Summary
We have defined a coordinate program of transcription of S-phase genes (DNA polymerase α, PCNA and the two ribonucleotide reductase subunits) that can be induced by the G1 cyclin, cyclin E. In Drosophila embryos, this program drives an intricate spatial and temporal pattern of gene expression that perfectly parallels the embryonic program of S-phase control. This dynamic pattern of expression is not disrupted by a mutation, string, that blocks the cell cycle. Thus, the transcriptional program is not a secondary consequence of cell cycle progression. We suggest that developmental signals control this transcriptional program and that its activation either directly or indirectly drives transition from G1 to S phase in the stereotyped embryonic pattern.
Keywords: Drosophila, G1-S phase, cell cycle, transcription, endoreduplication, cyclin E
Introduction
The cell cycle is not a relentless oscillator. It is regulated by a myriad of inputs such as cell size, nutritional status and, importantly, signals from other cells (O'Farrell, 1992; Pardee, 1989). Dramatic cell cycle regulation occurs during embryogenesis where precise and stereotyped programs of cell division guide orderly morphogenesis (Foe et al., 1993). In Drosophila, the mother supplies the egg with a plentiful reservoir of cell cycle components so that the earliest cycles are relatively unconstrained and lack the gap phases, G1 and G2. However, the early rapid divisions are soon curtailed as constraints are added. The constraints appear one at a time as developmental events remove or inhibit activities that were previously constitutive. Progress of subsequent cycles depends on reprovision of the limiting component. This reprovision is rigidly controlled by the developmental program (O'Farrell et al., 1989).
The first major constraint appears with the introduction of a G2 phase in cycle 14 (Edgar and O'Farrell, 1990). Like G2 of the fission yeast, Schizosaccharomyces pombe (Gould and Nurse, 1989; Lundgren et al., 1991; Moreno et al., 1990), this G2 is regulated by inhibitory phosphorylation of Cdc2 kinase. After the first 13 rapid cell cycles, destruction of the maternal supplies of a phosphatase allows inactivation of the Cdc2 kinase by inhibitory phosphorylation in cycle 14 (Edgar and O'Farrell, 1989; Edgar et al., 1994). Mitosis then awaits new expression of the phosphatase and reactivation of Cdc2 kinase. Expression of the phosphatase, which is encoded by the string gene (Edgar and O'Farrell, 1989; Gautier et al., 1991), is regulated in an intricate spatiotemporal program that guides the spatiotemporal program of mitoses throughout the embryo (Edgar and O'Farrell, 1990; Foe, 1989). But all regulation of the cell cycle does not occur at the transition from G2 to mitosis. After three string regulated cycles (14, 15 and 16), many of the cells acquire a G1 phase in cycle 17 (Edgar and O'Farrell, 1990). Thereafter, the length of the G1 phase is developmentally controlled in a stereotyped pattern. We have sought to define cell cycle events associated with the appearance of G1 and its subsequent developmental programming.
Numerous regulatory signals, including the mating pheromones of Saccharomyces cerevisiae and growth factors of mammalian cells, influence a decision whether to arrest the cell cycle prior to S phase (Pardee, 1989; Chang and Herskowitz, 1990; Ewen et al., 1993; Koff et al., 1993). These various signals appear to be integrated prior to the activation of a G1 regulatory decision which commits the cell to progress into S phase and divide (Pringle and Hartwell, 1981; Pardee, 1989). A transcriptional program that drives expression of a number of replication functions such as ribonucleotide reductase (RNR) accompanies the transition to S phase in diverse organisms (Andrews and Herskowitz, 1990; Lowndes et al., 1992b; Nevins, 1992).
We show here that transcription of four S phase genes that are constitutively expressed during the early cycles is extinguished upon appearance of a G1. Subsequent expression of each gene occurs in an identical, complex spatiotemporal pattern that is precisely coincident with the onset of S phase. This transcription program can be provoked by the G1 cyclin, cyclin E. We suggest that this coordinate expression of S phase genes is analogous to the START program of transcriptional control described in yeast and that, in Drosophila embryos, the program is developmentally regulated.
Materials and Methods
Fly stocks
Wild-type embryos and DNA were of the Oregon R strain. stg7B69/TM3 Sb and stgAR2/TM3 Sb (a transcription null allele; Bruce Edgar, personal communication) stocks were used for the analysis of homozygous embryos lacking string function. Mutant embryos were unambiguously identified from their altered morphology and from the reduced cell number as determined by staining nuclei with the DNA-binding dye Hoechst 33258. All embryos were fixed in a 1:1 mixture of heptane:7% formaldehyde/PBS for 25 minutes and devitellinized. Immunostained embryos were mounted for microscopy in Fluoromount G (Fisher).
Probes used for whole-mount in situ hybridization analysis
All probe sequences were obtained by PCR amplification of genomic DNA prepared from adult Oregon R flies. A degenerate 5′ DmRNR1 primer (5′AAGGTACCTA(T/C)TT(T/C)GGNTT(T/C)AA(A/G)ACN(C/T)TNGA3′) was derived from the conserved RNR large subunit amino acid sequence YFGFKTLE. A degenerate 3′ primer (5′-CCGAATTCCAT(A/G)AANA(A/G)(A/G)TCNGGNATCCA3′) was derived from the conserved RNR large subunit sequence WIPDLF. (N=A/C/T/G) The 5′ and 3′ primers for DmRNR2 were 5′AAGGTACCGA(T/C)GGNAT(T/C/A)GTNAA(T/C)GA(A/G)AA3′ and 5′CCGAATTC(C/T)TT(C/T)TC(A/G)AA(A/G)AA(A/G)TTNGT3′, derived from conserved RNR small subunit amino acid sequences DGIVNEN and TNFFEK, respectively. EcoRI + KpnI-digested PCR products were cloned into pBLUESCRIPT SKII–. PCNA probe sequences encompassing the entire open reading frame including the single intron (Yamaguchi et al., 1990) were obtained using the primers 5′CGGAATTCATGTTCGAGGCACGCCTGGGTCAA3′ (5′) and 5′CGTCTAGATTATGTCTCGTTGTCCTCGATCTT3′ (3′). DNA POLa probe sequences encompassing 900 bp of exon 2 (Hirose et al., 1991) were obtained using the primers 5′CGTCTAGAGGTTATGCAGAAGATCTTCGG3′ (5′) and 5′GCGAATTCTACTGGATCCTCATAGGCCTC3′ (3′). EcoRI + XbaI-digested PCNA and POLa PCR products were cloned into pBLUESCRIPT SKII–.
In situ hybridizations using digoxigenin-labeled DNA or RNA probes were performed as described (Tautz and Pfeifle, 1989; Lehner and O'Farrell, 1990b) except that xylene treatment was omitted. Digoxigenin-labeled DNA was made by either of two methods; random priming (Lehner and O'Farrell, 1990b) or inclusion of Dig-dUTP in PCR amplification of cloned DNA using the above primers. The final nucleotide concentrations in these reactions were 100 μM dATP, 100 μM dCTP, 100 μM dGTP, 60 μM dTTP, and 40 μM Dig-dUTP (Boehringer). Dig-PCR products were digested with HaeIII and AluI before hybridization to facilitate probe penetration into fixed embryos. Digoxigenin-labeled RNA (Boehringer kit) was made with 350 μM Dig-UTP, 650 μM TTP, and 1 mM each of ATP, CTP and GTP in T3 or T7 RNA polymerase transcription reactions using linearized pBLUESCRIPT clones as templates. RNA-RNA hybridizations were performed at 70°C in pH 4.5 hybridization solution (Lehner and O'Farrell, 1990b). Embryos were photographed using a Nikon 20× objective and Kodak Ektachrome 160T or Technical Pan film.
Detection of replicating nuclei
Dechorionated embryos were permeabilized with octane and pulse labeled with 1 mg/ml BrdU (Sigma) for 15-30 minutes in Schneider's medium and immediately fixed as described above. Permeabilization and BrdU detection was performed according to Edgar and O'Farrell (1990). Mouse anti-BrdU monoclonal antibody was purchase from Becton Dickinson. Texas Red- or rhodamine-conjugated goat anti-mouse secondary antibodies were from Jackson. For histochemistry, a biotin-conjugated donkey anti-mouse secondary (Jackson) and streptavidin conjugated to horse radish peroxidase (Vector) were used. Conjugates were detected by incubation in 0.5 mg/ml 3,3′-diaminobenzidine (DAB), 0.08% NiCl2 and 0.003% H2O2. Simultaneous BrdU labeling and in situ hybridization was performed exactly as in Richardson et al. (1993) except that incorporated BrdU was detected with fluorescently labeled secondary antibodies.
Heat-shock experiments
Overnight collections of embryos from hsp70-cyclin E/CyO or hsp70-cyclin E chromosome two homozygous stocks were heat shocked for 30 minutes by floating the small grape juice/agar egg collection plates on the surface of a 37°C water bath. After a 30-60 minute recovery period at room temperature (23°C), the embryos were prepared for in situ hybridization or labeled with BrdU for 15-45 minutes and immediately fixed as described above.
