Summary
The engrailed gene acts early in Drosophila embryogenesis and plays an essential role in the processes that establish and maintain the repeating segmental pattern. To begin molecular analysis of the role of the engrailed gene in embryonic pattern formation, we used a chromosomal walk to clone genomic sequences that encompass the locus, and have physically mapped the positions of 15 engrailed mutations. The positions of engrailed rearrangement mutations indicate that the engrailed complementation unit includes a minimum of 70 kb. The locus can be divided into two regions. Rearrangement mutations interrupting the centromere proximal 50 kb of the locus result in embryonic lethality while mutants altered in the distal 20 kb of the locus survive to show morphological abnormalities in several adult segments. It appears that long-range cis interactions play a role in the function of the engrailed gene.
Introduction
Genetic analysis has identified a number of genes that regulate key steps in Drosophila embryonic development (Lewis, 1978; Kaufman et al., 1980; Nusslein-Volhard and Wieschaus, 1980; Kornberg, 1981a; Nusslein-Volhard et al., 1984; Jurgens et al., 1984; Wieschaus et al., 1984). Mutations in some of these genes cause abnormal segmentation. For example, specific pattern elements are deleted in every segment of gooseberry embryos and in every alternate segment in hairy embryos. On the other hand, mutations in the homeotic genes do not affect the segment periodicity but rather alter their developmental fate (Lewis, 1978; Kaufman et al., 1980). This can result in striking transformations where, for example, Antennapedia mutants will grow legs where antennae are normally found. These mutant phenotypes suggest that segments are homologous units whose developmental pathway is under the control of these homeotic loci.
A segmental pattern of organization appears to be specified before it is visible. Positional values (Simcox and Sang, 1983), but not cell types (Garcia-Bellido et al., 1973; Morata and Ripall, 1975) are specified within the first 3 hr of embryogenesis. The formation of developmental compartments is an example of such a specification event. Segment anlagen are subdivided so that individual cells and their progeny are destined to contribute to either anterior or posterior parts of segments, the anterior and posterior compartments (Garcia-Bellido et al., 1973; Kornberg, 1981b, a; 1981b; Morata and Lawrence, 1979; Struhl, 1981; Wieschaus and Gehring, 1976). The compartment boundaries appear to define areas within which particular homeotic genes are expressed (Lawrence and Morata, 1983).
Assignment of cells to compartments plays an integral role in segmentation. In engrailed mutants assignment of cells to compartmental and segmental units eventually fails (Lawrence and Morata, 1976; Kornberg, 1981a). The aberrant form of engrailed mutant embryos indicates a profound effect on segmentation; pairs or larger groups of segments fuse together and the embryos die (Kornberg, 1981a). Analysis of mitotic clones lacking engrailed function has given important clues about its action. In anterior cells of each segment, absence of engrailed function is without apparent consequence. Posterior cells with a similar deficiency can acquire traits of anterior cells and can cross the borders that normally demark the posterior compartment. These observations can be summarized in the following model: positional information in the embryo defines a pattern of engrailed gene expression wherein groups of engrailed expressing cells alternate with groups of nonexpressing cells along the anterior/posterior axis (a zebra stripe pattern). In at least some cell lineages the state of engrailed expression, once established, is stably transmitted to daughter cells. Finally, engrailed product alters cell behavior and cell interaction so that expressing cells are defined as members of the posterior compartment.
Recent studies using cloned sequences have shown a remarkable evolutionary conservation among a number of genes that regulate Drosophila development, suggesting that these genes, and presumably the steps they control, are fundamental and universal (Scott and Weiner, 1984; McGinnis et al., 1984; Poole et al., 1985). The demonstration that these genes are expressed in a spatially restricted pattern suggests that their expression is spatially regulated so that function is expressed in the appropriate position (Hafen et al., 1984; Levine et al., 1983; Akam, 1983; Kornberg et al., 1985). Thus, it appears that much of early pattern formation can be addressed as an issue of spatial programming of the expression of these regulatory genes.
To pursue studies of how the engrailed gene is regulated and how the engrailed gene product acts as a regulator, we have undertaken molecular analysis of the locus. Using chromosome rearrangements as a guide (Kornberg, 1981a; Ali and Kornberg, unpublished) and following approaches pioneered by Bender et al. (1983b), we have isolated overlapping clones representing 225 kb of genomic DNA from a chromosomal region 48A–48B that encompasses the engrailed gene. This report describes the molecular structure of the locus and the physical mapping of 15 engrailed mutations.
Results
Chromosome Walking through the engrailed Locus
Cytological analysis localized the engrailed gene to position 48A on the polytene chromosome map (Kornberg, 1981a). Using tRNA met2 as a probe (Elder et al., 1980) we obtained from a λ phage bank (Maniatis et al., 1978) two genomic clones, E19 and E20, that hybridized to the 48B region.
We took advantage of a relatively small visible deletion that removes all of the cytological region 48A and part of 48B as an aid to the genomic cloning of the 48A region. This deleted chromosome lacks any engrailed function. We were able to establish the orientation of the entry point clones at 48B because the distal end of the enSF31 deletion lay within the E19 clone and could be detected by its altered pattern of DNA restriction fragments on Southern blots. A recombinant DNA bank prepared from enSF31/SM5 was screened with probes to detect clones homologous to E19. A single clone, E31, was isolated and shown to carry sequences from both sides of the deletion (Figure 1).
Figure 1. A Recombinant DNA Clone from Df(2R)enSF31 Spans the Polytene Region of 48A.
In situ hybridization (Pardue and Gall, 1975) has grains at 48A and 48B. The chromosomes are from a wild-type strain and the probe was from the clone E31, a clone containing the enSF31 breakpoint with sequences from 48A1 and 48B5.
