Abstract
Changes in gene expression in brain reward regions are thought to contribute to the pathogenesis and persistence of drug addiction. Recent studies have begun to focus on the molecular mechanisms by which drugs of abuse and related environmental stimuli, such as drug-associated cues or stress, converge on the genome to alter specific gene programs. Increasing evidence suggests that these stable gene expression changes in neurons are mediated in part by epigenetic mechanisms that alter chromatin structure on specific gene promoters. This review discusses recent findings from behavioral, molecular and bioinformatic approaches being used to understand the complex epigenetic regulation of gene expression by drugs of abuse. This novel mechanistic insight might open new avenues for improved treatments of drug addiction.
Mechanisms of drug addiction
Drug addiction is a debilitating psychiatric disorder that is characterized by compulsive drug seeking and taking despite severe adverse consequences [1-3]. Once an individual becomes addicted to a drug of abuse, there are few effective clinical options, and most addicts relapse within a short period of time. Thus, addiction research focuses on two major outstanding questions. First, what are the neural mechanisms underlying the transition from recreational drug use to a chronically addicted state? Second, what are the mechanisms responsible for the persistence of addictive behaviors even after prolonged drug abstinence? A better understanding of these mechanisms might provide clues into how we can block or even reverse the addicted state and thereby reduce the rate of relapse.
Drug-induced changes in gene expression in key brain reward regions, such as the nucleus accumbens (NAc), prefrontal cortex (PFC) and ventral tegmental area (VTA), represent one mechanism thought to contribute to both of these key questions [1-3]. For example, the transcription factor ΔFosB is induced several fold in the NAc by chronic drug exposure and has been implicated in the transition to an addicted state [4-6]. Altered expression of specific genes, such as activator of G-protein signaling 3 (AGS3) [7] and brain-derived neurotrophic factor (BDNF) [8], has been reported weeks after the last drug experience, and manipulation of these genes in rodents regulates drug relapse behavior [7,9,10]. Genome-wide mRNA analyses have identified many more potential gene targets for drugs of abuse in distinct brain reward regions, and these targets might also contribute to long-lasting behavioral effects [11-16]. Therefore, it has become of great interest to identify the underlying mechanisms by which chronic drug exposure promotes stable changes in gene expression and, ultimately, behavior. Recent evidence has suggested that epigenetic mechanisms – key cellular processes that integrate diverse environmental stimuli to exert potent and often long-lasting changes in gene expression through the regulation of chromatin structure – contribute to these drug-induced transcriptional and behavioral changes [17-19]. This review discusses current progress toward understanding how epigenetic mechanisms are regulated by drugs of abuse in brain reward regions and how such mechanisms might contribute to drug-related behaviors. Ultimately, better understanding of these mechanisms might reveal novel drug targets for the development of improved pharmaceutical interventions.
Epigenetic mechanisms
The word ‘epigenetic’ historically refers to a heritable phenotype not coded by DNA itself but by a cellular process ‘above the genome’. Cellular differentiation is a classic example where epigenetic phenomena have a critical role [20,21]. Because all cells in an organism contain the same genetic information, the ability to form clonal populations of distinct cell types with unique functions (e.g. neurons versus hepatocytes) is achieved by transmitting the correct transcriptional programs from parent to daughter cell. This epigenetic process is in large part coordinated through control of chromatin structure. Increasing evidence indicates that changes in chromatin structure not only mediate these heritable epigenetic phenomena [22] but also that the same types of changes in chromatin occur in mature, post-mitotic neurons [23,24].
Chromatin is made up of DNA and the histone proteins around which the DNA is wrapped. Histones are assembled into an octamer made up of two copies of H2A, H2B, H3 and H4 [25]. These histone proteins, together with DNA, undergo a complex supercoiling process, which results in a highly compact structure in which meters of extended DNA is condensed and organized. This highly condensed structure means that control over gene expression occurs partly by gating access of transcriptional activators to DNA [26,27]. The structure of chromatin, and hence access to the DNA sequence wrapped around it, is highly regulated by post-translational modifications of histones and the DNA itself [28]. Such modifications include acetylation, phosphorylation and methylation of histones, methylation of DNA, and copious others, with each modification either positively or negatively regulating the transcriptional activity of the underlying gene (Table 1). Ultimately, dozens of potential modifications that occur at many distinct histone residues are thought to summate to determine the final transcriptional output of a given gene [29].
Table 1.
Histone modifications and their functiona
| Modification | Transcriptional effectb | Enzyme (addition) | Enzyme (removal) |
|---|---|---|---|
| Acetylation |  |
HATs (CBP,p300) | HDACs (HDAC1−11) |
| Methylation |  |
KMTs (SUV39H1) | KDMs (JHDM2a) |
| Phosphorylation |  |
Kinases (MSK1) | Phosphatases (PP1) |
| Ubiquitination |  |
Ub ligases (RING2) | Ub protease (USP16) |
| Sumoylation |  |
SUMO E2s/E3s? (UBC9) | SUMO protease (SUSP1?) |
Abbreviations: HAT, histone acetyltransferase; HDAC, histone decacetylase; KMT, lysine methyltransferase; KDM, lysine demethylase; SUMO, small ubiquitin-related modifier; Ub, ubiquitin.
List of histone modifications, their known effects on transcription and examples of enzymes that catalyze their addition or removal from histones.
 = increased transcription;
= increased transcription;  = decreased transcription.
= decreased transcription.
The expansive number of combinatorial options possible enables epigenetic mechanisms to exert exquisite control over the transcriptional activity of each gene in the genome. One of the key hallmarks of epigenetic mechanisms is their potential stability within the cell. This is an important feature because maintaining the continuity of liver gene programs in hepatacytes and neural gene programs in neurons is necessary throughout the life of an individual. However, despite the stability of developmental epigenetic mechanisms in vivo, all types of chromatin modifications identified to date are potentially reversible and have specific enzymes or processes that mediate the addition or removal of each mark [28] (see Box 1). For example, histone acetyltransferases (HATs) add acetyl groups to histone proteins, whereas histone deacetylases (HDACs) remove them, and neuronal signaling appears to strongly regulate both the addition and removal of histone acetylation, histone methylation and DNA methylation in vivo, in some cases over relatively short time frames (e.g. hours) [24].
Box 1. Chromatin-modifying enzymes.
Chromatin modifications, or ‘marks’, such as histone acetylation and methylation are dynamic processes, controlled by enzymes that either add or remove the specific mark. Histone acetyltransferases (HATs) catalyze the addition of acetyl groups onto lysine residues of histone proteins. There are over a dozen known HATs, such as CBP, p300, Gcn5, and PCAF (p300/CBP-associated factor), many of which have been implicated in addiction-, stress- or memory-related behaviors [18,64,65]. Recently, several transcription factors [e.g. activating transcription factor 2 (ATF2), CLOCK] have also been shown to possess HAT activity [66,67].
Histone deacetylases (HDACs), which remove acetyl groups from histones, are divided into four classes. Class I HDACs (e.g. HDAC1, 2, 3) are ubiquitously expressed and probably mediate the majority of deacetylase activity within cells. Class II HDACs (e.g. HDAC4, 5, 9), enriched in specific tissues such as heart and brain, are larger proteins that contain both the class I deacetylase domain and an N-terminal regulatory domain that enables them to be shuttled in and out of the nucleus in a neural-activity-dependent manner [68]. Class III HDACs are NAD- (nicotinamide adenine dinucleotide) dependent and have been implicated in the regulation of life span and metabolism [69]. HDAC11 is a class IV HDAC and shares homology to both class I and class II enzymes [70].
