Abstract
An amended kinetic model for the self-assembly of empty capsids of brome mosaic virus is proposed. The model has been modified to account for a new feature in the assembly kinetics revealed by time-course light scattering experiments at higher temporal resolution than previously attempted. To be able to simulate the sharp takeoff from the initial lag phase to the growth phase in the kinetic curves, a monomer activation step was proposed.
Introduction
A significant fraction of viruses employ icosahedral protein capsids to protect and deliver their genomes. In vivo, nonenveloped icosahedral viruses assemble from protein subunits and nucleic acids. In some cases, the self-assembly process of infectious virions from protein and nucleic acid subunits can be reproduced in vitro.1 Moreover, in certain conditions of temperature, ionic strength and pH, the in vitro assembly of empty capsids is possible. The empty capsids of some viruses, e.g., Brome Mosaic Virus (BMV),2 papillomavirus,3 phage P22,4,5 Cowpea Chlorotic Mottle Virus (CCMV),6 and Hepatitis B Virus (HBV)7 are structurally indistinguishable from the capsids of infective viruses.
Understanding the kinetics of empty capsid assembly represents the first step in understanding whole-virion assembly and may provide new ways of developing antivirals and virus-based therapeutics.
Interestingly, a few common features of the icosahedral capsid assembly are conserved, independent of the type of virus:8-12
The time-dependent capsid growth is sigmoidal, with a lag phase followed by a rapid increase which then levels off.
Few intermediates are accumulated during the assembly;
Increasing protein concentration accelerates the reaction rate and there is a critical concentration below which no noticeable assembly occurs.
Malformations are occasionally observed under optimal assembly conditions but can be significant at other temperatures, concentrations, pH or ionic strength.
A significant amount of theoretical work has been done to rationalize these qualitative observations. The classic nucleation theory has logically been a big interest,13,14 and there have been attempts to seek hints from structural analysis15-17 concerning the specific forms of assembly nuclei. However, the predicted stable capsomers that were supposed to serve as the nuclei during capsid assembly did not agree well with experimental findings18 and capsid binding energies evaluated from structural analysis (from 15 kcal/mol to more than 100 kcal/mol)19,20 were usually much higher than assembly kinetics suggested (<10 kcal/mol).21
Although quantitative modeling of capsid assembly is still in its infancy, simulations have provided insights into the most general qualitative features. For example, a local rules model based upon subunits with a reduced number of degrees of freedom and generic interactions was proposed to explain the occurrence of icosahedral capsid out of several different possible outcomes.15,16 Molecular dynamics models employed protein subunits as blocks of well-defined shape and specific intersubunit interactions, which allowed studies of the temperature dependence of reaction rates and products.17,22 These models have suggested that the capsid grows mainly through monomer addition. Zlotnick et al. have described the assembly kinetics phenomenologically by using rate equations. Two models have emerged: the equilibrium model12 and the kinetically limiting (KL) model.11 Both are based on the assumption that the assembly follows a predefined cascade of monomer addition in agreement with molecular dynamics predictions. Both models are able to reproduce the observed sigmoidal kinetics of capsid growth and they predict only trace amounts of intermediates. Moreover, an analysis of early assembly extension and elongation rates can be used to infer the nature of the initial stable intermediate, the so-called “nucleus”.
In the present paper, a fast time-course light scattering study of assembly kinetics of empty BMV capsids is reported that reveals several new details of the assembly process:
(1) A more abrupt initial takeoff of the signal associated with capsids than could be accounted for by the KL models.
(2) A strong temperature dependence of the assembly rate.
(3) A bimodal size distribution observed by light scattering during the entire length of the process, indicating two dominant species: individual dimers and larger aggregates of size comparable to capsids.
These results lead us to propose a modification of the KL model to include contributions from an initial activated subunit state to account for the observed features.
Theory
The original KL model assumes a single-pathway for assembly. The process occurs through a cascade of monomer addition reactions, in which each reaction is reversible. The first few steps are slow and rate-limiting, being associated with nucleation. Subsequent faster steps correspond to elongation reactions:11,23
The original KL model includes many variables that count for each step of the assembly cascade. Therefore, it is difficult to use this model to quantitatively fit experimental data. To improve interpretability, we propose to simplify the KL model by omitting the intermediates in the elongation steps. Our reasoning is based upon two further approximations: unidirectional (forward only) elongation reactions and a steady-state approximation for the corresponding intermediates. The former is validated by the results of the original KL model simulations;23 the latter is applicable only if the intermediates associated with these steps accumulate to such small amount as to provide negligible detectable signal, as later shown in this paper. Under these approximations, the simplified KL model is described by the following formalism:
An assumed nucleus was designated as an nmer. The consequent simplified model (sKLn) followed a reduced and more tractable set of rate equations to calculate the intermediate and capsid concentrations as a function of time:
The rate constants, k1, k−1, k2, k−2,…kn−1 and kn, were used as fit parameters to model the experimental data. In addition to the rate constants, because we were simulating turbidity data, the molar scattering cross-sections of the assembly components were all parameters. Capsid scattering cross-section was treated as an adjustable variable, whereas that of a protein subunit (the monomer in the modeling and a protein dimer for BMV assembly) was measured from initial scattering intensity.
