Abstract
Protein biomarker discovery produces lengthy lists of candidates that must subsequently be verified in blood or other accessible biofluids. Use of targeted mass spectrometry (MS) to verify disease- or therapy-related changes in protein levels requires the selection of peptides that are quantifiable surrogates for proteins of interest. Peptides that produce the highest ion-current response (high-responding peptides) are likely to provide the best detection sensitivity. Identification of the most effective signature peptides, particularly in the absence of experimental data, remains a major resource constraint in developing targeted MS—based assays. Here we describe a computational method that uses protein physicochemical properties to select high-responding peptides and demonstrate its utility in identifying signature peptides in plasma, a complex proteome with a wide range of protein concentrations. Our method, which employs a Random Forest classifier, facilitates the development of targeted MS—based assays for biomarker verification or any application where protein levels need to be measured.
Proteomic discovery experiments in case-and-control comparisons of tissue or proximal fluids frequently generate lists comprising many tens to hundreds of candidate biomarkers1. Integrative genomic approaches incorporating microarray data and literature mining are also increasingly being used to guide identification of candidate protein biomarkers. To further credential biomarker candidates and move them toward possible clinical implementation, it is necessary to determine which of the proteins from lists of candidates differentially abundant in diseased versus healthy patients can be detected in body fluids, such as blood, that can be assayed with minimal invasiveness1.
This process, termed verification, has historically been approached using antibodies. High-quality, well-characterized collections of antibodies suitable for protein detection in tissue are now being developed2. But unfortunately, the required immunoassay-grade antibody pairs necessary for sensitive and specific detection in blood exist for only a tiny percentage of the proteome. Thus, for the majority of proteins, suitable reagents for their detection and quantification in blood (or other biofluids) do not yet exist and alternative technologies are needed to bridge the gap between discovery and clinical-assay development. This problem is an important aspect of the larger need in biology and medicine for quantitative methods to measure the presence and abundance of any protein of interest.
Targeted MS is emerging as an assay technology capable of selective and sensitive detection and quantification of potentially any protein of interest (or modification thereof) in the proteome3–6. In stable isotope dilution—multiple reaction monitoring (MRM)-MS, peptides (precursors) from candidate proteins of interest are selectively detected and caused to fragment (products) in the mass spectrometer. The resulting product ions are used to quantify the peptide, and therefore, the protein from which it was derived, by calculating the ratio of the signal response of the endogenous peptide to a stable isotope—labeled version of the peptide added as an internal standard3–6.
The first step in developing an MRM-MS—based assay involves selecting a subset of peptides to use as quantitative surrogates for each candidate protein. ‘Signature peptides’1 correspond to the subset of ‘proteotypic peptides’7 that, in addition to being sequence unique and detectable, are also the highest responding peptides for each protein. Current methods rely on selecting signature peptides based on detection in the initial MS discovery data3,5, identification in databases of MS experimental data8,9 or computational approaches to predict proteotypic peptides10–13. When multiple peptides are detected for a candidate protein for which experimental data are available, selection is primarily based on high peptide-response. Other considerations such as high-performance liquid chromatography (HPLC) retention time, amino acid composition, uniqueness in the genome and charge state also play a role. After selecting signature peptides, the targeted MRM-MS assay must be optimized for each peptide to select appropriate precursor-to-product ion transitions5,14. Because some peptides fail the optimization process due to poor chromatography, solubility problems, interference with matrix or failure to recover the peptide after digestion in plasma, it is common for laboratories to evaluate approximately five peptides per protein. This usually insures that at least one peptide per protein is suitable for developing a quantitative assay3,5.
Two key problems usually arise with the selection of signature peptides for assay development. First, only a fraction of peptides present in a complex sample are detected in discovery proteomic experiments. This undersampling problem is well known and leads to poor reproducibility of peptide and protein detection, even in replicate samples15. As a result, the best signature peptides for any given candidate may not be the ones observed in the discovery experiment. Second, it is of interest to quantify candidate proteins identified by methods other than proteomics, such as genomic experiments or literature mining. These candidate proteins may represent biomarkers or key components in signaling or metabolic pathways. In these situations, de novo prediction of signature peptides is required.
Here we describe the enhanced signature peptide (ESP) predictor, a computational method to predict high-responding peptides from a given protein. We (i) validate the method on ten diverse experimental data sets not used in training the ESP predictor, (ii) show that ESP predictions are significantly better at selecting high-responding peptides than existing computational methods10,12,13, (iii) demonstrate that the ESP predictor can be used to define the best peptides for targeted MRM-MS—based assay development in the absence of experimental proteomic data for the protein and (iv) identify the most relevant physicochemical properties used to predict high-responding peptides in the context of electrospray ionization (ESI)-MS.
RESULTS
Method overview
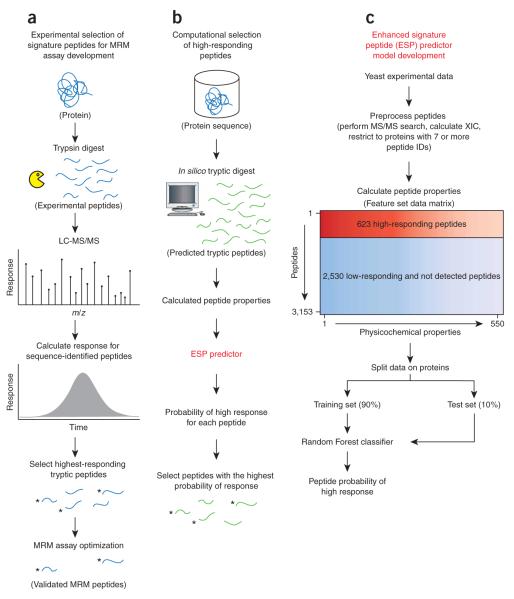
We developed a model to predict the probability that a peptide from a given protein will generate a high response in an ESI-MS experiment. We define peptide response as the sum of the extracted ion chromatogram (XIC) based on the monoisotopic peak for all charge states and modifications detected from sequence-identified peptides. This measure is more consistent with the intended application of the ESP predictor, which is to predict signature peptides from an in silico digest of a candidate protein (Fig. 1a,b).
Figure 1.
ESP application and model development overview. (a) A typical proteomic workflow to select signature peptides for targeted protein analysis using MRM. Candidate proteins are experimentally analyzed, and five signature peptides per protein are selected based primarily on high peptide-response and sequence composition, among other factors. After optimization, the remaining peptides are referred to as validated MRM peptides. (b) We computationally digest each candidate protein, in silico (no missed cleavages, 600–2,800 Da), to produce a set of predicted tryptic peptides. Peptide sequences are input into the ESP predictor and we select the five peptides with the highest probability of response for each protein. To validate the ESP predictions, we compare the top five predicted peptides to the experimentally determined five highest-responding peptides from a, denoted by asterisks (3 out of 5, in this example). (c) We developed the ESP predictor using peptides from a yeast lysate experimental analysis. We trained the ESP predictor using Random Forest on 90% of the peptides and held out 10% to test the model, referred to as Yeast test. We split the data at the protein level to avoid any bias in training and testing the model on peptides from the same protein and to keep the training and test data completely separated.
