Summary
The Drosophila developmental gene, engrailed, encodes a sequence-specific DNA binding activity. Using deletion constructs expressed as fusion proteins in E. coli, we localized this activity to the conserved homeodomain (HD). The binding site consensus, TCAATTAAAT, is found in clusters in the engrailed regulatory region. Weak binding of the En HD to one copy of a synthetic consensus is enhanced by adjacent copies. The distantly related HD encoded by fushi tarazu binds to the same sites as the En HD, but differs in its preference for related sites. Both HDs bind a second type of sequence, a repeat of TAA. The similarity in sequence specificity of En and Ftz HDs suggests that, within families of DNA binding proteins, close relatives will exhibit similar specificities. Competition among related regulatory proteins might govern which protein occupies a given binding site and consequently determine the ultimate effect of cis-acting regulatory sites.
Introduction
Transcriptional regulators of gene activity are expected to play important roles in directing embryonic development. Among the prime candidates for such regulators are a group of recently identified Drosophila developmental genes (Garcia-Bellido, 1975; Lewis, 1978; Nusslein-Volhard and Wieschaus, 1980). Mutations that affect these developmental regulators often alter the spatial patterns of expression of other regulatory genes (Hafen et al., 1984; Carroll and Scott, 1986; Harding et al., 1986; Howard and Ingham, 1986; DiNardo and O’Farrell, 1987). Extensive studies of this type have led to the proposal that the developmental genes interact in a complex regulatory network (reviewed in Scott and O’Farrell, 1986). It appears that this regulatory network progresses through a sequence of stages, each involving new regulators and each characterized by more detailed spatial distributions of the regulators involved. Apparently, the regulatory interactions of this network guide formation of embryonic pattern.
Many of the genes involved in this regulatory network are related by a region of sequence homology, the homeodomain (HD) (McGinnis et al., 1984b; Scott and Weiner, 1984). Consequently, gene duplication and divergence are thought to have played important roles in the evolutionary origin of the developmental genes (Lewis, 1951; McGinnis et al., 1984a). If functional constraints dictate the extraordinary evolutionary conservation of the 60 amino acid HD sequence, the genes encoding an HD must have functional similarities (McGinnis et al., 1984a). The presence of an HD in many of the Drosophila developmental genes suggests that reiteration of some fundamental interactions might underlie the regulatory network guiding pattern formation (O’Farrell et al., 1985).
Several lines of evidence have suggested that one of the functions of the HD is sequence-specific DNA binding. First, there is considerable homology of the HD with the yeast transcription factors, MATa1 and MATα2, and more distant homology to the helix-turn-helix structural motif of prokaryotic DNA binding proteins (Laughon and Scott, 1984; Shepherd et al., 1984). Second, HD-containing proteins are localized to the nucleus (for examples, see White and Wilcox, 1984; Beachy et al., 1985; Carroll and Scott, 1985; DiNardo et al., 1985). Third, a fusion protein containing part of the engrailed encoded protein, including the HD, has sequence-specific DNA binding activity in vitro (Desplan et al., 1985; see also Fainsod et al., 1986). Here we will provide further evidence that the DNA binding is specified by the HD sequences.
The demonstration that specificity of DNA binding is defined by such a highly conserved sequence provokes a new question. What is the relationship of the sequence specificity of the HDs found in various developmental regulators? We show here that the sequence specificities of the HDs encoded by the engrailed and fushi tarazu genes (En HD and Ftz HD) are very closely related. We suggest that evolution has created a family of regulators with related binding specificities. As in the case of the bacteriophage lambda regulators, repressor and cro, the HD-containing regulators might function in an interdependent fashion because of similarities in binding specificity (Ptashne, 1986).
Results
Specific DNA Binding Maps to the Homeodomain
To define the domain of the engrailed encoded protein (En protein) that specifies DNA binding, we examined the activity encoded by the constructs outlined in Figure 1. Various parts of the engrailed (en) coding sequence were fused to β-galactosidase (Figure 1, parts A, B, E, and F) or calcitonin (Figure 1, parts C and D) coding sequences. While the fusions are not likely to precisely mimic all the functions of the natural gene product, gene fusions have been used successfully to localize functional domains within proteins (Hall et al., 1984; Johnson and Herskowitz, 1985; Picard and Yamamoto, 1987). Here the fusions provide antigenic tags used in a convenient immunoprecipitation assay for DNA fragment binding activity (McKay, 1981). All constructs were tested in this fragment binding assay using antibodies to either β-galactosidase or calcitonin (Figures 1 and 5).
Figure 1. The En and Ftz Fusion Protein Constructs Tested for DNA Binding Activity.

The 60 amino acid homeodomain (HD) is indicated by the thick bold line, other En sequences by a thin line, and other Ftz sequences by a triple line. The numbers above constructs A and G indicate positions with respect to the intact En or Ftz proteins, and the numbers below the various constructs indicate positions with respect to the first amino acid of the HD. The position of the helix-turn-helix motif within the HD is also shown. The capacity of these constructs to bind DNA is indicated (+/−).
(A) Represents the En fusion protein used for experiments shown in Figures 2 and 5 (also see Desplan et al., 1985). The fusion contains 44 residues N-terminal to the HD and 39 residues extending from the end of the HD to the natural stop codon.
(B) Deletion of a C-terminal segment of en coding sequences replaces the last amino acid of the En HD (thr to ser, hatched line). Translation terminates at a TGA codon immediately after the altered amino acid.
(C) and (D) In these two constructs (Theis et al., unpublished data), smaller C-terminal parts of the En protein are fused to part of the preprocalcitonin rather than β-galactosidase. Fusion C contains 11 residues N-terminal to the HD, while D contains 41.
(E) Cleavage at the BgIII site, within the homeobox, and fusion to a different ORF (hatched line) results in an HD truncated beyond position 47 and lacking half of the putative recognition helix.
(F) This is an 11 residue deletion that removes amino acids 48 to 58, inclusively. This deletion removes half of the putative recognition helix.
(G) The Ftz construct includes 110 residues N-terminal to the HD and extends to the natural termination codon 97 residues C-terminal to the HD. The only homology between the En and Ftz proteins is within the HD (thick line). The Ftz protein expressed is derived from the Oregon R cDNA, which is proposed to encode a 410 amino acid protein (Laughon and Scott, 1984). The cloned ftz cDNA was generously provided by A. Laughon and M. P. Scott.
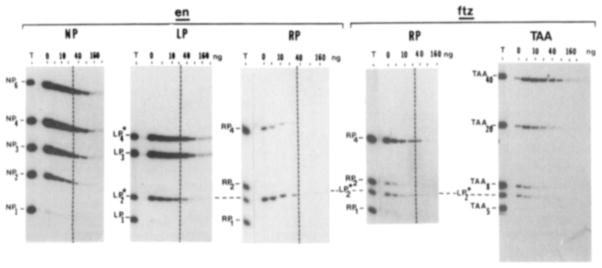
Figure 5. Binding of En and Ftz Fusion to DNA Fragments with Different Numbers of Copies of Synthetic Sites.