For intermediates, which were much smaller than the wavelength of light, Rayleigh scattering theory applied and cross-sections could be calculated from knowledge of subunit scattering and number of subunits.24 This is because, in the framework of the Rayleigh theory, the irradiance of the light scattered by any particle, regardless its shape, is proportional to the squared volume of the particle. For example, for an isotropic sphere, the polarizability is given by
whereas for an anisotropic sphere, the polarizability tensor reads
Assuming rapid random reorientation with respect to the detector response time, the anisotropic sphere ensemble is characterized by an effective average polarizability similar to the one for an isotropic sphere:
where the averages are taken over all possible orientations. Here α stands for polarizability, ε1 for the permittivity of the sphere material, and εm for the permittivity of the medium.
It will be shown later the contributions of the small intermediates to turbidity were vanishingly small. Therefore, considerations regarding their shapes were unimportant.
A further modification of the KL model was made by inserting a monomer activation step that contributed to the sharp takeoff from the initial lag phase to the growth phase in a scattering curve. The reaction equation corresponded to
This model with a specified nucleus, nmer, is referred to as sKL0n in the rest of the article. Accordingly, an additional rate equation was inserted and the first rate equation in the simplified KL model was changed to
Simulations were run in Excel by a least-squares fit procedure. At any reaction time, the reaction rates were calculated from the rate constants and concentrations. Then the concentrations at the next time step were computed; a time step of 0.01 s was taken for the initial 200 and 0.1 s for the rest. Iterating the variables was performed manually until a minimum χ2 value was found.
Experimental Section
The simplified model has been tested on data obtained from in vitro BMV capsid assembly by light scattering. The BMV capsid is a ~28 nm protein cage composed of 90 protein dimers arranged in a T = 3 icosahedral symmetry.25 Protein dimers were purified from intact virions and stored in a buffer of pH 7.4 containing 0.01 M Tris–HCl, 1 M KCl, 0.005 M MgCl2 and 0.02% DTT. Mixing 60 μL of this protein solution with 60 μL of an assembly buffer at pH 5.2 containing 0.2 M NaAc/HAc, 1 M KCl and 0.005 M MgCl2 led to the assembly of empty capsids.26,27 The empty capsid thus obtained was structurally indistinguishable from the native BMV capsid. The assembly was believed to use dimers of protein as the building block,28 which represented the monomer in the KL model.
BMV capsid assembly was monitored by two methods: (1) Continuous turbidity versus time was measured with a 10 ms step time using the scattered light intensity from a 2 mW 637 nm laser source detected at 90 degrees by a Si femtowatt photoreceiver (model 2151, New Focus). The photoreciever output was digitized by an oscilloscope (LeCroy Wavesurfer 432). Initial light scattering from protein dimers in the storage buffer was recorded for more than 50 s. Then the assembly buffer was injected quickly and mixed with the protein solution using a micropipette, when an immediate (less than half a second was necessary to recover from the disturbance of the pipet tip) drop of the scattering intensity to half of the original level was observed. This represented time 0 in the assembly curve. The whole scattering setup was enclosed in a thermal box (10 °C to room temperature), in which the protein solution and the assembly buffer were equilibrated at a designated temperature within ±0.1 °C. (2) Dynamic light scattering (DLS) taken at time steps of 18 s with each data point collected from 1 s scan was performed on a Zetasizer Nano-S. The protein solution had to be mixed with the assembly buffer outside the sample container in a cuvette, which was then inserted into the sample holder, when the Zetasizer performed the initial setup steps. During this period of time (~12 s), the assembly information was lost.
Results
DLS Observations
DLS was employed to monitor the assembly kinetic process. The initial stage was usually not resolved clearly due to the intrinsic limitation of DLS on analyzing a multicomponent system; usually, two types of particles need to differ in size by more than 1 order of magnitude to be distinguished clearly from each other. Furthermore, monitoring size distributions was complicated by concentration changes over time. However, we have observed that at 10 °C, an initial recording (at around 12 s from mixing the assembly solutions) of a single peak corresponding to BMV protein dimers was followed immediately (at an 18 s minimum scan interval) by the rise of a second peak close to capsid size (supplemental Figure 1, Supporting Information). Distinct peaks representing the two species were observed from time to time during assembly at various temperatures (Figure 1). We believe the two peak patterns reflect the nature of the assembly kinetic process, lacking intermediates. Eventually, the DLS spectra were dominated by a single capsid peak because scattering intensity scales proportionally with the scatter's radius to the sixth power.24 However, the onset depended on the temperature.