We used liquid chromatography (LC)-ESI-MS analyses of a yeast lysate sample, from three proteomic laboratories, to derive a training set to model peptide response (Fig. 1c). For each protein, we standardized the peptide response, using the z-score (z), and selected a threshold to define ‘high’ (z ≥ 0) and ‘low’ (z ≤ −1) responding peptides. We also derived a set of ‘not detected’ peptides from an in silico tryptic digest (no missed cleavages, mass 600–2,800 Da), but we considered only peptides not sequence identified in any form, including missed cleavages. Because we are only interested in detecting high-responding peptides, we combined the ‘low’ and ‘not detected’ peptides together to create the final training set of ‘high’ versus ‘low/not detected’.
To develop a predictive model, one must encode the peptides as an n-dimensional property vector. These properties represent specific characteristics of the peptides such as mass, hydrophobicity and gasphase basicity. We considered 550 physicochemical properties (Supplementary Table 1 online) to model peptide response16,17. For each physicochemical property, we computed the property value by averaging over all amino acids in each peptide. Thus, the training set comprised a matrix of ‘peptides by properties’ along with the class labels, ‘high’ or ‘low/not detected’.
We modeled peptide response using the Random Forest18 algorithm. Random Forest is a nonlinear ensemble classifier composed of many individual decision trees. We chose Random Forest because the algorithm, and its R implementation19, conveniently includes many features especially suited to this type of analysis. Specifically, Random Forest effectively handles data sets with large numbers of correlated features, provides insight into the model by determining the most relevant properties during training18–21 and exhibits better performance for this data set than using a Support Vector Machine22 does. Notably, the structure of the decision trees that make up the final model are learned using only the training set, and the model is fixed for subsequent testing and validation.
We also attempted to reduce the dimensionality of the training set by considering two feature-selection techniques, Fisher Criterion Score23 and the area under the receiver operator curve (ROC)24. We used the best features ranked by each of the feature selection methods to build Support Vector Machine models using three different kernels and Random Forest. Random Forest exhibited the best performance using all 550 properties, implying that feature selection is not helpful in this context (Supplementary Fig. 1 online).
Metrics to evaluate the ESP predictor
We created the ESP predictor to select high-responding peptides from candidate proteins, in the absence of MS experimental data, with the intention of developing an MRM-MS assay. Therefore, we developed metrics to assess the success of such predictions. When developing an MRM-MS assay, it is necessary to evaluate the assay performance of about five peptides per protein in the biological matrix of interest (typically plasma) to reliably obtain at least one peptide with suitable limits of detection and quantification. The expense and time associated with generating synthetic peptides and evaluating the assay performance of each for MRM quantification (typically involving generation of a ten-point concentration response curve for each peptide) make evaluation of more than five peptides per protein impractical.
We evaluated the ESP predictor on ten validation sets not used in training to assess its performance (Table 1). We experimentally analyzed each validation set using ESI-MS and selected the five highest-responding, fully tryptic (no missed cleavages) peptides from each protein (Fig. 1a). Then, using the ESP predictor, we ranked the predicted probability of high response for all tryptic peptides generated from an in silico digest of the same proteins and selected the top five peptides for each protein (Fig. 1b). We calculated two metrics designed to assess how well the ESP predictor selected the five highest-responding peptides for all proteins in each validation set. First, we calculated the protein sensitivity, which is the percent of proteins with one or more peptides predicted by the ESP predictor to be among the five highest responding. Second, we calculated a P-value to test the hypothesis that the ESP predictions are significantly better than random predictions, using a permutation test. In gauging the performance of the ESP predictor, a combination of high protein sensitivity and low P-value is desirable. A high protein sensitivity indicates that more proteins in the data set have at least one correctly predicted high-responding peptide, whereas statistical significance requires P < 0.05. We also compared the ESP predictor to three publicly available computational methods for predicting proteotypic peptides10,12,13.
Table 1.
Description of validation sets
| Validation seta | Experiment type (ESI) |
Proteinsb | Theoretical peptidesc |
PS ≥1d | PS ≥2e | Tsf | Mixture complexityg |
Database search |
Quantification |
|---|---|---|---|---|---|---|---|---|---|
| ISB-18 | LC-MS | 6 | 153 | 100% | 100% | 17h | Low | Spectrum Mill | XIC |
| Yeast test | LC-MS | 8 | 226 | 100% | 88% | 21h | Medium | Spectrum Mill | XIC |
| Plasma | LC-MS | 14 | 633 | 71% | 36% | 16i | Very High | Spectrum Mill | XIC |
| Sigma48 | LC-MS | 16 | 438 | 88% | 69% | 34h | Low | Spectrum Mill | XIC |
| Plasma Hu14 | LC-MS | 30 | 1,403 | 87% | 43% | 43h | Very high | Spectrum Mill | XIC |
| Yeast_2 | LC-MS | 94 | 1,930 | 97% | 82% | 242h | Medium | Spectrum Mill | XIC |
| HeLa_1 | LC-MS | 149 | 4,944 | 90% | 65% | 301h | High | Mascot | MSQuant |
| HeLa_2 | GeLC-MS | 300 | 15,172 | 86% | 54% | 498h | High | Mascot | MSQuant |
| Pull-down | GeLC-MS | 172 | 8,062 | 92% | 68% | 358h | Medium | Mascot | MSQuant |
| Plasma Hu14 SCX | SCX-LC-MS | 45 | 1,935 | 93% | 49% | 74h | High | Spectrum Mill | XIC |
All validation sets were analyzed using an LTQ-Orbitrap except the plasma and ISB-18 data, which were analyzed using an LTQ-FT.
Only proteins with six or more theoretical peptides (in silico digest) and at least five sequence-identified peptides were considered for validation.
In silico tryptic digest with no missed cleavages and a mass range of 600–2,800 Da.
Protein sensitivity (PS). The percent of proteins with one or more peptides predicted by the ESP predictor to be among the five highest responding. The weighted mean of all validation sets based on number of proteins is 89%.
The percent of proteins with two or more peptides predicted by the ESP predictor to be among the five highest responding. The weighted mean of all validation sets based on number of proteins is 60%.
Test statistic (Ts). The sum of correctly predicted peptides among the five highest-responding peptides for all proteins in the validation set.
Simple comparison of the number of proteins present in each sample mixture. For example, plasma has more than 10,000 proteins (very high) compared to sigma48, which has 48 proteins (low).
P < 0.0001
P = 0.0363 based on null distribution for the entire validation set, by permutation test.
Validation of the ESP predictor
We wanted to demonstrate the advantage of applying a single model to predict high-responding peptides in varied data spanning a wide range of different ESI experimental types, mixture complexities, database search algorithms and XIC quantification methods. For a fair assessment of how well the ESP predictor selects the five highest-responding peptides, we restricted the validation sets to proteins with six or more theoretical peptides and five or more sequence-identified peptides. The results indicate the ESP predictor performance is consistent across all ten validation sets despite very different types of proteomic data (Table 1). On average, the ESP predictor achieves a success rate of 89% at selecting one or more high-responding peptides per protein. Across all validation sets, the ESP predictor correctly selects approximately two out of five high-responding peptides from an average of 42 theoretical peptides per protein.