DNA sequencing and polytene chromosome in situ hybridization
Both strands of the cloned DmRNR1 and DmRNR2 PCR products were sequenced using nested oligonucleotide primers. Digoxigenin-labeled DmRNR1 and DmRNR2 probes were hybridized to salivary gland polytene chromosomes as follows. Salivary glands were dissected from late third instar larvae, fixed for 30 seconds in 45% acetic acid, transferred to a 1:2:3 mixture of lactic acid:water:acetic acid and squashed between a slide and a siliconized glass coverslip. The spread polytene chromosomes were subsequently dehydrated by soaking for 10 minutes in 95% ethanol and air drying. Dried chromosomes were denatured by rehydrating at 65°C in 2× SSC for 30 minutes and soaking in 70 mM NaOH for 3 minutes. After a brief wash in 2× SSC the denatured chromosomes were dehydrated with an ethanol series and air dried. Hybridization was performed overnight at 42°C with DIG-labelled DNA (prepared as described above) in 2× SSC, 50% deionized formamide, 12.3 mM Tris pH 7.5, 600 mM NaCl, 5× Denhardt's, 1 mM EDTA, 30 μg/ml salmon sperm DNA and 10% dextran sulfate. Hybridized chromosomes were washed 3× 20 minutes in 2× SSC at 53°C followed by 3× 5 minutes in PBS. After a 1 hour incubation with anti-DIG antibodies coupled to horse radish peroxidase (Boehringer) followed by three 5 minute PBS washes, hybrids were detected by incubation in 0.5 mg/ml DAB, 0.01% H2O2, 0.08% NiCl2. Polytene bands were visualized by Giemsa staining and chromosomes were mounted in Permount (Sigma).
Results
Developmental programming of G1
Visualization of mitotic spindles by tubulin staining and immunological detection of S-phase nuclei labeled with 5-bromodeoxyuridine (BrdU) have provided a detailed description of the cell cycle program in early Drosophila embryos (Edgar and O'Farrell, 1990; Foe, 1989). There is no detectable lag between mitosis and S phase during the first 16 cell cycles. However, following mitosis 16, most cells enter a prolonged G1 arrest in cycle 17. Many of these G1-arrested cells are destined to polytenize their genomes. Following S phase 17, these cells enter amitotic endoreduplication cycles in which rounds of DNA replication are interrupted by periods of quiescence. Although we will focus on the populations of cells that acquire a G1 and begin endoreduplication cycles, the still rapidly proliferating cells of the nervous system (Bodmer et al., 1989) and Malpighian tubules (Skaer, 1989; Skaer and Martinez-Arias, 1992) continue to label intensely with BrdU (e.g. see Fig. 3). It has not yet been determined if these more rapidly dividing cells have a G1 phase.
Fig. 3.
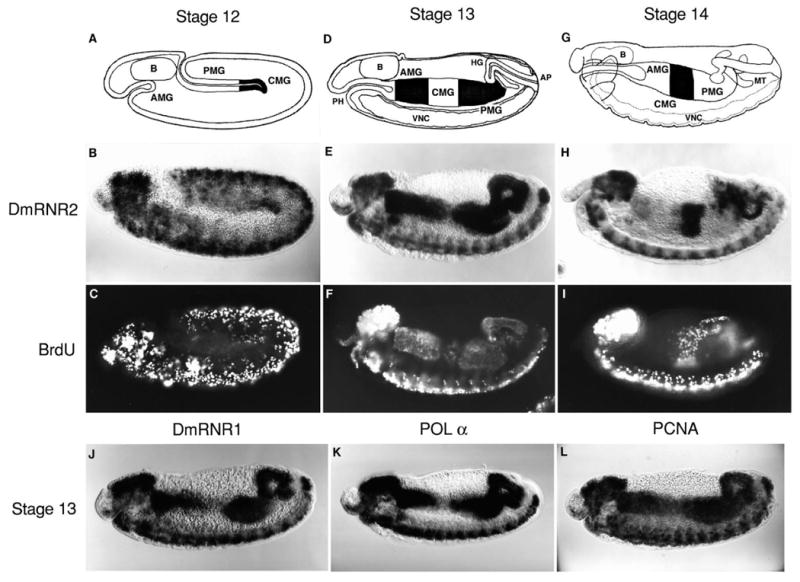
The patterned expression of S-phase genes correlates with the G1-S transition. In each panel, an embryo is displayed in lateral view with dorsal up and anterior to the left. (A) During early stage 12 of embryogenesis, the distinct anterior and posterior invaginations of the midgut primordia (AMG and PMG, respectively) remain separated while germ band retraction initiates. (B) DmRNR2 expression is extinguished in the G1-arrested epidermal cells but persists in neural tissue along the ventral midline and in the brain (B in diagram). At this stage, DmRNR2 expression can be detected in large endodermal cells at the leading edge of the migrating posterior midgut invagination (shaded region in A). These cells will occupy the central portion of the midgut (CMG) after fusion of the midgut primordia. (C) An embryo pulse labeled with BrdU at the same stage as B reveals that the cells of the CMG have entered S17. Replicating nuclei can also be detected in the nervous system. (D) By stage 13, germ band retraction is complete and the midgut primordia have fused on either side of the yolk resulting in three distinct regions of the midgut along the anterior-posterior axis: the AMG, CMG and PMG. (E) In stage 13, DmRNR2 expression has terminated in the CMG and initiated in the AMG and PMG. Several other tissues also accumulate DmRNR2 RNA at this time. Among those visible in this embryo are the hindgut (HG), the anal pads (AP), the pharynx (PH), and the brain (B) and ventral nerve cord (VNC). (F) The S-phase pattern of a stage 13 embryo is identical to the DmRNR2 expression pattern. Clearly visible is S17 in the AMG and PMG. Note that a few CMG cells near the end of S17 have incorporated BrdU (slightly out of focus). (G) The midgut cells begin migrating dorsally and ventrally on either side of a stage 14 embryo to eventually fuse and create a tube that envelopes the yolk. (H) The DmRNR2 expression pattern has changed substantially by stage 14 (this embryo is ∼1 hour older than the embryo in E). Expression has ceased in the AMG, PMG, HG, and PH and RNA reaccumulates in the CMG. Staining is also apparent in the Malpighian tubules (MT) and persists in the VNC. (I) The S-phase pattern at stage 14 changes in concert with the DmRNR2 expression pattern. The CMG has reentered S phase (S18), which is the first endoreduplication S in the midgut. The BrdU incorporation in the Malpighian tubules (MT) at this stage is due to endoreduplication S phases. The pattern of expression of DmRNR1 (J), DNA polymerase α (K), and PCNA (L) during stage 13 is identical to that of DmRNR2 (E).
S17 and the subsequent endoreduplication S phases occur in a stereotyped pattern (Smith and Orr-Weaver, 1991). The salient features of this pattern can be illustrated in the developing midgut. Three regions of the midgut, the anterior midgut (AMG), the central midgut (CMG) and the posterior midgut (PMG), initiate S phase at different times and thus define distinct ‘replication domains’ (see diagrams in Fig. 3). The CMG enters S17 early during germband retraction (stage 12; stages according to Campos-Ortega and Hartenstein, 1985), and is nearly completed by the time the AMG and PMG enter S17 (stage 13). Following S17 of the AMG and PMG, the CMG again enters S phase (stage 14), an endoreduplication S phase (S18). Although less well characterized, this alternating pattern of midgut S phases continues at least for one more cycle.
We define a program of coordinate transcription of four S-phase genes whose regulation parallels the three principle features of the S-phase program: (i) the initial appearance of a G1 in cycle 17, (ii) the arrest of some cells (e.g. the epidermal cells) in G1 throughout the remainder of embryogenesis and, most importantly, (iii) the re-entry of a number of the G1-arrested cells into S phase according to a complex developmental schedule.
Cloning of the Drosophila ribonucleotide reductase genes
Of the numerous S-phase genes whose expression is coupled to entry into S phase, only ribonucleotide reductase (RNR) has shown this coupling in all species examined, from bacteria to mammals (Bjorklund et al., 1990; Elledge and Davis, 1990; Standart et al., 1985). Active RNR is a tetramer composed of two large and two small subunits encoded by separate genes (Reichard, 1993). Degenerate primers were designed from highly conserved regions of each RNR subunit (see Materials and methods). PCR amplification of Drosophila genomic DNA yielded unique fragments representing each of the two genes. One product, a ∼500 bp fragment (DmRNR1), has an open reading frame whose sequence is 43% identical to the expected region of the large subunit of eukaryotic RNRs (Fig. 1A). A second product, a ∼650 bp fragment (DmRNR2), has an open reading frame whose sequence is 51% identical to the expected region of the small subunit of eukaryotic RNRs (Fig. 1B). Hybridization to genomic blots and salivary gland polytene chromosomes indicated that DmRNR1 and DmRNR2 are single copy genes mapping to 31D and 48D, respectively (data not shown).
Fig. 1.
