Abstract
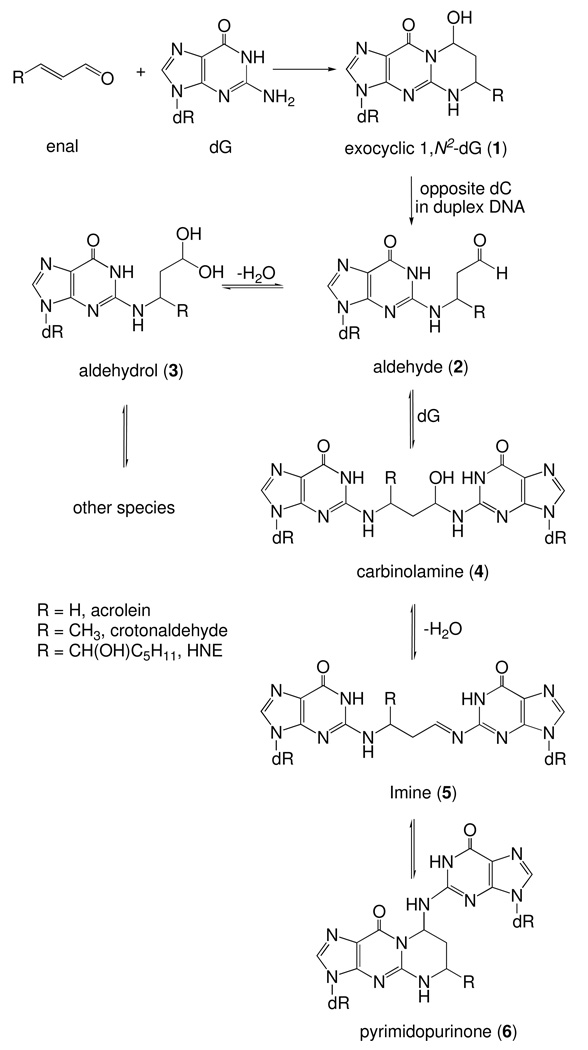
Acrolein reacts with dG to form hydroxylated 1,N2-propanodeoxyguanosine (OH-PdG) adducts. Most abundant are the epimeric 3-(2-deoxy-β-D-erythro-pentofuranosyl)-5,6,7,8-tetrahydro-8-hydroxypyrimido[1,2a] purin-10(3H)-ones, commonly referred to as the γ-OH-PdG adduct. When placed complementary to deoxycytosine in duplex DNA, these undergo rearrangment to the N2-(3-oxopropyl)-dG aldehyde. The latter forms diastereomeric interstrand N2-dG:N2-dG cross-links in the 5'-CpG-3' sequence. Here we report the structure of the stereochemically favored (R)-γ-hydroxytrimethylene N2-dG:N2-dG interstrand DNA cross-link in 5'-d(G1C2T3A4G5C6X7A8G9T10C11C12)-3'•5'-d(G13G14A15C16T17C18Y19C20T21A22G23C24)-3' (X7 is the dG adjacent to the α-carbon of the carbinolamine linkage and Y19 is the dG adjacent to the γ-carbon of the carbinolamine linkage; the cross-link is in the 5'-CpG-3' sequence). The structure was characterized using isotope-edited 15N NOESY-HSQC NMR, in which the exocyclic amines at X7 or Y19 were 15N-labeled. Analyses of NOE intensities involving Y19 N2H indicated that the (R)-γ-hydroxytrimethylene linkage was the major cross-link species, constituting 80–90% of the cross-link. The X7 and Y19 imino resonances were observed at 65 °C. Additionally, for the 5'-neighbor base pair G5•C20, the G5 imino resonance remained sharp at 55 °C, but broadened at 65 °C. In contrast, for the 3'-neighbor A8•T17 base pair, the T17 imino resonance was severely broadened at 55 °C. Structural refinement using NOE distance restraints obtained from isotope-edited 15N NOESY HSQC data indicated that the (R)-γ-hydroxytrimethylene linkage maintained the C6•Y19 and X7•C18 base pairs with minimal structural perturbations. The (R)-γ-hydroxytrimethylene linkage was located in the minor groove. The X7 N2 and Y19 N2 atoms were in the gauche-conformation with respect to the linkage, which maintained Watson-Crick hydrogen bonding of the cross-linked base pairs. The anti conformation of the hydroxyl group with respect to Cα of the tether minimized steric interaction, and more importantly, allowed the formation of a hydrogen bond between the hydroxyl group and C20 O2 located in the 5'-neighboring base pair G5•C20. The formation of this hydrogen bond may, in part, explain the thermal stability of this carbinolamine interstrand cross-link, and the stereochemical preference for the (R)-configuration of the cross-link.
Introduction
DNA interstrand cross-links represent a severe form of DNA damage due to their ability to block of DNA replication and transcription.1,2 The nucleotide excision repair (NER) pathway is involved in the repair of the interstrand cross-link lesion.3 Many chemicals cause interstrand cross-links, including bifunctional alkylating reagents, platinum compounds, and psoralen.3 α,β-Unsaturated aldehydes (enals) such as acrolein,4–6 crotonaldehyde,5 and trans-4-hydoxynonenal (HNE)7 represent a group of chemicals that produce DNA interstrand cross-links. They are produced from the incomplete combustion of organic matter such as fuels, wood, and tobacco;8,9 enals are also produced endogenously as a byproduct of lipid peroxidation.10–17
Enals are mutagenic.18–22 They are bis-electrophiles and undergo sequential reaction with two nucleophilic groups of DNA bases. Reaction of deoxyguanosine with enals produce exocyclic 1,N2-deoxyguanosine adduct 1 through an initial Michael addition.23–27 When placed opposite deoxycytosine in duplex DNA, these exocyclic 1,N2-dG adducts undergo ring-opening to N2-(3-oxopropyl) adduct 2, unveiling a reactive aldehyde functionality; it exists in equlibrium with the corresponding aldehydrols 3 or related species (Scheme 1).28–31 We have demonstrated that the N2-amino group of a neighboring deoxyguanosine on the complementary strand can react with the ring-opened aldehyde to form an interstrand N2-dG:N2-dG cross-links.4–7,32 The formation of the DNA interstrand cross-links by the exocyclic 1,N2-deoxyguanosine adducts is sequence dependent, forming in a local 5′-CpG-3′ sequence context but not in the 5′-GpC-3′ sequence context.5 This is attributed to the longer distance between the N2-dG atoms in the 3′-direction than in the 5′-direction. The formation of a 3-carbon tether in the 3′-direction greatly perturbs the cross-linked base pairs and destabilizes the DNA duplex.33,34 The formation of interstrand cross-links is also influenced by the stereochemistry of the exocyclic 1,N2-dG adducts derived from enal with longer carbon chains.5,7 The crotonaldehyde derived 1,N2-deoxyguanosine adduct of (6R) configuration induces interstrand cross-linking more efficiently than does the (6S) stereoisomer.5 Structure and modeling studies reveal that the orientations of the aldehyde group in the ring-opened (R)- and (S)-crotonaldehyde derived 1,N2-dG adducts are different.28 Steric interactions between the methyl group of the cross-link of the (6S) stereoisomer with the neighboring base pairs result in perturbations of the DNA duplex;28,35 these unfavorable steric interactions are not present in the (6R) crosslink. Similar considerations apply to the HNE-derived 1,N2-dG adducts,36 of which only the (6S,8R,11S) configuration forms interstrand cross-links in the 5′-CpG-3′ sequence.7
Scheme 1.
Chemistry of Enal-Derived Exocyclic 1,N2-Deoxyguanosine Adduct in the 5′-CpG-3′ Sequence When Placed Opposite Deoxycytosine in Duplex DNA
Possible structures for the DNA interstrand cross-links include carbinolamine 4, imine 5, and the ring-closed pyrimidopurinone 6, or possibly an equilibrium mixture of all three (Scheme 1). Pyrimidopurinone 6 is observed upon enzymatic digestion of the cross-linked DNA duplex5,7 but is undetectable in the duplex oligodeoxynucleotides.28,29,37 Although the existence of imine 5 in duplex DNA is surmised by the observation that the interstrand cross-link can be reduced by NaCNBH3,5,7 the interstrand cross-links involving the acrolein and crotonaldehyde derived 1,N2-dG adducts exist primarily as diastereomeric γ-hydroxytrimethylene (carbinolamine 4) tethers at N2-dG:N2-dG positions (Scheme 2).28,29,37 Both imine 2 or pyrimidopurinone 3 are predicted to perturb the Watson-Crick hydrogen bonding and stacking of the cross-linked base pairs, whereas the formation of the carbinolamine linkage has a minimum impact.28,29
Scheme 2.