Histone methyltransferases (HMTs) and demethylases (HDMs) are not only specific for the histone subunit and lysine residue (e.g. K4 versus K9) but also for the number of methyl groups they can add or remove [28]. For example, the HMT KMT1C (lysine methyltransferase 1C, formerly G9a), is specific for histone H3K9 but only adds one or two methyl groups, with the distinct HMT KMT1A (SUV39H1) catalyzing trimethylation of this site. Similarly, the HDM KDM3A [lysine demethylase, formerly jumonji domain-containing histone demethylase 2A (JHDM2a)], can demethylate one or two methyl groups on H3K9, requiring a distinct demethylase [e.g. KDM4D, formerly jumonji domain-containing protein 2D (JMJD2D)] to fully demethylate the trimethylated state. Thus, several enzymes are required to move between the unmethylated and fully trimethylated states, providing an additional level of regulation and information encoded by each methyl mark.
Methylation of DNA in brain is catalyzed by three main enzymes, DNA methyltransferase (DNMT)1, 3a and 3b. A role in adult neural plasticity is supported by the observation that DNMT inhibitors alter behavioral effects in learning and memory paradigms [71]. DNA demethylase enzymes are likely to exist based on reports that environmental stimuli induce the demethylation of particular genes in the brain, but the specific enzymes are not yet known [71].
Drug-induced changes in chromatin structure
Histone acetylation
Acetylation of histone lysine residues reduces the electrostatic interaction between histone proteins and DNA, which is thought to relax chromatin structure and make DNA more accessible to transcriptional regulators [28]. Histone acetylation is best characterized on histones H3 and H4: it can occur on lysines 9, 14, 18 and 23 on the N-terminal tail of H3 and at lysines 5, 8, 12 and 16 on the tail of H4. Genome-wide studies have shown that hyperacetylation in promoter regions is strongly associated with gene activation, whereas hypoacetylation is correlated with reduced gene expression [30,31]. This association also exists in the brain in vivo in response to drugs of abuse. Acute exposure to cocaine, for example, which is known to rapidly induce the immediate early genes c-fos and fosb in the NAc, increases histone H4 acetylation on their proximal gene promoters (Figure 1) [17]. Time-course analysis revealed that this modification occurs within 30 min and disappears by 3 h, consistent with the induction kinetics of these immediate early genes. At least for fosb, this increase in histone acetylation is dependent on the HAT cAMP response element-binding protein (CREB)-binding protein (CBP) [18]. Interestingly, despite several control gene promoters where acute cocaine does not affect histone acetylation (β-tubulin, tyrosine hydroxylase, histone H4), acute cocaine does increase global levels of histone H4 acetylation and histone H3 phospho-acetylation (see next section), but not H3 acetylation alone, within 30 min [17,32]. Thus, global changes in histone modifications, which have been observed not only in response to drugs of abuse [17], but also in learning models and in response to environmental enrichment [33,34], might be accounted for by acetylation, and strong induction, of a specific subset of genes.
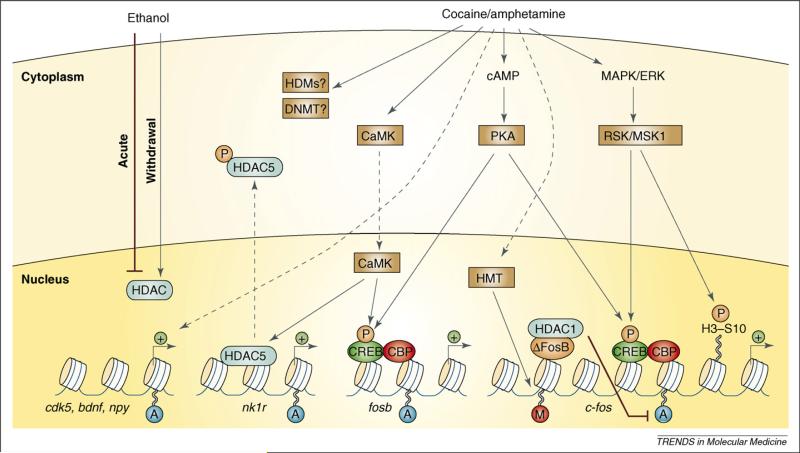
Figure 1.
Regulation of chromatin remodeling by drugs of abuse. Cocaine and amphetamine increase levels of cAMP in the nucleus accumbens (NAc) and activate protein kinase A (PKA). PKA then phosphorylates cAMP-response-element-binding protein (CREB), which allows for the recruitment of the histone acetyltransferase CREB-binding protein (CBP). Examples of this are shown on the fosb and c-fos genes. Chronic cocaine or amphetamine is also known to elevate levels of ΔFosB, which can recruit histone deacetylase 1 (HDAC1) to the c-fos promoter and inhibit subsequent induction of the gene (see also Figure 3). This desensitization of c-fos also involves increased repressive histone methylation, which is thought to occur via the induction of specific histone methyltransferases. It is not yet known how cocaine regulates histone demethylases (HDMs) or DNA methyltransferases (DNMTs). Cocaine also activates the mitogen-activated protein kinase (MAPK) cascade, which through MSK1 can phosphorylate CREB and histone H3 at serine 10. In addition, stimulant drugs regulate Ca2+ levels in NAc neurons (perhaps via regulation of glutamatergic synapses from cortical regions). This activates CaMK (calcium/calmodulin protein kinase) signaling, which, in addition to phosphorylating CREB, also phosphorylates HDAC5. This results in nuclear export of HDAC5 and increased histone acetylation on its target genes [e.g. the NK1 receptor (also known as the neurokinin 1 or substance P receptor)]. Several other genes have been shown to display increased acetylation on their promoters after cocaine or amphetamine exposure, including cdk5, bdnf and npy. In addition, acute ethanol has been shown to reduce histone acetylation by increasing HDAC activity, whereas withdrawal from chronic ethanol increases histone acetylation by reducing HDAC activity. Figure modified with permission from [24].
Repeated cocaine exposure, either forced (investigator) administration or self-administration, is known to induce a distinct set of genes in the NAc (e.g. cdk5 and bdnf), some of which remain elevated for days to weeks [7,8,12,15,35]. Consistent with such stable changes in gene expression, increased histone H3 acetylation was observed on the gene promoters of both cdk5 and bdnf for 1 to 7 days after the final dose of cocaine [17]. Stable changes in histone acetylation and gene expression have been observed for nearly two weeks after withdrawal from cocaine self-administration in the PFC as well. For example, npy (neuropeptide Y) expression was found to be upregulated and its gene promoter hyperacetylated while egr-1 (early growth response 1) was found to be downregulated and hypoacetylated after cocaine withdrawal [36].