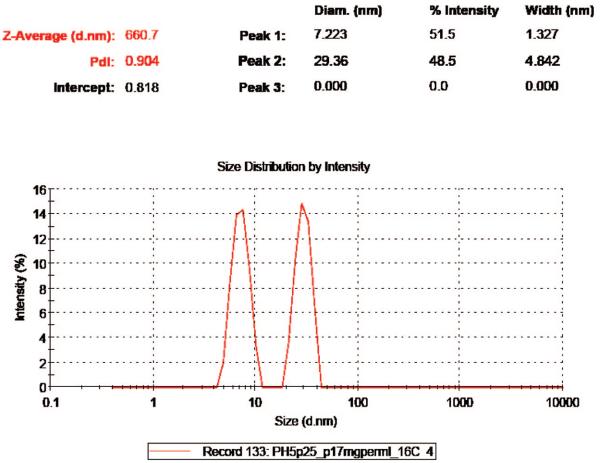
Figure 1.
An example DLS diagram during early stages of BMV capsid assembly. DLS was set for general fitting; 3 peaks were listed as default display setting, though there were only 2 components captured. The peak of ~7.22 nm represents protein dimers and the peak of 29.4 nm represents capsids. Notably, intermediates were barely observed. The diagram was recorded in 60 s at 16 °C and a concentration of 4.19 μM of protein dimers.
It was found by DLS that BMV capsid assembly strongly depended on temperature: At 22 °C, 4.2 μM of BMV protein dimers did not show any significant reaction for hours. However, when the solution was immersed in an ice bath for less than half-minute, a single strong peak emerged at ~29 nm and it remained constant in time, indicating a quick completion of the assembly process. Moreover, when the temperature was brought back up to 22 °C, the peak stayed unchanged in either shape or intensity even for hours, which indicated apparent irreversibility of the assembly reaction.
At 10 °C, assembly kinetics was slow enough to be followed by DLS. In this case, the scattering intensity peak representing BMV capsids increased steadily for more than 24 h of incubation, although the reaction rate decreased as the unassembled subunits became limiting.
Turbidity vs Time
The change in turbidity over time was measured to confirm the DLS results. As expected from previous reports, the reactions displayed sigmoidal character.8-12 Furthermore, because this experimental scheme provided access to much faster time scales than DLS, the initial lag phase of about 10 s was recorded with higher time resolution than previously. An initial flat line, followed by a rapid increase of the scattering intensity, characterized these reactions (Figure 2b and Figure 3). The lag phase shortened with increasing concentration of protein subunits. In addition, a steeper increase of intensity correlated with higher protein subunit concentration. Consistent with our previous observations, temperature had an inverse correlation with the change in turbidity (Figure 2a).
Figure 2.
Turbidity curves of BMV capsids assembly at various temperatures and protein concentration of 9.4 μM of protein dimers. The capsid assembly rate decreases with temperature (a). Turbidity curves are characterized by an initial flat line followed by a rapid increase of the scattering intensity (b).
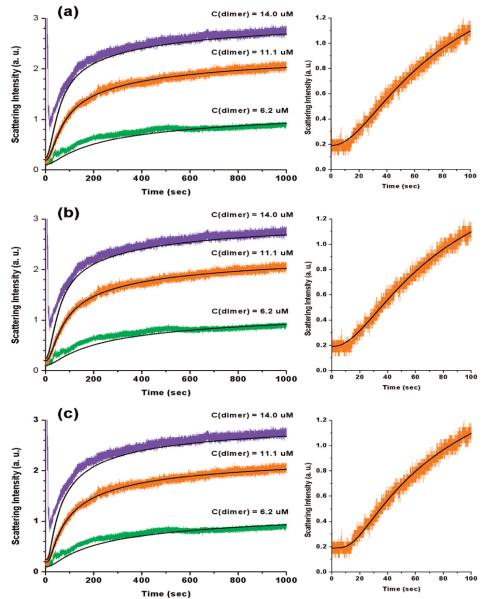
Figure 3.
Simulated turbidity curves (black lines) with the models of sKL3 (a), sKL4 (b) and sKL03 (c), and the experimental data (colored) for BMV capsid assembly at 10 °C. On the right column, the characteristic initial flat lag phase followed by a sharp turning into the rapid growth zone is illustrated for the assembly with 11.1 μM of protein dimers; sKL3 and sKL4 fail to produce the feature but sKL03 does.
