Abstract
A regulatory cascade, initiated during the syncytial stage of embryogenesis, culminates in the striped pattern of engrailed gene expression at the cellular blastoderm stage. The early regulatory genes, for example the pair-rule genes, are expressed transiently and as their products decay a distinct regulatory programme involving segment polarity genes takes over. This late programme maintains and perhaps modifies the striped pattern of engrailed expression through interactions that may involve cell communication.
An early step in the generation of the Drosophila body pattern is the determination of the segmental subdivisions of the embryo. This occurs quite rapidly. By the cellular blastoderm stage (2–2.5 h after fertilization) the segmental outlines of the embryo, although not morphologically visible, are represented by the precise expression patterns of the segmentation (or segment polarity) genes, such as engrailed1,2 and wingless3. The subsequent activity of genes such as these directs the morphological events leading to segmentation4,5.
The segmentation genes are expressed in their localized patterns as a consequence of an early regulatory cascade. Three gene classes (maternal genes6, and the zygotically acting gap and pair-rule genes4) act in sequence, each establishing the more refined spatial expression pattern of the next7–9. This takes place in a syncytial cytoplasm and is thought to rely on the diffusion and interaction of regulatory products encoded by these genes. The subdivision of the embryonic field by this cascade is rapid, occurring during the first thirteen nuclear divisions. After the thirteenth division, the nuclei are cellularized and the cellular blastoderm is formed.
One outcome of this regulatory hierarchy is the establishment of engrailed (en) expression at the cellular blastoderm stage in 14 single-cell-wide stripes transecting the antero-posterior axis of the embryo1,2,10,22. Previous work has shown that the activity of separate sets of pair-rule genes controls establishment of en expression in even- and in odd-numbered stripes11,12. The data presented here suggests further that the separate control of even-and odd-numbered stripes is reflected in the organization of the cis-regulatory sequences at en.
During the post-cellular blastoderm phase of development the expression pattern of en is maintained and modified1,10. It is unlikely that the pair-rule genes are involved in the post-blastoderm regulation of en, since the pair-rule genes characterized to date are only expressed transiently in their pair-rule blastoderm patterns13–16,31. Here we address the mechanism by which the en gene is regulated in later embryonic development.
We find that the striped expression pattern of en during post-blastoderm development is controlled by a cis-regulatory programme distinct from that controlling the establishment of expression at the cellular blastoderm stage. Control of this late-acting programme is exercised through the activity of various segment polarity genes. The activity of at least one of the segment polarity genes (wingless) is not cell autonomous1744. This leads us to a model for the control of post-blastoderm en gene expression involving cell interactions. We suggest that the late en regulatory programme allows for some plasticity in developmental programming.
An en-lacZ fusion gene
As part of an ongoing dissection of the en regulatory region we have constructed transgenic fly lines expressing β-galactosidase under the control of portions of the en regulatory region. Our dissection of the cis-regulatory sequences at en is in its initial stages, but the expression pattern of one transgenic construct is particularly instructive. It accurately expresses discrete portions of the en pattern and represents one line of evidence that there are distinct early and late regulatory programmes that control the expression of en in stripes.
Figure 1 shows the DNA construct, P(en/lac), in which expression of a β-galactosidase (β-gal) - engrailed fusion protein is directed by the en promoter and flanking sequences. The fusion gene is in a vector allowing the establishment of stably transformed ‘transgenic’ flies through P-element mediated integration into the germ line18. In addition to the promoter and 5′ flanking sequences, the construct includes 1099 base pairs (bp) of transcribed sequences from the en gene19, encompassing ~211 nucleotides of untranslated leader and the N-terminal 296 amino acids20 spliced in frame to the lacZ gene encoding β-galactosidase (β-gal) activity. Therefore, the expression of this construct could reflect control both at the transcriptional and post-transcriptional levels. The signal (β-gal activity or antigen) produced by P(en/lac) is unstable, thereby allowing us to detect transitions that occur in en expression patterns. The instability could be due to the inclusion of en leader and coding sequences in the fusion, as a relatively stable β-gal signal is produced by a ftz promoter-lacZ fusion, which does not contain coding sequences from the ftz gene21.
Fig. 1.
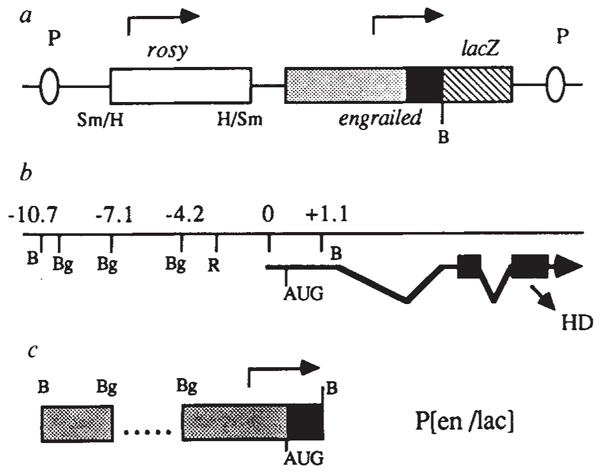
Construction of an en-lacZ fusion gene (not to scale), a, schematic diagram of P(en/lac) indicating sites for P-mediated integration (P); the rosy gene (open box); upstream and non-coding (stippled) and coding (filled) regions from engrailed; and lacZ coding sequences (hatched). An SV40 polyadenylation signal is at the end of lacZ. b, Map of en genomic region showing particular restriction site landmarks, the major transcription unit with start site (0) and presumed translation start site (AUG) and the portions of exons two and three (solid boxes) encoding the homoeodomain (HD)19,20,40. c, Specific genomic en regions used in constructing P(en/lac) (aligned with b). Dots represent the deletion of sequences between coordinates −4.2 and −7.1. Restriction sites: B, BamHI; Bg, BglII; H, HindIII; R, EcoRI; Sm, SmaI; symbol (/), joined by blunt-end ligation. Arrows indicate the direction of transcription.