Histone phosphorylation
Histone phosphorylation is generally associated with transcriptional activation; it can be observed on the promoters of immediate early genes such as c-fos when they are induced after cAMP induction or glutamate treatment in cultured striatal neurons [37,38]. One of the best characterized histone phosphorylation sites is serine 10 on histone H3 (H3S10). This modification stabilizes the HAT, Gcn5, on gene promoters while antagonizing the repressive modification – methylation of lysine 9 on histone H3 (H3K9) and its subsequent recruitment of heterochromatin protein 1 (HP1) [28]. Because phosphorylation at H3S10 recruits a HAT, the neighboring lysine residue at H3K9 is often acetylated in concert with phosphorylation (phospho-acetylation). In addition to the global induction of H3 phospho-acetylation in striatum by acute cocaine (as mentioned above), phospho-acetylation is also known to occur on the c-fos gene promoter at a time point when the gene is highly activated (Figure 1) [17,32]. Moreover, pretreatment of rats with an HDAC inhibitor before cocaine administration potentiates phospho-acetylation at the c-fos promoter and induction of c-fos mRNA, further illustrating the interplay between these two modifications. Interestingly, this interplay between histone phosphorylation and acetylation might occur only at certain genes in response to acute cocaine or other acute stimuli and has been implicated particularly in the induction of immediate early genes. Cocaine-induced H3S10 phosphorylation appears to be mediated by the downstream mitogen- and stress-activated protein kinase 1 (MSK1) (Figure 1) [32]. Additionally, cocaine-induced phosphorylation of dopamine- and cAMP-regulated phosphoprotein of 32 kDa (DARPP-32) has been shown recently to promote nuclear localization of this protein, which through inhibition of protein phosphatase 1 contributes to increases in H3S10 phosphorylation [72].
Histone methylation
Histone methylation is particularly complex and can exist in mono-, di- (me2) or tri-methylated (me3) states, enabling each state to recruit unique coregulators and exert distinct effects on transcriptional activity [28]. Histone methylation is also unique because each lysine residue has distinct, and often opposite, effects on transcription. For example, H3K4 is highly associated with gene activation, whereas H3K9 and H3K27 are usually associated with repression [27]. However, even this is an oversimplification, as H3K9 is often found downstream of the gene promoter in coding regions along with H3K36 and might be involved in transcriptional elongation [28,39]. Such complexity is reflected in the PFC of adolescent rats, where cocaine was found to induce significant reductions in global levels of both the activating H3K4me3 mark as well as the repressive H3K27me3 mark [40]. One likely explanation is that these global decreases in histone methylation are occurring on distinct gene promoters. Indeed, using the gene-specific technique, chromatin immunoprecipitation (Figure 2a), chronic cocaine exposure increases H3K9me2 on genes where histone acetylation, an activating mark, is not induced. Many of these hypermethylated genes are also downregulated by cocaine (see next section). There is also early evidence that specific histone methyltransferases and demethylases are themselves regulated by cocaine in the NAc [41].
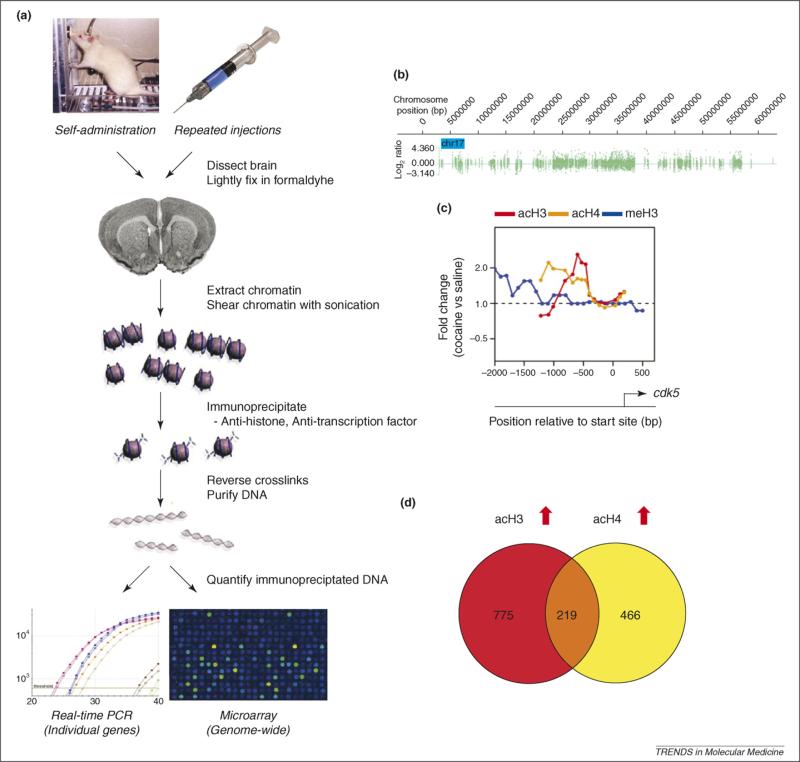
Figure 2.
Studying chromatin regulation in brain using chromatin immunoprecipitation. (a) Schematic of the chromatin immunoprecipitation (ChIP) protocol. ChIP is a technique used to quantify how much of a specific DNA sequence is occupied by a given histone modification or transcription factor. The technique involves lightly fixing the tissue or cells with formaldehyde to crosslink DNA with the associated histones and other associated proteins (chromatin). The crosslinked chromatin is then sonicated into ∼500 bp fragments and immunoprecipitated with an antibody raised against a specific histone modification or transcription factor. The immunoprecpitated chromatin is then reverse crosslinked from associated proteins and purified (pure DNA). Specific regions of this DNA can then be directly quantified by real time PCR (polymerase chain reaction) to determine how much of that DNA was immunoprecipitated in a drug-treated versus saline-treated animal. The final purified DNA can also be amplified for downstream use in genome-wide analysis techniques, such as microarrays (ChIP-chip) or next generation sequencing (ChIP-seq). (b) ChIP-chip data are typically displayed as an enrichment profile across each chromosome. For example, acetylated H3 (acH3)-binding on chromosome 17 from the nucleus accumbens (NAc) of a cocaine-treated mouse is displayed. One can then compare the enrichment profiles between cocaine- and saline-treated mice to determine the fold difference of acH3 on a specific chromosomal region. (c) The chromosome-wide data shown in (b) can be ‘zoomed in’ to display each gene in the genome. Displayed here is the cdk5 gene promoter in the NAc and the fold difference between cocaine- and saline-treated mice for acH3 (red), acH4 (orange) and methylated H3 (meH3) (blue). (d) The fold differences between cocaine- and saline-treated mice can be quantified for each gene and analyzed for statistical significance. The genes can then be compared and displayed, for example, using Venn diagrams to show how many genes are commonly regulated between two conditions. Shown here are the number of genes in the NAc on which cocaine commonly (overlapping area) or uniquely (non-overlapping areas) increases acH3 and acH4 binding.
Although enzymes exist to demethylate histones (Box 1), in a mouse model of depression (social defeat stress), increased H3K27me2 on the bdnf promoter was observed in the hippocampus for at least a month after the final stress [42]. This repressive mark correlated with a significant downregulation of BDNF expression. If the mice were treated with chronic antidepressants after the stress, however, increases in the activating mark, H3K4me2, occurred on the bdnf promoter and BDNF expression returned to normal levels, despite persistent increases in H3K27me2. Although the study of histone methylation in animal models of addiction is in its early phases, these studies together have shown that histone methylation is dynamically regulated by drugs of abuse and other emotional stimuli and has the potential to stably alter gene expression in vivo. It will be important to identify how such long-lasting changes in histone methylation are maintained and whether these mechanisms mediate persistent behavioral deficits.