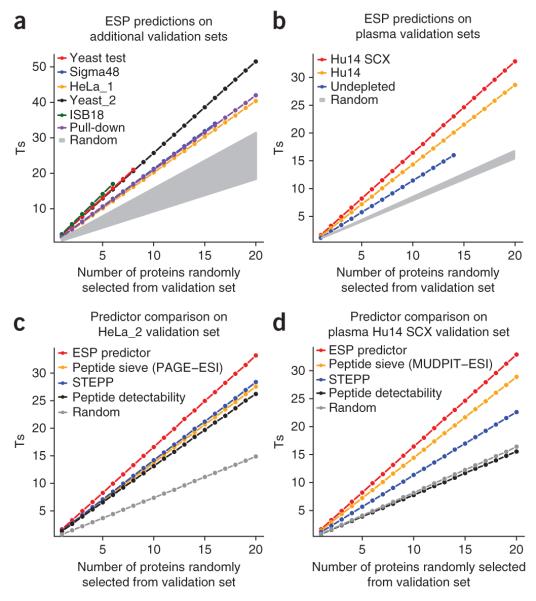
Next, we used a permutation test to confirm that the ESP predictions are statistically more significant, across multiple proteins, than random predictions and current computational methods (Fig. 2 and Supplementary Fig. 2 online). The predictions on nine of the ten validation sets tested were significantly better than random (P < 0.0001). Only the predictions on the most complex mixture, undepleted plasma, were less significant (P = 0.036). The predictions for the undepleted plasma are better understood in the context of predictions for the Plasma Hu14 (with the 14 most abundant proteins depleted) and Plasma Hu14 SCX (depleted and fractionated) validation sets (Fig. 2b). The number of correct peptides selected significantly increases (Table 1) as the mixture complexity decreases, suggesting less ion suppression and better quantification due to less interference.
Figure 2.
ESP predictor validation and method comparison. ESP predictions outperform existing computation models and are statistically significant for all validation data sets based on a random permutation test. We plotted the mean number of cumulative correctly predicted peptides (Ts) for random combinations of 1–20 proteins. We calculated the 95% confidence interval of the mean, but the error bars were too small to display. The null distribution for P-value calculation is derived using a predictor that randomly selects the top five high-responding peptides for a protein (Supplementary Fig. 2). (a) ESP predictor performance on multiple validation sets, with the performance of a random predictor shown in gray. Each validation set produces its own set of random distributions, depending on the number of peptides per protein. We grouped all random distributions into a single shaded area. (b) ESP predictions on plasma validation sets. The samples represent undepleted plasma, top 14 most-abundant proteins depleted, and depleted and then fractionated using SCX (also referred to as MUDPIT). Random selection of the top five peptides resulted in the gray area. (c) Comparison between the ESP predictor, proteotypic predictors and random predictions on a HeLa GeLC-MS cell lysate. (d) Comparison between the ESP predictor, proteotypic predictors and random predictions on a depleted and fractionated plasma sample. This is the sample type most commonly used for MRM biomarker verification. See Tables 1 and 2 for more details. STEPP, SVM technique for evaluating proteotypic peptides.
We also compared the performance of the ESP predictor on the HeLa_2 and Plasma Hu14 SCX validation sets to three computational methods designed to predict proteotypic peptides (Table 2). We demonstrate, using the HeLa_2 and Plasma Hu14 SCX validation sets, that our method for selecting high-responding peptides performs significantly better (based on Ts, Table 2) than methods designed to predict proteotypic peptides (Fig. 2c,d). Compared to the HeLa_2 validation set, these other methods exhibit more variability with the Plasma Hu14 SCX validation set, whereas ESP still performed well. Performance on fractionated plasma is especially important because it represents a sample type frequently used in MRM biomarker verification. It is relevant to note that these studies constitute the first evaluation of the performance of peptide response predictors in the context of plasma, the most difficult proteome of all with respect to complexity and dynamic range of protein abundance.
Table 2.
Comparison of computational methods
| Method | Validation set | PS ≥1a | PS ≥2b | Tsc |
|---|---|---|---|---|
| ESP Predictor | HeLa_2 (GeLC-MS) | 86% | 54% | 498d |
| STEPP13 | HeLa_2 (GeLC-MS) | 80% | 44% | 425d |
| Peptide sieve (PAGE-ESI)10 | HeLa_2 (GeLC-MS) | 77% | 43% | 413d |
| Peptide detectability12 | HeLa_2 (GeLC-MS) | 77% | 41% | 394d |
| ESP predictor | Plasma Hu14 SCX | 93% | 49% | 74d |
| Peptide sieve (MUDPIT-ESI) | Plasma Hu14 SCX | 82% | 46% | 65d |
| STEPP | Plasma Hu14 SCX | 69% | 36% | 51e |
| Peptide detectability | Plasma Hu14 SCX | 62% | 13% | 35f |
The ESP predictor demonstrates the best performance compared to existing computational methods. Refer to Table 1 for additional validation set information. STEPP, SVM technique for evaluating proteotypic peptides.
Protein sensitivity (PS): The percent of proteins with one or more peptides predicted by the ESP predictor to be among the five highest responding.
The percent of proteins with two or more peptides predicted by the ESP predictor to be among the five highest responding.
Test statistic (Ts). The sum of correct peptides among the five highest-responding peptides for all proteins in the validation set.
P < 0.0001
P = 0.0029
P = 0.6685 based on null distribution for the entire validation set, by permutation test.
To further demonstrate the robustness of the ESP predictor, we examined three quantification methods to calculate peptide response using the HeLa_1 validation set. In addition to MSQuant (Table 1 and Fig. 2a), we also searched the raw data using Spectrum Mill, which reports peptide intensity. We also calculated the XIC based on the monoisotopic peak from the raw data. All three methods exhibited similar performance (Supplementary Fig. 3 online). This suggests the ESP predictor is agnostic to the method of calculating peptide response, as long as it is done consistently.
The ESP predictor selects optimal signature peptides for MRM-MS assays
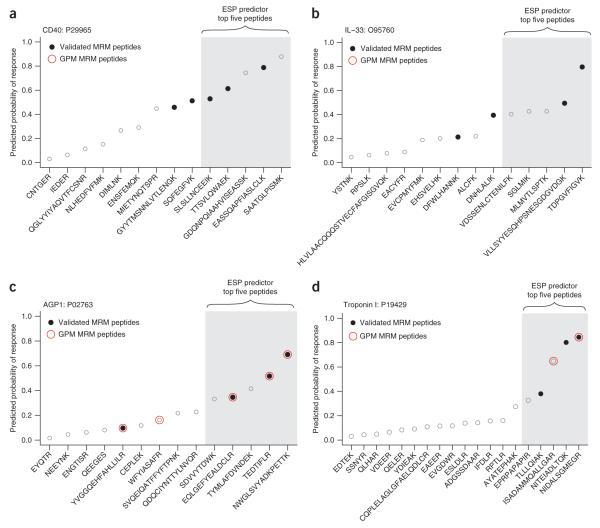
Having validated that the ESP predictor is successful at predicting high-responding peptides, we sought to determine if the predictions can be used to select signature peptides to configure MRM-MS assays in plasma. We tested the ability of the ESP predictor to select the correct signature peptides for a set of 14 proteins (9 cardiovascular biomarkers, 4 nonhuman proteins and prostate-specific antigen). For each of these proteins, we had previously experimentally defined the validated MRM peptides and then configured successful MRM-MS assays using these peptides. We used the ESP predictor to select five candidate signature-peptides and compared the results to the validated MRM peptides for each protein (Fig. 1a,b). The ESP predictor correctly selected two validated MRM peptides per protein, on average, yielding a protein sensitivity of 93% (Fig. 3 and Supplementary Data online for all plots).
