Abstract
Our studies in children with rheumatic diseases have led to the identification of two of the oldest cytokines, type I Interferon (IFN) and Interleukin 1 (IL-1), as important pathogenic players in Systemic Lupus Erythematosus (SLE) and Systemic onset Juvenile Arthritis (SoJIA) respectively. These findings were obtained by studying the transcriptional profiles of patient blood cells and by assessing the biological and transcriptional effect(s) of active patient sera on healthy blood cells. We also identified a signature which can be used to promptly diagnose SoJIA from other febrile conditions. Finally, our pilot clinical trials using IL-1 blockers have shown remarkable clinical benefits in SoJIA patients refractory to other medications.
INTRODUCTION
The immune system evolved to protect us from microbes (1, 2). The antigen (Ag)-nonspecific innate immunity and Ag-specific adaptive immunity synergize to eradicate the invading pathogen through cells, such as neutrophils, dendritic cells (DCs) and lymphocytes, and through their effector proteins, including antimicrobial peptides, complement, and antibodies (3). Through their unique capacity to present antigen and activate naïve T lymphocytes and their ability to secrete potent cytokines which impact both innate and adaptive immunity cells, DCs are the conductors of the immune system orchestra (4, 5). Under steady state conditions, DCs are not activated, so called immature, and are mostly prone to tolerance induction (6). Upon microbial invasion, inflammation leads to DC activation and maturation, which is thought to induce immunity. Under these inflammatory conditions a novel pool of DCs is generated through the maturation of monocytes. The intrinsic complexity of the immune system renders it prone to dysfunction, leading to cancer, autoimmunity, chronic inflammation, chronic infections and allergy (5, 7).
Rheumatology has been a central medical specialty leading to the development of clinical immunology. Most rheumatic diseases are diagnosed based on a combination of clinical and laboratory findings, as they characteristically lack specific diagnostic tests. Even though most of these diseases are chronic and involve remissions and relapses, reliable biomarkers of disease activity and predictors of flares are missing. Furthermore, many of these diseases continue to be treated in a non-specific manner with drugs that suppress broad inflammatory cascades.
Rheumatic diseases might be triggered by yet to be identified environmental factors in patients with a susceptible genetic background. Many years of genetic studies in humans and animal models have yielded a wealth of candidate genes whose mutations might contribute to disease development. None of these candidate genes however has led to the identification of novel therapeutic targets. Animal models supported, for example, a fundamental role for IL-1b in the pathogenesis of RA, as IL1b-deficient mice fail to develop chronic, erosive arthritis in response to arthritogenic stimuli (8) and IL-1Ra-deficient mice develop autoimmunity and arthritis spontaneously (9). However, clinical trials using IL-1 blockers in patients with this disease did not yield the expected beneficial results (10).
An important breakthrough in rheumatology occurred in the late 80s when Feldman and Maini proposed that a single cytokine, tumor necrosis factor (TNF), was of major importance in the pathogenesis of RA (11). Their prediction, which was based on experiments made with human synovial tissue explants, has been confirmed in multiple clinical trials using different modalities of TNF-alpha blockers (12, 13). This therapy has in fact changed the face of rheumatic diseases characterized by erosive joint disease including RA, psoriatic arthritis and the spondyloarthropaties. The beneficial effect of blocking a single cytokine in several diseases with common as well as unique clinical manifestations took the rheumatology community by surprise. We will next review how simple human studies have helped us extend this paradigm to other cytokines and to an even wider array of rheumatic diseases.
One of the most efficient tools that we have used in our studies is blood gene expression profiling. The use of this technology to discover diagnostic and prognostic biomarker signatures was first developed in the field of cancer (14, 15). These landmark studies, which focused on the analysis of tumor tissue, paved the way for a wider use of this technology in clinical research. We proposed to study the blood of patients with immune-mediated diseases (16) because the blood is the pipeline of the immune system. Indeed, every cell that circulates in the blood is associated to the immune system, including cells of the innate immune system (i.e. neutrophils and NK/NKT cells), the adaptive immune system (T and B cells), and cells at the interface of the two systems (monocytes and dendritic cells). Even red blood cells and platelets express molecules (i.e. complement receptors, co-stimulatory molecules etc) that contribute to the regulation of immune reactions. Thus, alterations in blood gene expression profiles reflect altered blood cell composition and/or signaling pathways (i.e. those induced by pathogen recognition, inflammatory cytokines etc.) that might originate in the tissues at sites of inflammation.
Two diseases have attracted our attention in the past ten years because they represented the most difficult and common challenges for the pediatric rheumatologist: Systemic Lupus Erythematosus (SLE) and Systemic onset Juvenile Arthritis (SoJIA) (Figure 1). The study of children offers several advantages over the study of adults, such as the possibility of analyzing samples at the time of disease diagnosis, before initiating treatments with drugs that for the most part have protean effects on the immune system. Children also tend to lack co-morbid conditions and display more aggressive disease, resulting in overall greater sample homogeneity.
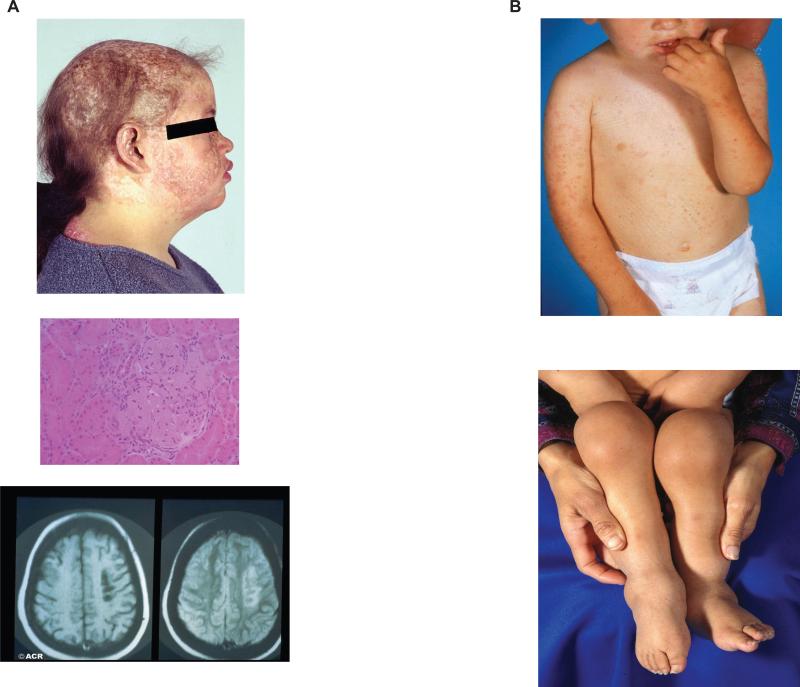
Figure 1. Clinical features of pediatric SLE and SoJIA.
A (top). Cutaneous involvement in a 12 year old girl with SLE; this patient also presented systemic manifestations including nephritis; (medium) light microscopy section showing lupus glomerulonephritis; (bottom) MRI showing lupus CNS involvement. B. (top) Salmon-colored rash typical of SoJIA in an 18 month old patient; (bottom) multiple joint swelling in the lower extremities of a 2 year old boy with SoJIA.
SLE and SOJIA, two unmet medical needs
Systemic Lupus Erythematosus (SLE) is the prototype systemic autoimmune disease. Thus, SLE patients present with the widest array of clinical manifestations affecting multiple organs, including the skin, kidneys, blood vessels, CNS etc. Although SLE typically affects young adults, especially females, up to 20% of all cases of lupus start in childhood. Children with SLE have more active disease at presentation and over time than adult SLE patients do, especially when renal disease is considered. Compared with adults with SLE, children require more intensive drug therapy and accrue more damage over time, often due to both disease severity and side effects of medications (17).
Diagnosing SLE in children requires the same 4/11 criteria established by the American College of Rheumatology to diagnose SLE in adults. Measuring disease activity at any age represents a challenge to the rheumatologist. At least 6 composite measures of SLE global disease activity are available (18-23). These instruments provide metrics to document and quantify disease activity. Current activity indexes not always coincide however with clinical impressions. Some of the included measures, for example, are not easy to obtain. Conversely, given the heterogeneous nature of the clinical disease, not all SLE manifestations are computed in these instruments, making the overall assessment of the patient condition difficult. Hence, there is an important need to develop efficient and objective systems to assess global SLE disease activity.
Juvenile idiopathic arthritis (JIA) is the most common rheumatic disease in childhood and an important cause of short and long-term disability. The term JIA includes a heterogeneous group of diseases, each of which may have a variety of causes and elicit a variety of host responses. All are characterized, however, by the development of idiopathic peripheral arthritis thought to be secondary to an immuno-inflammatory pathogenesis, possibly triggered by contact with external antigen(s). JIA is classified according to three major types of disease presentation: oligoarthritis, polyarthritis and systemic onset (SoJIA). Each of these groups is defined by a constellation of clinical signs and symptoms during the first six months of illness. Systemic Onset Juvenile Idiopathic Arthritis (SoJIA) represents up to 20% of all the cases of JIA (24). This disease is unique in terms of clinical manifestations, prognosis and lack of response to available therapies. High, spiking fever, a salmon-color rash that follows the fever spikes, anemia, leukocytosis and elevated ESR are the main initial features of the disease, sometimes lasting several months before the diagnosis can be established once arthritis appears. SoJIA patients usually lack autoantibodies and autoreactive T cells. As the symptoms are non specific and can mimic infections, malignancies, etc, patients undergo a series of very costly diagnostic tests and prolonged hospitalizations. The systemic manifestations may last from weeks to months and eventually tend to subside to be followed by the development of chronic arthritis. About 50% of patients will present oligoarticular involvement and will eventually recover. The other half will evolve into a polyarticular pattern, the prognosis of which correlates with the number of joints involved 6 months into the disease course. According to some studies, 48% of children with SoJIA will have active arthritis 10 years after the diagnosis is made (25, 26). These patients display an increased risk of developing hemophagocytic syndrome, also known as macrophage activation syndrome (MAS) (25). This disorder, which can occur as well in the context of infectious and neoplastic diseases, is associated with serious morbidity and/or death. Its etiology, especially in the context of SoJIA, is unknown.
Progresses in these diseases need to occur at multiple levels including: 1) understanding pathogenesis, 2) development of specific diagnostic tests, 3) identification of robust biomarkers of disease activity, 4) development of targeted therapies.
SLE AND TYPE I INTERFERON-MEDIATED DISEASES
Loss of tolerance to nuclear antigens and development of immune complexes that deposit in tissues and cause widespread inflammation are considered the hallmarks of lupus. Altered T/B cell interactions have been proposed to represent the pathogenic mechanism leading to disease (27). Indeed, we found that the peripheral B cell compartment is altered in SLE as reflected by an overall decrease in circulating naïve and conventional memory cells, an upregulation of germinal center markers like CD38, and an expansion of CD27- memory cells and plasma cell precursors (28, 29). B cell repertoire studies have shown that pediatric SLE patients fail to remove self-reactive and polyreactive antibody-expressing B cells from the naive blood repertoire (30). Studies on tonsils from adult SLE patients have also shown a defective tolerance checkpoint at the transition between follicular mantle and germinal center B cells (31).
Given that dendritic cells (DCs) are efficient stimulators of B and T lymphocytes as well as key controllers of immunity (4) and tolerance (6, 32), we hypothesized that SLE might actually be driven by unabated DC activation.
SLE Monocytes Display Functions of Dendritic Cells
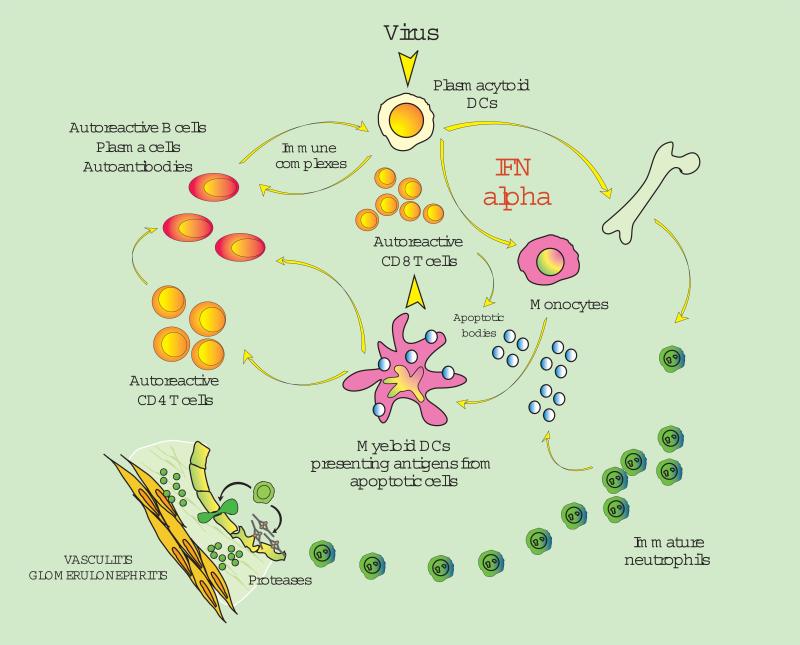
Monocytes represent circulating precursors of macrophages and DCs. Monocytes have limited antigen presenting capacity, as they cannot initiate primary immune responses, i.e. they cannot induce the proliferation of allogeneic naïve T cells in mixed lymphocyte reactions (MLR). However, blood monocytes from some SLE patients are able to induce potent MLR and thus behave like DCs (33). To further understand the mechanism responsible for this unusual property of lupus monocytes, Patrick Blanco exposed healthy monocytes to SLE serum and observed the rapid generation of cells with DC morphology and function, suggesting that SLE blood represents a DC-inducing environment (33) (Figure 2 A and B). This led us to hypothesize that unabated DC maturation could lead to the activation/expansion of autoreactive T cells which have escaped central tolerance in the thymus. Autoreactive T cells, which are silenced under steady state conditions by immature DCs, now expand and start attacking their targets (reviewed in (34)) (Figure 3).
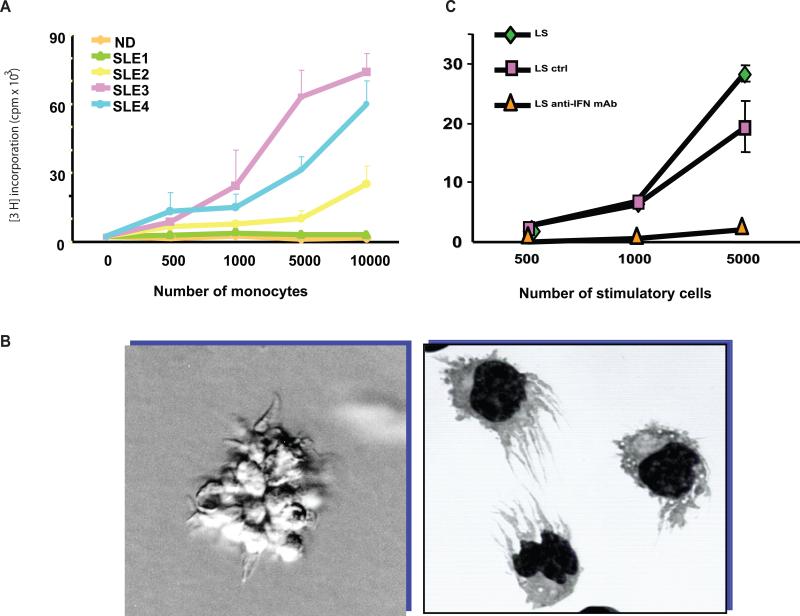
Figure 2. Dendritic cell alterations in SLE are driven by IFN-alpha.
A. Monocytes from ¾ pediatric SLE patients induce significant proliferation of allogeneic naïve CD4+ T cells. B. SLE serum induces healthy monocytes to differentiate into cells with DC properties. C. An IFN-alpha neutralizing antibody blocks the induction of DCs by SLE serum, as measured using the proliferation of allogeneic T cells.