Simulations
Simulations of the change in turbidity vs time were performed using the simplified KL model with and without an initial activation step (results are shown in Figure 3 and Table 1). In these reactions, 10 °C was used with varying initial capsid subunit concentrations. However, the least-squares procedure was only performed for the 11.1 μM concentration which provided the best signal. Nevertheless, kinetic data from other concentrations have been used for visual verification.
TABLE 1.
Simulation Results with the Models
| sKL3 |
capsid |
k1 = 1.44 × 10−4 μM−1 · s−1 k−1 = 4.65 × 10−2 s−1 k2 = 3.00 × 10−4 μM−1 · s−1 k3 = 3.47 × 10−2 μM−1 · s−1 χ2/ν = 1.4 |
| sKL4 |
capsid |
k1 = 1.45 × 10−4 μM−1 · s−1 k−1 = 4.52 × 10−2 s−1 k2 = 3.00 × 10−4 μM−1 · s−1 k−2 = 1.35 × 10−2 s−1 k3 = 7.56 × 10−2 μM−1 · s−1 k4 = 6.86 × 10−2 μM−1 · s−1 χ2/ ν = 1.4 |
| sKL03 |
capsid |
k0 = 7.65 × 10−2 s−1 k−0 = 1.21 × 10−1 s−1 k1 = 4.00 × 10−3 μM−1 · s−1 k−1 = 1.79 s−1 k2 = 7.50 × 10−3 μM−1 · s−1 k3 = 2.49 × 10−2 μM−1 · s−1 χ2/ ν = 1.4 |
The sKL3 model fit well the overall shape of the turbidity curves with a reduced chi-square of 1.4 (Figure 3a, Supporting Information). The greatest concern with this model was that it failed to produce a sharp turning from the initial lag phase to the rapid increase zone (Figure 3a, right images; this will be discussed later in detail).
In an attempt to reproduce the sharp takeoff, simulations with sKL4 and sKL03 were performed. Both models could only match the experimental data with a reduced chi-square of 1.4 - no improvement to the overall fitting in comparison with sKL3. The simulated curve from sKL4 overlapped everywhere with that from sKL3 and did not produce any sharper corner than sKL3 at the taking-off from the lag phase. The rate constants k1, k−1 and k2 in this case were very close to those from sKL3. Moreover, the simulated curves were not sensitive to the values of k−2, k3 and k4, which implied that the corresponding reaction steps are not rate-determining. An inspection of the reaction rates during the steady state revealed that 2mer + 1mer ⇔ 3mer was mainly a forward reaction with about 10% of backward reaction, which validated the initial unidirectional assumption of this step in sKL3.
Although sKL03 did not improve the overall curve fitting, it did improve the fitting of the initial phase by generating a flat lag phase followed by a sharp turning into the rapid growth zone. This could be explained by a monomer activation step at the beginning of the assembly process. The activation step introduced a nonlinear dependency of the assembly rate on the concentration of the intermediates, because the association of two activated monomers into a dimer would requires a second-order reaction rate, i.e., R = k1[1mer]2. Furthermore, activated monomers would also participate in each of the subsequent reaction steps. The influence of their accumulation should be manifested as an increase in the overall order of reaction. In sum, it is argued that the nonlinear effect can be seen in the turbidity-vs-time curve as a sudden takeoff from the lag phase.
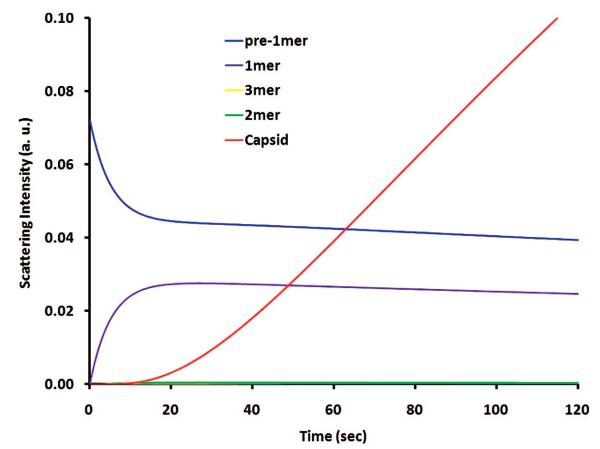
Figure 4 illustrate sKL03 predictions of scattering contributions from different components during BMV assembly for a protein concentration of 4.19 μM dimers at 10 °C. Apparently, the scattering from 2mer and 3mer is completely buried by the dominant signal from pre-1mer and 1mer (protein dimers) in the early stage and that from capsids, later. Hence, the approximation for the scattering cross-sections based on Rayleigh theory has a minor role in modeling. The prediction shows capsid signal rising up between 10 and 20 s, which corresponds well to the monomer activation period (Figure 4). Moreover, this timing for capsid occurrence from modeling agrees well with the DLS observation (supplemental Figure 1, Supporting Information).
Figure 4.
sKL03 prediction of scattering contributions of different components for the BMV assembly with a protein concentration of 4.19 μM dimers at 10 °C. The intermediates only make an ignorable contribution; the scattering from capsid rises up between 10 and 20 s, corresponding to the time needed for monomer activation, and in a fair agreement with DLS observation (supplemental Figure 1, Supporting Information).
Discussion
Kinetic simulations with the original KL model indicate that capsid assembly would reach a steady state whereas all intermediate concentrations change slowly after significant fluctuations during the early stage. Furthermore, inspection of the cascade of reactions at this stage revealed negligible back reactions in the elongation steps: every new nucleus entering the assembly line proceeds to form a capsid. The situation changes only when assembly approaches equilibrium.23 The simplified KL model takes the unidirectional approximation and extends it to the earlier assembly stage, as the system at this time is further away from equilibrium and closer to the reactant side than at the steady state and back-reactions are less pronounced. The comparison between sKL3 and sKL4 modeling results proved the validity of this approximation, because the extra bidirectional step in sKL4 turned out to be forward-reaction dominating.
Besides the unidirectional approximation of the elongation steps, the simplified KL model applies a steady-state approximation that treats the concentrations of the intermediates in elongation steps as infinitesimally small. Its validity is justified in the light of DLS observations, which show little scattering contribution from big intermediates throughout the entire assembly process. The modeling results are consistent with the approximation and further exclude the scattering contribution from small intermediates in the nucleation steps as well. Therefore, the light scattering mainly comes from the produced intact capsids and partially from individual protein dimers.
Modeling with sKL3 and sKL03 has achieved fair agreement with the experimental data in the overall sense, suggesting a trimer nucleus, in the assembly cascade. Moreover, a step-up from sKL3 to sKL4 indicates that the reaction 2mer + 1mer ⇔ 3mer is indeed forward-reaction dominating, whereas 1mer + 1mer ⇔ 2mer achieves equilibrium. This result is consistent with the concept of a nucleus: as soon as it is formed, it goes forward to form a capsid and there is no way back. Interestingly, CCMV, a similar virus, was found to assemble from a pentamer nucleus.29 It would be interesting to see if this conclusion would remain valid if tested by shorter time scale experiments similar to those discussed in the present article.
Protein association energies can be calculated from the derived rate constants in Table 1. For example, using sKL03 model,
The initial sudden takeoff in turbidity measurements of assembly cannot be reproduced by the simplified KL model without a monomer activation step. The sKL3 and sKL03 modeling results together with the experimental data are coplotted in Figure 5a. It can be seen that the deviation of sKL3 modeling from the data is apparently above the noise whereas the sKL03 result corners along with the data. As a further clarification, Figure 5b displays the local squared deviation averaged among an 8 s span of both models from the data. The sKL03 simulation deviates 3 times less than sKL3 result around the turning range (between 6 and 14 s).
Figure 5.
Noise analysis for the lag-phase takeoff ranges. (a) sKL3 and sKL03 modeling results together with the smoothed data (by average of 4 adjacent points, which reduces the time resolution to 40 ms). It is clear that sKL3 modeling rises over the noise during that range whereas sKL03 reproduces the sharp turning feature. (b) Local squared deviation averaged among an 8 s span of both modeling from the smoothed data. The sKL3 result shows significant larger values around the turning range.
A similar sharp turning feature from the initial lag phase into the fast growing phase has been observed on Papillomavirus9 and Salmonella typhimurium phage P228 as well. In the former case, it was suggested that a cooperative effect existed between assemblies of individual capsids; e.g., a growing intermediate could act as a template to help form new dimers.9 This could be a possible explanation for the observed nonlinearity in the BMV case, too.
As an alternative assumption, we propose here that a higher order influence of certain intermediates on the reaction rate could be responsible for the sharp takeoff; i.e., intermediates aggregate among themselves. However, any association reaction between two intermediates that are greater than a monomer would contradict the basic scheme of sequential addition of monomers in the KL model, unless the monomers themselves are intermediates. This implies a monomer activation step before they add up to build a capsid. The nature of the activated monomer is not clear at this time; the activation may be related to a protein conformational change occurring at the different pH required for assembly. Given that many viral processes can take place in environments with defined local conditions that are dramatically different than the rest of the cell, e.g., as vesicles and membrane invaginations with strict requirements for lipids,30,31 it is possible that the pH changes required in vitro reflect a change that takes place in vivo.
