Abstract
Phosphatidylinositol-specific phospholipase C (PI-PLC) enzymes simultaneously interact with the substrate, PI, and with non-substrate lipids such as phosphatidylcholine (PC). For Bacillus thuringiensis PI-PLC these interactions are synergistic with maximal catalytic activity observed at low to moderate mole fractions of PC (XPC) and maximal binding occurring at low mole fractions of anionic lipids. It has been proposed that residues in α helix B help modulate membrane binding and that dimerization on the membrane surface both increases affinity for PC and activates PI-PLC yielding the observed PI/PC synergy. Vesicle binding and activity measurements using a variety of PI-PLC mutants support many aspects of this model and reveal that while single mutations can disrupt anionic lipid binding and the anionic lipid/PC synergy, the residues important for PC binding are less localized. Interestingly, at high XPC mutations can both decrease membrane affinity and increase activity, supporting a model where reductions in wildtype activity at XPC>0.6 results from both dilution of the substrate and tight membrane binding of PI-PLC limiting enzyme hopping or scooting to the next substrate molecule. These results provide a direct analysis of vesicle binding and catalytic activity and shed light on how occupation of the activator site enhances enzymatic activity.
Keywords: peripheral membrane protein, interfacial activation, mixed phospholipid vesicles, fluorescence correlation spectroscopy
The conformation and activity of peripheral membrane proteins are often mediated by interactions with different types of phospholipids (1). Electrostatic attractions between proteins and anionic phospholipids may recruit proteins to the membrane, while interactions with these and other phospholipids can help anchor proteins to the membrane, target proteins to specific membranes or cellular compartments, and/or serve to regulate protein activity by inducing conformational changes. For phospholipases, which cleave an ester bond in target phospholipids, binding to non-substrate phospholipids both anchors the protein to the membrane and enhances phospholipase activity (2).
The numerous roles of lipid-protein interactions make it difficult to tease out how interactions with multiple types of phospholipids differentially affect protein binding, conformational changes, and activity. For example, do mutations in the active site also alter binding to non-substrate lipids? To address such questions for bacterial phosphatidylinositol-specific phospholipase C (PI-PLC), we have determined how mutations that should directly alter lipid binding or protein conformational changes affect both binding to and enzymatic activity towards multi-component lipid vesicles.
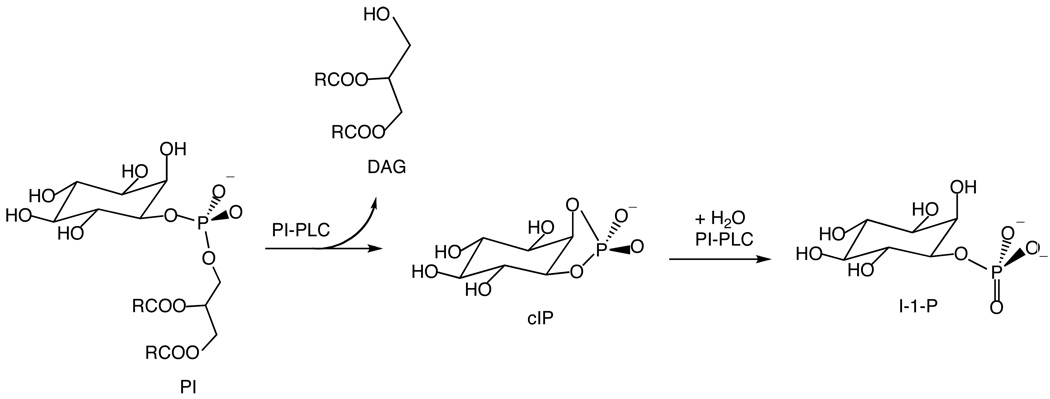
PI-PLC enzymes from Bacillus thuringiensis and related bacteria are secreted proteins that enhance bacterial virulence (3, 4), and several of these bacterial PI-PLC enzymes, including B. thuringiensis PI-PLC, are also able to release GPI anchored proteins from the surface of eukaryotic cells (5). B. thuringiensis PI-PLC catalyzes the specific cleavage of the sn-3-phosphodiester bond in phosphatidylinositol (PI) in two steps, an intramolecular phosphotransferase reaction at a phospholipid interface and a phosphodiesterase reaction where the substrate can be in the water phase (Scheme 1). Both of these steps are specifically activated by PI-PLC binding to phosphatidylcholine (PC) interfaces (6–8). This activation involves two critical tryptophan residues, Trp47 in the helix B region and Trp242 in a disordered loop shown in Figure 1 (9, 10). In addition, while wildtype B. thuringiensis and related bacterial PI-PLCs are monomeric in solution (6, 11), a B. thuringiensis PI-PLC mutant (Trp47Ala, Trp242Ala) is a homodimer suggesting that PI-PLC dimerization may play a role in PI-PLC activation (12).
Scheme 1.
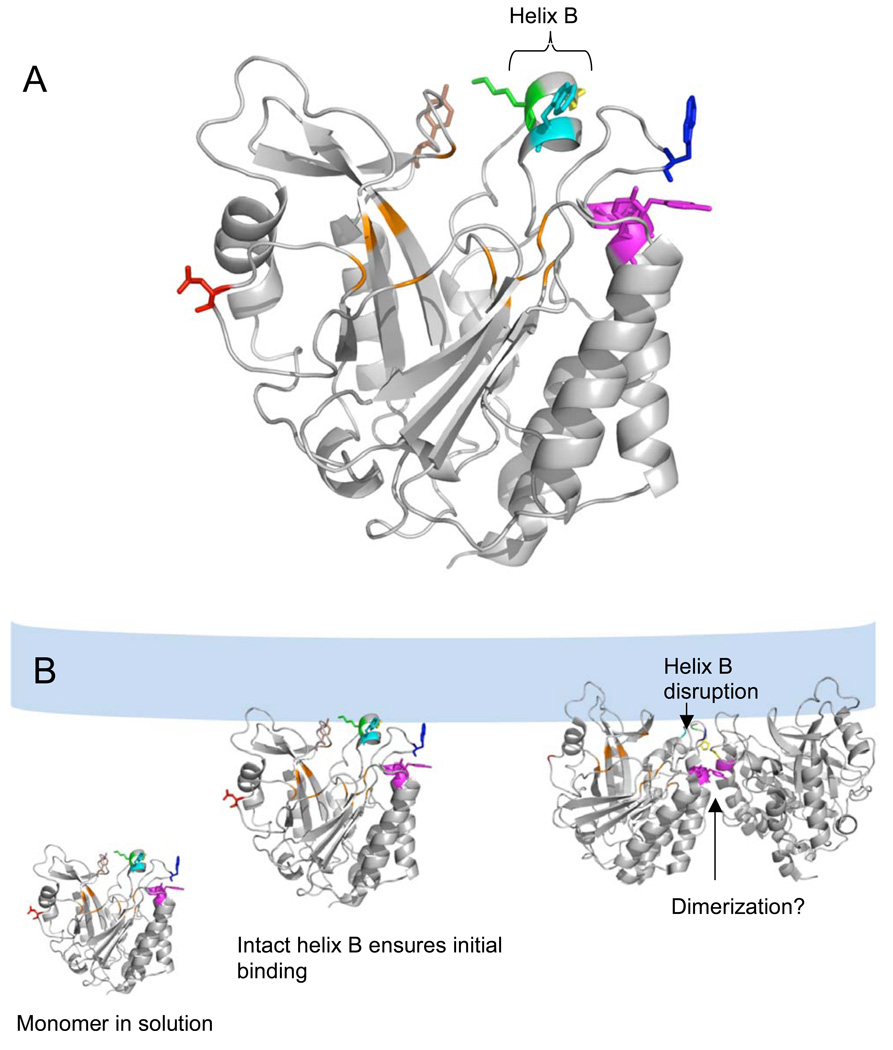
Figure 1.
(A) B. thuringiensis PI-PLC monomer structure (from the Y247S/Y251S crystal structure (14)) highlighting the active site pocket in orange and the surface residues altered in this work (Asn168, red; Pro42, yellow; Lys44, green; Tyr88, brown; Tyr246, Tyr247, Tyr248, magenta; Trp47, cyan and Trp242, blue). The short helix B contains Lys44 and Trp47 and is capped by Pro42 at the N-terminal end. (B) Proposed roles of the mutated residues in PI-PLC vesicle binding and activation (which may include dimerization). PI-PLC is a monomer in solution. After the initial Lys44 mediated attraction to the anionic membrane, an intact helix B orients Trp47 for membrane insertion. Subsequent disruption of helix B allows Pro42 to help stabilize the PI-PLC homodimer interface including a Tyr zipper involving Tyr residues 246–248. The dimer structure in this model is based on the W242A/W47A B. thuringiensis dimer crystal structure (12). The lipid bilayer is shown schematically in light blue.
The importance of helix B and the Trp residues for PI-PLC binding and activity form the basis of a recent model of PI-PLC membrane binding and activity (13). In this model (Figure 1B), membrane binding is mediated by a hydrophobic plug containing Trp47 in helix B and Trp242 on an adjacent loop as well as electrostatic interactions between Lys44 in helix B and anionic phospholipids. An intact helix B ensures that these residues are correctly oriented for membrane binding, and subsequent disruption of helix B may also allow Pro42 and Gln45 to form part of a PI-PLC homodimer interface that also includes a Tyr zipper involving Tyr residues 246–248, and 251 (12, 14). It has been proposed that such transient dimerization on the membrane surface activates PI-PLC towards both membrane bound and soluble substrates (13, 14).