Using the breakpoint clone E31 to make hybridization probes, we isolated a second entry point clone, E1, from the region proximal to the enSF31. The entry clones were then used to isolate a series of overlapping clones extending from the two ends toward the middle of the deletion. Comparison of restriction digests and hybridization analyses indicated when the two separate walks overlapped. A total of 225 kb of DNA was cloned from the 48AB region (Figure 2) and 205 kb were found to be deleted by enSF31.
Figure 2. Molecular Map of Polytene Region 48AB.
Coordinates are in kb, based on a zero point at the insertion site of the en1 transposition, and the map is orientated with the centromere to the left. Individual phage (E1-20) and cosmid (cos 189B, CH1A, and 190) clones are shown above the coordinate scale and below it are shown restriction maps that were determined for the individual phage. The arrowheads indicate the end points of the enSF31 deletion.
It is notable that in the course of this work a number of different chromosomes were analyzed without detecting any insertional polymorphisms. For comparison with analyses of other Drosophila chromosomal regions, see Table 1.
Table 1.
| Locusa | Length of Walk in kb | Insertionalb Polymorphisms |
|---|---|---|
| bithorax complex | 195 | 0 |
| rosy–Ace | 315 | 8 |
| Notch | 80 | 1 |
| y–achaete | 120 | 0 |
| Antennapedia | 290 | 2 |
| engrailed | 225 | 0 |
References: Bender et al., 1983a; 1983b; Artavanis-Tsakonas et al., 1983; Carramolino et al., 1982; Harald Biessmann, personal communication; Scott et al., 1983; Garber et al., 1983.
Polymorphisms are given for comparisons of Oregon vs. Canton only. Comparisons to additional chromosomes in some cases reveals additional insertional polymorphisms. Inclusion of results with additional chromosomes reinforces the apparent differences but comparable data is not available for all regions.
Localization of the engrailed Locus within the Cloned Segments
The engrailed gene can be localized within the 225 kb of cloned sequences by physically mapping DNA rearrangements that disrupt engrailed function. A number of chromosomal rearrangements with engrailed phenotypes have been isolated in screens for new engrailed alleles (Kornberg, 1981a; Eberlein and Russell, 1983). To map the positions of these rearrangements, we used in situ hybridization to polytene chromosomes and Southern analysis of genomic DNA to locate rearrangement breakpoints. To confirm these locations, we cloned the rear-ranged sequences.
To show that engrailed rearrangements had breakpoints within the cloned region, the entry point clones E1 and E19 were used as probes to hybridize to polytene chromosomes from selected rearrangement mutants. In all cases examined the E1 probe hybridized on the centromere proximal side of the rearrangement and the E19 probe hybridized to the distal side. Additional in situ hybridization experiments with several probes from the walk roughly located the breakpoints to the middle of the enSF31 deletion.
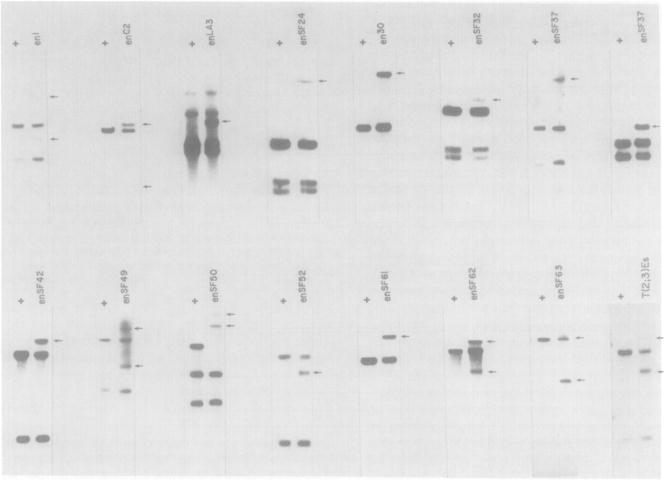
More accurate and convenient localization of the engrailed mutant breakpoints was accomplished by analyzing genomic Southern blots of restriction enzyme digests of mutant and parental DNA probed with phage DNA from the chromosomal walk. When a phage probe detected anomolous DNA fragments in digests with several different restriction enzymes (mostly Eco RI, Bam HI, Bgl II, and Xho I), it was taken to be a region of rearrangement. Determination of the particular wild-type fragment in which a break occurred was complicated by the presence of DNA from a en+ balancer chromosome in all of the engrailed mutant stocks. Thus, although new bands were detected in the mutant DNAs, the normal restriction fragments altered by breaks were not missing, but only reduced in intensity. However, evaluation of band intensity and use of partially overlapping probes or probes from small (1–6 kb) subclones localized the breakpoint lesions to within a few kilobases (Figure 3).
Figure 3. Demonstration of the Positions Altered by Chromosomal Rearrangements.
In each panel, DNA extracted from wild-type or parental flies (designated +) is compared to DNA extracted from an engrailed mutant (right lane). DNA was digested with a restriction endonuclease, transferred to nitrocellulose, and hybridized with a nick translated Eco RI fragment of DNA from the walk. Arrows indicate the novel restriction fragments created by the rearrangement. In some digests both the proximal and distal rearrangement fragments are seen, whereas in others only one of the new fragments is detected because of either limited sensitivity or resolution. Because of the presence of a wild-type allele of engrailed on the balancer chromosome, generally the DNA fragment broken by a DNA rearrangement mutation is still present in the mutant heterozygotes. However, in a few cases there is a polymorphism between the parental chromosome and the balancer; in these, the mutation causes a band to disappear (e.g., enSF50). Digestions of genome DNA and positions (see Figure 2) of the Eco RI fragments used for probes were: en1, Xho I (−0.2, +2.7); enc2, Bam HI (−0.2, +2.7); enLA3, Hind III (+25.3, +34.2); enSF24, Xho I (+2.7, +12.0); en30, Eco RI (+13, +20.5); en32, Xho I (−1.0, −4.7); enSF37, Xho I (−28.0, −33.9); enSF37, Bgl II (+2.7, +12.0); enSF42, Xho I (−10.6, −15.2); enSF49, Bam HI (−10.6, −15.2); enSF50, Xho I (−1.0, −4.7); enSF52, Xho I (−28.0, −33.9); enSF61, Eco RI (+13, +20.5); enSF82, Eco RI (−10.6, −15.2); enSF83, Bam HI (−5.4, −10.6); and enEs, Bgl II (+25.3, +34.2).