In each binding reaction, the En fusion protein or Ftz fusion protein is offered a mixture of fragments carrying different numbers of copies of NP, LP, RP, or TAA as indicated. A separation of each total mixture is shown (T), and subsequent lanes show the fragments immunoprecipitated with the En fusion protein in the absence (0) and presence of increasing amounts (5,10, 20, 40, 80,160, and 320 ng) of competitor DNA, a mixture of oligomerized synthetic double-stranded fragments, [(TAA)5]n. Note that the TAA competitor DNA competed out the binding of both TAA and LP* sequences. Competition with oligomerized NP sequences gives similar results. That is, the different competitor DNAs differed slightly in the concentration required to compete for binding; they gave the same order of competition of each of the labeled fragments. Fragments LP2* and LP4* contain 2 and 4 copies of the LP sequence, but modified (by a blunt-ending procedure) to remove two terminal nucleotides at each end. Cloning of the blunt-ended oligonucleotides in the Smal site of M13mp18 regenerates one of the two missing nucleotides from each end of the LP* fragments. Consequently, the LP* fragments differ in the position at which they are cloned (Smal versus BamHI) and are 4 bp shorter than the corresponding NP or RP fragments and differ from the LP sequence given in Figure 4B by lacking the leftmost T and rightmost A. Experiments using a perfect LP2 sequence cloned in the Smal site showed that the sequence difference was of little or no consequence to binding by En or Ftz fusions (data not shown). LP2* is included as an internal reference in all but the NP panel. The fragments named TAA5 and TAA8 contain five and eight tandem copies of the trinucleotide TAA, respectively. The other fragments, TAA20 and TAA40, contain four or eight copies of TAA5 ligated in various orientations. The dashed line is to aid alignment in making comparisons between experiments. Results similar to those shown for the En fusion are obtained using calcitonin constructs C and D.
The N-terminal 442 and the C-terminal 40 amino acid residues of the En protein were found to be dispensable for sequence-specific DNA binding. Taken together, these truncations suggest that the binding activity lies within a region beginning 11 amino acids N-terminal to the HD and extending through 59 residues of the HD. As expected from the predictions based on homology, constructs altered in the presumed recognition helix (Figure 1, parts E and F) show no DNA binding. Extracts from cells expressing inactive fusions served as controls demonstrating the specificity of the assays used (data not shown).
In the experiments presented here, we used construct A (Figure 1), the En fusion, or construct G (Figure 1), the Ftz fusion. Many of the described features have also been confirmed with protein constructs C or D as noted in the figure legends.
A DNA Sequence Recognized by the En Fusion Protein
Sites bound by the En fusion protein were located by DNAase I protection (Figure 2). We analyzed the fragments most efficiently bound in the immunoprecipitation assay: a 670 bp fragment upstream of en coding sequences, a 341 bp fragment in the first intron of en, and a fragment 3′ to the ftz coding region (Desplan et al., 1985; and Figure 3A). The left panel of Figure 2A shows two of the regions within the 670 bp fragment that were footprinted by the En fusion (the Ftz fusion will be discussed below). Flanking the positions of nuclease protection (−), we detected a number of DNAase I hypersensitive sites (+).
Figure 2. DNAase I Protection Patterns Produced by HD Fusions.
(A) DNAase I protection of regions 1 and 2 in en DNA. A Clal-Nael fragment from plasmid p615, 5′ end–labeled at the Clal site, was incubated for 30 min at 0°C without protein (0) or with 0.9 (1), 4.4 (2), or 22 μg (3) of bacterial extract containing the En HD fusion protein (construct A in Figure 1), partially digested with DNAase I as described in Experimental Procedures and electrophoresed on a 6% sequencing gel. The Ftz lanes represent protection obtained by 1.5 (1), 7.4 (2), or 37 (3) μg of bacterial extract containing the Ftz fusion protein (construct G in Figure 1). Protected (−) and enhanced (+) sites of DNAase I cleavage in and around sites 1 and 2 are indicated. The arrowhead indicates an enhanced band present only when the Ftz protein is used. No protection was observed in bacterial extracts producing truncated, inactive fusion proteins. A third protected region contained in this fragment is not visible in this separation. Protection by the En fusion spans 29,18, and 20 bp for regions 1,2, and 3, respectively.
(B) DNAase I protection of a fragment containing six copies of the NP sequence (NP6; see Figure 4A). Increasing amounts of the En fusion protein extract, no protein (0), 0.35 (1), 1.75 (2), 9 (3), or 44 μg (4) in 25 μl were incubated with the DNA fragment (end-labeled at its HindIII site). Digestion with 1 μg/ml of DNAase I is as in Experimental Procedures. The arrows indicate the positions and orientations of the six NP consensus sequences. The positions of the bands resulting from DNAase I cuts are indicated for the first copy of the NP sequence. These positions of cleavage are repeated in each subsequent copy of the NP sequence having the same orientation (copies 1 to 4). A characteristic pattern of protection/enhancement due to the En protein extract is observed in each of these copies. This pattern changes for copies 5 and 6, which are in the opposite polarity. Calcitonin fusions C and D (see Figure 1) exhibit a similar pattern of protection.
(C) Concentration of En HD fusion protein extract required to protect one or three copies of the LP sequence (see Figure 4B). Fragments LP1 and LP3 were footprinted with increasing amounts of the extract: no extract (0); 1.3 (1); 2.7 (2); 5.5 (3); 11 (4); 22 (5); or 44 μg (6) of total protein. The positions of the DNAase I cuts within the various LP sequences are indicated by comparison with the G/A Maxam–Gilbert sequencing lane (note that the positions of the bands in this ladder are shifted compared with the DNAase I lane because of the difference in the position of cleavage [Maxam and Gilbert, 1980]). Each palindrome and its center is indicated. The dashed lines represent the limits of each copy.
Figure 3. Consensus Sites near the engrailed Gene Are Clustered.

(A) The positions of regions protected from DNAase I by the En fusion (arrowheads) are clustered, and each protected region contains one or more sequences related to a consensus. Each footprinted region is designated with a number. The sequences of five footprinted regions are given, and positions matching the consensus (see B) are in upper case. Where a footprinted region (e.g., 1) includes more than one consensus site, these are distinguished with a prime (e.g., 1,1′, and 1″). Clal sites to the left of the illustrated sequences are the positions at which label was incorporated for analysis of DNAase I protection.
(B) Alignment of the sequences exhibiting footprints with the En fusion protein. Sequences of the footprinted regions are aligned based on their homology. Regions 1 to 6 are from the en gene of D. melanogaster (A). The sequences marked “en vir” are the corresponding regions in the en gene of D. virilis (Kassis et al., unpublished data). The ftz footprinted regions are located 3′ to the ftz gene (see Desplan et al., 1985). Each distinct footprint is designated by a number. Most of these footprinted regions contain several sequences that can be aligned, and each distinct alignment is indicated with the number of the region (e.g., sites 1, 1′, and 1″). All these aligned sites are present within regions protected from DNAase I digestion by the En fusion protein (e.g., Figure 2A).
The consensus is defined as the average between all these aligned sequences. The number of sites matching the consensus at each particular position as well as the score of each individual sequence matching the consensus are indicated. Three of the consensus sequences, en vir 2′, ftz 1′, and lambda 2′, are aligned on the strand opposite to the other represented sequences. The sites in lambda DNA are sequences present in fragments bound by the fusion protein. The sequences within these fragments that exhibit homology to the consensus are aligned. They have not been footprinted.
