Abstract
Biomarkers are needed to overcome critical roadblocks in the development of disease-modifying therapeutics for neurodegenerative diseases. Evolving genome-wide expression technologies can comprehensively search for molecular biomarkers and allow fascinating insights into the expanding complexity of the human transcriptome. The technology has matured to the point where some applications are deemed reliable enough for use in patient care. In the neurosciences, it has led to the discoveries of osteopontin in multiple sclerosis and SORL1/LR11 in Alzheimer's, and recent studies indicate its potential for identifying neurogenomic biomarkers. Advances in pre-analytical and analytical methods are improving search efficiency and reproducibility and may lead to a pipeline of biomarker candidates suitable for development into future neurologic diagnostics.
Keywords: gene expression, transcriptional profiling, microarray, biomarker, blood, biological fluids, variation, stability, reproducibility, validation, Parkinson's disease, Alzheimer's disease, multiple sclerosis, SORL1, LR11, α-synuclein
1. Introduction
Why does a neurologist need biomarkers?
Biological markers - biomarkers - are biologic indicators of disease or therapeutic effects, which can be measured through tests on blood and other biologic samples, and imaging tests. Biomarkers are needed to overcome critical roadblocks in the development of disease-modifying therapeutics for Parkinson's disease, Alzheimer's disease, and other common neurodegenerative diseases. Five major roadblocks involve early diagnosis, phase II and phase III clinical trials, and early drug development.
Roadblock 1
Diagnosis based on clinical exam delays detection until clinical deficits have already manifested, reflecting widespread underlying neuronal injury. Individuals at the earliest disease or at risk of disease with less complete neuronal damage, however, would be most responsive to a neuroprotective therapy. A simple laboratory biomarker could be used to identify individuals with high risk of developing neurodegeneration. These could then be prioritized for in-depth evaluation with time and cost intense, advanced clinical and neuroimaging instruments. Markers that identify high-risk individuals before a majority of neurons have been injured, combined with a risk-modifying or disease-modifying therapeutic could prevent the disease from ever manifesting (risk marker or early diagnostic).
Roadblock 2
In small phase II clinical trials, testing safety and tolerability of a compound is straightforward, however they lack power to detect slowing of disease progression based on clinical assessments alone. Therefore every compound has to go through large, costly and time-consuming phase III clinical trials to make decisions about its neuroprotective efficacy or failure. Markers that track the progression of neurodegenerative diseases in phase II clinical trials (progression marker) and that can serve as surrogates of therapeutic effect are needed to prioritize lead compounds for phase III clinical trials (response marker).
Roadblock 3
A second roadblock arises in phase III clinical trials. Entry diagnoses are based on a physician's assessment and clinical diagnostic scales, but even skilled neurologists misdiagnose common neurodegenerative diseases in up to 30% of cases. This dilutes power of phase III trials. A diagnostic that increases the specificity of the differential diagnosis or aids in defining disease subtypes would allow to enroll a more homogeneous population, increase power to detect disease modification, and reduce costs (diagnostic marker).
Roadblock 4
The pharmaceutical industry seeks markers that are a direct measure of modulation of the biologic drug target and measure quantitative changes in response to dose. These pharmacodynamic markers accelerate go/no go decisions in early drug development.
Roadblock 5
Markers that predict serious adverse medication effects would be of similar benefit in the early drug development of disease-modifying therapeutics (commonly referred to as pharmacogenomic marker).
2. Scientific rationale
Looking through the dumps: what is the biological basis of biomarkers in body fluids of patients with brain diseases?
Body fluids such as blood, urine (Muthukumar et al. 2005), saliva (Li et al. 2004), and very recently cerebrospinal fluid (Izzotti et al. 2008) haven been used for gene expression biomarker discovery. Non-neuronal cells and tissues such as fibroblasts, lymphoblasts, and muscle biopsies have also been explored. These analyses are based on the premise that biological fluids and non-neuronal cells can serve as surrogate substrates of select pathobiological and pharmacological processes in the brain. While clinical symptoms of Alzheimer's disease (AD), Parkinson's disease (PD), Huntington's disease (HD), and multiple sclerosis (MS) reflect preferential neuronal damage, DNA, RNA, and biochemical traits have been detected in peripheral cells. Genes causing familial AD are ubiquitously expressed (Schlossmacher et al. 1992; Citron et al. 1994), and skin fibroblasts from individuals carrying a familial AD mutation secrete excessive amounts of Aβ (Citron et al. 1994). Biochemical traits have been documented in blood cells of patients with AD (Schlossmacher et al. 1992; Ibarreta et al. 1998; Cecchi et al. 1999; Stieler et al. 2001; Tayebati et al. 2001; Scherzer et al. 2004; Hye et al. 2005; Blandini et al. 2006; Mhyre et al. 2007). Dopamine biosynthesis/signaling and mitochondrial function appear perturbed in blood cells of patients with PD (Yoshino et al. 1992; Schulz and Beal 1994; Nagai et al. 1996; Barbanti et al. 1999; Caronti et al. 1999; Caronti et al. 2001; Petrozzi et al. 2001). Lymphocytes of MS patients show biochemical changes (Kerlero de Rosbo et al. 1993; Mahad et al. 2003; Oki et al. 2004; Hallin et al. 2006; Vallittu et al. 2007). Blood cells of patients with Huntington's disease show perturbed mitochondrial function (Panov et al. 1999; Panov et al. 2005; Saft et al. 2005; Squitieri et al. 2006), apoptosis (Sawa et al. 1999; Maglione et al. 2006), adenosine receptor signaling (Maglione et al. 2005; Maglione et al. 2006), and tryptophan metabolism (Stoy et al. 2005). In addition, select transcriptional programs in blood cells and neurons overlap. The Parkinson's disease linked α-synuclein gene (SNCA) for example appears directly regulated by two members of the GATA family of transcription factors in erythroid blood cells and in neuronal dopamine cells (Scherzer et al. 2008).
Much progress has been made in developing neuroimaging biomarkers designed to visualize various aspects of the disease process in the brain (for example see companion review by David Eidelberg). In the future, we envision a toolbox of useful clinical, laboratory and imaging tests for neurodegenerative diseases, with distinct, but complementary, well-defined applications for accelerating drug trials and improving clinical care.
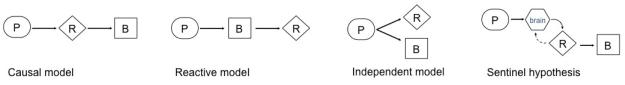
Four possible relations may link RNA biomarkers, biochemical traits, and neurodegenerative diseases in peripheral cells (Figure 1)
Figure 1.
Four possible relations may link gene expression biomarkers, biochemical traits, and neurodegenerative diseases in peripheral cells. P, primary cause of disease (e.g. genetic, toxic etc.); R, RNA trait; B, biochemical trait. The causal model (and more complex variations thereof) may yield disease-specific biomarkers causally linked to the disease locus. These will be ideal surrogate markers of therapeutic response. The reactive, independent and sentinel hypothesis models may yield disease-specific biomarkers reactively or independently linked to the disease etiology. Adapted from Schadt et al. 2005. See text for details.
(Hennecke and Scherzer 2008). In order to correctly interpret a gene expression biomarker experiment it is worth to consider the underlying scientific rationale. Standard gene-expression experiments cannot easily distinguish variations in RNA levels that are causal for complex traits from those that are reactive to complex traits. In most biomarker studies, the relation of the candidate RNA biomarker to the disease will be initially unclear. Then, detailed mechanistic analyses will be necessary to establish the relation of the candidate marker to the disease. Based on theoretical models a limited number of relationships between the traits are possible (Schadt et al. 2005). Four principal models are visualized in Figure 1 to illustrate these theoretical relationships (adapted from Schadt et al. 2005). A. Causal model. RNA expression changes in peripheral cells may be directly caused by copy number variations, transcriptional effects of single nucleotide polymorphisms in cis- or trans-acting regulatory regions, polymorphisms in regions controlling mRNA stability, as well as by epigenetic modification including histone modification, DNA methylation, and microRNAs. SNCA multiplications cause autosomal dominant PD through a dosage effect that is detectable in blood (and brain) with 50-100% increases in SNCA mRNA and SNCA protein (Miller et al. 2004). B. Reactive model. RNA expression changes may be reactive responses to primary genetic or toxic processes in peripheral cells. C. Independent model. Both biochemical traits and expression traits in peripheral cells may be independently induced by primary genetic and toxic etiologies; D. Sentinel hypothesis. Mononuclear white blood cells may transition between blood and brain compartments and initiate transcriptional and translational programs in response to brain pathology. In MS, activated T-cells from peripheral blood penetrate the blood brain barrier and initiate an inflammatory response that leads to demyelination (Prat et al. 2002; Prat et al. 2005).