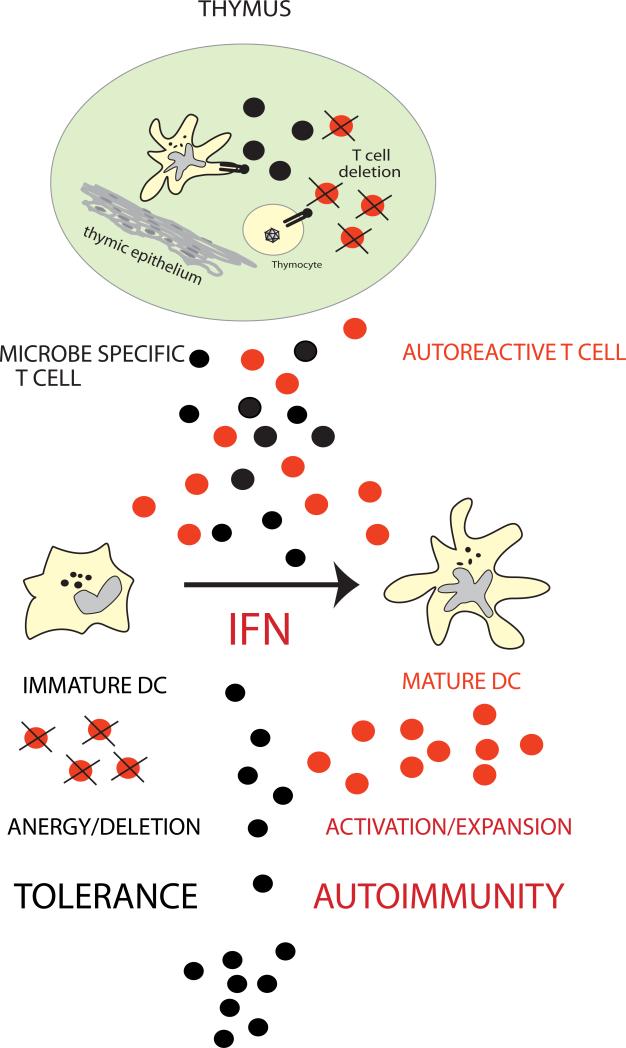
Figure 3. Fate of Autoreactive T Cells.
Autoreactive thymic escapees (in red) are normally silenced in the periphery through the recognition of autoantigens presented by immature DCs (peripheral tolerance). An excess of IFN-αβ, as observed in SLE, induces unabated DC maturation, which leads to activation/expansion of autoreactive T cells. (Original figure published in Immunity 2004;20:539-50)
IFN-alpha is responsible for monocyte activation in SLE
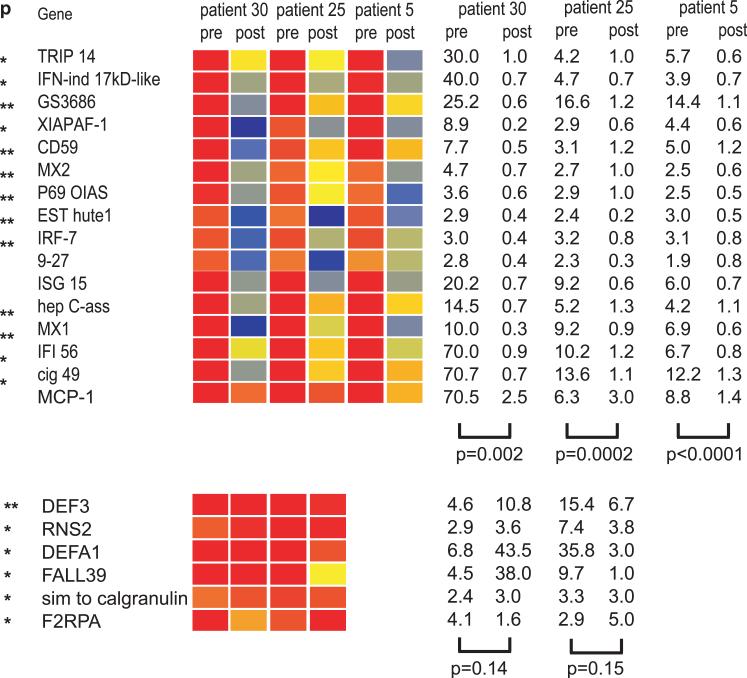
Several cytokines have been shown to allow the differentiation of precursor cells into DCs (7). In particular, IFN-α together with GM-CSF drives monocytes to become DCs (35-37). Accordingly, we found that the DC inducing property of SLE serum is dependent on type I IFN, as it can be blocked with a neutralizing anti-IFN α antibody (Figure 2C). Furthermore, the ability of the serum to induce DCs correlates with disease activity according to the SLEDAI (33). Thirteen different IFN-α subtypes together with IFN-beta, IFN-kappa, IFN-epsilon, and IFN-omega (reviewed in(38) compose the type I IFN family.~ Type I IFNs can be produced by numerous cell types. Plasmacytoid dendritic cells (pDCs), however, are equipped to quickly secrete large amounts of type I IFN (39, 40) and likely contribute type I IFN in SLE patients. Though pDC numbers are reduced in SLE blood (33, 41), these cells massively infiltrate inflamed lupus skin (42, 43). The decrease in SLE blood pDCs might thus result from their accelerated migration to inflammation sites, as demonstrated in allergen challenged nasal mucosa (44).
Studies in the 60's revealed that type I IFNs display immunoregulatory properties. Through their effect on B cells, type I IFNs directly enhance primary antibody responses to soluble proteins and induce the production of all subclasses of IgG in mice (45). In studies performed by Gaetan Jego, we found that pDCs triggered with virus induce CD40-activated B cells to differentiate into plasma cells through two pDC cytokines that act sequentially. First, type I IFN generates non-Ig-secreting plasma blasts, second, IL-6 induces the differentiation of these plasmablasts into Ig-secreting plasma cells (46) (Figure 4). The plasmablasts generated upon exposure to IFN-α in vitro are phenotypically identical to those that we had previously reported in the blood of pediatric lupus patients (28) as well as those found by others upon vaccination of healthy individuals (47). While the presence of these cells in the blood of vaccinated subjects is short-lived, they can be detected in SLE blood for extended periods of time (Arce, Pascual et al., submitted). Thus, type I IFN may contribute to increase autoantibody secretion and immune complex formation in SLE by directly activating B cells and inducing their differentiation into plasma cells. Murine studies indicate that type I IFN also acts directly on T cells, preventing activated T cell death during inflammatory responses (48). Exposure of CD8+ T cells to type I IFN appears critical for the generation of effector and memory cells in response to viral infection (49). Recent studies by Patrick Blanco showed that DCs generated with SLE patient sera promote the differentiation of CD8+ effector T lymphocytes. These CD8+ T cells can kill target cells and generate nucleosomes and SLE autoantigens in a granzyme-dependent manner (50). Nucleosomes may be uptaken by DCs and presented to T and B cells. Indeed, administration of DC loaded with apoptotic cells consistently triggers autoimmune responses in mice, although clinical autoimmunity only develops in genetically susceptible recipients (51, 52).
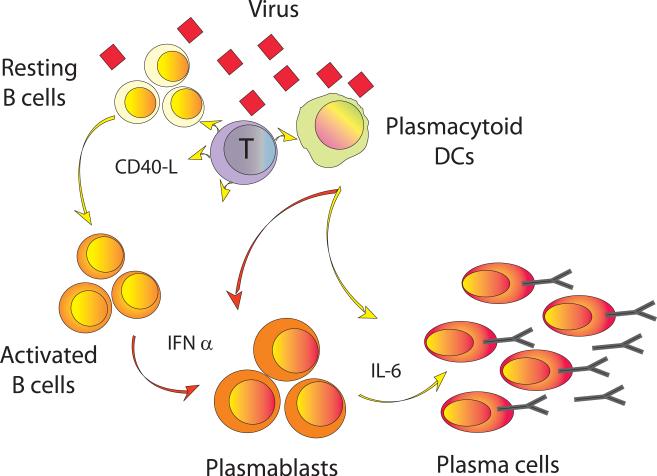
Figure 4. Plasmacytoid dendritic cells induce plasma cell differentiation through type I Interferon and Interleukin 6.
Upon virus encounter, pDCs promptly secrete type I IFN. The cells differentiate into DCs presenting viral antigens to T cells, which promptly secrete IL-2 and turns on CD40-L that signals pDCs to secrete IL-6 and activates B cells. Activated B cells differentiate into plasma blasts in response to type I IFN. The secretion of IL-6 further induces the plasmablasts to become plasma cells. The T cells also contribute importantly through their secretion of IL-2 and IL-10, the combination of all signals leading to the generation of CD38++ long-lived plasma cells. (Original figure published in Immunity, 2003, 19: 225-234)
We discussed above how the effects of type I IFN on the innate and adaptive immune system could explain many SLE features. Furthermore, the use of recombinant IFN-α to treat patients with cancer and chronic viral infections led to a wealth of clinical observations supporting the potential role of this cytokine in the development of autoimmunity. Thus, treatment with type I IFN induces autoantibody formation in 4%-19% of patients and a variety of SLE symptoms have been reported in 0.15%-0.7% of them (reviewed in (53)). Another important lead came from the group of Lars Ronnblom who showed that the numbers of natural IFN-producing cells (pDCs) were decreased in lupus blood and that DNA and/or nucleic acid-containing immune complexes, which are normally present in SLE serum, could induce these cells to secrete type I IFN (54, 55). Despite all this evidence, a certain skepticism in supporting a central role for this cytokine in the pathogenesis of human SLE drew from the following facts: i) not every SLE patient displays detectable serum IFN-α levels (56, 57), ii) most murine SLE-models do not seem to display type-I IFN dysregulation, and iii) genetic linkage and association studies failed to yield candidate lupus susceptibility genes within the IFN-pathway. We will describe next how addressing these concerns strengthened our original hypothesis.
SLE Leukocyte Gene Expression Profiling confirms the excess of type-I ifn
As type I IFN induces a remarkable cellular transcriptional program (58), we surmised that leukocyte gene expression profiling could be a sensitive test to detect in vivo IFN exposure. In spite of a previous report that failed to show any significant elevation of IFN-inducible genes in the blood of 24 adult SLE patients (59), we found that leukocytes from most (29/30) active pediatric SLE patients displayed a remarkable IFN signature (Figure 5). We were comforted in the value of this observation by the fact that treating SLE patients with high dose IV steroids, which are used to treat disease flares, abrogated this signature (16) (Figure 6). This effect, which might result from pDC depletion (60), further supports the role of pDCs and type I IFN in disease pathogenesis. At the time we described the almost universal expression of a type-I IFN signature in pediatric SLE patients, a similar signature was also reported in ~50% of adults with the disease (61, 62). The higher prevalence of the signature in children might be due to sample selection, as more patients with newly diagnosed, untreated disease were included in the pediatric study. They may also reflect the degree of disease severity and/or a genetic background responsible for the earlier appearance of disease manifestations.
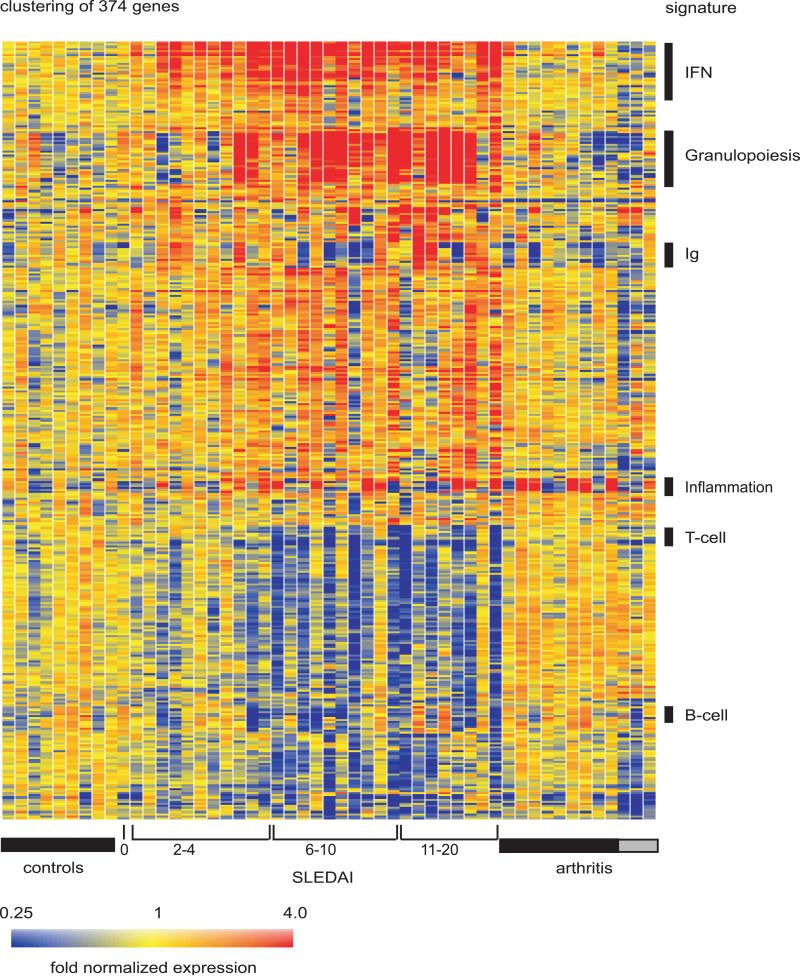
Figure 5. SLE signature.
Hierarchical clustering of gene expression data by blood leukocytes of 9 healthy children, 30 with SLE and 12 with juvenile chronic arthritis including 3 systemic arthritis. The SLE patients were ranked according to their SLEDAI at time of blood draw. Each row represents a separate gene and each column a separate patient. 374 transcript sequences were selected as being differentially expressed in SLE by comparison to healthy patients. The normalized expression index for each transcript sequence (rows) in each sample (columns) is indicated by a color code. Red, yellow and blue squares indicate that expression of the gene is greater than, equal to or less than the mean level of expression across 9 healthy controls. The scale extends from fluorescence ratios of 0.25 to 4.0. (Original figure published in J. Exp. Med. 2003 Mar 17;197(6):711-23)
Figure 6. High dose steroid intravenous pulse extinguishes the type I IFN signature in SLE blood.
Analysis of PBMCs from 3 pediatric SLE patients before and after treatment with high dose i.v. Methylprednisolone (1g/day for 3 days). All patients show down-regulation of IFN-regulated transcripts (upper panel) while expression of not type I IFN-inducible transcripts (lower panel) does not change significantly. p values on the right indicate significance of the gene expression level before and after steroid treatment (paired t-test). Original figure published in J. Exp. Med. 2003 Mar 17;197(6):711-23
One of the puzzles raised by these initial studies was the absence of type I IFN gene transcripts in spite of a plethora of IFN-inducible transcripts in SLE blood. A likely explanation is that the number of pDCs, the cells that transcribe the bulk of type-I IFN genes, is decreased in SLE blood as these cells migrate to tissues. Indeed, IFN-inducible genes and/or proteins can be detected in lupus kidney and skin biopsies (42, 63) . These organs, which are the target of inflammation in lupus, are indeed infiltrated with pDCs (64).
Microarray analyses on blood leukocytes have also shed light on the pathogenesis of drug-induced lupus. When treated with anti-TNF agents, patients with rheumatoid arthritis (RA) and Crohn's disease may develop anti-dsDNA antibodies and reversible SLE (13, 65). Accordingly, we observed that the PBMCs of these patients display an overt IFN signature. Furthermore, our in vitro studies revealed that pDCs exposed to viruses show increased IFN secretion by pDCs when TNF antagonists are added to the cultures (66).