Comparison of protected regions showed that all have at least one sequence approximating the 10 bp consensus TCAATTAAAT (Figure 3). Figure 3B aligns 21 sequences from which the consensus sequence was derived (the data from D. virilis en DNA will be described in detail elsewhere; J. Kassis, C. Desplan, D. Wright, and P. O’Farrell, unpublished data). In most cases, this consensus is repeated within the protected region. For example, protected region 1 contains three tandemly repeated sequences spaced by a single nulceotide.
In our earlier work (Desplan et al., 1985), we showed that a few restriction fragments of lambda DNA were specifically bound by the En fusion protein. If the consensus sequence represents the preferred binding site, it should also be found in these DNA fragments. Indeed, two of the lambda fragments contain sequences matching the consensus (Figure 3B). They have not been footprinted.
The En Fusion Protein Binds to Repeats of a Synthetic Consensus
The various footprinted regions identified in Drosophila DNA are clustered and some of these footprinted regions contain several copies of the consensus (Figure 3). To test the sufficiency of the consensus sequence for binding, we built DNA fragments containing single or multiple copies of a synthetic consensus, TCAATTAAATga (NP sequence). The G and A at the end of the 10 bp consensus were added primarily to create a restriction site between repeats of the sequence (but see consideration of symmetry below). We examined En fusion protein binding to fragments containing single or multiple copies of NP (Figure 4A). The relative efficiency of binding these fragments was compared in the presence of increasing amounts of competing DNA. A fragment carrying one copy of the consensus was bound only poorly, while fragments carrying two or more synthetic sites were bound very effectively (Figure 5, panel NP). The binding of a fragment containing three tandem copies of the NP sequence, NP3, surpassed the binding of any of the DNA fragments from the engrailed locus (data not shown). Thus, reiteration of sites produces a very effective binding site.
Figure 4. Synthetic Version of the Consensus Sequence.

(A) Different arrangements of a synthetic consensus sequence. A 12 bp nearly palindromic sequence (NP), TCAATTAAATGA, was synthesized. Positions 1 through 10 of this sequence represent the consensus sequence. The G and A (positions 11 and 12) were added in order to create a BcII site at the junction between two consensus sequences. The arrows indicate the orientations of the consensus sequences in cloned repeats. Note that the addition of the G and A at positions 11 and 12 creates a sequence in the opposite polarity that matches the consensus at 9 out of 10 positions. One or several copies of the NP sequence were cloned in various orientations within the BamHI site of the M13mp18 polylinker.
(B) The NP sequence is nearly palindromic, imperfect at positions 4 and 9, which are both A. Palindromic sequences were synthesized by duplicating in the opposite polarity the left six bases (left palindrome, LP) or duplicating the right six bases (right palindrome, RP) of the NP sequence. Various numbers of copies of the LP and RP sequences were cloned into the BamHI (LP1, LP3, and RP sequences) or Smal (LP2) sites of M13mp18.
A variety of spacing of the repeated sites is compatible with binding. Each of the three possible orientations of two NP sequences (tandem, head-to-head, and tail-to-tail) gives similar binding results (data not shown). Furthermore, the various engrailed DNA fragments showing binding have different spacings of sites.
Single Nucleotide Substitutions Influence Binding
Substitutions can be used to test whether individual base pairs of the binding site contribute to site recognition. Position 4 of the consensus sequence is A in 18 out of 21 footprinted sites and T in the remaining 3. A sequence containing the less preferred base at this position is referred to as right palindromic, or RP, since this alteration makes the site palindromic and related to the right half of the NP sequence (Figure 4B). Fragments carrying different numbers of RP sequences were bound much less well than fragments carrying the same number of copies of the NP sequence (Figure 5; see legend for description of the LP2* fragment that is used as an internal standard). Thus, as predicted by the consensus, A is clearly preferred at position 4.
Symmetry plays an important part in characterized protein DNA complexes. In these, symmetric dimers or tetramers make similar contacts on either side of a palindromic site. The consensus sequence defined by our analysis has only weak palindromic features. Positions 3, 5, and 6 and positions 7, 8, and 10 are symmetric with a dyad between positions 6 and 7. The synthetic consensus (NP) was made more symmetric by the addition of two nucleotides. The prokaryotic precedents and the effectiveness of the nearly palindromic NP sequence led us to test the importance of symmetry in the site. Since A is preferred at position 4 (see above), symmetry would predict a T at position 9 rather than the consensus A. We tested the effect of a T for A substitution on binding. We refer to this substituted sequence as the left palindromic, or LP sequence, because of its relationship to the left half of the NP sequence (Figure 4B). As shown in Figure 5, fragments carrying different numbers of copies of LP bind about as well as fragments carrying the same number of copies of NP. Consequently, there is no distinct preference of T over A at position 9, in contrast to the distinct preference for A at position 4. Thus, these symmetrically disposed positions appear to make different contributions to binding.
The En Fusion Binds Cooperatively to Repeated Sites
In order to more precisely define how the En protein recognizes repeated synthetic sequences, we performed DNAase I protection experiments at several concentrations of the En fusion protein. The NP6 sequence exhibited a periodic pattern of DNAase I protection (Figure 2B). A strong enhancement appeared between the 12th bp of the synthetic repeat and the 1st bp of the next repeat.
As expected from the weak binding of a fragment containing a single site, we did not see complete protection of an isolated NP site. However, high concentrations of extract resulted in partial protection and strong enhancements at positions similar to those described for the footprint of site 1 in the NP6 fragment (data not shown). We conclude that the En fusion binds to a single copy of the synthetic consensus, albeit weakly. Similarly, a single copy of LP was only protected at high concentrations of En fusion. The presence of adjacent sites dramatically reduced the concentration of fusion needed to produce protection (compare LP1 and LP3 in Figure 2C). This demonstrates a form of cooperativity between sites.
The DNA Binding Specificities of En and Ftz Fusions are Related
If sequence-specific DNA binding is one of the fundamental functions of HDs, we would expect this activity to be conserved among the family of proteins containing this element. Indeed, an HD-containing Ftz fusion protein had sequence-specific DNA binding activity (Figure 1). The sequence specificities of the Ftz and En fusion proteins were closely related. All sites footprinted by one fusion were also footprinted by the other (e.g., Figure 2A). The footprints produced by the two proteins differed somewhat in the strength of enhancements, the effectiveness of protection and the size of the protected region (Figure 2A). Both En and Ftz fusions bound to fragments carrying NP, LP, and RP sites and had higher affinities for fragments with increasing numbers of sites (data not shown). Despite close parallels in the sequence recognized and the influence of site repetition, the two proteins differed slightly in site preference. Figure 5 shows that the En fusion greatly favored LP sequences over RP sequences. While the Ftz fusion also preferred LP sites to RP sites, it did not discriminate between these sites as well as the En fusion did (Figure 5).