Previous studies (13, 14) using micelles or pure PC small unilamellar vesicles (SUVs) revealed that mutations in the helix B region or at the putative dimer interface both weaken binding to pure PC vesicles and impair activity (Table 1). However, for wildtype PI-PLC, membrane binding and activity are enhanced for vesicle systems containing both PC and anionic lipids, with the highest activity occurring between 0.2 and 0.5 mole fraction PC (XPC) and the tightest binding around XPC= 0.9 (15). This phospholipid "synergy", where highest activity and tightest binding require the presence of both anionic phospholipids and zwitterionic PC, suggests that the anionic lipid binding site(s) and the PC binding site(s) may communicate with each other. To rigorously test the model for PI-PLC membrane binding and activation as well as to determine the differential effects of substrate and non-substrate lipids, we have used fluorescence correlation spectroscopy (FCS) to monitor binding of these PI-PLC variants to multicomponent SUVs as well as 31P NMR to monitor PI-PLC activity.
Table 1.
Relative activities (Rel. Act.),a diC7PC activation of cIP hydrolysis, apparent Kd (determined by non-FCS methods) for pure PC SUVs, and kinetic defects of B. thuringiensis PI-PLC variants extracted from published work (13–15, 24).
| PI-PLC | Rel. Act. (PI/diC7PC) | PC activation of cIP hydrolysisb | Kd (mM), PC SUVs (non-FCS) | comment / kinetic defect |
|---|---|---|---|---|
| N168C | 1.00 | 22 | 0.064±0.010 c,d | both enzyme activities are enhanced by PC binding; synergistic effect of PG/PC on Kd for SUVs determined by FCS |
| P42G | 0.70 | 5.8 | 0.77±0.30 c | PC activation of cIP hydrolysis is impaired, while PI cleavage is close to wildtype; PC binding is impaired |
| K44A | 0.98 | 33 | 0.087±0.005 d | PI cleavage in PI/diC7PC micelles and PC activation of cIP hydrolysis like wildtype; binding to PC SUVs like wildtype |
| K44E | 0.44 | 4.5 | 0.5±0.2 d | PI cleavage in PI/diC7PC micelles and PC activation of cIP hydrolysis are significantly impaired; binding to PC vesicles is weaker |
| Y88A | 2.92 | 28 | 0.94±0.05 e | PI cleavage in PI/diC7PC micelles is 2-fold higher than wildtype; PC activation of cIP hydrolysis comparable to wildtype; binding to PC SUVs is weakened |
| 3YS f | 0.36 | 2.9 | 0.5±0.2 d | PI cleavage in PI/diC7PC micelles is about 30% of wildtype; PC activation of cIP hydrolysis is significantly reduced; binding to PC SUVs is much weaker than wildtype |
Standard assay systems include: 6 (or 8) mM PI dispersed in 24 (or 32) mM diC7PC. The specific activity of wildtype enzyme is 1630 µmol min−1 mg−1 for PI/diC7PC.
The extent of PC activation of cIP hydrolysis is measured as the ratio of the specific activity, vcIP, in the presence of 5 mM diC7PC over vcIP of the enzyme towards cIP alone. For 20 mM cIP in the absence of diC7PC vcIP was typically 2.8 µmol min−1 mg−1.
Measured by intrinsic fluorescence of PI-PLC when titrated with pure SUVs (13).
Measured by filtration assay, which briefly concentrates the sample (24).
Unpublished results from S. Guo, Boston College.
Data from (14).
To avoid vesicle fusion induced by the production of diacylglycerol (DAG) by active PI-PLC variants, FCS experiments were performed using SUVs containing the substrate analogue phosphatidylglycerol (PG) and PC while activity assays were performed using PI/PC vesicles. In the PG-rich region (low XPC), helix B mutations significantly increased the apparent Kd for the vesicles, although effects on Kd in the PC-rich region (high XPC) depended on the residue modified. In contrast, mutating the Tyr residues at the putative dimer interface significantly decreased PC binding with much smaller affects on binding to substrate analogue-rich bilayers. For XPC < 0.5, the specific activity of the PI-PLC variants towards PI/PC SUVs parallels the apparent Kd measured by FCS, and, interestingly, variants with poor PC binding are still fairly active towards PI indicating the important role of substrate/activator phospholipid synergy for this catalysis. Active site mutants were also examined, and these lost the pronounced synergistic vesicle binding behavior observed for wildtype and most of the other PI-PLC variants.
Materials & Methods
Chemicals
All phospholipid stock solutions in chloroform were purchased from Avanti Polar Lipids, Inc., and used without further purification. These included the long-chain lipids 1-palmitoyl-2-oleoylphosphatidylcholine (PC), dioleoylphosphatidylglycerol (PG), and L-α-phosphatidylinositol (PI) from bovine liver. Competent cells used in mutagenesis (XL1-Blue) and overexpression (BL21-Codonplus (DE3)-RIL) were obtained from Stratagene. The fluorescent dye Alexa Fluor 488 C5 maleimide (AF488) was purchased from Invitrogen. Most of the other chemicals, including bovine serum albumin (BSA), D2O, and Triton X-100, were purchased from Sigma. Q-Sepharose fast-flow anion-exchange resin and phenyl-Sepharose hydrophobic resin were purchased from GE Healthcare, and Micro Bio-spin6 columns were obtained from Bio-Rad Laboratories.
Preparation and purification of PI-PLC variants
Recombinant B. thuringiensis PI-PLC was overexpressed in E. coli and purified as described previously (8). Protein concentrations were measured by both Lowry assays and absorption at 280 nm using an extinction coefficient of 65.32 mM−1 cm−1 calculated by the web-based ProtParam software (16). PI-PLC mutants P42G, K44A, K44E, Y88A, and Y246/247/248S were constructed previously (13, 14), Y88A was constructed by S. Guo, Boston College, and the additional mutation N168C was introduced into these plasmids using QuikChange methodology (Stratagene). The active site PI-PLC mutation, H32A, was introduced into the N168C and wildtype background respectively using Quikchange. The altered gene sequences were confirmed by DNA sequencing (Genewiz). Each protein was specifically labeled at Cys168 with the hydrophilic dye AF488 according to the manufacturer's protocol. Prior to labeling, the proteins were incubated with 5 mM DTT for 20 min. The excess DTT was removed immediately before conjugation using Bio-Spin 6 columns (Bio-Rad) and the protein was incubated with AF488. The AF488 labeled protein was separated from free AF488 using a second spin column. All variants had a labeling ratio of 100±10%, determined by comparing the absorption of the protein at 280 nm to that of the probe at 495 nm. Labeling at N168C does not affect the enzyme activity or binding properties of wildtype PI-PLC (15), and this fluorescent protein is the control for the FCS binding experiments. The fluorescently labeled mutated proteins are denoted by the residue(s) changed and an asterisk, so that labeled K44A/N168C is indicated as *K44A; the control protein, N168C, is denoted as *N168C.
PI-PLC activity assays
Specific activities of PI-PLC enzymes toward PI in SUVs were measured by 31P NMR spectroscopy (17) using a Varian INOVA 500 as described previously (8, 10). The desired amount of POPC and PI in chloroform were mixed in a glass vial, the chloroform was removed under a stream of nitrogen gas, and the resultant film lyophilized overnight. The lipid film was rehydrated with 50 mM HEPES, pH 7.5. SUVs were prepared by sonication (Branson Sonifier Cell Disrupter) of lipid mixtures for 10 min on ice. The buffer used for the reaction was 50 mM HEPES, pH 7.5, with 1 mM EDTA, 5 mM dithiothreitol and 0.1 mg/ml BSA. Three types of experiments were carried out. (i) To assess the effect of the apparent Kd for vesicles on enzymatic activity, we carried out assays with fixed XPC = 0.2 and various total phospholipid concentrations: 10 mM (0.4 ml), 4 mM (1 ml) and 0.5 mM (8 ml). Wildtype or Y88A/N168C PI-PLC (0.1 to 0.15 µg) was added to the solution and incubated at 28°C for fixed times chosen so that less than 20% of the PI was cleaved. The reaction was quenched by incubating the sample in a boiling water bath for 4 min, and the sample was then lyophilized. The lyophilized powder was rehydrated in 30% D2O, and 1–2 drops of Triton X-100 were added (to solubilize the remaining phospholipids in detergent micelles) for acquisition of the 31P NMR spectrum. (ii) For “surface dilution” experiments, the PI concentration was 10 mM and increasing amounts of PC were incorporated in the vesicles. The reaction volume was 0.5 ml containing 0.1 to 0.4 µg PI-PLC. Samples (in duplicate) were incubated at 28°C, and the reaction was quenched by adding 30 µl acetic acid. Triton X-100 (50 µl) was added to solubilize the phospholipids for analysis by 31P NMR. (iii) Another variation used a fixed concentration of total phospholipid (10 mM) with XPC varied from 0.0 to 0.9 and workup as indicated above. In all of these assays, cIP was the only product and the specific activity was calculated from the integration of the cIP resonance compared to the total PI and PC resonance. In the figures, the error bars reflect the measured 15% variation in the specific activities.