PRE-AUTOIMMUNE MICE DEVELOP SLE UPON ADMINISTRATION OF IFN-ALPHA: THE “ACCELERATED” LUPUS MODEL
Numerous mouse models have been developed to attempt to understand the pathogenesis of SLE. They can be divided into three groups: (1) spontaneous, (2) congenic, and (3) engineered.
The best known spontaneous lupus-like models arise on New Zealand Black (NZB), New Zealand White (NZW), MRL, BXSB and SWR backgrounds. Hybrids of some of these background strains, like the NZB/W F1, NZM2410 and the SWR/NZB F1 develop anti-nuclear antibodies, glomerulonephritis and other features of human disease. The lpr (Fas) or gld (FasL) mutations on the MRL background also give rise to mice displaying many of the characteristics of human lupus (reviewed in (67). It is important to remember, however, that the massive degree of lymphoproliferation that occurs in these mice is not typical of human lupus. Additionally, although MRL-lpr/lpr mice develop a spontaneous arthritis, it shares more features with rheumatoid arthritis than with the arthropathy seen in human SLE, as it includes bone erosions and rheumatoid nodule-like lesions (68, 69) that are rarely seen in human SLE patients. Conversely, humans carrying mutations in the Fas/FasL genes do not develop SLE. Recently, the yaa mutation, that accelerates disease on the BXSB background, has been shown to be due to a translocation of the TLR7 gene into the y chromosome, explaining the predominantly male predisposition to disease in this particular model (70, 71), which is at striking difference with the 9:1 ratio of SLE in humans.
To better dissect the genetic basis of SLE, congenic mice bearing individual predisposing loci on lupus-resistant strains have been generated. Some of the best studied congenic models bear the NZM2410-derived Sle1, Sle2 and Sle3/5 intervals on the B6 background (72-75). These studies have revealed important epistatic interactions among different loci. Sle1, for example, which is a genetic interval responsible for breaking tolerance to chromatin, does not lead to disease manifestations unless in epistasis with other intervals encoding B cell hyperactivity (Sle2), antigenSle3/5 (APC hyperactivity), Yaa (TLR7 duplication) or lpr (Fas mutation).
Many lupus-like syndromes arise in mice deficient in single genes. They fall into two main categories: i) clearance of apoptotic cells and ii) lymphocyte activation and survival (reviewed in (67). Genes within the first (C1q, C2, C4 and Dnase I) and second groups (CD45, CTLA-4, PD1 and FcgRIIb) are also candidate in the human according to linkage and/or association studies (reviewed in (76).
Although none of the above described models predicted that type-I Interferon might play a central role in disease pathogenesis, mice harboring the Y-linked autoimmune accelerator (Yaa) locus have provided clues to support this hypothesis. These mice were recently found to display a duplication in the TLR 7 gene (70, 77). TLR 7 signaling, which can be triggered by immune complexes containing RNA and by RNA-associated autoantigens, induces type-I IFN production by pDCs and activation and pro-inflammatory cytokine production by B cells (78). Transgenic lines overexpressing TLR7 develop spontaneous autoimmunity beyond a 2-fold increase in expression of this gene. Whereas a modest increase in TLR7 gene dosage promoted autoreactive lymphocytes with RNA specificities and myeloid cell proliferation, a substantial increase in TLR7 expression caused fatal acute inflammatory pathology and profound dendritic cell dysregulation (79).
With Alexis Mathian and Sophie Koutouzov, we tested the role of type I IFN in vivo by delivering IFN-α to young (6-8 weeks old) preautoimmune NZB/W F(1) mice. This resulted in the rapid development of severe SLE. Anti-dsDNA Abs appeared as early as 10 days after initiation of IFN-α treatment. Proteinuria and glomerulonephritis-induced death occurred in all treated mice at 9 and 18 weeks respectively, a time when untreated mice did not show any sign of disease (80). Conversely, two independent groups have shown that the cross of both NZB and B6 lpr/lpr mice with a type-I IFN receptor KO strain significantly decreases morbidity and prolongs the survival of these animals (81, 82). Thus, these mouse experiments confirm the results of human studies.
Targeted genetics: how understanding biology helps discovering candidate genes
The genetic basis of human SLE has been the subject of extensive studies for more than 20 years. Indeed, genome-wide linkage studies have identified several genetic loci associated to SLE in multiplex families. Intriguingly, genetic alteration(s) leading to excessive IFN production and/or downstream signaling amplification had never been described as part of these studies. With the idea that this pathway lies at the center of lupus pathology, a focused analysis of 13 genes related to the type I IFN pathway was recently performed. This study revealed two novel strong associations with SLE: three polymorphisms within the flanking/intronic regions of the IFN regulatory factor 5 (IRF 5) gene and two within the coding region of the tyrosine kinase 2 (TYK2) gene (83).The transcription factor IRF5 is involved downstream of the TLR-MyD88 signaling pathway in the induction of pro-inflammatory cytokines (84). IRF5 is expressed in pDCs and B cells and seems to be a critical mediator of TLR7 signaling because i) it is activated by TLR7/8 ligation and ii) its ectopic expression enables type I IFN production in response to TLR7 ligands (85). Immune complexes and nucleic acids that activate pDCs and B cells through TLR-dependent and independent mechanisms can induce type-I IFN production and B cell activation (86-92) (88, 93). Mutations in IRF5 could amplify such responses. The association of SLE with IRF-5 polymorphisms giving rise to unique IRF-5 isoforms has been confirmed across different cohorts and ethnic backgrounds (94). Thus, these studies establish a causal role for type I IFN pathway genes in human SLE and represent a successful example of biology leading genetic studies. Further supporting the relevance of the type I IFN pathway in SLE development, a haplotype within the STAT-4 gene has been recently reported as strongly associated with susceptibility to SLE (95). STAT-4 encodes a transcription factor that transmits signals induced by several key cytokines, including type I IFN.
Using a combination of candidate gene-based to genome-wide low and high density SNP genotyping microarrays, a series of recent genetic association studies have identified 6 novel SLE susceptibility candidate loci (96-99). ITGAM, which encodes the α-chain of αMβ2-integrin (also known as Mac-1, CR3 and CD11b/CD18), displays the highest association. αMβ2 regulates leukocyte adhesion and emigration from the bloodstream via interactions with a wide range of ligands, including ICAM-1 and ICAM-2, C3bi, fibrinogen, GPIbα and othrs (100). αMβ2 levels were reported 20 years ago to be increased on neutrophils in SLE patients with active disease and postulated to possibly contribute to endothelial injury (101). Interesting, our initial gene expression profiling studies of blood mononuclear cells revealed a striking “neutrophil signature” in SLE patient samples (16). This signature, which derives from a fraction of low density neutrophils encompassing different stages of maturation, seems to correlate with the presence of end-organ disease, i.e. nephritis (Vega, Pascual et al., unpublished observations). Furthermore, increased urinary secretion of neutrophil-derived peptides, i.e. lipocalin, correlates with nephritis severity in children with SLE (102). Thus, more pieces of the lupus puzzle are finally fitting together.
A unified view of SLE pathogenesis
SLE flares are often associated to environmental triggers like viral infections. Thus, infection could trigger the unabated production of type I IFN in SLE patients. Our studies indicate that increased bioavailability of type I IFN is fundamental to SLE pathogenesis. It induces and maintains the generation of mature DCs, tilting the fate of autoreactive T lymphocytes which have escaped central tolerance from deletion to activation. These mature DCs activate cytotoxic CD8+ T cells to generate nucleosomes which can be captured and presented by IFN-DCs (50). Together with IL-6, type I IFN promotes the differentiation of mature B cells into plasma cells (103). Thus, the effects of type I IFN on DCs, B and T cells could explain the breakdown of tolerance to nuclear antigens, autoantibody secretion and IC formation characteristic of SLE. Chromatin-containing IC activate i) B cells through the co-engagement of BCR and TLRs and ii) pDCs to secrete more type I IFN through the co-engagement of FcγR and TLRs. How human nucleic acids within IC trigger type I IFN production by pDCs is starting to be understood. HMGB1, a nuclear DNA-binding protein released from necrotic cells, is an essential component of DNA-containing immune complexes that activate plasmacytoid dendritic cells and B cells in response to DNA and contribute to autoimmune pathogenesis. Furthermore, binding of HMGB1 to class A CpG oligodeoxynucleotides also increases cytokine production through the activation of TLR9 and RAGE. This results in an amplification of this pathogenic loop (104). A similar mechanism has been described for the antimicrobial peptide LL37 (also known as CAMP). This peptide, a product of keratinocytes and neutrophils, is the key factor that mediates pDC activation in psoriasis. LL37 converts inert self-DNA into a potent trigger of interferon production by binding the DNA to form aggregated and condensed structures that are delivered to and retained within early endocytic compartments in pDCs to trigger Toll-like receptor 9 (105).
As shown in different murine models of SLE, however, excessive IFN production only induces disease in certain genetic backgrounds (106). Polymorphisms in genes related to the IFN pathway may predispose to SLE by increasing the ability of PDCs to release type-I IFN and pro-inflammatory cytokines upon activation and enhancing B cell responses to these cytokines. We surmise that SLE patients display other genetic defects leading to alterations in autoreactive B cell checkpoints that may be independent from IFN and DCs. Such genetic alterations have been recently described in mice (107) and might allow the survival of autoreactive clones into the peripheral compartment, as it has been described in children with SLE (30). Finally, alterations in innate immunity cell-specific genes, i.e. ITGAM, may explain the predisposition to develop complications such as lupus nephritis. Whether there is a synergistic connection between neutrophil function and the IFN pathway remains to be elucidated (Figure 7).
Figure 7. A unified view of SLE pathogenesis.
SLE flares are often associated to environmental triggers like viral infections. Thus, infection could trigger the unabated production of type I IFN in SLE patients. Increased bioavailability of type I IFN induces and maintains the generation of mature DCs, tilting the fate of autoreactive T lymphocytes which have escaped central tolerance from deletion to activation. These mature DCs activate cytotoxic CD8+ T cells to generate nucleosomes which can be captured and presented by IFN-DCs. Together with IL-6, type I IFN promotes the differentiation of mature B cells into plasma cells. Thus, the effects of type I IFN on DCs, B and T cells could explain the breakdown of tolerance to nuclear antigens, autoantibody secretion and IC formation characteristic of SLE. Innate immunity cells such as neutrophils may also contribute to lupus pathogenesis and end organ damage. (Original figure published in J. Exp. Med. 2003 Mar 17;197(6):711-23)
Alterations of the type I IFN system in SLE have been described almost 30 years ago (56) but were forgotten. The study of SLE patients, however, has recently brought back to the spot light the role of this old cytokine family in the pathogenesis of the disease. Biological studies on blood monocytes and interferon producing cells together with blood gene expression profiling revived these earlier findings. Eventually, blocking IFN may bring relief to patients with this often devastating disease.
Type 1 IFN in other autoimmune diseases
Treatment of cancer and hepatitis patients with IFN-α may lead to thyroiditis, diabetes and other autoimmune manifestations. In dermatomyositis, an autoimmune disease targeting the skin and proximal muscle groups, muscle biopsies reveal infiltration with pDCs as well as IFN-α inducible gene and protein expression (108, 109). Furthermore, levels of IFN-inducible gene transcription in the blood seem to correlate with disease activity (110, 111). Sjogren's syndrome (SS), an autoimmune disease affecting mainly salivary and lacrymal glands, also seems to be associated to alterations in type I IFN production. Indeed, pDCs infiltrate SS salivary glands (112), IFN-inducible genes are overexpressed in minor salivary glands and ocular epithelial cells from patients, and ICs from the serum of patients activate pDCs to secrete type I IFN (113).
Although psoriasis patients are efficiently treated with TNF antagonists, several observations suggest the potential involvement of type I IFN in the pathogenesis of the disease. The induction of psoriasis after injection of recombinant type I IFN (114), the presence of an activated type I IFN signaling pathway in keratinocytes (115), and the development of psoriasiform inflammation in IRF-2-deficient mice (116) all point to the involvement of type I IFN. A functional role of type I IFN in the initiation of psoriasis has been recently demonstrated. Although IFN-α is not elevated in fully established psoriatic plaques, it is produced at early stages. Indeed, pDCs infiltrate the skin of psoriatic patients and become activated to produce IFN-α early during disease formation. In a xenograft model of human psoriasis, blocking type I IFN signaling or inhibiting the ability of pDCs to produce type I IFN prevented the T cell-dependent disease development (117). Thus, the initial stages of psoriasis development might be IFNa -dependent whereas the late stages are TNF-dependent. TNF may also play a role in late-stage end organ damage in SLE patients. Indeed, treatment with TNF blockers led to clinical improvement of arthritis and decreased proteinuria in an open label study including 6 SLE patients. As expected, autoantibodies to dsDNA and cardiolipin increased in the majority of them (118).
INTERLEUKIN 1-MEDIATED DISEASES
In 2002 we decided to study SoJIA because it was one of the most prevalent unmet medical needs in the pediatric rheumatology clinic. Thus, while children with other forms of juvenile arthritis tend to respond dramatically to anti-TNF therapy, the majority of SoJIA patients do not. As reviewed above, some of these patients display long term systemic and articular manifestations that leave them with profound sequelae.
From the early 1990s several pro-inflammatory cytokines, especially IL-6 and TNF, were postulated to play a role in SoJIA based on the detection of elevated levels in the serum or synovial fluid (SF) of these patients (119). Interleukin-6 (IL-6) levels are elevated in SoJIA serum and to correlate with disease activity, including the severity of joint involvement, platelet counts, CRP and fever spikes. Synovial fluid levels of IL-6 are markedly elevated in SoJIA and significantly higher than in patients with other types of JIA or adult rheumatoid arthritis. TNF levels are increased in all subtypes of JIA. In patients with active SoJIA, circulating levels of TNFa, sTNFR1 and sTNFR2 are significantly higher than in controls. The levels of sTNFR1 and sTNFR2, but not those of TNF, are associated with the persistence and severity of systemic symptoms. There are controversial reports regarding serum levels of IL-1 in SoJIA patients. IL-1b, however, may be difficult to detect in the serum as this cytokine binds to large proteins such as b-2-macroglobulin, complement, and the soluble type II IL-1 receptor (120).
We have used two different but complementary approaches based on microarray technology to help our understanding of the pathogenesis of SoJIA: 1) analysis of transcriptional patterns of freshly isolated blood mononuclear cells from active patients; 2) analysis of transcriptional changes induced upon culturing healthy blood mononuclear cells with serum from active patients. Transcriptional analysis of SoJIA PBMCs revealed significant alterations in the expression of >800 trancripts. The majority of these changes, however, were also found in children with systemic inflammation caused by bacterial and viral infections and did not point towards a specific cytokine pathway. The clue came from culture experiments, which revealed that SoJIA serum up-regulates IL-1 transcription in healthy cells after 6 hr incubation (Figure 8). As expected, IL-1 protein secretion was also induced in a disease-activity dependent manner (121). The serum effects included genes known to be induced by IL-1b (i.e. pentraxin 3), or potentially involved in IL-1b secretion (i.e. KCNJ15 or ATP-sensitive inward rectifier potassium channel 15). These transcripts turn out to also be upregulated in the PBMCs from the majority of patients. Furthermore, we found that mononuclear cells from active SoJIA patients secrete an excess of IL-1b protein upon activation. In the same cultures, IL-6 and TNF production is not significantly different from controls, suggesting a specific dysregulation of the IL-1 pathway in patients with SoJIA. Thus, simple experiments performed with active patient sera helped focus our attention on a specific cytokine pathway. This information would have otherwise been diluted among the many transcriptional changes that take place in the blood of patients in vivo (122-124).