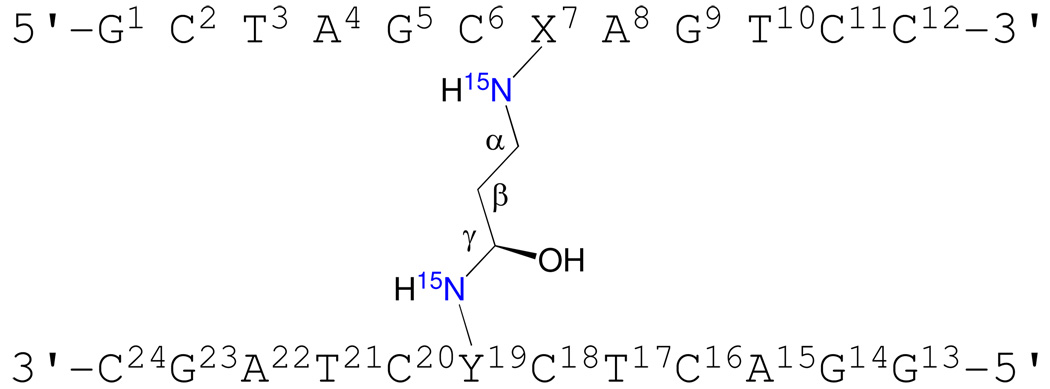
Numbering scheme of the oligodeoxynucleotides containing the (R)-γ-hydroxytrimethylene interstrand cross-link in the 5'-CpG-3' sequence. The two 15N isotopic labeling sites are indicated in blue.
While structures of reduced models of these cross-links, in which a trimethylene chain links the N2-positions have been reported,33,34 the native diastereomeric carbinolamine cross-links have not been examined. The observation of thermally stable carbinolamine interstrand cross-links in duplex DNA in aqueous solution28,29 suggests that additional interactions might stabilize the linkage. Presently, we show that the acrolein-derived diastereomeric (R)-γ-hydroxytrimethylene N2-dG:N2-dG interstrand DNA cross-link is favored in 5'-d(G1C2T3A4G5C6X7A8G9T10C11C12)-3'•5'-d(G13G14A15C16T17C18Y19C20T21A22G23C24)-3' (X7 is the dG adjacent to the α-carbon of the carbinolamine linkage and Y19 is the dG adjacent to the γ-carbon of the carbinolamine linkage; the cross-link is in the 5′-CpG-3′ sequence). The structure has been characterized using isotope-edited 15N NOESY-HSQC NMR. Analyses of NOE intensities involving the 15N-labeled Y19 N2H indicate that the (R-)-γ-hydroxytrimethylene linkage is the major interstrand cross-link species, constituting 80–90% of the total cross-link. Structural refinement using NOE distance restraints obtained from isotope-edited 15N NOESY HSQC data indicates that the (R-)-γ-hydroxytrimethylene linkage maintains the C6•Y19 and X7•C18 base pairs with minimal structural perturbations. The X7 N2 and Y19 N2 atoms are in the gauche-conformation with respect to the linkage, which maintains Watson-Crick hydrogen bonding of the cross-linked base pairs. The anti conformation of the hydroxyl group with respect to Cα of the tether minimizes steric interactions and allows the formation of a hydrogen bond between the hydroxyl group and C20 O2 located in the 5'-neighboring base pair G5•C20. The formation of this hydrogen bond may, in part, explain the thermal stability of this carbinolamine interstrand cross-link, and the stereochemical preference for the (R)-configuration of the cross-link.
Results
Stereochemistry of the Carbinolamine Cross-Link
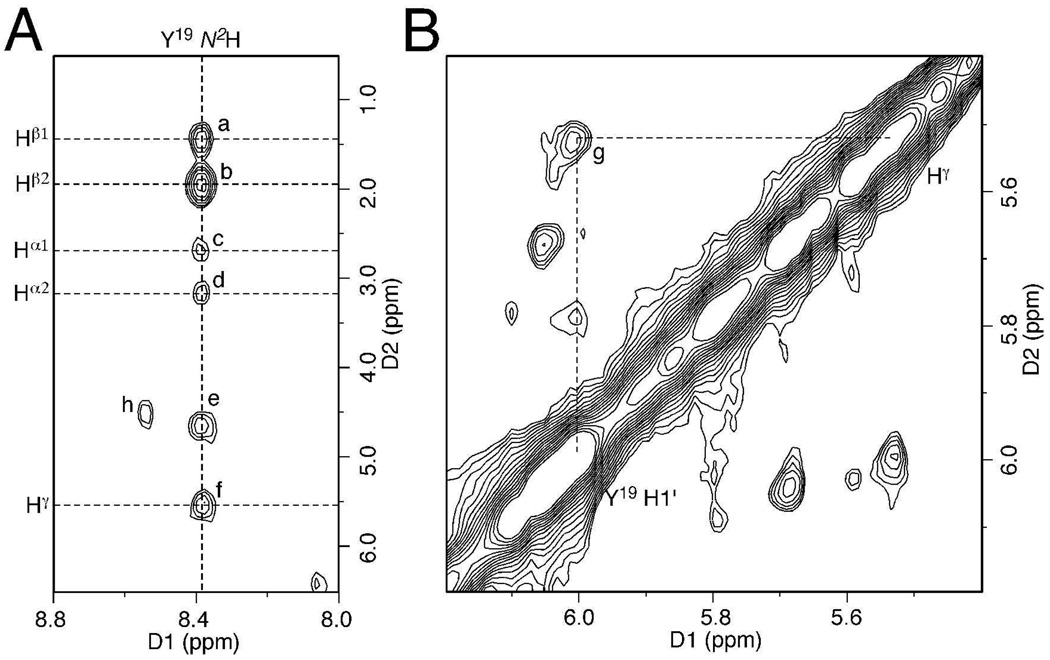
The carbinolamine interstrand cross-links of the acrolein adduct of dG were prepared as reported,5 and occurred as a mixture of (R) and (S) diastereomers at the carbinolamine carbon. An expansion of 15N HSQC filtered NOESY spectrum of the Y19 N2 15N-labeled interstrand cross-link is shown in Figure 1A. For the major diastereomeric cross-linked species, a complete set of NOEs between Y19 N2H and the γ-hydroxymethylene tether was observed. The Y19 N2H→Hγ NOE was weaker than the Y19 N2H→Hβ2 NOE and only slightly stronger than the two diastereotopic Y19 N2H→ Hα NOEs. This was consistent with modeling, in which the (R)-γ-hydroxytrimethylene tether placed Y19 N2H and Hγ at a distance of 2.9–3.1 Å. In contrast, the modeling suggested that the (S)-γ-hydroxytrimethylene tether placed Y19 N2H and Hγ at a closer distance of 2.2–2.4 Å, which would have resulted in a stronger NOE. In addition, a weak Y19 H1′→Hγ NOE was observed (Figure 1B), suggesting Hγ was oriented toward Y19. These data indicated that the (R)-γ-hydroxytrimethylene linkage (Scheme 2) was the major interstrand cross-link species. The relative peak intensities in the 15N HSQC spectrum suggested that the (R)-γ-hydroxytrimethylene cross-link constituted 80–90% of the overall interstrand cross-links.
Figure 1.
NOE cross peaks to identify the stereochemistry of the DNA interstrand cross-link. (A) 15N NOESY-HSQC of the Y19 N2 15N-labeled sample. (B) Non-isotope-edited NOESY. NOEs for the major diastereomer of the cross-link were assigned as: (a) Y19 N2H→Hβ1; (b) Y19 N2H→Hβ2; (c) Y19 N2H→Hα1; (d) Y19 N2H→Hα2; (e) water exchange peak of Y19 N2H; (f) Y19 N2H→Hγ; (g) Y19 H1′→Hγ; (h), Y19 N2H→Hγ of the minor diastereomer.