FCS
FCS based SUV binding experiments were performed using PI-PLC variants labeled at N168C and a home-built confocal setup based on an IX-70 inverted microscope (Olympus) as previously described (15, 18). In brief, FCS experiments were carried out at 22°C on 300 µL samples in PBS, pH 7.3, plus 1 mg/ml BSA to stabilize PI-PLC, in chambered coverglass wells (LabTek). Prior to use, the chambers were coated with 10 mg/ml BSA and rinsed with PBS to prevent protein adhesion to the sides of the wells. SUVs were prepared as described above. For vesicle binding experiments, 3.5 nM labeled PI-PLC was titrated with unlabeled vesicles, and the fraction bound to the vesicles, f, was determined from two species fits to the auto or cross correlation, G(τ), (19–21).
| (1) |
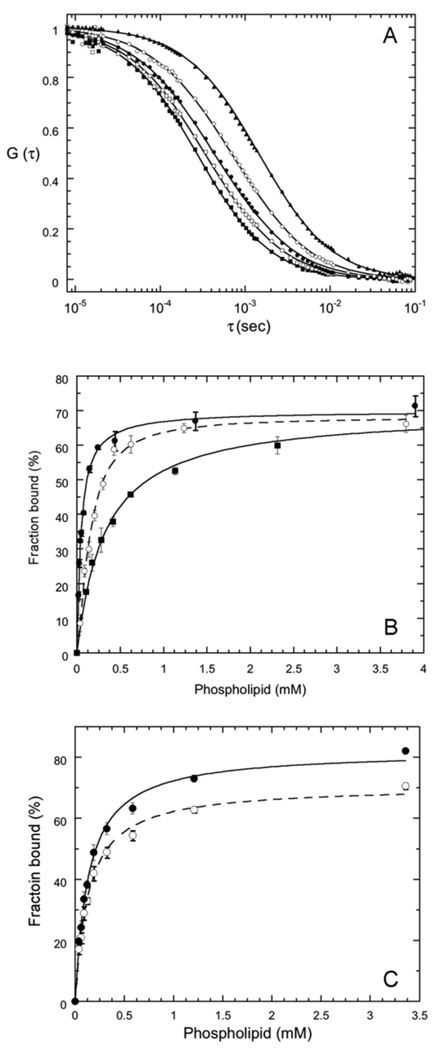
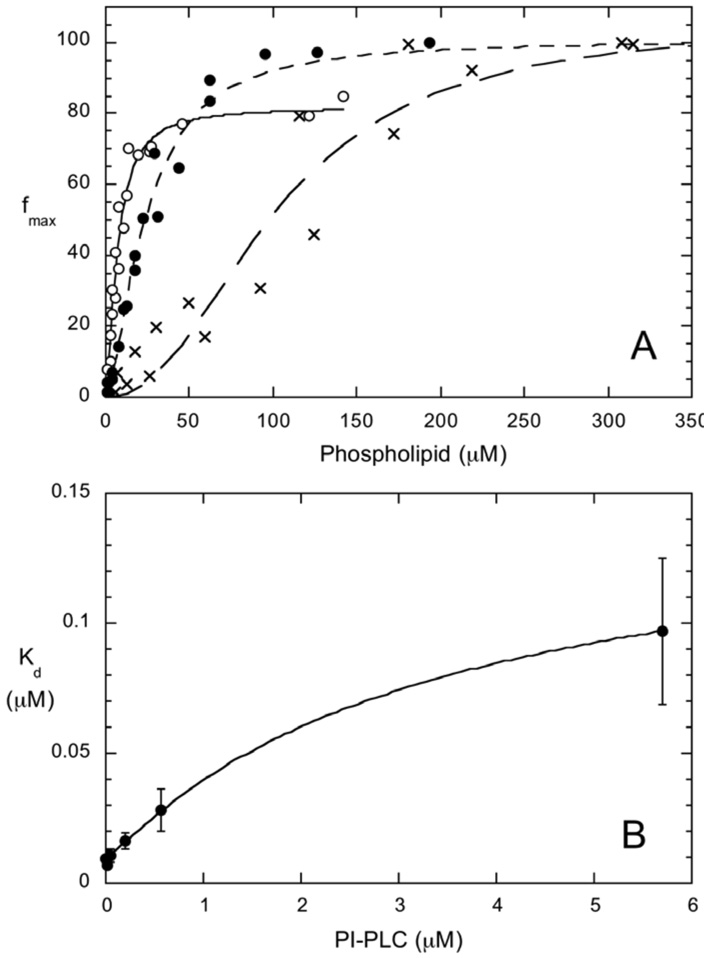
Here, <N> is the time averaged number of PI-PLC molecules in the effective volume, τfree and τbound are the diffusion times for free and vesicle-bound PI-PLC respectively, and S is the ratio of the axial (z) to the radial (x-y) dimension for the effective volume. The diffusion time is given by τspecies=ωo2/4Dspecies where Dspecies is the diffusion coefficient for each species (free or bound) and ωo is the radius of the observation volume in the x-y plane. The values of S and ωo were determined at the beginning and end of each day of experiments using the calibration dye rhodamine 110 with a reported D = 280 µm2/s (22). FCS experiments on labeled PI-PLC variants in the absence of SUVs showed that Dfree= 58±5 µm2/s for all of the variants, in agreement with previous work on *N168C (15). The values of Dfree and ωo were used to calculate τfree, which was fixed for fits to equation 1. The value of Dbound was determined from FCS experiments on vesicles containing fluorescent lipids. For fits to equation 1, τbound was either fixed to the diffusion time for free vesicles or globally floated (15). The maximum value of f, the fraction of protein bound to the vesicles, was typically 50–60%. Comparisons between independent datasets collected using different protein and phospholipid preparations for *Y88A, *H32A and *N168C, reveal that variations in the fraction bound between preparations, obtained from fits to equation 1, are approximately ±30% (Figure 2).
Figure 2.
(A) Crosscorrelation curves for free *Y88A (■), rhodamine-labeled PC SUVs (▲), and for 3.5 nM *Y88A binding to SUVs in the presence of 0.1 mM (❒), 0.4 mM (●) and 4.7 mM (❍) phospholipid, corresponding to 18%, 36% and 70% bound protein. The line through these data is the best fit to Equation 1 with Dbound =15 µm2/s. (B) Titration curves for *Y88A binding to pure PC (■), PC/PG (7:3) (●), and PC/PG (1:1) SUVs (❍). The lines through the data represent the best fits to equation 2. Error bars represent the standard deviations from three time points for the same titration. (C) Dependence of the fraction bound on the method used to fit equation 1. The fraction bound shown are for *Y88A and XPC = 0.9 either globally fitting Dbound (●) (resulting in Dbound=17 µm2/s) or fixing Dboundto 15 µm2/s (❍). The lines are hyperbolic (n=1) fits to equation 2.
The substrate, PI, was not used for FCS experiments (except for the inactive mutants, H32A) because PI cleavage by PI-PLC produces DAG leading to vesicle fusion (23). Instead, since *N168C binds PG tightly as long as some PC in present (15), PG was used as the anionic substrate analogue. Wildtype PI-PLC has very low activity towards PG cleaving the sn-3-phosphodiester bond on the time scale of days at µM protein concentrations (6); much longer times and much higher protein concentrations than used for the FCS experiments.
A single PI-PLC binding site may be formed by multiple lipids, and the apparent dissociation constant, Kd, representing the partitioning of the enzyme to the vesicle surface, as well as a cooperativity coefficient, n, were determined using the empirical equation:
| (2) |
where the fraction of PI-PLC bound, f, was determined from the FCS experiments for different total lipid concentrations, [PL] and fixed XPC, and fmzx is the apparent maximum fraction bound. For XPC>0.3, the binding curves have the expected hyperbolic shape and fits were performed with n fixed to 1. However, for XPC <0.3, plots of fversus [PL] are noticeably sigmoidal, particularly for the tighter binding PI-PLC variants, and n was a fitting parameter. This apparent cooperativity at low XPC likely results from PI-PLC's preference either for the subpopulation of vesicles with higher PC contents or for PC-rich regions in the vesicles, both of which are scarce when XPC<0.3 and the vesicle concentration is low (15). Typical hyperbolic binding curves for *Y88A are shown in Figure 2, and a typical sigmoidal binding curve is shown in the supplementary information (Figure S1).
To assess uncertainties in Kd values arising from differences in SUV preparation and/or protein purification, 2 different SUV and protein preparations were used for both the *N168C and *Y88A titration experiments. Experiments on these preparations were performed months apart. In the case of *N168C, the largest differences between preparations were observed for XPC<0.5, where the drop in Kd is steepest (Figure 3) and, in this region, Kd variations between the preparations ranged from 36 to 82% with an average variation of 61%. Higher values of XPC yielded less variation, with differences in Kd ranging from 9 to 53% with an average value of 27% for *N618C. For *Y88A, the dependence of Kd on XPC is less steep at low XPC and this is reflected in the smaller variation in Kd, 7 to 11% with an average value of 9% for XPC<0.5, while variations were similar for XPC≥0.5 ranging from 25 to 40% with an average value of 31%. For several of the other mutant enzymes specific XPC compositions were performed in duplicate to assess errors (Figure 3). Kd differences between *N168C and mutant enzymes are considered significant if they are an order of magnitude or larger, well outside any of the determined uncertainties. Kd was also determined for 3.5 nM labeled PI-PLC binding to PG/PC (1:1) SUVs in the presence of up to 0.20 mg/ml (5.7 µM) unlabeled PI-PLC as a way of estimating the maximum amount of protein bound to these vesicles.
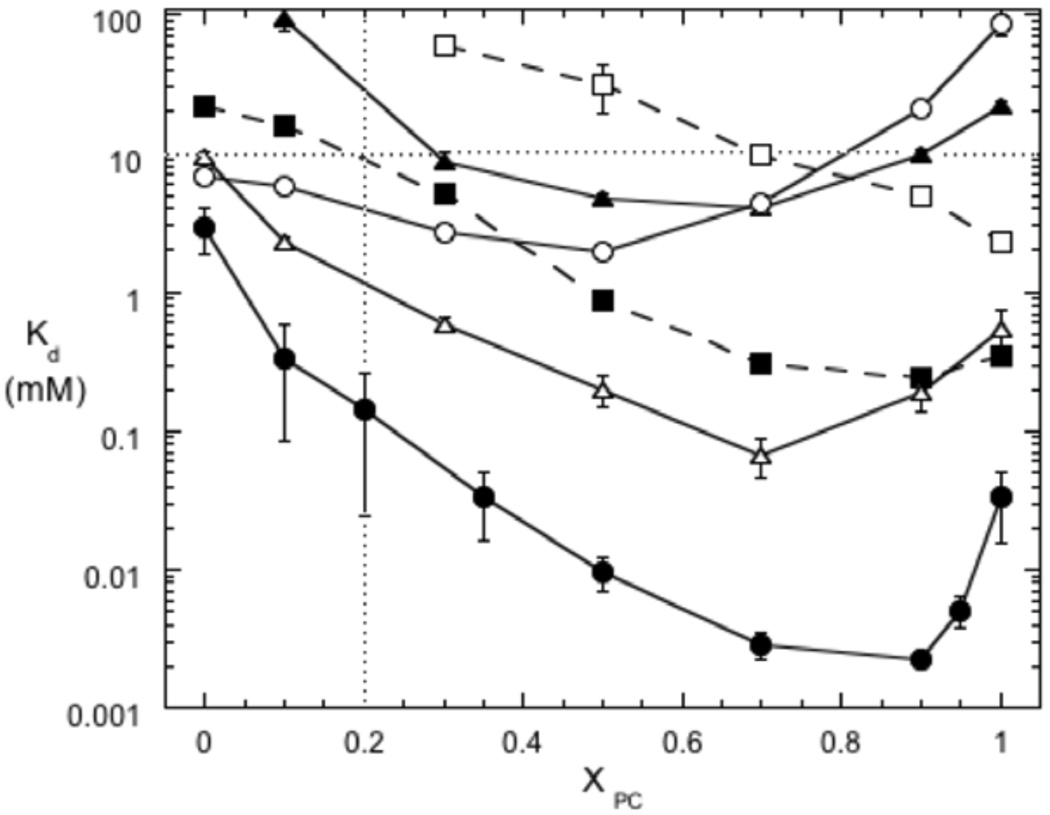
Figure 3.
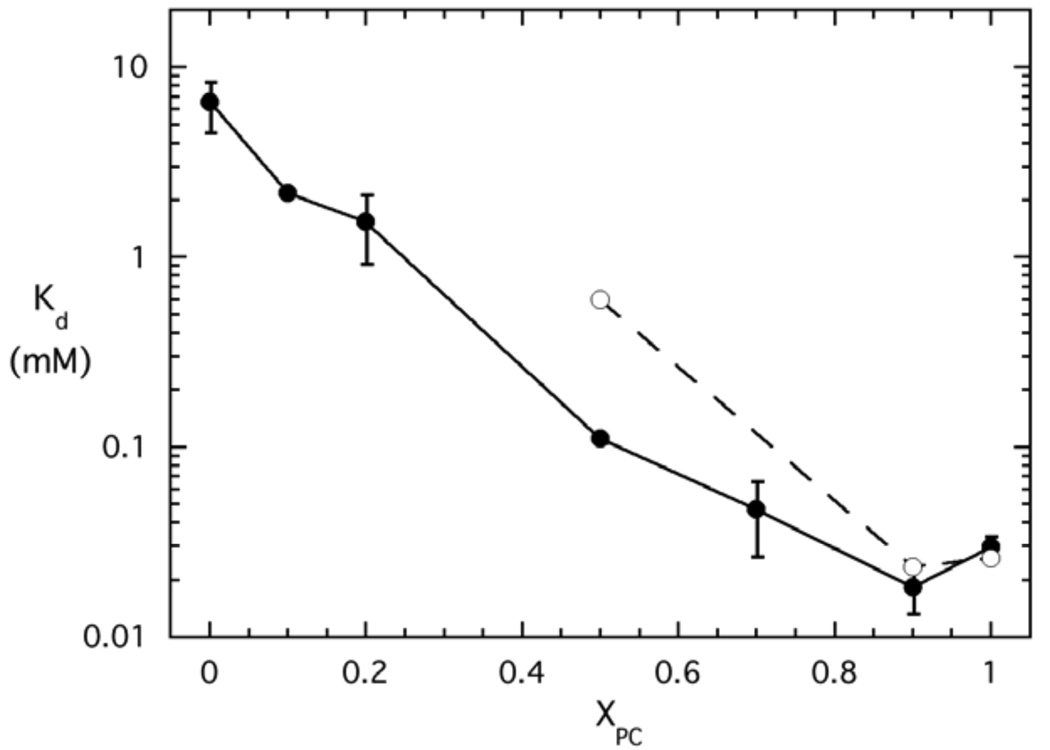
Binding of PI-PLC variants to PG/PC SUVs as a function of XPC. The apparent Kd values were measured by FCS for variants labeled with AF488 at N168C: *N168C (●), *P42G (▲), *K44A (■), *K44E (❒), *Y88A (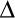 ), and *3YS (❍). The two dotted lines indicate XPC = 0.2 and 10 mM total phospholipids. The error bars are the standard deviations of Kd from two independent protein and SUV preparations.
), and *3YS (❍). The two dotted lines indicate XPC = 0.2 and 10 mM total phospholipids. The error bars are the standard deviations of Kd from two independent protein and SUV preparations.
Results
SUV size distributions and FCS fits
The fraction of PI-PLC bound to the vesicles, f, obtained from fits to the FCS data tend to asymptote at less than 100% binding (Figure 2B). Fits to the FCS data using equation 1 assume that only 2 species are present, free PI-PLC and SUV bound PI-PLC with 2 corresponding diffusion coefficients Dfree and Dbound. However, as observed in dynamic light scattering experiments (15), the SUVs have a variety of radii resulting in diffusion coefficients ranging from ~7 to ~35 µm2/s, corresponding to diffusion times of ~2 to ~0.5 ms. The SUV distributions are dominated by smaller vesicles, and SUVs with diffusion coefficients greater than 15 µm2/s (diffusion times less than 1 ms) constitute approximately 70% of the population (Figure S2). Modeling of the effects SUV size distributions on 2 species FCS fits reveals that Kd values are robust and do not significantly depend on the value of Dbound used for the 2 species fits. In contrast, the fraction bound, f, is very dependent Dbound and tends to asymptote at values well below 100% (Supplementary Information). This same trend, variations in f and fmax but robust values of Kd, is also observed when fitting the experimental data (Figure 2C).
For example, FCS data may be modeled using dynamic light scattering data for pure PG SUVs to approximate the SUV size distribution and f values of 0 (0% binding) to 1 (100% binding) (Supplemental Information, Figures S2 & S3). Fitting these model FCS curves using 2 species (Equation 1) with fixed values of Dbound from 10 to 15 µm2/s, yield fmax values ranging from 28% to 94% and Kd values ranging from 10.1 to 9.9 µM compared to modeled values of fmax=100% and Kd=10 µM (Figure S4 and Table S1). Globally fitting Dbound for the entire set of modeled titration data yield fmax=82% and Kd=10.3 µM. Thus, approximating the SUV distribution as a single species with a single, well-defined diffusion coefficient is likely to result in fmax values that are significantly less than 100% even in the presence of 100% binding. While the fmax values depend on the method used for the 2 species fit, the values of Kd are quite reliable.
These model results mirror the results of fitting the experimental data. As shown in Figure 2C, for *Y88A and XPC=0.90, globally fitting Dbound results in Dbound=17 µm2/s, and relatively high values for f while values of f decrease when Dbound is fixed to a value below the globally fitted value (fixed Dbound=15 µm2/s in Figure 2C). Despite the differences in f which lead to different values for fmax, 82 and 71% for the fitted and fixed Dbound data shown in Figure 2C, respectively, the values of Kd are similar, 135 µM and 134 µM, for fs determined using the fitted and fixed methods, respectively. The data in Figure 2C are typical and we see this trend of varying fmax but similar Kds for both experimental and model data.
As an alternative to using the 2 species fit, we could fit the FCS data based on the distribution of SUV sizes determined by dynamic light scattering. However, PI-PLC prefers to bind to smaller vesicles (15, 24) shifting the distribution of vesicle sizes observed in FCS binding experiments. The observed distribution is also affected by how well SUVs with different sizes and different fluorescence intensities are detected. For example, if PI-PLC prefers smaller SUVs, the smaller, faster moving SUVs in a population may bind multiple fluorescently labeled proteins and these brighter SUVs may be more easily detected than slower moving, larger SUVs with fewer bound fluorescent proteins. Since both of these factors can change the observed SUV distributions, we have chosen to use the 2 species fits which, while still an approximation, are easy to implement and provide reliable values for the apparent Kd.
Binding Wildtype PI-PLC
Wildtype PI-PLC is activated towards both PI and soluble substrates by binding to micelles or lipid vesicles (6, 8, 10), and the lipid composition affects both the extent of activation and the affinity for SUVs (15). For SUVs containing anionic lipids and PC, PI-PLC binding is strengthened by even small amounts of PC (XPC ~0.1) and the apparent Kd continues to decrease until XPC= 0.9 (Figure 3) (15). The relationship between vesicle binding and activity is complicated with an increase in activity at low XPC and the highest activity from XPC= 0.2 to 0.5. Above XPC= 0.5, enzymatic activity declines and the tightest vesicle binding is associated with reduced activity. The complicated relationships between binding and enzymatic activity as well as the synergistic effects of PC and anionic lipids make it difficult to predict how mutations will alter the binding and activity in the presence of multi-component lipid vesicles.