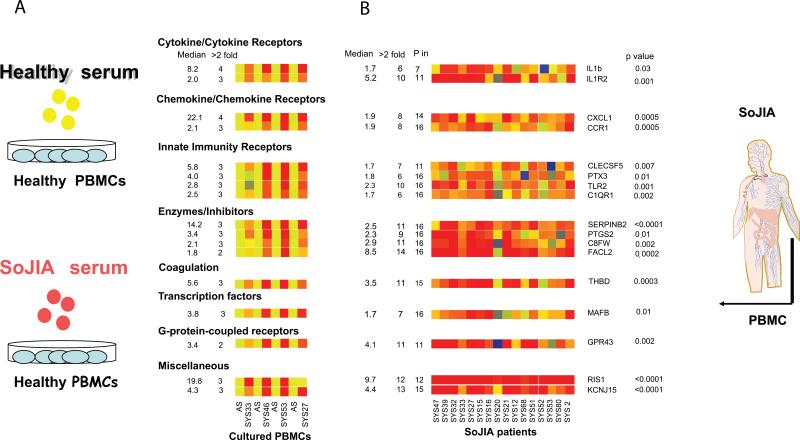
Figure 8. Gene expression profiling leads to the identification of IL-1 b in the pathogenesis of SoJIA.
A) Analysis of transcriptional changes induced upon incubation of healthy PBMCs with autologous sera (AS) or with sera from four patients with active SoJIA (SYS33, SYS46, SYS53, and SYS27). Sera from patients induced the up-regulation of 46 genes. Median fold up-regulation by the four SoJIA sera incubation is depicted on the left column. The number of SoJIA sera that induced greater than twofold up-regulation is shown in the next column. B) Expression of a set of gene probes from Fig.1a in the PBMCs of 16 active SoJIA patients. The patient PBMCs expression data were normalized to the median expression of the same gene probes in the PBMCs of 12 healthy children. Median gene expression and number of samples with greater than twofold up-regulation are depicted in the first two columns. The third column represents the number of samples with a P (present) flag according to Affymetrix MAS 5.0 scaled gene expression data. p-values (Mann-Whitney test) are given next to these genes. (Published in Curr Opin Immunol. 2007 Dec;19(6):623-32)
Most clinical and laboratory manifestations of SoJIA can be explained based on increased IL-1 production, but the origin of this dysregulation remains unknown. An unusual microorganism could target an otherwise normal innate immune system resulting in IL-1 overproduction. There is no epidemiological evidence however for clustering of SoJIA patients, which would be expected if this were to be the case. Alternatively, as described for inflammasome-mediated diseases, a common infectious or inflammatory trigger could lead to an excessive production of IL-1 in patients with underlying mutations in genes controlling IL-1 production. In favor of this hypothesis, non-specific activation of SoJIA PBMCs in vitro results in excessive IL-1 b secretion (121). Since IL-1b can upregulate its own transcription, IL-1b itself could also be responsible for the serum effects described above.
The IL-1 family and the inflammasome
Up to 11 members of the IL-1 family have been identified to date (125). Of those, only five have been thoroughly studied: IL-1a, IL-1b, IL-18, IL-1Ra and the most recently reported IL-33. The remaining six (IL-1F5; IL-1F6; IL-1F7; IL-1F8; IL-1F9; IL-1F10) have been shown to be expressed in various cell types or tissues, but their functions remain to be determined.
IL-1a and IL-1b are proinflammatory cytokines. Both are synthesized as precursor molecules (pro-IL1a and pro-IL1b) by many different cell types. Pro-IL1a is biologically active and needs to be cleaved by calpain to generate the smaller mature protein. By contrast, pro-IL1b is biologically inactive and requires enzymatic cleavage by caspase-1 in order to become active. IL-1a is primarily bound to the membrane whereas IL-1b is secreted and thus represents the predominant extracellular form of IL-1 (reviewed in (126)). IL-1Ra is an endogenous receptor antagonist. It exists as 3 intracellular isoforms and a secreted isoform (sIL-1Ra). IL-1Ra is predominantly produced by activated monocytes and macrophages. sIL-1Ra binds both type I and type II IL-1 receptors, thereby preventing binding and signal transduction by IL-1a and IL-1b (126).
The production and biological activity of IL-1 are regulated at multiple levels, including transcription, translation, cleavage and cellular release. IL-1a and IL-1b transcription is induced by a wide array of stimuli including bacterial and viral products, cytokines that include IL-1b itself, etc. The activation of caspase 1 is mediated by a multiprotein complex known as the “inflammasome”. Two main types of inflammasome have been described to date-the NALP1 inflammasome, composed of NALP1, the adaptor protein ASC, and caspases 1 and 5, and the NALP2/3 inflammasome that contains NALP2 or NALP3 (also known as CIAS1 or cryopyrin) as well as the caspase recruitment domain (CARD)-containing protein Cardinal, ASC and Caspase-1 (127). Until recently, the mechanism(s) of activation of this complex remained unknown. CIAS−/− mice have revealed however that the NALP3 inflammasome can be directly activated by bacteria (L. monocytogenes and S. aureus), the purine analogs R848 and R837, bacterial mRNA, double-stranded/viral RNA and uric acid crystals (128-131). Inflammasome activation leads to the conversion of pro-caspase-1 into caspase-1 and subsequent cleavage of pro-IL-1b into mature IL-1b (reviewed in (126)). Release of mature IL-1b depends on a second signal provided by the nucleotide P2X7 receptor, which can be activated by the human cathelicidin-derived peptide LL37 or by ATP, leading to an efflux of potassium from the cell (132). Potassium efflux is responsible for phosphatidylcholine-specific phospholipase C induction, which in turn allows the rise in intracellular free calcium concentration required for activation of phospholipase A(2). This activation is ultimately responsible for lysosome exocytosis and IL-1 beta secretion (132). Recently, a model in which K+ efflux is the common and specific trigger of NALP1 and NALP3 activation induced by all reported ligands through the involvement of K+ channel(s) has been proposed (133).
IL-1b mediated diseases
IL-1b is a major mediator of inflammation that plays a fundamental role in tissue injury repair as well as in the defense against microbial pathogens. In these situations, local (i.e. endothelial) and/or systemic (i.e. bone marrow) responses to this cytokine are responsible for beneficial effects, including among others cellular infiltration and neutrophil mobilization respectively. An excess of this cytokine, however, may have deleterious effects on a variety of cells and tissues. Injection of recombinant IL-1b in humans and in experimental animal models induces fever, anorexia and pain hypersensitivity through direct effects on the CNS. This cytokine also has important effects on endothelial cells that may lead to vasculitis and promote thrombosis. It plays a role in destructive joint and bone disease, and is toxic for insulin-producing β cells in the pancreas (120). Given these protean local and systemic effects, it would not be surprising that IL-1b also mediates pathological inflammatory cascades in a variety of human diseases. These could include febrile diseases of non-infectious origin, vasculitis, arthritis and diabetes. Conventional approaches such as cytokine measurement in the serum of patients, have failed however to establish such connection.
Blocking IL-1 is an effective therapy for SoJIA patients
When we saw that culturing healthy PBMCs with SoJIA serum induced IL-1 secretion, we felt compelled to try to block this cytokine in our patients. Interestingly, a recombinant soluble IL-1 receptor antagonist (Anakinra®) was already in the market to treat RA and JIA patients. Clinical trials in adult RA had yielded disappointing results (10, 134). A formal trial on SoJIA, especially on patients with systemic manifestations, had never been conducted before. With the hypothesis that blocking IL-1 would control the disease manifestations, we enrolled our first two patients in a pilot trial using (Anakinra®). Both patients had had SoJIA for years. The disease was refractory to a combination of steroids, methotrexate and anti-TNF agents. To our delight and that of our patients, two days after initiation of daily subcutaneous injections of Anakinra® the two patients were afebrile and felt subjectively a great improvement in their articular symptoms. The subjective improvement was reflected already within the first 2 weeks of treatment in a progressive substantial decrease in objective active joint count as well as laboratory parameters. In view of the spectacular results, we started 7 additional patients on Anakinra. Upon follow-up, 7 out of 9 patients entered in sustained full remission, one in partial sustained remission and the remaining patient had a transient partial response (121). These results were reproduced in a separate cohort of 9 patients. Overall, upon 3 year follow up more than 2/3 of the patients treated with Anakinra® were considered in remission (Punaro et al., manuscript in preparation).
Thus, blocking IL-1b results in remarkable clinical and hematological responses in SoJIA patients (121). Furthermore, IL-6 levels return to normal in patients after initiation of therapy, supporting that increased IL-6 production is a secondary event downstream of IL-1b (Allantaz, unpublished observations). Rapid responses to IL-1b blockade have also been described in patients with refractory adult-onset (Still's) disease (135). All of these findings point toward IL-1b as a mediator of SoJIA and therefore link this disease to the family of autoinflammatory disorders.
Tackling the next challenge: SoJIA diagnostic and activity biomarkers
A major remaining challenge is how to establish the prompt diagnosis of the disease to initiate effective therapy. We have therefore studied the gene expression patterns in blood leukocytes to find a diagnostic test for SoJIA. There is however a considerable degree of overlap between the blood gene signatures of SoJIA patients and those of other febrile inflammatory disease groups which represent a true differential diagnosis in the clinical setting. This is especially true for Gram (+) bacterial infections and autoinflammatory syndromes in which IL-1b production is also increased (136) (124).
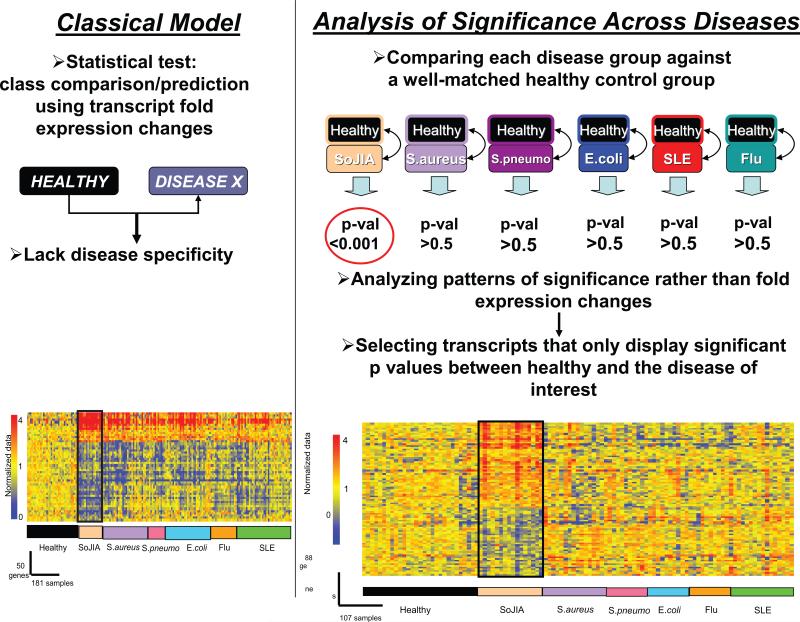
Analysis of significance patterns is a strategy that has been successfully applied to this type of situations (124, 137). First, statistical comparisons are performed between each group of patients and their respective control groups composed of age-matched and gender-matched healthy donors. The p-values obtained from each comparison are then subjected to selection criteria to identify genes significantly changed in the disease of interest versus its control group, and not in any of the other diseases versus their own control groups. The advantage of this analysis is that it permits normalization of each disease group to its own matched control group, therefore avoiding biological (i.e. age, gender) or technical (i.e. array runs) confounding factors. Using this approach, we studied samples from two groups of SoJIA patients: i) patients experiencing systemic symptoms and ii) patients who had resolved the systemic phase but continued having arthritis. We also included patients with systemic infectious diseases (S. aureus, S. pneumoniae and E. coli), SLE and PAPA syndrome, a hereditary autoinflammatory disease that results in excessive IL-1b production. A SoJIA-specific signature composed of 88 genes was identified (Figure 9). Furthermore, administration of IL-1 blockers resulted in the normalization of expression of the majority of these transcripts in the treated patients (Figure 10). Indeed, 12 highly significant genes from this analysis (p<0.0001 in SoJIA and >0.5 in all other groups) permitted accurate disease classification in 18/19 SoJIA patients (124). If validated with a larger number of patients in multi-centric studies, this “mini-signature” should permit to establish an early diagnosis and initiate specific therapy. Thus, the early diagnosis of SoJIA through genomic approaches might permit us to start treatment early after diagnosis and therefore avoid the subsequent development of long term disabilities.
Figure 9. Analysis of significance across diseases identifies 88 SoJIA-specific transcripts.
(A) Eight healthy and eight SoJIA samples were used as training set to generate a list of 50 classifier genes displaying the best ability to discriminate SoJIA patients from healthy controls. Those classifier genes were hierarchically clustered in a test set composed of 35 healthy controls, 16 SoJIA, 31 S. aureus, 12 S. pneumoniae, 31 E. coli, 18 Influenza A and 38 SLE patients. (B) Genes expressed at statistically different levels in SoJIA patients compared to healthy volunteers (p<0.01, Wilcoxon-Mann-Whitney test) were selected (4311 probe sets). Out of those, 88 were found expressed at statistically different levels in SoJIA patients compared to healthy volunteers (p<0.01, Wilcoxon-Mann-Whitney test) but not in all the other groups (p>0.5, Wilcoxon-Mann-Whitney test). The 88 genes are hierarchically clustered in the 107 samples from different disease groups used in (A). Expression values or the genes are normalized per-gene to the healthy group (Published in Curr Opin Immunol. 2007 Dec;19(6):623-32)
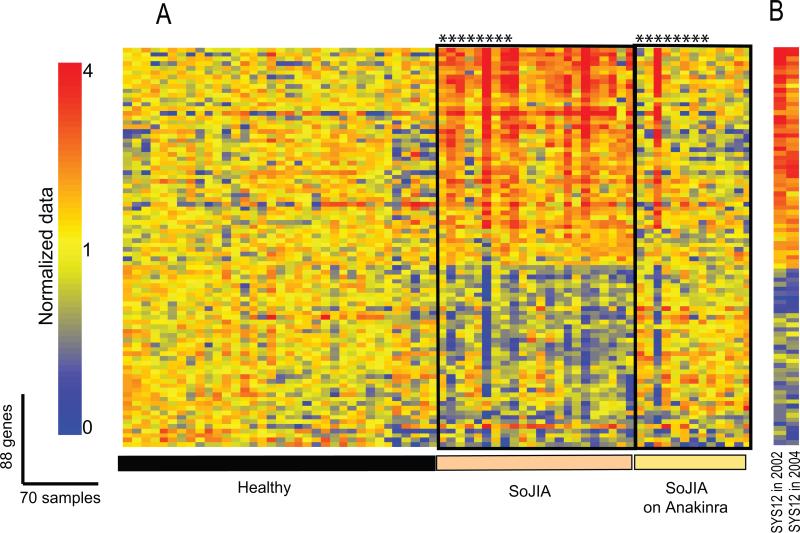
Figure 10. Treatment with IL-1Ra (Anakinra) extinguishes the SoJIA-specific signature.
(A). 88 SoJIA-specific genes were analyzed in 35 healthy, 22 SoJIA patients not receiving IL-1 blockers and 14 SoJIA patients after initiation of treatment with IL- blockers. *represents the same patients before and after initiation of the therapy. (B). The SoJIA signature is present in a patient on two occasions taken 2 years apart. On both occasions the patient was active and not receiving IL-1 blockers. (Published in Curr Opin Immunol. 2007 Dec;19(6):623-32)
An expanding spectrum of IL-1b mediated diseases resulting from inflammasome dysregulation
As discussed above, IL-1b production is tightly controlled at distinct steps. Most IL-1b mediated human diseases identified thus far have been linked to abnormal activation of this cytokine by the inflammasome (138). The clinical hallmark of these diseases, which are also known as “periodic fever” or “autoinflammatory” syndromes”, is the presence of recurrent symptoms which are easily explained by an excessive production of IL-1b: fever and inflammation predominantly affecting the serosal membranes, joints and skin. Their underlying genetic defects were identified through genome-wide linkage studies using multi-case families and controls. Among them, Familial Cold Autoinflammatory Syndrome (FCAS, MIM 120100), Muckle-Wells syndrome (MWS; MIM 19100), and Neonatal-Onset Multisystem Inflammatory Disease (NOMID, MIM 607115) result from mutations in the NALP3 or cryopyrin gene. Mutations in genes encoding proteins that interact with inflammasome components give rise to Familial Mediterranean Fever (FMF, MIM 249100), and the syndrome of Pyogenic Sterile Arthritis, Pyoderma gangrenosum and Acne (PAPA, MIM 604416) (139). Yet, many patients fulfilling diagnostic criteria for some of these diseases do not display mutations in the corresponding genes. For example, no mutations have been identified in up to 50% of patients with clinically diagnosed FCAS, MWS or CINCA (140).