Figure 3.
ESP predictions translate into experimentally validated MRM peptides. For each protein, we performed an in silico digest (600–2,800 Da) and ensured that the top five peptides predicted by the ESP predictor were unique in the Swiss-Prot human database. Although additional filtering criteria could easily be applied after analysis with the ESP predictor, we opted for no filtering (except top five uniqueness) to demonstrate the simplicity of using the ESP predictor to select candidate signature-peptides to configure an MRM-MS assay. For all plots, peptides are sorted by the ESP predicted probability of response (y-axis). The actual rank order of measured peptide response is shown in Supplementary Table 2. (a) The ESP predictor correctly selected all three validated MRM peptides (filled black circles) out of the five predicted candidate signature-peptides for troponin I. (b) The ESP predictor correctly selected two validated MRM peptides out of the five predicted candidate signature-peptides for IL-33. In a and b, two representative proteins not found in the GPM database are shown. (c) GPM correctly selected all four of the validated MRM peptides among the top five. Three peptides are common between the ESP predictor and GPM. (d) Only two peptides were suggested by GPM of which only one was a validated MRM peptide. In c and d, two representative proteins are shown where we overlaid the MRM peptides suggested by GPM (open red circles). Example d highlights the limitations of relying solely on database predictions because two validated MRM peptides would have been missed.
We then evaluated the usefulness of a proteomic database in defining signature peptides for MRM-MS assay configuration for these 14 proteins. Using the MRM feature of the Global Proteome Machine (GPM) respository8, a well-known and comprehensive database of proteomic experimental data, we obtained an average of only 0.8 validated MRM peptides per protein. Most importantly, for six of these proteins, no prior MS experimental data existed in GPM (CD40, BNP, HRP, IL-33, leptin, and MBP). For these six proteins, ESP correctly predicted 12 out of 18 validated MRM peptides. Across all 14 proteins, only 11 of the 39 validated MRM peptides were found in GPM, whereas ESP correctly predicted 29 of the 39 validated MRM peptides (Supplementary Table 2 online). For the eight proteins for which data were available in GPM, there was good agreement between the ESP predictor and GPM in predicting validated MRM peptides (Fig. 3c). These results point to potential issues in using proteomic data in databases for MRM-MS assay configuration, as recently noted by others25, and underscore the need for a computational approach to select signature peptides in the absence of MS experimental data.
Important physicochemical properties
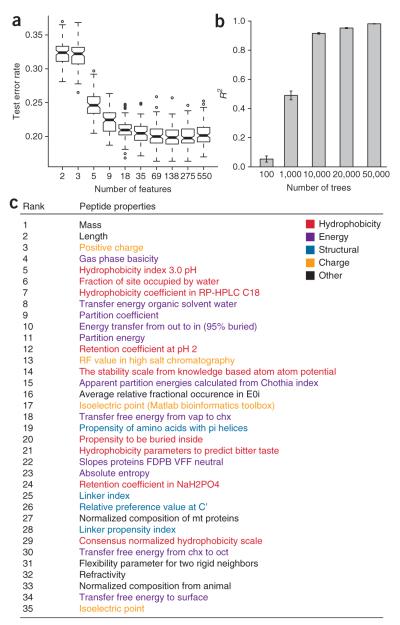
One major benefit of using Random Forest is that it facilitates model interpretation by determining an importance score for each physicochemical property. We followed a procedure similar to that described previously26 to determine the number of important properties. Briefly, we randomly split the yeast training data into train (80%) and test (20%) sets. We then trained a model using all 550 properties and recorded the test error using the variable importance measure to rank the properties. Note that the variable importance measure was calculated once using all properties to avoid overfitting. Next, we repeatedly removed the least important half of the properties and recorded the test-set error at each step. We repeated this entire process 100 times to produce an error distribution (Fig. 4a). Because the test error was distributed normally, we used a two-tailed t-test to determine the minimum number of properties at which the test error distributions were no longer significantly different (P < 0.05). We selected 35 properties as the most important and grouped them into five major categories (Fig. 4c and Supplementary Methods online for more information about the 35 properties). Even though we used all properties in the final Random Forest model, we selected these 35 properties to gain some insight into an interpretation of the model.
Figure 4.
Analysis of important physicochemical properties in predicting high-responding peptides. (a) The yeast training set was randomly split into training- (80%) and test- (20%) sets to produce 100 different Random Forest models (1,000 trees) at each step of halving the number of important properties. The box plot shows the test set error distribution. (b) The stability of property importance improves with increased number of trees in the Random Forest model. For a given number of trees, five models were built and the pairwise Spearman rank correlation coefficient of determination (R2) was calculated for the ranked list of important features (error bars ± 1 s.d). (c) The top 35 features from the ESP predictor using 50,000 trees are listed.
Stability of the ranking of important physicochemical properties is highly dependent on the number of trees used in Random Forest. We built five Random Forest models using 100, 1,000, 10,000, 20,000 and 50,000 trees and analyzed the pair-wise Spearman rank correlation (ten correlations with five models) of the property ranking for each Random Forest model. Not surprisingly, the pair-wise correlation for a Random Forest with 100 trees indicates almost no correlation of the property rank between models (R2 = 0.06 ± 0.02, mean ± s.d.). However, the correlation continues to improve as we increase the number of trees (for 50,000 trees, R2 = 0.98 ± 0.001, mean ± s.d.). With 50,000 trees, the list of important physicochemical properties becomes more stable and reproducible (Fig. 4b). We observed no indication of overfitting with 50,000 trees, which is consistent with the behavior of Random Forest (Supplementary Fig. 4 online).
DISCUSSION
The ESP predictor is more robust and performs significantly better than existing computational methods or random predictions across ten experimentally diverse validation sets. Based on our analyses, it provides a robust method to select candidate signature-peptides for MRM-MS protein quantification, especially in the absence of MS-based experimental data. When applied directly to MRM-MS—assay development for 14 proteins, our method achieved a success rate of 93%, and on average correctly selected two signature peptides per protein. In particular, we used the ESP predictor to successfully configure MRM-MS assays for six proteins in which MS discovery data were not found in a comprehensive proteomic database.
We showed, using two validation sets, that the ESP predictor performs significantly better than three previous methods designed to predict proteotypic peptides (Fig. 2c,d). We attribute the success of our method to the following two factors. First, a unique aspect of our study, relative to these prior studies, is the method used to determine the training set. Prior studies defined their training set based on peptides ‘detected’ or ‘not detected’ in an MS experiment. Our method focuses on predicting high-responding peptides. High-responding peptides are not only proteotypic (that is, detectable and unique) but constitute that subset of proteotypic peptides producing the highest MS response. Second, Random Forest is a committee of decision trees that vote on deciding a final classification, and each of these trees is based on random resampling in both feature and sample space. These characteristics of the Random Forest may be responsible for the ability of the model to generalize well beyond the training set (Supplementary Figs. 1 and 5 online).
