Summary
Cyclin proteins are thought to trigger entry into mitosis. During mitosis they are rapidly degraded. Therefore, mitosis and consequently cyclin degradation might be triggered at a time when cyclins have reaccumulated to a critical level. We cloned and sequenced a Drosophila cyclin A homolog and identified mutations in the corresponding gene. Immunofluorescent staining revealed that cyclin A accumulates in the interphase cytoplasm of cellularized embryos, but relocates to the nuclear region early in prophase and is completely degraded within metaphase. Cyclin A was expressed in dividing cells throughout development, and a functional cyclin A gene was required for continued division after exhaustion of maternally contributed cyclin A. Importantly, the timing of post cellularization divisions was not governed by the rate of accumulation or level of cyclin A.
Introduction
An extraordinary homology of some of the fundamental cell cycle regulators suggests that there are universal mechanisms governing cell cycle progression. While this invites comparison between different organisms, caution is needed because there is not a unique mode of regulation. In many organisms, the character of cell cycle regulation changes as development progresses.
Early embryonic cell cycles are generally extremely fast (8 min in Drosophila) and apparently uncoupled from regulatory events restricting later cell cycles (Blumenthal et al., 1973; Foe and Alberts, 1983; Edgar et al., 1986; Edgar and Schubiger, 1986). In Drosophila, these abbreviated cell cycles involve only the nuclei; they divide in a common syncytial cytoplasm. Here we will focus on the cell cycles occuring after cellularization of the syncytial nuclei following mitosis 13. In contrast to the first 13 mitoses, these mitoses are dependent on zygotic transcription (Edgar and Schubiger, 1986; Edgar et al., 1986). In addition, mitosis is no longer synchronous. Cells divide in an intricate spatial and temporal pattern (Foe and Alberts, 1983; Hartenstein and Campos-Ortega, 1985; Foe, 1988). This developmental switch from a simple to a compex cell cycle is common to many organisms, such as Xenopus, where it occurs during the midblastula transition (Kirschner et al., 1985; Kimelman et al., 1987).
Following three postcellularization cell cycles, further division is largely restricted to the nervous system and the germ cell lineage. Most cells forming the larval body never divide again, and many become polyploid. During larval life, however, a few diploid cells re-enter mitotic cell cycles and form the imaginal discs whose subsequent morphogenesis will form the adult body.
Molecular probes for proteins marking the progress of the cell cycle would help to further define the spatio-temporal program of embryonic cell divisions. Additionally, in Drosophila, mutations could be used to explicitly test the roles of such proteins in the regulation of these embryonic cell divisions. Here we describe a molecular and genetic analysis of the role of a Drosophila cyclin in the highly patterned postcellularization divisions.
Cyclin proteins were originally identified in eggs of the marine invertebrates clam, sea urchin, and starfish. They are continuously synthesized and accumulate throughout interphase, but are rapidly and completely degraded during each meiotic division and during each of the early cleavage divisions (Evans et al., 1983; Swenson et al., 1986; Standart et al., 1987). Evidence for a role in governing cell cycle progression came from experiments in which immature Xenopus oocytes were driven into meiosis by injection of synthetic mRNA made from either a clam or a sea urchin cyclin cDNA (Swenson et al., 1986; Pines and Hunt, 1987). The identification of cyclins having similar behaviors in several species suggests that these proteins have evolutionary conserved roles. Two highly homologous cyclin types, A and B, were originally described in the marine clam (Evans et al., 1983; Westendorf et al., 1989). The two cyclin types are also present in Xenopus (T. Hunt, personal communication) and Drosophila (this report; W. Whitfield and D. Glover, personal communication). Recently, the cdc13+ gene from the yeast Schizosaccharomyces pombe (S. pombe) has been shown to encode a cyclin homolog (Solomon et al., 1988; Goebl and Byers, 1988) which is required for entry into mitosis (Booher and Beach, 1987, 1988). Together these observations suggest that cyclins function universally in triggering mitosis (and meiosis).
Cyclin action is coupled to a conserved mitotic regulator, MPF (maturation or mitosis promoting factor). MPF is an experimentally defined activity that triggers, even in the absence of protein synthesis, either entry into meiosis after injection into immature Xenopus oocytes (Masui and Markert, 1971; Gerhart et al., 1984) or mitotic events in a cell free system (Lohka and Maller, 1985; Miake-Lye and Kirschner, 1985; Dunphy and Newport, 1988; Lohka et al., 1988). Recently, one component of Xenopus MPF has been shown to be homologous to the protein kinase encoded by the S. pombe cell cycle gene cdc2+ (Dunphy et al., 1988; Gautier et al., 1988). Two lines of evidence indicate a close relationship between cyclins and MPF. First, mRNA injection experiments with Xenopus oocytes imply that cyclins activate MPF (Swenson et al., 1986; Pines and Hunt, 1987). Second, in S. pombe, the cdc2+ gene product appears to interact with the cyclin encoded by the cdc13+ gene (Booher and Beach, 1987, 1988).
All of the above observations are consistent with the idea that cyclin accumulation to a critical level leads to MPF activation and consequently to entry into mitosis. Thus, the rate of cyclin accumulation could determine the time of mitosis. The degradation of cyclins during each division might play the role of resetting the cycle.
To analyze the role of cyclin in the regulation of embryonic cell cycles, we cloned a Drosophila cyclin A homolog and studied its expression in wild-type and mutant embryos. Cyclin A is expressed in dividing cells at all developmental stages. The phenotype of mutant alleles demonstrated that cyclin A is required for cell cycle progression. However, differential rates of cyclin A accumulation do not determine the temporal order of entry of cells into the first asynchronous cell division (mitosis 14).
Results
Cloning and Sequencing of a Drosophila Cyclin A Homolog
We used a degenerate oligonucleotide probe derived from a stretch of amino acids conserved between clam cyclin A and a sea urchin cyclin (Swenson et al., 1986; Pines and Hunt, 1987) to screen a λ phage cDNA library made from poly(A)+ RNA from Drosophila ovaries. Several clones were isolated, and the longest insert was sequenced (Figure 1). The sequence analysis revealed a long open reading frame. Its putative initiation codon is flanked by a sequence that fits the consensus sequence for translational start sites in Drosophila (Cavener, 1987). The sequences flanking the five ATG codons found upstream of the putative translational start site do not fit the consensus, and all these ATGs are followed by several stop codons. At the 3′ end of the cDNA, a polyadenylation signal followed by a poly(A) tract was found. As shown below, at least two different maternal transcripts were detected on Northern blots (Figure 4, lane 1). The predominant maternal transcript is about 300 bases longer than our longest insert. At present, we do not know whether this cDNA represents a full-length clone. Some or all of the observed size difference between cDNA and transcript might be accounted for by the presence of a longer poly(A) tail in the mRNA. The sequence of another cDNA clone revealed differences in the 5′ and 3′ untranslated regions (data not shown), arguing for alternative mRNA processing events. According to restriction mapping and partial sequencing, the coding sequence appears to be identical in all 18 cDNA clones.
Figure 1. Nucleotide Sequence and Predicted Amino Acid Sequence of the Drosophila Cyclin A cDNA.
The nucleotide sequence of a Drosophila cyclin A cDNA (2385 bp) has a long open reading frame encoding a protein of 491 amino acids. The predicted amino acid sequence is shown in three letter code above the nucleotide sequence. The putative initiating ATG is underlined along with flanking bases matching the consensus sequences for translational start sites in Drosophila (Cavener, 1987). Five additional upstream ATG codons and a putative polyadenylation signal are also indicated by underlining.
Figure 4. Cyclin A mRNA Levels during Embryonic Cell Cycle Progression.