Model A (and more complex variations thereof) may yield disease-specific biomarkers causally linked to the disease locus. These are ideal surrogate markers of therapeutic response. Model 2, 3, and 4 can yield disease-specific biomarkers reactively or independently linked to the disease locus or toxin. These can be useful diagnostic or progression biomarkers. Some of these biomarkers may also serve to measure surrogate responses to disease modifying-treatments (i.e. for example, if Coenzyme Q10 were to improve mitochondrial function in PD, this might lead to a “normalization” of compensatory, reactive expression changes of antioxidant genes).
3. Strengths, limitations, and unresolved aspects of the technology
Advances in the technology
From the first home made microarray to the first in vitro diagnostic multivariate index assay device
Gene expression microarray technology has matured to the point where some applications are deemed reliable enough for use in patient care. Eleven years after the initial report on microarry technology appeared in Science (Schena et al. 1995), the first microarray diagnostic, The MammaPrint assay, was approved by the US Food and Drug Administration on February 6, 2007. MammaPrint received the new designation of an “in vitro diagnostic multivariate index assay (IVDMIA) device” for predicting breast cancer recurrence within five to 10 years after a woman's initial cancer (van 't Veer et al. 2002; Bogaerts et al. 2006; Couzin 2007).
Reproducibility of microarray measurements: lessons from the Microarray Quality Control project
Enthusiasm for gene expression microarray technology has been curtailed by concerns about the reproducibility of this technique (Tan et al. 2003; Sotiriou and Piccart 2007). In view of the concerns raised on one hand and the great potential of this technology for personalized medicine on the other, the US Food and Drug Administration launched the Microarray Quality Control (MAQC) project (Shi et al. 2006). This consortium of 51 academic institutions and industrial partners systematically addressed the technical reproducibility of microarray measurements within and between laboratories, as well as across different gene expression platforms (Applied Biosystems, Affymetrix, Agilent, Eppendorf, GE Healthcare, Illumina, NCI_Operon, Taqman assays, Panomics, Gene Express) (Shi et al. 2006). Four different RNA samples were assayed in five replicates on seven microarray platforms by three laboratories each. The median coefficient of variation for the within-laboratory replicates ranged from 5–15% for the various microarray platforms and 10–20% for between-laboratory replicates (Shi et al. 2006). These results were in line with — or possibly better than — those reported for some traditional immunohistochemical assessments, for example of hormone receptors in breast tumors (Layfield et al. 2003; Sotiriou and Piccart 2007). There was an average 89% overlap of the differentially expressed genes between test sites using the same high-density microarray platform and a 74% overlap across different microarray platforms. Ranks of log ratios among all gene expression platforms were highly correlated. For example, gene lists rank-ordered by differential expression measured either by Taqman real-time PCR and Affymetrix or Illumina microarrays showed correlations of R = 0.92 or 0.91, respectively based on the comparison of ∼450-550 genes (Shi et al. 2006). The MAQC concluded that “… microarray results were generally repeatable within a test site, reproducible between test sites and comparable across platforms…” and sufficiently reliable to be used for clinical and regulatory purposes (Shi et al. 2006).
The expanding complexity and depth of the transcriptome: from chips to deep sequencing
At the same time, microarray technologies are evolving allowing fascinating insights into the expanding complexity of the human transcriptome, including microRNAs and mRNA splice variants. New digital transcript-counting approaches record the numerical frequency of a sequence in the library population and overcome many of the inherent limitations of array-based systems as well as limitations in detecting low-abundance transcripts (Marioni et al. 2008; Sultan et al. 2008). If (or perhaps when) the costs of deep sequencing are reduced, it may revolutionize - once more - transcriptome analyses and supersede many applications of gene expression microarray technology.
Limitations of the technology: blood, bias, overfitting, and power
The high-throughput capability of gene expression technology comes at a price. It is susceptible to biological noise inherent to the surrogate tissue, pre-analytical and analytical bias, overfitting of classifiers, and instability of gene lists.
Surrogate substrates are subject to biological noise inherent in the source tissue. Noninvasive biomarkers that can be measured in human blood would have many advantages over more invasive or cost-intense cerebrospinal fluid or imaging markers. Several confounding variables are specific to blood and need be dealt with in the experimental design and analysis. These are the type of blood cells assayed, the choice of blood RNA stabilizing reagent, and hemoglobin mRNA abundance. Two available whole blood RNA systems, PAXgene (PreAnalytiX) and Tempus (Applied Biosystems) are designed to isolate high-quality RNA from a diverse pool of blood cells including reticulocytes, neutrophils, eosinophils, basophils, lymphocytes, and monocytes. Large amounts of hemoglobin mRNA in blood tend to reduce the sensitivity of hybridization results on some platforms. It is however unclear whether the benefits of globin reduction protocols outweigh the limitation of introducing a new confounding variable (Burczynski and Dorner 2006; www.expressionanalysis.com October 2007).
Bias and overfitting pose critical threats to the validity of genomic and other multivariate biomarker studies - if they not carefully addressed in study design and conduct (Feng et al. 2004; Ransohoff 2004; Ludwig and Weinstein 2005; Ransohoff 2005). Bias poses a common and potentially fatal challenge to gene expression biomarker studies (Ransohoff 2005; Hennecke and Scherzer 2008). Many biomarker studies rely on banked convenience samples, use historic or convenience controls not drawn from the source population, enrolled at different times, from different populations, by different operators, using different procedures. These are susceptible to bias due to systematic differences in sample collection, processing, and storage. For example, if the experimental specimens had been processed immediately, while control samples were frozen and processed several months later, then a bias resulting from changes caused by storage could be ‘hard-wired’ into the data. Other potential sources of bias are baseline inequalities between groups (for example: if all experimental samples are recruited from clinic populations and most controls amongst paid or unpaid volunteers), sample run order (if most experimental samples are run first, most controls later), medication bias (if all cases are on cholinesterase inhibitors, but none of the controls), and hematological biases (if most cases were anemic, but most controls were not). Bias can be minimized by a prospective design, by enrolling controls from the source population, and by handling and analyzing specimens of cases and controls in parallel, in a uniform and blinded manner. Pre-analytical variables should be tightly controlled and monitored (Hennecke and Scherzer 2008). Controls should be comparable to the cases in that they are drawn from the same source population, are exposed to the same environmental variables, and could be identified as a case, if they had disease. Bias from sample processing can be minimized by collecting, handling and analyzing specimens of cases and controls in parallel, in a standardized manner. In addition, all steps of sample processing should be recorded in detail (e.g. time and operator performing the phlebotomy, duration of storage (Hennecke and Scherzer 2008). Monitoring these data points can further assist in identifying and statistically controlling for sources of bias (Ransohoff 2005).
Overfitting can occur when multivariate models show apparent discrimination that is actually caused by chance and is, therefore, not reproducible. Overfitting can occur when large numbers of genes are used to discriminate among a small number of outcome events. Lets assume that for 10 people with neurodegenerative disease and 10 without, 40,000 features with no relation to the disease are assessed, such as the number of movies they watched over the past month or the number of times they chew their food (Ransohoff 2004). If enough possible predictors are examined, even if nonsensical and random, a pattern could be found that perfectly discriminates among those 20 people with and without neurodegenerative disease (Ransohoff 2004). Such a pattern would ‘fit’ or discriminate among the group of individuals it was derived from in a training set, but it would not be able to correctly classify a second group of individuals. The solution to the threat of overfitting is to rigorously validate a multivariate classifier in independent validation studies using pre-specified parameters and outcome measures. Splitting of patients into separate training and validation sets should be an essential part of every multivariate biomarker publication (Feng et al. 2004). A survey of all microarray publications in cancer however found that independent validation has been carried out in only about 10% of reports about microarray research (Ntzani and Ioannidis 2003). In neurodegenerative diseases, the percentage of high-throughput biomarker studies that included an independent validation set is estimated to be even smaller.
It is important to realize that while such initial testing is a critical measure to guard against overfitting, it is just the first in a series of validation steps aimed at reducing uncertainty and increasing knowledge about a candidate biomarker. Successive steps in the biomarker development process are addressed below.