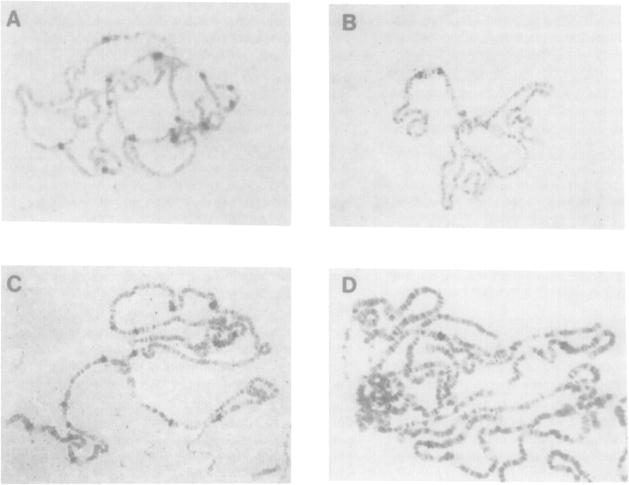
In order to characterize further the organization of the mutant DNA and to ensure that the detected anomalies were not due to polymorphisms, we cloned the rearranged sequences. Genomic clones containing either the novel fragment created by fusion of the distal sequences to a new region or the novel fragment generated by the proximal sequences were isolated from λ phage recombinant libraries prepared from engrailed mutant DNA. Breakpoint clones were isolated in this way for en1, enC2, enLA3, enSF24, enSF37, enSF42, enSF49, enSF52, and enEs. In situ hybridization to wild-type polytene chromosomes directly demonstrated that in these clones of rearranged sequences, the 48A region was fused with a site on either the second or the third chromosome (Figure 4).
Figure 4. In Situ Hybridization with Breakpoint Fragment Probes.
Wild-type polytene chromosomes from larval salivary glands were hybridized with nick translated probes from a subclone containing the en1 insertion element (A), the Bam HI breakpoint restriction fragment of the proximal enSF37 chromosome rearrangement (B), the Eco RI breakpoint restriction fragment of the distal enSF37 chromosome rearrangement (C) and the breakpoint restriction fragment of enSF24 (D). Note multiple sites of hybridization in (A) and (C), sites of hybridization at 48A and 46C in (B), and sites of hybridization at 48A and 65A in (D).
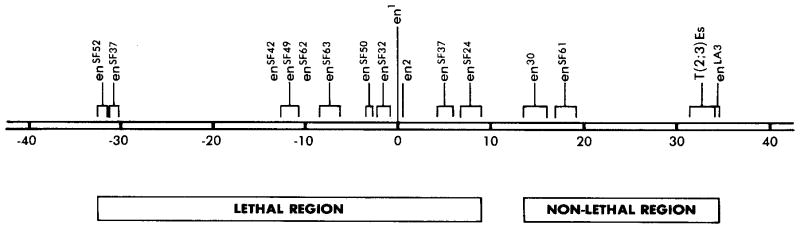
The breakpoint locations are shown in Figure 5. The mutation en1 has been arbitrarily designated as position 0 on this map. It is notable that the engrailed gene defined by these mutations is very large, at least 70 kb.
Figure 5. Physical Location of engrailed Breakpoint Mutations.
The locations of the engrailed breakpoint mutations on the restriction map of the region (see Figure 3) are given. The distances are measured in kb and the accuracy of localization of the breakpoints is indicated by brackets. Note the physical separation of the lethal and nonlethal rearrangement alleles. The two distinct breaks mapped for enSF37 are shown.
Features of engrailed Mutations
engrailed rearrangement mutations do not give null phenotypes. Most dramatically, the rearrangement alleles enLA3, enEs, en30, and enSF62 can complement the lethality of other engrailed alleles while failing to fully complement the engrailed morphological defects (Kornberg, 1981a; Eberlein and Russell, 1983; Epper and Sanchez, 1983). The lethal and nonlethal engrailed rearrangement break-points lie in distinct regions with the nonlethal alleles all lying distal to the lethal alleles (Figure 5). As defined by these chromosomal rearrangements, the size of the genomic region encoding the essential embryonic function is at least 40 kb.
The en1 allele arose spontaneously in 1926 (Eker, 1929). The en1lesion is associated with an insertion element of approximately 7 kb that is repeated about 16 times in the Oregon-R genome (Figure 4A). The phenotype of en1 mutants is unique and surprising. Although en1 flies are viable, the site of insertion is bracketed by lethal engrailed breakpoint mutations (Figure 5). In addition, because en1 homozygotes show severe morphological defects that are largely confined to the thoracic segments of the imago, the engrailed defect appears to be specific to stage and position. Finally, en1 gives a peculiar pattern of partial complementation with some other engrailed alleles (Kornberg, 1981a; Epper and Sanchez, 1983; Eberlein and Russell, 1983; see also below).