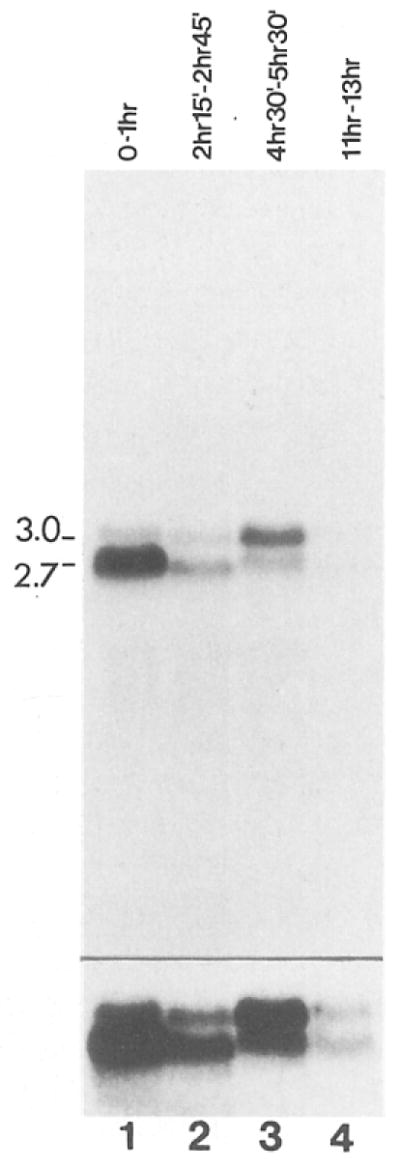
Total RNA from an equal number of early embryos, engaged in the rapid cleavage divisions (lane 1), early cycle 14 embryos in interphase (lane 2), embryos engaged in embryonic cell divisions 15 and 16 (lane 3), and older embryos where cell divisions are restricted to the nervous system (lane 4) were probed on Northern blots with the cyclin A cDNA. The estimated size (kb) of the cyclin mRNAs are indicated on the left side. Embryos were collected and aged at 25°C for the times indicated and staged under the microscope before isolation of RNA. In order to visualize the signals from cyclin A mRNAs in older embryos, a longer exposure is shown in the lower panel.
Amino acid sequence comparison between the 491 amino acid protein encoded by the Drosophila cDNA and the clam cyclins A and B (Swenson et al., 1986; Westendorf et al., 1989), a sea urchin cyclin (Pines and Hunt, 1987), and the S. pombe cdc13+ gene product (Booher and Beach, 1988) revealed extensive conservation. For example, within a 60 amino acid region (indicated by arrows in Figure 2) residues at 82% of the positions are similar and 72% are identical in the aligned Drosophila and clam cyclin A sequences. Within this same region, 47% of the positions are identical among all the cyclins, whether A or B type, or whether of yeast, mollusc, echinoderm, or insect origin (see residues marked with an asterisk). Conservation among this group of proteins extends throughout the carboxy-terminal two-thirds of these proteins (Figure 2). Based on this homology and on the cell cycle–dependent degradation of the encoded protein (see below), we conclude that the cloned sequence represents a Drosophila cyclin. In contrast to the sea urchin and the S. pombe cyclins, the Drosophila sequence is more similar to clam cyclin A than to clam cyclin B (50% identity as opposed to 34% identity in the region shown; see also boxes in Figure 2). The presence of amino acid clusters specifically conserved between Drosophila and clam A cyclins, and clusters specifically conserved among yeast, sea urchin, and clam B cyclins (see for instance Figure 2, residues marked with black dots), allows a tentative classification of these cyclins as A or B types.
Figure 2. Amino Acid Sequence Comparison of the Drosophila Cyclin A with Other Known Cyclin Proteins.
The amino acid sequences of the domain conserved in Drosophila cyclin A (D.A), clam cyclin A (C.A), clam cyclin B (C.B), a sea urchin cyclin (SU), and the S. pombe cdc13+ gene product (SP) are aligned. Residues identical in all the sequences are marked with asterisks (*). Bold letters indicate residues that are conserved in at least three sequences. The arrows delineate an especially highly conserved region (see text). Gaps, introduced for optimal alignment, are represented by dashes. Regions are boxed where the two A type sequences (D.A and C.A) and at least two of the B type sequences (C.B, SU, and SP) are similar (identical or conserved replacements using the following grouping: A,L,V,I,M; K,R; D,E; S,T; N,Q; Y,F). Black dots mark regions where distinctive differences distinguish A and B type cyclins.
Identification of Mutations in the Drosophila Cyclin A Gene
In situ hybridization to polytene chromosomes localized the cyclin A sequence to the genetically well characterized region 68 D/E on the left arm of chromosome three (data not shown). Hybridizations to a series of chromosomal deficiencies that subdivide this region (Akam et al., 1978) placed the cyclin A sequence between the proximal breakpoints of the deficiencies vin2 and vin3 (Figure 3A). An essentially saturating genetic screen had uncovered five lethal complementation groups (rsg11–rsg15) within this interval (Hoogwerf et al., 1988).
Figure 3. Identification of Mutations in the Cyclin A Gene.
(A) Cytogenetic map of the region on chromosome arm 3L containing the cyclin A gene. The regions deleted in the chromosomal deficiencies vin2 and vin3 (Akam et al., 1978) are indicated by black bars. In situ hybridization localized the cyclin A sequence within the two proximal breakpoints of vin2 and vin3 (arrow). Five lethal complementation groups (rsg11–rsg15) have been localized between these breakpoints (Hoogwerf et al., 1988). The EMS-induced allele I(3)183 of the complementation group rsg11 complements neither the EMS-induced allele I(3)v4-4 nor the allele neo114 isolated after P element insertion mutagenesis (Cooley et al., 1988)
(B) Mapping the P element insertion neo114. Southern analysis with a cyclin A cDNA probe was used to compare genomic restriction fragments from an rsg11 allele (neo114), a deletion of the region (vin3), and a control chromosome. The chromosome carrying the allele neo116 was chosen as control because it is derived from the same parental chromosome as neo114 and it also has a P element insert but in a different location. Since the chromosomes being compared carry recessive lethals, DNA was prepared from heterozygous flies. Novel fragments absent from the control DNA (neo116/TM3, lanes 1, 3, and 7) but apparent in neo114/TM3 DNA are marked with arrowheads. Lanes 1 and 2: Sall; lanes 3 and 4: EcoRI; lanes 5–8: HindIII. Despite the appearance of novel fragments, no loss of bands is observed in Sall and EcoRI digests of neo114/TM3 DNA because of the contribution of the balancer chromosome. However, one band present in the control (lane 7) cannot be detected in HindIII digests of neo114/TM3 DNA (lane 8, arrow). This disappearance indicates the presence of a restriction site polymorphism. In digests of DNA from flies carrying a deficiency deleting the cyclin A gene (vin3), the hybridizing fragments are uniquely derived from the balancer chromosome, and a comparison of the banding pattern derived from two different balancer chromosomes, In(3L)P (lane 5) and TM3 (lane 6), reveals a HindIII restriction site polymorphism. This polymorphism allows the unambiguos assignment of the hybridizing fragments to the different chromosomes present in neo116/TM3 flies or neo114/TM3 flies, respectively. Position and size (kbp) of molecular weight markers are indicated on the left side.
Independent work in the laboratory of Y.-N. Jan resulted in the cloning of the genomic region flanking the P element insert present in the fly line neo114. This P element insert is located in the chromosomal region 68D/E and the line neo114 does not complement the allele I(3)183 of the complementation group rsg11 (H. Vaessin and Y.-N. Jan, personal communication). On Southern blots, the cyclin A cDNA probe detected restriction fragments in neo114 fly DNA that were not found in control DNAs (Figure 3B). Additionally, the cyclin A cDNA hybridized to the cloned genomic sequences flanking the P element insert (H. Vaessin, Y.-N. Jan, C. F. Lehner, and P. H. O’Farrell, unpublished data). Southern analysis of the I(3)183 allele shows that this mutation is a small deletion that removes cyclin sequences (data not shown). Moreover, in embryos homozygous for either rsg11 allele, neo114, or I(3)183, no zygotic cyclin A was detected (see below). We conclude that neo114 and I(3)183 are mutant alleles of the cyclin A gene that disrupt the cloned sequence and interfere with expression of the encoded protein. The recessive lethality of these mutations suggests that the cyclin A gene encodes an essential function.