Methods. Transgenic lines were established18 using ‘wings-clipped’ helper, and an Sgs–4Ber–-1, cn; ry506 host. Lines were established from three independent G0 adults; the insertion mapped to chromosome II in one line and to chromosome III in the other two.
Early expression
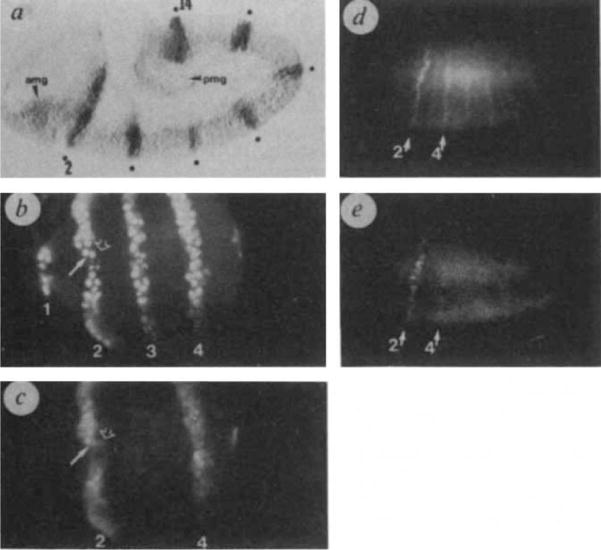
Three independent P(en/lac) lines have been analysed, and all exhibit a similar pattern of ectodermal expression. Seven stripes of expression from P(en/lac) are apparent along the extending germband (Fig. 2a), a stage at which en is expressed in fourteen stripes. The seven stripes expressing P(en/lac) precisely align with the even-numbered en stripes (Fig. 2b, c). Within these stripes P(en/lac) is faithfully expressed since all cells expressing en are also expressing β-gal. On occasion, an extra β-gal-expressing cell is observed ventrally at the trailing edge of even-numbered en stripes (Fig. 2b, c). The significance of this occasional non-correspondence is unclear.
Fig. 2.
Antibody-staining of P(en/lac) transgenic embryos show that regulatory sequences controlling establishment of even-numbered en stripes are functionally distinct from those controlling establishment of odd-numbered stripes. Anterior left and ventral down in all panels, unless indicated otherwise, a, P(en/lac) transformant stained with rabbit anti-β-galactosidase antibody to visualize the expression from P(en/lac), 3.5–4h after egg laying (AEL). Seven ectodermal stripes are evident (indicated by dots), corresponding to en stripes 2, 4, 6, 8, 10, 12 and 14, which mark the posterior portions of the maxillary, first and third thoracic, and second, fourth, sixth and eighth abdominal primordia. Variable expression seen in the anterior (amg) and posterior (pmg) midgut primordia is not part of the normal en expression program. This expression may be due to sequences from en, or may be spurious, since expression from similar vectors in mesodermal and gut primordia has been reported41. b, c: Magnified (ventral) view of a doubly-labelled embryo (mouse anti -en and rabbit anti-β-gal) reveals expression of endogenous en (b) and of P(en/lac) (c), 4.5 h AEL. The stripes of en expression are 1(Mn), 2(Max), 3(La) and 4(T1). Expression of β-gal is coincident with even-numbered en stripes 2(Max) and 4(T1); arrows indicate the same cells in b and c. There is no detectable β-gal expression at positions corresponding to odd-numbered en stripes. In even-numbered stripes there is virtually one-to-one correspondence between β-gal and en expressing cells. The occasional cell containing low levels of β-gal antigen but no detectable en is indicated (open arrow). The β-gal signal is not as discrete as the en signal since the en-β-gal fusion protein is not restricted to the nucleus, d, e: Embryos were doubly-labelled for en expression (d) and β-gal expression (e) at the onset of gastrulation, 3 h AEL. Expression from P(en/lac) is induced at about the same developmental stage as en, although there may be a slight lag in induction or accumulation. Arrows point to co-expression in 2(Max), and in 4(T1), where β-gal is just detectable.
Methods. Preparation of embryos for immunocytochemistry was as in refs 10, 12. Rabbit anti-β-gal was from Cappel, rabbit anti-en, (ref. 10); mouse monoclonal anti-en (and inv), a gift of K. Coleman, C. Goodman and T. Kornberg. Final magnification was ×130, except for b and c which were ×520.
The establishment of P(en/lac) expression at even-numbered en stripes is first evident at the same stage as endogenous en expression—the cellular blastoderm/early gastrula (Fig. 2d, e). As endogenous en expression is established, the odd-numbered stripes initially lag behind the development of even-numbered stripes10,22. However, en is equivalently expressed in the odd- and even-numbered stripes by the time the germband is elongating (compare panels d and b, Fig. 2). In contrast, during germband elongation or even after the germband fully extends, no β-gal expression (enzyme or antigenic activity) is detected in positions corresponding to the odd-numbered en stripes (Fig. 2a).