The description of the en30 allele (Russell and Eberlein, 1979; Eberlein and Russell, 1983) emphasized a cytologically evident deficiency, 48A3-4 to 48C6-8. Our molecular analysis detected a defect in the cloned region, but we have not directly demonstrated whether this is the proximal end point of the deficiency. On the basis of complementation it appears most reasonable to attribute the engrailed defect of en30 to the alteration that we have mapped within the cloned sequences. The phenotype of heterozygous combinations of en30 with other engrailed alleles is compatible with the observed location of the en30 sequence alteration in the nonlethal region.
Cytologically the enSF37 allele is an insertional translocation of 46C–48A to the heterochromatic base of chromosome 3. Our molecular analysis suggests that it is more complex. Two breakpoints were detected in the engrailed region, one at about −30 kb and one at about +5 kb. Thus, it appears that the translocated region was actually broken into two pieces that were inserted into chromosome 3 in a permuted order (see Figure 4b). It is, of course, uncertain whether the engrailed defect of this allele is due to the proximal, and/or the distal engrailed breakpoint.
Discussion
engrailed Is a Large Gene
The rearrangement mutations mapped here are all part of the engrailed complementation unit. They are dispersed over a 70 kb region. Although obvious uncertainties remain, we believe that 70 kb is a good approximation of the size of the genetic unit. Since mutant alleles mapping at great distances from the characterized transcription unit (see below) are as well represented as mutations in the immediate vicinity of the transcription unit, we argue that the distant lesions cannot be dismissed as unusual phenomena such as second site mutations or position effects. If mutant phenotypes were due to second site changes or position effects, we would not expect a correlation between the severity of mutant phenotype and breakpoint position (see below for discussion of nonlethal mutations).
The unusually large size of the engrailed locus has precedents among other Drosophila genes; two other loci involved in pattern formation, Antennapedia (Scott et al., 1983; Garber et al., 1983) and Ubx (Bender et al., 1983a), have primary transcription units of 105 kb and 70 kb respectively. It is notable that these sizes are a direct physical measure of the transcription unit. If the gene is defined by all mutations that fail to fully complement, the Ubx complementation unit is 30 kb larger than the transcription unit (vis. pbx and bxd mutations do not fully complement Ubx mutations). Thus, for the 100 kb Ubx complementation unit, the size of the transcription unit (70 kb) is a major, but not the exclusive, factor contributing to the large size of the genetic unit. Two features can contribute to the size of these genes, the transcription unit itself and the amount of flanking sequences required in cis for normal expression.
The phenotypes of rearrangement mutations divide both the engrailed and the Ubx complementation groups into lethal and nonlethal regions. Rearrangements within the 70 kb Ubx transcription unit give rise to lethal phenotypes whereas rearrangements within 30 kb upstream of this transcription unit give allele-specific nonlethal phenotypes (Lewis, 1978; Bender et al., 1983a; Beachy et al., 1985). Similarly, we have shown that engrailed rearrangements define distinct lethal regions of 50 kb and nonlethal regions of 20 kb. In contrast to the large primary transcription units of Ubx and Antp, a 2.7 kb engrailed transcript is derived from less than 5 kb of genomic DNA. Three criteria suggest that this transcript encodes en function: its time course of expression is appropriate to the times of engrailed action (Drees, O’Farrell, and Kornberg, unpublished); it is expressed in a position-specific fashion consistent with the pattern expected from genetic analyses (Kornberg et al., 1985; DiNardo, Kuner, Theis, and O’Farrell, unpublished); and, like the coding sequences of genes from the Bithorax and Antennapedia complex, it contains a homeo box sequence (Poole et al., 1985). As presently characterized, this transcript maps to genomic sequences located roughly at the center of the genetic unit (approximately position −13 to −18 on our chromosomal walk) and is transcribed in the distal to proximal direction (Poole et al., 1985; Drees, O’Farrell, and Kornberg, unpublished). Thus, it appears that the large size of the engrailed complementation unit is primarily due to a requirement for long range cis interactions for normal function (see below).
Structure of the engrailed Complementation Unit
The engrailed mutations belong to a single complementation unit. However, more detailed considerations suggest that the large region constituting the engrailed gene contains interacting elements. First, because engrailed mutations fail to fully complement en1, they are considered allelic; nonetheless, when the severities of the phenotypes are scored, the same engrailed alleles vary significantly in their ability to complement the en1 morphological defects. One aspect of the complementation shows no variation. The en1 allele provides an activity that complements the embryonic lethality of all engrailed lethal mutations. However, the nonrearrangement alleles show differing abilities to complement the adult morphological defects of en1. This complementation activity cannot be explained by proposing that these embryonic lethal alleles have normal adult function. Studies of mitotic clones (Kornberg, 1981a; Lawrence and Struhl, 1982) show that engrailed lethal alleles are unable to support normal development of adult pattern by themselves. Thus, the partial complementation between the nonrearrangement engrailed alleles and en1 suggests that these lethal alleles provide an activity that functions in collaboration with the en1 allele to promote more normal development of adult structures.
The nonlethal rearrangement mutations provide a second indicator of complexity of the engrailed locus. The existence of viable engrailed mutations that give allele-specific phenotypes suggests that some chromosome rearrangement mutations alter regulation rather than inactivate the encoded function. If so, the physical mapping of these mutations to sites 40 kb from the transcription unit raises the interesting possibility that regions far distant from the transcription unit are involved in the regulation that defines the normal pattern of engrailed expression. Similarly, it has been proposed that the regions upstream of Ubx transcript act in cis to regulate the Ubx unit (Ingham, 1984; Beachy et al., 1985).