Transcription of the Cyclin A Gene during Wild-Type Development
The relative cyclin A mRNA levels during different stages of embryogenesis were estimated by Northern blot experiments (Figure 4). During all stages, two bands (around 2.7 kb and 3.0 kb, respectively) were detected by the cyclin A cDNA probe. The lower band was especially prevalent in RNA from 0–1 hr embryos (Figure 4, lane 1). These embryos are transcriptionally inactive and therefore contain only maternal RNA.
In general, cyclin A mRNA levels appeared to be roughly correlated with the mitotic activity in the embryo. Consistent with the high frequency of clones found in the ovary cDNA library (0.1%), the maternal mRNAs appeared to be quite abundant in 0–1 hr embryos that are engaged in extremely rapid cleavage divisions (Figure 4, lane 1). Considerably lower levels of cyclin A mRNA were found in total RNA from early cycle 14 embryos (Figure 4, lane 2), suggesting that much of the maternal mRNA was degraded before gastrulation. The increase in cyclin A mRNAs during cycles 15 and 16 must have represented reaccumulation by zygotic transcription (Figure 4, lane 3). Only very low levels of mRNA were detected in older embryos, when only cells in the nervous system still divide (Figure 4, lane 4). The cyclin A gene might therefore be transcriptionally inactive in cells that have completed their limited number of mitotic divisions.
Characterization of Anti–Cyclin A Antibodies
In order to follow the accumulation of the cyclin A protein during development, an affinity purified rabbit antibody against bacterially produced cyclin A was used for immunoblotting and immunofluorescence experiments. A number of observations indicated that the affinity purified antibodies are monospecific for Drosophila cyclin A. Immunofluorescent labeling was reduced to background levels in embryos homozygous for either of the cyclin A mutant alleles, I(3)183 or neo114, at stages after the exhaustion of the maternal cyclin A supplies (see below). In addition, our anti–cyclin A antibodies did not react with either the sea urchin cyclin or the clam cyclin A made in a reticulocyte lysate (A. Murray and C. F. Lehner, unpublished data).
On immunoblots, two bands around 61 kd were detected in total embryo extracts from different developmental stages (Figure 5). These bands were not detected with a control antibody (data not shown). The lowest band corresponded in size to the product of in vitro translation reactions in which T7 RNA polymerase transcripts of the cyclin A cDNA were used to program a reticulocyte lysate (data not shown). Therefore, it is unlikely that the multiple bands are caused by proteolysis. More likely, they reflect modification of cyclin A. Modification of cyclin proteins has been reported in other organisms (Pines and Hunt, 1987; Standart et al., 1987). Interestingly, the ratios of the signal intensities in the different bands varied at different developmental stages (Figure 5, compare lanes 1 and 2 with lanes 3 and 4), suggesting that cyclin A modification might be developmentally regulated.
Figure 5. Cyclin A Abundance during Embryonic Development.
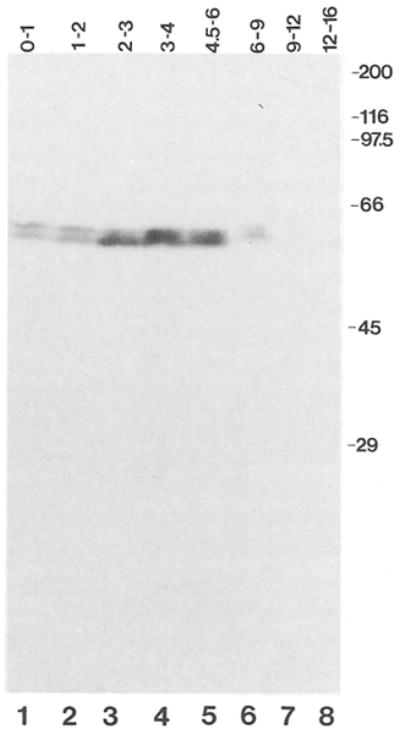
Total protein extracts from embryos collected and aged at 25°C for the times indicated were resolved on SDS gels, transferred to nitrocellulose, and probed with affinity purified antibodies against cyclin A followed by 125I iodinated protein A. The position and size (kd) of molecular weight markers are indicated on the right side.
Consistent with the results of Northern blot experiments, considerably lower signals were observed in older embryos, where only cells in the nervous system divide (Figure 5, lanes 6–8).
Subcellular Location and Degradation of Cyclin A during Mitotic Divisions
After egg activation, a fertilized Drosophila embryo undergoes 13 rapid and synchronous cleavage divisions during which only the nuclei divide (Foe and Alberts, 1983). When early embryos were stained with affinity purified anti–cyclin A antibodies in indirect immunofluorescence experiments, an almost uniformly distributed, bright signal was observed throughout the syncytial embryo. Surprisingly, only subtle differences in staining were observed between embryos in interphase and those in different mitotic stages (data not shown).
After the 13 cleavage divisions, nuclei become cellularized. In striking contrast to the early cleavage divisions, cyclin A was completely degraded during the subsequent cell divisions. Figure 6 shows cells from an embryo during mitosis 14 after double labeling with anti–cyclin A antibodies and a DNA stain. Cyclin A staining accumulated during interphase and was found exclusively in the cytoplasm (Figure 6, Interphase, see also Figure 7a). In contrast, during prophase the most intense cyclin A staining was localized in the region of the condensing chromatin (Figure 6, Prophase). Metaphase cells were either brightly stained (Figure 6, Metaphase A), intermediately stained (data not shown), or unstained (Figure 6, Metaphase B), indicating that cyclin A was completely and rapidly degraded within metaphase. Consistently, no cyclin A was detected in anaphase (Figure 6, Anaphase), telophase, and early in the following interphase (data not shown, but see Figure 7).
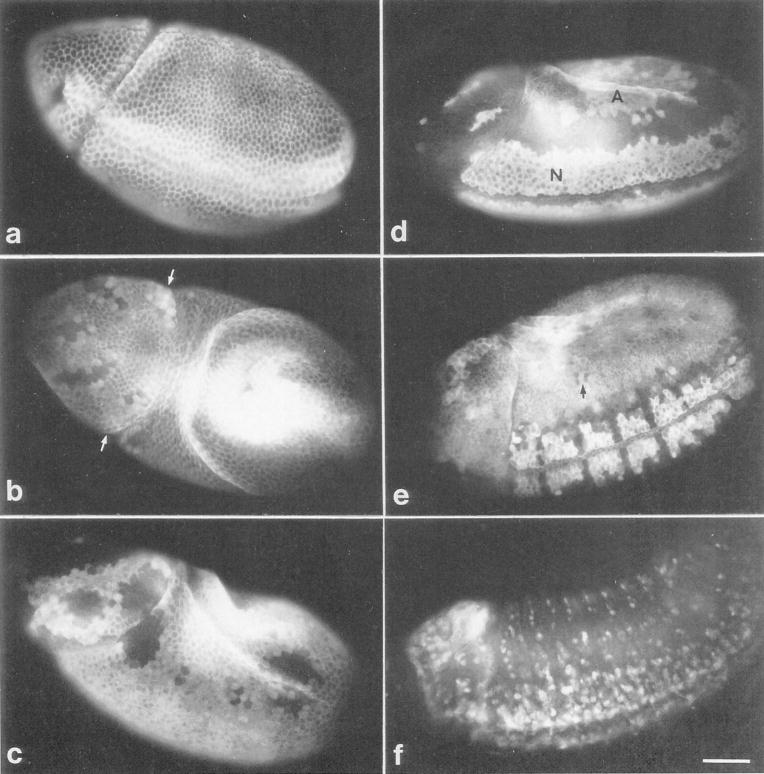
Figure 6. Intracellular Distribution of Cyclin A during Cell Division.