Genome-wide analysis of chromatin regulation in animal models of addiction
Beyond analyzing the effect of drugs of abuse on chromatin structure at specific genes, the next important step is to characterize these drug-induced histone modifications across every gene in the genome. Using genome-wide techniques involving hybridizing immunoprecipitated chromatin to genome-wide promoter microarrays (ChIP-chip) (see Figure 2a) or to high-throughput sequencing, a wealth of new information can been uncovered about epigenetic regulation in specific brain regions, as well as novel gene targets that control behavioral responses to drugs of abuse. Such analyses are just now getting underway for drug addiction models.
The data one gets from such analyses are illustrated in Figure 2b, where histograms represent the intensity of enrichment along each chromosome [43]. This can be ‘zoomed in’ for each gene in the genome, as shown in Figure 2c, where one can overlay cocaine-induced changes in histone H3 acetylation, H4 acetylation, H3K9me2 methylation, and so on. This high resolution map of histone modifications provides a new level of insight into basic transcriptional mechanisms occurring in the brain in vivo in response to chronic cocaine administration. One such insight directly addresses earlier findings where a few acutely induced genes showed selective H4 acetylation on their promoters while a few chronically induced genes displayed selective H3 acetylation [17]. Genome-wide ChIP-chip analysis reveals that although there are more H3 acetylated genes in the NAc of mice exposed to chronic cocaine, there is also a significant set of previously unrecognized, chronically-induced genes that are hyperacetylated only on H4 (Figure 2d) [43]. Interestingly, this analysis showed further that only a very small subset of genes contains both H3 and H4 acetylation. Previous findings in cocaine [17] and seizure models [44] found that, among the small number of genes studied, most that were induced after chronic treatments were hyperacetylated on histone H3, not H4. A subset of these genes, consisting mostly of immediate early genes, were also induced after an acute treatment and hyperacetylated primarily on H4. These findings suggested that most genes were activated acutely by acetylation at H4 and chronically by acetylation at H3. However, the identification by ChIP-chip of a significant number of chronically induced genes that are only acetylated on H4 suggests that the H4 to H3 switch previously observed in both cocaine and seizure models might be reserved for certain types of genes, such as immediate early genes.
Another striking finding from these first genome-wide studies in addiction models is that there are very few genes where cocaine induces hypoacetylation of either H3 or H4 [43]. This suggests that active histone deacetylation might not be the most common mechanism for downregulating gene expression by cocaine. One interpretation of these results is that HDACs more commonly serve to limit gene expression while an alternative mechanism serves to actually repress genes in response to cocaine. Histone methylation might be such a repressive mechanism because numerous genes were found to have elevated H3K9me2 methylation after chronic cocaine exposure, and many of these genes are known to be downregulated by cocaine [12,15,43]. Thus, histone methylation is an attractive candidate for a cocaine-induced mechanism to repress specific gene transcription.
Functions of chromatin remodeling in vivo
Despite the substantial progress being made in identifying chromatin modifications throughout the genome in a variety of in vivo model systems, many questions remain. For example, the tools are not yet available to determine whether histone modifications are the primary cause of changes in gene expression or whether they are simply a reflection of it. Even in simple biological systems this remains a challenging question because most genetic and pharmacological tools manipulate chromatin structure genome-wide. This makes it difficult to establish direct causal relationships between altered chromatin structure and observed transcriptional changes at a specific gene locus. Importantly, a recent breakthrough has demonstrated the proof of principle that zinc finger peptides fused to a DNA methyltransferase can target that enzyme to a specific gene in cultured cells and repress its activity [45,46], suggesting that we might soon be able to target chromatin-modifying enzymes to individual genes in the brain, for example, using viral-mediated gene transfer. Then, one could begin to determine whether stimulus (e.g. cocaine)-induced histone modifications on specific gene promoters are necessary and sufficient to alter the activity of that gene in vivo and, ultimately, whether such modifications play a part in behavioral responses to drugs of abuse.
Although these tools have not yet been used in brain, there are several examples in vivo that suggest certain histone modifications are more than a reflection of gene expression and might serve a function in gene priming. For example, bdnf is highly acetylated in the NAc within 24 h after a rat self-administers cocaine [17]; however, the steady-state levels of BNDF protein are not significantly elevated until a week of cocaine withdrawal [8]. This is an example where histone acetylation on a gene promoter precedes the induction of gene expression and supports the idea that, at least for a subset of genes, histone acetylation might play a role in priming genes for subsequent induction. Therefore, investigating histone modifications genome-wide might provide unique insight into cocaine-induced gene regulation beyond a reflection of steady-state mRNA levels. Additionally, the persistent increase in levels of BDNF during cocaine withdrawal is particularly interesting from an epigenetic perspective, especially because BDNF has been implicated in cocaine relapse [9,10]. Although it is not known if changes in chromatin structure persist for as long as BDNF is upregulated, histone H3 acetylation has been observed on the bdnf promoter for at least a week of cocaine withdrawal. Further research is required to better characterize the mechanisms mediating this long-lasting transcriptional change [17].
Role of epigenetic mechanisms in drug-related behaviors
The identification of cocaine-induced alterations in histone acetylation, phosphorylation and methylation in the NAc and other brain areas suggests that such modifications might be involved in regulating behavioral responses to drugs of abuse. Indeed, the first evidence for this came from studies that demonstrated that the pharmacological and genetic manipulation of certain HDACs in the NAc alters levels of histone acetylation in vivo and profoundly affects behavioral sensitivity to cocaine [17]. In the conditioned place preference test, in which an animal learns to associate the rewarding effects of cocaine with a specific environment, either systemic administration of sodium butyrate or trichostatin A, both non-specific HDAC inhibitors, significantly potentiates the rewarding effects of cocaine [17]. Delivery of the more-specific HDAC inhibitor suberoylanilide hydroxamic acid (SAHA) directly into the NAc is sufficient to increase cocaine reward [19]. Similar potentiating effects of HDAC inhibition on drug-related behavior were observed with amphetamine and D1 agonists [47,48]. Consistent with the hypothesis that increased histone acetylation potentiates behavioral sensitivity to cocaine, mice that are deficient in CBP, a HAT, exhibit reduced histone acetylation on the fosb promoter, as well as reduced sensitivity to cocaine [18]. Similarly, reducing histone acetylation in the NAc by virally overexpressing certain HDACs (HDAC4 or HDAC5, but not HDAC9) in the NAc significantly decreases cocaine place conditioning and, at least for HDAC5, this effect requires the C-terminal catalytic deacetylase domain [19]. However, class II HDACs such as HDAC5 associate with class I HDACs (e.g. HDAC3) at this same C-terminal domain [49], so the relative contribution of this interaction, versus any catalytic activity of HDAC5, to its full effects on cocaine reward remains unclear.