Spectral Assignments of the (R)-γ-Hydroxytrimethylene Cross-Link
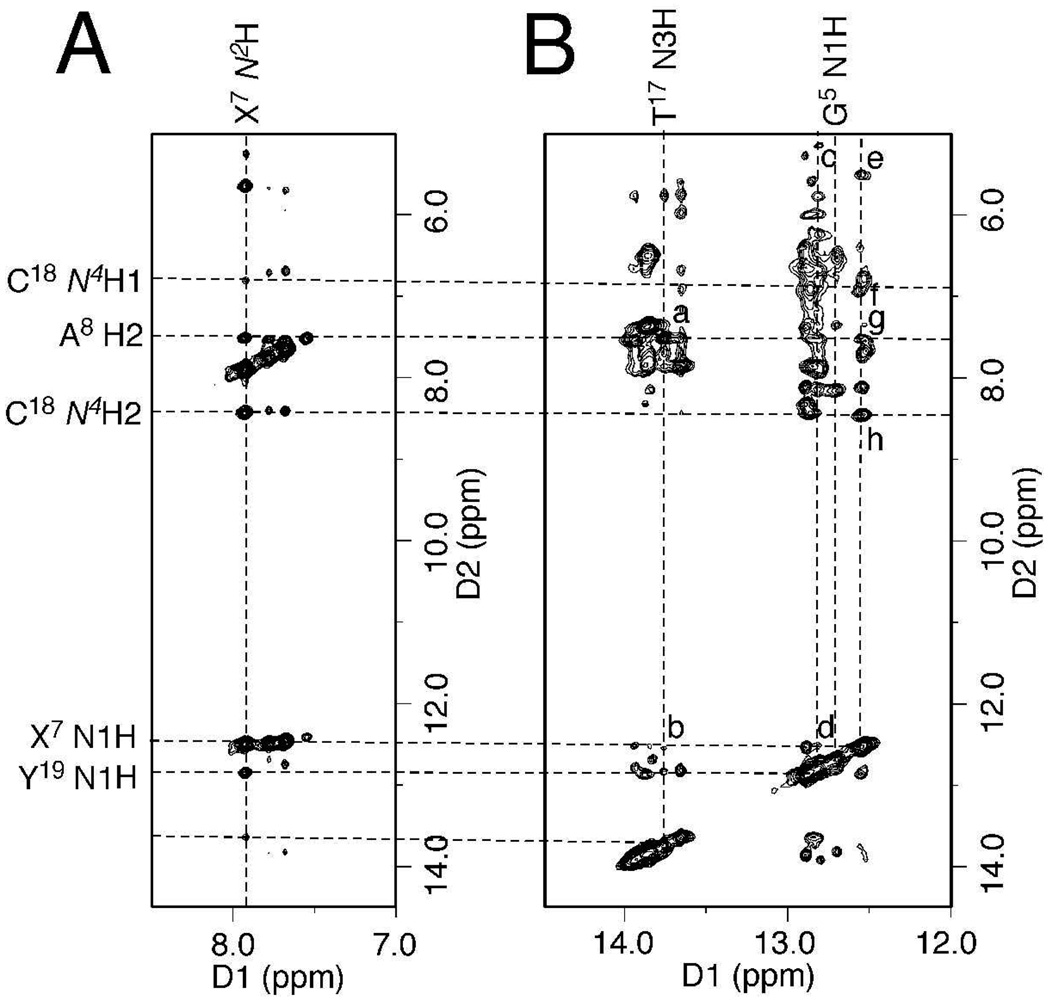
By comparison of the non-isotope-edited NOESY spectrum with the 15N NOESY-HSQC spectrum of the X7 N2 15N-labeled interstrand cross-link, the G5, X7, T17, and Y19 imino protons were assigned (Figure 2). The G5 N1H→C20 N4Hs, X7 N1H→C18 N4Hs, T17 N3H→A8 H2, and Y19 N1H→C6 N4Hs NOE interactions arising from Watson-Crick hydrogen bonding were observed, indicating the Watson-Crick hydrogen bonds at the cross-linked base pairs were not significantly perturbed.
Figure 2.
Expansions of NOESY spectra indicating G5•C20, C6•Y19, X7•C18 and A8•T17 base pairs of the (R)-γ-hydroxytrimethylene cross-link maintained Watson-Crick base pairing. (A) 15N NOESY-HSQC of the X7 N2 15N-labeled sample. (B) The non-isotopically edited NOESY spectrum. NOEs were assigned as: (a) T17 N3H→A8 H2; (b) T17 N3H→X7 N1H; (c) Y19 N1H→C6 H5; (d) Y19 N1H→X7 N1H; (e) X7 N1H→C18 H5; (f) X7 N1H→C18 N4H1; (g) X7 N1H→A8 H2; (h) X7 N1H→C18 N4H2.
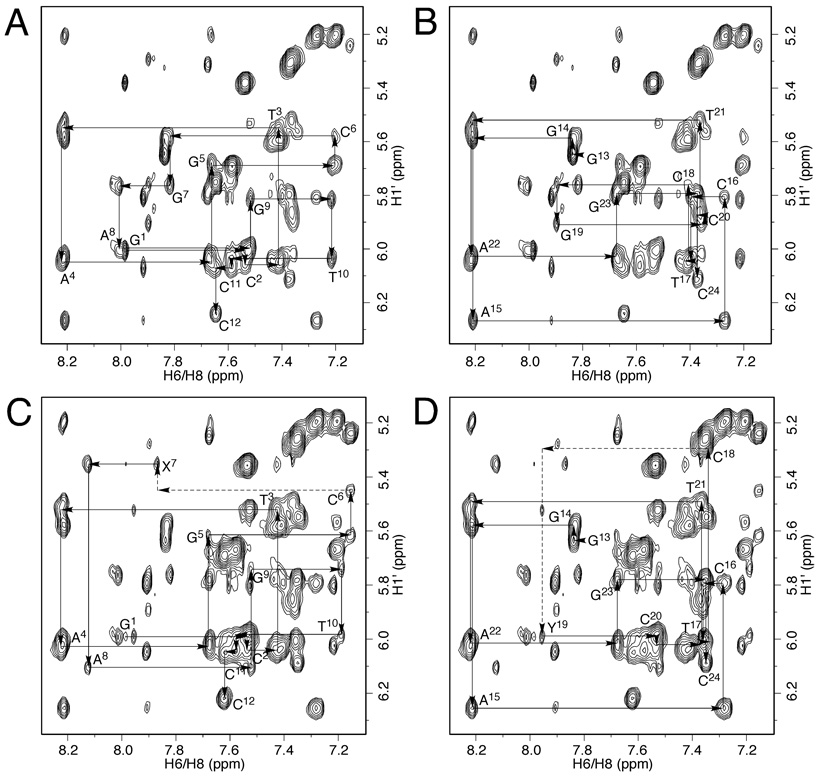
The base aromatic protons and the deoxyribose H1′ protons were assigned by comparing non-isotopically edited NOESY spectra of the modified duplex before and after cross-linking (Figure 3). For the freshly prepared duplex, aldehydrol 3 was the predominant species present in neutral solution.29,30 The major NOE cross peaks in Figures 3A and 3B, which were sequentially connected,38,39 were therefore assigned to the correlations of aromatic H6/H8 protons with H1′ protons of aldehydrol 3. The sample was then incubated for a week at 37 °C, during which time the interstrand cross-link 4 formed, and was eventually present at 50% of the duplex5,29 with the (R)-γ-hydroxytrimethylene predominant. Figures 3C and 3D display the same region of the NOESY spectrum of the cross-linked sample. Except for the nucleotides close to the ends of the duplex, two sets of NOEs were sequentially assigned. The first set of NOEs were the same as those in Figures 3A and 3B (not demonstrated in Figures 3C and 3D) and arose from aldehydrol 3, whereas the second set of NOEs were assigned to the (R)-γ-hydroxytrimethylene of cross-link 4. These NOEs arising from cross-link 4 were sequentially connected with interruptions from C6 to X7 and from C18 to Y19 (Figures 3C and 3D), indicating the (R)-γ-hydroxytrimethylene interstrand cross-link maintained a B-DNA geometry. The X7 H8→C6 H1′ and Y19 H8→C18 H1′ correlations were missing and the A8 H8→X7 H1′ interaction was stronger than corresponding others, indicating structural perturbation at the cross-linked base pairs. The assignments were supported by other NOEs such as H6/H8→H3′, H6/H8→H2′(′), H1′→H3′, and H1′→H2′ (′) etc.
Figure 3.
NOESY spectra of the duplex containing the acrolein-derived 1,N2-dG exocyclic adduct and the resulting interstrand cross-links. (A, B) Duplex before cross-linking. The major NOEs, which were sequentially connected by solid lines, were assigned to the aldehydrol 3. (C, D) Duplex after cross-linking. Two sets of NOE connectivities were assigned. One set arose from aldehydrol 3, the other was assigned to (R)-γ-hydroxytrimethylene cross-link 4. The NOEs of the (R)-γ-hydroxytrimethylene cross-link 4 were sequentially connected (solid and dashed lines). The X7 H8→C6 H1′ and Y19 H8→C18 H1′ NOEs were missing and the A8 H8→X7 H1′ correlation was stronger than corresponding others.