The en1 Mutation Is Associated with an Insertion Element
Only chromosomes carrying the en1 mutation contained a detectable insertion in the engrailed region. Although the parental chromosome from which the spontaneous en1 mutation was isolated (Eker, 1929) is not available as a control, we believe that this 7 kb insertion is responsible for the mutant phenotype. Among all the chromosomes we analyzed, only the en1 chromosome contains an insertion within the cloned region and thus it seems unlikely that it is a polymorphism coincidentally associated with the mutation. Furthermore, this conclusion is consistent with earlier demonstrations that spontaneous mutations at bithorax (Bender et al., 1983a), white (Rubin, 1983), Notch (Artavanis-Tsakonas et al., 1983; Kidd et al., 1983), scute (Carramolino et al., 1982), and Antennapedia (Scott et al., 1983; Garber et al., 1983) loci are generally associated with an insertion event.
Sequences Governing Complex Developmental Programs of Expression
The function of the engrailed locus in the production of embryonic pattern may rely on the spatial control of its expression (Kornberg et al., 1985). Much of what is fundamental to the establishment of pattern might then lie in the sequences that control engrailed expression. The mapping of engrailed mutations suggests that extensive flanking sequences are involved in the spatial and temporal regulation of expression. Two other loci having complex spatial patterns of activity, the Bithorax complex and scute, have rearrangement alleles resembling those of engrailed: the positions of these rearrangement alleles are dispersed over a large region of the genome; these alleles do not have null phenotypes; and, they give rise to allele-specific spatially restricted defects (Lewis, 1978; Campuzano et al., 1985). Perhaps this represents a general feature of spatial and temporal control and genes exhibiting such complex patterns of regulation will frequently be associated with an extended regulatory region.
Experimental Procedures
Fly Strains and Culture
All crosses were carried out in standard culture medium at 25°C. engrailed mutant strains were isolated as alleles of en1, enLA4, or Df(2R)enSF31 after X-ray or EMS mutagenesis. enLA4, Df(2R)enSF31, enC2 (in [2R] 478,48A), enSF24 (T[2;3] 48A;90C), enSF32 (T[Y;2]48A), and enSF37 (T[2;3] 46C;48A;80) are lethal engrailed alleles (Kornberg, 1981a), as are enSF42 (T[2;3] 48A; 65F), enSF49 (in [2R] 47F;48A), enSF50 (T[2;3] 48A;57A;81A), enSF52 (T[2;3] 48A; 57B;88F), and enSF61 (T[2;3] 48A;89A) (All and Kornberg, unpublished). Nonlethal alleles are enLA3 (T[2;3] 48A;96C; Kornberg, 1981), en30 (Df[2R] 48A 3–4;48C 6–8; Eberlein and Russell, 1983), enSF82 (T[2;3] 48A;84D; All and Kornberg, unpublished), and enES (T[2;3] 48A;84D; Lindsley et al., 1972). Descriptions of all other strains can be found in Lindsley and Grell (1968).
Recombinant DNA Libraries
An amplified library of Charon 4A clones carrying inserts from wild-type (Canton S) Drosophila melanogaster (Maniatis et al., 1978) was obtained from D. Hogness and W. Bender. A cosmid library constructed by E. Meyerowitz (1980) was obtained from D. Hogness and S. Artavanis-Tsakonis.
Strategy for the Chromosome Walk
Once a rough restriction map for a particular phage was determined, a restriction fragment near the most advanced end of the insert was chosen as the primary probe for the next step. In addition, two fragments, one slightly behind the most advanced, and another behind that, were used as auxiliary probes. 32P-labeled DNA fragments were prepared either by nick translation (Rigby et al., 1977) with DNA polymerase I or by the chewback-fill-in procedure with T4 DNA polymerase (O’Farrell, 1981; O’Farrell et al., 1980). Screening phage libraries was as described in Maniatis et al. (1982). Probing three replicas of the same plate with the three probes identified plaques that were positive for the primary probe and negative for the auxiliary probes, yielding steps that extended farthest in the desired direction.
Restriction maps were determined for a few selected phages, and comparisons among them and with the previous step revealed those that had actually advanced the walk the farthest.
The same plates and replicas could be reused for several successive steps. Bound labeled probe was removed by washing the replicas for 1 hr at 70°C in a prehybridization mixture before re-use. Comparisons could then be made with plaques that were positive in the previous step to help guide the selection of plaques.
Clones were also isolated from a cosmid library made by Meyerowitz (1980) or one made with the pJB8 vector (M. Nakanishi and P. O’Farrell, unpublished). Although the cosmid blanks provided some helpful large steps, they proved to be difficult and inefficient to use because of the instability of the cloned fragments. Phage clones were therefore principally used for the chromosome walk, and only those phage clones with minimal neighbor overlap are described here. Preparation of phage stocks and isolation phage DNA was as in Maniatis et al. (1982).
Purification of Drosophila DNA
Two procedures were used. With the first, 1 g of adult flies was homogenized on ice with a teflon homogenizer in 30 ml of buffer H (0.32 M sucrose, 100 mM Tris, pH 7.8, 50 mM NaCl, 5 mM CaCl2, 1% Triton X 100). Debris was removed by filtering through four layers of cheesecloth and a Nitex screen mesh. Nuclei in the filtrate were pelleted at 2000 × g for 5 min and resuspended in 5 ml of the buffer H. To a 15 ml corex tube, 5 ml of buffer F (10% sucrose, 0.75 M NaCl, 3.3 mM EDTA, 5 mM Tris, pH 8.1, 0.2% Titron X 100) was added and the nuclear suspension was layered on top. The nuclei were pelleted through the buffer F layer in a swinging bucket rotor at 16,000 × g for 6 min; this step removes nucleases, RNA, and mitochondria. The pellet was resuspended in buffer P (50 mM Tris, pH 8, 10 mM EDTA). To this, 3 ml of buffer P containing 2 mg of proteinase K was added (the proteinase K solution had previously been autodigested for 15 min at 37°C to reduce nuclease contaminants). Then 0.5 ml of 10% SDS was added and mixed on ice, followed by incubation at 37°C for 2 hr. Debris was removed by centrifugation at 16,000 × g for 5 min. To the supernatant 1 ml of 6 M NaClO4 was added and mixed, followed by 3 ml of CIA (CHCl3 [24 parts]: isoamyl alcohol [1 part]). Then 3 ml of phenol was added and gently mixed for 10 min. After centrifugation, the aqueous phase was collected and extracted twice with CIA. DNA was precipitated with ethanol, spooled, washed in 70% ethanol, and dissolved in TE. The yield was approximately 200 μg per gram of flies.
The second protocol was that of R. Lifton (personal communication). Two hundred adult flies were homogenized in 0.125 M Tris-HCl (pH 8.5), 0.08 M NaCl, 0.06 M EDTA, and 0.16 M sucrose, 0.5% SDS, and incubated for 30 min at 65°C. With the addition of potassium acetate to 1 M, the mixture was chilled to 0°C for 1 hr. The supernatant from a 5 rain centrifugation at 10 K was phenol extracted, ethanol precipitated, and resuspended in TE (0.01 M Tris, pH 8, 0.001 M EDTA). These preparations were used for Southern blot analysis and for construction of genomic libraries.
Lambda Libraries from Mutant Flies
Two methods were employed. The λ vector 1059 was used to clone Sau 3a partial digests as described by Karn et al. (1980). The extent of digestion was monitored by electrophoretic separation in agarose and the appropriate size fraction (15–20 kb) obtained by centrifugation of 100 μg of DNA through a gradient of 5%–20% NaCl in TE (5 hr at 35,000 RPM in a SW40 Beckman rotor at 20°C).
In the second method, the Charon 34 vector was digested with either Eco RI or Bam HI restriction enzyme and, after annealing of the cohesive ends, the arms were purified through agarose, electroeluted, extracted with phenol–chloroform, and precipitated with ethanol. Fly DNA was digested to completion, ligated to the purified λ arms, and packaged in vitro (Maniatis et al., 1982). Libraries were plated on C600 for screening or for amplification. Phage carrying insert sequences were purified and amplified, and their DNA was extracted and subjected to digestion with a restriction endonuclease to distinguish between phage with inserts of wild-type or mutant origin. Mutant restriction fragments were subcloned into the plasmid pUC8 (Vieira and Messing, 1982) or pEMBL8 (Dente et al., 1983), mapped for sites of restriction enzyme cleavage, and nick translated for use in genomic Southern blots and in situ hybridization.
Acknowledgments
We thank Robert Elder and Olke Uhlenbeck for the tRNA met2, Joyce Lauer, Welcome Bender, and Spyros Artavanis-Tsakonas for genomic libraries, our colleagues for their support, and Judy Kassis, Steve DiNardo, Elizabeth Sher, and Claude Desplan for their comments on the manuscript. This work was supported by National Science Foundation (P. H. O’F,) and National Institutes of Health (T. K.) grants, American Cancer Society and Giannini fellowships (J. M. K.), Weingart Foundation scholarship (L M. K.), and predoctoral training grants (J. T., E. G., and B. D.). We also thank Douglass Forbes, Louise Liao, Eliane Mohier, and Crawford Harris for their helpful contributions at early stages of this project.
References
- Akam ME. The location of Ultrabithorax transcripts in Drosophila tissue sections. EMBO J. 1983;2:2075–2084. doi: 10.1002/j.1460-2075.1983.tb01703.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Artavanis-Tsakonas S, Muskavitch MAT, Yedvobnick B. Molecular cloning of Notch, a locus affecting neurogenesis in Drosophila melanogaster. Proc Natl Acad Sci USA. 1983;80:1977–1981. doi: 10.1073/pnas.80.7.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beachy PA, Helfand SL, Hogness DS. Segmental distribution of bithorax complex proteins during Drosophila development. Nature. 1985;313:545–551. doi: 10.1038/313545a0. [DOI] [PubMed] [Google Scholar]
- Bender W, Akam M, Karch F, Beachy PA, Peifer M, Spierer P, Lewis EB, Hogness DS. Molecular genetics of the bithorax complex in Drosophila melanogaster. Science. 1983a;221:23–29. doi: 10.1126/science.221.4605.23. [DOI] [PubMed] [Google Scholar]
- Bender W, Spierer P, Hogness DS. Chromosomal walking and jumping to isolate DNA from the Ace and rose loci and the bithorax complex in Drosophila melanogaster. J Mol Biol. 1983b;168:17–33. doi: 10.1016/s0022-2836(83)80320-9. [DOI] [PubMed] [Google Scholar]
- Campuzano S, Carramolino L, Cabrera CV, Ruiz-Gomez M, Villares R, Boronat A, Modolell J. Molecular genetics of the achaete-scute gene complex of D. melanogaster. Cell . 1985;40:327–338. doi: 10.1016/0092-8674(85)90147-3. [DOI] [PubMed] [Google Scholar]
- Carramolino L, Ruiz-Gomez M, Carmen-Guerrero M, Campuzano S, Modolell J. DNA map of mutations at the scute locus of Drosophila melanogaster. EMBO J. 1982;1:1185–1191. doi: 10.1002/j.1460-2075.1982.tb00011.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dente L, Cesareni G, Cortese R. pEMBL: a new family of single stranded plasmids. Nucl Acids Res. 1983;11:1645–1655. doi: 10.1093/nar/11.6.1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eberlein S, Russell M. Effects of deficiencies in the engrailed region of Drosophila melanogaster. Dev Biol. 1983;100:227–237. doi: 10.1016/0012-1606(83)90215-4. [DOI] [PubMed] [Google Scholar]
- Eker R. The recessive mutant engrailed in Drosophila melanogaster. Hereditas. 1929;12:217–222. [Google Scholar]
- Elder R, Szabo P, Uhlenbeck OC. In: Transfer RNA: Biological Aspects. Soll D, Abelson J, Schunmel P, editors. Cold Spring Harbor, New York: Cold Spring Harbor Laboratory; 1980. [Google Scholar]
- Epper F, Sanchez L. Effects of engrailed in the genital disc of Drosophila melanogaster. Dev Biol. 1983;100:387–398. doi: 10.1016/0012-1606(83)90233-6. [DOI] [PubMed] [Google Scholar]
- Garber RL, Kuroiwa A, Gehring WJ. Genomic and cDNA clones of the homeotic locus Antennapedia in Drosophila. EMBO J. 1983;2:2027–2036. doi: 10.1002/j.1460-2075.1983.tb01696.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Bellido A. Genetic control of wing disc development in Drosophila. Cell Patterning, CIBA Foundation Symp. 1975;29:161–182. doi: 10.1002/9780470720110.ch8. [DOI] [PubMed] [Google Scholar]
- Garcia-Bellido A, Santamaria P. Developmental analysis of the wing disc in the mutant engrailed of Drosophila melanogaster. Genetics. 1972;72:87–104. doi: 10.1093/genetics/72.1.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Bellido A, Ripoll P, Morata G. Developmental compartmentalization of the wing disc of Drosophila. Nature New Biol. 1973;245:251–253. doi: 10.1038/newbio245251a0. [DOI] [PubMed] [Google Scholar]
- Hafen E, Kuroiwa A, Gehring WJ. Spatial distribution of transcripts from the segmentation gene fushi tarazu during Drosophila embryonic development. Cell. 1984;37:833–841. doi: 10.1016/0092-8674(84)90418-5. [DOI] [PubMed] [Google Scholar]
- Ingham PW. A gene that regulates the bithorax complex differentially in larval and adult cells of Drosophila. Cell. 1984;37:815–823. doi: 10.1016/0092-8674(84)90416-1. [DOI] [PubMed] [Google Scholar]
- Jurgens G, Wieschaus E, Nusslein-Volhard C, Kluding M. Mutations affecting the pattern of the larval cuticle in Drosophila melanogaster. II Zygotic loci on the third chromosome Wilhelm Roux’s. Arch Dev Biol. 1984;193:283–295. doi: 10.1007/BF00848157. [DOI] [PubMed] [Google Scholar]
- Karn J, Brenner S, Barnett L, Cesarini G. Novel bacteriophage λ cloning vector. Proc Natl Acad Sci USA. 1980;77:5172–5176. doi: 10.1073/pnas.77.9.5172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufman TC, Lewis R, Wakimoto B. Cytogenetic analysis of chromosome 3 in Drosophila melanogaster: the homeotic gene complex in polytene chromosome interval 84A–B. Genetics . 1980;94:115–133. doi: 10.1093/genetics/94.1.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kidd S, Lockett TJ, Young MW. The Notch locus of Drosophila melanogaster. Cell. 1983;34:421–433. doi: 10.1016/0092-8674(83)90376-8. [DOI] [PubMed] [Google Scholar]
- Kornberg T. engrailed: a gene controling compartment and segment formation in Drosophila. Proc Natl Acad Sci USA. 1981a;78:1095–1099. doi: 10.1073/pnas.78.2.1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornberg T. Compartments in the abdomen of Drosophila and the role of the engrailed locus. Dev Biol. 1981b;86:363–372. doi: 10.1016/0012-1606(81)90194-9. [DOI] [PubMed] [Google Scholar]
- Kornberg T, Siden I, O’Farrell P, Simon M. The engrailed locus of Drosophila: in situ localization of transcripts reveals compartment-specific expression. Cell. 1985;40:45–53. doi: 10.1016/0092-8674(85)90307-1. [DOI] [PubMed] [Google Scholar]
- Lawrence PA, Morata G. Compartments in the wing of Drosophila: a study of the engrailed gene. Dev Biol. 1976;50:321–337. doi: 10.1016/0012-1606(76)90155-x. [DOI] [PubMed] [Google Scholar]
- Lawrence PA, Morata G. The elements of the bithorax complex. Cell. 1983;35:595–601. doi: 10.1016/0092-8674(83)90091-0. [DOI] [PubMed] [Google Scholar]
- Lawrence PA, Struhl G. Further studies on the engrailed phenotype in Drosophila. EMBO J. 1982;1:827–833. doi: 10.1002/j.1460-2075.1982.tb01255.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine M, Hafen E, Garber RL, Gehring WJ. Spatial distribution of Anatennapedia transcripts during Drosophila development. EMBO J. 1983;2:2037–2046. doi: 10.1002/j.1460-2075.1983.tb01697.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis EB. A gene complex controlling segmentation in Drosophila. Nature. 1978;276:565–570. doi: 10.1038/276565a0. [DOI] [PubMed] [Google Scholar]
- Lindsley D, Grell E. Genetic Variations of Drosophila melanogaster. Carnegie Inst Wash. 1968 Publ. No. 627. [Google Scholar]