The observation that chronic cocaine treatment regulates HDAC5 in the NAc raises the exciting possibility that this class II HDAC is involved in the behavioral transitions that occur between acute and chronic cocaine exposure (e.g. between drug experimentation and compulsive drug use). Specifically, chronic, but not acute, cocaine administration induces HDAC5 phosphorylation and nuclear export in the NAc, actions that block the enzyme's effects on histones. Nuclear export of HDAC5 results in histone hyperacetylation and increased mRNA expression of specific HDAC5 target genes, which would then contribute to sensitized behavioral responses to the drug [19]. An example of such a target gene is the NK1 (neurokinin 1 or substance P) receptor. Consistent with this model, naïve HDAC5-knockout mice display normal rewarding responses to initial cocaine exposures, but become hypersensitive if they are previously exposed to a chronic course of cocaine [19]. Importantly, HDAC5-knockout mice also hypersensitize to other chronic, but not acute, stimuli, including chronic social defeat stress, chronic cardiac stress and chronic neuropathic pain [19,50,51]. These findings, taken together with HDAC inhibitor studies in learning and memory and depression models, suggest that histone acetylation controls the saliency of a wide variety of environmental stimuli [34,42,52]. Whether it is cocaine, stress, pain or memory, pharmacological and genetic manipulations that result in elevated histone acetylation appear to potentiate the respective behavioral responses.
Histone H3 phosphorylation and phospho-acetylation also appear to play key roles in drug-regulated behaviors. As discussed earlier, cocaine rapidly increases global levels of histone H3 phosphorylation and phospho-acetylation in the striatum with similar kinetics to c-fos mRNA induction [17,32]. Moreover, c-fos seems to be highly linked to these modifications because c-fos induction by acute cocaine is potentiated by HDAC inhibitors and entirely blocked by loss of the histone H3 kinase MSK1 [17,32]. MSK1 is a downstream member of the MAP kinase cascade and, as mentioned earlier, is necessary for cocaine-induced H3 phosphorylation. Loss of MSK1 also reduces locomotor responses to cocaine [32], which is consistent with a mechanism involving dysregulation of c-fos [53].
Histone methylation is also regulated by cocaine; however, the behavioral significance of this modification is still under investigation. New inhibitors of histone methyltransferases, in addition to viral-mediated gene transfer, are allowing this question to be directly addressed. For example, preliminary findings suggest that inhibition of a particular H3K9 histone methyltransferase, the expression of which is regulated in the NAc by chronic cocaine, potentiates behavioral responses to the drug [41]. Because inhibition of H3K9 methylation would be expected to enhance gene activity, these results are consistent with studies of histone acetylation [17,19] and indicate that manipulations that increase gene transcription generally promote behavioral sensitivity to drugs of abuse.
The strong behavioral consequences of pharmacological or genetic manipulation of epigenetic mechanisms in the brain in vivo suggests that chromatin-modifying enzymes, such as histone deacetylases or methyltransferases, might be useful targets for the development of new psychiatric treatments. Histone deacetylase inhibitors, for example, have antidepressant-like effects in animal models [42,52]. Consistent with this idea is the recent finding that tranylcypromine, a monoamine oxidase inhibitor (MAOI) used to treat depression, is a much stronger inhibitor of the histone demethylase KDM1 (lysine demethylase 1, formerly LSD1) than it is of either monoamine oxidase AorB [54].
Interplay between transcription factors and epigenetic mechanisms
In order for environmental stimuli to regulate chromatin structure on the correct set of genes, mechanisms exist to guide the proper chromatin-remodeling enzymes and transcriptional regulators to the right gene locus. Transcription factors serve as a key mechanism by which distinct gene programs are controlled because they bind to highly specific DNA regulatory sequences. These regulatory sequences – termed response elements – serve as an address, so the cell can rapidly initiate specific gene programs that are influenced by a given set of transcription factors. Whereas some chromatin-remodeling enzymes can bind directly to a chromatin mark through specialized protein domains [28], many others get targeted to chromatin by interacting with specific transcription factors. The transcription factor CREB, which plays an essential role in behavioral responses to cocaine [55], was one of the first transcription factors known to direct a chromatin-modifying enzyme to gene promoters. When CREB is phosphorylated, it interacts with CBP, a HAT that helps facilitate target gene activation by acetylating neighboring histones [56].
ΔFosB, mentioned earlier, is another key transcription factor involved in behavioral responses to cocaine. ΔFosB is a stable, truncated splice product of the fosB gene that binds to AP-1 (activator protein 1) sites in the promoter regions of responsive genes [5,6]. Interestingly, ΔFosB activates certain genes (e.g. cdk5) [12,17,35] and represses others (e.g. c-fos under certain conditions) [12,57]. Recent studies have found that at the cdk5 gene, which is upregulated after chronic cocaine [35], ΔFosB binds to the promoter and recruits transcriptional activators, such as the SWI/SNF (switch/sucrose nonfermentable)-remodeling protein BRG1 (BRM/SWI2-related gene 1) (Figure 3a) [17].
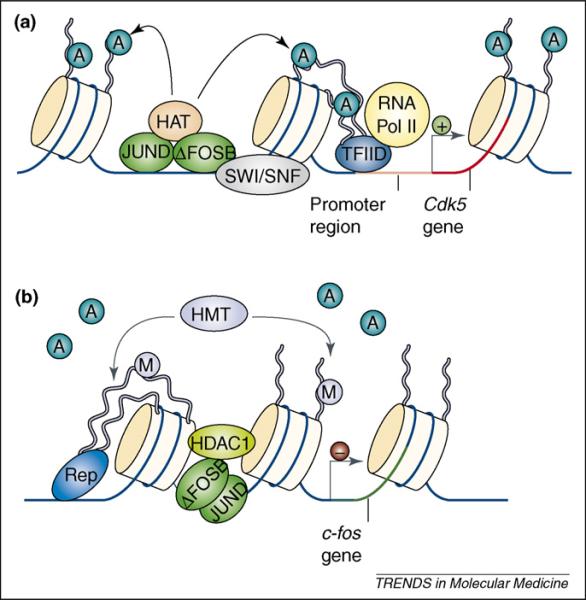
Figure 3.
Gene-dependent recruitment of chromatin-remodeling enzymes. (a) Cocaine- and amphetamine-induced increases in ΔFosB in the nucleus accumbens (NAc) are known to activate transcription of the cdk5 gene. This involves binding of the ΔFosB/JunD heterodimer to the cdk5 promoter and recruitment of the SWI/SNF ATP-dependent chromatin-remodeling complexes and histone acetyltransferases (HATs). This allows for the assembly of RNA polymerase II (RNA Pol II), Transcription Factor IID (TFIID) and several other protein complexes (not shown) involved in the initiation and elongation of transcription. Histone deacetylases (HDACs) are not present on the cdk5 promoter, which permits significantly higher levels of acetylated histone H3 after chronic cocaine exposure. (b) Cocaine- and amphetamine-induced increases in ΔFosB also act as a transcriptional repressor at a different gene locus, c-fos. After repeated stimulant exposure, the c-fos gene is desensitized in the NAc and much more weakly induced by subsequent drug exposures. This involves the binding of ΔFosB to the c-fos gene promoter and recruitment of HDAC1 to reduce histone acetylation and gene activity. In concert with HDAC1, chronic drug exposure increases the levels of the repressive histone methyltransferase (HMT); KMT1a/SUV39H1 and levels of histone H3K9 methylation on the c-fos promoter. Together, these enzymes and histone modifications serve to repress c-fos gene activity through a mechanism involving ΔFosB and other corepressors (Rep). Figure modified with permission from [24].