To test whether the establishment of P(en/lac) expression is under the control of the same regulatory factors that control the establishment of even-numbered en stripes, we have analysed the expression of P(en/lac) in particular pair-rule mutants. Two pair-rule mutants known to affect the expression of even-numbered en stripes, fushi tarazu11,12 and odd-skipped12, have identical effects on the establishment of P(en/lac) expression and on endogenous en expression (data not shown).
We conclude that P(en/lac) expression is regulated faithfully at positions corresponding to even-numbered en stripes, and that this regulation involves the same trans-acting activities as the endogenous en gene. Therefore, the regulatory region present in P(en/lac) includes cis-acting sequences that direct the establishment of even-numbered en stripes. Presumably, the sequences required for expression in odd-numbered stripes are not present, or are functionally disrupted in this construct.
Late expression
As development continues, there is a striking transition in the expression of P(en/lac). The changes in expression provide evidence that the expression of en stripes is separable into two temporally and mechanistically distinct programmes.
After the germband is fully extended, the expression of P(en/lac) in stripes corresponding to even-numbered en stripes begins to fade. (Note the spotty appearance of the stripes in Fig. 3a where the embryo is about 1.5h older than the stage exhibited in Fig. 2a.) At this late stage, individual cells located dorso-laterally at positions corresponding to odd-numbered en stripes begin to accumulate β-gal antigen (Fig. 3a). Over the next hour or two, β-gal staining is lost ventrally from even-numbered stripes, but intense expression is induced dorso-laterally at positions corresponding to both even- and odd-numbered en stripes (in the dorsal ridge, T1 through A7, and to some extent A8; Fig. 3b). The eleven dorso-lateral stripes expressed from P(en/lac) at this late developmental stage correspond cell-for-cell with en expressing cells (Fig. 3c, d).
Fig. 3.
P(en/lac) uncovers a distinct late program of control over en, which is regulated by segment polarity genes, a, staining for β-gal, 5 h AEL, anterior left and ventral down (×130). The seven β-gal stripes (dots) are beginning to fade. Note the spotty appearance laterally (open arrows). As development proceeds this signal fades ventrally. In addition, note that some cells at dorso-lateral positions are beginning to accumulate β-gal signal (solid arrows). The signal in the dorsal ectoderm will be heavily induced during subsequent stages. Though restricted to dorsal regions, the position of this signal corresponds accurately to even-and odd-numbered en stripes, b, Staining for β-gal at the onset of germband retraction, 7–8 h AEL, ventro-lateral view (×130). Expression of P(en/lac) in ventral areas is low, but expression at dorso-lateral positions is quite high. Expression in both ventral and dorsal positions occurs in the posterior portions of 11 segment primordia: Labial, T1–T3, and A1–A7. Significant expression of P(en/lac) in Mandibular and Maxillary primordia and the terminalia during this late programme is usually not observed. Expression in dorsal-most A8 is sometimes observed. Bracketed area is magnified (from a different embryo) in c and d. VM, ventral midline. c, d: Magnified view (×520) of a doubly-labelled embryo at the turn of the germband (A2, A3 and A4 region) showing one to one correspondence between en (c) and β-gal (d) expressing cells, e, f: Same as c, d but in ptc mutant background, with arrow showing that P(en/lac) is ectopically induced (f) as is the endogenous en gene (e).
As the germband retracts the P(en/lac) signal appears to extend ventrally, although it usually does not completely fill the en-expressing region. Often, the level of P(en/lac) expression on the ventral surface is higher in cells at the posterior edge of the en stripes, and lower in other cells of the en stripe (data not shown). In contrast, dorso-lateral expression of P(en/lac) remains uniformly coextensive with en-expressing cells. We conclude that the P(en/lac) construct contains at least some of the regulatory sequences necessary for position specific expression in ventro-lateral regions, whereas it contains all sequences necessary for dorso-lateral expression. The discrimination between dorso-lateral and ventro-lateral regions in the embryo is not unique to this construct. Recent data have shown that there is also a sharp ventral/dorsal distinction in neurogenic potential and in the mitotic division schedule23 (V. Foe, personal communication).
At this later developmental stage, the expression of P(en/lac) in positions corresponding to odd-numbered en stripes is in striking contrast to the absence of detectable β-gal expression in these positions at early stages. Therefore, P(en/lac) responds later in development to regulatory signals that control expression in both even- and odd-numbered positions. These data show that two programmes of striped en expression are distinguishable by character and time of onset. The first programme results in the initiation of stripes at the cellular blastoderm, and involves the separate regulation of even- and odd-numbered stripes by pair-rule gene action. The second programme becomes evident after the germband has elongated, as initial expression fades and de novo expression becomes visible at both even- and odd-numbered positions. Since late P(en/lac) expression is not present in all cells that express the endogenous en gene (for example, ventral P(en/lac) stripes do not fill the en domain), it is unlikely that P(en/lac) expression is being regulated simply by the presence of endogenous en protein. Such a positive autoregulatory loop may be one of the components of late regulation, but en must also be responding to other signals. We next address the nature of these other inputs by investigating the effects that particular segmentation genes have on the pattern of expression of the endogenous en gene.