A Sequence Unlike the Consensus Is Bound by the En and Ftz Fusion Proteins
It is possible that En and Ftz HDs can also recognize other specific sequences that did not occur within the DNA we have analyzed. We were led to suspect this because a different consensus binding sequence has been described for the Ubx encoded protein (Ubx protein), which has an HD closely related to the Ftz HD (P. Beachy, M. Krasnow, L. Gavis, and D. Hogness, personal communication; also see Robertson, 1987). This consensus sequence, deduced from Ubx protein binding to sites in the putative regulatory regions of the Ubx and Antp genes, consists of repetitions of the trinucleotide TAA, with an apparent preference for five copies, (TAA)5. We found that the En fusion could also bind the (TAA)5-like sequences present in Ubx and Antp DNA, although these sites were bound more weakly than sites matching the NP consensus in en DNA (data not shown). Both the En fusion and the Ftz fusion bound to synthetic versions of the (TAA)5 sequence, but, relative to LP*2 (see legend of Figure 5), the Ftz fusion bound the TAA type of sequence better than the En fusion. Consequently, it appears that both of these HD-containing proteins can bind two different types of sequences but with differing preferences.
To probe the relationship of the activities responsible for binding TAA and NP sequences, we tested whether the two types of sequences compete with each other for binding. Synthetic oligonucleotides representing NP and (TAA)5 were ligated and used as competitors. The binding of one type of oligonucleotide prevented binding of the other (Figure 5 and data not shown). Thus, the extract contains a single activity that binds both sequences. Furthermore, binding of the two sequences must rely on interdependent sites or even the same site on the protein.
Discussion
Molecular characterization of eukaryotic transcription factors has increasingly supported the generalization that these regulators are organized in families of evolutionary related members (Chowdbury et al., 1987; Evans and Hollenberg, 1988; Jones et al., 1988). Recently, it has become apparent that some regulators are related not only by sequence homology but also by similarities in their DNA binding specificity. This is particularly well documented for the hormone receptors for mineralocorticoids, glucocorticoids, and progesterone, all of which can activate transcription from the same enhancer region (the MMTV LTR, Chandler et al., 1983; Cato et al., 1986; Arriza et al., 1987). Progesterone and glucocorticoid receptors bind to the same sequences with subtle differences that can influence the relative strength of binding to different sites (Chalepakis et al., 1988). The more diverged estrogen receptor has a distinct specificity (Green and Chambon, 1987). Other examples of regulators with overlapping sequence specificity include members of the Jun–Ap1 family (Struhl, 1987; Franza et al., 1988) and a number of CAAT binding proteins (Jones et al., 1988).
The highly conserved homeodomain (HD) sequence identifies a large family of related regulators (Gehring and Hiromi, 1986). The Drosophila members of this family function together in a regulatory network guiding embryonic pattern formation. Because many of the Drosophila genes encoding HD-containing proteins have been identified by mutation, these regulators might be particularly amenable to analyses exploring the functional interrelationships that tie a family of regulators together. Our data suggest the possibility that one such tie might be overlapping sequence specificities of different HD-containing proteins.
The Homeodomain Is Responsible for Sequence-Specific DNA Binding
As had been predicted on the basis of homology with the prokaryotic helix-turn-helix proteins, our results demonstrate that sequences within the HD are responsible for DNA binding (Laughon and Scott, 1984). Deletions of the En fusion delimit the region essential for DNA binding activity to 70 amino acids, beginning 11 amino acids N-terminal to the HD and extending through the first 59 amino acids of the HD (Figure 1). Since the Ftz protein has no homology to the 11 residues N-terminal to the En HD and yet binds the same DNA sequences, we conclude that the binding activity is specified by conserved amino acid residues in the HD. Consistent with this, other proteins, whose only homology to the En protein is within the HD, bind DNA with specificities related to the En HD (Hoey and Levine, 1988; R. Kostriken, personal communication; P. Beachy, M. Krasnow, L. Gavis, and D. Hogness, personal communication). In addition, mutational analysis of the yeast MATα2 protein has roughly located its DNA binding activity to the HD (Hall and Johnson, 1987).
Our deletion analysis suggests that the in vitro DNA binding specificity that we observed is intrinsic to the HD without influence from the remainder of the protein. In the natural gene products, this intrinsic binding specificity could be modified by interactions outside the HD (Sauer et al., 1979).
Site Sequence and Repetition Contribute to En Fusion Protein Binding to DNA
The clustering of consensus sequences in tightly bound natural DNA fragments suggested that both primary sequence recognition and site reiteration might be important for binding. Indeed, analysis of two synthetic versions of the consensus sequence differing by a single base pair shows that in vitro binding of the En fusion protein depends on primary sequence of the sites (compare NP and RP; first and third panels of Figure 5) and also on the number of repetitions of the site (e.g., compare NP and NP3 in Figure 5).
The improvement in binding seen with site repetition is not simply additive; the concentration of fusion protein required to protect an individual site from DNAase I is decreased 10- to 25-fold by the presence of an adjacent site. This type of result, enhancement of binding by the presence of adjacent sites, has been used as an assay for cooperative interactions in binding (Hochschild et al., 1986; Brenowitz et al., 1986). Unfortunately, the assay is ambiguous unless the form of the binding protein is known. Oligomerization or aggregation of the binding protein could result in preferential interaction with fragments having multiple sites. Our lacZ fusions might exhibit preferential binding to multiple sites because β-galactosidase is a tetramer.
The clustering of binding sites near the engrailed coding region is conserved in the distantly related D. virilis genome (Figure 3; and J. Kassis, C. Desplan, D. Wright and P. O’Farrell, unpublished data). Because chance occurrence and conservation of such clusters is implausible, we believe that the clustering of sites is important and suggest that it is because regulators acting at this site bind cooperatively.
The DNA Binding Specificities of En and Ftz HD Are Related
Here we have shown that a Ftz fusion protein binds to the same sites as the En fusion protein. This applies to “natural” and to synthetic sites. Even in a screen of more than one hundred “natural” and sites of high and low affinity, we failed to detect sites uniquely recognized by one of these fusion proteins (D. Wright, J. Kassis, C. Desplan, and P. O’Farrell, unpublished data).
The En and Ftz HD sequences differ by 52%. Similarly, the HD of eve has diverged from both En and Ftz HDs by about 50% while also retaining a sequence specificity related to that of the En HD (Hoey and Levine, 1988; J. Treisman and C. Desplan, unpublished data). On the other hand, the sequence specificity of yeast MATα2, which has a more distantly related HD (32% identity with the En HD), is not obviously related to that described here (Johnson and Herskowitz, 1985). Consequently, we propose that the HD family of regulators will shown similarities in DNA binding specificity that parallel their similarities in amino acid sequence. From this we expect that HDs exhibiting higher sequence identity than the En:Ftz pair will exhibit very similar binding specificity. For example, the HD of invected (88% sequence identity with the En HD, Coleman et al., 1987) ought to have a specificity extremely similar to that of en. Perhaps differences in binding specificity of HDs will parallel differences in the putative recognition residues (Laughon and Scott, 1984; Table 1). Such a correlation would support the widely accepted but not yet tested view that HD containing proteins bind to DNA in a fashion analogous to helix-turn-helix proteins.
Table 1.