- Lindsley D, Sandler L, Baker B, Carpenter A, Denell R, Hall J, Jacobs P, Miklos G, Davis B, Getthman R, Hardy R, Hessler A, Miller S, Nozawa H, Parry D, Gould-Somero M. Segmental aneuploidy and the genetic gross structure of the Drosophila genome. Genetics. 1972;71:157–184. doi: 10.1093/genetics/71.1.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maniatis T, Hardison RC, Lacy E, Lauer J, O’Connell C, Quon D, Sim GK, Efstradiadis A. The isolation of structural genes from libraries of eucaryotic DNA. Cell. 1978;15:687–701. doi: 10.1016/0092-8674(78)90036-3. [DOI] [PubMed] [Google Scholar]
- Maniatis T, Fritsch EF, Sambrook J. Molecular Cloning. Cold Spring Harbor, New York: Cold Spring Harbor Laboratory; 1982. [Google Scholar]
- McGinnis W, Levine MS, Hafen E, Kuroiwa A, Gehring WJ. A conserved DNA sequence in homeotic genes of the Drosophila Antennapedia and bithorax complexes. Nature. 1984;308:428–433. doi: 10.1038/308428a0. [DOI] [PubMed] [Google Scholar]
- Meyerowitz E, Guild G, Prestidge L, Hogness D. A new high-capacity cosmid vector and its use. Gene. 1980;11:271–282. doi: 10.1016/0378-1119(80)90067-0. [DOI] [PubMed] [Google Scholar]
- Morata G, Lawrence P. Development of the eye-antennal imaginal disc of Drosophila. Dev Biol. 1979;70:355–371. doi: 10.1016/0012-1606(79)90033-2. [DOI] [PubMed] [Google Scholar]
- Morata G, Ripoll P. Minutes: mutants of Drosophila autonomously affecting cell division rate. Dev Biol. 1975;42:211–221. doi: 10.1016/0012-1606(75)90330-9. [DOI] [PubMed] [Google Scholar]
- Nusslein-Volhard C, Wieschaus E. Mutations affecting segment number and polarity in Drosophila. Nature. 1980;287:795–801. doi: 10.1038/287795a0. [DOI] [PubMed] [Google Scholar]
- Nusslein-Volhard C, Weischaus E, Kluding H. Mutations affecting the pattern of the larval cuticle in Drosophila melanogaster. I Zygotic loci on the second chromosome Wilhelm Roux’s. Arch Dev Biol. 1984;193:267–282. doi: 10.1007/BF00848156. [DOI] [PubMed] [Google Scholar]
- O’Farrell PH. Replacement synthesis methods of labeling DNA fragments. Bathesda Research Lab Focus. 1981;3:1–2. [Google Scholar]
- O’Farrell PH, Kutter E, Nakanishi M. A restriction map of the bacteriophage T4 genome. Mol Gen Genet. 1980;179:421–435. doi: 10.1007/BF00425473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pardue ML, Gall JG. Nucleic acid hybridization to the DNA of cytological preparations. Methods Cell Biol. 1975;10:1. doi: 10.1016/s0091-679x(08)60727-x. [DOI] [PubMed] [Google Scholar]
- Poole SJ, Kauvar LM, Drees B, Kornberg T. The engrailed locus of Drosophila: structural analysis of an embryonic transcript. Cell. 1985;40:37–43. doi: 10.1016/0092-8674(85)90306-x. [DOI] [PubMed] [Google Scholar]
- Rigby P, Dieckmanen M, Rhodes C, Berg P. Labeling DNA to high specific activity in vitro by nick translation and DNA polymerase I. J Mol Biol. 1977;113:237–245. doi: 10.1016/0022-2836(77)90052-3. [DOI] [PubMed] [Google Scholar]
- Rubin GM. Dispersed repetitive DNA’s in Drosophila. In: Shapiro JA, editor. Mobile Genetic Elements. New York: Academic Press; 1983. [Google Scholar]
- Russell M, Eberlein S. New mutants of engrailed in Drosophila melanogaster. Genetics. 1979;591:109. [Google Scholar]
- Scott MP, Weiner AJ. Structural relationships among genes that control development: sequence homology between the Antennapedia, Ultrabithorax, and fushi tarazu loci of Drosophila. Proc Natl Acad Sci USA. 1984;81:4115–4119. doi: 10.1073/pnas.81.13.4115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott MP, Weiner AJ, Hazelrigg TI, Polisky BA, Pirrotta V, Scalenghe F, Kaufman TC. The molecular organization of the Antennapedia locus of Drosophila. Cell. 1983;35:763–776. doi: 10.1016/0092-8674(83)90109-5. [DOI] [PubMed] [Google Scholar]
- Simcox AA, Sang JH. When does determination occur in Drosophila embryos? Dev Biol. 1983;97:212–221. doi: 10.1016/0012-1606(83)90078-7. [DOI] [PubMed] [Google Scholar]
- Struhl G. Anterior and posterior compartments in the proboseis of Drosophila. Dev Biol. 1981;84:372–385. doi: 10.1016/0012-1606(81)90406-1. [DOI] [PubMed] [Google Scholar]
- Vieria J, Messing J. The pUC plasmids, an M13 mp7-derived system for insertion mutagen and sequencing with synthesis universal primers. Gene. 1982;19:259–268. doi: 10.1016/0378-1119(82)90015-4. [DOI] [PubMed] [Google Scholar]
- Wieschaus E, Gehring W. Clonal analysis of primordal disc cells in the early embryo of Drosophila. Dev Biol. 1976;50:249–265. doi: 10.1016/0012-1606(76)90150-0. [DOI] [PubMed] [Google Scholar]
- Wieschaus E, Nusslein-Volhard C, Jurgens G. Mutations affecting the pattern of the larval cuticle in Drosophila melanogaster. III Zygotic loci on the X-chromosome and fourth chromosome Wilhelm Roux’s. Arch Dev Biol. 1984;193:296–307. doi: 10.1007/BF00848158. [DOI] [PubMed] [Google Scholar]