Regulation of late programme
It is unlikely that the pair-rule genes directly regulate the late programme of en expression, since products encoded by these genes are no longer detectable at this stage13–16,24,25,31. Analysing the en expression pattern in particular segment polarity mutants provides evidence that these genes are involved in regulating the second programme.
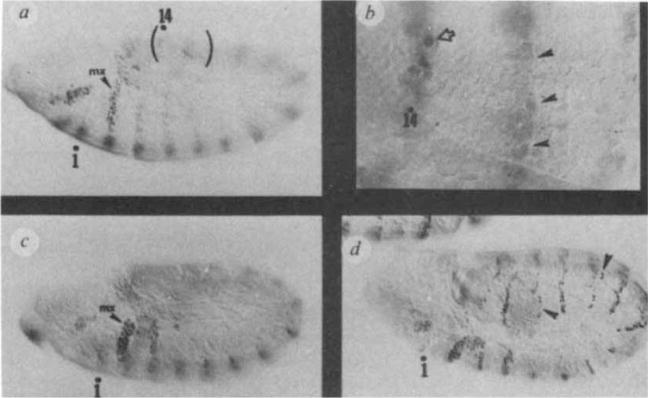
In wg null mutants en expression is established normally at the cellular blastoderm in fourteen roughly single-cell-wide stripes. This pattern remains wild-type through gastrulation and germband elongation, en stripes becoming 2–3 cells wide at this stage as a result of both cell movements and cell division. However, after the germband is fully extended, ectodermal en expression decays prematurely. Two developmental stages are shown in Fig. 4a, c, revealing the progress of the decay (summary, Fig. 6). The only ectodermal expression that remains is in the maxillary (Mx) stripe, a portion of the labial (La) stripe, and in a cluster of dorsally located cells in developing T1 (Fig. 4c). The effect of strong, but probably not null, alleles of hh, arm and fu are similar to the effects of the wg mutations, except that the en stripes do not disappear fully. Rather, a variable but small number of cells retain en expression. An example is shown for hh6N (Fig. 4d) (note the spotty and ‘thin’ appearance of en stripes). Weak alleles of wg, such as wgIL114ts grown at semi-permissive temperature, similarly leave some en-expressing cells unaffected (data not shown). This raises the possibility that the true null condition for hh, fu and arm (including removal of any maternal contribution), would result in the total loss of en expression as in wg nulls.
Fig. 4.
Premature decay of en expression in wg and hh mutants. Embryos, anterior left and ventral down, were stained with anti-en-antibody, a, Mutant wgcx4: 4–5 h AEL(×130). Most en stripes are faint, but still visible above background. The Max and La stripes are affected least. Similar results have been obtained in wgL5, wgIG22, and also in wgIL114ts when aged at non-permissive temperatures (25–28 °C). In wild-type embryos en antigen can be visualized in cells until the onset of cuticular differentiation, 12–14h AEL10. b, Mutant wgcx4: magnified (×520) view (in a different embryo) of bracketed region shown in (a) as signal from en expression decays. Particular en stripes lose signal at varying times; thus, these two adjacent en stripes clearly show that the decay in en expression occurs while cells are still part of the ectodermal cell sheet (arrowhead, decaying signal), c, Mutant wgcx4: 6.5 h AEL (×130). The remaining en signal is lost from ectodermal cells, except in Max and La regions. The blurred ventral signal is due to en expression in cells (out of this focal plane) of the developing central nervous system, d, hh6N: 6 h AEL (×130). Note the spotty and thin appearance of en stripes. Similar results are found for armxk22, fu113, fu113/fu94, hh6n/hh9k, and in wgIL114ts (raised at 16–18 °C). The segment polarity mutants gooseberry and cubitus interruptsD have not been studied.
Fig. 6.