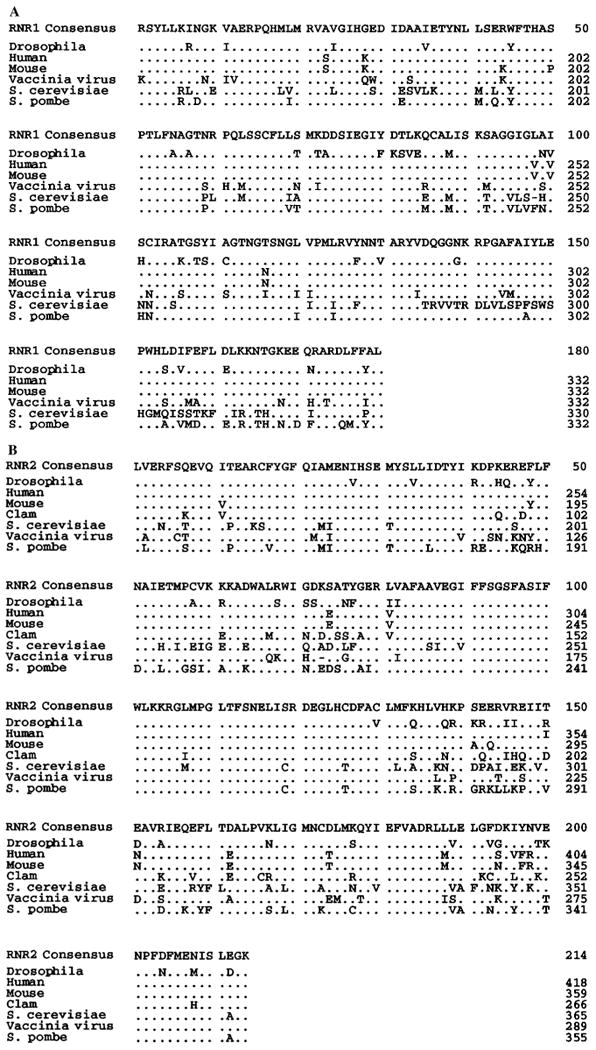
Predicted amino acid sequence of the DmRNR1 and DmRNR2 cloned PCR products with comparison to other eukaryotic and viral ribonucleotide reductases. Only sequence between and not including the PCR primer sites is shown. The alignments were performed using Geneworks. ‘Consensus’ amino acids appear in ≥2 of the sequences. Dots indicate identities with the consensus. Dashes indicate gaps introduced in a sequence to maximize the alignment. (A) RNR large subunit (RNR1) alignment. The S. cerevisiae genome contains two large subunit genes, RNR1 and RNR3 (Elledge and Davis, 1990), and sequence from the latter is shown in this alignment. (B) RNR small subunit (RNR2) alignment.
Oscillations in DmRNR2 RNA mark the onset of G1 cycles
In situ hybridization of whole embryos revealed the spatiotemporal pattern of DmRNR2 RNA accumulation during embryogenesis. A strong signal detected prior to initiation of zygotic transcription indicates that there is a maternal supply of the DmRNR2 message (Fig. 2A). The maternal RNA declines steadily during cycles 10-13 to near background levels early in cycle 14 (blastoderm stage; Fig. 2B). About 30 minutes into cycle 14 (mid cellularization) zygotic transcription drives reaccumulation of DmRNR2 RNA. Initial accumulation of DmRNR2 RNA appears faintly striped along the anterior-posterior axis, perhaps suggesting that the promoter is sensitive to regulators of early pattern formation (data not shown). Through the remainder of cycle 14, all of cycle 15 (data not shown) and into early cycle 16 (Fig. 2E) DmRNR2 RNA is ubiquitous and nearly uniform. The faint patterns evident at these stages reflect only quantitative differences in accumulation of transcript in different cells, and these differences bare no apparent relation to cell cycle events.
Fig. 2.
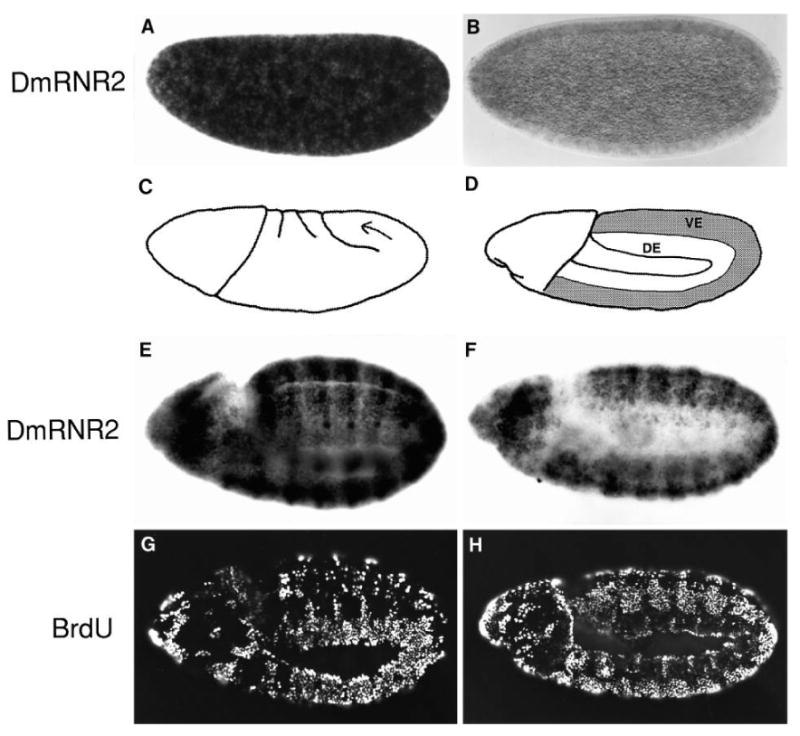
Embryonic expression of DmRNR2 prior to the appearance of G1-S control. DmRNR2 mRNA was detected by whole embryo in situ hybridizations using a digoxigenin-labeled probe. (A) A preblastoderm embryo with high levels of maternal DmRNR2 message. (B) The maternal message nearly disappears by blastoderm cellularization during early cycle 14. (C) Gastrulation begins shortly after cellularization is complete. Invagination of cells along the ventral midline creates a three-layered germ band. The arrow indicates the movement of cells of the germ band around the posterior tip of the embryo during germ band extension. (D) In a fully germ band extended embryo, the ventral epidermis (VE) extends around to the dorsal side of the embryo as well (shaded region), while the dorsal epidermis (DE) is located laterally. The embryos in E-H are at the germ band extended stage. They have been pulse labeled with BrdU and then hybridized with a DmRNR2 probe prior to immunofluorescent detection of incorporated BrdU (G and H). (E) The constitutive and ubiquitous levels of DmRNR2 that characterize the postblastoderm divisions are beginning to decline in the dorsal epidermis. This same embryo viewed with fluorescent illumination is shown in G. The heavily labeled nuclei in the dorsal epidermis are in early S16, while the unlabeled cells in the ventral epidermis have just completed S15. (F) DmRNR2 expression in a slightly older embryo. Note that DmRNR2 message is significantly reduced in the dorsal epidermis, while the level remains high in ventral epidermis. (H) Fluorescence view of the embryo in F shows the S-phase pattern. The ventral epidermal cells have finished G2 and mitosis of cycle 15 and entered S16 without an intervening G1. Cells in the dorsal epidermis have completed or nearly completed S16 and will enter G1 after progressing through a short G2 and mitosis. In each panel, dorsal is to the top and anterior is to the left. The embryo in E and G is rotated slightly on its side relative to the embryo in F and H such that the ventral midline is apparent.
Double label experiments reveal that DmRNR2 RNA begins to disappear as cells complete cycle 15. Although the elaborate spatial and temporal program of cell cycles 15 and 16 have not been characterized throughout the embryo, the timing of these cell cycles within the dorsal epidermis is relatively simple, and S phases 15 and 16 can be recognized by the pattern of labeling in precisely staged embryos (Foe, 1989; Edgar and O'Farrell, 1990; Lehner and O'Farrell, 1990a; Hartenstein and Campos-Ortega, 1985). BrdU labeling shows that dorsal epidermis of the embryo in Fig. 2G is in S phase 16, and hybridization with DmRNR2 probe shows that the RNA has begun to decline (Fig. 2E). In a slightly older embryo, the cells of the dorsal epidermis have completed S phase 16, and are in G2 (Fig. 2H and data not shown). DmRNR2 RNA is no longer detectable in these cells (Fig. 2F). Ventral epidermal cells enter S phase 16 later (Edgar and O'Farrell, 1990; also compare Fig. 2G and H) and lose DmRNR2 RNA correspondingly later (Fig. 2E and F). A strong signal is retained in rapidly proliferating tissues, such as the nervous system (Fig. 3B,E,H). Thus, DmRNR2 RNA declines towards the end of S phase 16 in those cells destined to arrest in G1 in the next cell cycle.
In contrast to early ubiquitous expression, the levels of DmRNR2 RNA are patterned from late cycle 16 onward. DmRNR2 RNA is undetectable in G1-arrested cells (e.g. see WT embryo in Fig. 5), except for brief bursts of accumulation in cells that re-enter S phase. The bursts of DmRNR2 expression occur in a spatiotemporal pattern that precisely parallels the pattern of S phases in these tissues. For example, DmRNR2 RNA signal seen in the CMG early in stage 12 embryos, corresponds to the early S phase 17 characteristic of this region of the midgut (Fig. 3A-C). In an older embryo (stage 13), the central region of the midgut has no signal while the AMG and PMG have a strong signal, corresponding to the later S17 in these regions (Fig. 3D-F). During stage 14, the CMG enters its first endoreduplication S phase (S18), and the signals reverse again: the CMG has a strong signal and the flanking AMG and PMG are again unstained (Fig. 3G-I). This correlation of patterns of DmRNR2 RNA expression and BrdU incorporation holds throughout the embryo. Thus, onset of a G1 phase is anticipated by extinction of DmRNR2 expression, and there is pulse of RNA expression associated with each subsequent S phase.