Relationship among Homeodomain Sequences in the Putative Recognition Helix
| Residue Number | *1 | *2 | 3 | 4 | *5 | *6 | 7 | 8 | *9 |
|---|---|---|---|---|---|---|---|---|---|
| Common Residues | |||||||||
| - in all HDs | x | x | x | I/V | x | x | W | F | x |
| - in classes I, II, & III | E | x | Q | I/V | K | I | W | F | Q |
| Variable Residues | |||||||||
| - in class I (Antp, Ubx, ftz, Scr, Dfd AbdB, zen, zen2, cad) | - | R | - | - | - | - | - | - | - |
| - in class II (en & inv) | - | A | - | - | - | - | - | - | - |
| - in class III (99B, labial, rough) | - | T | - | - | - | - | - | - | - |
| - in class IV (prd, gsbl, gsb2) | - | A | R | - | Q | V | - | - | S |
| - in eve | - | S | T | - | - | V | - | - | - |
| - in bcd | T | A | - | - | - | - | - | - | K |
Nine residues corresponding to positions 42 through 50 in the homeodomain are predicted to constitute the recognition helix, based on the alignment proposed by Laughon and Scott (1984) of the homeodomain sequences with the helix-turn-helix motif of prokaryotic DNA binding proteins. Position 4 in the helix is always I or V. Since both of these amino acids are seen in prokaryotic bacteriophage proteins, yeast transcriptional regulators, MATa1 and MATα2, and also both Drosophila and vertebrate HDs, they appear to be equivalent alternatives. All 19 HD sequences compiled here are conserved at positions 4, 7, and 8, while the majority of the presently identified HD sequences differ only at position 2. The sources of the sequence are as follows: Antp, Ubx, and ftz (McGinnis et al., 1984b; Scott and Weiner, 1984); Scr (Kuroiwa et al., 1985); Dfd and AbdB (Regulski et al., 1985); zen (Doyle et al., 1986); zen2 (C. Rushlow and M. Levine, personal communication); cad (Mlodzik et al., 1985, 1987; Macdonald and Struhl, 1986); en (Poole et al., 1985; Fjose et al., 1985); inv (Coleman et al., 1987); 99B (B. Jacq, A. Fjose, and W. Gehring, personal communication); F 90-2, homeodomain of the lab gene (Hoey et al., 1986; and personal communication from A. Mahowald); rough (B. Kalionis and R. Saint, personal communication); prd, gsb1, and gsb2 (Bopp et al., 1986); eve (Macdonald et al., 1986); and bcd (Frigerio et al., 1986).
A Second Sequence Is Recognized by Homeodomains
Studies of the DNA binding activity of the Ubx protein suggested that it binds specifically to a simple trinucleotide repeat, (TAA)5, unrelated to NP (P. Beachy, M. Krasnow, L. Gavis, and D. Hogness, personal communication; see Robertson, 1987). Superficially, this seemed inconsistent with the interpretation made from our results, that Ubx and Ftz proteins should have similar sequence specificity because they have closely homologous HDs (77% identity). However, precedents exist for dual sequence specificity of DNA binding proteins (Ross and Landy, 1982; Pfeifer et al., 1987) and indeed, binding experiments with the En and Ftz fusion proteins show that they are also able to bind this second sequence. Again, though showing related binding specificities, the two fusion proteins exhibit different site preferences. The Ftz fusion seems to bind to TAA repeats about as well as it binds the NP class of sequences, while the En fusion shows a preference for binding to the NP class.
A Network of Related Regulators
The observations here suggest that HD-containing regulators might compete for binding to sites. Since a number of eukaryotic regulators have been found to share overlapping binding specificities (Von der Ahe et al., 1985; Cato et al., 1986; Struhl, 1987; Franza et al., 1988), we suggest the generalization that evolutionary duplication and divergence have created families of regulators with varying levels of functional homology. Consequently, it seems likely that many DNA binding sites will not have unique cognate transcription factors. Rather, competition among related binding proteins would govern which protein occupies a site and thus determine the ultimate effect of the site. Thus, the relative affinities of different proteins for different sites would play a major role in defining their regulatory specificity. This behavior would be analogous to the regulatory behavior of lambda repressor and cro. These related proteins compete for binding to the bacteriophage rightward operator and have opposing regulatory consequences (Ptashne, 1986).
Many of the HD-containing proteins function to guide embryonic pattern formation. These related developmental regulators act in an elaborate network that proceeds through a cascade of steps. At each step, regulators are expressed in overlapping spatial distributions. These act in combinatorial codes to control the spatial pattern of expression of subsequent regulators. Accordingly, competition and cooperation among HDs might provide a tie that interconnects the component regulators in an integrated network.
Experimental Procedures
Plasmid Constructions
The engrailed HD-lacZ fusion construct (A in Figure 1) is described in Desplan et al. (1985): briefly, a BamHI-HindIII fragment from the en cDNA (Poole et al., 1985), containing the homeobox plus flanking sequences, was fused in-frame with the lacZ gene in a pUR290 vector (Ruther and Muller-Hill, 1983) opened at the BamHI and HindIII sites of its polylinker. Construct B derived from construct A by splicing out a 32 bp SaII (cuts between codons 58 and 59 of the HD) to PstI fragment (see Poole et al., 1985). The two sites were blunt-ended with T4 polymerase and ligated. The resulting open reading frame regenerates codon 59 of the HD, replaces the last codon of the HD (thr to ser), and immediately terminates.
To create the fusions to the calcitonin (CT) gene (construct C and D in Figure 1), a BgIII-BsmI (BsmI site blunted with mung bean nuclease) fragment encoding part of preprocalcitonin (Le Moullec et al., 1984) was cloned into pUC8 opened at the AccI (blunt-ended with mung bean nuclease) and BamHI sites. In the resulting construct, pLac.Ct, a calcitonin-containing peptide is expressed under the control of the lac promoter. To create new fusion junctions in the en sequences, construct A was cut at the unique BamHI site (upstream of the homeobox), digested with BaI31 to resect the ends, then digested with EcoRI and blunt-ended by filling in with Klenow polymerase. The various fragments were then cloned into the filled-in (with Klenow polymerase) HindIII site of pLac.CT. Clones were screened by sizing the fusion proteins produced. The CT-En junctions of several plasmids were sequenced (Chen and Seeburg, 1985). In construct C, the fusion occurs 11 codons prior to the homeobox. In construct D, the fusion is located 41 codons upstream of the homeobox. The C-terminal part of these molecules is the same as in construct A.
Construct E (Figure 1) is a deletion encompassing the C-terminal part of the En HD. The sequence coding for the HD was interrupted at the BgIII site (codon 47 of the HD), blunt-ended by filling in with Klenow polymerase, and fused with the filled-in XhoI site located nine codons prior to the stop codon of the en cDNA (Poole et al., 1985). The resulting open reading frame (ORF) differs after codon 47 of the HD. Construct F is a deletion of amino acids 48 to 58 of the HD, inclusive. A BgIII–SaII fragment was spliced out of the construct A, and the plasmid was recircularized, after filling in the two sites with Klenow polymerase. The reading frame, after the deletion, is conserved to the end of the En protein.