Stabilities of the Base Pairs for the (R)-γ-Hydroxytrimethylene Interstrand Cross-link
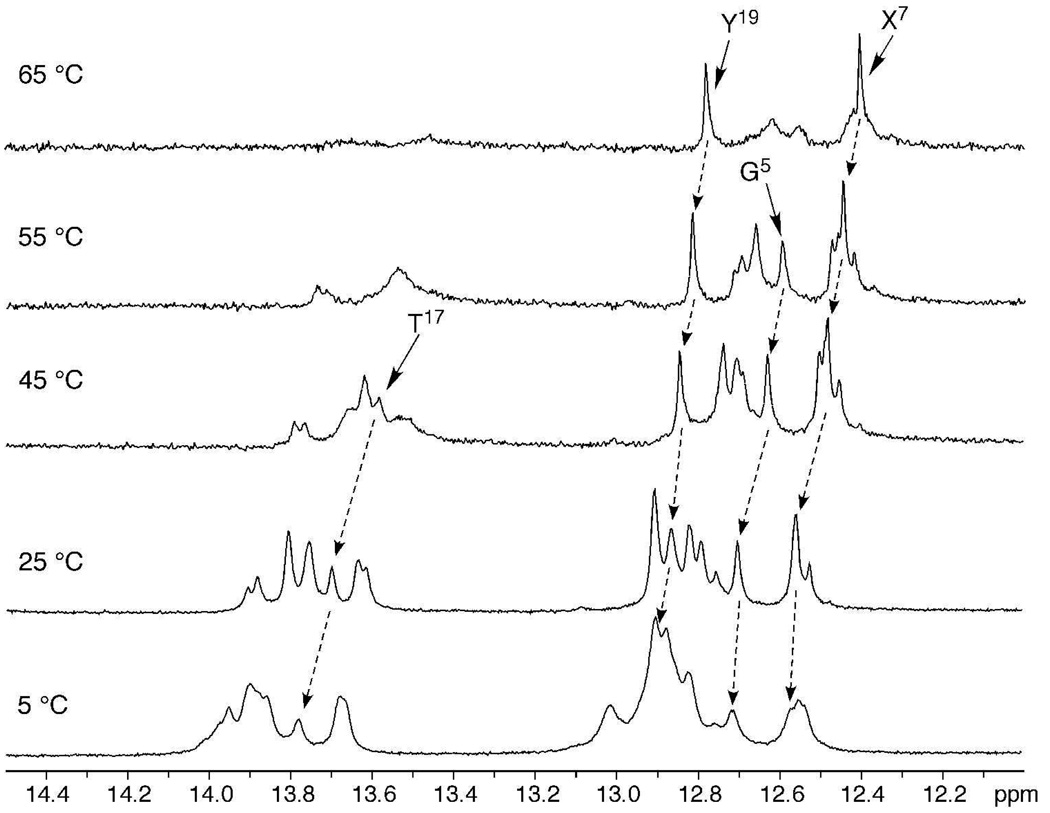
The resonances of pyrimidine N3H and purine N1H imino protons at different temperatures are shown in Figure 4. The melting temperature (Tm) of the DNA interstrand cross-links was 91 °C in 1.0 M NaCl buffer.5 The X7 and Y19 imino resonances were still observable at 65 °C, although starting to broaden, indicating the cross-linked base pairs were stabilized; this observation is consistent with a high melting structure. For the 5'-neighbor base pair G5•C20, the G5 imino resonance remained sharp at 55 °C, but was broadened at 65 °C. At the 3'-neighbor A8•T17 base pair, the T17 imino resonance was severely broadened at 55 °C. For the corresponding unmodified duplex these imino resonances were all broadened at 40 °C.
Figure 4.
NMR melting studies of the (R)-γ-hydroxytrimethylene interstrand cross-link showing basepairs G5•C20, C6•Y19, and X7•C18 at the cross-linked region were stabilized.
Chemical Shift Perturbations of the (R)-γ-Hydroxytrimethylene Cross-Link
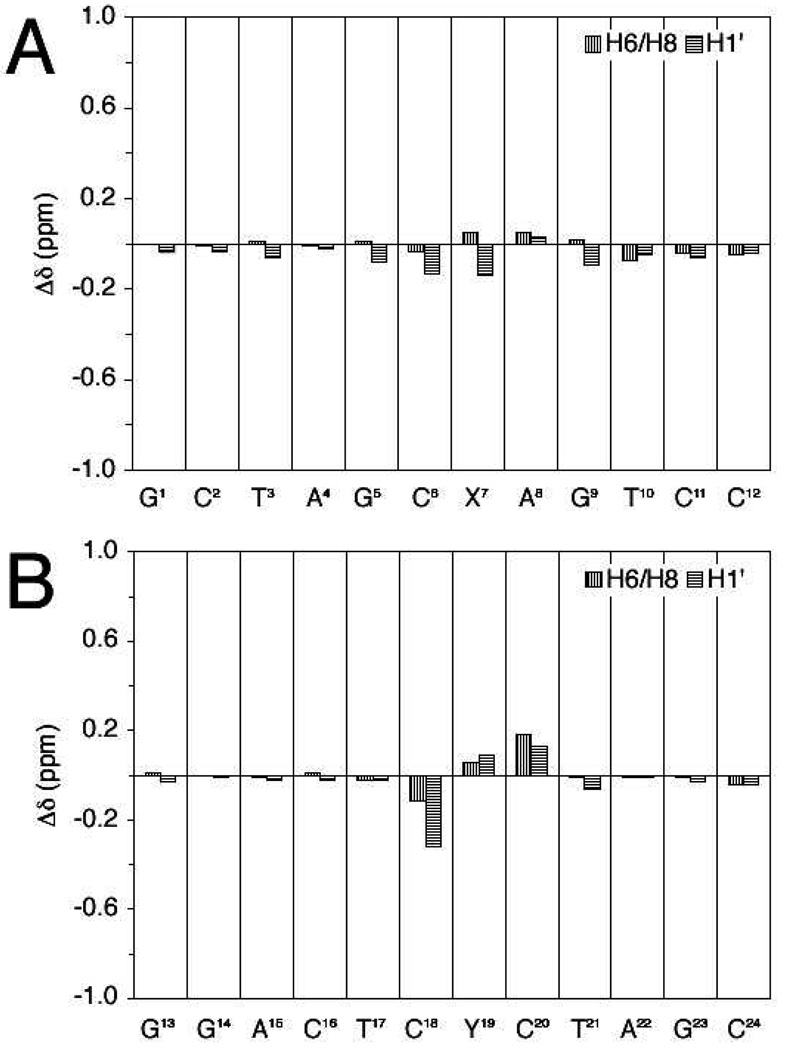
The chemical shifts were compared with the corresponding unmodified duplex (Figure 5). Small chemical shift perturbations were observed for most of the nucleotides. The X7•C18 base pair was minimally perturbed, and the C18 H6 and C18 H1′ resonances shifted upfield 0.11 and 0.32 ppm, respectively. The chemical shift perturbations for C6•Y19 base pair were not remarkable. The C6 H6, C6 H1′, Y19 H8, and Y19 H1′ resonances shifted −0.03, −0.13, 0.06, and 0.09 ppm, respectively. Significantly, the 5'-neighboring C20 H6 had the most significant perturbation among the aromatic protons, 0.18 ppm downfield from the related proton in the unmodified sequence. C20 H5 and C20 H1′ also shifted downfield, 0.22 and 0.13 ppm, respectively, indicating the neighboring C20 was involved in the cross-link formation.
Figure 5.
Chemical shift perturbations of the (R)-γ-hydroxytrimethylene interstrand cross-link compared to the unmodified duplex.
The backbone conformation was evaluated by the distribution of phosphorus resonances. The assignments of the C6 and C18 H3′ protons of the cross-link were based upon their H1′→H3′ and H6→H3′ NOEs, respectively. They exhibited 31P→H3′ correlations in the HMBC spectrum. Compared with other nucleotides, the C6 and C18 phosphates were shifted downfield, indicating that the backbone at the cross-linked base pairs was perturbed from ideal B-DNA.
Structural Refinement of the (R)-γ-hydroxytrimethylene Cross-Link
The cross-link was simulated in explicit water at 300 K. The nineteen NOE cross peaks in the 15N NOESY-HSQC spectra assigned to X7 N2H and Y19 N2H were converted to distance restraints to refine the cross-link tether. In addition, 52 empirical distance restraints and 200 empirical torsion angle restraints were used to maintain Watson-Crick hydrogen bonding and B-type DNA, respectively.
A fully solvated molecular dynamics simulation of 8 ns was carried out starting from both A- and B-type DNAs. The all-atom mass-weighted root-mean-square deviations (RMSDs) referenced to the starting structures were used to determine the time for the simulations to reach equilibrium, and in turn, the range of the trajectories to be used for the sequence analysis. The simulation equilibrated within 500–600 ps starting from A-type DNA, and 200–300 ps starting from B-type DNA.
The rMD trajectories after simulating for 1 ns were used to analyze the potential hydrogen bonding of the carbinol hydroxyl group of the tether. Its occupancy by hydrogen bond acceptors in the rMD trajectories revealed that it was likely to form a hydrogen bond in solution. Starting from both A- and B-type DNA, the simulations predicted the same hydrogen bond trends (Table 1). The carbinol hydroxyl group did not form a hydrogen bond with the solvent throughout the simulations. The duplex tended to adopt a conformation in which the hydroxyl group formed a hydrogen bond with C20 O2, although the potential for a hydrogen bond to C20 O4′ was also observed. No hydrogen bonds with C6 O2 or Y19 N3 were formed.