Inflammasome-mediated diseases, however, do not necessarily present with fever or systemic inflammation and do not always have a well-defined genetic basis. A combination of biochemical approaches and inflammasome gene knock-out in mice demonstrated that activation of the NALP3 inflammasome with contact sensitizers or uric acid crystals triggers the inflammatory cascades underlying contact hypersensitivity and gout/pseudogout respectively (131, 141-143). These diseases affect considerably larger patient populations than the rare familial periodic fever syndromes.
In agreement with the notion that the inflammasome ultimately regulates the secretion of IL-1, blocking this cytokine with a recombinant IL-1 receptor antagonist has emerged as a successful form of therapy for many of these disorders.
Blood microarrays support the role of IL-1 b in the pathogenesis of type 1 and type 2 diabetes
Early studies reported the presence of Type I IFN in the islets of patients with recently diagnosed type 1 diabetes (144). Furthermore, treatment of cancer and hepatitis C with recombinant IFN-α led to the development of diabetes in a group of patients. We thus hypothesized that blood gene expression profiling of newly diagnosed diabetes patients might show an IFN signature. We also hypothesized that this signature might be discrete, as the autoimmune attack remains localized to the islets in contrast to the most systemic attack of SLE. Our hypothesis proved to be only partly correct in that a blood signature indeed was found in diabetes patients. This signature turned out however not to be type I IFN-induced. Rather, the blood of children with newly diagnosed type 1 and type 2 diabetes showed that 5 of the 10 most highly overexpressed genes in these patients overlap with those found overexpressed in patients with SoJIA. These genes are also found expressed in healthy PBMCs incubated with SoJIA serum. Among them, one of the most significantly upregulated is IL-1b (145). These data would support that type 1 and type 2 diabetes share a common pathway for beta cell dysfunction that includes secretion of IL-1b and downstream proteins that may exacerbate preexisting beta cell dysfunction and contribute to further hyperglycemia. A recent study that recapitulates our own study using SoJIA serum to culture healthy PBMCs found that sera from recent onset type 1 diabetes patients induced an expression signature that included IL-1 cytokine family members (146). These results support the value of this type of studies in helping understand disease processes associated with an altered immune system.
This finding does not come as a total surprise because IL-1b had been implicated as an effector molecule in the process of inflammatory beta-cell destruction leading to diabetes (147). Perhaps the most compelling data to support the role of IL-1b in diabetes pathogenesis is that the administration of recombinant soluble IL-1 receptor antagonist results inclinical improvement. In particular, treatment of patients with type 2 diabetes results in improved glycemia and beta-cell secretory function and reduced markers of systemic inflammation (148). Given the overall safety clinical record of IL-1 blockers, similar studies should be conducted in newly diagnosed type 1 diabetes patients to evaluate the value of this form of therapy in preserving beta cell function and reduce exogenous insulin requirement in this disease.
CONCLUSION
Animal models of rheumatic diseases have not been very helpful in predicting therapeutic targets for human disease. As of today, the most successful treatments for these diseases have been identified either by serendipity; i;e: methotrexate in RA and anti-malarials in SLE or by direct examination of human samples. The successful treatment of RA patients with anti-TNF agents and SoJIA patients with IL-1 antagonists represent examples of how simple experiments using human samples can yield fundamental clues regarding disease pathogenesis.
Microarray analyses of patients with rheumatic diseases have shown that diseases with diverse pathogenesis and clinical manifestations may share common immune mediators, which represent therapeutic targets for intervention. Since our initial reports(16, 61, 149) , blood transcriptional signatures have now been described in patients with other autoimmune diseases, including multiple sclerosis (150-152), rheumatoid arthritis (153-156), inflammatory bowel disease (157), psoriasis (158) and dermatomyositis (108, 110). Blood profiling studies extended rapidly to other diseases also characterized by a strong inflammatory component, especially infections caused by viruses like HIV (159), adenovirus (160), influenza (161), or dengue (162); bacteria like Staphylococcus aureus, Streptococcus pneumoniae or Escherichia coli (161); Mycobacterium tuberculosis (163); parasites (Plasmodium) (164); as well as sepsis (165). The blood of transplant recipients has also been profiled, in kidney (166), liver (167), heart (168), and hematopoeitic cell transplant recipients (169). Furthermore, disease signatures have been detected in the blood of patients with diabetes (145, 170), cardio vascular diseases (171), and non-hematological malignancies (172, 173).
Microarray technology, however, raises significant challenges. Patient selection, for example, is very important when searching for disease-specific signatures, and therefore collaborations with skilled clinicians are fundamental. Sample homogeneity with respect to time since diagnosis might be also important, as immune alterations as well as drugs used at the time of disease initiation might be different from those in later stages of disease. Challenges inherent to system-wide studies, such as noise and technical variability, also need to be considered.
Blood microarrays can also be used to diagnose disease and follow clinical activity as well as response to therapy in the clinical setting. Our own studies, for example, have identified transcriptional biomarkers of SLE disease activity and prediction of nephritis (Chaussabel et al., submitted). These biomarkers, which are being validated in large longitudinal cohorts, might also offer in the future the possibility of predicting end-organ involvement (i.e. lupus nephritis) before overt clinical symptoms appear, thus allowing the initiation of therapy before the damage might be irreversible. They may also allow to understand disease heterogeneity and to predict whether individual patients might respond to targeted therapies or not.
We foresee that blood microarray analysis will soon become an integral part of clinical medicine and an essential component of Personalized Medicine (for a description of the HSS Personalized Health Care Initiative go to: http://www.hhs.gov/myhealthcare/). They will be used to monitor health and early disease development as an alteration in transcriptional activity will likely precede the appearance of clinical symptoms.
ACKNOWLEDGEMENTS
Supported by Baylor Health Care System Foundation, the National Institutes of Health: AR054083, AR055503 and AR050770 (Chandra Mohan PI) to VP, CA78846, AI068842 and U19A1057234 to JB, the Alliance for Lupus Research (VP), the DANA Foundation (DC) and the Mary Kirkland Foundation (VP). JB holds the Caruth Chair for Transplantation Immunology. KP holds the Michael A. E. Ramsay Chair for Cancer Immunology Research. We thank Drs. Michael Ramsay and Bill Duncan for continuous support. We also thank Drs. Edsel Arce, Patrick Blanco, Lynda Bennett, Gaetan Jego, Ellen Kaizer, Sophie Koutouzov, Alexis Mathian, Michel Nussenzweig, Octavio Ramilo, Dorothee Stichweh, Barbara Vega, Perrin White and Sergey Yurasov for collaborating in the studies described in this review. We thank Drs. Lynn Punaro and Katherine Madson and our patients at the Pediatric Rheumatology Clinic over Texas Scottish Rite Hospital in Dallas for their willingness to participate in our studies.
REFERENCES
- 1.Pulendran B, Ahmed R. Translating innate immunity into immunological memory: implications for vaccine development. Cell. 2006;124:849–63. doi: 10.1016/j.cell.2006.02.019. [DOI] [PubMed] [Google Scholar]
- 2.Germain RN, Bajenoff M, Castellino F, Chieppa M, Egen JG, Huang AY, Ishii M, Koo LY, Qi H. Making friends in out-of-the-way places: how cells of the immune system get together and how they conduct their business as revealed by intravital imaging. Immunol Rev. 2008;221:163–81. doi: 10.1111/j.1600-065X.2008.00591.x. [DOI] [PubMed] [Google Scholar]
- 3.Palucka AK, Banchereau J. How dendritic cells and microbes interact to elicit or subvert protective immune responses. Curr Opin Immunol. 2002;14:420–31. doi: 10.1016/s0952-7915(02)00365-5. [DOI] [PubMed] [Google Scholar]
- 4.Banchereau J, Steinman RM. Dendritic cells and the control of immunity. Nature. 1998;392:245–52. doi: 10.1038/32588. [DOI] [PubMed] [Google Scholar]
- 5.Steinman RM, Banchereau J. Taking dendritic cells into medicine. Nature. 2007;449:419–26. doi: 10.1038/nature06175. [DOI] [PubMed] [Google Scholar]
- 6.Steinman RM, Hawiger D, Nussenzweig MC. Tolerogenic dendritic cells. Annu Rev Immunol. 2003;21:685–711. doi: 10.1146/annurev.immunol.21.120601.141040. [DOI] [PubMed] [Google Scholar]
- 7.Ueno H, Klechevsky E, Morita R, Aspord C, Cao T, Matsui T, Di Pucchio T, Connolly J, Fay JW, Pascual V, Palucka AK, Banchereau J. Dendritic cell subsets in health and disease. Immunol Rev. 2007;219:118–42. doi: 10.1111/j.1600-065X.2007.00551.x. [DOI] [PubMed] [Google Scholar]
- 8.Saijo S, Asano M, Horai R, Yamamoto H, Iwakura Y. Suppression of autoimmune arthritis in interleukin-1-deficient mice in which T cell activation is impaired due to low levels of CD40 ligand and OX40 expression on T cells. Arthritis Rheum. 2002;46:533–44. doi: 10.1002/art.10172. [DOI] [PubMed] [Google Scholar]
- 9.Horai R, Saijo S, Tanioka H, Nakae S, Sudo K, Okahara A, Ikuse T, Asano M, Iwakura Y. Development of chronic inflammatory arthropathy resembling rheumatoid arthritis in interleukin 1 receptor antagonist-deficient mice. J Exp Med. 2000;191:313–20. doi: 10.1084/jem.191.2.313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Genovese MC, Cohen S, Moreland L, Lium D, Robbins S, Newmark R, Bekker P. Combination therapy with etanercept and anakinra in the treatment of patients with rheumatoid arthritis who have been treated unsuccessfully with methotrexate. Arthritis Rheum. 2004;50:1412–9. doi: 10.1002/art.20221. [DOI] [PubMed] [Google Scholar]
- 11.Brennan FM, Chantry D, Jackson A, Maini R, Feldmann M. Inhibitory effect of TNF alpha antibodies on synovial cell interleukin-1 production in rheumatoid arthritis. Lancet. 1989;2:244–7. doi: 10.1016/s0140-6736(89)90430-3. [DOI] [PubMed] [Google Scholar]
- 12.Maini RN, Feldmann M. The pitfalls in the development of biologic therapy. Nat Clin Pract Rheumatol. 2007;3:1. doi: 10.1038/ncprheum0373. [DOI] [PubMed] [Google Scholar]
- 13.Feldmann M, Maini RN. Anti-TNF alpha therapy of rheumatoid arthritis: what have we learned? Annu Rev Immunol. 2001;19:163–96. doi: 10.1146/annurev.immunol.19.1.163. [DOI] [PubMed] [Google Scholar]
- 14.Alizadeh AA, Eisen MB, Davis RE, Ma C, Lossos IS, Rosenwald A, Boldrick JC, Sabet H, Tran T, Yu X, Powell JI, Yang L, Marti GE, Moore T, Hudson J, Jr., Lu L, Lewis DB, Tibshirani R, Sherlock G, Chan WC, Greiner TC, Weisenburger DD, Armitage JO, Warnke R, Levy R, Wilson W, Grever MR, Byrd JC, Botstein D, Brown PO, Staudt LM. Distinct types of diffuse large B-cell lymphoma identified by gene expression profiling. Nature. 2000;403:503–11. doi: 10.1038/35000501. [DOI] [PubMed] [Google Scholar]
- 15.Bittner M, Meltzer P, Chen Y, Jiang Y, Seftor E, Hendrix M, Radmacher M, Simon R, Yakhini Z, Ben-Dor A, Sampas N, Dougherty E, Wang E, Marincola F, Gooden C, Lueders J, Glatfelter A, Pollock P, Carpten J, Gillanders E, Leja D, Dietrich K, Beaudry C, Berens M, Alberts D, Sondak V. Molecular classification of cutaneous malignant melanoma by gene expression profiling. Nature. 2000;406:536–40. doi: 10.1038/35020115. [DOI] [PubMed] [Google Scholar]
- 16.Bennett L, Palucka AK, Arce E, Cantrell V, Borvak J, Banchereau J, Pascual V. Interferon and granulopoiesis signatures in systemic lupus erythematosus blood. J Exp Med. 2003;197:711–23. doi: 10.1084/jem.20021553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brunner HI, Gladman DD, Ibanez D, Urowitz MD, Silverman ED. Difference in disease features between childhood-onset and adult-onset systemic lupus erythematosus. Arthritis Rheum. 2008;58:556–62. doi: 10.1002/art.23204. [DOI] [PubMed] [Google Scholar]
- 18.Bencivelli W, Vitali C, Isenberg DA, Smolen JS, Snaith ML, Sciuto M, Bombardieri S. Disease activity in systemic lupus erythematosus: report of the Consensus Study Group of the European Workshop for Rheumatology Research. III. Development of a computerised clinical chart and its application to the comparison of different indices of disease activity. The European Consensus Study Group for Disease Activity in SLE. Clin Exp Rheumatol. 1992;10:549–54. [PubMed] [Google Scholar]
- 19.Hay EM, Bacon PA, Gordon C, Isenberg DA, Maddison P, Snaith ML, Symmons DP, Viner N, Zoma A. The BILAG index: a reliable and valid instrument for measuring clinical disease activity in systemic lupus erythematosus. Q J Med. 1993;86:447–58. [PubMed] [Google Scholar]