Power
Insufficient power for detecting small true positive differences in gene expression is another important limitation of many microarray studies in neurodegenerative diseases. Small sample sizes might both hinder the identification and replication of truly important genes. It has been estimated that thousands of samples are needed to achieve sufficient power for replicating individual genes on a gene lists (Ein-Dor et al. 2006). The challenges facing neurologic research in this regard may even exceed the challenges in cancer research, where autopsied tumor tissue is directly available for analysis and the magnitude of gene expression changes exceeds the magnitude typically seen in neurodegenerative diseases. For molecular medicine to fulfill its arrays of promises, we should aim for studies with thousands of patients, a hundred-fold more than the current standard (Ioannidis 2005).
Unresolved questions on the clinical utility of gene expression classifiers
Replication of gene expression classifiers in independent populations
The MAQC study provided strong evidence that gene expression platforms can be used to reliably measure gene expression in technical replicates of the same RNA specimen in different laboratories. Technical replicates serve to estimate the retest reliability or technical precision of the platform. In clinical practice, a second source of variation, biologic variation is of critical importance. The MAQC study however did not address biological variation in gene expression. Biologic variation is introduced by differences in gene expression between any two humans. Thus for clinical applications, the key question arises whether a gene expression signature or gene list can be replicated in biospecimens from different individuals of one clinical phenotype (biological replicates).
Recent progress in cancer research suggests that gene expression classifiers can indeed be replicated in independent populations. TRANSBIG, the translational research network of the Breast International Group, examined two breast cancer gene signatures (a 70-gene and a 76-gene predictor) in large replication studies involving 307 and 198 patients, respectively, not included in the original studies (Buyse et al. 2006; Desmedt et al. 2007). Although there was only a 3-gene overlap between the two signatures, the clinical utility of both predictors was validated (Buyse et al. 2006; Desmedt et al. 2007). By contrast in Huntington's disease, 12 candidate mRNA markers (Borovecki et al. 2005) were not replicated in an independent cohort of 65 subjects (Runne et al. 2007).
These studies address the question whether a given gene expression classifier has predictive value in independent populations. A distinct concern has been raised by several studies that have each attempted to re-discover a classifier for the same clinical phenotype in different patient cohorts. These studies have yielded highly divergent lists of genes differentially expressed in experimental versus control groups.
Stability of gene lists
Five different groups have reported five different gene lists (intrinsic subtypes classifier, 70-gene profile, wound response classifier, 21-gene recurrence score, and 2-gene ratio) that appear to predict survival in breast cancer. Much confusion has been generated by the observation that the five gene lists do not appreciably overlap. Nevertheless, a head-to-head comparison of these five non-overlapping gene-expression predictors showed significant agreement in the outcome predictions for individual patients for four of the five predictors tested (Fan et al. 2006). How is it possible for five different groups to detect five non-overlapping gene lists that when applied to a common data set show highly concordant results? This is a puzzling question that has not been convincingly resolved. Understanding the underlying scientific reasons for this apparent puzzle may, however, offer important guidance for the future selection of robust and clinically reliable gene lists.
One possible explanation is suggested by emerging insights into the structure of gene expression data. Gene expression changes in a molecular pathway tend to be highly coordinated and correlated. It is possible that distinct - but correlated - genes were being discovered as predictors by different groups that all tag the same underlying molecular information or pathway (Ein-Dor et al. 2005; Sotiriou and Piccart 2007). The relative ranking of individual genes on the basis of their association with the clinical phenotype (either by correlation coefficient or by P-value) is unstable and changes dramatically when different patient populations are used. Different genes then fluctuate to the top of rank-ordered lists resulting in the selection of apparently different gene lists.
According to this view assessing the overlap in gene identity among gene-expression profiles may not be the most efficient measure of reproducibility. Rather than simply counting the overlaps of the gene lists from different studies for a complex disease, novel metrics are needed to characterize and tag the underlying molecular pathways in a standardized manner.
4. Highlights of gene expression biomarker analyses in neurodegenerative diseases
Differential gene expression has been detected in peripheral blood as well as other peripheral tissues of patients with multiple sclerosis, Alzheimer's disease (Kalman et al. 2005; Maes et al. 2007), Parkinson's disease (addressed below), Huntington's disease (Borovecki et al. 2005; Strand et al. 2005), spinal muscular atrophy (Sumner et al. 2006), stroke (for review (Sharp et al. 2006; Baird 2007), neurodevelopmental, and neuropsychiatric disorders (reviewed in (Mohr and Liew 2007). Here I will highlight a few - out of many remarkable - recent developments that illustrate important approaches and applications.
1. Brain-to-body fluid biomarker discovery (Figure 2a)
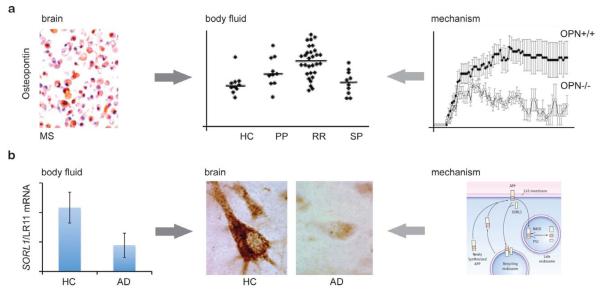
Figure 2.
Gene expression analyses in neurodegenerative diseases. a, Brain-to-body fluids biomarker discovery in MS. Gene expression scans discovered high levels of the proinflammatory gene osteopontin in plaques dissected from brains of patients with multiple sclerosis and spinal cords from rats with experimental autoimmune encephalomyelitis (Chabas et al. 2001). Osteopontin expression in the center of an actively demyelinating MS plaque is confirmed by immunostaining in macrophages (Chabas et al. 2001). Elevated levels of osteoponin were detected in plasma of patients with active relapsing-remitting MS (RR), but not in primary (PP) or secondary progressive MS (SP) (Vogt et al. 2003). Osteopontin-deficient mice were resistant to progressive experimental autoimmune encephalomyelitis (Chabas et al. 2001) suggesting a mechanistic role in the disease process. Osteopontin knock-out mice (OPN−/−; open circles) had milder disease with lower severity scores (y-axis) than OPN+/+ controls (closed circles) (Chabas et al. 2001). Right and left top panels from Chabas et al. 2001, reprinted with permission from AAAS. Middle panel from Vogt et al. 2003, reprinted with pending permission from Annals of Neurology.
b, Body-fluids-to-brain disease gene discovery. A microarray scan discovered low transcript levels of SORL1/LR11 (for sortilin-related receptor LDLR class A repeats-containing gene) in lymphoblasts of patients with AD (bottom left panel; Scherzer et al. 2004). Low expression levels of SORL1/LR11 were confirmed in brains of patients with sporadic AD (middle panel) and of individuals with mild cognitive impairment. Genetic and molecular studies have suggested a mechanistic model linking SORL1/LR11 to increased Ab production. In this model, SORL1 directs trafficking of APP into recycling pathways. When SORL1 is underexpressed, APP instead is sorted into the late endosome, where the enzymes BACE and presenilin 1 (PS1) generate neurotoxic Ab. Middle panel from Scherzer et al. 2004, reproduced with pending permission from Archives of Neurology; right panel from Marx 2007, reprinted with permission from AAAS.
In 2001, Larry Steinman and colleagues performed a seminal high-throughput transcriptional analysis of MS plaques. They found elevated levels of osteopontin transcripts, encoding an integrin binding protein involved in immunity and inflammation, in plaques dissected from brains of patients with multiple sclerosis and in spinal cords from rats paralyzed by experimental autoimmune encephalomyelitis (Chabas et al. 2001). Osteopontin-deficient mice were resistant to progressive experimental autoimmune encephalomyelitis and had frequent remissions (Chabas et al. 2001). Recently, elevated levels of osteoponin were also found in CSF (Braitch et al. 2008) and plasma of patients with MS (Vogt et al. 2003; Comabella et al. 2005). This emerging research illustrates that a CNS expression scan can yield a valuable candidate circulating biomarker.