A major advantage of the ESP predictor is that a single model performs well across all common ESI experimental types. Unlike existing methods, which developed separate models for different ESI platforms10 or even data set—specific models11, we observe very consistent performance with a single model, indicating the model does not need to be retrained. We show this by testing the ESP predictor against validation sets from multiple database-search algorithms, quantification methods, mass spectrometers and experimental conditions. The ESP predictor would probably need to be retrained to be used on data produced using matrix-assisted laser desorption ionization (MALDI) MS, if a protease other than trypsin was used, or if other sample preparation procedures differ significantly from those used for the training set (e.g., not reducing and alkylating cysteines before digestion or using different LC solvent buffers).
Use of the Random Forest classifier provides insight into the model by calculating the most important physicochemical properties used to predict high-responding peptides. Because Random Forest is a nonlinear model, it is not possible to determine the direction of response from each property (Supplementary Fig. 6 online). It is difficult to compare relevant physicochemical properties (which are heavily influenced by the underlying experimental data) across previous studies because each study used different training sets, properties and computational models. For example, previously13 cysteine was shown to be important in classifying a peptide as ‘not proteotypic’ because the sample was not alkylated, making cysteine-containing peptides unlikely to be detected by MS. To further illustrate this point, in our study, we applied two different feature-selection techniques and found minimal overlap with the top 35 properties reported by the final Random Forest model (Supplementary Fig. 7 online). However, there is broad agreement that hydrophobicity, positive charge and energy terms are critical for predicting high-responding peptides and proteotypic peptides10–13. We grouped the top 35 properties into five categories: hydrophobicity, energy, structural, charge and other (Fig. 4c). In previous studies examining ESI response, it was observed that Gibbs free-energy transfer between amino acids has led to an increased response in peptides with nonpolar regions27. This supports our findings that hydrophobicity and energy properties influence peptide response. The structural properties may indicate likely cleavage sites during protein digestion, and we know peptides must carry a charge to be detected in a mass spectrometer28. It is worth mentioning that, although many of the properties appear similar in name (that is, hydrophobicity), often the amino acid values were determined under different experimental conditions. For example, a mathematical model has been developed29 to calculate amino acid hydrophobicity based on HPLC performance of synthetic amino acids (rank 5 in Fig. 4c). On the other hand, a model of retention time (that is, hydrophobicity; rank 12 inFig. 4c)30 was developed based on HPLC performance using a synthetic 5-mer peptide in which individual amino acids were sequentially added in the middle. This suggests Random Forest is able to leverage subtle differences in amino acid property values to appropriately calculate peptide response.
In summary, we have shown that the ESP predictor is a robust method to predict high-responding peptides from a given protein based entirely on the peptide sequence. The ESP predictor greatly facilitates selection of optimal candidate signature-peptides for developing targeted assays to detect and quantify any protein of interest in the proteome. The ESP predictor fills a critical gap, enabling selection of candidate signature-peptides for proteins of interest in the absence of high-quality MS-based experimental evidence. Its use should improve the efficiency of biomarker verification, currently one of the most significant resource constraints in the development of biomarkers for early detection of disease, and the development of pharmacodynamic markers of therapeutic efficacy1,31,32.
METHODS
Defining empiric peptide classification training set
The National Cancer Institute Clinical Proteomic Technology Assessment in Cancer Program (NCI-CPTAC) prepared a tryptic digest of a yeast lysate sample and sent it to three proteomic laboratories: Vanderbilt University, New York University (NYU) and the Broad Institute. All laboratories were expected to follow the same MS protocol on an LTQ-Orbitrap mass spectrometer. Vanderbilt analyzed the sample in duplicate on two instruments, NYU analyzed the sample in duplicate, and the Broad Institute performed six replicates. Thus, the yeast lysate was analyzed 12 times across four LTQ-Orbitraps. The raw files were searched using Spectrum Mill v3.4 beta with a precursor mass tolerance of 0.05 Da and fragment mass tolerance of 0.7 Da, specifying up to two missed cleavages and the following modifications: cysteine carbamidomethylation, carbamylation of N termini and lysine, oxidized methionine and pyroglutamic acid. The tandem MS (MS/MS) data were autovalidated at the protein level with a protein score of 25 and at the peptide level using a score of 13, percent similarity of 70%, forward-reverse score of 2, and rank 1-2 score difference of 2, for all charge states. In total, 4,230 peptides (570 proteins) were identified. The peptide identities, m/z, and retention time were exported to calculate the XIC for the monoisotopic peak.
The XIC for each peptide (in a given charge state) was calculated by determining the location (m/z and retention time) of the peptide peak. If a peptide was sequenced multiple times (that is, has many MS/MS spectra), the peptide with the best Spectrum Mill score on a per charge basis was used for this purpose. Peptides with the highest score indicate the highest confidence in matching the fragment spectra compared to spectra with lower scores for the same peptide.
In each LC-MS/MS run, different sets of peptides were sequence identified owing to the stochastic behavior of the mass spectrometer. Therefore, retention times were propagated across different LC-MS/MS runs using a quadratic regression model (R2 = 0.99 for all LC-MS/MS runs). This yielded an approximate elution time, and allowed us to ‘hunt’ for peptides not sequence identified in a particular LC-MS/MS run. The XIC was calculated using a combination of retention time and m/z for each peptide.
An in-house program was developed to automatically calculate the XIC using the Thermo Software Development Kit. The XIC was calculated using a retention time tolerance of ± 2.5 min and m/z tolerance of ± 15 p.p.m. A summary table was created where the response for each peptide was obtained by summing the XIC values for all peptide variations (that is, peptides with multiple charge states and common modifications). This reduced the list to 3,637 peptides.
The yeast LC-MS/MS runs from each institute (Vanderbilt, NYU, Broad Institute) were then median normalized to account for any instrument or processing differences (which were expected to be minor because all samples were processed following the same protocol). The median normalization divides each LC-MS/MS run by its median XIC value and then multiplies it by the common median XIC (the median of the median of all 12 LC-MS/MS runs). A table of identified peptides was created, with their corresponding XIC (if present) in all 12 LC-MS/MS runs. The median of all 12 LC-MS/MS runs was selected as the ‘official’ XIC value for each peptide. Peptides with a coefficient of variance (s.d./mean×100%) >100% were rejected. In addition, any peptide with a median XIC of zero was rejected, indicating that it was not reliably detected in all LC-MS/MS runs.
Next, a set of peptides ‘not detected’ in the mass spectrometer was created. An in silico tryptic digest was performed for all sequence-identified proteins. A substring search was used to remove any in silico peptide where we had evidence of a sequence-identified peptide. For example, if the in silico peptide was LQTISALPK and the sequence-identified peptide was LQTISALPKGDELR, the in silico peptide was rejected because it is a substring of the sequence-identified peptide. Thus, the ‘not detected’ set of peptides was not seen in any form of the sequence-identified peptides. In addition, any peptide sequence that was not unique and any N- or C-terminal peptides (~4% of the peptides) were removed. The final peptide set contained a list of sequence-identified peptides (with their corresponding XIC) and peptides that were not sequence identified in any form.