- 20.Bombardier C, Gladman DD, Urowitz MB, Caron D, Chang CH. Derivation of the SLEDAI. A disease activity index for lupus patients. The Committee on Prognosis Studies in SLE. Arthritis Rheum. 1992;35:630–40. doi: 10.1002/art.1780350606. [DOI] [PubMed] [Google Scholar]
- 21.Liang MH, Socher SA, Larson MG, Schur PH. Reliability and validity of six systems for the clinical assessment of disease activity in systemic lupus erythematosus. Arthritis Rheum. 1989;32:1107–18. doi: 10.1002/anr.1780320909. [DOI] [PubMed] [Google Scholar]
- 22.Bae SC, Koh HK, Chang DK, Kim MH, Park JK, Kim SY. Reliability and validity of systemic lupus activity measure-revised (SLAM-R) for measuring clinical disease activity in systemic lupus erythematosus. Lupus. 2001;10:405–9. doi: 10.1191/096120301678646146. [DOI] [PubMed] [Google Scholar]
- 23.Petri M, Buyon J, Kim M. Classification and definition of major flares in SLE clinical trials. Lupus. 1999;8:685–91. doi: 10.1191/096120399680411281. [DOI] [PubMed] [Google Scholar]
- 24.Petty RE, Southwood TR, Manners P, Baum J, Glass DN, Goldenberg J, He X, Maldonado-Cocco J, Orozco-Alcala J, Prieur AM, Suarez-Almazor ME, Woo P. International League of Associations for Rheumatology classification of juvenile idiopathic arthritis: second revision, Edmonton, 2001. J Rheumatol. 2004;31:390–2. [PubMed] [Google Scholar]
- 25.Cassidy JT, Ross E. Textbook of Pediatric Rheumatology. 4th Edition 2001. Juvenile Rheumatoid Arthritis. pp. 218–321. [Google Scholar]
- 26.Lomater C, Gerloni V, Gattinara M, Mazzotti J, Cimaz R, Fantini F. Systemic onset juvenile idiopathic arthritis: a retrospective study of 80 consecutive patients followed for 10 years. J Rheumatol. 2000;27:491–6. [PubMed] [Google Scholar]
- 27.Shlomchik MJ, Craft JE, Mamula MJ. From T to B and back again: positive feedback in systemic autoimmune disease. Nat Rev Immunol. 2001;1:147–53. doi: 10.1038/35100573. [DOI] [PubMed] [Google Scholar]
- 28.Arce E, Jackson DG, Gill MA, Bennett LB, Banchereau J, Pascual V. Increased frequency of pre-germinal center b cells and plasma cell precursors in the blood of children with systemic lupus erythematosus. J Immunol. 2001;167:2361–9. doi: 10.4049/jimmunol.167.4.2361. [DOI] [PubMed] [Google Scholar]
- 29.Nicholas MW, Dooley MA, Hogan SL, Anolik J, Looney J, Sanz I, Clarke SH. A novel subset of memory B cells is enriched in autoreactivity and correlates with adverse outcomes in SLE. Clin Immunol. 2008;126:189–201. doi: 10.1016/j.clim.2007.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yurasov S, Wardemann H, Hammersen J, Tsuiji M, Meffre E, Pascual V, Nussenzweig MC. Defective B cell tolerance checkpoints in systemic lupus erythematosus. J Exp Med. 2005 doi: 10.1084/jem.20042251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cappione A, 3rd, Anolik JH, Pugh-Bernard A, Barnard J, Dutcher P, Silverman G, Sanz I. Germinal center exclusion of autoreactive B cells is defective in human systemic lupus erythematosus. J Clin Invest. 2005;115:3205–16. doi: 10.1172/JCI24179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hawiger D, Inaba K, Dorsett Y, Guo M, Mahnke K, Rivera M, Ravetch JV, Steinman RM, Nussenzweig MC. Dendritic cells induce peripheral T cell unresponsiveness under steady state conditions in vivo. J Exp Med. 2001;194:769–79. doi: 10.1084/jem.194.6.769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Blanco P, Palucka AK, Gill M, Pascual V, Banchereau J. Induction of dendritic cell differentiation by IFN-alpha in systemic lupus erythematosus. Science. 2001;294:1540–3. doi: 10.1126/science.1064890. [DOI] [PubMed] [Google Scholar]
- 34.Banchereau J, Pascual V, Palucka AK. Autoimmunity through cytokine-induced dendritic cell activation. Immunity. 2004;20:539–50. doi: 10.1016/s1074-7613(04)00108-6. [DOI] [PubMed] [Google Scholar]
- 35.Paquette RL, Hsu NC, Kiertscher SM, Park AN, Tran L, Roth MD, Glaspy JA. Interferon-alpha and granulocyte-macrophage colony-stimulating factor differentiate peripheral blood monocytes into potent antigen-presenting cells. J Leukoc Biol. 1998;64:358–67. doi: 10.1002/jlb.64.3.358. [DOI] [PubMed] [Google Scholar]
- 36.Santini SM, Lapenta C, Logozzi M, Parlato S, Spada M, Di Pucchio T, Belardelli F. Type I interferon as a powerful adjuvant for monocyte-derived dendritic cell development and activity in vitro and in Hu-PBL-SCID mice. J Exp Med. 2000;191:1777–88. doi: 10.1084/jem.191.10.1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Luft T, Pang KC, Thomas E, Hertzog P, Hart DN, Trapani J, Cebon J. Type I IFNs enhance the terminal differentiation of dendritic cells. J Immunol. 1998;161:1947–53. [PubMed] [Google Scholar]
- 38.Pestka S, Krause CD, Walter MR. Interferons, interferon-like cytokines, and their receptors. Immunol Rev. 2004;202:8–32. doi: 10.1111/j.0105-2896.2004.00204.x. [DOI] [PubMed] [Google Scholar]
- 39.Siegal FP, Kadowaki N, Shodell M, Fitzgerald-Bocarsly PA, Shah K, Ho S, Antonenko S, Liu YJ. The nature of the principal type 1 interferon-producing cells in human blood [In Process Citation]. Science. 1999;284:1835–7. doi: 10.1126/science.284.5421.1835. [DOI] [PubMed] [Google Scholar]
- 40.Liu YJ. IPC: professional type 1 interferon-producing cells and plasmacytoid dendritic cell precursors. Annu Rev Immunol. 2005;23:275–306. doi: 10.1146/annurev.immunol.23.021704.115633. [DOI] [PubMed] [Google Scholar]
- 41.Gill MA, Blanco P, Arce E, Pascual V, Banchereau J, Palucka AK. Blood dendritic cells and DC-poietins in systemic lupus erythematosus. Hum Immunol. 2002;63:1172–80. doi: 10.1016/s0198-8859(02)00756-5. [DOI] [PubMed] [Google Scholar]
- 42.Farkas L, Beiske K, Lund-Johansen F, Brandtzaeg P, Jahnsen FL. Plasmacytoid Dendritic Cells (Natural Interferon- alpha/beta-Producing Cells) Accumulate in Cutaneous Lupus Erythematosus Lesions. Am J Pathol. 2001;159:237–43. doi: 10.1016/s0002-9440(10)61689-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Blomberg S, Eloranta ML, Cederblad B, Nordlin K, Alm GV, Ronnblom L. Presence of cutaneous interferon-alpha producing cells in patients with systemic lupus erythematosus. Lupus. 2001;10:484–90. doi: 10.1191/096120301678416042. [DOI] [PubMed] [Google Scholar]
- 44.Jahnsen FL, Lund-Johansen F, Dunne JF, Farkas L, Haye R, Brandtzaeg P. Experimentally induced recruitment of plasmacytoid (CD123high) dendritic cells in human nasal allergy. J Immunol. 2000;165:4062–8. doi: 10.4049/jimmunol.165.7.4062. [DOI] [PubMed] [Google Scholar]
- 45.Le Bon A, Schiavoni G, D'Agostino G, Gresser I, Belardelli F, Tough DF. Type i interferons potently enhance humoral immunity and can promote isotype switching by stimulating dendritic cells in vivo. Immunity. 2001;14:461–70. doi: 10.1016/s1074-7613(01)00126-1. [DOI] [PubMed] [Google Scholar]
- 46.Jego G, Palucka AK, Blanck JP, Chalouni C, Pascual V, Banchereau J. Plasmacytoid dendritic cells induce plasma cell differentiation through type I interferon and interleukin 6. Immunity. 2003;19:225–34. doi: 10.1016/s1074-7613(03)00208-5. [DOI] [PubMed] [Google Scholar]
- 47.Medina F, Segundo C, Campos-Caro A, Gonzalez-Garcia I, Brieva JA. The heterogeneity shown by human plasma cells from tonsil, blood, and bone marrow reveals graded stages of increasing maturity, but local profiles of adhesion molecule expression. Blood. 2002;99:2154–61. doi: 10.1182/blood.v99.6.2154. [DOI] [PubMed] [Google Scholar]
- 48.Marrack P, Kappler J, Mitchell T. Type I interferons keep activated T cells alive. J Exp Med. 1999;189:521–30. doi: 10.1084/jem.189.3.521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kolumam GA, Thomas S, Thompson LJ, Sprent J, Murali-Krishna K. Type I interferons act directly on CD8 T cells to allow clonal expansion and memory formation in response to viral infection. J Exp Med. 2005;202:637–50. doi: 10.1084/jem.20050821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Blanco P, Pitard V, Viallard JF, Taupin JL, Pellegrin JL, Moreau JF. Increase in activated CD8+ T lymphocytes expressing perforin and granzyme B correlates with disease activity in patients with systemic lupus erythematosus. Arthritis Rheum. 2005;52:201–11. doi: 10.1002/art.20745. [DOI] [PubMed] [Google Scholar]
- 51.Bondanza A, Zimmermann VS, Dell'Antonio G, Dal Cin E, Capobianco A, Sabbadini MG, Manfredi AA, Rovere-Querini P. Cutting edge: dissociation between autoimmune response and clinical disease after vaccination with dendritic cells. J Immunol. 2003;170:24–7. doi: 10.4049/jimmunol.170.1.24. [DOI] [PubMed] [Google Scholar]
- 52.Georgiev M, Agle LM, Chu JL, Elkon KB, Ashany D. Mature dendritic cells readily break tolerance in normal mice but do not lead to disease expression. Arthritis Rheum. 2005;52:225–38. doi: 10.1002/art.20759. [DOI] [PubMed] [Google Scholar]
- 53.Stewart TA. Neutralizing interferon alpha as a therapeutic approach to autoimmune diseases. Cytokine Growth Factor Rev. 2003;14:139–54. doi: 10.1016/s1359-6101(02)00088-6. [DOI] [PubMed] [Google Scholar]
- 54.Cederblad B, Blomberg S, Vallin H, Perers A, Alm GV, Ronnblom L. Patients with systemic lupus erythematosus have reduced numbers of circulating natural interferon-alpha- producing cells. J Autoimmun. 1998;11:465–70. doi: 10.1006/jaut.1998.0215. [DOI] [PubMed] [Google Scholar]
- 55.Vallin H, Perers A, Alm GV, Ronnblom L. Anti-double-stranded DNA antibodies and immunostimulatory plasmid DNA in combination mimic the endogenous IFN-alpha inducer in systemic lupus erythematosus. J Immunol. 1999;163:6306–13. [PubMed] [Google Scholar]
- 56.Hooks JJ, Moutsopoulos HM, Geis SA, Stahl NI, Decker JL, Notkins AL. Immune interferon in the circulation of patients with autoimmune disease. N Engl J Med. 1979;301:5–8. doi: 10.1056/NEJM197907053010102. [DOI] [PubMed] [Google Scholar]
- 57.Preble OT, Black RJ, Friedman RM, Klippel JH, Vilcek J. Systemic lupus erythematosus: presence in human serum of an unusual acid-labile leukocyte interferon. Science. 1982;216:429–31. doi: 10.1126/science.6176024. [DOI] [PubMed] [Google Scholar]
- 58.Samanta H, Engel DA, Chao HM, Thakur A, Garcia-Blanco MA, Lengyel P. Interferons as gene activators. Cloning of the 5' terminus and the control segment of an interferon activated gene. J Biol Chem. 1986;261:11849–58. [PubMed] [Google Scholar]
- 59.Maas K, Chan S, Parker J, Slater A, Moore J, Olsen N, Aune TM. Cutting edge: molecular portrait of human autoimmune disease. J Immunol. 2002;169:5–9. doi: 10.4049/jimmunol.169.1.5. [DOI] [PubMed] [Google Scholar]
- 60.Shodell M, Shah K, Siegal FP. Circulating human plasmacytoid dendritic cells are highly sensitive to corticosteroid administration. Lupus. 2003;12:222–30. doi: 10.1191/0961203303lu362xx. [DOI] [PubMed] [Google Scholar]
- 61.Baechler EC, Batliwalla FM, Karypis G, Gaffney PM, Ortmann WA, Espe KJ, Shark KB, Grande WJ, Hughes KM, Kapur V, Gregersen PK, Behrens TW. Interferon-inducible gene expression signature in peripheral blood cells of patients with severe lupus. Proc Natl Acad Sci U S A. 2003;100:2610–5. doi: 10.1073/pnas.0337679100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kirou KA, Lee C, George S, Louca K, Papagiannis IG, Peterson MG, Ly N, Woodward RN, Fry KE, Lau AY, Prentice JG, Wohlgemuth JG, Crow MK. Coordinate overexpression of interferon-alpha-induced genes in systemic lupus erythematosus. Arthritis Rheum. 2004;50:3958–67. doi: 10.1002/art.20798. [DOI] [PubMed] [Google Scholar]
- 63.Peterson KS, Huang JF, Zhu J, D'Agati V, Liu X, Miller N, Erlander MG, Jackson MR, Winchester RJ. Characterization of heterogeneity in the molecular pathogenesis of lupus nephritis from transcriptional profiles of laser-captured glomeruli. J Clin Invest. 2004;113:1722–33. doi: 10.1172/JCI19139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tucci M, Quatraro C, Lombardi L, Pellegrino C, Dammacco F, Silvestris F. Glomerular accumulation of plasmacytoid dendritic cells in active lupus nephritis: role of interleukin-18. Arthritis Rheum. 2008;58:251–62. doi: 10.1002/art.23186. [DOI] [PubMed] [Google Scholar]
- 65.Cush JJ. Unusual toxicities with TNF inhibition: heart failure and drug-induced lupus. Clin Exp Rheumatol. 2004;22:S141–7. [PubMed] [Google Scholar]
- 66.Palucka AK, Blanck JP, Bennett L, Pascual V, Banchereau J. Cross-regulation of TNF and IFN-alpha in autoimmune diseases. Proc Natl Acad Sci U S A. 2005;102:3372–7. doi: 10.1073/pnas.0408506102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Liu K, Mohan C. What do mouse models teach us about human SLE? Clin Immunol. 2006;119:123–30. doi: 10.1016/j.clim.2006.01.014. [DOI] [PubMed] [Google Scholar]
- 68.Hang L, Theofilopoulos AN, Dixon FJ. A spontaneous rheumatoid arthritis-like disease in MRL/l mice. J Exp Med. 1982;155:1690–701. doi: 10.1084/jem.155.6.1690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Koopman WJ, Gay S. The MRL-lpr/lpr mouse. A model for the study of rheumatoid arthritis. Scand J Rheumatol Suppl. 1988;75:284–9. doi: 10.3109/03009748809096780. [DOI] [PubMed] [Google Scholar]
- 70.Pisitkun P, Deane JA, Difilippantonio MJ, Tarasenko T, Satterthwaite AB, Bolland S. Autoreactive B Cell Responses to RNA-Related Antigens Due to TLR7 Gene Duplication. Science. 2006 doi: 10.1126/science.1124978. [DOI] [PubMed] [Google Scholar]
- 71.Subramanian S, Tus K, Li QZ, Wang A, Tian XH, Zhou J, Liang C, Bartov G, McDaniel LD, Zhou XJ, Schultz RA, Wakeland EK. From the Cover: A Tlr7 translocation accelerates systemic autoimmunity in murine lupus. Proc Natl Acad Sci U S A. 2006;103:9970–5. doi: 10.1073/pnas.0603912103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Morel L, Mohan C, Yu Y, Croker BP, Tian N, Deng A, Wakeland EK. Functional dissection of systemic lupus erythematosus using congenic mouse strains. J Immunol. 1997;158:6019–28. [PubMed] [Google Scholar]