The apparent irreversibility when the completed assembly in an ice bath is brought back to 22 °C implies the assembly process is kinetically controlled. However, an alternative explanation could be the hysteresis due to the complexity of a multistep reaction system.32 Whether a capsid assembly would reach equilibrium or not in a long time range has been controversial,33 and there has been no way to rigorously examine it so far.34 Fortunately, in the case of our simulations, the assembly reactions takes at least for 24 h; hence the first 1000 s of data reflect the system status being far away from equilibrium and should be safe to be used in kinetic simulations.
BMV capsid assembly rate accelerates with lowering the temperature. The formal interpretation of this result corresponds to an apparent negative activation energy for the assembly reaction.35 Expressing the apparent activation energy in terms of individual steps' activation energies can help to identify the most probable origin of the temperature dependence. In the case of sKL03 model, the assembly achieves a steady state and the formation of capsids follows:
The apparent third order of the reaction rate is the result of 3mer being the nucleus. The apparent rate constant is kapp = (k1 · k2/k−1) · (k0/k0 + k−0) and the apparent activation energy can be calculated according to the Arrhenius equation:
In the above equations, Ea is the activation energy and ΔH is the enthalpy change of a reaction; the subscripts 0, 1, and 2 correspond to the monomer activation step and the reactions leading to a 2mer and 3mer, respectively. Because the activation energy of an elementary reaction can only be positive, the overall negative activation energy must result from negative ΔH0 or ΔH1. A negative ΔH1 would mean exothermic binding of protein subunits in contradiction of the commonly accepted idea that the process is entropy driven and endothermic.36,37 Therefore, the first activation step is probably very exothermic and responsible for the negative value of the overall activation energy.
Conclusion
Faster time-course light scattering studies of assembly kinetics of empty BMV capsids have revealed a more abrupt initial takeoff of the signal at the end of the assembly lag phase. In light of this new experimental observation, it is shown here that the accepted kinetically limited model needs to be amended to quantitatively fit the data. Direct computations of protein binding energies (for the initial association steps) from simulations has confirmed Zlotnick et al.'s conclusion of relatively weak association between protein subunits.
In addition, a negative dependence of the assembly rate on temperature has been found, which is somewhat unusual when compared to other known virus capsid assemblies such as the Hepatitis B virus.38 One possible explanation is the apparent negative activation energy due to a hypothetical initial monomer activation step. The question of the nature of the negative activation energy and the nature of the monomer activation need further exploration.
Although the model examined here has been only applied to the BMV capsid assembly at one specific pH, ionic strength and temperature, it offers the possibility of obtaining more insight into capsid assemblies when applied to wider ranges of pH, ionic strength and temperatures. For example, entropy and enthalpy change from protein binding could be calculated using the Van't Hoff approach.
Supplementary Material
Acknowledgment
We thank Adam Zlotnick for many helpful discussions. We gratefully acknowledge partial support for this work from the National Science Foundation (grant 0631982), the MetaCYT endowment fund from the Eli Lilly Co., and the National Institutes of Health (grant GM081029-01).
Footnotes
Part of the “Stephen R. Leone Festschrift”.
Supporting Information Available: Description of reduced χ2 and of the variable iteration procedure. DLS spectra. This material is available free of charge via the Internet at http://pubs.acs.org.