Fig. 5.
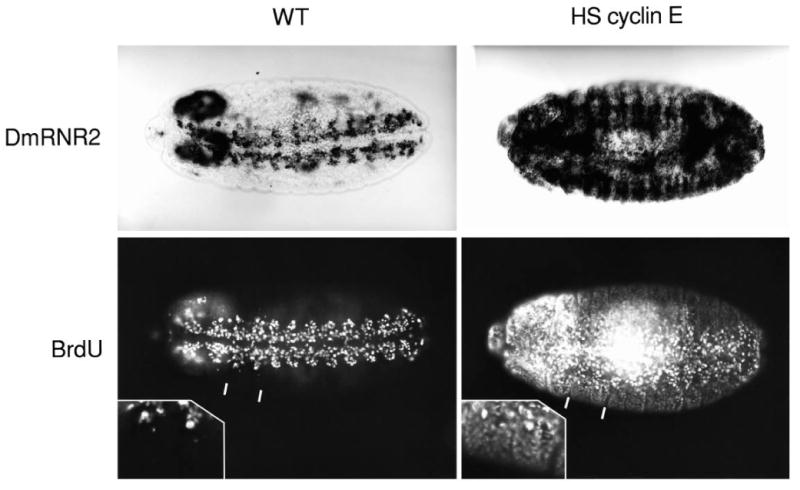
Ectopic production of Drosophila cyclin E induces the G1-S transcriptional program and drives G1 cells into S phase. All panels show a ventral view of a stage 14 embryo either stained for DmRNR2 expression or BrdU incorporation. The VNC is clearly visible in the wild-type (WT) embryos. Note that the G1-arrested epidermal cells on either side of the VNC neither incorporate BrdU nor express DmRNR2. Embryos homozygous for a hsp70-cyclin E transgene (HS cyclin E) were heat shocked at 37°C for 30 minutes. After a subsequent 45 minute recovery period at room temperature, they were either fixed and stained for DmRNR2 expression or pulse labeled with BrdU. The insets represent enlargements of the two segments included between the white markings in the BrdU-labeled embryos.
The relative timing of the pulse of DmRNR2 expression and DNA synthesis is illustrated by the cells of the anal pads located at the end of the retracting germ band. To verify that these cells are in G1 during the early stages of germ band shortening, we showed that they fail to incorporate BrdU (i.e. they are not in S phase; Fig. 4C, see inset), and that they fail to stain with antibodies to cyclin A or cyclin B, indicating recent destruction of these proteins in mitosis 16 (data not shown). Early during this G1 phase there is no DmRNR2 signal (Fig. 4B). In slightly older embryos that have not yet begun incorporating BrdU into these cells (Fig. 4F) a DmRNR2 RNA signal is detected (Fig. 4E). DmRNR2 RNA remains at high levels during early S phase after the completion of germ band retraction (Fig. 4H,I). Thus, the burst of expression precedes and overlaps S phase. Expression declines before the completion of S phase (data for the anal pads are not shown, but see CMG in Fig. 3E,F).
Fig. 4.
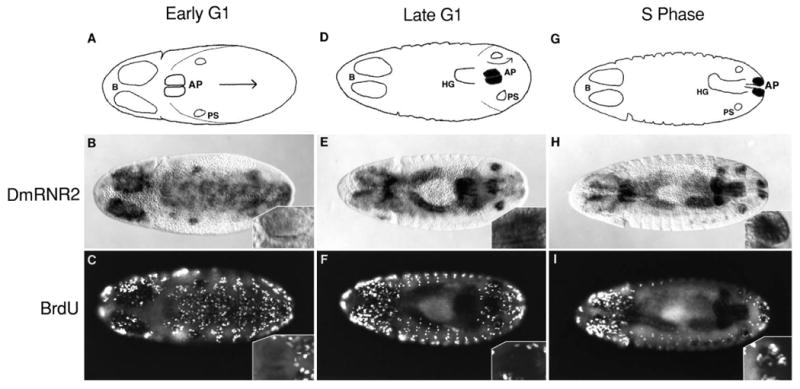
DmRNR2 message accumulation occurs late in G1 prior to the initiation of DNA replication. Embryos were pulse labeled with BrdU and then hybridized with a DmRNR2 probe prior to immunofluorescent detection of incorporated BrdU. All embryos are viewed from the dorsal perspective and anterior is to the left. The schematics indicate the changing position of the anal pads (AP), which occupy the tip of the germ band. In stage 12, the germ band retracts along the same pathway as when it extended. The arrow indicates the direction of cell movement during this process. PS indicates the bilaterally symmetric groups of cells that will form the posterior spiracles, which are already in S phase and express DmRNR2 in the embryos shown. The B in the schematics indicates the two lobes of the brain. In the leftmost three panels (A-C), the cells of the AP are in early G1, and they neither express DmRNR2 (B) nor incorporate BrdU (C). The insets show a close-up view of the right (towards the top) anal pad. In the central panels (D-F), the AP cells are in late G1. At this stage, segmentation is obvious and retraction of the germ band (75%) has revealed the hindgut (HG). The movements of the germ band (arrow in D) are such that the AP will eventually appear more posterior than the PS. The AP now expresses DmRNR2 RNA (E, and indicated by shading in the diagrams), but still fails to incorporate BrdU (F). In the rightmost three panels (G-I), the cells of the AP are in S phase. At this stage, germ band retraction is complete, placing the AP at the end of the hindgut tube. DmRNR2 RNA is still detected (H, and indicated by the shading in G). The fluorescence signal from BrdU labeling of the AP cells is partially quenched by the histochemical staining of the DmRNR2 RNA, but it is nonetheless evident that these cells are in S phase (I, see inset). Note that most of the numerous isolated cells that incorporate BrdU and stain in C, F and I are rapidly dividing cells of the peripheral and central nervous systems.
Together these data reveal three important features of the dynamic DmRNR2 expression pattern. First, early zygotic transcription is constitutive in cell cycles that lack a G1 phase. Second, DmRNR2 expression ceases prior to, and is thus not a consequence of, entry into the first G1. Third, re-accumulation of DmRNR2 RNA occurs shortly before DNA synthesis begins, presumably in response to signals promoting the onset of S phase.
A coordinate program of G1-S transcriptional control
The START transcriptional program of S. cerevisiae includes a number of genes encoding enzymes involved in DNA replication (Andrews and Herskowitz, 1990). To test if this is also the case in Drosophila, we examined expression of the other subunit of ribonucleotide reductase, DmRNR1, as well as DNA polymerase α(Hirose et al., 1991) and the DNA polymerase δ accessory subunit, PCNA (Yamaguchi et al., 1990). Each of these genes has a spatiotemporal program of expression that is identical to that of DmRNR2, except for minor differences in the extent of the decline in maternal signal prior to zygotic cycle 14 expression (data not shown). Fig. 3E,J,K,L illustrates the patterns of expression observed in stage 13 embryos and provides a striking example of the parallel transcriptional activation of these genes. This coincidence holds for the entirety of the detailed program of G1 to S control at least through stage 16 (the last stage we have examined). The coordination of transcription of DmRNR1, DmRNR2, PCNA and POLα presumably involves common transactivation mechanisms, since these genes map to widely separated sites: 31D, 48D, 56EF and 93E, respectively (this work and Yamaguchi et al., 1990; Melov et al., 1992).
Cyclin E will activate the G1-S transcriptional program
G1 cyclins activate the START transcriptional program of S. cerevisiae (Dirick et al., 1992; Marini and Reed, 1992; Nasmyth and Dirick, 1991; Ogas et al., 1991). Several observations suggested that cyclin E might play an analogous role in metazoans (Cao et al., 1992; Hinds et al., 1992; Lees et al., 1992). Since induction of cyclin E expression in Drosophila can drive G1 cells into S phase (H. Richardson and R. Saint, personal communication; Knoblich et al., 1994), we tested whether it would activate transcription of the S-phase genes. Embryos were collected from a stock carrying a homozygous cyclin E construct under the control of a heat-shock promoter (provided by H. Richardson and R. Saint). A 30 minute heat shock (37°C) resulted in induction of S phase in cells arrested in G1 as revealed by BrdU incorporation into the epidermis of a stage 14 embryo (Fig. 5). Parallel heat shocks of embryos lacking the cyclin E transgene showed no alteration in S-phase pattern (Fig. 5 WT panels). Heat-shock-induced cyclin E expression also activated widespread expression of DmRNR1, DmRNR2, PCNA and Pol α (shown for DmRNR2 in Fig. 5). This activation includes tissues that are normally silent for the remainder of embryogenesis after entry into G1 (e.g. see the epidermis in Fig. 5). However, the induced expression is not uniform and the responsiveness of given tissues changes at different times. Heat treatment of embryos collected from a stock heterozygous for the cyclin E transgene caused more pronounced patterns (not shown), suggesting that the degree of the response is sensitive to the dosage of cyclin E. Apparently, the ability of cyclin E to activate the G1-S transcriptional program is modulated by additional factors that we have not yet characterized.