To classify peptides as high- or low-responding, we considered only proteins with seven or more sequence-identified peptides. The peptide response within each protein was log transformed (excluding peptides ‘not detected’) to create a normal distribution and is justified by the Box Cox transformation33. The log-transformed data were then standardized, using the z-score (z), within each protein. High-responding peptides were selected with a z ≥ 0 whereas low-responding peptides were selected with a z ≤ −1. This procedure was used only to create the training set and does not apply to the validation sets, where we examined only the five highest-responding peptides. The ‘not detected’ peptides were then appended to the low-responding peptides to create a binary high (n = 623) versus low/not detected (n = 2,530) classifier.
Calculation of physicochemical properties for peptides
A diverse set of 550 physicochemical properties was used to calculate the peptide feature set. Properties such as length, number of acidic (glutamic acid, asparagine) and basic (arginine, lysine, histidine) residues were calculated by counting the number of amino acids in each peptide. The Bioinformatics package in Matlab was used to calculate the peptide mass and pI. The gas phase basicity was calculated from Zhang’s model17. The remaining 544 physicochemical properties contained individual values for each amino acid. For each peptide and a given property, the constituent amino acid numerical values were averaged to produce a single value. Missing values were ignored. The average (rather than median or sum) was chosen because it is sensitive to outliers and normalizes for peptide length. It was assumed that the average physicochemical property across each peptide was sufficient to capture relevant information about peptide response. The model does not incorporate protein context such as flanking amino acids or protein information (e.g., protein molecular weight or protein pI). We view this as a separate problem from predicting high-responding peptides34,35. Calculations of the peptide feature set were performed in Matlab R2006b (MathWorks).
Random Forest classifier for predicting high-responding peptides
Random Forest is a nonlinear ensemble algorithm composed of many individual decision trees. Each tree is grown using a randomized tree-building algorithm. For each tree (num_tree), a bootstrap sample (that is, random data subset sampled with replacement) is selected from the training set. At each decision branch in the tree, the best spilt is chosen from a randomly selected subset of properties (rather than all properties), num_feature. With these two random steps each tree is different. Predictions result from the ensemble of all trees by taking the majority vote. Instead of relying on this binary classification, a probabilistic output (the fraction of trees that vote high) was used and referred to as probability of response.
The peptide training data were imbalanced. High-responding peptides, the class of interest, comprised only ~20% of the data. Most classifiers focus on optimizing overall accuracy at the expense of misclassifying the minority class (high-responding peptides). Down sampling is a common technique to handle imbalanced data sets36. In Random Forest, the number of training samples for each class was set to the size of the minority class (n = 623), and samples were selected via bootstrapping with replacement from both the minority and majority classes. This process was repeated for each tree and exhibits a significant improvement in performance and generalization36.
Balanced class sizes were used to optimize num_tree and num_feature parameters in Random Forest. The num_feature parameter was optimized by setting num_tree to 1,000 and varying num_feature between 2 and 550 features. The optimal value for num_feature was determined to be 90 (Supplementary Fig. 8 online). The num_tree parameter was optimized by increasing the number of trees until the variable importance measure was consistent and reproducible (Fig. 4b). The num_tree parameter was set to 50,000 trees.
The training data were used to calculate a no call region in order to judge the model performance on peptides confidently classified as either high or low/not detected. Peptides with a predicted probability of response between 0.38–0.65 were labeled as no call and the model was not penalized. Peptides with a predicted probability greater than or equal to 0.65 were classified as high and peptides with a predicted probability less than or equal to 0.38 were classified as low/not detected. The reject region was selected based on a false positive rate (1 — specificity) of 10%. This choice of reject region yielded calls on 74% of the training data.
The weighted accuracy was used to account for the imbalanced class size. The weighted accuracy is calculated as: Aw = 0.5 * (sensitivity + specificity) where sensitivity is the percent of true positives and specificity is the percent of true negatives. The yeast training data were split into training (90%) and test (10%) sets. The training and test set weighted accuracies were 81% and 76%, respectively. We also examined the area under the curve (AUC) for a receiver operating characteristic (ROC) plot24 on the test data. The AUC is a standard measure of performance where a perfect classification would have an AUC of 1 and random classification would have an AUC of 0.5. The AUC for the test set was 83% (P = 9.4e-9) indicating the predictions are significantly better than random (Supplementary Fig. 9 online). Random Forest and ROC calculations were performed in R (http://CRAN.R-project.org/) using the Random Forest package v. 4.5-18 (ref. 19) and ROCR library v. 1.0-2 (ref. 37), respectively.
Random Forest variable importance score
A measure of how each property contributes to the overall model performance is determined during Random Forest training. When the values for an important property are permuted there should be a noticeable decrease in model accuracy. Likewise, when the values for an irrelevant property are permuted there should be little change in model accuracy. The difference in the two accuracies are then averaged for all trees and normalized by the standard error to produce an importance measure, referred to as the variable importance score.
Permutation test to evaluate the significance of the ESP predictions
All proteins were required to contain at least six or more predicted tryptic peptides (from an in silico digest) and at least five or more sequence-identified peptides. For each protein, the five highest-responding peptides were selected (based on experimental data, Fig. 1a down to ‘MRM-MS assay optimization’). Then, using the same protein, five peptides with the highest probability of response were selected using the ESP predictor (Fig. 1b). For each validation set, the actual test statistic (Ts) was calculated as the sum of the number of peptides in common between the top five peptides from the experimental and computational methods for each protein. Next, a random test statistic (Trs) was calculated by randomly sampling five peptides and taking the sum of the number of peptides in common with the top five experimentally derived peptides for each protein in the validation set. This process was repeated 10,000 times to produce a null distribution for each validation set. The resulting distribution was used to estimate a one-tailed P-value. Using this procedure, the statistical significance of the predictions made by the ESP predictor was calculated as the number of proteins (also selected at random from the respective validation set) increased. The permutation test implicitly accounts for differences in the number of peptides from each protein. The permutation test calculations were performed in R.
Analysis and MS summary for all validation sets
All protein mixtures were digested using trypsin and analyzed using reversed-phased nano LC-ESI-MS/MS on multiple LTQ Oribtrap and LTQ-FT mass spectrometers (Thermo). Specific conditions concerning chromatography, buffers, injection volume and MS analysis settings varied according to each validation set (full details for all validation sets are provided in the Supplementary Methods). Validation sets were subsequently processed using either Spectrum Mill 3.4 beta (Agilent Technologies) or Mascot v. 2.1.0.3 (Matrix Science) to determine sequence-identified peptides from the collected MS/MS spectra. Peptide response was calculated using either an in-house developed program to calculate the XIC, MSQuant v. 1.4.2 b5 (http://msquant.sourceforge.net/), or Spectrum Mill. The total peptide response was calculated by summing all forms of a given peptide (that is, multiple charge states and the following modifications: carbamidomethylation, carbamylated lysine, oxidized methionine and pyroglutamic acid). The following is a brief summary of each validation set:
ISB-18 is a publicly available standard protein mix consisting of 18 proteins provided by the Institute for Systems Biology (ISB)38. Only the LTQ-FT data were considered.