References and Notes
- 1.Hiebert E, Bancroft JB. Factors Affecting the Assembly of Some Spherical Viruses. Virology. 1969;39:296–311. doi: 10.1016/0042-6822(69)90050-6. [DOI] [PubMed] [Google Scholar]
- 2.Pfeiffer P, Hirth L. Aggregation States of Brome Mosaic Virus Protein. Virology. 1974;61:160–167. doi: 10.1016/0042-6822(74)90251-7. [DOI] [PubMed] [Google Scholar]
- 3.Chen XJS, Garcea RL, Goldberg I, Casini G, Harrison SC. Structure of Small Virus-Like Particles Assembled from the L1 Protein of Human Papillomavirus 16. Mol. Cell. 2000;5(3):557–567. doi: 10.1016/s1097-2765(00)80449-9. [DOI] [PubMed] [Google Scholar]
- 4.Fuller MT, King J. Assembly Invitro of Bacteriophage-P22 Procapsids from Purified Coat and Scaffolding Subunits. J. Mol. Biol. 1982;156(3):633–665. doi: 10.1016/0022-2836(82)90270-4. [DOI] [PubMed] [Google Scholar]
- 5.Prevelige PE, Thomas D, King J. Scaffolding Protein Regulates the Polymerization of P22 Coat Subunits into Icosahedral Shells Invitro. J. Mol. Biol. 1988;202(4):743–757. doi: 10.1016/0022-2836(88)90555-4. [DOI] [PubMed] [Google Scholar]
- 6.Bancroft JB, Wagner GW, Bracker CE. The Self-Assembly of a Nucleic-Acid Free Pseudo-Top Component for a Small Spherical Virus. Short Commun. 1968:146–149. doi: 10.1016/0042-6822(68)90126-8. [DOI] [PubMed] [Google Scholar]
- 7.Kenney JM, Vonbonsdorff CH, Nassal M, Fuller SD. Evolutionary Conservation in the Hepatitis-B Virus Core Structure Comparison of Human and Duck Cores. Structure. 1995;3(10):1009–1019. doi: 10.1016/s0969-2126(01)00237-4. [DOI] [PubMed] [Google Scholar]
- 8.Prevelige PE, Jr., Thomas D, King J. Nucleation and Growth Phases in the Polymerization of Coat and Scaffolding Subunits into Icosahedral Procapsid Shells. Biophys. J. 1993;64:824–835. doi: 10.1016/S0006-3495(93)81443-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Casini GL, Graham D, Heine D, Garcea RL, Wu DT. In Vitro Papillomavirus Capsid Assembly Analyzed by Light Scattering. Virology. 2004;325(2):320–327. doi: 10.1016/j.virol.2004.04.034. [DOI] [PubMed] [Google Scholar]
- 10.Cuillel M, Berthet-Colominas C, Krop B, Tardieu A, Vachette P, Jacrot B. Self-Assembly of Brome Mosaic Virus Capsids - Kinetic Study Using Neutron and X-Ray Solution Scattering. J. Mol. Biol. 1983;164:645–650. doi: 10.1016/0022-2836(83)90055-4. [DOI] [PubMed] [Google Scholar]
- 11.Zlotnick A, Johnson JM, Wingfield PW, Stahl SJ, Endres D. A Theoretical Model Successfully Identifies Features of Hepatitis B Virus Capsid Assembly. Biochemistry. 1999;38(44):14644–14652. doi: 10.1021/bi991611a. [DOI] [PubMed] [Google Scholar]
- 12.Zlotnick A. To Build a Virus Capsid - an Equilibrium-Model of the Self-Assembly of Polyhedral Protein Complexes. J. Mol. Biol. 1994;241(1):59–67. doi: 10.1006/jmbi.1994.1473. [DOI] [PubMed] [Google Scholar]
- 13.Prevelige PE, Thomas D, King J. Nucleation and Growth Phases in the Polymerization of Coat and Scaffolding Subunits into Icosahedral Procapsid Shells. Biophys. J. 1993;64(3):824–835. doi: 10.1016/S0006-3495(93)81443-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zandi R, van der Schoot P, Reguera D, Kegel W, Reiss H. Classical Nucleation Theory of Virus Capsids. Biophys. J. 2006;90(6):1939–1948. doi: 10.1529/biophysj.105.072975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schwartz R, Shor PW, Prevelige PE, Berger B. Local Rules Simulation of the Kinetics of Virus Capsid Self-Assembly. Biophys. J. 1998;75(6):2626–2636. doi: 10.1016/S0006-3495(98)77708-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Berger B, Shor PW, Tuckerkellogg L, King J. Local Rule-Based Theory of Virus Shell Assembly. Proc. Nat.l Acad. Sci. U.S.A. 1994;91(16):7732–7736. doi: 10.1073/pnas.91.16.7732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rapaport DC. Self-Assembly of Polyhedral Shells: A Molecular Dynamics Study. Phys. Rev. E. 2004;70(5) doi: 10.1103/PhysRevE.70.051905. [DOI] [PubMed] [Google Scholar]