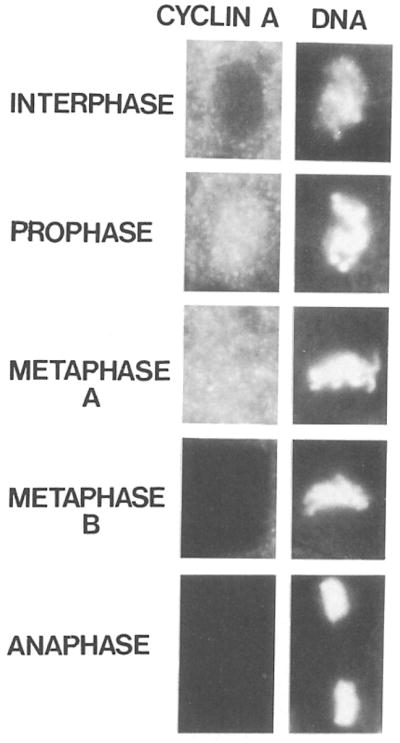
Embryos were double labeled with affinity purified antibodies against cyclin A followed by rhodamine conjugated secondary antibodies (left column) and with the DNA stain Hoechst 33258 (right column). Cells in interphase, prophase, metaphase, and anaphase are shown. Cyclin A is degraded within metaphase and therefore metaphases with cyclin A staining (Metaphase A) as well as without (Metaphase B) can be detected.
Figure 7. Accumulation and Degradation of Cyclin A during Embryonic Cell Cycle Progression.
Embryos were stained with affinity purified antibodies against cyclin A and rhodamine conjugated secondary antibodies. The following developmental stages are shown (the age of the embryos is given as the time after onset of mitosis 14, which is about 200 min after egg deposition):
(a) immediately before onset of mitosis 14.
(b) 5 min after onset of mitosis 14. The cells in the unstained domains in the head have already completed mitosis. Arrows point to a domain where mitosis is initiated but not yet completed.
(c) 20 min divisions in the lateral regions posterior to the cephalic furrow have started.
(d) 30 min most cells have completed mitosis 14 except cells in the amnioserosa (A), the neurogenic region (N), and a few cells in the head region.
(e) 70 min onset of mitosis 15. Cyclin A staining is again present in cells unstained in the 330 min embryo (d), and a few cells (arrow) in the thoracic region have already completed mitosis 15 and are unstained.
(f) 7 hr cell divisions are restricted to the peripheral and central nervous system.
Assignment of the developmental ages was according to Foe (1989). Embryos are oriented anterior to the left, posterior to the right, dorsal up and ventral down, except the embryo in (b), which shows a dorsal view. The bar in (f) corresponds to 50 μm.
Cyclin A and the Temporal and Spatial Program of Embryonic Cell Divisions
The divisions following cellularization (mitoses 14–16) are no longer synchronous: the cells enter mitosis in a spatially and temporally defined, bilaterally symmetrical pattern (Foe and Alberts, 1983; Hartenstein and Campos-Ortega, 1985; Foe, 1988). Anti–cyclin A labeling permitted dramatic visualization of this spatio-temporal program of embyronic cell divisions. The first cells to enter the 14th mitosis were found in a cluster in the head region of the embryo. Shortly thereafter, cells in another cluster in the head region underwent mitosis 14. In the embryo shown in Figure 7b, most of the cells in these first two clusters had already passed metaphase and therefore cyclin A was no longer detectable. In addition, divisions in another domain were anticipated by cyclin A staining in nuclei, a distribution characteristic of early mitotic stages (Figure 7b, arrows). In a slightly older embryo (Figure 7c), the cells in this domain, just anterior to the cephalic furrow, had completed mitosis and were therefore no longer stained. With time, existing domains expanded as more cells divided and additional domains appeared in the head and lateral region (Figures 7c, and 7d).
Cyclin A staining persisted in cells of the amnioserosa (Figure 7d, region A) and in the neurogenic region (Figure 7d, region N). The cells in the neurogenic region eventually enter mitosis 14, some of them synchronously in a segmentally repeated pattern (Figure 7e). Cell cycle 14 in the neurogenic region can be up to three times longer than in the head where cells enter mitosis 14 first. In contrast, amnioserosa cells never go into mitosis 14. Rather than disappear abruptly, the cyclin A in these cells appeared to be slowly degraded. Except for the amnioserosa, the patterned loss of cyclin A labeling was strictly correlated with the pattern of mitotic divisions revealed by double labeling using a DNA stain (data not shown).
In contrast, to degradation, the accumulation of cyclin A did not reflect the pattern of mitosis 14. Cyclin A accumulated very uniformly in all cells throughout the embryo (Figure 7a). The rate of accumulation appeared to be identical in cells that complete mitosis 14 very early (head region), very late (neurogenic region), or never (amnioserosa).
Cyclin A cycled during all embryonic cell divisions after cellularization. Following degradation during mitosis 14, cyclin A reaccumulated during interphase 15. This can be seen in Figure 7e, where the cells in the head and the lateral regions of the embryo were again labeled. In cycle 15, the cells never stained as intensely as those cells that enter mitosis 14 very late (Figure 7e). In fact, the first cells to divide in the thoracic region of the embryo shown in Figure 7e (arrow) had already passed metaphase 15 and were therefore no longer labeled. Cyclin A staining in a slightly older embryo demonstrated that not only mitosis 14 but also mitosis 15 is highly patterned (Figure 8c). Note that the segmentally repeated pattern of the 15th division in the lateral region of the embryo (Figure 8c) was not predictable from the pattern of reaccumulation (Figure 7e). Thus, like mitosis 14, the timing of mitosis 15 is not correlated with the rate of cyclin accumulation.
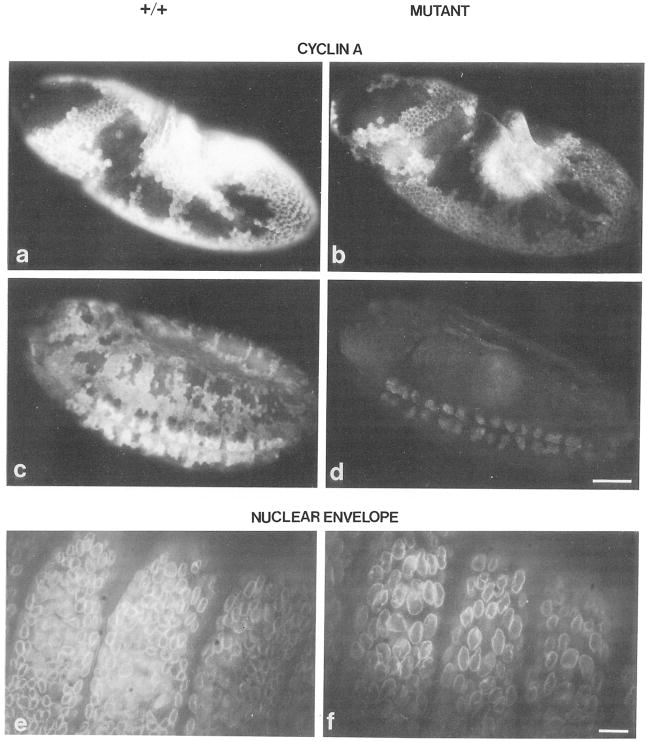
Figure 8. Cyclin A Distribution and Nuclear Density in Mutant Embryos.