Yeast test refers to the 10% of proteins held-out from the training set in order to evaluate the model performance.
Plasma refers to neat plasma (that is, undepleted plasma).
Sigma48 refers to a set of 48 equimolar proteins (Universal Proteomics Standard Set, Sigma). The samples were digested using a trifluoroethanol-assisted digestion protocol39.
Plasma Hu14 SCX refers to a plasma sample with the 14 most abundant proteins removed using a MARS Hu-14 column (Agilent Technologies) and then fractionated using strong cation exchange (SCX). Eleven fractions were collected and analyzed.
Yeast_2 refers to a separate independent analysis of a yeast mixture. Importantly, proteins in common with the yeast training set were removed.
HeLa_1 refers to HeLaS3 cell lysate digested in-solution.
HeLa_2 refers to HeLaS3 cell lysate analyzed by GeLC-MS (Supplementary Methods).
Pull-Down refers to a GeLC-MS affinity pull-down experiment from a HeLaS3 cell lysate.
Plasma Hu14 refers to a plasma sample with the 14 most abundant proteins removed using a MARS Hu-14 column.
MRM-MS assay development
The validated MRM peptides were defined from single protein digests for each of the 14 proteins. Peptide selection for the 14 target proteins was based upon experimental observation using commercially available protein standards. Briefly, the proteins were individually digested with trypsin and analyzed by nano LC-MS/MS in positive-ion electrospray on an LTQ linear ion trap mass spectrometer (Thermo) with data-dependent acquisition. Peptide-sequence identity was determined using Spectrum Mill on the collected MS/MS spectra. Approximately five candidate peptide standards per protein were chosen based primarily on high relative response. Exclusion criteria included large hydrophobic or small hydrophilic peptides, flanking tryptic ends with dibasic amino acids (KK, RR, KR, RK) at the N or C terminus and peptide identity corresponding to multiple endogenous plasma proteins. Peptide standards containing methionine and cysteine were avoided if possible. Stable isotope—labeled versions of each candidate peptide were synthesized for quantification and MRM response curves were optimized in plasma for each protein over a wide concentration range. All peptides that performed satisfactorily over the response curves are referred to as “validated MRM peptides.”
All MRM experiments were performed on a 4000 Q Trap Hybrid triple quadrupole/linear ion trap mass spectrometer coupled to a Tempo LC system (Applied Biosystems). Data analysis was done using MultiQuant software (Applied Biosystems).
The GPM database was searched (December 19, 2008) by entering the protein Ensembl accession number and then selecting the ‘MRM’ link. For some proteins, a large number of peptides were listed. Only the top five peptides were considered based on the number of times observed in the GPM database.
Data and software availability
The yeast MS data used to develop the model are publicly available from Tranche (http://tranche.proteomecommons.org/). The ESP predictor is freely available as a module in the GenePattern integrative genomics software package (http://www.genepattern.org/) under the category ‘proteomics’. The automated script to calculate the XIC using the Thermo Software Development Kit is available upon request. Source code and examples are available as Supplementary Source Code online. The data associated with this manuscript may be downloaded from the ProteomeCommons.org Tranche system ⟨/http://www.proteomecommons.org/data-downloader.jsp?fileName=90MaGKV4KHKHOyOvNGSXxtDhAEQbJA3KbZap6ruHxvUFDk%2BvOFyhawX%2BhSQa%2Bxa/KvG6oQCYON4nsZ/uDw55FfNDAU0AAAAAAAAMLw==S⟩.
Supplementary Material
ACKNOWLEDGMENTS
We thank the National Cancer Institute (NCI) Clinical Proteomic Technology Assessment in Cancer Program (NCI-CPTAC, http://proteomics.cancer.gov/programs/CPTAC/) for providing samples of yeast lysate and raw MS data generated by the CPTAC centers. We thank Rushdy Ahmad, Kathy Do, Amy Ham, Emily Rudomin, and Shao-En Ong for MS data generation, and Hasmik Keshishian and Terri Addona for generating the lists of validated MRM peptides. We also thank Shao-En Ong, Jacob Jaffe, Karl Clauser, Eric Kuhn, Pablo Tamayo, and Nick Patterson for helpful discussions. We would like to thank the reviewers for their insightful comments. This work was supported in part by grants to S.A.C. from the National Institutes of Health Grants 1U24 CA126476 as part of the NCI’s Clinical Proteomic Technologies Assessment in Cancer Program, the National Heart, Lung, and Blood Institute, U01-HL081341 and The Women’s Cancer Research Fund; to J.P.M. from the National Science Foundation and NIGMS the National Institutes of Health (NIGMS and NCI); to D.R.M. from the National Institutes of Health grant R01 CA126219, as part of NCI’s Clinical Proteomic Technologies for Cancer Program.
Footnotes
Reprints and permissions information is available online at http://npg.nature.com/reprintsandpermissions/
Note: Supplementary information is available on the Nature Biotechnology website.