Table 1.
Occupancy of the Hydroxyl Group by the Hydrogen Bond Acceptors During Molecular Dynamics Simulations, and the Distances and Angles of the Corresponding Atoms in the Average Structure of the (R)-γ-Hydroxytrimethylene Cross-link.
| potential hydrogen bond |
occupancy of OH (%) a | distance (Å) | angle (°) | |
|---|---|---|---|---|
| from A-DNA | from B-DNA | |||
| Carbinol OH→C20 O2 | 62 | 74 | 2.1 | 177 |
| Carbinol OH→C20 O4′ | 22 | 14 | 3.0 | 97 |
| Carbinol OH→Y19 N3 | 2.7 | 1.7 | 3.2 | 103 |
| Carbinol OH→C6 O2 | 1.0 | 0.70 | 3.6 | 93 |
| Carbinol OH→solvent | 0.20 | 0 | ||
The criteria for a hydrogen bond formation: distance ≤ 3.5 Å and angle ≥ 120°.
The trajectories of the molecular dynamics simulations from 1–8 ns starting from A- and B-type DNAs were averaged. The RMSD value of these two refined structures was 0.54 Å, reflecting the similarities of the converged structures. The atom distances agreed with the NOE intensities (Table 1) and suggested that the refined structures obtained from the molecular dynamics simulations were accurate. Significantly, for the refined structures, the Y19 N2H→ Hγ distance was 3.0 Å, which corroborated the observation of a weak NOE between Y19 N2H and Hγ. The NOEs in Figures 3C and 3D were not used as the distance restraints. The distances of X7 H8→C6 H1′, A8 H8→X7 H1′ and Y19 H8→C18 H1′ in the average structure were 6.0, 2.9, and 5.5 Å, respectively. Consistently, the X7 H8→C6 H1′ and Y19 H8→C18 H1′ NOEs were missing and A8 H8→X7 H1′ NOE were stronger than other NOEs of aromatic protons with the 5′-neighbor H1′ protons (Figures 3C and 3D). Other isolated NOEs of the cross-link in Figures 3C and 3D also agreed with the average structure. These indicated that the average structure provided an accurate depiction of the NMR data.
Refined Structure of the (R)-γ-hydroxytrimethylene Cross-Link
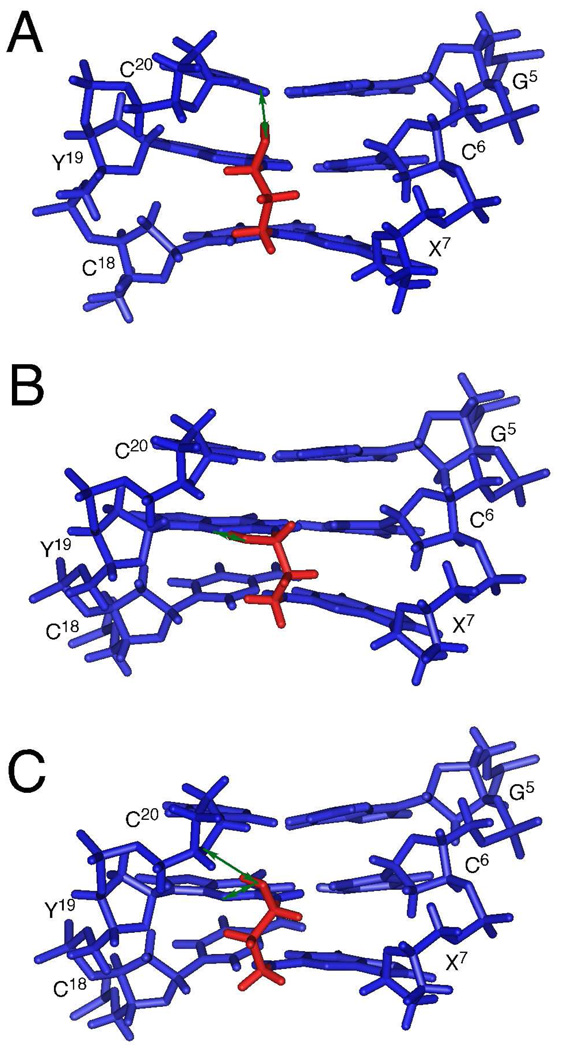
Starting from either A- or B-type DNA, the cross-link converged to B-type with the tether located in the minor groove. Figure 6A provides an expanded view of the average structure observed from the minor groove. Consistent with Y19 N1H→C6 N4Hs and X7 N1H→C18 N4Hs NOEs (Figure 2), the C6•Y19 and X7•C18 base pairs conserved Watson-Crick hydrogen bonding with small distortions. They were closer to each other as compared to other base pairs. The backbone torsion angles of C6, X7, C18, and Y19 were not ideal B-form geometry and agreed with the downfield shifts of C6 and C18 phosphates.
Figure 6.
Expanded views, observed from the minor groove, of the diastereomeric carbinolamine interstrand cross-links. (A) Average structure of the (R)-γ-hydroxytrimethylene cross-link. (B) Predicted structure of the (S)-γ-hydroxytrimethylene cross-link from molecular dynamics simulations starting from A-DNA. (C) Predicted structure of the (S)-γ-hydroxytrimethylene cross-link from molecular dynamics simulations starting from B-DNA. Blue and red sticks represent nucleotides and the γ-hydroxytrimethylene tether, respectively. The green arrows indicate putative hydrogen bonds involving the hydroxyl groups.
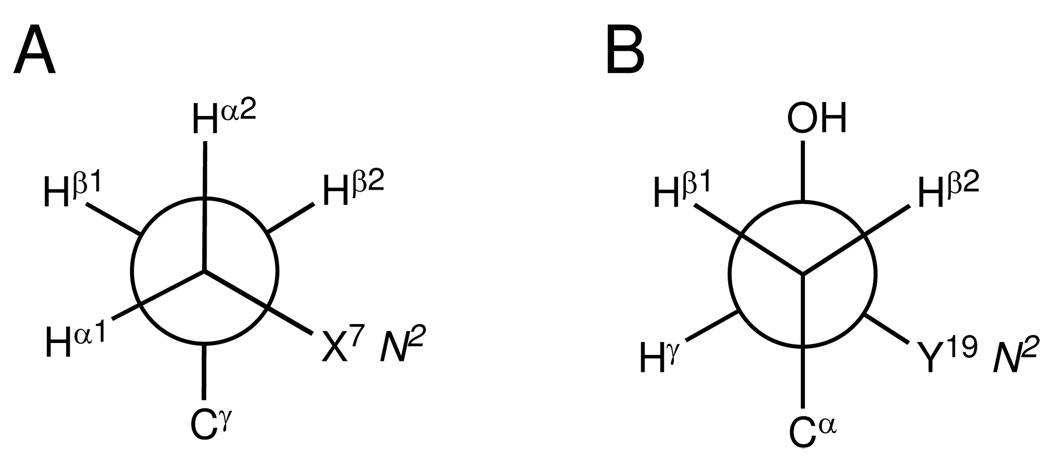
The (R)-γ-hydroxytrimethylene tether was located in the minor groove (Figure 6A). The β-carbon oriented towards C6 and X7. Figure 7 provides Newman projections of the tether viewed along the Cα-Cβ and Cβ-Cγ bonds. Both X7 N2 and Y19 N2 were in the gauche-conformation with respect to the (R)-γ-hydroxytrimethylene tether, which facilitated the Watson-Crick hydrogen bonding of the C6•Y19 and X7•C18 base pairs. The trans-configuration of the hydroxyl group with Cα minimized their steric interaction.
Figure 7.
Newman projections showing conformation of the (R)-γ-hydroxytrimethylene tether. (A) Viewed along Cα-Cβ bond. (B) Viewed along Cβ-Cγ bond. Both X7 N2 and Y19 N2 were in the gauche-conformation with the tether to maintain the Watson-Crick hydrogen bonding. The trans-conformation of hydroxyl group with Cα minimizes the steric interaction and allows the formation of the carbinol OH→C20 O2 hydrogen bond.
As shown in Figure 6A, the hydroxyl group of the (R)-γ-hydroxytrimethylene tether oriented towards C20 O2 and formed a hydrogen bond. The distance and angle of the carbinol OH→C20 O2 in the average structure were 2.1 Å and 177°, respectively (Table 2). No hydrogen bond interaction existed between the hydroxyl group and C6 O2, Y19 N3, or C20 O4′. The orientation of the hydroxyl group towards C20 O2 agreed with its occupancy by the hydrogen bond acceptors in the trajectory analysis (Table 1). The hydrogen bond was also consistent with the NMR data. The G5•C20 base pair was stabilized by the cross-link (Figure 4). Chemical shift perturbations of C20 H5, H6, and H1′ were observed (Figure 5). In contrast, chemical shifts of the corresponding cytosine H5, H6, and H1′ protons in the interstrand cross-links bridged by either trimethylene34 or α-methyltrimethylene tether,35 in which a hydrogen bond is not possible, underwent small perturbations.