- 73.Mohan C, Alas E, Morel L, Yang P, Wakeland EK. Genetic dissection of SLE pathogenesis. Sle1 on murine chromosome 1 leads to a selective loss of tolerance to H2A/H2B/DNA subnucleosomes. J Clin Invest. 1998;101:1362–72. doi: 10.1172/JCI728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Mohan C, Yu Y, Morel L, Yang P, Wakeland EK. Genetic dissection of Sle pathogenesis: Sle3 on murine chromosome 7 impacts T cell activation, differentiation, and cell death. J Immunol. 1999;162:6492–502. [PubMed] [Google Scholar]
- 75.Mohan C, Morel L, Yang P, Wakeland EK. Genetic dissection of systemic lupus erythematosus pathogenesis: Sle2 on murine chromosome 4 leads to B cell hyperactivity. J Immunol. 1997;159:454–65. [PubMed] [Google Scholar]
- 76.Alarcon-Riquelme ME. The genetics of systemic lupus erythematosus. J Autoimmun. 2005;25(Suppl):46–8. doi: 10.1016/j.jaut.2005.09.012. [DOI] [PubMed] [Google Scholar]
- 77.Subramanian S, Tus K, Li QZ, Wang A, Tian XH, Zhou J, Liang C, Bartov G, McDaniel LD, Zhou XJ, Schultz RA, Wakeland EK. A Tlr7 translocation accelerates systemic autoimmunity in murine lupus. Proc Natl Acad Sci U S A. 2006;103:9970–5. doi: 10.1073/pnas.0603912103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Lau CM, Broughton C, Tabor AS, Akira S, Flavell RA, Mamula MJ, Christensen SR, Shlomchik MJ, Viglianti GA, Rifkin IR, Marshak-Rothstein A. RNA-associated autoantigens activate B cells by combined B cell antigen receptor/Toll-like receptor 7 engagement. J Exp Med. 2005;202:1171–7. doi: 10.1084/jem.20050630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Deane JA, Pisitkun P, Barrett RS, Feigenbaum L, Town T, Ward JM, Flavell RA, Bolland S. Control of toll-like receptor 7 expression is essential to restrict autoimmunity and dendritic cell proliferation. Immunity. 2007;27:801–10. doi: 10.1016/j.immuni.2007.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Mathian A, Weinberg A, Gallegos M, Banchereau J, Koutouzov S. IFN-alpha induces early lethal lupus in preautoimmune (New Zealand Black x New Zealand White) F1 but not in BALB/c mice. J Immunol. 2005;174:2499–506. doi: 10.4049/jimmunol.174.5.2499. [DOI] [PubMed] [Google Scholar]
- 81.Santiago-Raber ML, Baccala R, Haraldsson KM, Choubey D, Stewart TA, Kono DH, Theofilopoulos AN. Type-I Interferon Receptor Deficiency Reduces Lupus-like Disease in NZB Mice. J Exp Med. 2003;197:777–88. doi: 10.1084/jem.20021996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Braun D, Geraldes P, Demengeot J. Type I Interferon controls the onset and severity of autoimmune manifestations in lpr mice. J Autoimmun. 2003;20:15–25. doi: 10.1016/s0896-8411(02)00109-9. [DOI] [PubMed] [Google Scholar]
- 83.Sigurdsson S, Nordmark G, Goring HH, Lindroos K, Wiman AC, Sturfelt G, Jonsen A, Rantapaa-Dahlqvist S, Moller B, Kere J, Koskenmies S, Widen E, Eloranta ML, Julkunen H, Kristjansdottir H, Steinsson K, Alm G, Ronnblom L, Syvanen AC. Polymorphisms in the tyrosine kinase 2 and interferon regulatory factor 5 genes are associated with systemic lupus erythematosus. Am J Hum Genet. 2005;76:528–37. doi: 10.1086/428480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Takaoka A, Yanai H, Kondo S, Duncan G, Negishi H, Mizutani T, Kano S, Honda K, Ohba Y, Mak TW, Taniguchi T. Integral role of IRF-5 in the gene induction programme activated by Toll-like receptors. Nature. 2005;434:243–9. doi: 10.1038/nature03308. [DOI] [PubMed] [Google Scholar]
- 85.Schoenemeyer A, Barnes BJ, Mancl ME, Latz E, Goutagny N, Pitha PM, Fitzgerald KA, Golenbock DT. The interferon regulatory factor, IRF5, is a central mediator of toll-like receptor 7 signaling. J Biol Chem. 2005;280:17005–12. doi: 10.1074/jbc.M412584200. [DOI] [PubMed] [Google Scholar]
- 86.Bave U, Magnusson M, Eloranta ML, Perers A, Alm GV, Ronnblom L. Fc gamma RIIa is expressed on natural IFN-alpha-producing cells (plasmacytoid dendritic cells) and is required for the IFN-alpha production induced by apoptotic cells combined with lupus IgG. J Immunol. 2003;171:3296–302. doi: 10.4049/jimmunol.171.6.3296. [DOI] [PubMed] [Google Scholar]
- 87.Means TK, Latz E, Hayashi F, Murali MR, Golenbock DT, Luster AD. Human lupus autoantibody-DNA complexes activate DCs through cooperation of CD32 and TLR9. J Clin Invest. 2005;115:407–17. doi: 10.1172/JCI23025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Barrat FJ, Meeker T, Gregorio J, Chan JH, Uematsu S, Akira S, Chang B, Duramad O, Coffman RL. Nucleic acids of mammalian origin can act as endogenous ligands for Toll-like receptors and may promote systemic lupus erythematosus. J Exp Med. 2005;202:1131–9. doi: 10.1084/jem.20050914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Boule MW, Broughton C, Mackay F, Akira S, Marshak-Rothstein A, Rifkin IR. Toll-like receptor 9-dependent and -independent dendritic cell activation by chromatin-immunoglobulin G complexes. J Exp Med. 2004;199:1631–40. doi: 10.1084/jem.20031942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Honda K, Yanai H, Negishi H, Asagiri M, Sato M, Mizutani T, Shimada N, Ohba Y, Takaoka A, Yoshida N, Taniguchi T. IRF-7 is the master regulator of type-I interferon-dependent immune responses. Nature. 2005;434:772–7. doi: 10.1038/nature03464. [DOI] [PubMed] [Google Scholar]
- 91.Bave U, Alm GV, Ronnblom L. The combination of apoptotic U937 cells and lupus IgG is a potent IFN-alpha inducer [In Process Citation]. J Immunol. 2000;165:3519–26. doi: 10.4049/jimmunol.165.6.3519. [DOI] [PubMed] [Google Scholar]
- 92.Lovgren T, Eloranta ML, Bave U, Alm GV, Ronnblom L. Induction of interferon-alpha production in plasmacytoid dendritic cells by immune complexes containing nucleic acid released by necrotic or late apoptotic cells and lupus IgG. Arthritis Rheum. 2004;50:1861–72. doi: 10.1002/art.20254. [DOI] [PubMed] [Google Scholar]
- 93.Vollmer J, Tluk S, Schmitz C, Hamm S, Jurk M, Forsbach A, Akira S, Kelly KM, Reeves WH, Bauer S, Krieg AM. Immune stimulation mediated by autoantigen binding sites within small nuclear RNAs involves Toll-like receptors 7 and 8. J Exp Med. 2005;202:1575–85. doi: 10.1084/jem.20051696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Graham RR, Kozyrev SV, Baechler EC, Reddy MV, Plenge RM, Bauer JW, Ortmann WA, Koeuth T, Escribano MF, Collaborative Groups TA. Pons-Estel B, Petri M, Daly M, Gregersen PK, Martin J, Altshuler D, Behrens TW, Alarcon-Riquelme ME. A common haplotype of interferon regulatory factor 5 (IRF5) regulates splicing and expression and is associated with increased risk of systemic lupus erythematosus. Nat Genet. 2006;38:550–5. doi: 10.1038/ng1782. [DOI] [PubMed] [Google Scholar]
- 95.Remmers EF, Plenge RM, Lee AT, Graham RR, Hom G, Behrens TW, de Bakker PI, Le JM, Lee HS, Batliwalla F, Li W, Masters SL, Booty MG, Carulli JP, Padyukov L, Alfredsson L, Klareskog L, Chen WV, Amos CI, Criswell LA, Seldin MF, Kastner DL, Gregersen PK. STAT4 and the risk of rheumatoid arthritis and systemic lupus erythematosus. N Engl J Med. 2007;357:977–86. doi: 10.1056/NEJMoa073003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Nath SK, Han S, Kim-Howard X, Kelly JA, Viswanathan P, Gilkeson GS, Chen W, Zhu C, McEver RP, Kimberly RP, Alarcon-Riquelme ME, Vyse TJ, Li QZ, Wakeland EK, Merrill JT, James JA, Kaufman KM, Guthridge JM, Harley JB. A nonsynonymous functional variant in integrin-alpha(M) (encoded by ITGAM) is associated with systemic lupus erythematosus. Nat Genet. 2008;40:152–4. doi: 10.1038/ng.71. [DOI] [PubMed] [Google Scholar]
- 97.Harley JB, Alarcon-Riquelme ME, Criswell LA, Jacob CO, Kimberly RP, Moser KL, Tsao BP, Vyse TJ, Langefeld CD. Genome-wide association scan in women with systemic lupus erythematosus identifies susceptibility variants in ITGAM, PXK, KIAA1542 and other loci. Nat Genet. 2008;40:204–10. doi: 10.1038/ng.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Kozyrev SV, Abelson AK, Wojcik J, Zaghlool A, Linga Reddy MV, Sanchez E, Gunnarsson I, Svenungsson E, Sturfelt G, Jonsen A, Truedsson L, Pons-Estel BA, Witte T, D'Alfonso S, Barrizzone N, Danieli MG, Gutierrez C, Suarez A, Junker P, Laustrup H, Gonzalez-Escribano MF, Martin J, Abderrahim H, Alarcon-Riquelme ME. Functional variants in the B-cell gene BANK1 are associated with systemic lupus erythematosus. Nat Genet. 2008;40:211–6. doi: 10.1038/ng.79. [DOI] [PubMed] [Google Scholar]
- 99.Hom G, Graham RR, Modrek B, Taylor KE, Ortmann W, Garnier S, Lee AT, Chung SA, Ferreira RC, Pant PV, Ballinger DG, Kosoy R, Demirci FY, Kamboh MI, Kao AH, Tian C, Gunnarsson I, Bengtsson AA, Rantapaa-Dahlqvist S, Petri M, Manzi S, Seldin MF, Ronnblom L, Syvanen AC, Criswell LA, Gregersen PK, Behrens TW. Association of Systemic Lupus Erythematosus with C8orf13-BLK and ITGAM-ITGAX. N Engl J Med. 2008 doi: 10.1056/NEJMoa0707865. [DOI] [PubMed] [Google Scholar]
- 100.Yakubenko VP, Lishko VK, Lam SC, Ugarova TP. A molecular basis for integrin alphaMbeta 2 ligand binding promiscuity. J Biol Chem. 2002;277:48635–42. doi: 10.1074/jbc.M208877200. [DOI] [PubMed] [Google Scholar]
- 101.Buyon JP, Shadick N, Berkman R, Hopkins P, Dalton J, Weissmann G, Winchester R, Abramson SB. Surface expression of Gp 165/95, the complement receptor CR3, as a marker of disease activity in systemic Lupus erythematosus. Clin Immunol Immunopathol. 1988;46:141–9. doi: 10.1016/0090-1229(88)90014-1. [DOI] [PubMed] [Google Scholar]
- 102.Brunner HI, Mueller M, Rutherford C, Passo MH, Witte D, Grom A, Mishra J, Devarajan P. Urinary neutrophil gelatinase-associated lipocalin as a biomarker of nephritis in childhood-onset systemic lupus erythematosus. Arthritis Rheum. 2006;54:2577–84. doi: 10.1002/art.22008. [DOI] [PubMed] [Google Scholar]
- 103.Jego G, Pascual V, Palucka AK, Banchereau J. Dendritic cells control B cell growth and differentiation. Curr Dir Autoimmun. 2005;8:124–39. doi: 10.1159/000082101. [DOI] [PubMed] [Google Scholar]
- 104.Tian J, Avalos AM, Mao SY, Chen B, Senthil K, Wu H, Parroche P, Drabic S, Golenbock D, Sirois C, Hua J, An LL, Audoly L, La Rosa G, Bierhaus A, Naworth P, Marshak-Rothstein A, Crow MK, Fitzgerald KA, Latz E, Kiener PA, Coyle AJ. Toll-like receptor 9-dependent activation by DNA-containing immune complexes is mediated by HMGB1 and RAGE. Nat. Immunol. 2007;8:487–96. doi: 10.1038/ni1457. [DOI] [PubMed] [Google Scholar]
- 105.Lande R, Gregorio J, Facchinetti V, Chatterjee B, Wang YH, Homey B, Cao W, Wang YH, Su B, Nestle FO, Zal T, Mellman I, Schroder JM, Liu YJ, Gilliet M. Plasmacytoid dendritic cells sense self-DNA coupled with antimicrobial peptide. Nature. 2007;449:564–9. doi: 10.1038/nature06116. [DOI] [PubMed] [Google Scholar]
- 106.Lauwerys BR, Wakeland EK. Genetics of lupus nephritis. Lupus. 2005;14:2–12. doi: 10.1191/0961203305lu2052oa. [DOI] [PubMed] [Google Scholar]
- 107.Kumar KR, Li L, Yan M, Bhaskarabhatla M, Mobley AB, Nguyen C, Mooney JM, Schatzle JD, Wakeland EK, Mohan C. Regulation of B cell tolerance by the lupus susceptibility gene Ly108. Science. 2006;312:1665–9. doi: 10.1126/science.1125893. [DOI] [PubMed] [Google Scholar]
- 108.Greenberg SA, Pinkus JL, Pinkus GS, Burleson T, Sanoudou D, Tawil R, Barohn RJ, Saperstein DS, Briemberg HR, Ericsson M, Park P, Amato AA. Interferon-alpha/beta-mediated innate immune mechanisms in dermatomyositis. Ann Neurol. 2005;57:664–78. doi: 10.1002/ana.20464. [DOI] [PubMed] [Google Scholar]
- 109.Tezak Z, Hoffman EP, Lutz JL, Fedczyna TO, Stephan D, Bremer EG, Krasnoselska-Riz I, Kumar A, Pachman LM. Gene expression profiling in DQA1*0501+ children with untreated dermatomyositis: a novel model of pathogenesis. J Immunol. 2002;168:4154–63. doi: 10.4049/jimmunol.168.8.4154. [DOI] [PubMed] [Google Scholar]
- 110.Baechler EC, Bauer JW, Slattery CA, Ortmann WA, Espe KJ, Novitzke J, Ytterberg SR, Gregersen PK, Behrens TW, Reed AM. An interferon signature in the peripheral blood of dermatomyositis patients is associated with disease activity. Mol Med. 2007;13:59–68. doi: 10.2119/2006-00085.Baechler. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Walsh RJ, Kong SW, Yao Y, Jallal B, Kiener PA, Pinkus JL, Beggs AH, Amato AA, Greenberg SA. Type I interferon-inducible gene expression in blood is present and reflects disease activity in dermatomyositis and polymyositis. Arthritis Rheum. 2007;56:3784–92. doi: 10.1002/art.22928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Gottenberg JE, Cagnard N, Lucchesi C, Letourneur F, Mistou S, Lazure T, Jacques S, Ba N, Ittah M, Lepajolec C, Labetoulle M, Ardizzone M, Sibilia J, Fournier C, Chiocchia G, Mariette X. Activation of IFN pathways and plasmacytoid dendritic cell recruitment in target organs of primary Sjogren's syndrome. Proc Natl Acad Sci U S A. 2006;103:2770–5. doi: 10.1073/pnas.0510837103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Bave U, Nordmark G, Lovgren T, Ronnelid J, Cajander S, Eloranta ML, Alm GV, Ronnblom L. Activation of the type I interferon system in primary Sjogren's syndrome: a possible etiopathogenic mechanism. Arthritis Rheum. 2005;52:1185–95. doi: 10.1002/art.20998. [DOI] [PubMed] [Google Scholar]
- 114.Funk J, Langeland T, Schrumpf E, Hanssen LE. Psoriasis induced by interferon-alpha. Br J Dermatol. 1991;125:463–5. doi: 10.1111/j.1365-2133.1991.tb14774.x. [DOI] [PubMed] [Google Scholar]