By contrast, at c-fos, which is repressed after chronic cocaine injections in the animal's homecage [4], ΔFosB binds to the c-fos promoter and recruits HDAC1 to deacetylate nearby histones (Figure 3b) [57]. Importantly, over-expression of ΔFosB alone is sufficient both to upregulate cdk5 and to repress c-fos, nicely illustrating how a single transcription factor can direct distinct chromatin-modifying enzymes to specific genes. The exact mechanisms that mediate the gene-specific effects of ΔFosB remain unclear, but neighboring promoter regulatory elements that recruit distinct transcription factors or unique post-translational modifications might contribute. Moreover, the activating versus repressive effects of ΔFosB might be influenced by drug-associated environmental cues (e.g. via glutamatergic inputs from cerebral cortex) because c-fos induction is higher in rats repeatedly injected with cocaine in a novel environment [58]. Such interplay between transcription factors and chromatin-remodeling enzymes suggests that developing small molecules that stabilize or disrupt these complexes could provide a new avenue for addiction treatment.
Future studies
Drug-induced alterations in chromatin structure have now been implicated in both the pathogenesis and maintenance of the addicted state. An important area for future research is to translate these findings from simple behavioral models, such as conditioned place preference and locomotor responses, to self-administration and relapse paradigms, which better model the human syndrome. Moreover, cocaine and other related stimulant addiction makes up only a small part of the substance abuse problem, making it equally important to focus on other drugs of abuse, such as opiates, nicotine, ethanol, cannabinoids and inhalants (see Box 2). Indeed, progress is being made, as several recent findings have begun to explore other drugs of abuse. In the amygdala, for example, regulation of histone acetylation was observed in ethanol-treated rats, where acute ethanol exposure reduces HDAC activity and increases global levels of histone H3 and H4 acetylation, and withdrawal from chronic ethanol has the opposite effect (increased HDAC activity and reduced histone acetylation) (Figure 1) [59]. Changes in acetylation have also been implicated in the molecular and behavioral responses to an inhalant in Drosophila [60].
Another area for future research is to dissect the many types of potential changes in chromatin structure beyond histone acetylation that also occur after drug exposure. Perhaps a place to start is to explore those marks known to be regulated in brain in other behavioral models, such as the long-lasting histone H3K27 methylation seen in a mouse model of depression [42] or DNA methylation seen in rats with poor maternal care [61]. These mechanisms might contribute to the longer-lasting components of addiction, such as withdrawal symptoms and relapse, because they are generally more stable in cell-culture models than histone acetylation and might persist in vivo for as long as the addicted state. In addition to numerous types of histone modifications and DNA methylation, other mechanisms of chromatin regulation, such as ATP-dependent chromatin remodeling by SWI/SNF proteins and histone replacement (e.g. H3.1,3.3, H2A.Z), might be important pathways for drugs of abuse as well. However, regardless of the epigenetic mechanism studied, we must ultimately understand what is happening to the underlying expression of specific gene targets. In theory, any epigenetic change should be manifested by a change in gene expression or, as in the case of BDNF during cocaine withdrawal, the transcriptional potential of a gene that ultimately alters neural function [17].
Box 2. Outstanding questions.
How long after chronic cocaine exposure do changes in chromatin structure persist?
On which genes do cocaine-induced changes in chromatin structure occur? How is the activity of these genes regulated?
What other types of epigenetic mechanisms (e.g. methylation, ubiquitination, sumoylation, ATP-dependent nucleosome remodeling, etc.) are important in drug addiction?
Does pharmacological or genetic manipulation of chromatin-remodeling enzymes alter how an animal self-administers cocaine?
Do epigenetic mechanisms control drug relapse after prolonged withdrawal?
How does cocaine regulate chromatin structure in specific cell types (e.g. neurons, glia, dopamine D1R-positive, dopamine D2R-positive, etc.)?
How is chromatin regulated in other brain regions involved in drug addiction?
How is chromatin regulated by other drugs of abuse?
Concluding remarks
There is now growing evidence that epigenetic mechanisms, such as histone acetylation, are involved in the regulation of the saliency of environmental stimuli in several behavioral models [17-19,24,33,34,42,51,62]. This has important implications for the pathogenesis of drug addiction and other neuropsychiatric disorders because novel therapeutics could target such mechanisms to block or even reverse the transition from recreational drug use to a chronically addicted state. Likewise, epigenetic mechanisms are attractive candidates for molecular substrates that mediate long-lived changes in brain, such as drug addiction and relapse. General support for this idea comes from rats subjected to poor maternal care, which is associated with changes in chromatin structure that persist into adulthood and with behavioral abnormalities that can be reversed by HDAC inhibitors [63]. However, it has yet to be determined whether similar long-lasting changes in chromatin structure are involved in the maintenance of drug addiction. If so, the ability to reverse the epigenetic signature of an addicted state would offer a fundamentally new approach for more effective treatments of drug relapse.
Acknowledgements
Preparation of this review was supported by grants from the National Insitute on Drug Abuse (E.J.N.) and the University of Texas Southwestern Medical Scientist Training Program (W.R.).
References
- 1.Hyman SE, et al. Neural mechanisms of addiction: the role of reward-related learning and memory. Annu. Rev. Neurosci. 2006;29:565–598. doi: 10.1146/annurev.neuro.29.051605.113009. [DOI] [PubMed] [Google Scholar]
- 2.Kalivas PW, et al. Unmanageable motivation in addiction: a pathology in prefrontal-accumbens glutamate transmission. Neuron. 2005;45:647–650. doi: 10.1016/j.neuron.2005.02.005. [DOI] [PubMed] [Google Scholar]
- 3.Koob G, Kreek MJ. Stress, dysregulation of drug reward pathways, and the transition to drug dependence. Am. J. Psychiatry. 2007;164:1149–1159. doi: 10.1176/appi.ajp.2007.05030503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hope BT, et al. Induction of a long-lasting AP-1 complex composed of altered Fos-like proteins in brain by chronic cocaine and other chronic treatments. Neuron. 1994;13:1235–1244. doi: 10.1016/0896-6273(94)90061-2. [DOI] [PubMed] [Google Scholar]
- 5.Kelz MB, et al. Expression of the transcription factor deltaFosB in the brain controls sensitivity to cocaine. Nature. 1999;401:272–276. doi: 10.1038/45790. [DOI] [PubMed] [Google Scholar]