The time course of the cyclin E response is consistent with an involvement of the transcription program in progress to S phase. In stage 14 and older embryos, short BrdU pulses (15 minutes) begin detecting ectopic S phase between 45 and 60 minutes after the heat-shock treatment. In contrast, ectopic DmRNR2 RNA accumulation has already occurred by 45 minutes after heat shock (not shown). Therefore, the transcription program may be required to restore sufficient levels of S-phase functions after prolonged G1 arrest (see Discussion).
Developmental signals rather than cell cycle progress activates the G1-S transcriptional program
To examine whether the transcriptional program that we have characterized is only a secondary outcome of cell cycle progress toward S phase, we tested whether the program operates following cell cycle arrest. The cell cycle of string mutant embryos is blocked in G2 of cycle 14 (Edgar and O'Farrell, 1989). The patterns of expression revealed by probes for DmRNR2 (Fig. 6) and PCNA (not shown) were virtually identical in wild-type and string mutant embryos. Constitutive early expression terminates normally, and subsequent bursts of expression mimic the wild-type pattern (cf. Fig. 6A with Fig. 3E and Fig. 6C with Fig. 3H). Note that S17 is blocked in the string mutant (Fig. 6B), while the programmed transcription of DmRNR2 is unabated (Fig. 6A). Thus, the program of G1- to S-phase transcription operates independently of cell cycle events. Apparently, developmental controls trigger this program at the appropriate times and in the appropriate cells.
Fig. 6.
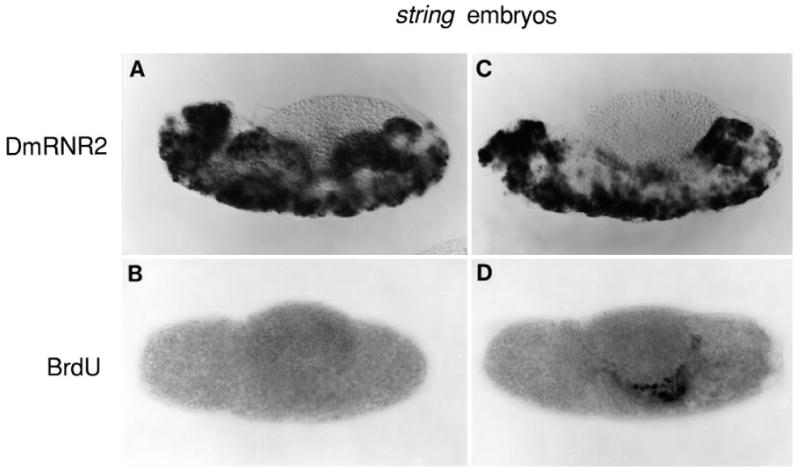
The G1-S transcriptional program operates independently of cell cycle progress. Expression of DmRNR2 (A,C) and BrdU incorporation (B,D) are shown in approximately stage 13 (A,B) and stage 14 (C,D) string mutant embryos. See Fig. 3 panels E, F, H and I for analogous staining of wild-type embryos (note that BrdU incorporation was detected histochemically in this experiment but immunofluorescently in the experiment shown in Fig. 3). In stage 13 string embryos, DmRNR2 is expressed in a near normal pattern (A) despite the lack of detectable DNA replication (B). In stage 14 string embryos, DmRNR2 is also expressed in a near normal pattern (C), and DNA replication is confined to a few cells (note the large nuclei in the center of the midgut in D). These replicating cells are apparently those that would undergo an endoreduplication S phase at this stage (see text).
A developmental program that overcomes the block to re-replication
Endoreduplicating cells must bypass the usual requirement of S phase for a preceding mitosis. Since S phase 18 is the first S phase that occurs without a preceding mitosis, we expect the bypass to be induced in those cells undergoing this round of DNA replication. This bypass was detected in our analysis of DNA synthesis in string mutant embryos. As expected, BrdU labeling failed to detect DNA synthesis in string embryos at the times of normal S phase 15 or 16 (not shown). Similarly, despite the accurate timing of the bursts of DmRNR2 expression, string null mutant embryos exhibited no BrdU incorporation in the S phase 17 pattern (Fig. 6B). However, BrdU incorporation was detected in string embryos at the time and position of the first endoreduplication S phase (S18 in the CMG, Fig. 6D). Note that, while our results are not unlike those of Smith and Orr-Weaver (1991), we discriminate between S17 (which follows mitosis) and S18 (an endoreduplication S phase). Only those S phases that normally occur without a preceding mitosis occur in a string mutant. Thus, an ability to enter S phase without passing through mitosis (checkpoint bypass) is acquired in particular cells (those exhibiting endoreduplication) just before they progress to S phase 18. The observed renewal of replication in a string mutant suggests that an event leading to checkpoint bypass is programmed by developmental controls independent of cell cycle processes.
Discussion
We have shown that coordinate transcription of genes involved in DNA replication occurs prior to the G1- to S-phase transition in Drosophila embryos. This transcriptional program is governed by developmental signals, as expected if its regulation were a means of developmental control of cell cycle progress.
Introduction of developmental constraints on the cell cycle
Development begins with abundant supplies of maternally provided cell cycle components and few cell cycle constraints. Constraints are added by changes that make previously constitutive components rate limiting. The introduction of a delay between mitosis and S phase (a G1 phase) at cycle 17 requires introduction of a constraint that prevents immediate progress from mitosis to S phase. Reduction in one or more activities required for DNA replication could provide such a constraint. It is our premise that it is a change in gene expression, from constitutive to regulated, that introduces a G1 phase into the cell cycle and that regulates its length.
In diverse species, transcriptional activation of a number of genes accompanies entry into S phase. Our analysis of the expression of the small subunit of ribonucleotide reductase, DmRNR2, uncovered such a G1 to S phase program of transcription in Drosophila embryos. DmRNR2 RNA is expressed constitutively prior to cycle 17, extinguished in anticipation of appearance of a G1, and re-expressed in abrupt bursts at subsequent transitions from G1 to S phase.
Ribonucleotide reductase is, perhaps, an unlikely candidate for the key regulator of the G1- to S-phase transition. Indeed, it is a marker of a larger regulatory program rather than the focus of it. In addition to DmRNR2, we have examined three other genes encoding products involved in DNA synthesis, DmRNR1, polymerase α and PCNA. Expression of each of these genes, but not other genes less directly involved in DNA synthesis such as topoisomerase II (R. J. D and P. H. O'F., unpublished observations), parallels that of DmRNR2. In addition, cyclin E appears to be similarly regulated (Knoblich et al., 1994; R. J. D and P. H. O'F., unpublished observations). Thus, a coordinate program of G1- to S-phase transcription drives expression of at least five genes. Upon introduction of a G1, we suggest that one or more of the products under the control of the coordinate G1- to S-phase transcriptional program becomes limiting. But which if any of the regulated genes limits progress to S phase?
In S. cerevisiae, the products of all CDC genes, except for CDC4, are sufficient for more than one cell cycle, and overexpression of Cdc4 provides a supply sufficient for several cell cycles without disturbing the cycle (Breck Byers, personal communication). This suggests that periodic expression of these genes is not crucial to the cell cycle. In contrast, periodic expression of the G1 cyclins encoded by the CLN genes appears to necessary for the S. cerevisiae cell cycle (Nasmyth and Dirick, 1991; Ogas et al., 1991), and constitutive expression disturbs the cycle and drives premature S phase (Hadwiger et al., 1989). Similarly, in Drosophila Cyclin E mutant embryos S phase is blocked once maternal cyclin E stores are depleted (Knoblich et al. 1994), and heat-shock induction of cyclin E can induce S phase in G1-arrested cells (H. Richardson and R. Saint, personal communication; Knoblick et al., 1994; and the present work). Thus, cyclin E limits progress to S phase, but it is not clear whether it acts directly.
Our work shows that cyclin E expression induces a coordinate program of gene expression of S-phase genes. This introduces, but does not resolve the question of whether or not this transcriptional induction is instrumental in driving progress from G1 to S phase. Analysis of mutations in the induced genes (e.g. RNR2) will determine whether new expression of these genes is required for the progress to S phase. Additionally, analysis of RNR expression in cyclin E mutant embryos will reveal whether or not the transcriptional program requires cyclin E as might be suggested by the induction of this program by heat-shock-induced expression of cyclin E. Although we must await these experiments to resolve these issues, analogies with yeast suggest that the essential role of the S-phase transcriptional program is to induce G1 cyclins, and that the remainder of the transcriptional program is of secondary importance (Koch et al., 1993).