- 115.van der Fits L, van der Wel LI, Laman JD, Prens EP, Verschuren MC. In psoriasis lesional skin the type I interferon signaling pathway is activated, whereas interferon-alpha sensitivity is unaltered. J Invest Dermatol. 2004;122:51–60. doi: 10.1046/j.0022-202X.2003.22113.x. [DOI] [PubMed] [Google Scholar]
- 116.Hida S, Ogasawara K, Sato K, Abe M, Takayanagi H, Yokochi T, Sato T, Hirose S, Shirai T, Taki S, Taniguchi T. CD8(+) T cell-mediated skin disease in mice lacking IRF-2, the transcriptional attenuator of interferon-alpha/beta signaling. Immunity. 2000;13:643–55. doi: 10.1016/s1074-7613(00)00064-9. [DOI] [PubMed] [Google Scholar]
- 117.Nestle FO, Conrad C, Tun-Kyi A, Homey B, Gombert M, Boyman O, Burg G, Liu YJ, Gilliet M. Plasmacytoid predendritic cells initiate psoriasis through interferon-alpha production. J Exp Med. 2005;202:135–43. doi: 10.1084/jem.20050500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Aringer M, Graninger WB, Steiner G, Smolen JS. Safety and efficacy of tumor necrosis factor alpha blockade in systemic lupus erythematosus: an open-label study. Arthritis Rheum. 2004;50:3161–9. doi: 10.1002/art.20576. [DOI] [PubMed] [Google Scholar]
- 119.Adams A, Lehman TJ. Update on the pathogenesis and treatment of systemic onset juvenile rheumatoid arthritis. Curr Opin Rheumatol. 2005;17:612–6. doi: 10.1097/01.bor.0000169363.69066.d0. [DOI] [PubMed] [Google Scholar]
- 120.Dinarello CA. Biologic basis for interleukin-1 in disease. Blood. 1996;87:2095–147. [PubMed] [Google Scholar]
- 121.Pascual V, Allantaz F, Arce E, Punaro M, Banchereau J. Role of interleukin-1 (IL-1) in the pathogenesis of systemic onset juvenile idiopathic arthritis and clinical response to IL-1 blockade. J Exp Med. 2005;201:1479–86. doi: 10.1084/jem.20050473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Pascual V, Allantaz F, Arce E, Punaro M, Banchereau J. Role of interleukin-1 (IL-1) in the pathogenesis of systemic onset juvenile idiopathic arthritis and clinical response to IL-1 blockade. J Exp Med. 2005 doi: 10.1084/jem.20050473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Fall N, Barnes M, Thornton S, Luyrink L, Olson J, Ilowite NT, Gottlieb BS, Griffin T, Sherry DD, Thompson S, Glass DN, Colbert RA, Grom AA. Gene expression profiling of peripheral blood from patients with untreated new-onset systemic juvenile idiopathic arthritis reveals molecular heterogeneity that may predict macrophage activation syndrome. Arthritis Rheum. 2007;56:3793–804. doi: 10.1002/art.22981. [DOI] [PubMed] [Google Scholar]
- 124.Allantaz F, Chaussabel D, Stichweh D, Bennett L, Allman W, Mejias A, Ardura M, Chung W, Wise C, Palucka K, Ramilo O, Punaro M, Banchereau J, Pascual V. Blood leukocyte microarrays to diagnose systemic onset juvenile idiopathic arthritis and follow the response to IL-1 blockade. J Exp Med. 2007;204:2131–44. doi: 10.1084/jem.20070070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Smith DE, Renshaw BR, Ketchem RR, Kubin M, Garka KE, Sims JE. Four new members expand the interleukin-1 superfamily. J Biol Chem. 2000;275:1169–75. doi: 10.1074/jbc.275.2.1169. [DOI] [PubMed] [Google Scholar]
- 126.Dinarello CA. Blocking IL-1 in systemic inflammation. J Exp Med. 2005;201:1355–9. doi: 10.1084/jem.20050640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Martinon F, Tschopp J. Inflammatory caspases and inflammasomes: master switches of inflammation. Cell Death Differ. 2007;14:10–22. doi: 10.1038/sj.cdd.4402038. [DOI] [PubMed] [Google Scholar]
- 128.Kanneganti TD, Body-Malapel M, Amer A, Park JH, Whitfield J, Franchi L, Taraporewala ZF, Miller D, Patton JT, Inohara N, Nunez G. Critical Role for Cryopyrin/Nalp3 in Activation of Caspase-1 in Response to Viral Infection and Double-stranded RNA. J Biol Chem. 2006;281:36560–8. doi: 10.1074/jbc.M607594200. [DOI] [PubMed] [Google Scholar]
- 129.Kanneganti TD, Ozoren N, Body-Malapel M, Amer A, Park JH, Franchi L, Whitfield J, Barchet W, Colonna M, Vandenabeele P, Bertin J, Coyle A, Grant EP, Akira S, Nunez G. Bacterial RNA and small antiviral compounds activate caspase-1 through cryopyrin/Nalp3. Nature. 2006;440:233–6. doi: 10.1038/nature04517. [DOI] [PubMed] [Google Scholar]
- 130.Mariathasan S, Weiss DS, Newton K, McBride J, O'Rourke K, Roose-Girma M, Lee WP, Weinrauch Y, Monack DM, Dixit VM. Cryopyrin activates the inflammasome in response to toxins and ATP. Nature. 2006;440:228–32. doi: 10.1038/nature04515. [DOI] [PubMed] [Google Scholar]
- 131.Martinon F, Petrilli V, Mayor A, Tardivel A, Tschopp J. Gout-associated uric acid crystals activate the NALP3 inflammasome. Nature. 2006;440:237–41. doi: 10.1038/nature04516. [DOI] [PubMed] [Google Scholar]
- 132.Ferrari D, Pizzirani C, Adinolfi E, Lemoli RM, Curti A, Idzko M, Panther E, Di Virgilio F. The P2X7 receptor: a key player in IL-1 processing and release. J Immunol. 2006;176:3877–83. doi: 10.4049/jimmunol.176.7.3877. [DOI] [PubMed] [Google Scholar]
- 133.Petrilli V, Papin S, Dostert C, Mayor A, Martinon F, Tschopp J. Activation of the NALP3 inflammasome is triggered by low intracellular potassium concentration. Cell Death Differ. 2007;14:1583–9. doi: 10.1038/sj.cdd.4402195. [DOI] [PubMed] [Google Scholar]
- 134.Buch MH, Bingham SJ, Seto Y, McGonagle D, Bejarano V, White J, Emery P. Lack of response to anakinra in rheumatoid arthritis following failure of tumor necrosis factor alpha blockade. Arthritis Rheum. 2004;50:725–8. doi: 10.1002/art.20115. [DOI] [PubMed] [Google Scholar]
- 135.Fitzgerald AA, Leclercq SA, Yan A, Homik JE, Dinarello CA. Rapid responses to anakinra in patients with refractory adult-onset Still's disease. Arthritis Rheum. 2005;52:1794–803. doi: 10.1002/art.21061. [DOI] [PubMed] [Google Scholar]
- 136.Gamero AM, Oppenheim JJ. IL-1 can act as number one. Immunity. 2006;24:16–7. doi: 10.1016/j.immuni.2005.12.007. [DOI] [PubMed] [Google Scholar]
- 137.Chaussabel D, Allman W, Mejias A, Chung W, Bennett L, Ramilo O, Pascual V, Palucka AK, Banchereau J. Analysis of significance patterns identifies ubiquitous and disease-specific gene-expression signatures in patient peripheral blood leukocytes. Ann N Y Acad Sci. 2005;1062:146–54. doi: 10.1196/annals.1358.017. [DOI] [PubMed] [Google Scholar]
- 138.Martinon F, Gaide O, Petrilli V, Mayor A, Tschopp J. NALP Inflammasomes: a central role in innate immunity. Semin Immunopathol. 2007 doi: 10.1007/s00281-007-0079-y. [DOI] [PubMed] [Google Scholar]
- 139.Brydges S, Kastner DL. The systemic autoinflammatory diseases: inborn errors of the innate immune system. Curr Top Microbiol Immunol. 2006;305:127–60. doi: 10.1007/3-540-29714-6_7. [DOI] [PubMed] [Google Scholar]
- 140.Aksentijevich I, C DP, Remmers EF, Mueller JL, Le J, Kolodner RD, Moak Z, Chuang M, Austin F, Goldbach-Mansky R, Hoffman HM, Kastner DL. The clinical continuum of cryopyrinopathies: novel CIAS1 mutations in North American patients and a new cryopyrin model. Arthritis Rheum. 2007;56:1273–85. doi: 10.1002/art.22491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Watanabe H, Gaide O, Petrilli V, Martinon F, Contassot E, Roques S, Kummer JA, Tschopp J, French LE. Activation of the IL-1beta-Processing Inflammasome Is Involved in Contact Hypersensitivity. J Invest Dermatol. 2007 doi: 10.1038/sj.jid.5700819. [DOI] [PubMed] [Google Scholar]
- 142.McDermott MF, Tschopp J. From inflammasomes to fevers, crystals and hypertension: how basic research explains inflammatory diseases. Trends Mol Med. 2007 doi: 10.1016/j.molmed.2007.07.005. [DOI] [PubMed] [Google Scholar]
- 143.Sutterwala FS, Ogura Y, Szczepanik M, Lara-Tejero M, Lichtenberger GS, Grant EP, Bertin J, Coyle AJ, Galan JE, Askenase PW, Flavell RA. Critical role for NALP3/CIAS1/Cryopyrin in innate and adaptive immunity through its regulation of caspase-1. Immunity. 2006;24:317–27. doi: 10.1016/j.immuni.2006.02.004. [DOI] [PubMed] [Google Scholar]
- 144.Huang X, Yuang J, Goddard A, Foulis A, James RF, Lernmark A, Pujol-Borrell R, Rabinovitch A, Somoza N, Stewart TA. Interferon expression in the pancreases of patients with type I diabetes. Diabetes. 1995;44:658–64. doi: 10.2337/diab.44.6.658. [DOI] [PubMed] [Google Scholar]
- 145.Kaizer EC, Glaser CL, Chaussabel D, Banchereau J, Pascual V, White PC. Gene expression in peripheral blood mononuclear cells from children with diabetes. J Clin Endocrinol Metab. 2007;92:3705–11. doi: 10.1210/jc.2007-0979. [DOI] [PubMed] [Google Scholar]
- 146.Wang X, Jia S, Geoffrey R, Alemzadeh R, Ghosh S, Hessner MJ. Identification of a molecular signature in human type 1 diabetes mellitus using serum and functional genomics. J Immunol. 2008;180:1929–37. doi: 10.4049/jimmunol.180.3.1929. [DOI] [PubMed] [Google Scholar]
- 147.Mandrup-Poulsen T, Spinas GA, Prowse SJ, Hansen BS, Jorgensen DW, Bendtzen K, Nielsen JH, Nerup J. Islet cytotoxicity of interleukin 1. Influence of culture conditions and islet donor characteristics. Diabetes. 1987;36:641–7. doi: 10.2337/diab.36.5.641. [DOI] [PubMed] [Google Scholar]
- 148.Larsen CM, Faulenbach M, Vaag A, Volund A, Ehses JA, Seifert B, Mandrup-Poulsen T, Donath MY. Interleukin-1-receptor antagonist in type 2 diabetes mellitus. N Engl J Med. 2007;356:1517–26. doi: 10.1056/NEJMoa065213. [DOI] [PubMed] [Google Scholar]
- 149.Crow MK, Kirou KA, Wohlgemuth J. Microarray analysis of interferon-regulated genes in SLE. Autoimmunity. 2003;36:481–90. doi: 10.1080/08916930310001625952. [DOI] [PubMed] [Google Scholar]
- 150.Achiron A, Feldman A, Mandel M, Gurevich M. Impaired expression of peripheral blood apoptotic-related gene transcripts in acute multiple sclerosis relapse. Ann N Y Acad Sci. 2007;1107:155–67. doi: 10.1196/annals.1381.017. [DOI] [PubMed] [Google Scholar]
- 151.Achiron A, Gurevich M, Snir Y, Segal E, Mandel M. Zinc-ion binding and cytokine activity regulation pathways predicts outcome in relapsing-remitting multiple sclerosis. Clin Exp Immunol. 2007;149:235–42. doi: 10.1111/j.1365-2249.2007.03405.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Singh MK, Scott TF, LaFramboise WA, Hu FZ, Post JC, Ehrlich GD. Gene expression changes in peripheral blood mononuclear cells from multiple sclerosis patients undergoing beta-interferon therapy. J Neurol Sci. 2007;258:52–9. doi: 10.1016/j.jns.2007.02.034. [DOI] [PubMed] [Google Scholar]
- 153.Edwards CJ, Feldman JL, Beech J, Shields KM, Stover JA, Trepicchio WL, Larsen G, Foxwell BM, Brennan FM, Feldmann M, Pittman DD. Molecular profile of peripheral blood mononuclear cells from patients with rheumatoid arthritis. Mol Med. 2007;13:40–58. doi: 10.2119/2006-000056.Edwards. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.van der Pouw Kraan TC, Wijbrandts CA, van Baarsen LG, Voskuyl AE, Rustenburg F, Baggen JM, Ibrahim SM, Fero M, Dijkmans BA, Tak PP, Verweij CL. Rheumatoid arthritis subtypes identified by genomic profiling of peripheral blood cells: assignment of a type I interferon signature in a subpopulation of patients. Ann Rheum Dis. 2007;66:1008–14. doi: 10.1136/ard.2006.063412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Lequerre T, Gauthier-Jauneau AC, Bansard C, Derambure C, Hiron M, Vittecoq O, Daveau M, Mejjad O, Daragon A, Tron F, Le Loet X, Salier JP. Gene profiling in white blood cells predicts infliximab responsiveness in rheumatoid arthritis. Arthritis Res Ther. 2006;8:R105. doi: 10.1186/ar1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Batliwalla FM, Baechler EC, Xiao X, Li W, Balasubramanian S, Khalili H, Damle A, Ortmann WA, Perrone A, Kantor AB, Gulko PS, Kern M, Furie R, Behrens TW, Gregersen PK. Peripheral blood gene expression profiling in rheumatoid arthritis. Genes Immun. 2005;6:388–97. doi: 10.1038/sj.gene.6364209. [DOI] [PubMed] [Google Scholar]
- 157.Burczynski ME, Peterson RL, Twine NC, Zuberek KA, Brodeur BJ, Casciotti L, Maganti V, Reddy PS, Strahs A, Immermann F, Spinelli W, Schwertschlag U, Slager AM, Cotreau MM, Dorner AJ. Molecular classification of Crohn's disease and ulcerative colitis patients using transcriptional profiles in peripheral blood mononuclear cells. J Mol Diagn. 2006;8:51–61. doi: 10.2353/jmoldx.2006.050079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Stoeckman AK, Baechler EC, Ortmann WA, Behrens TW, Michet CJ, Peterson EJ. A distinct inflammatory gene expression profile in patients with psoriatic arthritis. Genes Immun. 2006;7:583–91. doi: 10.1038/sj.gene.6364334. [DOI] [PubMed] [Google Scholar]
- 159.Lempicki RA, Polis MA, Yang J, McLaughlin M, Koratich C, Huang DW, Fullmer B, Wu L, Rehm CA, Masur H, Lane HC, Sherman KE, Fauci AS, Kottilil S. Gene expression profiles in hepatitis C virus (HCV) and HIV coinfection: class prediction analyses before treatment predict the outcome of anti-HCV therapy among HIV-coinfected persons. J Infect Dis. 2006;193:1172–7. doi: 10.1086/501365. [DOI] [PubMed] [Google Scholar]
- 160.Thach DC, Agan BK, Olsen C, Diao J, Lin B, Gomez J, Jesse M, Jenkins M, Rowley R, Hanson E, Tibbetts C, Stenger DA, Walter E. Surveillance of transcriptomes in basic military trainees with normal, febrile respiratory illness, and convalescent phenotypes. Genes Immun. 2005 doi: 10.1038/sj.gene.6364244. [DOI] [PubMed] [Google Scholar]
- 161.Ramilo O, Allman W, Chung W, Mejias A, Ardura M, Glaser C, Wittkowski KM, Piqueras B, Banchereau J, Palucka AK, Chaussabel D. Gene expression patterns in blood leukocytes discriminate patients with acute infections. Blood. 2007;109:2066–77. doi: 10.1182/blood-2006-02-002477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Simmons CP, Popper S, Dolocek C, Chau TN, Griffiths M, Dung NT, Long TH, Hoang DM, Chau NV, Thao le TT, