Abstract
Background
Hepatitis E virus (HEV) is a common cause of acute viral hepatitis (AVH) in many developing countries. In Egypt, HEV seroprevalence is among the highest in the world; however, only a very limited number of Egyptian HEV sequences are currently available.
Objectives
The objectives were to determine the HEV genotype(s) currently circulating in Egypt.
Study Design
AVH patients without serologic evidence of hepatitis A, B, and C viruses were evaluated for possible HEV infection using serologic assays for anti-HEV IgM and anti-HEV IgG and real-time PCR for HEV RNA. Stool suspensions from suspected cases were inoculated into rhesus macaques to confirm the presence of HEV. Sequence analysis was utilized to determine HEV genotype.
Results
Of 287 subjects with AVH enrolled, 58 had serologic evidence of acute HEV infection. Stool samples for two of these patients were repeatedly positive for HEV RNA by real-time PCR. Macaques experimentally inoculated with these human stools also developed viremia. Sequence analysis of open reading frame (ORF) 1 demonstrated that these isolates belonged to HEV genotype 1 and were 3.9% – 9.5% divergent from other genotype 1 isolates. ORF2 was 5.3% – 8.7% divergent from previously reported Egyptian isolates.
Conclusions
This study strongly suggests that genotype 1 HEV related to other North African isolates is circulating in acute symptomatic patients in Egypt. Further evaluation of genotypic variability is underway in this highly endemic cohort and is considered an important component of our increased understanding of HEV pathogenesis.
Keywords: Hepatitis E virus (HEV), Egypt, symptomatic, genotype, diversity
Background
Hepatitis E virus (HEV) is an enterically transmitted virus that represents a common cause of acute viral hepatitis (AVH) in many developing countries 1. Although most patients with HEV recover fully, mortality rates of ~1% in the general population and up to 20% in pregnant women have been reported 2. Based on genomic variability observed among HEV isolates, four genotypes have been identified 3. As with other hepatitis viruses, genotype may play a role in disease severity 1, 4.
In Egypt, the prevalence of anti-HEV is among the highest of any country in the world, approaching 80% 5–10; yet, outbreaks have not been reported. A prospective study also estimated the HEV incidence at 41.6/1000 person-years 11; however, none of the seroconverting individuals gave a history compatible with AVH during the observation period. Similarly, HEV infection among pregnant women was not associated with history of jaundice or liver disease as reported elsewhere 11. Reasons for the infrequent clinical hepatitis remain unclear but could be the result of early childhood exposure leading to inapparent infection. Alternatively, the predominant HEV genotype(s) in Egypt could be less virulent than those circulating in Asia.
Objectives
Only a very limited number of HEV sequences are currently available from North Africa; therefore, we investigated HEV infection and genotypes in two Egyptian communities.
Methods
Study participants and sample collection
In Egypt, patients with acute viral hepatitis (AVH) are routinely referred to specialty fever hospitals. Suspected acute hepatitis patients were enrolled after providing informed consent. Clinical data were obtained by the treating physician and recorded in a pre-designed clinical research datasheet. From 2006–08, we enrolled 287 acute hepatitis subjects. Patients were serologically screened for hepatitis A virus (HAV), hepatitis B virus (HBV), and hepatitis C virus (HCV) using rapid tests for anti-HAV IgM (CTK Bioteck Inc, San Diego, CA), anti-HBV core IgM (IND Diagnostic, Canada), and anti-HCV IgG (Clinpro International, Union City, CA).
Serologic testing for HEV infection
Sera from patients with no evidence of HAV, HBV, or HCV infection were tested for possible HEV infection using the commercial anti-HEV IgM kit (HEV-IgM ELISA 3.0, MP Diagnostics, formerly Genelabs Diagnostic, Singapore). Anti-HEV IgG was measured as described previously using an in-house ELISA 12. Using the manufacturer’s criteria for positive anti-HEV IgM, 58 acute HEV infections were identified; 44 were subjects with acute symptomatic hepatitis, while 14 were contacts (mostly asymptomatic) of symptomatic cases.
Experimental HEV infection
Rhesus macaques derived from a hepatitis virus-free colony were housed at Bioqual Inc. (Rockville, Maryland, USA) and maintained according to the American Association of Accreditation of Laboratory Animal Care guidelines. For the current study, one animal each was inoculated with serum or a 20% suspension of stool derived from HEV-infected Egyptian patient TS00223 or TS00226 to confirm the presence of HEV. This study was approved by the Animal Care and Use Committees of Bioqual Inc. and the National Institute of Allergy and Infectious Diseases (Bethesda, Maryland, USA).
Real-time RT-PCR for HEV infection
Viral RNA was extracted from 20% stool suspensions from humans or macaques using TriZol and resuspended in DEPC-treated water. Viral RNA was extracted from 140 uL of patient serum using the QIAamp Viral RNA kit (QIAGEN, Valencia, CA). To determine the presence of HEV RNA in patient stool samples, two real-time RT-PCR methods were employed. The method of Jothikumar et al. amplifies ORF 3 and detects as few as 4 genome equivalent copies of HEV 13, while the method of Gyarmati et al. amplifies ORF 2 and detects 1–20 genome equivalents of HEV 14. Positive controls included HEV-infected rhesus stool samples corresponding to genotypes 1–4. A negative control with no input RNA was included in each run. 1:100 and 1:1000 dilutions of stool-derived RNA were also tested to reduce the effects of PCR inhibitory substances.
Reverse transcriptase (RT)-PCR amplification of HEV
To analyze genotype diversity, qualitative RT-PCR assays for ORF 1, ORF 2, and the ORF2/3 overlap region were employed. Primer sequences for ORF1 were CTG GCA TYA CTA CTG CYA TTG AGC (outer sense; nts 64 – 87), CCA TCR ARR CAG TAA GTG CGG TC (outer antisense; nucleotides [nts] 459 – 581), CTG CCY TKG CGA ATG CTG TGG (inner sense; nts 112 – 132), and GGC AGW RTA CCA RCG CTG AAC ATC (inner antisense; nts 375 – 398) 15, 16. Primer sequences for ORF2 were CTG TTT AAY CTT GCT GAC AC (outer sense; nts 6342 – 6361), TGA AAG CCA WAG CAC ATC (6633 – 6650), GAC AGA ATT RAT TTC GTC GGC TGG (inner sense; nts 6380 – 6403), and CTT GTT CRT GYT GGT TRT CAT AAT C (inner antisense; nts 6552 – 6576) 15. Primer sequences for ORF2/3 were GCR GTG GTT TCT GGG GTG AC (outer sense; nts 5279 – 5298), CTG GGM TYG GTC DCG CCA AG (outer antisense; nts 5485 – 5321), GYT GAT TCT CAG CCC TTC GC (inner sense; nts 5302 – 5321), and GMY TGG TCD CGC CAA GHG GA (inner antisense; nts 5419 – 5428) 17. One-step RT-PCR conditions were 50°C for 60 minutes, 94°C for 10 minutes, and 40 cycles of 94°C for 1 minute, 53°C for 45 seconds, and 72°C for 1 minute, followed by an additional 10 minutes at 72°C. Nested PCR conditions were 94°C for 2 minutes, 40 cycles of 94°C for 1 minute, 53°C for 45 seconds, and 72°C for 1 minute, followed by an additional 10 minutes at 72°C. Negative controls included one reaction with no reverse transcriptase and a separate reaction with no RNA. PCR products were gel purified and directly sequenced in both directions.
Phylogenetic analyses
The HEV genotype was determined by direct sequencing of the PCR fragment. All alignments were performed using Clustal X version 1.64b 18. GenBank references used to identify the HEV genotype included genotypes 1 (AF076239, M80581, AF051830, D11093, AY230202, AY204877, and X98292), 2 (M74506), 3 (AB246676, AF082843, AF060668, and AB189075) and 4 (AJ272108, AB220975, AB220978, and AB220979). Sequences from Egypt and Algeria - AF051350, AF051351, and AF051352 19 -were also included.
Results
Demographic and clinical characterization
58 HEV-infected subjects were identified. 52% of the population was female. The average age was 8 years. Median ALT and AST values were 94 (interquartile range [IR]: 60, 112) and 89 (IR: 51, 920), respectively. These levels were 5.4 (0.3, 52.4) and 3.6 (0.3, 89.7) fold above the upper limit of normal (ULN), respectively. 35 (60%) individuals reported jaundice, while 36 (62%) reported fever. 18 (30%) individuals reported contact with animals. The source of water was public/outside the residence for 24 (41%), residential tap water for 18 (31%), or wells for 16 (28%).
Virologic detection of HEV infection by real-time RT-PCR
Stool suspensions from 56 individuals were negative for HEV RNA by real-time PCR but were repeatedly positive for two patients – TS00223 and TS00226. TS00223 was a 24 year-old female from the Assuit area who was 4 months pregnant at the time of diagnosis. She presented with acute onset of fatigue, dark urine, yellow sclera, and abdominal pain for 3 days. Serum and stool was collected three days after disease onset. She was negative for HAV, HBV, and HCV. ALT and AST values were 94 and 96, respectively; these values are 7.8 and 8.0 fold above the ULN. She recovered completely within 2 weeks and was followed for 9 months with no morbidity. She had a normal delivery of a healthy baby. TS00226 was a 10 year-old female from a distinct village also in the Assuit area. TS00226 was not related to TS00223, although they did share the same well water source. She presented with acute onset of fatigue, loss of appetite, diarrhea, dark urine, and yellow sclera for 4 days. Serum and stool were collected four days after disease onset. She was negative for HAV, HBV, and HCV. ALT and AST values were 89 and 85, respectively; these values are 7.4 and 7.1 fold above the ULN. She recovered completely within 2 weeks and was followed for 9 months with no morbidity.
While HEV RNA was detectable in stool samples for TS00223 and TS00226, the corresponding serum samples did not have detectable HEV RNA. Similarly, a subset of individuals with no detectable stool HEV RNA was also negative for HEV RNA in their corresponding sera. For TS00223 and TS00226, there was insufficient RNA to determine the HEV genotype by qualitative RT-PCR; therefore, experimental inoculation of macaques with human serum and/or stool samples was attempted. Both animals inoculated with stool suspensions from these patients became positive for HEV RNA by real-time PCR, although neither corresponding serum sample transmitted HEV to the macaques.
HEV genotype variability
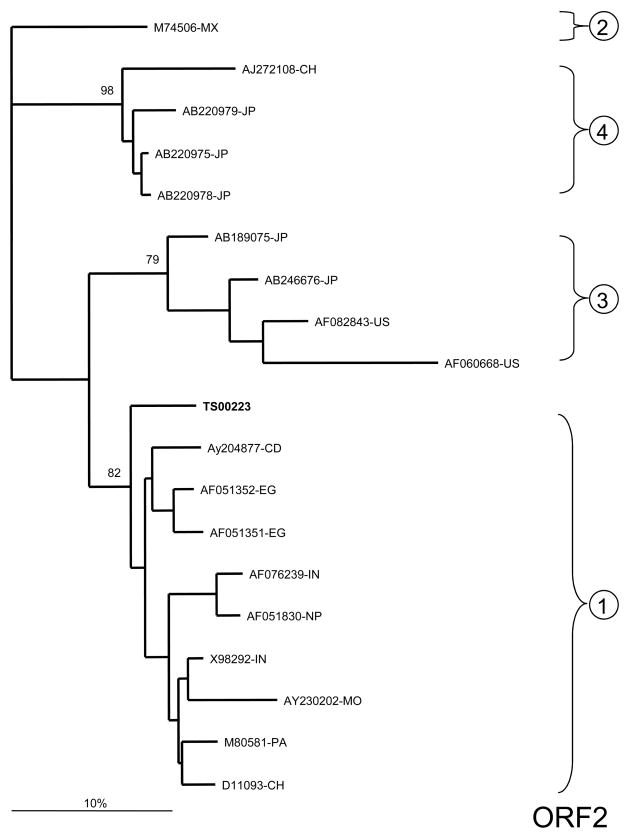
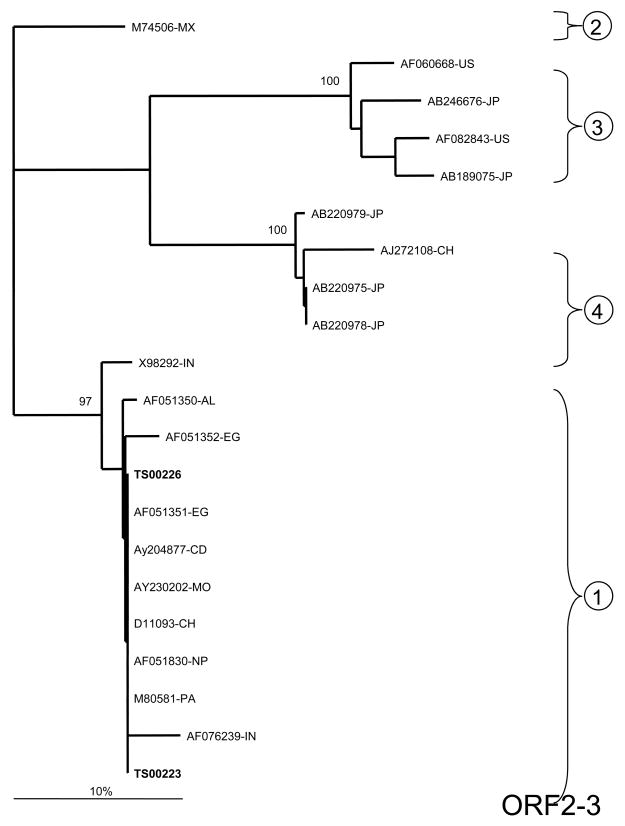
Stool samples from animals that were inoculated with stool suspensions from TS00223 and TS00226 were utilized in qualitative RT-PCR assays to determine the HEV genotype. Analysis of ORF1 demonstrated that TS00223 and TS00226 grouped with other African isolates representing HEV genotype 1 (Figure 1A). While they were only 0.4% divergent from one another at the nucleotide level, they were 3.9%–9.5% divergent from other genotype 1 isolates. No data from other Egyptian isolates were available for this ORF; however, TS00223 and TS00226 were divergent from isolates from Chad and Morocco.
Figure 1.
Phylogenetic analysis of HEV sequences from two symptomatic Egyptian patients. A) ORF 1 (242 base pairs [bp]; B) ORF2/3 overlap region (97 bp); C) ORF 2 (98 bp). Reference sequences are denoted by their GenBank accession numbers and the following country-specific abbreviations: Mexico (MX), United States (US), Japan (JP), China (CH), Chad (CD), Morocco (MO), India (IN), Pakistan (PA), Nepal (NP), Egypt (EG), and Algeria (AL). The two Egyptian samples characterized in the current study are highlighted in bold. The statistical robustness and reliability of the branching order within each phylogenetic tree were confirmed by bootstrap analysis using 100 replicates 25. Sequences from the current study – with primer sequences removed – have been submitted to GenBank under accession numbers FJ423076 – FJ423080.
For ORF2, only TS00223 could be amplified. This sample belonged to HEV genotype 1 and was 7.6%–11.2% divergent from other previously reported isolates. TS00223 was also 8.7% and 5.3% divergent from isolates AF051351-EG and AF051352-EG, respectively, previously collected from Egypt in 1993–94 (Figure 1B).
Analysis of the highly conserved ORF2/3 overlap region also confirmed that TS00223 and TS00226 belonged to HEV genotype 1 (Figure 1C). Both TS00223 and TS00226 were identical to one another and were 0.0%–3.2% divergent from other genotype 1 isolates. TS00223 and TS00226 were identical to Egyptian isolate AF051351-EG but 2.1% divergent from isolate AF051352-EG.
Discussion
Universal community exposure is likely responsible for the high seroprevalence of HEV in Egypt 5, 8, 11. However, HEV-associated AVH is relatively uncommon, leading some investigators to suggest that HEV strains in Egypt may be less virulent than in other geographic locations 6, 11. Tsarev et al. previously reported on two sporadic cases of HEV infection recovered from male army recruits living in the Cairo area in 1993–94 19. These isolates were closely related to other African isolates. Our finding of genotype 1 further supports the circulation of HEV genotype 1 throughout Egypt. Importantly, because our samples were collected in 2006–08 as part of a large prospective study of HEV infection in the cities of Assuit (Upper Egypt) and Mansoura (in the Delta), they represent the HEV isolates currently circulating in the central region of Egypt. While the intragenotypic variability observed suggests geographic diversity and/or viral evolution with time in this population, continued evaluation of HEV variation in this and other endemic settings is clearly warranted.
The detection of HEV RNA in only two stool samples in the current study is somewhat surprising given that HEV RNA has been detected in the stool in epidemic settings 20–22. However, the presence of viremia in the serum may be less frequent 23, 24. Furthermore, variable detection rates could reflect differences in the viral extraction methodology, assay sensitivity, and/or a highly divergent HEV strain that is not readily detected by current PCR-based assays. However, in the current analysis, we utilized two highly sensitive, real-time PCR assays that readily detected HEV RNA in human serum and stool in previous studies 13, 14. Additionally, HEV genotype 1 strains related to other North African isolates were detected, suggesting that assay sensitivity and viral divergence were unlikely to explain our findings. Importantly, the low frequency of HEV RNA detection in Egypt could also reflect true low-level viremia in HEV-infected individuals that merits additional investigation. This is further supported by the fact that HEV viremia was detected but only after experimental inoculation of human stool suspensions into macaques. Collectively, these data suggest that an attenuated genotype 1 may be circulating in Egypt and could be responsible for the low rate of HEV mortality observed.
Acknowledgments
This work was presented at the 13th International Symposium on Viral Hepatitis and Liver Disease held in Washington, DC in March 2009. This work was supported by an NIAID R21 (AI 067868) award to MTS, NIDDK K24 (DK 070528) to KES, and the Division of Intramural Research at NIAID.
Abbreviations
- HEV
hepatitis E virus
- HAV
hepatitis A virus
- HBV
hepatitis B virus
- HCV
hepatitis C virus
- AVH
acute viral hepatitis
- ORF
open reading frame
- RT
reverse transcriptase
- ULN
upper limit of normal
- SD
standard deviation
- ALT
alanine transaminase
- AST
aspartate transaminase
- S/C
sample/cut off ratio
- IU
international units
- nts
nucleotides
- bp
base pair
Footnotes
Competing interests: None declared
Approval for human studies was provided by the University of Cincinnati IRB. Animal studies were approved by the Animal Care and Use Committees of Bioqual Inc. and the National Institute of Allergy and Infectious Diseases (Bethesda, Maryland, USA).
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Emerson SU, Purcell RH. Hepatitis E virus. Reviews in Medical Virology. 2003 May–Jun;13(3):145–154. doi: 10.1002/rmv.384. [DOI] [PubMed] [Google Scholar]
- 2.Mushahwar I. Hepatitis E virus: molecular virology, clinical features, diagnosis, transmission, epidemiology, and prevention. Journal of Medical Virology. 2008;80(4):646–658. doi: 10.1002/jmv.21116. [DOI] [PubMed] [Google Scholar]
- 3.Okamoto H. Genetic variability and evolution of hepatitis E virus. Virus Research. 2007;127(2):216–228. doi: 10.1016/j.virusres.2007.02.002. [DOI] [PubMed] [Google Scholar]
- 4.Mizuo H, Yazaki Y, Sugawara K, et al. Possible risk factors for the transmission of hepatitis E virus and for the severe form of hepatitis E acquired locally in Hokkaido, Japan. J Med Virol. 2005 Jul;76(3):341–349. doi: 10.1002/jmv.20364. [DOI] [PubMed] [Google Scholar]
- 5.Fix AD, Abdel-Hamid M, Purcell RH, et al. Prevalence of antibodies to hepatitis E in two rural Egyptian communities. American Journal of Tropical Medicine and Hygiene. 2000;62(4):519–523. doi: 10.4269/ajtmh.2000.62.519. [DOI] [PubMed] [Google Scholar]
- 6.Stoszek SK, Abdel-Hamid M, Saleh DA, et al. High prevalence of hepatitis E antibodies in pregnant Egyptian women. Transactions of the Royal Society of Tropical Medicine and Hygiene. 2006;100(2):95–101. doi: 10.1016/j.trstmh.2004.12.005. [DOI] [PubMed] [Google Scholar]
- 7.Divizia M, Gabrieli R, Stefanoni ML, et al. HAV and HEV infection in hospitalised hepatitis patients in Alexandria, Egypt. European Journal of Epidemiology. 1999;15(7):603–609. doi: 10.1023/a:1007514030062. [DOI] [PubMed] [Google Scholar]
- 8.Abe K, Li TC, Ding X, et al. International collaborative survey on epidemiology of hepatitis E virus in 11 countries. Southeast Asian Journal of Tropical Medicine and Public Health. 2006;37(1):90–95. [PubMed] [Google Scholar]
- 9.Abdel Hady SI, El-Din MS, El-Din M. A high hepatitis E virus (HEV) seroprevalence among unpaid blood donors and haemodialysis patients in Egypt. Journal of the Egyptian Public Health Association. 1998;73(3–4):165–179. [PubMed] [Google Scholar]
- 10.El-Esnawy N. Examination for hepatitis E virus in wastewater treatment plants and workers by nested RT-PCR and ELISA. Journal of the Egyptian Public Health Association. 2000;75(1–2):219–231. [PubMed] [Google Scholar]
- 11.Stoszek SK, Engle RE, Abdel-Hamid M, et al. Hepatitis E antibody seroconversion without disease in highly endemic rural Egyptian communities. Transactions of the Royal Society of Tropical Medicine and Hygiene. 2006;100 (2):89–94. doi: 10.1016/j.trstmh.2005.05.019. [DOI] [PubMed] [Google Scholar]
- 12.Shata MT, Barrett A, Shire NJ, et al. Characterization of hepatitis E-specific cell-mediated immune response using IFN-gamma ELISPOT assay. Journal of Immunological Methods. 2007;328(1–2):152–161. doi: 10.1016/j.jim.2007.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jothikumar N, Cromeans TL, Robertson BH, Meng XJ, Hill V. A broadly reactive one-step real-time RT-PCR assay for rapid and sensitive detection of hepatitis E virus. Journal of Virological Methods. 2006;131(1):65–71. doi: 10.1016/j.jviromet.2005.07.004. [DOI] [PubMed] [Google Scholar]
- 14.Gyarmati P, Mohammed N, Norder H, Blomberg J, Belák S, Widén F. Universal detection of hepatitis E virus by two real-time PCR assays: TaqMan and Primer-Probe Energy Transfer. Journal of Virological Methods. 2007;146(1–2):226–235. doi: 10.1016/j.jviromet.2007.07.014. [DOI] [PubMed] [Google Scholar]
- 15.Schlauder GG, Desai SM, Zanetti AR, Tassopoulos NC, Mushahwar IK. Novel hepatitis E virus (HEV) isolates from Europe: evidence for additional genotypes of HEV. Journal of Medical Virology. 1999 Mar;57(3):243–251. doi: 10.1002/(sici)1096-9071(199903)57:3<243::aid-jmv6>3.0.co;2-r. [DOI] [PubMed] [Google Scholar]
- 16.Wang Y, Zhang H, Ling R, Li H, Harrison TJ. The complete sequence of hepatitis E virus genotype 4 reveals an alternative strategy for translation of open reading frames 2 and 3. Journal of General Virology. 2000 Jul;81(Pt 7):1675–1686. doi: 10.1099/0022-1317-81-7-1675. [DOI] [PubMed] [Google Scholar]
- 17.Inoue J, Takahashi M, Yazaki Y, Tsuda F, Okamoto H. Development and validation of an improved RT-PCR assay with nested universal primers for detection of hepatitis E virus strains with significant sequence divergence. Journal of Virological Methods. 2006 Nov;137(2):325–333. doi: 10.1016/j.jviromet.2006.07.004. [DOI] [PubMed] [Google Scholar]
- 18.Thompson J, Gibson T, Plewniak F, Jeanmougin F, Higgins D. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Research. 1997;25(24):4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tsarev SA, Binn LN, Gomatos PJ, et al. Phylogenetic analysis of hepatitis E virus isolates from Egypt. Journal of Medical Virology. 1999 Jan;57(1):68–74. doi: 10.1002/(sici)1096-9071(199901)57:1<68::aid-jmv10>3.0.co;2-e. [DOI] [PubMed] [Google Scholar]
- 20.Boccia D, Guthmann JP, Klovstad H, et al. High mortality associated with an outbreak of hepatitis E among displaced persons in Darfur, Sudan. Clinical Infectious Diseases. 2006;42(12):1679–1684. doi: 10.1086/504322. [DOI] [PubMed] [Google Scholar]
- 21.Isaäcson M, Frean J, He J, Seriwatana J, Innis B. An outbreak of hepatitis E in Northern Namibia, 1983. American Journal of Tropical Medicine and Hygiene. 2000;62(5):619–625. doi: 10.4269/ajtmh.2000.62.619. [DOI] [PubMed] [Google Scholar]
- 22.Chobe LP, Chadha MS, Banerjee K, Arankalle V. Detection of HEV RNA in faeces, by RT-PCR during the epidemics of hepatitis E in India (1976–1995) Journal of Viral Hepatitis. 1997;4(2):129–133. doi: 10.1111/j.1365-2893.1997.tb00215.x. [DOI] [PubMed] [Google Scholar]
- 23.Kantala T, Maunula L, von Bonsdorff CH, Peltomaa J, Lappalainen M. Hepatitis E virus in patients with unexplained hepatitis in Finland. Journal of Clinical Virology. 2009 doi: 10.1016/j.jcv.2009.03.006. Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- 24.Zhu G, Qu Y, Jin N, et al. Seroepidemiology and molecular characterization of hepatitis E virus in Jilin, China. Infection. 2008;36(2):140–146. doi: 10.1007/s15010-007-7130-8. [DOI] [PubMed] [Google Scholar]
- 25.Felsenstein J. Confidence limits on phylogenies: an approach using the bootstrap. Evolution. 1985;39:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]