Cyclin E and an S-phase trigger
Entry into S phase is a decisive event for a cell because it usually represents a commitment to progress through an entire cycle and to divide. The control system governing this commitment step ought to generate an unambiguous on/off signal. In S. cerevisiae, this type of signalling is accomplished via a positive feedback loop. The S. cerevisiae G1 cyclins, Cln1 and Cln2, stimulate the Cdc28 kinase to activate two heteromeric transcription factors that each include a Swi6 subunit (Swi4/Swi6 and Mbp1/Swi6) (Andrews and Herskowitz, 1989; Dirick et al., 1992; Koch et al., 1993; Lowndes et al., 1992a; Primig et al., 1992; Verma et al., 1992). These factors drive the program of G1-S transcription which includes expression of CLN1 and CLN2. Activation of this self re-enforcing transcriptional loop appears to be the pivotal event in the START decision (Cross and Tinkelenberg, 1991; Dirick and Nasmyth, 1991; Koch et al., 1993; Nasmyth and Dirick, 1991; Ogas et al., 1991).
G1 cyclins also play an important role in advancing animal cells to S phase (Baldin et al., 1993; Ohtsubo and Roberts, 1993; Sherr, 1993), and increasing evidence suggests that G1 cyclins are involved in transcriptional activation. For instance, cyclin E complexes with the cdk2 kinase (Dulic et al., 1992; Koff et al., 1992), promoting phosphorylation of the tumor suppressor gene product Rb (Hinds et al., 1992). Phosphorylation of Rb is thought to induce transcription by disrupting the inhibitory interaction of Rb with E2F-type transcription factors (Cao et al., 1992; Chellappan et al., 1991; Devoto et al., 1992; Mudryj et al., 1991; Shirodkar et al., 1992; Weintraub et al., 1992; Zamanian and La Thangue, 1993; Zhu et al., 1993). Complexes containing E2F have been implicated in transcriptional regulation of replication functions at G1-S (Blake and Azizkhan, 1989; Mudryj et al., 1990; Pearson et al., 1991; Slansky et al., 1993). Furthermore, overexpression of transcriptionally active forms of E2F can drive quiescent mammalian cells into S phase (Johnson et al., 1993).
Consistent with the suggestions from mammalian systems, cyclin E in Drosophila induces both S phase and the G1-S transcriptional program. If the transcriptional program includes activation of cyclin E expression, then a switch-like positive feedback loop analogous to S. cerevisiae may drive the G1-S transition in Drosophila embryos. While no analog of Rb has yet been identified in flies, other components of the presumed positive feedback loop have been defined. Drosophila Cdc2c appears to be the analog of the mammalian cdk2 (Lehner and O'Farrell, 1990a; Stern et al., 1993; and C. Lehner personal communication) and a Drosophila homolog of E2F has recently been cloned (Dynlacht et al., 1994). Thus, based on our demonstration that cyclin E induces a transcriptional program, together with the analogies to yeast and mammals, we suggest that cyclin E induces a positive feedback loop to turn on its own expression and consequently progress to S phase.
While G1 cyclins might play the major role in the G1-S transition, evolutionary conservation suggests that the broader program of coordinate expression of replication functions is also important. We suggest that new expression of genes such as RNR and DNA polymerase adopts increasing importance during growth arrest. Early in a G1 arrest only the more unstable gene products (perhaps only cyclin E) will have decayed to a point requiring renewed expression for entry into S phase. However, with increasing time during a G1 arrest the more stable gene products will decline, eventually falling below a threshold required for S phase. Consequently, re-entry into S phase from a prolonged G1 would require activation of the full coordinate program of gene expression.
Developmental programming of G1
Whatever the mechanistic basis of the G1- to S-phase transition, the timing of the transition is regulated during development. Orderly morphogenesis is associated with rigid developmental programs and stereotyped times of mitosis and S phase in each tissue (O'Farrell, 1992; O'Farrell et al., 1989). While developmental inputs control earlier cycles at the G2 to M transition, once a G1 appears in cycle 17, its length is regulated, and the stereotyped pattern of endoreduplication S phases that follow must be controlled by developmental inputs governing entry into S phase.
The G1 to S transcriptional program might represent part of the signaling system that drives the programmed S phases, or it might be coupled to and a consequence of transit through a step in the cell cycle. To address this, we tested the influence of cell cycle arrest on the G1- to S-phase transcriptional program. In embryos mutant for string, the cell cycle arrests in G2 of cycle 14, but development continues with few defects beyond those attributable to the shortage of cells (Edgar and O'Farrell, 1989; Gould et al., 1990; Hartenstein and Posakony, 1990). Analysis of the expression of genes involved in embryonic pattern formation, such as Ubx, show a fairly normal spatiotemporal program (Gould et al., 1990). Similarly, analysis of the expression of DmRNR2 and PCNA revealed that these genes also followed their normal spatiotemporal program of expression. For example, at the time that S phase 17 would ordinarily occur, string embryos exhibit the normal pattern of pulses of RNR2 expression without an associated S phase. The failure of the string mutation to disrupt the G1- to S-phase transcriptional program demonstrates that it is not driven by cell cycle events, and the detailed spatiotemporal pattern of this program indicates that it is controlled by developmental signals. We suggest that this transcriptional program is coupled to the developmental signal that triggers reactivation of the cycle in G1 cells.
Modifying the cycle
In a typical cell cycle, DNA replication follows mitosis. Indeed, work in several systems has suggested that a ubiquitous checkpoint mechanism prevents re-entry into S phase until completion of mitosis, and thus prevents re-replication (Hartwell and Weinert, 1989; Murray, 1992; Li and Deshaies, 1993). In Drosophila such a dependency of DNA synthesis on mitosis also seems to operate, as suggested by the finding that string, which is required for entry into mitosis, prevents S phases after the mitotic block (Edgar and O'Farrell, 1989, 1990).
Following S phase 17, many of the larval cells evade this checkpoint and re-replicate their genomes in a series of ‘endocycles’. These endocycles generate polytene nuclei, such as the well-known salivary gland nuclei, which have an especially high degree of polytenization. To initiate endocycles, the checkpoint must be bypassed. We have detected this ‘checkpoint bypass’ by examining DNA replication in string mutants that are blocked in G2 of cycle 14. Such G2-arrested mutants cannot initiate S phase until the checkpoint is bypassed. At the time and positions at which the first endocycles S phases begin, replication is again detected in the mutant. Thus, the spatial and temporal program of checkpoint bypass occurs independent of cycle progression. We suggest that developmental programs direct the spatial and temporal pattern of expression of a ‘checkpoint bypass gene’, that directs this modification of the cell cycle (e.g. see Moreno and Nurse, 1994).
Initiation of each endocycle S phase is associated with a pulse of expression of DmRNR2 RNA as well as expression of the other genes of the G1- to S-phase transcriptional program. These endocycle S phases also require cyclin E function (Knoblich et al., 1994). Thus, it seems likely that initiation of endocycle S phases is controlled in manner similar to a typical G1 to S transition, and that the checkpoint bypass is only a permissive modification of the cycle that removes a constraint on the normal progress to S phase without modifying the mechanism that triggers this progress.
Acknowledgments
We would like to thank Helena Richardson and Rob Saint for the gracious provision of the hsp70-cyclin E stock and Christian Lehner for communication of results prior to publication. We also thank Bruce Edgar for valuable discussions at the initiation of this work and all the members of the O'Farrell laboratory for helpful comments on the manuscript. This work was supported by the Cancer Research Fund of the Damon Runyon-Walter Winchell Foundation (DRG-1161 to R. J. D.) and NIH-PO1 HL 43821 to P. H. O.
References
- Andrews BJ, Herskowitz I. Identification of a DNA binding factor involved in cell-cycle control of the yeast HO gene. Cell. 1989;57:21–29. doi: 10.1016/0092-8674(89)90168-2. [DOI] [PubMed] [Google Scholar]
- Andrews BJ, Herskowitz I. Regulation of cell cycle-dependent gene expression in yeast. J Biol Chem. 1990;265:14057–14060. [PubMed] [Google Scholar]
- Baldin V, Lukas J, Marcote MJ, Pagano M, Draetta G. Cyclin D1 is a nuclear protein required for cell cycle progression in G1. Genes Dev. 1993;7:812–821. doi: 10.1101/gad.7.5.812. [DOI] [PubMed] [Google Scholar]
- Bjorklund S, Skog S, Tribukait B, Thelander L. S-phase-specific expression of mammalian ribonucleotide reductase R1 and R2 subunit mRNAs. Biochemistry. 1990;29:5452–5458. doi: 10.1021/bi00475a007. [DOI] [PubMed] [Google Scholar]
- Blake MC, Azizkhan JC. Transcription factor E2F is required for efficient expression of the hamster dihydrofolate reductase gene in vitro and in vivo. Mol Cell Biol. 1989;9:4994–5002. doi: 10.1128/mcb.9.11.4994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodmer R, Carretto R, Jan YN. Neurogenesis of the peripheral nervous system in Drosophila embryos, DNA replication patterns and cell lineages. Neuron. 1989;3:21–32. doi: 10.1016/0896-6273(89)90112-8. [DOI] [PubMed] [Google Scholar]
- Campos-Ortega JA, Hartenstein V. The Embryonic Development of Drosophila melanogaster. Berlin: Springer-Verlag; 1985. [Google Scholar]
- Cao L, Faha B, Dembski M, Tsai LH, Harlow E, Dyson N. Independent binding of the retinoblastoma protein and p107 to the transcription factor E2F. Nature. 1992;355:176–179. doi: 10.1038/355176a0. [DOI] [PubMed] [Google Scholar]
- Chang F, Herskowitz I. Identification of a gene necessary for cell cycle arrest by a negative growth factor of yeast, FAR1 is an inhibitor of a G1 cyclin, CLN2. Cell. 1990;63:999–1011. doi: 10.1016/0092-8674(90)90503-7. [DOI] [PubMed] [Google Scholar]
- Chellappan SP, Hiebert S, Mudryj M, Horowitz JM, Nevins JR. The E2F transcription factor is a cellular target for the RB protein. Cell. 1991;65:1053–1061. doi: 10.1016/0092-8674(91)90557-f. [DOI] [PubMed] [Google Scholar]
- Cross FR, Tinkelenberg AH. A Potential Positive Feedback Loop Controlling CLN1 and CLN2 Gene Expression at the Start of the Yeast Cell Cycle. Cell. 1991;65:875–883. doi: 10.1016/0092-8674(91)90394-e. [DOI] [PubMed] [Google Scholar]
- Devoto SH, Mudryj M, Pines J, Hunter T, Nevins JR. A cyclin A-protein kinase complex possesses sequence-specific DNA binding activity, p33cdk2 is a component of the E2F-cyclin A complex. Cell. 1992;68:167–176. doi: 10.1016/0092-8674(92)90215-x. [DOI] [PubMed] [Google Scholar]
- Dirick L, Moll T, Auer H, Nasmyth K. A central role for SWI6 in modulating cell cycle Start-specific transcription in yeast. Nature. 1992;357:508–513. doi: 10.1038/357508a0. [DOI] [PubMed] [Google Scholar]
- Dirick L, Nasmyth K. Positive feedback in the activation of G1 cyclins in yeast. Nature. 1991;351:754–757. doi: 10.1038/351754a0. [DOI] [PubMed] [Google Scholar]
- Dulic V, Lees E, Reed SI. Association of human cyclin E with a periodic G1-S phase protein kinase. Science. 1992;257:1958–1961. doi: 10.1126/science.1329201. [DOI] [PubMed] [Google Scholar]
- Dynlacht BD, Brook A, Dembski M, Yenush L, Dyson N. DNA-binding and trans-activation properties of Drosophila E2F and DP proteins. Proc Natl Acad Sci USA. 1994 doi: 10.1073/pnas.91.14.6359. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar BA, O'Farrell PH. Genetic control of cell division patterns in the Drosophila embryo. Cell. 1989;57:177–187. doi: 10.1016/0092-8674(89)90183-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar BA, O'Farrell PH. The three postblastoderm cell cycles of Drosophila embryogenesis are regulated in G2 by string. Cell. 1990;62:469–480. doi: 10.1016/0092-8674(90)90012-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar BA, Sprenger F, Duronio RJ, Leopold P, O'Farrell P. Distinct molecular mechanisms regulate the cell cycle at successive stages of Drosophila embryogenesis. Genes Dev. 1994 doi: 10.1101/gad.8.4.440. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elledge SJ, Davis RW. Two genes differentially regulated in the cell cycle and by DNA-damaging agents encode alternative regulatory subunits of ribonucleotide reductase. Genes Dev. 1990;4:740–751. doi: 10.1101/gad.4.5.740. [DOI] [PubMed] [Google Scholar]
- Ewen ME, Sluss HK, Whitehouse LL, Livingston DM. TGFβ Inhibition of Cdk4 Synthesis is Linked to Cell Cycle Arrest. Cell. 1993;74:1009–1020. doi: 10.1016/0092-8674(93)90723-4. [DOI] [PubMed] [Google Scholar]
- Foe VE. Mitotic domains reveal early commitment of cells in Drosophila embryos. Development. 1989;107:1–22. [PubMed] [Google Scholar]
- Foe VE, Odell GM, Edgar BA. Mitosis and morphogenesis in the Drosophila embryo, Point and counterpoint. In: Bate M, Martinez Arias A, editors. The Development of Drosophila melanogaster. Vol. 1. Cold Spring Harbor, New York: Cold Spring Harbor Laboratory Press; 1993. pp. 149–300. [Google Scholar]
- Gautier J, Solomon MJ, Booher RN, Bazan JF, Kirschner MW. cdc25 is a specific tyrosine phosphatase that directly activates p34cdc2. Cell. 1991;67:197–211. doi: 10.1016/0092-8674(91)90583-k. [DOI] [PubMed] [Google Scholar]
- Gould AP, Lai RY, Green MJ, White RA. Blocking cell division does not remove the requirement for Polycomb function in Drosophila embryogenesis. Development. 1990;110:1319–1325. doi: 10.1242/dev.110.4.1319. [DOI] [PubMed] [Google Scholar]
- Gould KL, Nurse P. Tyrosine phosphorylation of the fission yeast cdc2+ protein kinase regulates entry into mitosis. Nature. 1989;342:39–45. doi: 10.1038/342039a0. [DOI] [PubMed] [Google Scholar]
- Hadwiger JA, Wittenberg C, Richardson HE, de Barros Lopes M, Reed SI. A family of cyclin homologs that control the G1 phase in yeast. Proc Nat Acad Sci USA. 1989;86:6255–6259. doi: 10.1073/pnas.86.16.6255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartenstein V, Campos-Ortega JA. The Embryonic Development of Drosophila melanogaster. New York: Springer-Verlag; 1985. [Google Scholar]
- Hartenstein V, Posakony JW. Sensillum development in the absence of cell division, the sensillum phenotype of the Drosophila mutant string. Dev Biol. 1990;138:147–158. doi: 10.1016/0012-1606(90)90184-k. [DOI] [PubMed] [Google Scholar]
- Hartwell LH, Weinert TA. Checkpoints, controls that ensure the order of cell cycle events. Science. 1989;246:629–634. doi: 10.1126/science.2683079. [DOI] [PubMed] [Google Scholar]
- Hinds PW, Mittnacht S, Dulic V, Arnold A, Reed SI, Weinberg RA. Regulation of retinoblastoma protein functions by ectopic expression of human cyclins. Cell. 1992;70:993–1006. doi: 10.1016/0092-8674(92)90249-c. [DOI] [PubMed] [Google Scholar]
- Hirose F, Yamaguchi M, Nishida Y, Masutani M, Miyazawa H, Hanaoka F, Matsukage A. Structure and expression during development of Drosophila melanogaster gene for DNA polymerase alpha. Nucleic Acids Res. 1991;19:4991–4998. doi: 10.1093/nar/19.18.4991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson DG, Schwarz JK, Cress WD, Nevins JR. Expression of transcription factor E2F1 induces quiescent cells to enter S phase. Nature. 1993;365:349–352. doi: 10.1038/365349a0. [DOI] [PubMed] [Google Scholar]
- Knoblich JA, Sauer K, Jones L, Richardson H, Saint R, Lehner CF. Cyclin E controls S-phase progression and its down-regulation during Drosophila embryogenesis is required for the arrest of cell proliferation. Cell. 1994 doi: 10.1016/0092-8674(94)90239-9. in press. [DOI] [PubMed] [Google Scholar]
- Koch C, Moll T, Neuberg M, Ahorn H, Nasmyth K. A role for the transcription factors Mbp1 and Swi4 in progression from G1 to S phase. Science. 1993;261:1551–1557. doi: 10.1126/science.8372350. [DOI] [PubMed] [Google Scholar]
- Koff A, Giordano A, Desai D, Yamashita K, Harper JW, Elledge S, Nishimoto T, Morgan DO, Franza BR, Roberts JM. Formation and activation of a cyclin E-cdk2 complex during the G1 phase of the human cell cycle. Science. 1992;257:1689–1694. doi: 10.1126/science.1388288. [DOI] [PubMed] [Google Scholar]
- Koff A, Ohtsuki M, Polyak K, Roberts JM, Massague J. Negative regulation of G1 in mammalian cells, inhibition of cyclin E-dependent kinase by TGF-beta. Science. 1993;260:536–539. doi: 10.1126/science.8475385. [DOI] [PubMed] [Google Scholar]
- Lees E, Faha B, Dulic V, Reed SI, Harlow E. Cyclin E/cdk2 and cyclin A/cdk2 kinases associate with p107 and E2F in a temporally distinct manner. Genes Dev. 1992;6:1874–1885. doi: 10.1101/gad.6.10.1874. [DOI] [PubMed] [Google Scholar]