The Ftz fusion protein (G in Figure 1) was constructed by fusing a BstEII (filled-in with Klenow polymerase) to HindIII fragment of the ftz cDNA (Laughon and Scott, 1984) to the lacZ gene of a pUR290 vector (Ruther and Muller-Hill, 1983) opened at the BamHI (filled-in with Klenow) and at the HindIII sites. The resulting plasmid expresses a fusion protein that includes the 144 amino acids N-terminal to the HD, the 60 amino acid HD, and the C-terminal 97 residues of the Ftz protein.
For all the constructions, the fusion proteins were extracted as described in Desplan et al. (1985).
The synthetic version of the consensus sequence (NP) was cloned into the BamHI site of M13mp18 as one or several copies (see Figure 4A). The LP1, LP3, and all RP constructs were cloned in the BamHI site, while LP2, LP2*, LP4, and LP4* were cloned in the Smal site. LP2* and LP4* are distinct from LP2 and LP4, respectively, as described in the legend of Figure 4.
Immunoprecipitation of DNA Fragments
The technique is described in Desplan et al. (1985). Each of the various M13mp18 DNAs containing the different versions of the consensus was digested with HindIII, labeled with T4 kinase, and redigested with EcoRI. The excised fragments were purified on a 5% polyacrylamide gel. Various labeled purified fragments were mixed and incubated for 30 min at 0°C with En or Ftz fusion protein extracts in 25 μl of binding buffer (50 mM NaCl, 20 mM Tris-HCl [pH 7.6], 0.25 mM EDTA, 1 mM DTT, 10% glycerol) with differing amounts of competitor DNA (in Figure 5, the competitor is a mixture of oligomerized double-stranded oligonucleotides prepared from the sequence (TAA)5 and its complement; oliogomers of the NP sequence gave comparable results). The fragments complexed to the fusion protein were immunoprecipitated by addition of 0.5 μl of partially purified polyclonal anti-β-galactosidase antiserum (Cappel) adsorbed on 10 μl of fixed Staphylococcus (Pansorbin, Calbiochem). The pellets were phenol extracted, and the DNA was ethanol precipitated and electrophoresed on 8% sequencing gels. The amount of protein extract used in this experiment was 2.2 μg per 25 μl for the En fusion and 3.7 μg per 25 μl for the Ftz fusion.
DNAase I Protection Assays
5′ end-labeled DNA was incubated, for 30 min at 0°C, with the bacterial extract (0–44 μg/sample for the En protein, 0–37 μg/sample for the Ftz protein) in 25 μl of binding buffer with 1 mM EDTA. The mixture was then diluted to 200 μl with 10 mM Tris-HCl (pH 7.5), 12 mM MgCl2, 2.5 mM CaCl2, 1 mM DTT, 10% glycerol, and 10 μg/ml of carrier DNA (Calf thymus) and immediately incubated for 5 min at 0°C in the presence of 250 ng/ml (for footprints of the en and ftz fragments) or 1 μg/ml (for footprints of the fragments containing the NP or LP sequences) of DNAase I (BRL). The reaction was stopped by addition of 200 μl of 40 mM Tris-HCl (pH 8.0), 20 mM EDTA, and 600 mM NaCl and then 400 μl of 1:1 phenol-chloroform mixture. The DNA was ethanol precipitated from the aqueous phase and electrophoresed on 6% or 8% sequencing gels. Parallel lanes containing similar DNA treated with the chemical sequencing reactions of Maxam and Gilbert (1980) were also run on the same gels.
Acknowledgments
We are grateful to the people in the O’Farrell lab for their support. In particular, we thank Steve DiNardo, Jim Jaynes, Jill Jongens, Bill Kalionis, Delia Lakich, Christian Lehner, and John Little for their valuable comments on the manuscript, and Judy Piccini for handling its editing. We also thank people in the C. Guthrie and K. Yamamoto labs, in particular Shelly Haltiner Jones. We would like to acknowledge open discussions with Matt Scott, Alan Laughon, Tim Hoey, and Mike Levine, who have been very generous with material, information, and encouragement. We also thank Phil Beachy, Liz Qavis, Mark Krasnow, and David Hogness for sharing their data on the Ubx protein and its binding sites, Jessica Treisman for the data on the binding of various classes of homeodomains, and Annick Jullienne and M. S. Moukhtar for the calcitonin cDNA and constant encouragement. A special thank you is addressed to Steve DiNardo and Judy Kassis who have been invaluable colleagues and friends who have contributed many ideas during challenging and stimulating discussions. This work was supported by National Institutes of Health and National Science Foundation grants (to P. H. O.). C. D. was an EMBO fellow, and J. T. was supported by a National Institutes of Health training grant.
References
- Arriza JL, Weinberger C, Cerelli G, Glaser TM, Handelin BL, Housman DE, Evans RM. Cloning of human mineralocorticoid receptor complimentary DNA: structural and functional kinship with the glucocorticoid receptor. Science. 1987;237:268–274. doi: 10.1126/science.3037703. [DOI] [PubMed] [Google Scholar]
- Beachy PA, Helfand SL, Hogness DS. Segmental distribution of bithorax complex proteins during Drosophila development. Nature. 1985;313:545–551. doi: 10.1038/313545a0. [DOI] [PubMed] [Google Scholar]
- Bopp D, Burri M, Baumgartner S, Frigerio G, Noll M. Conservation of a large protein domain in the segmentation gene paired and in functionally related genes in Drosophila. Cell. 1986;47:1033–1049. doi: 10.1016/0092-8674(86)90818-4. [DOI] [PubMed] [Google Scholar]
- Brenowitz M, Senear DF, Shea MA, Ackers GK. “Footprint” titrations yield valid thermodynamic isotherms. Proc Natl Acad Sci USA. 1986;83:8462–8466. doi: 10.1073/pnas.83.22.8462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll SB, Scott MP. Localization of the fushi tarazu protein during Drosophila embryogenesis. Cell. 1985;43:47–57. doi: 10.1016/0092-8674(85)90011-x. [DOI] [PubMed] [Google Scholar]
- Carroll SB, Scott MP. Zygotically active genes that affect the spatial expression of the fushi tarazu segmentation gene during early Drosophila embryogenesis. Cell. 1986;45:113–126. doi: 10.1016/0092-8674(86)90543-x. [DOI] [PubMed] [Google Scholar]
- Cato ACB, Miksicek R, Schutz G, Arnemann J, Beato M. The hormone regulatory element of mouse mammary tumor virus mediates progesterone induction. EMBO J. 1986;5:2237–2240. doi: 10.1002/j.1460-2075.1986.tb04490.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalepakis G, Arnemann J, Slater E, Brüller HJ, Gross B, Beato M. Differential gene activation by glucocorticoids and progestins through the hormone regulatory element of mouse mammary tumor virus. Cell. 1988;53:371–382. doi: 10.1016/0092-8674(88)90157-2. [DOI] [PubMed] [Google Scholar]