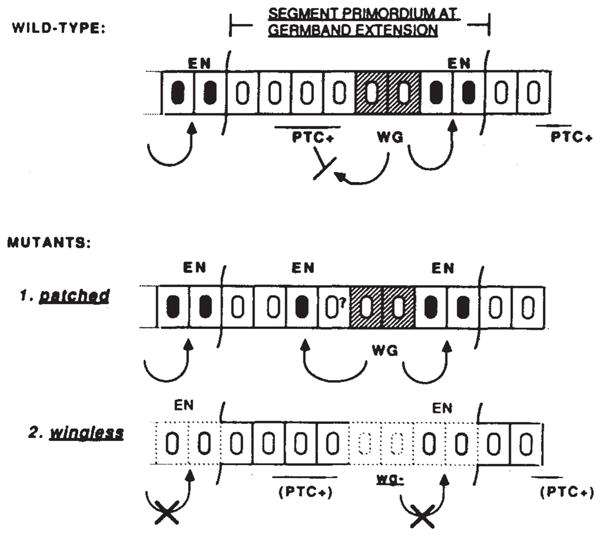
Interactions regulating en during post-blastoderm development. Wild-type: The segment primordium at germband extension stage (anterior to left) is represented as eight cells wide—the roughly four-cell wide primordia having expanded due to a post-cellular blastoderm cell division and some interdigitation of cells. Several domains within each primordium appear to exist, each ‘marked’ by the expression of, or by the requirement for the activity of, particular genes. Cells expressing en and wg account for two of the domains: the en domain comprises the last two cells of the primordium4–6 (blackened nuclei); the anteriorly adjacent two cells constitute the wg domain3 (hatching). The analysis of ptc mutations suggests that the remaining cells of the primordium are divided into at least two domains: those that ectopically induce en in the absence of ptc activity, and those that do not. The placement of the ptc-requiring domain is based on the position of ectopic en expression in ptc mutants. It has been proposed that the activity of another gene, naked, is required for the anterior-most cells of the primordium35. Each of the domains is presumably established at the cellular blastoderm stage by the action of pair-rule segmentation genes, as has been shown for en11,12 and wg domains43. The establishment of en expression constitutes the early regulatory programme of en. The second regulatory programme acts during post-blastoderm development as wg-expressing cells induce (or maintain) en expression in posteriorly located cells (arrow). Data presented here do not rule out other interactions. Mutants: (1) In the absence of ptc activity, en is induced in cells anterior to the wg domain. The induction requires wg (arrow to anterior), as no induction is detected in the absence of wg activity. Thus, in wild-type, wg induces en posterior to the wg domain, but is inhibited, directly or indirectly, from doing so anteriorly by ptc activity (blocking arrow shown in ‘wild-type’ panel). A cell (marked ‘?’) lies between the wg domain and the cell ectopically inducing en (see text). (2) the wg mutation leads to the premature loss of en expression (stippled nuclei), as a result of the loss of a morpho-genetic signal from wg expressing cells, perhaps the wg product itself. The hh, fu and/or arm products may play some role in the transduction of this signal. Cuticle pattern normally produced by en and wg-dependent cell types is missing3,4 either due to cell death or transformation of fate (dashed cell outlines). It is not known if the wg mutation affects cells located anteriorly to the wg domain.
Some cell death occurs in these mutants during germband retraction26,27. Thus, it is possible that cell death leads to the observed loss of en signal. However, in wg mutants, there is no evidence of cell death at the stage when cells are in the process of losing en antigen (Fig. 4b). This suggests that the loss of en expression is an early effect in wg mutants and not secondary to cell death.
The results indicate that the segment polarity loci wg, fu, hh and arm act in some manner to reinforce or maintain en expression once it has been established. The segment polarity mutant patched (ptc)4 has qualitatively different effects on en expression, providing further evidence for the existence of a distinct regulatory programme controlling en expression during post-blastoderm development.
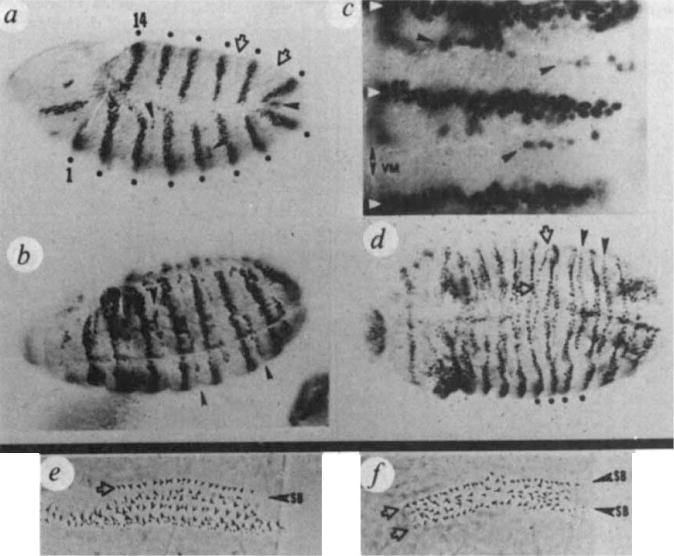
In patched mutants, en expression is established normally. However, at full germband extension in ptc mutants, the program of en expression is altered as en is induced in cells lying a few cell diameters posterior to each normal en stripe (Fig. 5a). The cells that are inducing en lie near and/or within an abnormal furrow that develops in the ectodermal cell sheet (Fig. 5a, b, c). As development proceeds more cells express en (Fig. 5b). By the onset of germband retraction the ectopic expression forms an incomplete transverse stripe positioned eccentrically between each pair of normal en stripes (Fig. 5d; summary, Fig. 6).
Fig. 5.
Reprogramming of cells to express en in patched mutants. Mutant embryos ptcIN108were stained with anti-en-antibody, a, Germband extended embryo (4.5 h AEL), anterior left and ventral down (×130). The normal 14 en stripes, (established at the cellular blastoderm stage) are now two to three cells wide and are indicated by dots. The patches and stripe of en-expressing cells anterior to stripe 1 resemble wild-type. Cells newly accumulating en antigen are indicated (arrowheads). The stage at which ectopic expression is first detectable cannot be due to leakiness of the ptc allele because the same delay is observed in ptcIN108 homozygotes as in the deficiency/ptcIN108 heteroallelic combination. A furrow in the ectodermal cell sheet is becoming visible between each pair of en stripes at the approximate position of ectopic induction (open arrows). The asterisk marks a cell group that induces en at this stage in wild-type also, b, Ventral view, 6 h AEL (×130). Eight en stripes are visible. More cells now express en between each pair of normal stripes, giving the appearance of an eccentrically positioned, incomplete stripe (arrowheads), c, Magnified (×520) ventro-lateral view (6h AEL) showing three normal en stripes (white arrowheads) and cells that have induced en, located near and often within the deep furrows, d, Germband retracted (9 h AEL) anterior left ventral view (×130). A few normal stripes (dots) and the intervening ectopic stripes are indicated (arrowheads). A given ectopic stripe has a wavy appearance, fusing along part of its length alternately with a posteriorly located en stripe, and then ventro-laterally with an anteriorly located stripe (open arrows). This fusion may be the result of shifts in cell position during germband retraction. Such fusions and reorganization of en stripes are frequently seen at late developmental stages in many of the segmentation mutants, e, Wild-type fourth abdominal denticle belt with border (SB) between third (upper) and fourth (lower) segment indicated. f, Fourth abdominal denticle belt in mutant ptc1N108/Df(2R)ptc showing duplication of segment border (SB), and anterior row of denticles (open arrows). In ptc6p43, enIM99 double mutants (raised at 18–20 °C) both normal and duplicate anterior row denticles do not appear, showing dependence on en function.
Methods. Stocks were obtained from C. Nusslein-Volhard and E. Wieschaus, from N. Baker (wgcx4), and J. Hooper and M. Scott (Df(2R)ptc44C–E). Crosses were performed in standard fashion, at 25 °C on molasses-cornmeal-agar. Embryos were prepared for immunocytochemical detection of en protein by affinity-purified rabbit anti-en antibody10,12 Cuticles were prepared as in ref. 42.
The ptc mutant phenotype suggests that the novel en stripes have a functional consequence on development. The ptc mutants develop duplicated cuticular structures within each segment4. Among these duplicated structures are an anterior row of denticle hairs and a segment border, which are characteristic of en-expressing cells28 (S. D., unpublished data; Fig. 5e,f). In a ptc, en double mutant, whereas other duplicated structures are still produced, those dependent on en activity are not produced. We conclude that the ectopic induction of en has a consequence on the final body pattern. Therefore, in ptc mutants particular cells that are initially specified at the cellular blastoderm stage to be negative for en expression are reprogrammed later in development.
wg and ectopic induction
The absence of the ptc product leads to a late programme of en expression in novel positions within a developing segment. Although pair-rule gene activity is unlikely to control late en expression in wild-type embryos, ectopic induction might in principle involve the same regulatory products that establish early en expression, such as ftz and eve, if their expression patterns are correspondingly altered in a ptc mutant. We consider this unlikely since we and others find no changes or increase in the lifetime of the ftz8 and eve29 patterns, nor evidence for renewed expression just before, or concommitant with, the induction of ectopic stripes. Furthermore, we can experimentally induce late expression of ftz in transgenic flies that express ftz from the inducible heat shock promoter30 and such late ftz expression has no consequence on the late en pattern. The observations suggest that these pair-rule gene products are neither present nor competent to induce ectopic en expression at the time that ectopic en stripes appear in ptc mutants.
In fact, we find that ectopic induction of en in ptc mutants is dependent on wg activity. Specifically, in wg, ptc double mutants, en expression is established normally, but decays prematurely, as it does in wg single mutants. There is no subsequent ectopic induction as seen in ptc single mutants (data not shown).
The ectopic induction of en in positions where there was no previously detectable en accumulation raises the possibility that wg activity has the capacity to induce en expression de novo. Therefore, what we have been referring to as ‘maintenance’ of en expression by wg, and other segment polarity genes, may be a re-induction of en expression. This observation complements the conclusion drawn from the analysis of P(en/lac) expression, which suggested that the en program has a distinct, late regulatory component that is independent of the establishment of early expression.
Late control of en-lacZ
Interestingly, the developmental stage at which segment polarity gene mutations affect en expression coincides with the transition between early and late en regulatory programmes as determined from the analysis of P(en/lac) expression (see above). The temporal link implies that the late programme of induction from P(en/lac) is due to the action of segment polarity genes. This has been tested by examining P(en/lac) expression in wg and ptc mutant embryos. In wg mutants, early P(en/lac) expression is identical to that in wild-type embryos, but the late programme does not initiate (data not shown). In ptc mutants, the early programme is also normal, but when the late programme is induced, P(en/lac) is expressed ectopically, as is the endogenous en gene (Fig. 3e,f). In summary, the effects of wg or ptc mutations on expression from P(en/lac) correlate with the effect of these mutations on endogenous en expression.
Cell communication
The data show that there are two programmes regulating en gene expression in stripes during embryogenesis. The first regulatory programme involves the independent control of even- and odd-numbered en stripes by pair-rule gene action11,12,15, and may involve direct transcriptional regulation by some of the pair-rule products.
The second regulatory programme operates after cellularization, and involves segment polarity gene action. Recent molecular analysis of wg, coupled with the data presented here, suggests that cell interactions are involved in the late control of en striped expression.
The wg gene, like en, is expressed in stripes, but wg transcripts accumulate mainly in cells that are anteriorly adjacent to those that express en, and probably not in the cells that express en (ref. 3, Fig. 6). Similarities noted between the predicted Drosophila wg protein and the product of the mammalian oncogene int-1 have led to the suggestion that the wg product acts as a signal in cell communication32. This suggestion is consistent with genetic analysis showing that wg is not cell-autonomous17,33,44. Our results identify one set of target cells for wg activity—the neighbouring en-expressing cells (Fig. 6)—and we suggest that en gene expression may be a target of a signal that is initiated at the cell surface by the wg protein. Since hh, arm and fu have phenotypes similar to wg with respect to their effects on en expression, it is tempting to speculate that some of these genes may encode other components involved in the transduction of the wg signal. The existence of genes in the mouse that are homologous to en34 and wg32 suggests that the circuitry proposed here for the control of en by wg may be phylogenetically conserved.