Molecular Dynamics Simulations of the (S)-γ-Hydroxytrimethylene Tether
It was not possible to obtain NOE-based distance restraints for the (S)-γ-hydroxytrimethylene tether, which was present in solution at only low levels (Figure 1). Consequently, to explore the conformational possibilities for this stereoisomeric of the cross-link, non-restrained molecular dynamics simulations were carried out in explicit solvent, starting from either A- or B-DNAs, respectively. Figures 6B and 6C show expanded views of the refined structures starting from A- and B-type DNAs, respectively. In both cases, the (S)-γ-hydroxytrimethylene tether converged to similar structures, but which were different from the refined structures of the (R)-γ-hydroxymethylene tether. For the (S)-γ-hydroxytrimethylene tether, the calculations predicted that the carbinol hydroxyl might form a hydrogen bond with Y19 N3 (Figure 6B) or a 3-point hydrogen bond with Y19 N3 and C20 O4′ (Figure 6C). The distance for the Y19 N2H to the Hγ for the (S)-γ-hydroxytrimethylene tether, was 2.2 or 2.3 Å, respectively.
Discussion
Interstrand DNA cross-links block DNA replication and transcription, leading to cytotoxic and mutagenic effects if not repaired.1,2 A DNA interstrand cross-link that covalently connects both strands represents a challenge for the cell and an elaborate mechanism is required for its repair.3 The DNA interstrand cross-link induced by the exocyclic 1,N2-dG adduct 1 exists as an equilibrium mixture of carbinolamine 4, imine 5, and pyrimidopurinone 6. We have demonstrated that carbinolamine 4 is the only observable cross-linked species in duplex DNA.29 Imine 5 and pyrimidopurinone 6 are predicted to cause significant perturbations to the DNA duplex and are thereby disfavored. Carbinolamine 4 might be anticipated to be readily reversible,40–42 yet it stabilizes the DNA duplex with respect to thermal denaturation (Tm > 90° C); thus, the forces involved in stabilizing these stereoisomeric carbinolamine interstrand cross-links are of interest.
(R)-γ-Hydroxytrimethylene Linkage as the Major Interstrand Cross-Link
The weak Y19 N2H→Hγ and Y19 H1′→Hγ NOE interactions (Figure 1) suggest that the (R)-γ-hydroxytrimethylene linkage is the major form of the interstrand cross-link. The (R)-γ-hydroxytrimethylene interstrand cross-link conserves B-type DNA geometry. The (R)-γ-hydroxytrimethylene tether has a minor groove orientation and adopts the U-conformation.34 Watson-Crick hydrogen bonding of the cross-linked C6•Y19 and X7•C18 base pairs is conserved (Figure 6A). X7 N2 and Y19 N2 are in the gauche-orientation with respect to the α- or γ-carbons, respectively, in the extended chain conformation (Figure 7). The C6•Y19 and X7•C18 base pairs are placed closer as compared to others.
Role of Hydrogen Bonding in Interstrand Cross-Link Formation
The orientation of the (R)-γ-hydroxytrimethylene tether towards C6 and X7 minimizes the steric interaction of the bulky substituents (Figure 7) and more importantly, facilitates the formation of a hydrogen bond between the hydroxyl group and C20 O2 (Figure 6A), such as has been observed for the active sites of type I aldolases in which carbinolamine is stabilized in the active site.43–46 Thermal denaturation of the DNA duplex results in the loss of this hydrogen bond, allowing the (R)-γ-hydroxytrimethylene tether to quickly dissociate.5 The formation of this hydrogen bond to C20 O2 augments the Watson-Crick hydrogen bonding (Figure 4) and base stacking of the cross-linked base pairs.
The presence of this hydrogen bond to C20 O2 may explain the favored (R) stereochemistry of the carbinolamine linkage. These calculations suggested that for the (S) stereochemistry, the hydroxyl group might form a hydrogen bond with Y19 N3 (Figure 6B) or a 3-point hydrogen bond with Y19 N3 and C20 O4′ (Figure 6C), neither of which would be expected to stabilize the duplex in the same manner as the cross-link to C20 O2. Overall, we suggest that formation of the carbinolamine linkage favors the (R) stereochemistry because of the favorable energetics associated with hydrogen bonding to C20 O2.
Comparison to Reduced Trimethylene Cross-link Structures
Dooley et al.33,34 cross-linked DNA duplexes with a trimethylene tether to simulate the DNA interstrand cross-links induced by the malondialdehyde and acrolein derived 1,N2-dG adducts. In contrast to the present results, they concluded that the reduced interstrand cross-link badly perturbed the Watson-Crick hydrogen bonding of the cross-linked base pairs.34 In their structures, the trimethylene tether adopted an extended chain W-conformation and was accommodated by unwinding of the duplex at the cross-linking base pairs to produce a bulge. In the extended chain conformation of the tether, the exocyclic nitrogens of each deoxyguanosine were in the anti-orientation with respect to the α- or γ-carbons of the reduced tether, respectively.
Crotonaldehyde-Derived DNA Interstrand Cross-links
The α-methyl group of the crotonaldehyde lesion can assume either the R or S configuration. In this 5′-CpG-3′ sequence the crotonaldehyde derived 1,N2-dG adduct with the R configuration of the α-methyl group induces DNA interstrand cross-links more efficiently than does the adduct with the S-configuration of the α-methyl group. The R configuration of the α-methyl group orients the reactive aldehyde proximal to the N2-dG cross-linking site in the complementary strand, and, moreover, the resulting carbinolamine cross-link results in less structural perturbation to the DNA duplex.28,35
The stereochemistry of the carbinolamine carbon of the native carbinolamine cross-links is not available. The orientations of the stereoisomeric α-methyltrimethylene tethers of the reduced analogs are different. 35 For both, the steric interaction of the methyl group was minimized by the anti-conformation of the methyl group with respect to Cγ. The orientation of the (S)-α-methyltrimethylene tether also reduced the steric interference of the α-methyl group with the neighbor A8•T17 base pair. Extrapolating this structural information to the native cross-link suggests that the orientation of the (R)-stereochemistry of the carbinolamine would not be positioned to form the same hydrogen bond with C20 O2 at the 5'-neighbor base pair as presently observed for the acrolein-derived 1,N2-dG exocyclic adduct. 35 Instead, formation of a hydrogen bond to C20 O2 by the (R)-γ-hydroxytrimethylene linkage arising from the crotonaldehyde-derived R-α-methyltrimethylene tether would require re-orientation of the α-CH3 group and Cγ into the gauche-conformation, which would be predicted to be less stable than the conformation of the (R)-γ-hydroxytrimethylene tether induced by acrolein derived 1,N2-dG adduct. This might explain, in part, why the crotonaldehyde-derived 1,N2-dG lesion having the R configuration of the α-methyl group ultimately produces less of the interstrand cross-links in the 5′-CpG-3′ sequence than does the acrolein-derived 1,N2-dG adduct.5
Summary
When placed opposite deoxycytosine in the 5′-CpG-3′duplex, the acrolein-derived exocyclic 1,N2-dG adduct induces stereoisomeric carbinolamine interstrand cross-links. The R-configuration of the carbinolamine linkage is favored. The formation of a hydrogen bond between the hydroxyl group and C20 O2 located in the 5'-neighboring base pair G5•C20, may in part explain the stereochemical preference for the formation of the (R)-γ-hydroxytrimethylene linkage and its thermal stability in duplex DNA. The cross-link causes minimal structural perturbations to the DNA duplex, and allows maintenance of Watson-Crick hydrogen bonding and base stacking at the tandem cross-linked base pairs.
Experimental Section
Materials
The oligodeoxynucleotide 5'-d(GGACTCGCTAGC)-3' was synthesized and purified by anion-exchange chromatography by the Midland Certified Reagent Co. (Midland, TX). The G19 N2 15N-labeled 5′-d(GGACTCGCTAGC)-3′ and the oligodeoxynucleotides containing the acrolein-derived exocyclic 1,N2-dG adduct in the dodecamer 5'-d(GCTAGCXAGTCC)-3' (X=1,N2-dG adduct 1, whether N2 15N-labeled or not), were synthesized, purified, and characterized as reported.29,47 The purities of the oligodeoxynucleotides were assessed by capillary gel electrophoresis, HPLC, and MALDI-TOF mass spectroscopy. Oligodeoxynucleotides were desalted by chromatography on Sephadex G-25. The stereoisomeric carbinolamine interstrand cross-links were prepared at pH 7.0 at 37°C, as reported.5 Reaction was monitored by 15N HSQC experiments.