- 18.Willits D, Zhao X, Olson N, Baker TS, Zlotnick A, Johnson JE, Douglas T, Young MJ. Effects of the Cowpea Chlorotic Mottle Bromovirus Beta-Hexamer Structure on Virion Assembly. Virology. 2003;306(2):280–288. doi: 10.1016/s0042-6822(02)00054-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Reddy VS, Giesing HA, Morton RT, Kumar A, Post CB, Brooks CL, Johnson JE. Energetics of Quasiequivalence: Computational Analysis of Protein-Protein Interactions in Icosahedral Viruses. Biophys. J. 1998;74(1):546–558. doi: 10.1016/S0006-3495(98)77813-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Horton N, Lewis M. Calculation of the Free-Energy of Association for Protein Complexes. Protein Sci. 1992;1(1):169–181. doi: 10.1002/pro.5560010117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zlotnick A. Are Weak Protein-Protein Interactions the General Rule in Capsid Assembly. Virology. 2003;315(2):269–274. doi: 10.1016/s0042-6822(03)00586-5. [DOI] [PubMed] [Google Scholar]
- 22.Nguyen HD, Reddy VS, Brooks CL. Deciphering the Kinetic Mechanism of Spontaneous Self-Assembly of Icosahedral Capsids. Nano Lett. 2007;7(2):338–344. doi: 10.1021/nl062449h. [DOI] [PubMed] [Google Scholar]
- 23.Zlotnick A. Theoretical Aspects of Virus Capsid Assembly. J. Mol. Recognit. 2005;18(6):479–490. doi: 10.1002/jmr.754. [DOI] [PubMed] [Google Scholar]
- 24.Bohren CF, Huffman DR. Absorption and Scattering of Light by Small Particles. Wiley Science; NY: 1983. [Google Scholar]
- 25.Lucas RW, Larson SB, McPherson A. The Crystallographic Structure of Brome Mosaic Virus. J. Mol. Biol. 2002;317(1):95–108. doi: 10.1006/jmbi.2001.5389. [DOI] [PubMed] [Google Scholar]
- 26.Pfeiffer P, Hirth L. Aggregation States of Brome Mosaic-Virus Protein. Virology. 1974;61(1):160–167. doi: 10.1016/0042-6822(74)90251-7. [DOI] [PubMed] [Google Scholar]
- 27.Pfeiffer P, Herzog M, Hirth L. Stabilization of Brome Mosaic-Virus. Philos. Trans. R. Soc. London Ser. B-Biol. Sci. 1976;276(943):99–107. doi: 10.1098/rstb.1976.0100. [DOI] [PubMed] [Google Scholar]
- 28.Cuillel M, Zulauf M, Jacrot B. Self-Assembly of Brome Mosaic Virus Protein into Capsids - Initial and Final States of Aggregation. J. Mol. Biol. 1983;164:589–603. doi: 10.1016/0022-2836(83)90052-9. [DOI] [PubMed] [Google Scholar]
- 29.Zlotnick A, Aldrich R, Johnson JM, Ceres P, Young MJ. Mechanism of Capsid Assembly for an Icosahedral Plant Virus. Virology. 2000;277(2):450–456. doi: 10.1006/viro.2000.0619. [DOI] [PubMed] [Google Scholar]
- 30.Taylor MP, Kirkegaard K. Potential Subversion of Autophagosomal Pathway by Picornaviruses. Autophagy. 2008;4(3):286–9. doi: 10.4161/auto.5377. [DOI] [PubMed] [Google Scholar]
- 31.Kopek BG, Perkins G, Miller DJ, Ellisman MH, Ahlquist P. Three-Dimensional Analysis of a Viral RNA Replication Complex Reveals a Virus-Induced Mini-Organelle. Plos. Biol. 2007;5:2022–2034. doi: 10.1371/journal.pbio.0050220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Singh S, Zlotnick A. Observed Hysteresis of Virus Capsid Disassembly Is Implicit in Kinetic Models of Assembly. J. Biol. Chem. 2003;278(20):18249–18255. doi: 10.1074/jbc.M211408200. [DOI] [PubMed] [Google Scholar]
- 33.Parent KN, Suhanovsky MM, Teschke CM. Phage P22 Procapsids Equilibrate with Free Coat Protein Subunits. J. Mol. Biol. 2007;365(2):513–522. doi: 10.1016/j.jmb.2006.09.088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zlotnick A. Distinguishing Reversible from Irreversible Virus Capsid Assembly. J. Mol. Biol. 2007;366(1):14–18. doi: 10.1016/j.jmb.2006.11.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Oosawa F, Asakura S. Thermodynamics of the Polymerization of Protein. Academic Press; London: 1975. [Google Scholar]
- 36.Kegel WK, van der Schoot P. Competing Hydrophobic and Screened-Coulomb Interactions in Hepatitis B Virus Capsid Assembly. Biophys. J. 2004;86(6):3905–3913. doi: 10.1529/biophysj.104.040055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nemethy G. Hydrophobic Interactions. Angew. Chem. Int. Ed. 1967;6(3):195–280. doi: 10.1002/anie.196701951. [DOI] [PubMed] [Google Scholar]
- 38.Ceres P, Zlotnick A. Weak Protein-Protein Interactions Are Sufficient to Drive Assembly of Hepatitis B Virus Capsids. Biochemistry. 2002;41(39):11525–11531. doi: 10.1021/bi0261645. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.