In formal terms ptc activity is required to keep en expression repressed in particular cell rows within each developing segment. Interestingly, the derepression of en in a ptc mutant requires a signal from wg-expressing cells. Consequently, one function of ptc may be to prohibit, directly or indirectly, wg from inducing en ectopically (Fig. 6). Thus, ptc activity may impart a polarity on patterning within segments, since the induction of en is restricted to cells located posterior to the wg domain (Fig. 6).
Ectopic induction of en in ptc mutants occurs in cells which do not abut the normal wg domain. Either wg can act over several cell diameters, or there are other genetic interactions missing from the scheme. Indeed, Martinez-Arias and co-workers have found that the wg expression domain is expanded in ptc mutants and that this change precedes the induction of en35 Thus, the defects in a ptc mutant may lead to a cascade of compensatory changes, at least in part involving wg, that lead to the induction of en.
Separate regulatory programmes
A teleological rationale for the existence of two discrete programmes for striped en expression can be offered. The observation that cells are respecified in ptc mutants suggests that there is still considerable plasticity in developmental programming at the cellular blastoderm. The later programme, involving the action of segment polarity genes, may represent an editing function that corrects subtle mistakes made in the establishment of en expression. In this scenario, the late programme is an important accessory programme that improves the fidelity of the system. In fact, although the establishment of en expression at blastoderm is remarkably precise—>95% of the cells in even-numbered en stripes map precisely to the anterior-most cells of ftz stripes36,37 —the remaining 2–5% of cells may represent true ‘mistakes’37, where en expression is initiated in cells internal to the anterior edge of the ftz stripe. Perhaps these mistakes are corrected during the post-blastoderm phase of development. In addition, this editing function may be what governs the classical ‘regulative response’, in which adjustments are made in body pattern in response to physical perturbations38,39.
A second possibility, not incompatible with the first, is that the existence of a separate early programme is an evolutionary adaptation for the very rapid embryonic development observed in Drosophila. The early programme, which establishes striped en expression at cellular blastoderm, represents the successful conclusion of a regulatory cascade that subdivides the embryonic field very rapidly9 and operates largely in a syncytium. Such a rapid and syncytial developmental plan is rather peculiar. Perhaps the late regulatory programme, involving cell interactions, is sufficient to achieve the gradual and sequential subdivision of the germband seen in most organisms. In this scenario, the late programme would be phylogenetically conserved, and the rapid cascade of regulatory interactions that culminates in the early program would be an accessory programme used by Drosophila and its relatives to rapidly establish spatial subdivisions.
We are indebted to Gus del Puerto for help in establishing transgenic lines and immunocytochemistry; P. Ingham, A. Martinez-Arias and N. Baker for communicating unpublished data and members of the laboratory for comments. We thank S. Carroll and M. Frasch for antibodies against ftz and eve, respectively; J. Hooper and M. Scott for Df(2R) ptc stock; and V. Finkel. Research was supported by NSF and NIH (P. O’F.), a Lucille P. Markey Scholar Award (S.D.), Damon Runyon-Walter Winchell (E.S.), NIH training grant (J.H.-J.), and the Giannini Foundation (J.A.K.).
References
- 1.Kornberg T, et al. Cell. 1985;40:45–53. doi: 10.1016/0092-8674(85)90307-1. [DOI] [PubMed] [Google Scholar]
- 2.Fjose A, McGinnis WJ, Gehring WJ. Nature. 1985;313:284–289. doi: 10.1038/313284a0. [DOI] [PubMed] [Google Scholar]
- 3.Baker N. EMBO J. 1987;6:1765–1773. doi: 10.1002/j.1460-2075.1987.tb02429.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nüsslein-Volhard C, Wieschaus E. Nature. 1980;287:795–801. doi: 10.1038/287795a0. [DOI] [PubMed] [Google Scholar]
- 5.Kornberg T. Proc natn Acad Sci USA. 1981;78:1095–1099. doi: 10.1073/pnas.78.2.1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nüsslein-Volhard C, Frohnhöfer HG, Lehmann R. Science. 1987;238:1675–1681. doi: 10.1126/science.3686007. [DOI] [PubMed] [Google Scholar]