NMR
Samples for the NMR experiments were at 1.0 mM strand concentration. Samples for the non-exchangeable protons were dissolved in 280 µL of a buffer containing 10 mM NaH2PO4, 100 mM NaCl, 50 µM Na2EDTA (pH 7.0). They were exchanged with D2O and suspended in 280 µL 99.996% D2O. The pH was adjusted to 7.0 with dilute DCl or NaOD solutions. Samples for the observation of exchangeable protons were dissolved in 280 µL of 10 mM NaH2PO4, 100 mM NaCl, 50 µM EDTA, (pH 7.0) containing 9:1 H2O/D2O (v/v) and the pH was adjusted to 7.0. 31P experiments were carried out on a Bruker Avance 600 spectrometer. Other NMR experiments were performed on a Bruker Avance 500 spectrometer with a cryogenic probe (Bruker Instruments). The temperature was 25 °C for observation of the non-exchangeable protons and 5 °C for observation of the exchangeable protons. Chemical shifts for 1H were referenced to water, and chemical shifts for 13C, 15N and 31P were referenced indirectly. Data were processed using FELIX 2000 (Accelrys Inc.). For all NMR experiments, a relaxation delay of 1.5 s was used. Two-dimensional homonuclear NMR spectra were recorded with 512 real data in the t2 dimension and 2048 real data in the t1 dimension. NOESY spectra were zero-filled during processing to create a matrix of 1024 × 1024 real points, other spectra were zero-filled to create a matrix of 1024 × 512 real points. For assignment of exchangeable protons, NOESY experiments used the WATERGATE sequence.48 The mixing time was 250 ms. For assignment of non-exchangeable protons and the derivation of distance restraints, NOESY experiments used TPPI quadrature detection and a mixing time of 250 ms were used. 2D 15N NOESY-HSQC pulse programs were modified from corresponding 3D pulse program49,50 with 128 points centered at 81ppm. The water signal was suppressed with the WATERGATE method. The mixing time was 250 ms. The spectrum was recorded with 512 real data in the t2 dimension and 2048 real data in the t1 dimension and was zero-filled during process to create 1024×512 matrix.
Experimental Restraints
NOE-derived distance restraints used for the refinement of the (R)-γ-hydroxytrimethylene structure were calculated from NOE cross-peak volumes in the 15N NOESY-HSQC spectra using MARDIGRAS v3.2.51,52 Isotropic correlation times 2, 3 and 4 ns were used. The relaxation time for the analyses was 1.5 s. The volume error was one-half the volume of the smallest peak. Volumes were normalized based on all the peak intensities. The RANDMARDI algorithm carried out 50 iterations for each set of data, randomizing peak volumes within limits specified by the input noise level.53 The distances were averaged. The standard deviation in a particular distance served as the error bond for the distance. Empirical restraints preserving Watson-Crick hydrogen bonding and preventing propeller twisting between basepairs were used.54 The backbone and sugar pucker torsion angle restraints were also derived using empirical data derived from B-DNA.55
Structural Refinement of the (R)-γ-Hydroxytrimethylene Cross-Link
Molecular dynamics simulations used the AMBER force field (Vol. 9, parm99 force field).56 Partial charges of the carbinolamine linkage were obtained by DFT calculation at B3LYP/6-31G* level with Gaussian 03.57 Starting structures were constructed from A- and B-type DNA using Insight II. The duplexes were energy minimized by conjugate gradients for 1,000 iterations without experimental and empirical restraints. Molecular dynamics simulations were carried out in explicit water using the particle mesh Ewald (PME) method. DNA duplexes were placed in a 10 Å cubic TIP3P water box. 22 Na+ ions were added to neutralize the DNA interstrand cross-link. The cutoff radius for nonbonding interactions was 10 Å. The restraint energy function contained terms describing distance and torsion angle restraints, both in the form of square well potentials. Bond lengths involving hydrogens were fixed with the SHAKE algorithm.58
Firstly, a 1,000-step energy minimization was performed with an integrator time of 1 fs with the heavy atoms of the cross-link restrained. A 10,000-iteration simulation was carried out with constant volume at 300 K. The heavy atoms of the cross-link were restrained. Then, molecular dynamics simulation at constant pressure was performed at 300 K for 8 ns with an integrator time of 1 fs. Experimental and empirical distance and torsion angle restraints with force constants of 32.0 kcal·mol−1·Å−1 were applied. One trajectory was saved in each 1,000-iteration during simulation. The trajectories after reaching simulation equilibrium were averaged. The mean structures were filtered out from water box and energy-minimized for 250 iterations without experimental and empirical restraints.
The PTRAJ program from AMBER 9 package was used to analyze the trajectories. The RMSD values of the trajectories were referenced to the starting structures. A distance of less than 3.5 Å, and an angle of greater than 120° between the potential hydrogen donor and acceptor, were used as criteria for hydrogen bond formation. The backbone conformation of the average structure was analyzed with the program 3DNA.59
Structural Refinement of the (S)-γ-Hydroxytrimethylene Cross-Link
Similar molecular dynamics simulations were performed to refine the structure of the (S)-γ-hydroxytrimethylene cross-link. Because this was the minor species in solution, it was not possible to obtain experimental NOEs, and thus molecular dynamics simulations were carried out without experimental distance restraints.
Supplementary Material
Acknowledgement
Dr. Markus Voehler assisted with NMR experiments. This work was supported by NIH grant PO1 ES-05355 (C.J.R., T.M.H., and M.P.S.).
Footnotes
Supporting Information Available
Table S1, NOEs used in the rMD simulations and the corresponding atom distances in the average refined structure; complete references 56 and 57. This material is available free of charge via Internet at http://pubs.acs.org.
References
- 1.Lawley PD, Brookes P. Nature. 1965;206:480–483. doi: 10.1038/206480a0. [DOI] [PubMed] [Google Scholar]
- 2.Lawley PD, Brookes P. J. Mol. Biol. 1967;25:143–160. doi: 10.1016/0022-2836(67)90285-9. [DOI] [PubMed] [Google Scholar]
- 3.Noll DM, Mason TM, Miller PS. Chem. Rev. 2006;106:277–301. doi: 10.1021/cr040478b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kozekov ID, Nechev LV, Sanchez A, Harris CM, Lloyd RS, Harris TM. Chem. Res. Toxicol. 2001;14:1482–1485. doi: 10.1021/tx010127h. [DOI] [PubMed] [Google Scholar]
- 5.Kozekov ID, Nechev LV, Moseley MS, Harris CM, Rizzo CJ, Stone MP, Harris TM. J. Am. Chem. Soc. 2003;125:50–61. doi: 10.1021/ja020778f. [DOI] [PubMed] [Google Scholar]
- 6.Sanchez AM, Kozekov ID, Harris TM, Lloyd RS. Chem. Res. Toxicol. 2005;18:1683–1690. doi: 10.1021/tx0500528. [DOI] [PubMed] [Google Scholar]
- 7.Wang H, Kozekov ID, Harris TM, Rizzo CJ. J. Am. Chem. Soc. 2003;125:5687–5700. doi: 10.1021/ja0288800. [DOI] [PubMed] [Google Scholar]
- 8.Witz G. Free Radic. Biol. Med. 1989;7:333–349. doi: 10.1016/0891-5849(89)90137-8. [DOI] [PubMed] [Google Scholar]