2. Body-fluid-to-brain disease gene discovery (Figure 2b)
While the analysis of circulating gene expression markers for AD is in its infancy, such an approach in this neurodegenerative disease has led to remarkable progress in elucidating a novel molecular pathobiological pathway. In 2003, we originally discovered an association between low transcript levels of SORL1 (also known as LR11) and AD in two gene expression scans of > 7270 transcripts in lymphoblasts (Scherzer et al. 2004). Low expression levels of SORL1 were confirmed in brain of patients with sporadic AD (Scherzer et al. 2004) and of individuals with mild cognitive impairment (Sager et al. 2007). SORL1 directs trafficking of APP into recycling pathways and when SORL1 is underexpressed, APP is sorted into Aβ-generating compartments (Andersen et al. 2005; Offe et al. 2006; Rogaeva et al. 2007). In 2007, variants in the SORL1 gene that appear to interfere with mRNA processing were found to be associated with increased susceptibility for AD (Rogaeva et al. 2007) and this has been replicated in Belgium (Bettens et al. 2008), China (Tan et al. 2007), autopsy-confirmed AD (Lee et al. 2008), and an urban community-based cohort (Lee et al. 2007). Other studies led to marginal or controversial (Webster et al. 2007; Li et al. 2008; Shibata et al. 2008), or negative results (Minster et al. 2008). While the discovery of SORL1 has yielded tangible insights into the disease process and is a promising novel drug target (Ma et al. 2007), its potential as a clinical biomarker is unclear and awaits characterization.
3. Pharmacodynamic and response marker
Interferon beta (INFb) is a standard disease-modifying therapy for relapsing-remitting MS. Transcript levels of MxA measure pharmacologic response (Gandhi et al. 2008), correlate with the presence of IFNb neutralizing antibodies, and predicted time to next relapse (Malucchi et al. 2008). During two years, pharmacologic nonresponders had shorter times to the next relapse and fewer nonresponders were relapse free (Malucchi et al. 2008). Given the cost of IFNb therapy, benefits of early effective treatment for modifying disease progression, and the lack of sensitive clinical or imaging methods to determine therapeutic response to IFNb, biomarkers such as MxA mRNA may aid in developing tailored – personalized - treatment regimens for individual patients.
4. Pharmacogenomic markers
Wyeth Research and Elan incorporated gene expression analysis into a phase IIa, double-blind, placebo-controlled, multi-center study conducted to evaluate the safety and tolerability, and pilot efficacy of immunization with β-amyloid AN1792 in 372 patients with mild to moderate AD (O'Toole et al. 2005). Six immunizations were planned but were halted when meningoencephalities developed in 6% of immunized patients. In what anticipates the future of microarray prediction of toxicogenomic responses in neurological trials, high pre-immunization levels in blood of STAT1 a critical gene in the proinflammatory signal transduction pathway, were strongly associated with meningoencephalities (odds ratio for menigoencephalities = 230, false discovery rate = 0.004).
5. Risk markers
Risk markers for complex diseases are not simply present or absent (Manolio 2003). Rather, they have a wide range of values that overlap in persons with a disease and in those without it (Manolio 2003). The risk typically increases progressively with increasing levels (Manolio 2003). PD is such a complex, etiologically, genetically, pathologically and clinically heterogeneous neurodegenerative disease (Scherzer and Feany 2004; Scherzer et al. 2004; Forman et al. 2005). We proposed a continuous score to identify individuals with higher or lower risk of PD that may account for the complex biology of the disease (Scherzer et al. 2007). In a cross-sectional case control study analysis of a training set (n = 66) identified an 8-gene set score that was associated with risk of PD. Leave-one-out cross-validation revealed that the tertile of individuals with the highest risk scores were over five times more likely to have PD than were the lowest tertile (P = 0.005 for trend); a similar risk was seen after validation of the score in 39 additional individuals. In a second analysis we identified 22 additional genes in pathobiologically relevant processes with markedly different expression in patients with PD relative to healthy controls.
5. From discovery to clinical trials: three clinical phases of biomarker development
The biomarker validation process involves several validation studies, each adding information and confidence to the biomarkers. In principle the scientific assessment of a biomarker progresses through different phases of assay development and study design advancing from discovery to small to medium scale cross-sectional studies (here termed phase I biomarker studies) to large-scale prospective studies (phase II biomarker studies) to clinical phase III trials of biomarker & disease-modifying drug combinations (combined phase III clinical & biomarker trial (Figure 3). This involves a careful and step-wise process of biomarker development similar to the phases of drug development. Difficult choices regarding the “go-to-the-clinic” platform need also be made. Options include to stay with a modification of the discovery platform or to leap to other platforms (for example from microarray to real-time PCR to ELISA).
Figure 3.
Phases of molecular biomarker development. Biomarker development advances from discovery studies to small to medium scale cross-sectional studies (designated phase I biomarker studies) to large-scale prospective studies (phase II biomarker studies) to clinical phase III randomized, placebo-controlled trials of biomarker & disease-modifying drug combinations (combined phase III clinical & biomarker trial). Each phase reduces uncertainty about the candidate molecular biomarker.
Ultimately, validation of a biomarker as surrogate of the clinical outcome will require that it be used in a clinical trial of a therapeutic intervention, which is found to favorably impact the true clinical endpoint (Alonso et al. 2006). The feasibility of this process is precedented by ongoing, randomized combination gene expression/drug trials for secondary prevention of breast cancer recurrence, MINDACT and TAILORx. The Microarray In Node-Negative Disease May Avoid Chemotherapy Trial (MINDACT) is a European 6,000-patient randomized, multicentric study (Cardoso et al. 2008). The Trial Assigning IndividuaLized Options for Treatment (TAILORx) is conducted by the US National Cancer Institute and examines in 10,000 women whether a gene expression marker that is frequently associated with risk of breast cancer recurrence can be used to assign patients to the most appropriate and effective treatment (Sparano 2006). In neurodegeneration, the process is further complicated. On the one hand, the absence of a surrogate response marker is a major roadblock in the development of a disease-modifying therapeutic, on the other hand definite validation of a response marker requires an effective therapeutic. This no-win situation can be tackled by co-developing disease-modifying therapeutics and response markers akin to the co-evolution of cholesterol and statins in cardiovascular disease.
6. Look to the future
Emerging studies of gene expression markers in neurologic diseases provide starting points for the long and difficult journey of developing mature neurological biomarkers. While these signals are encouraging, it is important to frame them correctly. All published studies are in the discovery phase of biomarker studies – necessarily prone to false positives and false negatives - and will need to go through phase I to III clinical biomarker studies. This process replaces the idea of absolute “validation” with a scientific understanding of degrees of certainty in various dimensions. Many microarray studies published up to date in neurology lack an explicit and rigorous biospecimen collection, and an independent validation set, thus making them potentially susceptible to bias as well as overfitting. Furthermore, a majority of published gene expression studies in neurodegenerative diseases involve very small samples sizes (< 50) and are insufficiently powered for identifying true positives with small effect sizes. Most candidate biomarkers are expected to fail along this development process, just as most drugs fail at various stages of drug development. A rigorous three-phase clinical biomarker development process (Figure 3) will increase the odds for success and yield clearly defined results for each step of the process.
Rigorously designed, large and collaborative molecular biomarker studies consistent with this approach are on the way. The Harvard NeuroDiscovery Biomarker Program is conducting a longitudinal case-control study of 2,000 individuals with PD, AD, and controls focused on markers of disease progression (http://www.neurodiscovery.harvard.edu/research/biomarkers.html). The Smell Testing as Diagnostic and Prognostic Biomarkers in Parkinson's disease (PROBE) Study is an ongoing case-control study of 200 individuals recruited at 20 sites in North America funded by the U.S. Department of Defense (http://clinicaltrials.gov). PROBE is designed to evaluate the feasibility and utility of protein, RNA, and olfactory biomarkers for PD in a multi-center trial. TRACK-HD, is an ongoing European, multi-center, two-year, longitudinal biomarker study of premanifest and early stage HD (http://www.track-hd.net/) funded by the HighQ Foundation. It will examine gene expression and other candidate biomarkers of progression in 360 individuals with premanifest, early stage HD, and controls. A longitudinal study designed to discover and validate metabolomic, genomic, and imaging biomarkers of disease progression in HD is on the way at Massachusetts General Hospital funded by the National Institute of Neurological Disorders and Stroke. The next years will undoubtedly bring informative results from large and rationally designed clinical biomarker studies.
In the near future a pipeline of candidate neurologic biomarkers will emerge suitable for further development towards the bedside. This will be a laborious and difficult, but essential process.
ACKNOWLEDGEMENTS
Dr. Scherzer is Co-Director of the Harvard NeuroDiscovery Biomarker Program and member of the Steering Committee of PROBE. He is Co-Investigator of a biomarker study funded by the Michael J. Fox Foundation conducted in collaboration with Diagenic of Norway and is listed as Co-Inventor on a patent application on biomarkers for neurologic diseases held by the Brigham & Women's Hospital. He declares no competing financial interests in Diagenic or other companies.
Dr. Steven Hersch and Dr. Peter Lansbury provided insightful comments and analyses of critical roadblocks in the development of disease-modifying therapeutics for HD and PD. I thank the members of the Scherzer lab for their excellent contributions. I am particularly grateful to Drs. John Growdon, Bernard Ravina, and Alberto Ascherio for their expertise. Dr. Scherzer's work is supported by a Paul B. Beeson K08AG024816 from the NIA & the American Federation for Aging Research, NINDS grants NS064155, NS060227, and NS058793, the Harvard NeuroDiscovery Center, the Department of Defense, the M.E.M.O. Hoffman Foundation, the RJG Foundation, and the Michael J. Fox Foundation.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Alonso A, Molenberghs G, Geys H, Buyse M, Vangeneugden T. A unifying approach for surrogate marker validation based on Prentice's criteria. Stat Med. 2006;25(2):205–21. doi: 10.1002/sim.2315. [DOI] [PubMed] [Google Scholar]
- Andersen OM, Reiche J, Schmidt V, Gotthardt M, Spoelgen R, Behlke J, von Arnim CA, Breiderhoff T, Jansen P, Wu X, et al. Neuronal sorting protein-related receptor sorLA/LR11 regulates processing of the amyloid precursor protein. Proc Natl Acad Sci U S A. 2005;102(38):13461–6. doi: 10.1073/pnas.0503689102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baird AE. Blood genomics in human stroke. Stroke. 2007;38(2 Suppl):694–8. doi: 10.1161/01.STR.0000250431.99687.7b. [DOI] [PubMed] [Google Scholar]
- Barbanti P, Fabbrini G, Ricci A, Cerbo R, Bronzetti E, Caronti B, Calderaro C, Felici L, Stocchi F, Meco G, et al. Increased expression of dopamine receptors on lymphocytes in Parkinson's disease. Mov Disord. 1999;14(5):764–71. doi: 10.1002/1531-8257(199909)14:5<764::aid-mds1008>3.0.co;2-w. [DOI] [PubMed] [Google Scholar]
- Bettens K, Brouwers N, Engelborghs S, De Deyn PP, Van Broeckhoven C, Sleegers K. SORL1 is genetically associated with increased risk for late-onset Alzheimer disease in the Belgian population. Hum Mutat. 2008;29(5):769–70. doi: 10.1002/humu.20725. [DOI] [PubMed] [Google Scholar]
- Blandini F, Sinforiani E, Pacchetti C, Samuele A, Bazzini E, Zangaglia R, Nappi G, Martignoni E. Peripheral proteasome and caspase activity in Parkinson disease and Alzheimer disease. Neurology. 2006;66(4):529–34. doi: 10.1212/01.wnl.0000198511.09968.b3. [DOI] [PubMed] [Google Scholar]
- Bogaerts J, Cardoso F, Buyse M, Braga S, Loi S, Harrison JA, Bines J, Mook S, Decker N, Ravdin P, et al. Gene signature evaluation as a prognostic tool: challenges in the design of the MINDACT trial. Nat Clin Pract Oncol. 2006;3(10):540–51. doi: 10.1038/ncponc0591. [DOI] [PubMed] [Google Scholar]
- Borovecki F, Lovrecic L, Zhou J, Jeong H, Then F, Rosas HD, Hersch SM, Hogarth P, Bouzou B, Jensen RV, et al. Genome-wide expression profiling of human blood reveals biomarkers for Huntington's disease. Proc Natl Acad Sci U S A. 2005;102(31):11023–8. doi: 10.1073/pnas.0504921102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braitch M, Nunan R, Niepel G, Edwards LJ, Constantinescu CS. Increased osteopontin levels in the cerebrospinal fluid of patients with multiple sclerosis. Arch Neurol. 2008;65(5):633–5. doi: 10.1001/archneur.65.5.633. [DOI] [PubMed] [Google Scholar]
- Burczynski ME, Dorner AJ. Transcriptional profiling of peripheral blood cells in clinical pharmacogenomic studies. Pharmacogenomics. 2006;7:187–202. doi: 10.2217/14622416.7.2.187. [DOI] [PubMed] [Google Scholar]
- Buyse M, Loi S, van't Veer L, Viale G, Delorenzi M, Glas AM, d'Assignies MS, Bergh J, Lidereau R, Ellis P, et al. Validation and clinical utility of a 70-gene prognostic signature for women with node-negative breast cancer. J Natl Cancer Inst. 2006;98(17):1183–92. doi: 10.1093/jnci/djj329. [DOI] [PubMed] [Google Scholar]
- Cardoso F, Van't Veer L, Rutgers E, Loi S, Mook S, Piccart-Gebhart MJ. Clinical application of the 70-gene profile: the MINDACT trial. J Clin Oncol. 2008;26(5):729–35. doi: 10.1200/JCO.2007.14.3222. [DOI] [PubMed] [Google Scholar]
- Caronti B, Antonini G, Calderaro C, Ruggieri S, Palladini G, Pontieri FE, Colosimo C. Dopamine transporter immunoreactivity in peripheral blood lymphocytes in Parkinson's disease. J Neural Transm. 2001;108(7):803–7. doi: 10.1007/s007020170030. [DOI] [PubMed] [Google Scholar]
- Caronti B, Tanda G, Colosimo C, Ruggieri S, Calderaro C, Palladini G, Pontieri FE, Di Chiara G. Reduced dopamine in peripheral blood lymphocytes in Parkinson's disease. Neuroreport. 1999;10(14):2907–10. doi: 10.1097/00001756-199909290-00006. [DOI] [PubMed] [Google Scholar]
- Cecchi C, Latorraca S, Sorbi S, Iantomasi T, Favilli F, Vincenzini MT, Liguri G. Gluthatione level is altered in lymphoblasts from patients with familial Alzheimer's disease. Neurosci Lett. 1999;275(2):152–4. doi: 10.1016/s0304-3940(99)00751-x. [DOI] [PubMed] [Google Scholar]
- Chabas D, Baranzini SE, Mitchell D, Bernard CC, Rittling SR, Denhardt DT, Sobel RA, Lock C, Karpuj M, Pedotti R, et al. The influence of the proinflammatory cytokine, osteopontin, on autoimmune demyelinating disease. Science. 2001;294(5547):1731–5. doi: 10.1126/science.1062960. [DOI] [PubMed] [Google Scholar]
- Citron M, Vigo-Pelfrey C, Teplow DB, Miller C, Schenk D, Johnston J, Winblad B, Venizelos N, Lannfelt L, Selkoe DJ. Excessive production of amyloid beta-protein by peripheral cells of symptomatic and presymptomatic patients carrying the Swedish familial Alzheimer disease mutation. Proc Natl Acad Sci U S A. 1994;91(25):11993–7. doi: 10.1073/pnas.91.25.11993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comabella M, Pericot I, Goertsches R, Nos C, Castillo M, Blas Navarro J, Rio J, Montalban X. Plasma osteopontin levels in multiple sclerosis. J Neuroimmunol. 2005;158(12):231–9. doi: 10.1016/j.jneuroim.2004.09.004. [DOI] [PubMed] [Google Scholar]
- Couzin J. Diagnostics. Amid debate, gene-based cancer test approved. Science. 2007;315(5814):924. doi: 10.1126/science.315.5814.924. [DOI] [PubMed] [Google Scholar]
- Desmedt C, Piette F, Loi S, Wang Y, Lallemand F, Haibe-Kains B, Viale G, Delorenzi M, Zhang Y, d'Assignies MS, et al. Strong time dependence of the 76-gene prognostic signature for node-negative breast cancer patients in the TRANSBIG multicenter independent validation series. Clin Cancer Res. 2007;13(11):3207–14. doi: 10.1158/1078-0432.CCR-06-2765. [DOI] [PubMed] [Google Scholar]
- Ein-Dor L, Kela I, Getz G, Givol D, Domany E. Outcome signature genes in breast cancer: is there a unique set? Bioinformatics. 2005;21(2):171–8. doi: 10.1093/bioinformatics/bth469. [DOI] [PubMed] [Google Scholar]
- Ein-Dor L, Zuk O, Domany E. Thousands of samples are needed to generate a robust gene list for predicting outcome in cancer. Proc Natl Acad Sci U S A. 2006;103(15):5923–8. doi: 10.1073/pnas.0601231103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan C, Oh DS, Wessels L, Weigelt B, Nuyten DS, Nobel AB, van't Veer LJ, Perou CM. Concordance among gene-expression-based predictors for breast cancer. N Engl J Med. 2006;355(6):560–9. doi: 10.1056/NEJMoa052933. [DOI] [PubMed] [Google Scholar]