- 7.Gaul U, Jäckle H. Cell. 1987;51:549–555. doi: 10.1016/0092-8674(87)90124-3. [DOI] [PubMed] [Google Scholar]
- 8.Carroll SB, Scott MP. Cell. 1986;45:113–126. doi: 10.1016/0092-8674(86)90543-x. [DOI] [PubMed] [Google Scholar]
- 9.Scott MP, O’Farrell PH. A Rev Cell Biol. 1986;2:49–80. doi: 10.1146/annurev.cb.02.110186.000405. [DOI] [PubMed] [Google Scholar]
- 10.DiNardo S, et al. Cell. 1985;43:59–69. doi: 10.1016/0092-8674(85)90012-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Howard K, Ingham P. Cell. 1986;44:949–957. doi: 10.1016/0092-8674(86)90018-8. [DOI] [PubMed] [Google Scholar]
- 12.DiNardo S, O’Farrell PH. Genes & Dev. 1987;1:1212–1225. doi: 10.1101/gad.1.10.1212. [DOI] [PubMed] [Google Scholar]
- 13.Hafen E, Kuroiwa A, Gehring WJ. Cell. 1984;37:833–841. doi: 10.1016/0092-8674(84)90418-5. [DOI] [PubMed] [Google Scholar]
- 14.Macdonald PM, Ingham P, Struhl G. Cell. 1986;47:721–734. doi: 10.1016/0092-8674(86)90515-5. [DOI] [PubMed] [Google Scholar]
- 15.Harding K, et al. Science. 1986;233:953–959. doi: 10.1126/science.3755551. [DOI] [PubMed] [Google Scholar]
- 16.Kilcherr F, Baumgartner S, Bopp D, Frei E, Noll M. Nature. 1986;321:493–499. [Google Scholar]
- 17.Morata G, Lawrence PA. Devl Biol. 1977;56:227–240. doi: 10.1016/0012-1606(77)90266-4. [DOI] [PubMed] [Google Scholar]
- 18.Spradling AC, Rubin GM. Science. 1982;218:341–347. doi: 10.1126/science.6289435. [DOI] [PubMed] [Google Scholar]
- 19.Drees B, et al. EMBO J. 1987;6:2803–2809. doi: 10.1002/j.1460-2075.1987.tb02576.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Poole SJ, et al. Cell. 1985;40:37–43. doi: 10.1016/0092-8674(85)90306-x. [DOI] [PubMed] [Google Scholar]
- 21.Hiromi Y, Kuroiwa A, Gehring WJ. Cell. 1985;43:603–613. doi: 10.1016/0092-8674(85)90232-6. [DOI] [PubMed] [Google Scholar]
- 22.Weir M, Kornberg T. Nature. 1985;318:433–439. doi: 10.1038/318433a0. [DOI] [PubMed] [Google Scholar]
- 23.Campos-Ortega JA, Hartenstein V. The Embryonic Development of Drosophila melanogaster. Springer; Berlin: 1985. [Google Scholar]
- 24.Carroll SB, Scott MP. Cell. 1985;43:47–57. doi: 10.1016/0092-8674(85)90011-x. [DOI] [PubMed] [Google Scholar]
- 25.Frasch M, et al. EMBO J. 1987;6:749–759. doi: 10.1002/j.1460-2075.1987.tb04817.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Martinez-Arias A. J Embryol exp Morph. 1985;87:99–114. [PubMed] [Google Scholar]
- 27.Perrimon N, Mahowald AP. Devl Biol. 1987;119:587–600. doi: 10.1016/0012-1606(87)90061-3. [DOI] [PubMed] [Google Scholar]
- 28.Gergen JP, Coulter D, Wieschaus E. In: Gametogenesis and the Early Embryo. Gall J, editor. New York: 1986. pp. 195–220. [Google Scholar]
- 29.Frasch M, Levine M. Genes & Dev. 1987;1:981–995. doi: 10.1101/gad.1.9.981. [DOI] [PubMed] [Google Scholar]
- 30.Struhl G. Nature. 1985;318:677–680. doi: 10.1038/318677a0. [DOI] [PubMed] [Google Scholar]
- 31.Ingham PW, Howard KR, Ish-Horowicz D. Nature. 1985;318:439–445. [Google Scholar]
- 32.Rijsewijk F, et al. Cell. 1987;50:649–657. doi: 10.1016/0092-8674(87)90038-9. [DOI] [PubMed] [Google Scholar]
- 33.Wieschaus E, Riggleman R. Cell. 1987;49:177–184. doi: 10.1016/0092-8674(87)90558-7. [DOI] [PubMed] [Google Scholar]
- 34.Joyner AL, Martin GR. Genes & Dev. 1987;1:29–38. doi: 10.1101/gad.1.1.29. [DOI] [PubMed] [Google Scholar]
- 35.Martinez-Arias A, Baker N, Ingham PW. Development. (submitted) [DOI] [PubMed] [Google Scholar]
- 36.Lawrence PA, Johnston P, Macdonald P, Struhl G. Nature. 1987;328:440–442. doi: 10.1038/328440a0. [DOI] [PubMed] [Google Scholar]
- 37.Carroll SB, et al. Genes & Dev. 2 (in the press) [Google Scholar]
- 38.French V, Bryant PJ, Bryant S. Science. 1976;193:969–981. doi: 10.1126/science.948762. [DOI] [PubMed] [Google Scholar]
- 39.Wright DA, Lawrence PA. Devl Biol. 1981;85:317–327. doi: 10.1016/0012-1606(81)90263-3. [DOI] [PubMed] [Google Scholar]
- 40.Kuner JM, et al. Cell. 1985;42:309–316. doi: 10.1016/s0092-8674(85)80126-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hiromi Y, Gehring W. Cell. 1987;50:963–974. doi: 10.1016/0092-8674(87)90523-x. [DOI] [PubMed] [Google Scholar]
- 42.Van der Meer S. Drosoph Inf Serv. 1977;52:160. [Google Scholar]
- 43.Ingham PW, Baker N, Martinez-Arias A. Nature. 1988;331:73–75. doi: 10.1038/331073a0. [DOI] [PubMed] [Google Scholar]