References
- 1.Rifai N, Gillette MA, Carr SA. Protein biomarker discovery and validation: the long and uncertain path to clinical utility. Nat. Biotechnol. 2006;24:971–983. doi: 10.1038/nbt1235. [DOI] [PubMed] [Google Scholar]
- 2.Uhlen M, Hober S. Generation and validation of affinity reagents on a proteome-wide level. J. Mol. Recognit. 2008 doi: 10.1002/jmr.891. [DOI] [PubMed] [Google Scholar]
- 3.Anderson L, Hunter CL. Quantitative mass spectrometric multiple reaction monitoring assays for major plasma proteins. Mol. Cell. Proteomics. 2006;5:573–588. doi: 10.1074/mcp.M500331-MCP200. [DOI] [PubMed] [Google Scholar]
- 4.Gerber SA, Rush J, Stemman O, Kirschner MW, Gygi SP. Absolute quantification of proteins and phosphoproteins from cell lysates by tandem MS. Proc. Natl. Acad. Sci. USA. 2003;100:6940–6945. doi: 10.1073/pnas.0832254100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Keshishian H, Addona T, Burgess M, Kuhn E, Carr SA. Quantitative, multiplexed assays for low abundance proteins in plasma by targeted mass spectrometry and stable isotope dilution. Mol. Cell. Proteomics. 2007;6:2212–2229. doi: 10.1074/mcp.M700354-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stahl-Zeng J, et al. High sensitivity detection of plasma proteins by multiple reaction monitoring of N-glycosites. Mol. Cell. Proteomics. 2007;6:1809–1817. doi: 10.1074/mcp.M700132-MCP200. [DOI] [PubMed] [Google Scholar]
- 7.Kuster B, Schirle M, Mallick P, Aebersold R. Scoring proteomes with proteotypic peptide probes. Nat. Rev. Mol. Cell Biol. 2005;6:577–583. doi: 10.1038/nrm1683. [DOI] [PubMed] [Google Scholar]
- 8.Craig R, Cortens JP, Beavis RC. Open source system for analyzing, validating, and storing protein identification data. J. Proteome Res. 2004;3:1234–1242. doi: 10.1021/pr049882h. [DOI] [PubMed] [Google Scholar]
- 9.Deutsch EW, Lam H, Aebersold R. PeptideAtlas: a resource for target selection for emerging targeted proteomics workflows. EBMO reports. 2008;9:429–434. doi: 10.1038/embor.2008.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mallick P, et al. Computational prediction of proteotypic peptides for quantitative proteomics. Nat. Biotechnol. 2007;25:125–131. doi: 10.1038/nbt1275. [DOI] [PubMed] [Google Scholar]
- 11.Sanders WS, Bridges SM, McCarthy FM, Nanduri B, Burgess SC. Prediction of peptides observable by mass spectrometry applied at the experimental set level. BMC Bioinformatics. 2007;8(Suppl 7):S23. doi: 10.1186/1471-2105-8-S7-S23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tang H, et al. A computational approach toward label-free protein quantification using predicted peptide detectability. Bioinformatics. 2006;22:e481–e488. doi: 10.1093/bioinformatics/btl237. [DOI] [PubMed] [Google Scholar]
- 13.Webb-Robertson BJ, et al. A support vector machine model for the prediction of proteotypic peptides for accurate mass and time proteomics. Bioinformatics. 2008;24:1503–1509. doi: 10.1093/bioinformatics/btn218. [DOI] [PubMed] [Google Scholar]
- 14.Jaffe JD, et al. Accurate inclusion mass screening: a bridge from unbiased discovery to targeted assay development for biomarker verification. Mol. Cell. Proteomics. 2008;7:1952–1962. doi: 10.1074/mcp.M800218-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Malmstrom J, Lee H, Aebersold R. Advances in proteomic workflows for systems biology. Curr. Opin. Biotechnol. 2007;18:378–384. doi: 10.1016/j.copbio.2007.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kawashima S, Kanehisa M. AAindex: amino acid index database. Nucleic Acids Res. 2000;28:374. doi: 10.1093/nar/28.1.374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang Z. Prediction of low-energy collision-induced dissociation spectra of peptides. Anal. Chem. 2004;76:3908–3922. doi: 10.1021/ac049951b. [DOI] [PubMed] [Google Scholar]
- 18.Breiman L. Random forest. Mach. Learn. 2001;45:5–32. [Google Scholar]
- 19.Liaw A, Wiener M. ClassificatIon and Regression by randomForest. R News. 2002;2:18–22. [Google Scholar]
- 20.Diaz-Uriarte R, de Andres S. Alvarez. Gene selection and classification of microarray data using random forest. BMC Bioinformatics. 2006;7:3. doi: 10.1186/1471-2105-7-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Enot DP, Beckmann M, Overy D, Draper J. Predicting interpretability of metabolome models based on behavior, putative identity, and biological relevance of explanatory signals. Proc. Natl. Acad. Sci. USA. 2006;103:14865–14870. doi: 10.1073/pnas.0605152103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vapnik V. The Nature of Statistical Learning Theory. Springer; New York: 1995. [Google Scholar]
- 23.Bishop C. Neural Networks for Pattern Recognition. Oxford University Press; Oxford: 1995. [Google Scholar]
- 24.Fawcett T. ROC Graphs: Notes and Practical Considerations for Researchers. HP Laboratories; Palo Alto, CA, USA: 2004. Technical report. [Google Scholar]
- 25.Lange V, Picotti P, Domon B, Aebersold R. Selected reaction monitoring for quantitative proteomics: a tutorial. Mol. Syst. Biol. 2008;4:222. doi: 10.1038/msb.2008.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Svetnik V, et al. Random forest: a classification and regression tool for compound classification and QSAR modeling. J. Chem. Inf. Comput. Sci. 2003;43:1947–1958. doi: 10.1021/ci034160g. [DOI] [PubMed] [Google Scholar]
- 27.Cech NB, Enke CG. Relating electrospray ionization response to nonpolar character of small peptides. Anal. Chem. 2000;72:2717–2723. doi: 10.1021/ac9914869. [DOI] [PubMed] [Google Scholar]
- 28.Cech NB, Enke CG. Practical implications of some recent studies in electrospray ionization fundamentals. Mass Spectrom. Rev. 2001;20:362–387. doi: 10.1002/mas.10008. [DOI] [PubMed] [Google Scholar]
- 29.Cowan R, Whittaker RG. Hydrophobicity indices for amino acid residues as determined by high-performance liquid chromatography. Pept. Res. 1990;3:75–80. [PubMed] [Google Scholar]
- 30.Parker JM, Guo D, Hodges RS. New hydrophilicity scale derived from high-performance liquid chromatography peptide retention data: correlation of predicted surface residues with antigenicity and X-ray-derived accessible sites. Biochemistry. 1986;25:5425–5432. doi: 10.1021/bi00367a013. [DOI] [PubMed] [Google Scholar]
- 31.Whiteaker JR, et al. Integrated pipeline for mass spectrometry-based discovery and confirmation of biomarkers demonstrated in a mouse model of breast cancer. J. Proteome Res. 2007;6:3962–3975. doi: 10.1021/pr070202v. [DOI] [PubMed] [Google Scholar]
- 32.Zolg JW, Langen H. How industry is approaching the search for new diagnostic markers and biomarkers. Mol. Cell. Proteomics. 2004;3:345–354. doi: 10.1074/mcp.M400007-MCP200. [DOI] [PubMed] [Google Scholar]
- 33.Sokal RR, Rohlf FJ. Biometry the Principles and Practice of Statistics in Biological Research. edn. 3 W.H. Freeman and Company; 1995. [Google Scholar]
- 34.Thomson R, Hodgman TC, Yang ZR, Doyle AK. Characterizing proteolytic cleavage site activity using bio-basis function neural networks. Bioinformatics. 2003;19:1741–1747. doi: 10.1093/bioinformatics/btg237. [DOI] [PubMed] [Google Scholar]
- 35.Yen CY, et al. Improving sensitivity in shotgun proteomics using a peptide-centric database with reduced complexity: protease cleavage and SCX elution rules from data mining of MS/MS spectra. Anal. Chem. 2006;78:1071–1084. doi: 10.1021/ac051127f. [DOI] [PubMed] [Google Scholar]
- 36.Chen C, Liaw A, Breiman L. Using Random Forest to Learn Imbalanced Data. Statistics Department of University of California at Berkeley; Berkeley: 2004. Technical Report 666. [Google Scholar]
- 37.Sing T, Sander O, Beerenwinkel N, Lengauer T. ROCR: visualizing classifier performance in R. Bioinformatics. 2005;21:3940–3941. doi: 10.1093/bioinformatics/bti623. [DOI] [PubMed] [Google Scholar]
- 38.Klimek J, et al. The standard protein mix database: a diverse data set to assist in the production of improved peptide and protein identification software tools. J. Proteome Res. 2008;7:96–103. doi: 10.1021/pr070244j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang H, et al. Development and evaluation of a micro- and nanoscale proteomic sample preparation method. J. Proteome Res. 2005;4:2397–2403. doi: 10.1021/pr050160f. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.