- 6.McClung CA, et al. ΔFosB: a molecular switch for long-term adaptation in the brain. Brain Res. Mol. Brain Res. 2004;132:146–154. doi: 10.1016/j.molbrainres.2004.05.014. [DOI] [PubMed] [Google Scholar]
- 7.Bowers MS, et al. Activator of G protein signaling 3: a gatekeeper of cocaine sensitization and drug seeking. Neuron. 2004;42:269–281. doi: 10.1016/s0896-6273(04)00159-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Grimm JW, et al. Time-dependent increases in brain-derived neurotrophic factor protein levels within the mesolimbic dopamine system after withdrawal from cocaine: implications for incubation of cocaine craving. J. Neurosci. 2003;23:742–747. doi: 10.1523/JNEUROSCI.23-03-00742.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Graham DL, et al. Dynamic BDNF activity in nucleus accumbens with cocaine use increases self-administration and relapse. Nat. Neurosci. 2007;10:1029–1037. doi: 10.1038/nn1929. [DOI] [PubMed] [Google Scholar]
- 10.Lu L, et al. A single infusion of brain-derived neurotrophic factor into the ventral tegmental area induces long-lasting potentiation of cocaine seeking after withdrawal. J. Neurosci. 2004;24:1604–1611. doi: 10.1523/JNEUROSCI.5124-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Freeman WM, et al. Chronic cocaine-mediated changes in non-human primate nucleus accumbens gene expression. J. Neurochem. 2001;77:542–549. doi: 10.1046/j.1471-4159.2001.00252.x. [DOI] [PubMed] [Google Scholar]
- 12.McClung CA, Nestler EJ. Regulation of gene expression and cocaine reward by CREB and ΔFosB. Nat. Neurosci. 2003;6:1208–1215. doi: 10.1038/nn1143. [DOI] [PubMed] [Google Scholar]
- 13.McClung CA, et al. Regulation of gene expression by chronic morphine and morphine withdrawal in the locus ceruleus and ventral tegmental area. J. Neurosci. 2005;25:6005–6015. doi: 10.1523/JNEUROSCI.0062-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Winstanley CA, et al. ΔFosB induction in orbitofrontal cortex mediates tolerance to cocaine-induced cognitive dysfunction. J. Neurosci. 2007;27:10497–10507. doi: 10.1523/JNEUROSCI.2566-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yao WD, et al. Identification of PSD-95 as a regulator of dopamine-mediated synaptic and behavioral plasticity. Neuron. 2004;41:625–638. doi: 10.1016/s0896-6273(04)00048-0. [DOI] [PubMed] [Google Scholar]
- 16.Yuferov V, et al. Differential gene expression in the rat caudate putamen after ‘binge’ cocaine administration: advantage of triplicate microarray analysis. Synapse. 2003;48:157–169. doi: 10.1002/syn.10198. [DOI] [PubMed] [Google Scholar]
- 17.Kumar A, et al. Chromatin remodeling is a key mechanism underlying cocaine-induced plasticity in striatum. Neuron. 2005;48:303–314. doi: 10.1016/j.neuron.2005.09.023. [DOI] [PubMed] [Google Scholar]
- 18.Levine AA, et al. CREB-binding protein controls response to cocaine by acetylating histones at the fosB promoter in the mouse striatum. Proc. Natl. Acad. Sci. U. S. A. 2005;102:19186–19191. doi: 10.1073/pnas.0509735102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Renthal W, et al. Histone deacetylase 5 epigenetically controls behavioral adaptations to chronic emotional stimuli. Neuron. 2007;56:517–529. doi: 10.1016/j.neuron.2007.09.032. [DOI] [PubMed] [Google Scholar]
- 20.Feinberg AP. Phenotypic plasticity and the epigenetics of human disease. Nature. 2007;447:433–440. doi: 10.1038/nature05919. [DOI] [PubMed] [Google Scholar]
- 21.Schuettengruber B, et al. Genome regulation by polycomb and trithorax proteins. Cell. 2007;128:735–745. doi: 10.1016/j.cell.2007.02.009. [DOI] [PubMed] [Google Scholar]
- 22.Grewal SI, Moazed D. Heterochromatin and epigenetic control of gene expression. Science. 2003;301:798–802. doi: 10.1126/science.1086887. [DOI] [PubMed] [Google Scholar]
- 23.Siegmund KD, et al. DNA methylation in the human cerebral cortex is dynamically regulated throughout the life span and involves differentiated neurons. PLoS ONE. 2007;2:e895. doi: 10.1371/journal.pone.0000895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tsankova N, et al. Epigenetic regulation in psychiatric disorders. Nat. Rev. Neurosci. 2007;8:355–367. doi: 10.1038/nrn2132. [DOI] [PubMed] [Google Scholar]
- 25.Luger K, Richmond TJ. The histone tails of the nucleosome. Curr. Opin. Genet. Dev. 1998;8:140–146. doi: 10.1016/s0959-437x(98)80134-2. [DOI] [PubMed] [Google Scholar]
- 26.Felsenfeld G, Groudine M. Controlling the double helix. Nature. 2003;421:448–453. doi: 10.1038/nature01411. [DOI] [PubMed] [Google Scholar]
- 27.Li B, et al. The role of chromatin during transcription. Cell. 2007;128:707–719. doi: 10.1016/j.cell.2007.01.015. [DOI] [PubMed] [Google Scholar]
- 28.Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 29.Strahl BD, Allis CD. The language of covalent histone modifications. Nature. 2000;403:41–45. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- 30.Kurdistani SK, et al. Mapping global histone acetylation patterns to gene expression. Cell. 2004;117:721–733. doi: 10.1016/j.cell.2004.05.023. [DOI] [PubMed] [Google Scholar]
- 31.Pokholok DK, et al. Genome-wide map of nucleosome acetylation and methylation in yeast. Cell. 2005;122:517–527. doi: 10.1016/j.cell.2005.06.026. [DOI] [PubMed] [Google Scholar]
- 32.Brami-Cherrier K, et al. Parsing molecular and behavioral effects of cocaine in mitogen- and stress-activated protein kinase-1-deficient mice. J. Neurosci. 2005;25:11444–11454. doi: 10.1523/JNEUROSCI.1711-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fischer A, et al. Recovery of learning and memory is associated with chromatin remodelling. Nature. 2007;447:178–182. doi: 10.1038/nature05772. [DOI] [PubMed] [Google Scholar]
- 34.Levenson JM, et al. Regulation of histone acetylation during memory formation in the hippocampus. J. Biol. Chem. 2004;279:40545–40559. doi: 10.1074/jbc.M402229200. [DOI] [PubMed] [Google Scholar]
- 35.Bibb JA, et al. Effects of chronic exposure to cocaine are regulated by the neuronal protein Cdk5. Nature. 2001;410:376–380. doi: 10.1038/35066591. [DOI] [PubMed] [Google Scholar]
- 36.Freeman WM, et al. Persistent alterations in mesolimbic gene expression with abstinence from cocaine self-administration. Neuropsychopharmacology. 2007;33:1807–1817. doi: 10.1038/sj.npp.1301577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Brami-Cherrier K, et al. Glutamate induces histone H3 phosphorylation but not acetylation in striatal neurons: role of mitogen- and stress-activated kinase-1. J. Neurochem. 2007;101:697–708. doi: 10.1111/j.1471-4159.2006.04352.x. [DOI] [PubMed] [Google Scholar]
- 38.Li J, et al. Dopamine D2-like antagonists induce chromatin remodeling in striatal neurons through cyclic AMP-protein kinase A and NMDA receptor signaling. J. Neurochem. 2004;90:1117–1131. doi: 10.1111/j.1471-4159.2004.02569.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vakoc CR, et al. Histone H3 lysine 9 methylation and HP1gamma are associated with transcription elongation through mammalian chromatin. Mol. Cell. 2005;19:381–391. doi: 10.1016/j.molcel.2005.06.011. [DOI] [PubMed] [Google Scholar]