- Feng Z, Prentice R, Srivastava S. Research issues and strategies for genomic and proteomic biomarker discovery and validation: a statistical perspective. Pharmacogenomics. 2004;5(6):709–19. doi: 10.1517/14622416.5.6.709. [DOI] [PubMed] [Google Scholar]
- Forman MS, Lee VM, Trojanowski JQ. Nosology of Parkinson's disease: looking for the way out of a quagmire. Neuron. 2005;47(4):479–82. doi: 10.1016/j.neuron.2005.07.021. [DOI] [PubMed] [Google Scholar]
- Gandhi KS, McKay FC, Schibeci SD, Arthur JW, Heard RN, Stewart GJ, Booth DR. BAFF is a biological response marker to IFN-beta treatment in multiple sclerosis. J Interferon Cytokine Res. 2008;28(9):529–39. doi: 10.1089/jir.2008.0007. [DOI] [PubMed] [Google Scholar]
- Hallin E, Mellergard J, Vrethem M, Ernerudh J, Ekerfelt C. In vitro Th2 deviation of myelin-specific peripheral blood lymphocytes from patients with multiple sclerosis. J Neuroimmunol. 2006;171(12):156–62. doi: 10.1016/j.jneuroim.2005.09.013. [DOI] [PubMed] [Google Scholar]
- Hennecke G, Scherzer CR. RNA biomarkers of Parkinson's disease: developing tools for novel therapies. Biomarkers in Medicine. 2008;2:41–53. doi: 10.2217/17520363.2.1.41. [DOI] [PubMed] [Google Scholar]
- Hye A, Kerr F, Archer N, Foy C, Poppe M, Brown R, Hamilton G, Powell J, Anderton B, Lovestone S. Glycogen synthase kinase-3 is increased in white cells early in Alzheimer's disease. Neurosci Lett. 2005;373(1):1–4. doi: 10.1016/j.neulet.2004.10.031. [DOI] [PubMed] [Google Scholar]
- Ibarreta D, Urcelay E, Parrilla R, Ayuso MS. Distinct pH homeostatic features in lymphoblasts from Alzheimer's disease patients. Ann Neurol. 1998;44(2):216–22. doi: 10.1002/ana.410440212. [DOI] [PubMed] [Google Scholar]
- Ioannidis JP. Microarrays and molecular research: noise discovery? Lancet. 2005;365(9458):454–5. doi: 10.1016/S0140-6736(05)17878-7. [DOI] [PubMed] [Google Scholar]
- Izzotti A, Fazzi E, Orcesi S, Cartiglia C, Longobardi M, Capra V, Lebon P, Cama A, Pulliero A, La Piana R, et al. Brain damage as detected by cDNA-microarray in the spinal fluid of patients with Aicardi-Goutieres syndrome. Neurology. 2008;71(8):610–2. doi: 10.1212/01.wnl.0000313934.05965.71. [DOI] [PubMed] [Google Scholar]
- Kalman J, Kitajka K, Pakaski M, Zvara A, Juhasz A, Vincze G, Janka Z, Puskas LG. Gene expression profile analysis of lymphocytes from Alzheimer's patients. Psychiatr Genet. 2005;15(1):1–6. doi: 10.1097/00041444-200503000-00001. [DOI] [PubMed] [Google Scholar]
- Kerlero de Rosbo N, Milo R, Lees MB, Burger D, Bernard CC, Ben-Nun A. Reactivity to myelin antigens in multiple sclerosis. Peripheral blood lymphocytes respond predominantly to myelin oligodendrocyte glycoprotein. J Clin Invest. 1993;92(6):2602–8. doi: 10.1172/JCI116875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Layfield LJ, Goldstein N, Perkinson KR, Proia AD. Interlaboratory variation in results from immunohistochemical assessment of estrogen receptor status. Breast J. 2003;9(3):257–9. doi: 10.1046/j.1524-4741.2003.09325.x. [DOI] [PubMed] [Google Scholar]
- Lee JH, Cheng R, Honig LS, Vonsattel JP, Clark L, Mayeux R. Association between genetic variants in SORL1 and autopsy-confirmed Alzheimer disease. Neurology. 2008;70(11):887–9. doi: 10.1212/01.wnl.0000280581.39755.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JH, Cheng R, Schupf N, Manly J, Lantigua R, Stern Y, Rogaeva E, Wakutani Y, Farrer L, St George-Hyslop P, et al. The association between genetic variants in SORL1 and Alzheimer disease in an urban, multiethnic, community-based cohort. Arch Neurol. 2007;64(4):501–6. doi: 10.1001/archneur.64.4.501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Rowland C, Catanese J, Morris J, Lovestone S, O'Donovan MC, Goate A, Owen M, Williams J, Grupe A. SORL1 variants and risk of late-onset Alzheimer's disease. Neurobiol Dis. 2008;29(2):293–6. doi: 10.1016/j.nbd.2007.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, St John MA, Zhou X, Kim Y, Sinha U, Jordan RC, Eisele D, Abemayor E, Elashoff D, Park NH, et al. Salivary transcriptome diagnostics for oral cancer detection. Clin Cancer Res. 2004;10(24):8442–50. doi: 10.1158/1078-0432.CCR-04-1167. [DOI] [PubMed] [Google Scholar]
- Ludwig JA, Weinstein JN. Biomarkers in cancer staging, prognosis and treatment selection. Nat Rev Cancer. 2005;5(11):845–56. doi: 10.1038/nrc1739. [DOI] [PubMed] [Google Scholar]
- Ma QL, Teter B, Ubeda OJ, Morihara T, Dhoot D, Nyby MD, Tuck ML, Frautschy SA, Cole GM. Omega-3 fatty acid docosahexaenoic acid increases SorLA/LR11, a sorting protein with reduced expression in sporadic Alzheimer's disease (AD): relevance to AD prevention. J Neurosci. 2007;27(52):14299–307. doi: 10.1523/JNEUROSCI.3593-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maes OC, Xu S, Yu B, Chertkow HM, Wang E, Schipper HM. Transcriptional profiling of Alzheimer blood mononuclear cells by microarray. Neurobiol Aging. 2007;28(12):1795–809. doi: 10.1016/j.neurobiolaging.2006.08.004. [DOI] [PubMed] [Google Scholar]
- Maglione V, Cannella M, Gradini R, Cislaghi G, Squitieri F. Huntingtin fragmentation and increased caspase 3, 8 and 9 activities in lymphoblasts with heterozygous and homozygous Huntington's disease mutation. Mech Ageing Dev. 2006;127(2):213–6. doi: 10.1016/j.mad.2005.09.011. [DOI] [PubMed] [Google Scholar]
- Maglione V, Cannella M, Martino T, De Blasi A, Frati L, Squitieri F. The platelet maximum number of A2A-receptor binding sites (Bmax) linearly correlates with age at onset and CAG repeat expansion in Huntington's disease patients with predominant chorea. Neurosci Lett. 2006;393(1):27–30. doi: 10.1016/j.neulet.2005.09.037. [DOI] [PubMed] [Google Scholar]
- Maglione V, Giallonardo P, Cannella M, Martino T, Frati L, Squitieri F. Adenosine A2A receptor dysfunction correlates with age at onset anticipation in blood platelets of subjects with Huntington's disease. Am J Med Genet B Neuropsychiatr Genet. 2005;139(1):101–5. doi: 10.1002/ajmg.b.30223. [DOI] [PubMed] [Google Scholar]
- Mahad DJ, Lawry J, Howell SJ, Woodroofe MN. Longitudinal study of chemokine receptor expression on peripheral lymphocytes in multiple sclerosis: CXCR3 upregulation is associated with relapse. Mult Scler. 2003;9(2):189–98. doi: 10.1191/1352458503ms899oa. [DOI] [PubMed] [Google Scholar]
- Malucchi S, Gilli F, Caldano M, Marnetto F, Valentino P, Granieri L, Sala A, Capobianco M, Bertolotto A. Predictive markers for response to interferon therapy in patients with multiple sclerosis. Neurology. 2008;70(13 Pt 2):1119–27. doi: 10.1212/01.wnl.0000304040.29080.7b. [DOI] [PubMed] [Google Scholar]
- Manolio T. Novel risk markers and clinical practice. N Engl J Med. 2003;349(17):1587–9. doi: 10.1056/NEJMp038136. [DOI] [PubMed] [Google Scholar]