Embryos were stained with affinity purified antibodies against cyclin A followed by rhodamine conjugated goat anti-rabbit antibodies (a–d) and with a monoclonal antibody against a nuclear envelope component followed by fluorescein conjugated goat anti-mouse antibodies (e and f). (a, c, and e) Normal embryos, (b, d, and f) Mutant embryos. Embryos are at the time of mitosis 14 (a and b), mitosis 15 (c and d), or after mitosis 16 (9 hr, e and f). The level of cyclin A staining in the progeny of the cross neo114/TM3 × vin3/TM3 is wild type (a and c) in 25%, intermediate (data not shown) in 50%, and clearly lower (b and d) in 25% of the embryos. Double labeling with anti–cyclin A antibodies (data not shown) and antinuclear envelope antibodies (e and f) revealed that embryos, which have no detectable cyclin A (f), have less than half the number of nuclei than in embryos where cyclin A was readily detected (e). The bar in (d) corresponds to 50 μm. Only the dorsolateral epidermis of three embryonic segments is shown in (e) and (f). The bar in (f) corresponds to 5 μm.
A similar accumulation and degradation accompanied cycle 16 (data not shown). Subsequently, mitotic divisions in the embryo stop except for cells in the central and peripheral nervous system and cells in the germ cell lineage. At these stages, cyclin A staining was detected exclusively in these cells (Figure 7f). Cyclin A expression was not restricted to embryonic cells; it was detected in imaginal disc cells and by immunoprecipitation in Schneider G2 tissue culture cells (data not shown). Thus, cyclin A appears to be expressed in all dividing cells, but not in nondividing cells (except for the nondividing aminoserosa cells where cyclin A is initially present but later disappears).
Cyclin A Accumulation and Cell Divisions in Mutant Embryos
It is clear that early embryos in Drosophila and other species have large stores of maternal cyclin mRNA. The amount of cyclin A produced from this maternal mRNA can be estimated by immunofluorescent labeling of embryos that are unable to produce cyclin A zygotically. The consequence of a zygotic defect can be examined after exhaustion of the maternal contribution. We analyzed mutant embyros homozygous for either neo114 or I(3)183 or carrying these alleles over the deficiency vin3. No clear difference between wild-type and mutant embryos was detectable during the early cleavage stages (data not shown). In older mutant embryos, no anti–cyclin A staining was detected (data not shown, but see below), suggesting that the maternal cyclin A mRNA pool was exhausted.
We analyzed cyclin A expression and the cell division program by double labeling timed embryo collections with anti–cyclin A antibodies and a DNA stain. The mutant embryos proceeded normally through the first 13 divisions. In the cell cycle preceding mitosis 14, they never accumulated cyclin A to the normal level (compare Figures 8a and 8b). Nevertheless, the pattern of mitosis 14 was normal, as visualized by DNA staining (data not shown) and by the spatial pattern of cyclin A (compare Figures 8a and 8b). Comparing normal and mutant embyros of the same age demonstrated that not only the pattern but also the timing of mitosis 14 was unaffected in mutant embryos despite the lower levels of cyclin A (Figures 8a and 8b).
After mitosis 14, cyclin A accumulated only to barely detectable levels in mutant embryos (compare Figures 8c and 8d). Nevertheless, at least some cells went through division 15 (data not shown). During the stage when embyros would normally be engaged in mitosis 16, no cyclin A was detected in mutant embryos. Interestingly, no mitotic figures were detected in these mutant embryos, clearly indicating that cell divisions were blocked in the absence of cyclin A. Consistently, at stages when the ectodermal division program was completed, the nuclear density revealed by labeling of nuclear envelopes was clearly lower in mutant embryos (Figures 8e and 8f). The number of nuclei in the ectoderm of mutant embryos was slightly less than half of the number of normal embryos.
Discussion
Here we report the cloning and sequencing of a cDNA encoding a Drosophila homolog of cyclin A. In addition, we identified mutations in the corresponding gene and analyzed its expression during embryonic cell cycle progression in wild-type and mutant embryos.
The Drosophila Cyclin A Gene Is Not Redundant
Cyclin A and cyclin B genes have been identified in Drosophila, as well as in clam and Xenopus (Evans et al., 1983; Westendorf et al., 1989; W. Whitfield and D. Glover, personal communication; T. Hunt, personal communication). Despite the presence of a cyclin B gene, the alleles identified in the Drosophila cyclin A gene are lethal. Interestingly, disruption of the cdc13+ gene encoding a B type cyclin in S. pombe is also lethal (Booher and Beach, 1988). These observations and the fact that the two cyclin types have been independently conserved in evolution strongly suggest that these two homologous proteins have at least subtly different functions and that the distinctions are essential. So far, however, no functional differences have been found and both, the clam cyclin A and the B type sea urchin cyclin, can induce Xenopus oocyte maturation (Swenson et al., 1986; Pines and Hunt, 1987).
Function of Cyclin A
We have analyzed cyclin A accumulation and cell cycle progression in wild-type and mutant embryos. Whereas no aberrant phenotype could be detected in early mutant embryos that still express cyclin A from maternal mRNA, cell divisions were found to be blocked in older mutant embryos at a time when cyclin A levels have declined below detectability. The results of Northern blot experiments are consistent with the interpretation that the decline of cyclin A reflects the exhaustion of maternal mRNA. Although we have only directly demonstrated a requirement for the cyclin A gene in mitosis 16, we have observed an extraordinary spatial and temporal pattern of cyclin A expression whose tight correlation with the pattern of cell divisions throughout development suggests that cyclin A is required for every cell division.
We would like to emphasize that the mutant embryos do not stop cell divisions as a consequence of cell death. They continue to develop and differentiate many structures, including a normal cuticle (Jürgens et al., 1984), after they have stopped dividing.
Cyclin A Accumulation and the Timing of Entry into Mitosis
Cyclin accumulation, starting anew after each division, might be sufficient to activate MPF and consequently trigger mitosis once it has reached a critical level (Swenson et al., 1986; Pines and Hunt, 1987; see also Murray, 1987). Therefore, the rate of cyclin accumulation might determine the time of entry into mitosis.
The above model predicts that cells entering mitosis at different times should have correspondingly different rates of cyclin accumulation in order to attain the critical cyclin level at different times. We have tested this prediction for the first asynchronous mitosis in Drosophila embryos (mitosis 14). Cyclin A accumulates at the same rate in cells that go into mitosis 14 very early, very late, or never. In addition, the spatial pattern of cell division 14 is not delayed in mutant embryos despite the reduced rates of cyclin A accumulation. Therefore, the temporal order of entry into mitosis 14 is not determined by differential rates of cyclin A accumulation, and another regulatory mechanism must time this division. The same conclusion holds also for mitosis 15, but we cannot exclude that the rate of cyclin A accumulation determines the time of entry into mitosis during other developmental stages.
Intracellular Distribution of Cyclin A during Mitosis
The immunofluorescent localization of cyclin A has not only allowed a dramatic visualization of cyclin degradation but has also revealed a surprising redistribution early in prophase. In contrast to its exclusive extranuclear distribution during interphase, cyclin A appears to be concentrated over the region of the condensing chromatin in prophase cells. Perhaps cyclin A redistribution in early prophase is required to initiate mitotic events like nuclear envelope breakdown and chromosome condensation. Alternatively, the tight coupling of mitosis and cyclin A degradation might require an interaction with the mitotic apparatus.
Cyclins are periodically degraded in distantly related organisms (Evans et al., 1983; Swenson et al., 1986; Standart et al., 1987; this report). Surprisingly, according to the results of immunofluorescent experiments, Drosophila cyclin A is continuously present in cleavage embryos. However, we have observed subtle, cell cycle–dependent redistributions around syncytial nuclei, and consequently, we suspect that cyclin A levels oscillate locally. (The early pattern of cyclin localization and degradation will be the subject of a separate communication.) In contrast to the nuclear cleavage cycles, cyclin A is completely and rapidly degraded during each of the subsequent cell divisions.