- Chandler VL, Maler BA, Yamamoto KR. DNA sequences bound specifically by glucocorticoid receptor in vitro render a heterologous promoter hormone responsive in vivo. Cell. 1983;33:489–499. doi: 10.1016/0092-8674(83)90430-0. [DOI] [PubMed] [Google Scholar]
- Chen EY, Seeburg PH. Supercoil sequencing: a fast and simple method for sequencing plasmid DNA. DNA. 1985;4:165–170. doi: 10.1089/dna.1985.4.165. [DOI] [PubMed] [Google Scholar]
- Chowdhury K, Deutsch U, Gruss P. A multigene family encoding several “fingers” structures is present and differentially active in mammalian genomes. Cell. 1987;48:771–778. doi: 10.1016/0092-8674(87)90074-2. [DOI] [PubMed] [Google Scholar]
- Coleman KG, Poole SJ, Weir MP, Soeller WC, Kornberg T. The invected gene of Drosophila: sequence analysis and expression studies reveal a close kinship to the engrailed gene. Genes Dev. 1987;1:19–28. doi: 10.1101/gad.1.1.19. [DOI] [PubMed] [Google Scholar]
- Desplan C, Theis J, O’Farrell PH. The Drosophila developmental gene engrailed encodes a sequence specific DNA binding activity. Nature. 1985;318:630–635. doi: 10.1038/318630a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiNardo S, O’Farrell PH. Establishment and refinement of segmental pattern in the Drosophila embryo: spatial control of engrailed expression by pair rule genes. Genes Dev. 1987;1:1212–1225. doi: 10.1101/gad.1.10.1212. [DOI] [PubMed] [Google Scholar]
- DiNardo S, Kuner JM, Theis J, O’Farrell PH. Development of embryonic pattern in D. melanogaster as revealed by accumulation of the nuclear engrailed protein. Cell. 1985;43:59–69. doi: 10.1016/0092-8674(85)90012-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle HJ, Harding K, Hoey T, Levine M. Transcripts encoded by a homeo box gene are restricted to dorsal tissues of Drosophila embryos. Nature. 1986;323:76–79. doi: 10.1038/323076a0. [DOI] [PubMed] [Google Scholar]
- Evans RM, Hollenberg SM. Zinc fingers: gilt by association. Cell. 1988;52:1–3. doi: 10.1016/0092-8674(88)90522-3. [DOI] [PubMed] [Google Scholar]
- Fainsod A, Bogarad LD, Ruusala T, Lubin M, Crothers DM, Ruddle FH. The homeodomain of a murine protein binds 5′ to its own homeo box. Proc Natl Acad Sci USA. 1986;83:9532–9536. doi: 10.1073/pnas.83.24.9532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fjose A, McGinnis WJ, Gehring WJ. Isolation of a homeo box–containing gene from the engrailed region of Drosophila and the spatial distribution of its transcripts. Nature. 1985;313:284–289. doi: 10.1038/313284a0. [DOI] [PubMed] [Google Scholar]
- Franza BR, Jr, Rauscher FJ, III, Josephs SF, Curran T. The fos complex and fos-related antigens recognize sequence elements that contain AP-1 binding sites. Science. 1988;239:1150–1153. doi: 10.1126/science.2964084. [DOI] [PubMed] [Google Scholar]
- Frigerio G, Burri M, Bopp D, Baumgartner S, NoII M. Structure of the segmentation of gene paired and the Drosophila PRD gene set as part of a gene network. Cell. 1986;47:735–746. doi: 10.1016/0092-8674(86)90516-7. [DOI] [PubMed] [Google Scholar]
- Garcia-Bellido A. Genetic control of wing disc development in Drosophila. “Cell Patterning,” Ciba Foundation Symp. 1975;29:161–182. doi: 10.1002/9780470720110.ch8. [DOI] [PubMed] [Google Scholar]
- Gehring WJ, Hiromi Y. Homeotic genes and the homeobox. Annu Rev Genet. 1986;20:147–173. doi: 10.1146/annurev.ge.20.120186.001051. [DOI] [PubMed] [Google Scholar]
- Green S, Chambon P. Oestradiol induction of a glucocorticoid-responsive gene by a chimaeric receptor. Nature. 1987;325:75–78. doi: 10.1038/325075a0. [DOI] [PubMed] [Google Scholar]
- Hafen E, Levine M, Gehring W. Regulation of Antennapedia transcript distribution by the bithorax complex in Drosophila. Nature. 1984;307:287–289. doi: 10.1038/307287a0. [DOI] [PubMed] [Google Scholar]
- Hall MN, Johnson AD. Homeo domain of the yeast repressor α2 is a sequence-specific DNA-binding domain but is not sufficient for repression. Science. 1987;237:1007–1012. doi: 10.1126/science.2887035. [DOI] [PubMed] [Google Scholar]
- Hall MN, Hereford L, Herskowitz I. Targeting of E. coli β-galactosidase to the nucleus in yeast. Cell. 1984;36:1057–1065. doi: 10.1016/0092-8674(84)90055-2. [DOI] [PubMed] [Google Scholar]
- Harding K, Rushlow C, Doyle H, Hoey T, Levine M. Cross-regulatory interactions among pair-rule genes in Drosophila. Science. 1986;233:953–959. doi: 10.1126/science.3755551. [DOI] [PubMed] [Google Scholar]
- Hochschild A, Douhan J, III, Ptashne M. How λ repressor and λ cro distinguish between OR1 and OR3. Cell. 1986;47:807–816. doi: 10.1016/0092-8674(86)90523-4. [DOI] [PubMed] [Google Scholar]
- Hoey T, Levine M. Divergent homeo box proteins recognize similar DNA sequences in Drosophila. Nature. 1988;332:858–861. doi: 10.1038/332858a0. [DOI] [PubMed] [Google Scholar]
- Hoey T, Rushlow C, Doyle H, Levine M. Homeo box gene expression in anterior and posterior regions of the Drosophila embryo. Proc Natl Acad Sci USA. 1986;83:4809–4813. doi: 10.1073/pnas.83.13.4809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howard K, Ingham P. Regulatory interactions between the segmentation genes fushi tarazu, hairy, and engrailed in the Drosophila blastoderm. Cell. 1986;44:949–957. doi: 10.1016/0092-8674(86)90018-8. [DOI] [PubMed] [Google Scholar]
- Johnson AD, Herskowitz I. A repressor (MATα2 product) and its operator control expression of a set of cell type specific genes in yeast. Cell. 1985;42:237–247. doi: 10.1016/s0092-8674(85)80119-7. [DOI] [PubMed] [Google Scholar]
- Jones NC, Rigby PWJ, Ziff EB. Trans acting protein factors and the regulation of eukaryotic transcription: lessons from studies on DNA tumor viruses. Genes Dev. 1988;2:267–281. doi: 10.1101/gad.2.3.267. [DOI] [PubMed] [Google Scholar]
- Kuroiwa A, Kloter U, Baumgartner P, Gehring WJ. Cloning the homeotic Sex combs reduced in gene in Drosophila and in situ localization of its transcripts. EMBO J. 1985;4:3757–3764. doi: 10.1002/j.1460-2075.1985.tb04145.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laughon A, Scott MP. Sequence of a Drosophila segmentation gene: protein structure homology with DNA binding proteins. Nature. 1984;310:25–31. doi: 10.1038/310025a0. [DOI] [PubMed] [Google Scholar]