- Marioni JC, Mason CE, Mane SM, Stephens M, Gilad Y. RNA-seq: An assessment of technical reproducibility and comparison with gene expression arrays. Genome Res. 2008;18(9):1509–17. doi: 10.1101/gr.079558.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marx J. Trafficking protein suspected in Alzheimer's disease. Science. 2007;315:314. doi: 10.1126/science.315.5810.314. [DOI] [PubMed] [Google Scholar]
- Mhyre TR, Loy R, Tariot PN, Profenno LA, Maguire-Zeiss KA, Zhang D, Coleman PD, Federoff HJ. Proteomic analysis of peripheral leukocytes in Alzheimer's disease patients treated with divalproex sodium. Neurobiol Aging. 2007 doi: 10.1016/j.neurobiolaging.2007.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller DW, Hague SM, Clarimon J, Baptista M, Gwinn-Hardy K, Cookson MR, Singleton AB. Alpha-synuclein in blood and brain from familial Parkinson disease with SNCA locus triplication. Neurology. 2004;62(10):1835–8. doi: 10.1212/01.wnl.0000127517.33208.f4. [DOI] [PubMed] [Google Scholar]
- Minster RL, DeKosky ST, Kamboh MI. No association of SORL1 SNPs with Alzheimer's disease. Neurosci Lett. 2008;440(2):190–2. doi: 10.1016/j.neulet.2008.05.082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohr S, Liew CC. The peripheral-blood transcriptome: new insights into disease and risk assessment. Trends Mol Med. 2007;13(10):422–32. doi: 10.1016/j.molmed.2007.08.003. [DOI] [PubMed] [Google Scholar]
- Muthukumar T, Dadhania D, Ding R, Snopkowski C, Naqvi R, Lee JB, Hartono C, Li B, Sharma VK, Seshan SV, et al. Messenger RNA for FOXP3 in the urine of renal-allograft recipients. N Engl J Med. 2005;353(22):2342–51. doi: 10.1056/NEJMoa051907. [DOI] [PubMed] [Google Scholar]
- Nagai Y, Ueno S, Saeki Y, Soga F, Hirano M, Yanagihara T. Decrease of the D3 dopamine receptor mRNA expression in lymphocytes from patients with Parkinson's disease. Neurology. 1996;46(3):791–5. doi: 10.1212/wnl.46.3.791. [DOI] [PubMed] [Google Scholar]
- Ntzani EE, Ioannidis JP. Predictive ability of DNA microarrays for cancer outcomes and correlates: an empirical assessment. Lancet. 2003;362(9394):1439–44. doi: 10.1016/S0140-6736(03)14686-7. [DOI] [PubMed] [Google Scholar]
- O'Toole M, Janszen DB, Slonim DK, Reddy PS, Ellis DK, Legault HM, Hill AA, Whitley MZ, Mounts WM, Zuberek K, et al. Risk factors associated with beta-amyloid(1-42) immunotherapy in preimmunization gene expression patterns of blood cells. Arch Neurol. 2005;62(10):1531–6. doi: 10.1001/archneur.62.10.1531. [DOI] [PubMed] [Google Scholar]
- Offe K, Dodson SE, Shoemaker JT, Fritz JJ, Gearing M, Levey AI, Lah JJ. The lipoprotein receptor LR11 regulates amyloid beta production and amyloid precursor protein traffic in endosomal compartments. J Neurosci. 2006;26(5):1596–603. doi: 10.1523/JNEUROSCI.4946-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oki T, Takahashi S, Kuwabara S, Yoshiyama Y, Mori M, Hattori T, Suzuki N. Increased ability of peripheral blood lymphocytes to degrade laminin in multiple sclerosis. J Neurol Sci. 2004;222(12):7–11. doi: 10.1016/j.jns.2004.03.026. [DOI] [PubMed] [Google Scholar]
- Panov A, Obertone T, Bennett-Desmelik J, Greenamyre JT. Ca(2+)-dependent permeability transition and complex I activity in lymphoblast mitochondria from normal individuals and patients with Huntington's or Alzheimer's disease. Ann N Y Acad Sci. 1999;893:365–8. doi: 10.1111/j.1749-6632.1999.tb07856.x. [DOI] [PubMed] [Google Scholar]
- Panov AV, Lund S, Greenamyre JT. Ca2+-induced permeability transition in human lymphoblastoid cell mitochondria from normal and Huntington's disease individuals. Mol Cell Biochem. 2005;269(12):143–52. doi: 10.1007/s11010-005-3454-9. [DOI] [PubMed] [Google Scholar]
- Petrozzi L, Lucetti C, Gambaccini G, Bernardini S, Del Dotto P, Migliore L, Scarpato R, Bonuccelli U. Cytogenetic analysis oxidative damage in lymphocytes of Parkinson's disease patients. Neurol Sci. 2001;22(1):83–4. doi: 10.1007/s100720170058. [DOI] [PubMed] [Google Scholar]
- Prat A, Biernacki K, Antel JP. Th1 and Th2 lymphocyte migration across the human BBB is specifically regulated by interferon beta and copolymer-1. J Autoimmun. 2005;24(2):119–24. doi: 10.1016/j.jaut.2005.01.004. [DOI] [PubMed] [Google Scholar]
- Prat A, Biernacki K, Lavoie JF, Poirier J, Duquette P, Antel JP. Migration of multiple sclerosis lymphocytes through brain endothelium. Arch Neurol. 2002;59(3):391–7. doi: 10.1001/archneur.59.3.391. [DOI] [PubMed] [Google Scholar]
- Ransohoff DF. Rules of evidence for cancer molecular-marker discovery and validation. Nat Rev Cancer. 2004;4(4):309–14. doi: 10.1038/nrc1322. [DOI] [PubMed] [Google Scholar]
- Ransohoff DF. Bias as a threat to the validity of cancer molecular-marker research. Nat Rev Cancer. 2005;5(2):142–9. doi: 10.1038/nrc1550. [DOI] [PubMed] [Google Scholar]
- Rogaeva E, Meng Y, Lee JH, Gu Y, Kawarai T, Zou F, Katayama T, Baldwin CT, Cheng R, Hasegawa H, et al. The neuronal sortilin-related receptor SORL1 is genetically associated with Alzheimer disease. Nat Genet. 2007 doi: 10.1038/ng1943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Runne H, Kuhn A, Wild EJ, Pratyaksha W, Kristiansen M, Isaacs JD, Regulier E, Delorenzi M, Tabrizi SJ, Luthi-Carter R. Analysis of potential transcriptomic biomarkers for Huntington's disease in peripheral blood. Proc Natl Acad Sci U S A. 2007;104(36):14424–9. doi: 10.1073/pnas.0703652104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saft C, Zange J, Andrich J, Muller K, Lindenberg K, Landwehrmeyer B, Vorgerd M, Kraus PH, Przuntek H, Schols L. Mitochondrial impairment in patients and asymptomatic mutation carriers of Huntington's disease. Mov Disord. 2005;20(6):674–9. doi: 10.1002/mds.20373. [DOI] [PubMed] [Google Scholar]
- Sager KL, Wuu J, Leurgans SE, Rees HD, Gearing M, Mufson EJ, Levey AI, Lah JJ. Neuronal LR11/sorLA expression is reduced in mild cognitive impairment. Ann Neurol. 2007;62(6):640–7. doi: 10.1002/ana.21190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawa A, Wiegand GW, Cooper J, Margolis RL, Sharp AH, Lawler JF, Jr., Greenamyre JT, Snyder SH, Ross CA. Increased apoptosis of Huntington disease lymphoblasts associated with repeat length-dependent mitochondrial depolarization. Nat Med. 1999;5(10):1194–8. doi: 10.1038/13518. [DOI] [PubMed] [Google Scholar]
- Schadt EE, Lamb J, Yang X, Zhu J, Edwards S, Guhathakurta D, Sieberts SK, Monks S, Reitman M, Zhang C, et al. An integrative genomics approach to infer causal associations between gene expression and disease. Nat Genet. 2005;37(7):710–7. doi: 10.1038/ng1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schena M, Shalon D, Davis RW, Brown PO. Quantitative monitoring of gene expression patterns with a complementary DNA microarray. Science. 1995;270(5235):467–70. doi: 10.1126/science.270.5235.467. [DOI] [PubMed] [Google Scholar]