- 9.Stevens JF, Maier CS. Mol. Nutr. Food Res. 2008;52:7–25. doi: 10.1002/mnfr.200700412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pan J, Chung FL. Chem. Res. Toxicol. 2002;15:367–372. doi: 10.1021/tx010136q. [DOI] [PubMed] [Google Scholar]
- 11.Chung FL, Nath RG, Nagao M, Nishikawa A, Zhou G-D, Randerath K. Mutat. Res. 1999;424:71–81. doi: 10.1016/s0027-5107(99)00009-3. [DOI] [PubMed] [Google Scholar]
- 12.Wilson VL, Foiles PG, Chung FL, Povey AC, Frank AA, Harris CC. Carcinogenesis. 1991;12:1483–1490. doi: 10.1093/carcin/12.8.1483. [DOI] [PubMed] [Google Scholar]
- 13.Nath RG, Ocando JE, Chung FL. Cancer Res. 1996;56:452–456. [PubMed] [Google Scholar]
- 14.Nath RG, Chung F-L. Proc. Natl. Acad. Sci. USA. 1994;91:7491–7495. doi: 10.1073/pnas.91.16.7491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Benedetti A, Comporti M, Esterbauer H. Biochim. Biophys. Acta. 1980;620:281–296. doi: 10.1016/0005-2760(80)90209-x. [DOI] [PubMed] [Google Scholar]
- 16.Esterbauer H, Schaur RJ, Zollner H. Free Radic. Biol. Med. 1991;11:81–128. doi: 10.1016/0891-5849(91)90192-6. [DOI] [PubMed] [Google Scholar]
- 17.Burcham PC. Mutagenesis. 1998;13:287–305. doi: 10.1093/mutage/13.3.287. [DOI] [PubMed] [Google Scholar]
- 18.Sierra LM, Barros AR, García M, Ferreiro JA, Comendador MA. Mutat. Res. 1991;260:247–256. doi: 10.1016/0165-1218(91)90033-i. [DOI] [PubMed] [Google Scholar]
- 19.Yang IY, Johnson F, Grollman AP, Moriya M. Chem. Res. Toxicol. 2002;15:160–164. doi: 10.1021/tx010123c. [DOI] [PubMed] [Google Scholar]
- 20.Czerny C, Eder E, Rünger TM. Mutat. Res. 1998;407:125–134. doi: 10.1016/s0921-8777(97)00069-4. [DOI] [PubMed] [Google Scholar]
- 21.Jha AM, Singh AC, Sinha U, Kumar M. Mutat .Res. 2007;632:69–77. doi: 10.1016/j.mrgentox.2007.04.008. [DOI] [PubMed] [Google Scholar]
- 22.Eckl PM. Mol. Aspects Med. 2003;24:161–165. doi: 10.1016/s0098-2997(03)00010-4. [DOI] [PubMed] [Google Scholar]
- 23.Chung F-L, R Y, Hecht SS. Cancer Res. 1984;44:990–995. [PubMed] [Google Scholar]
- 24.Gölzer P, Janzowski C, Pool-Zobel BL, Eisenbrand G. Chem. Res. Toxicol. 1996;9:1207–1213. doi: 10.1021/tx9600107. [DOI] [PubMed] [Google Scholar]
- 25.Chung FL, Young R, Hecht SS. Carcinogenesis. 1989;10:1291–1297. doi: 10.1093/carcin/10.7.1291. [DOI] [PubMed] [Google Scholar]
- 26.Chung FL, Chen HJ, Nath RG. Carcinogenesis. 1996;17:2105–2111. doi: 10.1093/carcin/17.10.2105. [DOI] [PubMed] [Google Scholar]
- 27.Chung FL, Nath RG, Ocando J, Nishikawa A, Zhang L. Cancer Res. 2000;60:1507–1511. [PubMed] [Google Scholar]
- 28.Cho Y-J, Wang H, Kozekov ID, Kurtz AJ, Jacob J, Voehler M, Smith J, Harris TM, Lloyd RS, Rizzo CJ, Stone MP. Chem. Res. Toxicol. 2006;19:195–208. doi: 10.1021/tx050239z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cho Y-J, Kim H-Y, Huang H, Slutsky A, Minko IG, Wang H, Nechev LV, Kozekov ID, Kozekova A, Tamura P, Jacob J, Voehler M, Harris TM, Lloyd RS, Rizzo CJ, Stone MP. J. Am. Chem. Soc. 2005;127:17686–17696. doi: 10.1021/ja053897e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.De los Santos C, Zaliznyak T, Johnson F. J. Biol. Chem. 2001;276:9077–9082. doi: 10.1074/jbc.M009028200. [DOI] [PubMed] [Google Scholar]
- 31.Huang H, Wang H, Qi N, Kozekova A, Rizzo CJ, Stone MP. J. Am. Chem. Soc. 2008;130:10898–10906. doi: 10.1021/ja801824b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stone MP, Cho YJ, Huang H, Kim HY, Kozekov ID, Kozekova A, Wang H, Minko IG, Lloyd RS, Harris TM, Rizzo CJ. Acc. Chem. Res. 2008 doi: 10.1021/ar700246x. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dooley PA, Zhang M, Korbel GA, Nechev LV, Harris CM, Stone MP, Harris TM. J. Am. Chem. Soc. 2003;125:62–72. doi: 10.1021/ja0207798. [DOI] [PubMed] [Google Scholar]
- 34.Dooley PA, Tsarouhtsis D, Korbel GA, Nechev LV, Shearer J, Zegar IS, Harris CM, Stone MP, Harris TM. J. Am. Chem. Soc. 2001;123:1730–1739. doi: 10.1021/ja003163w. [DOI] [PubMed] [Google Scholar]
- 35.Cho YJ, Kozekov ID, Harris TM, Rizzo CJ, Stone MP. Biochemistry. 2007;46:2608–2621. doi: 10.1021/bi061381h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Huang H, Wang H, Qi N, Lloyd RS, Rizzo CJ, Stone MP. Biochemistry. 2008;47:11457–11472. doi: 10.1021/bi8011143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kim HY, Voehler M, Harris TM, Stone MP. J. Am. Chem. Soc. 2002;124:9324–9325. doi: 10.1021/ja020333r. [DOI] [PubMed] [Google Scholar]
- 38.Patel DJ, Shapiro L, Hare D. Q. Rev. Biophys. 1987;20:35–112. doi: 10.1017/s0033583500004224. [DOI] [PubMed] [Google Scholar]
- 39.Reid BR. Q. Rev. Biophys. 1987;20:2–28. doi: 10.1017/s0033583500004212. [DOI] [PubMed] [Google Scholar]
- 40.Jencks WP. Catalysis in Chemistry and Enzymology. New York: McGraw-Hill; 1969. [Google Scholar]
- 41.Bruylants A, Feytmants-de Medicis E. In: Chemistry of Carbon-Nitrogen Double Bonds. Patai S, editor. London: Interscience; 1970. pp. 465–504. [Google Scholar]
- 42.Bruckner R. Advanced Organic Chemistry: Reaction Mechanisms. San Diego: Harcourt/Academic Press; 2002. [Google Scholar]
- 43.Allard J, Grochulski P, Sygusch J. Proc. Natl. Acad. Sci. USA. 2001;98:3679–3684. doi: 10.1073/pnas.071380898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Erskine PT, Newbold R, Brindley AA, Wood SP, Shoolingin-Jordan PM, Warren MJ, Cooper JB. J. Mol. Biol. 2001;312:133–141. doi: 10.1006/jmbi.2001.4947. [DOI] [PubMed] [Google Scholar]
- 45.Thorell S, Schürmann M, Sprenger GA, Schneider G. J. Mol. Biol. 2002;319:161–171. doi: 10.1016/S0022-2836(02)00258-9. [DOI] [PubMed] [Google Scholar]
- 46.Heine A, DeSantis G, Luz JG, Mitchell M, Wong C-H, Wilson IA. Science. 2001;294:369–374. doi: 10.1126/science.1063601. [DOI] [PubMed] [Google Scholar]
- 47.Nechev LV, Harris CM, Harris TM. Chem. Res. Toxicol. 2000;13:421–429. doi: 10.1021/tx990167+. [DOI] [PubMed] [Google Scholar]
- 48.Piotto M, Saudek V, Sklenar V. J. Biomol. NMR. 1992;2:661–665. doi: 10.1007/BF02192855. [DOI] [PubMed] [Google Scholar]
- 49.Mori S, Abeygunawardana C, Johnson M, vanZul PCM. J. Magn. reson. B. 1995;108:94–98. doi: 10.1006/jmrb.1995.1109. [DOI] [PubMed] [Google Scholar]
- 50.Talluri S, Wagner G. J. Magn. Reson. B. 1996;112:200–205. doi: 10.1006/jmrb.1996.0132. [DOI] [PubMed] [Google Scholar]
- 51.Borgias BA, James TL. J. Magn. Reson. 1990;87:475–487. [Google Scholar]
- 52.Liu H, Tonelli M, James TL. J. Magn. Reson. B. 1996;111:85–89. doi: 10.1006/jmrb.1996.0064. [DOI] [PubMed] [Google Scholar]
- 53.Liu H, Spielmann HP, Ulyanov NB, Wemmer DE, James TL. J. Biomol. NMR. 1995;6:390–402. doi: 10.1007/BF00197638. [DOI] [PubMed] [Google Scholar]
- 54.Kouchakdjian M, Marinelli E, Gao X, Johnson F, Grollman A, Patel D. Biochemistry. 1989;28:5647–5657. doi: 10.1021/bi00439a047. [DOI] [PubMed] [Google Scholar]
- 55.Blackburn GM, Gait MJ. Nucleic Acids in Chemistry and Biology. New York: Oxford University Press; 1996. [Google Scholar]
- 56.Case DA, et al. AMBER 9. San Francisco: San Francisco, CA: University of California; 2006. [Google Scholar]
- 57.Frisch MJ, et al. GAUSSIAN 03. Wallingford, CT: Gaussian, Inc.; 2004. [Google Scholar]
- 58.Ryckaert J-P, Ciccotti G, Berendsen HJC. J. Comput. Phys. 1977;23:327–341. [Google Scholar]
- 59.Lu X-J, Olson WK. Nucleic Acids Res. 2003;31:5108–5121. doi: 10.1093/nar/gkg680. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.