- Le Moullec JM, Jullienne A, Chenais J, Lasmoles F, Guliana JM, Milhaud G, Moukhtar MS. The complete sequence of human preprocalcitonin. FEBS Lett. 1984;167:93–97. doi: 10.1016/0014-5793(84)80839-x. [DOI] [PubMed] [Google Scholar]
- Lewis EB. Pseudoallelism and genome evolution. Cold Spring Harbor Symp Quant Biol. 1951;16:159–174. doi: 10.1101/sqb.1951.016.01.014. [DOI] [PubMed] [Google Scholar]
- Lewis EB. A gene complex controlling segmentation in Drosophila. Nature. 1978;276:565–570. doi: 10.1038/276565a0. [DOI] [PubMed] [Google Scholar]
- Macdonald PM, Struhl G. A molecular gradient in early Drosophila embryos and its role in specifying the body pattern. Nature. 1986;324:537–545. doi: 10.1038/324537a0. [DOI] [PubMed] [Google Scholar]
- Macdonald PM, Ingham P, Struhl G. Isolation, structure, and expression of even-skipped: a second pair-rule gene of Drosophila containing a homeo box. Cell. 1986;47:721–734. doi: 10.1016/0092-8674(86)90515-5. [DOI] [PubMed] [Google Scholar]
- Maxam AM, Gilbert W. Sequencing end labeled DNA with base specific chemical cleavages. Meth Enzymol. 1980;65:499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- McGinnis W, Garber RL, Wirz J, Kuroiwa A, Gehring WJ. A homologous protein-coding sequence in Drosophila homeotic genes and its conservation in other metazoans. Cell. 1984a;37:403–408. doi: 10.1016/0092-8674(84)90370-2. [DOI] [PubMed] [Google Scholar]
- McGinnis W, Levine MS, Hafen E, Kuroiwa A, Gehring WJ. A conserved DNA sequence in homeotic genes of the Drosophila Antennapedia and Bithorax complexes. Nature. 1984b;308:428–433. doi: 10.1038/308428a0. [DOI] [PubMed] [Google Scholar]
- McKay R. Binding of a simian virus 40 T-antigen related protein to DNA. J Mol Biol. 1981;145:471–488. doi: 10.1016/0022-2836(81)90540-4. [DOI] [PubMed] [Google Scholar]
- Mlodzik M, Gehring WJ. Expression of the caudal gene in the germ line of Drosophila: formation of an RNA and a protein gradient during early embryogenesis. Cell. 1987;48:465–478. doi: 10.1016/0092-8674(87)90197-8. [DOI] [PubMed] [Google Scholar]
- Mlodzik M, Fjose A, Gehring WJ. Isolation of caudal, a Drosophila homeo box-containing gene with maternal expression, whose transcripts form a concentration gradient at the pre-blastoderm stage. EMBO J. 1985;4:2961–2969. doi: 10.1002/j.1460-2075.1985.tb04030.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nusslein-Volhard C, Wieschaus E. Mutations affecting segment number and polarity in Drosophila. Nature. 1980;287:795–801. doi: 10.1038/287795a0. [DOI] [PubMed] [Google Scholar]
- O’Farrell PH, Desplan C, DiNardo S, Kassis JA, Kuner J, Lim E, Sher E, Theis J, Wright D. Molecular analysis of the involvement of the Drosophila engrailed gene in embryonic pattern formation. In: Edelman GM, editor. Molecular Determinants of Animal Form, UCLA Symposium on Molecular and Cellular Biology, New Series. Vol. 31. New York: Alan R. Liss; 1985. pp. 489–519. [Google Scholar]
- Pabo CO, Sauer RT. Protein DNA recognition. Annu Rev Biochem. 1984;53:293–321. doi: 10.1146/annurev.bi.53.070184.001453. [DOI] [PubMed] [Google Scholar]
- Pfeifer K, Prezant T, Guarente L. Yeast HAP1 activator binds to two upstream activation sites of different sequence. Cell. 1987;49:19–27. doi: 10.1016/0092-8674(87)90751-3. [DOI] [PubMed] [Google Scholar]
- Picard D, Yamamoto KR. Two signals mediate hormone-dependent nuclear localization of the glucocorticoid receptor. EMBO J. 1987;6:3333–3340. doi: 10.1002/j.1460-2075.1987.tb02654.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poole SJ, Kauvar LM, Drees B, Kornberg T. The engrailed locus of Drosophila: structural analysis of an embryonic transcript. Cell. 1985;40:37–43. doi: 10.1016/0092-8674(85)90306-x. [DOI] [PubMed] [Google Scholar]
- Ptashne M. A genetic switch. Cambridge, Massachusetts and Palo Alto, California: Cell Press and Blackwell Scientific Publications; 1986. [Google Scholar]
- Regulski M, Harding K, Kostriken R, Karch F, Levine M, McGinnis W. Homeo box genes of the Antennapedia and Bithorax complexes of Drosophila. Cell. 1985;43:71–80. doi: 10.1016/0092-8674(85)90013-3. [DOI] [PubMed] [Google Scholar]
- Robertson M. A genetic switch in Drosophila morphogenesis. Nature. 1987;327:556–557. [Google Scholar]
- Ross W, Landy A. Bacteriophage lambda Int protein recognized two classes of sequence in the phage att site: characterization of arm-type sites. Proc Natl Acad Sci USA. 1982;79:7724–7728. doi: 10.1073/pnas.79.24.7724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruther U, Muller-Hill B. Easy identification of cDNA clones. EMBO J. 1983;2:1791–1794. doi: 10.1002/j.1460-2075.1983.tb01659.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauer RT, Pabo CO, Meyer BJ, Ptashne M, Backman KD. Regulatory functions of lambda repressor reside in the amino-terminal domain. Nature. 1979;279:396–400. doi: 10.1038/279396a0. [DOI] [PubMed] [Google Scholar]
- Scott MP, O’Farrell PH. Spatial programming of gene expression in early Drosophila embryogenesis. Annu Rev Cell Biol. 1986;2:49–80. doi: 10.1146/annurev.cb.02.110186.000405. [DOI] [PubMed] [Google Scholar]
- Scott MP, Weiner AJ. Structural relationships among genes that control development: sequence homology between the Antennapedia, Ultrabithorax and fushi tarazu loci of Drosophila. Proc Natl Acad Sci USA. 1984;81:4115–4119. doi: 10.1073/pnas.81.13.4115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shepherd JCW, McGinnis W, Carrasco AE, DeRobertis EM, Gehring WJ. Fly and frog homoeo domains show homologies with yeast mating type regulatory proteins. Nature. 1984;370:70–71. doi: 10.1038/310070a0. [DOI] [PubMed] [Google Scholar]
- Struhl K. The DNA-binding domains of the jun oncoprotein and the yeast GCN4 transcriptional activator protein are functionally homologous. Cell. 1987;50:841–846. doi: 10.1016/0092-8674(87)90511-3. [DOI] [PubMed] [Google Scholar]
- Von der Ahe D, Janich S, Scheidereit C, Renkawitz R, Schutz G, Beato M. Glucocorticoid and progesterone receptors bind to the same sites in two hormonally regulated promoters. Nature. 1985;373:706–709. doi: 10.1038/313706a0. [DOI] [PubMed] [Google Scholar]
- White RAH, Wilcox M. Protein products of the Bithorax complex in Drosophila. Cell. 1984;39:163–171. doi: 10.1016/0092-8674(84)90202-2. [DOI] [PubMed] [Google Scholar]