Effects of specific mutations on PI-PLC binding to SUVs
1. Mutations in helix B
The rigid residue Pro42 at the N terminus of helix B is likely important for orienting the helix for membrane binding. In the mutant dimer structure Pro42 also forms part of the dimer interface (12) and it may therefore be involved in the conformational changes associated with interfacial activation. Replacing Pro42 with a flexible Gly only slightly reduced phosphotransferase activity but severely impaired the cyclic phosphodiesterase step (Table 1, (13)). FCS experiments reveal that the binding of *P42G to pure PG or pure PC SUVs is much weaker than *N168C (Figure 3). In fact, the interaction with pure PG SUVs is so weak that an apparent Kd could not be measured, while the interaction with pure PC SUVs increased Kd 40-fold compared to *N168C. As indicated in Table 1, this low affinity is consistent with the failure of pure diC7PC micelles to activate P42G towards the soluble substrate 1,2-cyclic inositol phosphate (cIP) (13). However, despite the low affinity for PC, tightest binding is still only observed in the presence of both PC and PG with Kd reaching a minimum of ~4 mM at XPC ~ 0.7. The Kd for pure PC SUVs is 5-fold greater than this minimum Kd, well outside the uncertainties in the Kd values. More importantly, the improved binding in the presence of both PC and PG suggests that as long as the total phospholipid concentration is above Kd, the phosphotransferase reaction is still efficient and specific activity will be close to that of the wildtype enzyme.
Lys44 is the only charged group in both helix B and the adjacent 240s loop region. In the monomer crystal structures of a B. thuringiensis PI-PLC variant (Y247S/Y251S) (kinetically similar to wildtype PI-PLC) (14) and the nearly identical B. cereus PI-PLC (11), the Trp47 side chain, which interacts with the membrane (9, 10), points outward into solution and Lys44 is nearby but directed away from Trp47. However, high B factors for this region in both structures suggest that helix B may be flexible. If Lys44 is important for electrostatic interactions with anionic membranes, then mutations that alter its charge should reduce PI-PLC affinity for anionic surfaces. Mutation of Lys to Ala does not change the enzyme activity towards PI dispersed in diC7PC micelles or, in the presence of diC7PC, towards the soluble substrate cIP, and binding to pure PC SUVs is similar to that of wildtype PI-PLC (24). However, if the cationic Lys is replaced by anionic Glu, the specific activity in the PI/diC7PC system and the PC activation of cIP are significantly impaired (Table 1, (13)). FCS experiments reveal that affinity of the Lys44 variants for anionic SUVs is decreased significantly, as expected if Lys44 mediates interactions with anionic lipids (Figure 3). For the charge reversal mutant, the apparent Kd could not even be measured for XPC below ~0.3, and above this XPC it decreased continuously with increasing XPC (Figure 3). Neither Lys44 variant shows the significant increase in Kd between 0.9 and 1 XPC observed for *N168C, indicating that anionic lipids and PC no longer have synergistic effects on Kd when Lys44 is mutated.
These results confirm the importance of this cationic residue for protein binding to surfaces rich in negatively charged phospholipids. Clearly, PC facilitates interaction of K44A protein with bilayers resulting in activities comparable to wildtype PI-PLC in PC-rich assay systems such as PI/diC7PC and cIP/diC7PC (13). The weaker binding of the Glu variant suggests that like-charge repulsion hinders the initial interaction between the helix B region and the substrate-containing vesicle (or micelle) as well as the subsequent promotion of the protein conformation optimized for both binding and catalysis. This leads to the observed lower phosphotransferase (and cyclic phosphodiesterase) activity for K44E.
2. Mutating the putative dimer interface
Tyr residues 246–248 are key components of the dimer interface formed by the interfacially impaired PI-PLC W47A/W242A (12), and mutation of these Tyr residues to Ser should disrupt the dimer interface. The Y246S/Y247S/Y248S (3YS) mutant has reduced affinity for pure PC bilayers and lower phosphotransferase and phosphodiesterase activities (14). However, it is not known whether removing these residues alters the initial interactions of the protein with anionic phospholipids. From Figure 3, we can see that the Y246S/Y247S/Y248S/*N168C (*3YS) variant and *N168C have similar affinities for pure PG vesicles; while the apparent Kd of *3YS for pure PC vesicles is ~3 orders of magnitude higher than that of *N168C. As shown in Table 1, previous estimates of 3YS binding to pure PC SUVs yielded much lower apparent Kd values (0.6-1.0 mM (14)) than those obtained by FCS (64±20 mM). Those earlier experiments used a filtration/centrifugation assay that requires much higher protein concentrations. The filtration step, which separates free and SUV-bound PI-PLC, likely concentrates both PI-PLC and the vesicles (24). These increased concentrations enhance the affinity of PI-PLC for the membranes. In contrast, FCS requires only nanomolar protein concentrations, similar to the concentrations used in the activity assays, avoiding the complications resulting from filtration.
What about multicomponent vesicles? There is a 3-fold drop in the apparent Kd up to XPC = 0.5, suggesting that the presence of both phospholipids still improves binding. However, as the bilayer becomes more PC-rich, protein affinity is significantly reduced. The apparent Kd of *3YS towards SUVs with XPC = 0.9, is about 1000-fold higher than that of the control, *N168C. These results indicate that tight binding to PC rich vesicles requires the protein conformation stabilized by these Tyr residues. In *3YS the poor PC binding could be consistent with three or more of the Tyr residues inserting into the membrane. However, these Tyr residues are located on helix G, and proper orientation of the active site opening towards the membrane, allowing substrate diffusion into the active site, along with Tyr insertion into the membrane would require significant unwinding of helix G. A better possibility is that these Tyr residues form a specific binding site for an individual PC molecule – perhaps providing a π-cation interaction for a choline moiety. Alternatively, the *3YS results are also consistent with surface-induced dimerization of PI-PLC via the Tyr zipper observed in the mutant dimer structure (12).
3. Y88A, a mutation near the lipid binding region
Tyr88 is at the top of the barrel rim and could directly interact with the bilayer (Figure 1). The Y88A variant is an extremely active enzyme whose specific activity is 3-fold higher than recombinant PI-PLC using a PI/diC7PC (1:4) assay system (S. Guo, unpublished results). In the presence of diC7PC, it has a cyclic phosphodiesterase activity comparable to wildtype. Its apparent Kd for pure PC SUVs as determined by a filtration/centrifugation assay (24) is approximately an order of magnitude higher than that of wildtype PI-PLC, consistent with this side chain partitioning into a PC bilayer (13). Since two types of phospholipids are involved in anchoring PI-PLC to surfaces, it is extremely useful to examine a more complete binding profile for this mutant protein in order to understand why the enzyme is more active than wildtype PI-PLC. Therefore, the variation of the apparent Kd with XPC for *Y88A was determined by FCS (Figure 3). The profile is qualitatively similar to that of *N168C with similar affinity for pure PG SUVs but with a smaller and more gradual drop in the apparent Kd as XPC increases. From 0 to 0.7 XPC, Kd decreased 880-fold for *N168C but only 140-fold for *Y88A. The minimum Kd also decreased from XPC = 0.9 for *N168C to XPC = 0.7 for *Y88A. Such behavior is consistent with interactions between this surface aromatic group and PC, but not PG, molecules in the bilayer (potentially via the π-cation as well as partial insertion). The shape of the Kd versus XPC curves for *Y88A closely resembles that of the *P42G mutant. However, *Y88A has much higher affinity for lipid surfaces with the lowest Kd values of all the studied mutants. Thus, as long as the total phospholipid concentration is high (typically 4–8 mM in the phosphotransferase micelle system, and 5 mM PC in cIP activation kinetics experiments), the specific activities of Y88A and wildtype PI-PLC should be similar.
4. Active site PI-PLC mutations and the loss of synergistic binding
While Lys44 is important for initial interactions between anionic lipids and PI-PLC, PI and anionic substrate analogue lipids must also bind to the active site. Mutations in the active site could perturb anionic lipid binding as well as the synergistic effects of binding to anionic lipids and PC. Alternatively, mutating residues that are crucial for catalysis could degrade activity with only modest effects on binding.
His32 acts as a general base in PI cleavage, and mutating this residue abolishes catalytic activity (25, 26). *H32A has negligible activity, but binds to pure PG and pure PC SUVs with essentially the same affinities as *N168C. However, the binding affinity of *H32A for multicomponent vesicles is strikingly different than wildtype (Figure 4). Unlike *N168C binding (Figure 3) with its concave dependence on XPC and minimum Kd around XPC=0.9, for *H32A the dependence of Kd on XPC gradually decreases as XPC increases. The apparent lack of a significant minimum in the Kd vs. XPC curves (Figure 4) demonstrates that an active site mutation can destroy the synergistic effects of anionic lipid and PC binding. However, unlike mutations to Lys44 which degrade both synergy and binding to pure PC vesicles, this active site mutant has Kd values for single component SUVs that are comparable to wildtype protein.
Figure 4.
Binding of inactive PI-PLC variants to SUVs as a function of XPC. The apparent Kd values were measured by FCS for *H32A binding to PG/PC (●) or PI/PC (❍) SUVs. The error bars are the standard deviations of Kd from two independent protein and SUV preparations.
FCS studies on active PI-PLC variants are limited to SUVs containing PC and substrate lipid analogues due to the production of DAG (Scheme 1) which leads to vesicle fusion (23). This is not a problem for the inactive H32A; thus, we used FCS to examine the interactions of this inactive variant with SUVs containing the substrate PI dispersed in PC (Figure 4). Interactions between *H32A and pure PI SUVs are essentially the same as those with pure PG SUVs. However, *H32A binding to 1:1 PI/PC SUVs is noticeably weaker than that to 1:1 PG/PC SUVs (Figure 4). This difference between PI and PG, a substrate analogue, indicates that at intermediate XPC, PI-PLC may bind more tightly to substrate analogues than to substrate, but this difference in binding affinity does not alter the observed trends.