- Lehner CF, O'Farrell PH. Drosophila cdc2 homologs, a functional homolog is coexpressed with a cognate variant. EMBO J. 1990a;9:3573–3581. doi: 10.1002/j.1460-2075.1990.tb07568.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehner CF, O'Farrell PH. The roles of Drosophila cyclins A and B in mitotic control. Cell. 1990b;61:535–547. doi: 10.1016/0092-8674(90)90535-m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li JJ, Deshaies RJ. Exercising self-restraint, discouraging illicit acts of S and M in eukaryotes. Cell. 1993;74:223–226. doi: 10.1016/0092-8674(93)90413-k. [DOI] [PubMed] [Google Scholar]
- Lowndes NF, Johnson AL, Breeden L, Johnston LH. SWI6 protein is required for transcription of the periodically expressed DNA synthesis genes in budding yeast. Nature. 1992a;357:505–508. doi: 10.1038/357505a0. [DOI] [PubMed] [Google Scholar]
- Lowndes NF, McInerny CJ, Johnson AL, Fantes PA, Johnston LH. Control of DNA synthesis genes in fission yeast by the cell-cycle gene cdc10+ Nature. 1992b;355:449–453. doi: 10.1038/355449a0. [DOI] [PubMed] [Google Scholar]
- Lundgren K, Walworth N, Booher R, Dembski M, Kirschner M, Beach D. mik1 and wee1 cooperate in the inhibitory tyrosine phosphorylation of cdc2. Cell. 1991;64:1111–1122. doi: 10.1016/0092-8674(91)90266-2. [DOI] [PubMed] [Google Scholar]
- Marini NJ, Reed SI. Direct Induction of G1-Specific Transcripts Following Reactivation of the Cdc28 Kinase in the Absence of de novo Protein Synthesis. Genes Dev. 1992;6:557–567. doi: 10.1101/gad.6.4.557. [DOI] [PubMed] [Google Scholar]
- Melov S, Vaughan H, Cotterill S. Molecular characterization of the gene for the 180 subunit of the DNA polymerase-primase of Drosophila melanogaster. J Cell Sci. 1992;102:847–856. doi: 10.1242/jcs.102.4.847. [DOI] [PubMed] [Google Scholar]
- Moreno S, Nurse P, Russell P. Regulation of mitosis by cyclic accumulation of p80cdc25 mitotic inducer in fission yeast. Nature. 1990;344:549–552. doi: 10.1038/344549a0. [DOI] [PubMed] [Google Scholar]
- Moreno S, Nurse P. Regulation of progression through the G1 phase of the cell cycle by the rum1+ gene. Nature. 1994;367:236–242. doi: 10.1038/367236a0. [DOI] [PubMed] [Google Scholar]
- Mudryj M, Devoto SH, Hiebert SW, Hunter T, Pines J, Nevins JR. Cell cycle regulation of the E2F transcription factor involves an interaction with cyclin A. Cell. 1991;65:1243–1253. doi: 10.1016/0092-8674(91)90019-u. [DOI] [PubMed] [Google Scholar]
- Mudryj M, Hiebert SW, Nevins JR. A role for the adenovirus inducible E2F transcription factor in a proliferation dependent signal transduction pathway. EMBO J. 1990;9:2179–2184. doi: 10.1002/j.1460-2075.1990.tb07387.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray AW. Creative blocks, cell-cycle checkpoints and feedback controls. Nature. 1992;359:599–604. doi: 10.1038/359599a0. [DOI] [PubMed] [Google Scholar]
- Nasmyth K, Dirick L. The role of SWI4 and SWI6 in the activity of G1 cyclins in yeast. Cell. 1991;66:995–1013. doi: 10.1016/0092-8674(91)90444-4. [DOI] [PubMed] [Google Scholar]
- Nevins JR. E2F, a link between the Rb tumor suppressor protein and viral oncoproteins. Science. 1992;258:424–429. doi: 10.1126/science.1411535. [DOI] [PubMed] [Google Scholar]
- O'Farrell PH. Cell cycle control, many ways to skin a cat. Trends in Cell Biology. 1992;2:159–163. doi: 10.1016/0962-8924(92)90034-k. [DOI] [PubMed] [Google Scholar]
- O'Farrell PH, Edgar BA, Lakich D, Lehner CF. Directing cell division during development. Science. 1989;246:635–640. doi: 10.1126/science.2683080. [DOI] [PubMed] [Google Scholar]
- Ogas J, Andrew BJ, Herskowitz I. Transcriptional Activation of CLN1, CLN2, and a Putative New G1 Cyclin (HCS26) by SWI4, a Positive Regulator of G1-Specific Transcription. Cell. 1991;66:1015–1026. doi: 10.1016/0092-8674(91)90445-5. [DOI] [PubMed] [Google Scholar]
- Ohtsubo M, Roberts JM. Cyclin-dependent regulation of G1 in mammalian fibroblasts. Science. 1993;259:1908–1912. doi: 10.1126/science.8384376. [DOI] [PubMed] [Google Scholar]
- Pardee AB. G1 events and regulation of cell proliferation. Science. 1989;246:603–608. doi: 10.1126/science.2683075. [DOI] [PubMed] [Google Scholar]
- Pearson BE, Nasheuer HP, Wang TS. Human DNA polymerase alpha gene, sequences controlling expression in cycling and serum-stimulated cells. Mol Cell Biol. 1991;11:2081–2095. doi: 10.1128/mcb.11.4.2081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Primig M, Sockanathan S, Auer H, Nasmyth K. Anatomy of a transcription factor important for the start of the cell cycle in Saccharomyces cerevisiae. Nature. 1992;358:593–597. doi: 10.1038/358593a0. [DOI] [PubMed] [Google Scholar]
- Pringle JP, Hartwell LH. The Saccharomyces cerevisiae cell cycle. In: Strathern JD, Jones EW, Broach JR, editors. The Molecular Biology of the Yeast Saccharomyces. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, New York: 1981. [Google Scholar]
- Reichard P. From RNR to DNA, Why So Many Ribonucleotide Reductases? Science. 1993;260:1773–1777. doi: 10.1126/science.8511586. [DOI] [PubMed] [Google Scholar]
- Richardson HE, O'Keefe LV, Reed SI, Saint R. A Drosophila G1-specific cyclin E homolog exhibits different modes of expression during embryogenesis. Development. 1993;119:673–690. doi: 10.1242/dev.119.3.673. [DOI] [PubMed] [Google Scholar]
- Sherr CJ. Mammalian G1 cyclins. Cell. 1993;73:1059–1065. doi: 10.1016/0092-8674(93)90636-5. [DOI] [PubMed] [Google Scholar]
- Shirodkar S, Ewen M, DeCaprio JA, Morgan J, Livingston DM, Chittenden T. The transcription factor E2F interacts with the retinoblastoma product and a p107-cyclin A complex in a cell cycle-regulated manner. Cell. 1992;68:157–166. doi: 10.1016/0092-8674(92)90214-w. [DOI] [PubMed] [Google Scholar]
- Skaer H. Cell division in Malpighian tubule development in D. melanogaster is regulated by a single tip cell. Nature. 1989;342:566–569. [Google Scholar]
- Skaer H, Martinez-Arias A. The wingless product is required for cell proliferation in the Malpighian tubule anlage of Drosophila melanogaster. Development. 1992;116:745–754. [Google Scholar]
- Slansky JE, Li Y, Kaelin WG, Farnham PJ. A protein synthesis-dependent increase in E2F1 mRNA correlates with growth regulation of the dihydrofolate reductase promoter. Mol Cell Biol. 1993;13:1610–1618. doi: 10.1128/mcb.13.3.1610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith AV, Orr-Weaver TL. The regulation of the cell cycle during Drosophila embryogenesis, the transition to polyteny. Development. 1991;112:997–1008. doi: 10.1242/dev.112.4.997. [DOI] [PubMed] [Google Scholar]
- Standart NM, Bray SJ, George EL, Hunt T, Ruderman JV. The small subunit of ribonucleotide reductase is encoded by one of the most abundant translationally regulated maternal RNAs in clam and sea urchin eggs. J Cell Biol. 1985;100:1968–1976. doi: 10.1083/jcb.100.6.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stern B, Ried G, Clegg NJ, Grigliatti TA, Lehner CF. Genetic analysis of the Drosophila cdc2 homolog. Development. 1993;117:219–232. doi: 10.1242/dev.117.1.219. [DOI] [PubMed] [Google Scholar]
- Tautz D, Pfeifle C. A non-radioactive in situ hybridization method for the localization of specific RNAs in the Drosophila embryo reveals translational control of the segmentation gene hunchback. Chromosoma. 1989;98:81–85. doi: 10.1007/BF00291041. [DOI] [PubMed] [Google Scholar]
- Verma R, Smiley J, Andrews B, Campbell JL. Regulation of the yeast DNA replication genes through the Mlu I cell cycle box is dependent on SWI6. Proc Natl Acad Sci U S A. 1992;89:9479–9483. doi: 10.1073/pnas.89.20.9479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weintraub SJ, Prater CA, Dean DC. Retinoblastoma protein switches the E2F site from positive to negative element. Nature. 1992;358:259–261. doi: 10.1038/358259a0. [DOI] [PubMed] [Google Scholar]
- Yamaguchi M, Nishida Y, Moriuchi T, Hirose F, Hui CC, Suzuki Y, Matsukage A. Drosophila proliferating cell nuclear antigen (cyclin) gene, structure, expression during development, and specific binding of homeodomain proteins to its 5′-flanking region. Mol Cell Biol. 1990;10:872–879. doi: 10.1128/mcb.10.3.872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zamanian M, La Thangue NB. Transcriptional repression by the Rb-related protein p107. Mol Biol Cell. 1993;4:389–396. doi: 10.1091/mbc.4.4.389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu L, van den Heuvel S, Helin K, Fattaey A, Ewen M, Livingston D, Dyson N, Harlow E. Inhibition of cell proliferation by p107, a relative of the retinoblastoma protein. Genes Dev. 1993;7:1111–25. doi: 10.1101/gad.7.7a.1111. [DOI] [PubMed] [Google Scholar]