- 40.Black YD, et al. Altered attention and prefrontal cortex gene expression in rats after binge-like exposure to cocaine during adolescence. J. Neurosci. 2006;26:9656–9665. doi: 10.1523/JNEUROSCI.2391-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Maze I, et al. Histone methylation in the nucleus accumbens controls behavioral responses to cocaine. Soc. Neurosci. Abs. (in press) [Google Scholar]
- 42.Tsankova NM, et al. Sustained hippocampal chromatin regulation in a mouse model of depression and antidepressant action. Nat. Neurosci. 2006;9:519–525. doi: 10.1038/nn1659. [DOI] [PubMed] [Google Scholar]
- 43.Kumar A, et al. Genome-wide epigenetic changes underlying chronic cocaine-induced neuroadaptations in the mouse nucleus accumbens. Soc. Neurosci. Abs. 2007;767:5. [Google Scholar]
- 44.Tsankova NM, et al. Histone modifications at gene promoter regions in rat hippocampus after acute and chronic electroconvulsive seizures. J. Neurosci. 2004;24:5603–5610. doi: 10.1523/JNEUROSCI.0589-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li F, et al. Chimeric DNA methyltransferases target DNA methylation to specific DNA sequences and repress expression of target genes. Nucleic Acids Res. 2007;35:100–112. doi: 10.1093/nar/gkl1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Smith AE, et al. Heritable gene repression through the action of a directed DNA methyltransferase at a chromosomal locus. J. Biol. Chem. 2008;283:9878–9885. doi: 10.1074/jbc.M710393200. [DOI] [PubMed] [Google Scholar]
- 47.Kalda A, et al. Histone deacetylase inhibitors modulates the induction and expression of amphetamine-induced behavioral sensitization partially through an associated learning of the environment in mice. Behav. Brain Res. 2007;181:76–84. doi: 10.1016/j.bbr.2007.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schroeder FA, et al. Drug-induced activation of dopamine D(1) receptor signaling and inhibition of class I/II histone deacetylase induce chromatin remodeling in reward circuitry and modulate cocaine-related behaviors. Neuropsychopharmacology. 2008 doi: 10.1038/npp.2008.15. DOI: 10.1038/npp.2008.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fischle W, et al. Enzymatic activity associated with class II HDACs is dependent on a multiprotein complex containing HDAC3 and SMRT/N-CoR. Mol. Cell. 2002;9:45–57. doi: 10.1016/s1097-2765(01)00429-4. [DOI] [PubMed] [Google Scholar]
- 50.Chang S, et al. Histone deacetylases 5 and 9 govern responsiveness of the heart to a subset of stress signals and play redundant roles in heart development. Mol. Cell. Biol. 2004;24:8467–8476. doi: 10.1128/MCB.24.19.8467-8476.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Renthal W, et al. Histone deacetylase 5 controls analgesic responsiveness to tricyclic antidepressants in a mouse model of neurophatic pain. Soc. Neurosci. Abs. (in press) [Google Scholar]
- 52.Schroeder FA, et al. Antidepressant-like effects of the histone deacetylase inhibitor, sodium butyrate, in the mouse. Biol. Psychiatry. 2007;62:55–64. doi: 10.1016/j.biopsych.2006.06.036. [DOI] [PubMed] [Google Scholar]
- 53.Zhang J, et al. c-Fos facilitates the acquisition and extinction of cocaine-induced persistent changes. J. Neurosci. 2006;26:13287–13296. doi: 10.1523/JNEUROSCI.3795-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lee MG, et al. Histone H3 lysine 4 demethylation is a target of nonselective antidepressive medications. Chem. Biol. 2006;13:563–567. doi: 10.1016/j.chembiol.2006.05.004. [DOI] [PubMed] [Google Scholar]
- 55.Carlezon WA, Jr, et al. Regulation of cocaine reward by CREB. Science. 1998;282:2272–2275. doi: 10.1126/science.282.5397.2272. [DOI] [PubMed] [Google Scholar]
- 56.Mayr B, Montminy M. Transcriptional regulation by the phosphorylation-dependent factor CREB. Nat. Rev. Mol. Cell Biol. 2001;2:599–609. doi: 10.1038/35085068. [DOI] [PubMed] [Google Scholar]
- 57.Renthal W, et al. ΔFosB mediates epigenetic desensitization of the cfos gene after chronic amphetamine exposure. J. Neurosci. doi: 10.1523/JNEUROSCI.1043-08.2008. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hope BT, et al. Cocaine-induced locomotor activity and Fos expression in nucleus accumbens are sensitized for 6 months after repeated cocaine administration outside the home cage. Eur. J. Neurosci. 2006;24:867–875. doi: 10.1111/j.1460-9568.2006.04969.x. [DOI] [PubMed] [Google Scholar]
- 59.Pandey SC, et al. Brain chromatin remodeling: a novel mechanism of alcoholism. J. Neurosci. 2008;28:3729–3737. doi: 10.1523/JNEUROSCI.5731-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wang Y, et al. Drug-induced epigenetic changes produce drug tolerance. PLoS Biol. 2007;5:2342–2353. doi: 10.1371/journal.pbio.0050265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Meaney MJ, Szyf M. Maternal care as a model for experience-dependent chromatin plasticity? Trends Neurosci. 2005;28:456–463. doi: 10.1016/j.tins.2005.07.006. [DOI] [PubMed] [Google Scholar]
- 62.Zhang CL, et al. Class II histone deacetylases act as signal-responsive repressors of cardiac hypertrophy. Cell. 2002;110:479–488. doi: 10.1016/s0092-8674(02)00861-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Weaver IC, et al. Epigenetic programming by maternal behavior. Nat. Neurosci. 2004;7:847–854. doi: 10.1038/nn1276. [DOI] [PubMed] [Google Scholar]
- 64.Maurice T, et al. Altered memory capacities and response to stress in p300/CBP-associated cactor (PCAF) histone acetylase knockout mice. Neuropsychopharmacology. 2008;33:1584–1602. doi: 10.1038/sj.npp.1301551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Oliveira AM, et al. Transgenic mice expressing an inhibitory truncated form of p300 exhibit long-term memory deficits. Learn. Mem. 2007;14:564–572. doi: 10.1101/lm.656907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Doi M, et al. Circadian regulator CLOCK is a histone acetyltransferase. Cell. 2006;125:497–508. doi: 10.1016/j.cell.2006.03.033. [DOI] [PubMed] [Google Scholar]
- 67.Kawasaki H, et al. ATF-2 has intrinsic histone acetyltransferase activity which is modulated by phosphorylation. Nature. 2000;405:195–200. doi: 10.1038/35012097. [DOI] [PubMed] [Google Scholar]
- 68.Chawla S, et al. Neuronal activity-dependent nucleocytoplasmic shuttling of HDAC4 and HDAC5. J. Neurochem. 2003;85:151–159. doi: 10.1046/j.1471-4159.2003.01648.x. [DOI] [PubMed] [Google Scholar]
- 69.Haigis MC, Guarente LP. Mammalian sirtuins – emerging roles in physiology, aging, and calorie restriction. Genes Dev. 2006;20:2913–2921. doi: 10.1101/gad.1467506. [DOI] [PubMed] [Google Scholar]
- 70.Yang XJ, Seto E. The Rpd3/Hda1 family of lysine deacetylases: from bacteria and yeast to mice and men. Nat. Rev. Mol. Cell Biol. 2008;9:206–218. doi: 10.1038/nrm2346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Miller CA, Sweatt JD. Covalent modification of DNA regulates memory formation. Neuron. 2007;53:857–869. doi: 10.1016/j.neuron.2007.02.022. [DOI] [PubMed] [Google Scholar]
- 72.Stipanovich A, et al. A phosphatase cascade by which rewarding stimuli control nucleosomal response. Nature. 2008;453:879–884. doi: 10.1038/nature06994. [DOI] [PMC free article] [PubMed] [Google Scholar]