- Scherzer CR, Eklund AC, Morse LJ, Fefer D, Locascio JJ, Schwarzschild M, Schlossmacher MG, Hauser MA, Vance JM, Sudarsky L, et al. Molecular markers of early Parkinson's disease based on gene expression in blood. Proc Natl Acad Sci U S A. 2007;104:955–960. doi: 10.1073/pnas.0610204104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scherzer CR, Feany MB. Yeast genetics targets lipids in Parkinson's disease. Trends Genet. 2004;20(7):273–7. doi: 10.1016/j.tig.2004.05.005. [DOI] [PubMed] [Google Scholar]
- Scherzer CR, Jensen RV, Gullans SR, Freese A, Simeone FA, Leone P, Janson C. Principles of Molecular Neurosurgery. Vol. 18. Karger; Basel: 2004. Simplifying complex neurodegenerative diseases by gene chip analysis; pp. 246–57. [Google Scholar]
- Scherzer CR, Offe K, Gearing M, Rees HD, Fang G, Heilman CJ, Schaller C, Bujo H, Levey AI, Lah JJ. Loss of apolipoprotein E receptor LR11 in Alzheimer disease. Arch Neurol. 2004;61(8):1200–5. doi: 10.1001/archneur.61.8.1200. [DOI] [PubMed] [Google Scholar]
- Schlossmacher MG, Ostaszewski BL, Hecker LI, Celi A, Haass C, Chin D, Lieberburg I, Furie BC, Furie B, Selkoe DJ. Detection of distinct isoform patterns of the beta-amyloid precursor protein in human platelets and lymphocytes. Neurobiol Aging. 1992;13(3):421–34. doi: 10.1016/0197-4580(92)90117-g. [DOI] [PubMed] [Google Scholar]
- Schulz JB, Beal MF. Mitochondrial dysfunction in movement disorders. Curr Opin Neurol. 1994;7(4):333–9. doi: 10.1097/00019052-199408000-00010. [DOI] [PubMed] [Google Scholar]
- Sharp FR, Xu H, Lit L, Walker W, Apperson M, Gilbert DL, Glauser TA, Wong B, Hershey A, Liu DZ, et al. The future of genomic profiling of neurological diseases using blood. Arch Neurol. 2006;63(11):1529–36. doi: 10.1001/archneur.63.11.1529. [DOI] [PubMed] [Google Scholar]
- Shi L, Reid LH, Jones WD, Shippy R, Warrington JA, Baker SC, Collins PJ, de Longueville F, Kawasaki ES, Lee KY, et al. The MicroArray Quality Control (MAQC) project shows inter- and intraplatform reproducibility of gene expression measurements. Nat Biotechnol. 2006;24(9):1151–1161. doi: 10.1038/nbt1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibata N, Ohnuma T, Baba H, Higashi S, Nishioka K, Arai H. Genetic association between SORL1 polymorphisms and Alzheimer's disease in a Japanese population. Dement Geriatr Cogn Disord. 2008;26(2):161–4. doi: 10.1159/000149821. [DOI] [PubMed] [Google Scholar]
- Sotiriou C, Piccart MJ. Taking gene-expression profiling to the clinic: when will molecular signatures become relevant to patient care? Nat Rev Cancer. 2007;7(7):545–53. doi: 10.1038/nrc2173. [DOI] [PubMed] [Google Scholar]
- Sparano JA. TAILORx: trial assigning individualized options for treatment (Rx) Clin Breast Cancer. 2006;7(4):347–50. doi: 10.3816/CBC.2006.n.051. [DOI] [PubMed] [Google Scholar]
- Squitieri F, Cannella M, Sgarbi G, Maglione V, Falleni A, Lenzi P, Baracca A, Cislaghi G, Saft C, Ragona G, et al. Severe ultrastructural mitochondrial changes in lymphoblasts homozygous for Huntington disease mutation. Mech Ageing Dev. 2006;127(2):217–20. doi: 10.1016/j.mad.2005.09.010. [DOI] [PubMed] [Google Scholar]
- Stieler JT, Lederer C, Bruckner MK, Wolf H, Holzer M, Gertz HJ, Arendt T. Impairment of mitogenic activation of peripheral blood lymphocytes in Alzheimer's disease. Neuroreport. 2001;12(18):3969–72. doi: 10.1097/00001756-200112210-00023. [DOI] [PubMed] [Google Scholar]
- Stoy N, Mackay GM, Forrest CM, Christofides J, Egerton M, Stone TW, Darlington LG. Tryptophan metabolism and oxidative stress in patients with Huntington's disease. J Neurochem. 2005;93(3):611–23. doi: 10.1111/j.1471-4159.2005.03070.x. [DOI] [PubMed] [Google Scholar]
- Strand AD, Aragaki AK, Shaw D, Bird T, Holton J, Turner C, Tapscott SJ, Tabrizi SJ, Schapira AH, Kooperberg C, et al. Gene expression in Huntington's disease skeletal muscle: a potential biomarker. Hum Mol Genet. 2005;14(13):1863–76. doi: 10.1093/hmg/ddi192. [DOI] [PubMed] [Google Scholar]
- Sultan M, Schulz MH, Richard H, Magen A, Klingenhoff A, Scherf M, Seifert M, Borodina T, Soldatov A, Parkhomchuk D, et al. A global view of gene activity and alternative splicing by deep sequencing of the human transcriptome. Science. 2008;321(5891):956–60. doi: 10.1126/science.1160342. [DOI] [PubMed] [Google Scholar]
- Sumner CJ, Kolb SJ, Harmison GG, Jeffries NO, Schadt K, Finkel RS, Dreyfuss G, Fischbeck KH. SMN mRNA and protein levels in peripheral blood: biomarkers for SMA clinical trials. Neurology. 2006;66(7):1067–73. doi: 10.1212/01.wnl.0000201929.56928.13. [DOI] [PubMed] [Google Scholar]
- Tan EK, Lee J, Chen CP, Teo YY, Zhao Y, Lee WL. SORL1 haplotypes modulate risk of Alzheimer's disease in Chinese. Neurobiol Aging. 2007 doi: 10.1016/j.neurobiolaging.2007.10.013. [DOI] [PubMed] [Google Scholar]
- Tan PK, Downey TJ, Spitznagel EL, Jr., Xu P, Fu D, Dimitrov DS, Lempicki RA, Raaka BM, Cam MC. Evaluation of gene expression measurements from commercial microarray platforms. Nucleic Acids Res. 2003;31(19):5676–84. doi: 10.1093/nar/gkg763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tayebati SK, Amenta F, Amici S, El-Assouad D, Gallai V, Ricci A, Parnetti L. Peripheral blood lymphocytes muscarinic cholinergic receptor subtypes in Alzheimer's disease: a marker of cholinergic dysfunction? J Neuroimmunol. 2001;121(12):126–31. doi: 10.1016/s0165-5728(01)00435-0. [DOI] [PubMed] [Google Scholar]
- Vallittu AM, Saraste M, Airas L. CCR7 expression on peripheral blood lymphocytes is up-regulated following treatment of multiple sclerosis with interferon-beta. Neurol Res. 2007;29(8):763–6. doi: 10.1179/016164107X228633. [DOI] [PubMed] [Google Scholar]
- van 't Veer LJ, Dai H, van de Vijver MJ, He YD, Hart AA, Mao M, Peterse HL, van der Kooy K, Marton MJ, Witteveen AT, et al. Gene expression profiling predicts clinical outcome of breast cancer. Nature. 2002;415(6871):530–6. doi: 10.1038/415530a. [DOI] [PubMed] [Google Scholar]
- Vogt MH, Lopatinskaya L, Smits M, Polman CH, Nagelkerken L. Elevated osteopontin levels in active relapsing-remitting multiple sclerosis. Ann Neurol. 2003;53(6):819–22. doi: 10.1002/ana.10606. [DOI] [PubMed] [Google Scholar]
- Webster JA, Myers AJ, Pearson JV, Craig DW, Hu-Lince D, Coon KD, Zismann VL, Beach T, Leung D, Bryden L, et al. Sorl1 as an Alzheimer's Disease Predisposition Gene? Neurodegener Dis. 2007 doi: 10.1159/000110789. [DOI] [PubMed] [Google Scholar]
- www.expressionanalysis.com Expression Profiling of Whole Blood Specimens on Illumina BeadChips. Expression Analysis Tech Note. 2007 October; www.expressionanalysis.com D., NC.
- Yoshino H, Nakagawa-Hattori Y, Kondo T, Mizuno Y. Mitochondrial complex I and II activities of lymphocytes and platelets in Parkinson's disease. J Neural Transm Park Dis Dement Sect. 1992;4(1):27–34. doi: 10.1007/BF02257619. [DOI] [PubMed] [Google Scholar]