Correlation of vesicle binding and observed phosphotransferase activity towards PI in SUVs
For a better understanding of the relationships between vesicle binding affinity and activity, we need to measure enzyme kinetics towards PI presented in SUVs. Two types of activity experiments were performed. In the first type of kinetic experiment, XPC is kept constant and the total phospholipid concentration is varied. In the second type of experiment, ‘surface dilution’, XPC is varied by conducting experiments at constant PI concentration with increasing amounts of PC, thus increasing XPC.
1. PI-PLC activity at constant XPC
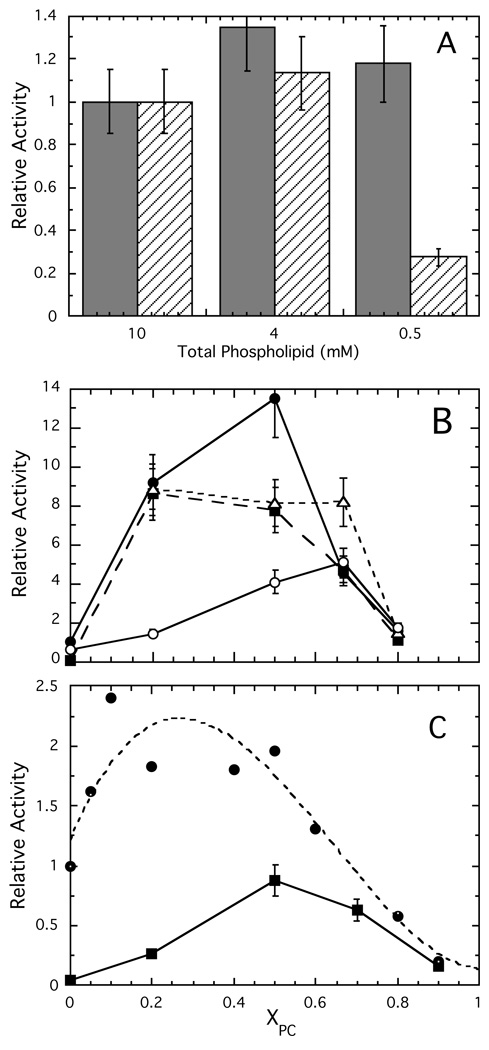
In the presence of PC, all of the mutants have higher Kds than does *N168C. This reduced affinity for membranes should result in decreased activity at lipid concentrations below the mutant Kds. We therefore compared the enzyme activity of wildtype PI-PLC and Y88A (Y88A/N168C) at fixed XPC = 0.2 and total phospholipid concentrations of 10, 4 or 0.5 mM (Figure 5A). Only wildtype and Y88A PI-PLC were used for these experiments due to the high apparent Kd values of the other variants at low XPC. At XPC = 0.2, the chosen total lipid concentrations are all above the 0.14±0.11 mM apparent Kd determined for *N168C. In contrast, 0.5 mM phospholipid is less than the ~1 mM apparent Kd of *Y88A. Above the dissociation constant, the protein should bind well to the lipids but the binding should drop dramatically when the total lipid concentration is below Kd. Indeed, the specific activity (Figure 5A) parallels the binding. The unaltered PI-PLC has similar specific activities at each of the three concentrations. In contrast, the Y88A specific activities at 10 and 4 mM are similar but there is a pronounced drop for the 0.5 mM phospholipid sample. This confirms that PG is a good substrate analogue in this anionic lipid-rich region, and indicates that vesicle binding controls the enzyme activity towards these PI-rich vesicles.
Figure 5.
Relative activities of PI-PLC enzymes toward PI/PC SUVs. (A) Activities at fixed XPC = 0.2 with different total phospholipid concentrations for wildtype ( ) and Y88A (///) enzymes. (B) Relative activities, compared to wildtype activity towards pure PI SUVs, of wildtype (●), K44A (■), Y88A (
) and Y88A (///) enzymes. (B) Relative activities, compared to wildtype activity towards pure PI SUVs, of wildtype (●), K44A (■), Y88A ( ), and 3YS (❍) towards 10 mM PI in the presence of increasing concentrations of PC. (C) Relative activity of wildtype (●), data taken from reference (15), and K44A (■) at fixed total phospholipid (10 mM) with varying XPC. Assays for K44A were carried out in duplicate; the 15% error bars represent the maximum error for these assays.
), and 3YS (❍) towards 10 mM PI in the presence of increasing concentrations of PC. (C) Relative activity of wildtype (●), data taken from reference (15), and K44A (■) at fixed total phospholipid (10 mM) with varying XPC. Assays for K44A were carried out in duplicate; the 15% error bars represent the maximum error for these assays.
2. Diluting the substrate and PI-PLC activity
While low Kd values can be advantageous at high substrate concentrations, at low substrate concentrations where the enzyme may need to move in order to find the next substrate molecule, tight binding may be a disadvantage. To investigate this, we performed surface dilution experiments in which the PI concentration is fixed at 10 mM and XPC is raised by increasing the PC concentration (Figure 5B). Wildtype PI-PLC showed a large (~9-fold) increase in activity with a small amount of PC (XPC= 0.2). The activity increased a bit further, then decreased steeply by XPC = 0.67. The increase with a small amount of PC and the decrease in activity in PC-rich bilayers were similar to what has been observed using SUVs with a fixed total phospholipid concentration and varying PC (15). At high XPC, this decreased enzyme activity was tentatively explained as sequestration of the enzyme in the PC-rich regions (15).
As expected from the FCS vesicle binding experiments (Figure 3), the activity of Y88A more or less parallels that of the wildtype enzyme (Figure 5B). However, Y88A PI-PLC is substantially more active than wildtype PI-PLC against SUVs with XPC = 0.67. Interestingly, at this XPC, Y88A binds more weakly to SUVs and also exhibits higher specific activity in the PI/diC7PC (1:4) mixed micelle system than does the wildtype enzyme. These results support the idea that PI-PLC's ability to hop and/or scoot around or between vesicles is particularly important at lower substrate surface concentrations.
In the surface dilution experiment, the specific activity of K44A (K44A/N168C) for pure PI SUVs was very low (~26 fold less than wildtype), in agreement with the very high apparent Kd for pure PG SUVs. The dependence of the relative activity on XPC was, however, similar to that seen for wildtype PI-PLC (Figure 5B). With fixed PI, increasing PC caused a substantial increase in activity followed by a decrease at high XPC (Figure 5B). A different comparison of the two enzymes is provided by keeping the total phospholipid concentration fixed and altering XPC (Figure 5C). K44A activity increased more gradually than wildtype enzyme with increasing XPC. This likely reflects the poorer binding of this variant to PC-poor vesicles as observed by FCS. However, the activity decreases at high PC are similar to those observed for wildtype PI-PLC. Recent field cycling NMR studies of wildtype PI-PLC interacting with phosphatidylmethanol (PMe)/PC vesicles (15) suggest that such a decrease is likely to be the result of enzyme-induced demixing of PMe (substrate analogue) and PC (activator) leading to sequestration of the enzyme in PC-rich regions resulting in reduced accessibility to substrate.
The interaction of vesicle binding and activity as a function of XPC is particularly interesting for the putative dimer interface mutant, 3YS (Y246S/Y247S/Y248S/N168C). The fluorescently labeled mutant has apparent Kd values for SUVs that decrease slightly as PC increases, with a possible minimum at XPC = 0.5 followed by dramatic increases in Kd and very weak binding above XPC = 0.8 (Figure 3). In contrast, the specific activity in the surface dilution experiment increased gradually as PC was added, reaching a maximum at XPC ~ 0.7 (Figure 5B). The gradual 8-fold increase in activity, in a regime where Kd varies only 2–3 fold and is at least 1 order of magnitude higher than that of Y88A or wildtype PI-PLC indicates that PC must do more than just bind the protein to the surface. It has been proposed that PI-PLC activation on PC containing surfaces is driven by dimerization, and the three mutated Tyr residues form part of the mutant homo-dimer interface; thus, these Tyr residues may promote transient dimerization on the surface (12, 14). Alternatively, interactions between PC and these Tyr residues may help drive conformational changes in surface bound, monomeric PI-PLC. In either case, the gradually increasing specific activity likely reflects not just partitioning of the enzyme to the vesicles but the surface active conformation that is promoted by PC.
At XPC= 0.8 with a total phospholipid concentration of 50 mM, there was a decrease in 3YS activity similar to that seen for other variants at slightly lower XPC. Since 50 mM is considerably above the apparent Kd for high XPC, the protein would be expected to bind to these SUVs. Therefore, the observed decrease cannot reflect poor partitioning of the enzyme to the vesicles. More likely this reflects either a reduced affinity for or reduced accessibility to the PI in the PC-rich interface.
Pro42 is also associated with the putative dimer interface, and previous kinetic characterization of P42G showed that while cleavage of PI (8 mM) in 32 mM diC7PC was about 70% that of wildtype enzyme, diC7PC (8 mM) activation of cIP hydrolysis was severely reduced (13). The FCS binding studies clearly indicate that binding of *P42G to both anionic phospholipids and PC is impaired, but with high enough PC, activities may approach that of the unaltered enzyme. To test this, we examined the kinetics of cIP hydrolysis with increased diC7PC to see if increased PC binding would promote the water-soluble reaction. With 5 mM and 25 mM diC7PC present, the specific activities toward 20 mM cIP were similar. Increasing the diC7PC to 50 mM roughly doubled the enzyme specific activity to 21.4 µmol˙min−1˙mg−1, 7- fold higher than the value in the absence of PC. However, this level of activation is still far lower than what is observed for wildtype enzyme (20- to 30-fold activation). These results are consistent with very weak binding of this variant to pure PC aggregates. It also emphasizes that the synergistic effects of anionic lipid and PC binding, presumably mediated by communication between the active and activator lipid binding sites, requires the anionic substrate analogue to have acyl chains.
Dependence of Kd on PI-PLC concentration – estimation of protein binding footprint
For a simple monomer binding model, the addition of unlabeled protein to the FCS experiments will lead to competition between labeled and unlabeled protein for SUV binding sites. If, at all lipid concentrations, there are adequate binding sites for the unlabeled protein the fraction of bound, labeled protein will be unaffected. However, if the unlabeled protein competes for available binding sites, the apparent Kd determined by FCS will increase. When 0.02 or 0.57 µM unlabeled PI-PLC was added to 3.5 nM *N168C, the apparent Kd values were essentially the same (Figure 6B). Further additions of unlabeled PI-PLC leads to small increases in Kd. Kd increases by approximately an order of magnitude in the presence of 5.7 µM unlabeled PI-PLC indicating that there is significant competition for binding sites and most of the sites are occupied by unlabeled protein at this concentration. Even at these high PI-PLC concentrations, there are unlikely to be interactions between monomers in solution, since the protein crystallizes as a monomer at even higher concentrations and the diffusion times of free *N168C are essentially the same in the absence and presence of 5.7 µM unlabeled protein indicating that no large PI-PLC aggregates, i.e. tetramer or larger, are present. Under these conditions, the fraction bound for the fluorescently labeled protein reaches a maximum around 0.3 mM phospholipid (Figure 6A). If we assume that all of the protein is bound at 0.3 mM phospholipid then we can estimate how many phospholipids comprise a PI-PLC binding site on the vesicle. Assuming 2/3 of the lipids are in the outer leaflet of the small vesicle, when the binding is saturated the outer leaflet lipid to protein ratio is around 35 to 1, suggesting that on 1:1 PG/PC SUVs, a single protein binding site requires around 35 phospholipids. This estimate assumes that the maximum of the titration curve corresponds to 100% binding; however, while we have shown that 2 species fits to the correlations tend to underestimate fmax (Supplementary Information) we do not know that the asymptote in the titration curve represents 100% binding. We can, therefore, estimate an upper limit for the PI-PLC footprint by assuming that the fraction bound at 0.3 mM phospholipid, i.e. f=50%, is the actual fraction bound rather than an underestimate. In this case, with the same small vesicles, the outer leaflet phospholipid to protein ratio is 70 to 1. Previous crude estimates of the number of phospholipids required to form the PI-PLC binding site, based on a Langmuir binding analysis using much higher PI-PLC concentrations and anionic vesicles (24), suggested 50–150 lipids per PI-PLC. The determination, by FCS, of 35–70 lipids per PI-PLC provides a better estimate of this parameter.
Figure 6.
Fluorescently labeled PI-PLC protein (3.5 nM) bound to vesicles as a function of total lipid concentration with (1:1) PG/PC SUVs: *N168C with 0.02 µM (❍), 0.57 µM (●) or 5.7 µM (X) unlabeled wildtype PI-PLC added. The points shown are from two different titrations with different enzyme preparations and different vesicle preparations. (B) Variation in apparent Kd for 3.5 nM *N168C binding to PG/PC (1:1) SUVs with unlabeled PI-PLC concentration.
Discussion
The model for PI-PLC membrane binding and activity predicts that helix B is crucial for binding PI-PLC monomers to the membrane while Tyr residues 246–248 and 251 as well as Pro42 in helix B help mediate dimerization on the membrane surface. For interfaces containing even small amounts of PC, dimerization or other conformational changes activate B. thuringiensis PI-PLC towards both phosphotransferase and phosphodiesterase substrates (6–8). The combination of FCS to measure vesicle binding and NMR to measure activity allow us to test this model and to determine correlations between binding and activity.
FCS experiments on wildtype enzyme clearly show that small amounts of PC greatly increase the affinity of PI-PLC for membranes (Figure 3, (15)). Low XPC also enhances PI-PLC activity, presumably because PC assists in anchoring the protein to the surface, allowing it to access the lipid substrate. While small amounts of PC decrease the apparent Kd, the tightest binding is observed not in pure PC vesicles but rather for PG/PC vesicles containing approximately XPC = 0.9 (15); while for PI/PC vesicles, optimal enzymatic activity is found from 0.2 to 0.5 XPC. Thus, the activator PC and the substrate PI or substrate analogue PG have synergistic effects on membrane binding and activity, and the model must account for both membrane binding and synergism between substrate and non-substrate phospholipids.
Helix B stability and lability
Helix B is a short, 7 amino acid helix with the sequence PIKQVWG (11), and replacement of the N terminal Pro helix cap with a flexible Gly should unravel the helix (27). As expected, this structural disruption greatly reduces the affinity of PI-PLC for PG/PC SUVs no matter what the XPC while preserving the PG/PC synergy (Figure 3). This implies that efficient interactions of PI-PLC with bilayers are facilitated by the rigidity of the proline residue which keeps the helix B hydrophobic groups solvent exposed and properly oriented for membrane insertion. However, Pro42 could have multiple roles both stabilizing the monomer conformation and contributing to the homodimer interface as it does in the dimeric W47A/W242A PI-PLC mutant (12). Thus, the reduced binding of P42G to PC vesicles could also reflect changes in the monomer-dimer equilibrium in solution and/or on the membrane surface.
Electrostatic attraction between anionic lipids and cationic amino acids also facilitate membrane binding (1). Mutation of Lys44, the only cationic residue in the helix B region, has a pronounced effect on the affinity of PI-PLC for anionic surfaces. In fact, the affinity of the charge switch mutant, K44E, for pure PG SUVs is too low to measure by FCS. The K44A and K44E mutations also severely impair PG/PC synergy. The importance of Lys44 for binding to anionic surfaces, along with higher crystallographic B factors observed for helix B but not for the surrounding loops (11), suggest that this short helix may be labile allowing Lys44 to orient itself towards the membrane.
The results for the Pro42G and Lys44 mutations seem to be contradictory with the former arguing for helix B stability while the latter argues for helix B lability. This suggests that for the short B helix there may a balance between too much flexibility, which could alter the interaction of the hydrophobic residues with the membrane and too little flexibility, which could reduce electrostatic attractions. While our results are suggestive, this hypothesis needs to be tested by directly measuring the dynamics of helix B and its surroundings.
Synergism between active and activator sites
Only two classes of PI-PLC mutants failed to show the pronounced synergistic effect of both lipids on PI-PLC binding: (i) Lys44 mutants and (ii) the active site mutant H32A. Both K44A and K44E have dramatically diminished affinity for PG-rich vesicles and slightly reduced affinity for PC-rich vesicles. However, the K44 mutations should not alter the active site and in high PC assay systems, PI-PLC cleavage of PI is reasonably efficient. This is consistent with the role of Lys44 in driving the protein to anionic surfaces, and this initial interaction appears to be crucial for binding to PG-rich vesicles.
The active site mutant behaves differently with unaltered PC sites and binding affinities for pure PG and pure PC SUVs that are comparable to wildtype. What H32A fails to show is the dramatic increase in binding affinity at low XPC and tightest binding at an XPC <0.9, implying that both substrate and PC aid in anchoring the enzyme. This strongly suggests that a fully functional active site is needed for the synergism between the PC and substrate binding sites. Such an interaction also hints that communication between both sites results in PC interfacial activation of the enzyme.
Interestingly these single site mutations both impair binding to anionic vesicles and abrogate anionic lipid/PC synergy. In contrast, mutants with seriously impaired PC binding, e.g. P42G and 3YS, still bind most tightly to mixed lipid vesicles. This suggests that either we have yet to identify the key residues that bind to PC and/or mediate PI/PC crosstalk from the PC end or that PC binding as well as the PC contribution to communication between lipid binding sites is more distributed than anionic lipid binding and thus more difficult to disrupt.
Tight lipid binding versus catalytic activity
All of these PI-PLC mutant enzymes show dramatically decreased specific activity at XPC > 0.8. This surface dilution inhibition is often attributed to the reduced two-dimensional concentration of the substrate PI. However, at that XPC the substrate concentrations are still quite high (for example, at XPC=0.8 there are still 20 substrate lipids per every 100 lipids), and if two-dimensional scooting about the vesicle or three-dimensional hopping around or between vesicles is efficient, the drop in activity should be more gradual assuming that the lipids are randomly distributed. What then happens to PI-PLC at XPC>0.7? Recent field-cycling NMR experiments suggest that PC is not randomly distributed in PC-rich vesicles due to enzyme-enhanced demixing of PI and PC, and that PI-PLC is sequestered in these PC rich regions (15). This demixing and sequestration would increase the distance PI-PLC has to move in order to find the next substrate molecule. In this work, the late onset of surface dilution inhibition for Y88A is associated with higher Kds, indicating that tight binding of PC hinders efficient movement of wildtype PI-PLC, again making it harder to find the next PI. Thus, at moderate XPC (0.6<XPC<0.8) the tighter binding of wildtype PI-PLC to PC-rich vesicles, likely associated with PI-PLC sequestration in PC-rich regions, enhances the effects of diluting PI in the membrane resulting in a precipitous rather than gradual decline in activity.
In summary, our results support many of the steps previously proposed for activation of B. thuringiensis PI-PLC and provide details on how some key residues interact with mixed phospholipid surfaces. For highly anionic membranes, but not PC-rich vesicles, binding correlates well with relative activity. The mutant PI-PLC binding studies suggest that Lys44 mediates the initial electrostatic interaction of the protein with substrate, orienting PI-PLC at the vesicle surface followed by Trp47 (and likely Trp242) insertion (13), which requires PC interaction with the protein. PI-PLC binding to both types of phospholipids then promotes conformational activation, possibly due to transient dimerization. Strikingly, this work also shows that very tight binding of the protein to PC-rich vesicles can actually reduce catalytic efficiency. Optimization of PI-PLC binding to substrate-containing vesicles is a balancing act between anchoring the protein in the correct conformation and orientation while also allowing it to dissociate in order to find substrate phospholipids or GPI anchored proteins by scooting and/or hopping.
Supplementary Material
Abbreviations
- DAG
Diacylglycerol
- FCS
Fluorescence Correlation Spectroscopy
- XPC
Mole Fraction Phosphatidylcholine
- PC
Phosphatidylcholine
- PG
Phosphatidylglycerol
- PI
Phosphatidylinositol
- PI-PLC
Phosphatidylinositol Specific Phospholipase C
- PMe
Phosphatidylmethanol
- SUV
Small Unilamellar Vesicle
Footnotes
This work was supported by N.I.H. GM60418 (M.F.R.)
Supporting Information
A titration curve for low XPC as well as a detailed description of the effects on fmax and Kd of using a single diffusion coefficient to describe a distribution of SUV sizes. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Johnson JE, Cornell RB. Amphitropic proteins: regulation by reversible membrane interactions. Mol. Membr. Biol. 1999;16:217–235. doi: 10.1080/096876899294544. [DOI] [PubMed] [Google Scholar]
- 2.Gelb MH, Cho W, Wilton DC. Interfacial binding of secreted phospholipases A2: more than electrostatics and a major role for tryptophan. Curr. Opin. Struct. Biol. 1999;9:428–432. doi: 10.1016/S0959-440X(99)80059-1. [DOI] [PubMed] [Google Scholar]
- 3.Daugherty S, Low MG. Cloning, expression, and mutagenesis of phosphatidylinositol-specific phospholipase C from Staphylococcus aureus: a potential staphylococcal virulence factor. Infect. Immun. 1993;61:5078–5089. doi: 10.1128/iai.61.12.5078-5089.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mengaud J, Braun-Breton C, Cossart P. Identification of phosphatidylinositol-specific phospholipase C activity in Listeria monocytogenes: a novel type of virulence factor? Mol. Microbiol. 1991;5:367–372. doi: 10.1111/j.1365-2958.1991.tb02118.x. [DOI] [PubMed] [Google Scholar]
- 5.Hirose S, Knez JJ, Edward Medof M, Patrick JC, Janice EB. Mammalian glycosylphosphatidylinositol-anchored proteins and intracellular precursors. Methods Enzymol. 1995;250:582–614. doi: 10.1016/0076-6879(95)50099-5. [DOI] [PubMed] [Google Scholar]
- 6.Zhou C, Qian X, Roberts MF. Allosteric activation of phosphatidylinositol-specific phospholipase C: Specific phospholipid binding anchors the enzyme to the interface. Biochemistry. 1997;36:10089–10097. doi: 10.1021/bi970846o. [DOI] [PubMed] [Google Scholar]
- 7.Zhou C, Roberts MF. Nonessential activation and competitive inhibition of bacterial phosphatidylinositol-specific phospholipase C by short-chain phospholipids and analogues. Biochemistry. 1998;37:16430–16439. doi: 10.1021/bi980601r. [DOI] [PubMed] [Google Scholar]
- 8.Zhou C, Wu Y, Roberts MF. Activation of phosphatidylinositol-specific phospholipase C toward inositol 1,2-(cyclic)-phosphate. Biochemistry. 1997;36:347–355. doi: 10.1021/bi960601w. [DOI] [PubMed] [Google Scholar]
- 9.Feng J, Bradley WD, Roberts MF. Optimizing the interfacial binding and activity of a bacterial phosphatidylinositol-specific phospholipase C. J. Biol. Chem. 2003;278:24651–24657. doi: 10.1074/jbc.M301207200. [DOI] [PubMed] [Google Scholar]
- 10.Feng J, Wehbi H, Roberts MF. Role of tryptophan residues in interfacial binding of phosphatidylinositol-specific phospholipase C. J. Biol. Chem. 2002;277:19867–19875. doi: 10.1074/jbc.M200938200. [DOI] [PubMed] [Google Scholar]
- 11.Heinz DW, Ryan M, Bullock TL, Griffith OH. Crystal structure of the phosphatidylinositol-specific phospholipase C from Bacillus cereus in complex with myo-inositol. EMBO J. 1995;14:3855–3863. doi: 10.1002/j.1460-2075.1995.tb00057.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shao C, Shi X, Wehbi H, Zambonelli C, Head JF, Seaton BA, Roberts MF. Dimer structure of an interfacially impaired phosphatidylinositol-specific phospholipase C. J. Biol. Chem. 2007;282:9228–9235. doi: 10.1074/jbc.M610918200. [DOI] [PubMed] [Google Scholar]
- 13.Guo S, Zhang X, Seaton BA, Roberts MF. Role of helix B residues in interfacial activation of a bacterial phosphatidylinositol-specific phospholipase C. Biochemistry. 2008;47:4201–4210. doi: 10.1021/bi702269u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shi X, Shao C, Zhang X, Zambonelli C, Redfield AG, Head JF, Seaton BA, Roberts MF. Modulation of Bacillus thuringiensis phosphatidylinositol-specific phospholipase C activity by mutations in the putative dimerization interface. J. Biol. Chem. 2009;284:15607–15618. doi: 10.1074/jbc.M901601200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pu M, Fang X, Redfield AG, Gershenson A, Roberts MF. Correlation of vesicle binding and phospholipid dynamics with phospholipase C activity: insights into phosphatidylcholine activation and surface dilution inhibition. J. Biol. Chem. 2009;284:16099–16107. doi: 10.1074/jbc.M809600200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gasteiger E, Hoogland C, Gattiker A, Duvaud S, Wilkins MR, Appel RD, Bairoch A. Protein identification and analysis tools on the ExPASy server. In: Walker JM, editor. The Proteomics Protocols Handbook. Totowa, NJ: Humana Press; 2005. pp. 571–607. [Google Scholar]
- 17.Volwerk JJ, Shashidhar MS, Kuppe A, Griffith OH. Phosphatidylinositol-specific phospholipase C from Bacillus cereus combines intrinsic phosphotransferase and cyclic phosphodiesterase activities: a phosphorus-31 NMR study. Biochemistry. 1990;29:8056–8062. doi: 10.1021/bi00487a010. [DOI] [PubMed] [Google Scholar]
- 18.Liu L, Mushero N, Hedstrom L, Gershenson A. Conformational distributions of protease-serpin complexes: a partially translocated complex. Biochemistry. 2006;45:10865–10872. doi: 10.1021/bi0609568. [DOI] [PubMed] [Google Scholar]
- 19.Elson EL, Magde D. Fluorescence correlation spectroscopy. I. Conceptual basis and theory. Biopolymers. 1974;13:1–27. doi: 10.1002/bip.1974.360130103. [DOI] [PubMed] [Google Scholar]
- 20.Takakuwa Y, Pack CG, An XL, Manno S, Ito E, Kinjo M. Fluorescence correlation spectroscopy analysis of the hydrophobic interactions of protein 4.1 with phosphatidyl serine liposomes. Biophys. Chem. 1999;82:149–155. doi: 10.1016/s0301-4622(99)00114-3. [DOI] [PubMed] [Google Scholar]
- 21.Thompson NL. In: Fluorescence correlation spectroscopy, in Topics in Fluorescence Spectroscopy. Lakowicz JR, editor. New York: Plenum Press; 1991. pp. 337–378. [Google Scholar]
- 22.Magde D, Elson EL, Webb WW. Fluorescence correlation spectroscopy. II. An experimental realization. Biopolymers. 1974;13:29–61. doi: 10.1002/bip.1974.360130103. [DOI] [PubMed] [Google Scholar]
- 23.Goñi F, Alonso A. Membrane fusion induced by phospholipase C and sphingomyelinases. Biosci. Rep. 2000;20:443–463. doi: 10.1023/a:1010450702670. [DOI] [PubMed] [Google Scholar]
- 24.Wehbi H, Feng J, Kolbeck J, Ananthanarayanan B, Cho W, Roberts MF. Investigating the interfacial binding of bacterial phosphatidylinositol-specific phospholipase C. Biochemistry. 2003;42:9374–9382. doi: 10.1021/bi034195+. [DOI] [PubMed] [Google Scholar]
- 25.Gässler CS, Ryan M, Liu T, Griffith OH, Heinz DW. Probing the roles of active site residues in phosphatidylinositol-specific phospholipase C from Bacillus cereus by site-directed mutagenesis. Biochemistry. 1997;36:12802–12813. doi: 10.1021/bi971102d. [DOI] [PubMed] [Google Scholar]
- 26.Hondal RJ, Zhao Z, Kravchuk AV, Liao H, Riddle SR, Yue X, Bruzik KS, Tsai M-D. Mechanism of phosphatidylinositol-specific phospholipase C: a unified view of the mechanism of catalysis. Biochemistry. 1998;37:4568–4580. doi: 10.1021/bi972646i. [DOI] [PubMed] [Google Scholar]
- 27.Viguera AR, Serrano L. Stable proline box motif at the N-terminal end of α-helices. Prot. Sci. 1999;8:1733–1742. doi: 10.1110/ps.8.9.1733. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.