Several observations argue that cyclin degradation is important for the metaphase–anaphase transition. First, the S. pombe cyclin encoded by the cdc13+ gene is not only involved in the G2/M transition but also in the metaphase–anaphase transition. Whereas null mutations result in a G2 arrest, a temperature-sensitive allele of cdc13+ has been isolated that leads to a metaphase arrest at the restricted temperature. Second, protein degradation has been implicated in the regulation of the metaphase–anaphase transition (Picard et al., 1985; Shoji-Kasai et al., 1988; Schollmeyer, 1988). The recently reported, complex fluctuations in the intracellular Ca2+ concentration during metaphase (Ratan et al., 1988) might modulate the Ca2+-activated proteases that appear to regulate the metaphase–anaphase transition (Schollmeyer, 1988), perhaps by degrading cyclin proteins.
In our immunofluorescence experiments using a DNA stain for double labeling, we were able to accurately identify the mitotic stage during which cyclin A is degraded. Based on the roughly equivalent frequency of stained and unstained metaphase cells, we conclude that degradation of cyclin A must occur over a period occupying only a fraction of metaphase, which lasts for about 1 min in Drosophila (Foe and Alberts, 1983). In addition, the degradation appears to be completed before the metaphase–anaphase transition, suggesting that cyclin A degradation is not immediately triggering this event. Interestingly, in clam embryos, cyclin B degradation is delayed relative to cyclin A degradation (Evans et al., 1983).
The temporal correlation of the dynamic changes in subcellular distribution and concentration of cyclin proteins with progression through mitosis, suggests that these processes might be part of a regulatory cascade controlling the sequence of the complex mitotic events.
Conclusions
Our results demonstrate that the cyclin A gene is essential and that its expression is in general restricted to dividing cells. Immunofluorescent localization of cyclin A reveals striking changes in subcellular distribution and stability, precisely correlated with progression through mitotic stages. In addition, the phenotype of mutations in the cyclin A gene demonstrates that cyclin A is required for mitosis. However, the rate of cyclin A accumulation does not necessarily determine the time of entry into mitosis.
Experimental Procedures
Fly Strains
Wild-type Drosophila melanogaster embryos were collected from flies of the Sevelen strain. The fly strains carrying the deficiency vin2 balanced over TM3,Sb,Ser or the deficiency vin3 balanced over In(3L)P,Me,h,D3 were obtained from D. Roberts, Oxford University, Oxford. A fly strain carrying the deficiency vin3 balanced over TM3,Sb,Ser was obtained from E. M. Meyerowitz, California Institute of Technology, Pasadena, CA. The two deficiencies vin2 and vin3 were originally isolated by Akam et al. (1978) and their breakpoints have been mapped to 67F2–3;68D6 (vin2) and 68C5–6;68E3–4 (vin3). The fly strains, I(3)v4-4A TM3,Sb,Ser and I(3)183/TM3,Sb,Ser (Hoogwerf et al., 1988), carrying EMS-induced lethals, and the fly strain neo114/TM3,Sb (Cooley et al., 1988), carrying a P element insertion at 68D/E, were obtained from Y.-N. Jan, Howard Hughes Institute, San Francisco, CA. The line neo116/TM3,Sb (Cooley et al., 1988), having a P element insertion at 82D, was used as a control in Southern blot experiments.
cDNA Cloning and Sequencing
A degenerate oligonucleotide probe was derived from a conserved stretch of amino acids found in clam cyclin A and sea urchin cyclin B corresponding to the amino acids 291–299 in Drosophila cyclin A. Some codons were selected based on the codon usage in Drosophila (O’Connell and Rosbash, 1984), and others were made degenerate resulting in a mixture of 48 26-mers with the following sequence: 5′-TC(GT)GG(GT)GG(AG)TA(CGT)ATCTCCTC(AG)TACTT-3′. The oligonucleotides were labeled with 32P by polynucleotide kinase (Maxam and Gilbert, 1977), and 50,000 plaques from a λzap cDNA library (a kind gift of B. Hay and L. Jan, Howard Hughes Institute, San Francisco, CA), made from poly(A)+ RNA of Drosophila ovaries, were screened. Hybridizations were carried out in 18% formamide, 6× SSC, 4× Denhardt’s solution, 0.1% SDS, 0.2 mg/ml of salmon sperm DNA, and 0.05 M HEPES (pH 7.0) at 42°C. After hybridization, filters were washed in 2× SSC, 0.1% SDS twice at 22°C for 5 min, three times at 42°C for 5 min, and once at 50°C for 1 min. After autoradiography, 24 plaques giving rise to the strongest signals were isolated and plaque purified. Bluescript plasmids containing the cDNA inserts were rescued from the zap phages according to the instructions of the manufacturer (Stratagene). By restriction mapping experiments using HaeIII and Sau3A, 18 cDNA clones were found to be related, and the largest insert found among these was cloned into M13mp18 and M13mp19 vectors (Norrander et al., 1983). A series of unidirectional deletions was generated from both ends of the insert (Dale et al., 1985) and sequenced using a Sequenase kit (United States Biochemical Corporation).
In Situ Hybridizations, Southern and Northern Blots
Hybridizations to polytene chromosomes from salivary glands of wandering third instar larvae were carried out using biotinylated DNA probes (Langer-Safer et al., 1982) and a biotin alkaline phosphatase detection system (ENZO Biochem, Inc.).
DNA for Southern blot analysis was isolated as follows. About 450 adult flies from stock bottles were frozen in liquid N2 and stored at −70°C. They were ground into a fine powder in a mortar on dry ice. The powder was extracted at 4°C in 0.5 ml of homogenization buffer (0.03 M Tris-HCl [pH 8.0], 0.1 M NaCl, 0.02 M Na2EDTA, 0.05% Triton X-100, 0.01 M 2-mercaptoethanol) for 30 sec on a vortex mixer. A crude nuclear pellet was sedimented during 1 min at 17,000 × g, washed once in 0.5 ml of extraction buffer (0.1 M Tris-HCl [pH 8.4], 0.1 M NaCl, 0,02 M Na2EDTA), and resuspended in 0.3 ml of extraction buffer. Nuclei were lysed by adding sodium sarcosyl (10%) to a final concentration of 1%, and proteins were degraded by protease K (0.1 mg/ml) for 1 hr at 50°C. After sequential extractions with phenol:chloroform (1:1) and chloroform:isoamylalcohol (24:1), DNA was precipitated with ethanol. After dissolving the pellet in TE (0.01 M Tris-HCl [pH 8.0], 0.001 M Na2EDTA), residual RNA was digested with RNAase A (1 μg/ml) for 30 min at 37°C and the extractions with organic solvents were repeated followed again by ethanol precipitation. DNA blots were prepared as described by Maniatis et al. (1982).
Total RNA from different embryonic stages was isolated as described by Edgar et al. (1986) using embryos collected after defined times after oviposition and staged under the microscope. RNA was resolved on formaldehyde-agarose gels, transferred to nylon membranes, and hybridized by standard procedures. Probes for northern and southern blots were prepared by random primer labeling (Hodgson and Fisk, 1987).
Production and Purification of Anti–Cyclin A Antibodies
The following plasmid construction was made in order to allow overproduction of a fusion protein that was used as an immunogen. The large XhoII–EcoRI fragment of the Drosophila cyclin A cDNA was cloned into a pAR3038 vector (Studier and Moffat, 1986) cut with BamHI and EcoRI. The resulting construct directed the synthesis of a cyclin A protein in which the 5 N-terminal amino acids are replaced by the 11 N-terminal amino acids of the coat protein encoded by the T7 phage gene 10. The fusion protein was produced as described by Studier and Moffat (1986) and purified by preparative SDS gel electrophoresis. After dialysis against PBS, 0.2 mg of purified protein emulsified in Freund’s Complete Adjuvant was subcutaneously injected into a male New Zealand rabbit. After booster injections (0.05 mg of protein in Freund’s Incomplete Adjuvant) were applied twice in 3 week intervals, the immune serum was isolated.
In order to construct an affinity matrix to purify anti–cyclin A antibodies, a β-galactosidase–cyclin A fusion protein was isolated as follows. The XhoII–HindIII fragment of the cyclin A cDNA was cloned into the pUR292 vector (Rüther and Müller-Hill, 1983) cut with BamHI and HindIII. The resulting construct directed the synthesis of the same part of cyclin A that was present in the immunogen (amino acids 6–491), but in this case fused to β-galactosidase. This fusion protein was produced in the E. coli strain DG101 as described by Carroll and Laughon (1987) and adsorbed to an anti-β-galactosidase antibody–Sepharose 4B matrix (Cappel). Crosslinking of the fusion protein to the antibody matrix and affinity purification of anti–cyclin A antibodies was done according to Carroll and Laughon (1987), except that bound antibodies were eluted in 10% dioxane, 0.2 M HCl adjusted to pH 2.2 with 2 M glycine. Eluted antibodies were neutralized by addition of 0.33 vol of 1 M K2HPO4 and dialyzed against PBS. After addition of 10% normal goat serum (Vector Laboratories, Inc.) and 0.02% NaN3, antibodies were stored at 4°C.
Immunoblotting and Immunofluorescence Experiments
Total protein extracts were made from timed collections of embryos by Dounce homogenization in SDS gel sample buffer after dechorionation in 2.5% sodium hypochlorite. Extracts were boiled for 3 min, centrifuged for 5 min at 17,000 × g, and the supernatants were loaded onto 10% SDS polyacrylamide gels (Laemmli, 1970). After resolution, proteins were transferred to nitrocellulose using a polyblot TM electroblotting system (American Bionetics, Inc.) according to the manufacturer’s instructions. The membranes were probed with affinity purified anti–cyclin A antibodies (using a concentration corresponding to a 1:400 dilution of the immune serum) followed by 125I iodinated protein A (Amersham) as previously described (Lehner et al., 1987).
For immunofluorescence experiments, embryos were dechorionated and fixed according to Mitchison and Sedat (1983). After blocking nonspecific binding in PBS containing 2% normal goat serum and 0.1% Triton X-100 for 1 hr at 22°C, embryos were incubated overnight at 4°C in the same buffer containing affinity purified anti–cyclin A antibodies diluted 1:400. After washing (3 times for 20 min at 22°C), embryos were incubated with rhodamine-conjugated goat anti-rabbit antibodies (Cappel) preabsorbed with fixed Drosophila embryos. Excess antibodies were again washed off in PBS containing 0.1% Triton X-100 as described above. During the last wash, the DNA stain Hoechst 33258 was included (1 μg/ml). After an additional 5 min wash in PBS, embryos were mounted in Fluoromount (Southern Biotechnology Associates, Inc.) and viewed in a Nikon Optiphot fluorescence microscope equipped with a 25× plan Neofluar lens (Zeiss) and a 100× Fluor lens (Nikon). Photomicrographs were taken on Kodak Technical Pan film. In the experiments where mutant embryos were compared with normal embryos, exposure times were kept constant in order to allow a comparison of signal intensities.
For double labeling experiments, rabbit anti–cyclin A antibodies were diluted in hybridoma supernatants containing mouse monoclonal antibodies against a nuclear envelope antigen (kind gift of D. Kellog and B. Alberts, University of California, San Francisco, CA). Secondary antibodies (rhodamine coupled goat anti-rabbit antibodies and fluorescein coupled goat anti-mouse antibodies) were also applied as a mixture.
Acknowledgments
We are indebted to H. Vaessin, E. Giniger, and Y.-N. Jan for their help with the identification of cyclin A mutant fly strains, for their helpful discussions of unpublished data, and for providing genomic clones and fly strains. We would like to thank D. Roberts and E. Meyerowitz for providing fly strains, B. Edgar for the Northern blot, T. Hunt, J. Ruderman, and D. Glover for communicating results prior to publication, and B. Edgar, J. Jongens, B. Kalionis, D. Lakich, J. Little, A. Murray, and J.-P. Vincent for critical reading of the manuscript. This work was supported by a fellowship of the Swiss National Science Foundation (to C. F. L.) and by a National Science Foundation grant (to P. H. O.).
References
- Akam ME, Roberts DB, Richards GP, Ashburner M. Drosophila: the genetics of two major larval proteins. Cell. 1978;13:215–225. doi: 10.1016/0092-8674(78)90190-3. [DOI] [PubMed] [Google Scholar]
- Blumenthal AB, Kriegstein HJ, Hogness DS. The units of DNA replication in Drosophila melanogaster chromosomes. Cold Spring Harbor Symp Quant Biol. 1973;38:205–233. doi: 10.1101/sqb.1974.038.01.024. [DOI] [PubMed] [Google Scholar]
- Booher R, Beach D. Interaction between cdc13+ and cdc2+ in the control of mitosis in fission yeast; dissociation of the G1 and G2 roles of the cdc2+ protein kinase. EMBO J. 1987;6:3441–3447. doi: 10.1002/j.1460-2075.1987.tb02667.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Booher R, Beach D. Involvement of cdc13+ in mitotic control in Schizosaccharomyces pombe: possible interaction of the gene product with microtubules. EMBO J. 1988;7:2321–2327. doi: 10.1002/j.1460-2075.1988.tb03075.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll SB, Laughon A. Production and purification of polyclonal antibodies to the foreign segment of (β-galactosidase fusion proteins. In: Glover D, editor. DNA Cloning: A Practical Approach. Oxford: IRL Press; 1987. pp. 89–111. [Google Scholar]
- Cavener DR. Comparison of the consensus sequence flanking translational start sites in Drosophila and vertebrates. Nucl Acids Res. 1987;15:1353–1361. doi: 10.1093/nar/15.4.1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooley L, Kelley R, Spradling A. Insertional mutagenesis of the Drosophila genome with single P elements. Science. 1988;239:1121–1128. doi: 10.1126/science.2830671. [DOI] [PubMed] [Google Scholar]
- Dale RMK, McClure BA, Houchins JP. A rapid single-stranded cloning strategy for producing a sequential series of overlapping clones for use in DNA sequencing: application to sequencing the corn mitochondrial 18S rDNA. Plasmid. 1985;13:31–40. doi: 10.1016/0147-619x(85)90053-8. [DOI] [PubMed] [Google Scholar]
- Dunphy WG, Newport JW. Mitosis-inducing factors are present in a latent form during interphase in Xenopus embryos. J Cell Biol. 1988;106:2047–2056. doi: 10.1083/jcb.106.6.2047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunphy WG, Brizuela L, Beach D, Newport J. The Xenopus cdc2 protein is a component of MPF a cytoplasmic regulator of mitosis. Cell. 1988;54:423–431. doi: 10.1016/0092-8674(88)90205-x. [DOI] [PubMed] [Google Scholar]
- Edgar BA, Schubiger G. Parameters controlling transcriptional activation during early Drosophila development. Cell. 1986;44:871–877. doi: 10.1016/0092-8674(86)90009-7. [DOI] [PubMed] [Google Scholar]
- Edgar BA, Kiehle CP, Schubiger G. Cell cycle control by the nucleo-cytoplasmic ratio in early Drosophila development. Cell. 1986;44:365–372. doi: 10.1016/0092-8674(86)90771-3. [DOI] [PubMed] [Google Scholar]
- Evans T, Rosenthal ET, Youngblom J, Distel D, Hunt T. Cyclin: a protein specified by maternal mRNA in sea urchin eggs that is destroyed at each cleavage division. Cell. 1983;33:389–396. doi: 10.1016/0092-8674(83)90420-8. [DOI] [PubMed] [Google Scholar]
- Foe VE. Mitotic domains reveal early commitment of cells in Drosophila embryos. Development. 1989 in press. [PubMed] [Google Scholar]
- Foe VE, Alberts BM. Studies of nuclear and cytoplasmic behaviour during five mitotic cycles that precede gastrulation in Drosophila embryogenesis. J Cell Sci. 1983;61:31–70. doi: 10.1242/jcs.61.1.31. [DOI] [PubMed] [Google Scholar]
- Gautier J, Norbury C, Lohka M, Nurse P, Maller J. Purified maturation-promoting factor contains the product of a Xenopus homolog of the fission yeast cell cycle control gene cdc2+ Cell. 1988;54:433–439. doi: 10.1016/0092-8674(88)90206-1. [DOI] [PubMed] [Google Scholar]
- Gerhart J, Wu M, Kirschner M. Cell cycle dynamics of an M-phase-specific cytoplasmic factor in Xenopus laevis oocytes and eggs. J Cell Biol. 1984;98:1247–1255. doi: 10.1083/jcb.98.4.1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goebl M, Byers B. Cyclin in fission yeast. Cell. 1988;54:739–740. [Google Scholar]
- Hartenstein V, Campos-Ortega JA. Fate-mapping in wildtype Drosophila melanogaster. I The spatio-temporal pattern of embryonic cell divisions. Roux’s Arch Dev Biol. 1985;184:181–195. [Google Scholar]
- Hodgson CP, Fisk RZ. Hybridization probe size control: optimized ‘oligolabeling’. Nucl Acids Res. 1987;15:6295. doi: 10.1093/nar/15.15.6295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoogwerf AM, Akam M, Roberts D. A genetic analysis of the rose-gespleten region (68C8-69B5) of Drosophila melanogaster. Genetics. 1988;118:665–670. doi: 10.1093/genetics/118.4.665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jürgens G, Wieschaus E, Nüsslein-Volhard C, Kluding H. Mutations affecting the pattern of the larval cuticle in Drosophila melanogaster. II Zygotic loci on the third chromosome. Roux’s Arch Dev Biol. 1984;193:283–295. doi: 10.1007/BF00848157. [DOI] [PubMed] [Google Scholar]
- Kimelman D, Kirschner M, Scherson T. The events of the midblastula transition in Xenopus are regulated by changes in the cell cycle. Cell. 1987;48:399–407. doi: 10.1016/0092-8674(87)90191-7. [DOI] [PubMed] [Google Scholar]
- Kirschner M, Newport J, Gerhart J. The timing of early developmental events in Xenopus. Trends Genet. 1985;1:41–47. [Google Scholar]
- Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Langer-Safer P, Levine M, Ward D. Immunological method for mapping genes on Drosophila polytene chromosomes. Proc Natl Acad Sci USA. 1982;79:4381–4385. doi: 10.1073/pnas.79.14.4381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehner CF, Stick R, Eppenberger HM, Nigg EA. Differential expression of nuclear lamin proteins during chicken development. J Cell Biol. 1987;105:577–587. doi: 10.1083/jcb.105.1.577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohka MJ, Maller JL. Induction of nuclear envelope breakdown, chromosome condensation and spindle formation in cell free extracts. J Cell Biol. 1985;101:518–523. doi: 10.1083/jcb.101.2.518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohka MJ, Hayes MK, Maller JL. Purification of maturation-promoting factor, an intracellular regulator of early mitotic events. Proc Natl Acad Sci USA. 1988;85:3009–3013. doi: 10.1073/pnas.85.9.3009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maniatis T, Fritsch EF, Sambrook J. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor, New York: Cold Spring Harbor Laboratory; 1982. [Google Scholar]
- Masui Y, Markert CL. Cytoplasmic control of nuclear behavior during meiotic maturation of frog oocytes. J Exp Zool. 1971;177:129–146. doi: 10.1002/jez.1401770202. [DOI] [PubMed] [Google Scholar]
- Maxam AM, Gilbert W. A new method for sequencing DNA. Proc Natl Acad Sci USA. 1977;74:560–564. doi: 10.1073/pnas.74.2.560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miake-Lye R, Kirschner MW. Induction of early mitotic events in a cell-free system. Cell. 1985;41:165–175. doi: 10.1016/0092-8674(85)90071-6. [DOI] [PubMed] [Google Scholar]
- Mitchison TJ, Sedat J. Localization of antigenic determinants in whole Drosophila embryos. Dev Biol. 1983;99:261–264. doi: 10.1016/0012-1606(83)90275-0. [DOI] [PubMed] [Google Scholar]
- Murray AW. Cyclins in meiosis and mitosis. Nature. 1987;326:542–543. doi: 10.1038/326542a0. [DOI] [PubMed] [Google Scholar]
- Norrander J, Kempe T, Messing J. Construction of improved M13 vectors using oligonucleotide directed mutagenesis. Gene. 1983;26:101–106. doi: 10.1016/0378-1119(83)90040-9. [DOI] [PubMed] [Google Scholar]
- O’Connell P, Rosbash M. Sequence, structure, and codon preference of the Drosophila ribosomal protein 49 gene. Nucl Acids Res. 1984;12:5495–5513. doi: 10.1093/nar/12.13.5495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picard A, Peaucellier G, Le Bouffant F, Le Peuch C, Doree M. Role of protein synthesis and proteases in production and inactivation of maturation-promoting activity during meiotic maturation of starfish oocytes. Dev Biol. 1985;109:311–320. doi: 10.1016/0012-1606(85)90458-0. [DOI] [PubMed] [Google Scholar]
- Pines J, Hunt T. Molecular cloning and characterization of the mRNA for cyclin from sea urchin eggs. EMBO J. 1987;6:2987–2995. doi: 10.1002/j.1460-2075.1987.tb02604.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratan RR, Maxfield FR, Shelanski ML. Long-lasting and rapid calcium changes during mitosis. J Cell Biol. 1988;107:993–999. doi: 10.1083/jcb.107.3.993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rüther U, Müller-Hill B. Easy identification of cDNA Clones. EMBO J. 1983;2:1791–1794. doi: 10.1002/j.1460-2075.1983.tb01659.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schollmeyer JE. Calpain II involvement in mitosis. Science. 1988;240:911–913. doi: 10.1126/science.2834825. [DOI] [PubMed] [Google Scholar]
- Shoji-Kasai Y, Senshu M, Iwashita S, Imahori K. Thiol protease-specific inhibitor E-64 arrests human epidermoid carcinoma A431 cells at mitotic metaphase. Proc Natl Acad Sci USA. 1988;85:146–150. doi: 10.1073/pnas.85.1.146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solomon M, Booher R, Kirschner M, Beach D. Cyclin in fission yeast. Cell. 1988;54:738–739. [Google Scholar]
- Standart N, Minshull J, Pines J, Hunt T. Cyclin synthesis, modification and destruction during meiotic maturation of the starfish oocyte. Dev Biol. 1987;124:248–258. doi: 10.1016/0012-1606(87)90476-3. [DOI] [PubMed] [Google Scholar]
- Studier FW, Moffatt BA. Use of bacteriophage T7 RNA polymerase to direct selective high-level expression of cloned genes. J Mol Biol. 1986;189:113–130. doi: 10.1016/0022-2836(86)90385-2. [DOI] [PubMed] [Google Scholar]
- Swenson KI, Farrell KM, Ruderman JV. The clam embryo protein cyclin A induces entry into M phase and the resumption of meiosis in Xenopus oocytes. Cell. 1986;47:861–870. doi: 10.1016/0092-8674(86)90801-9. [DOI] [PubMed] [Google Scholar]
- Westendorf JM, Swenson KI, Ruderman JV. The role of cyclin B in meiosis I. J Cell Biol. 1989 doi: 10.1083/jcb.108.4.1431. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]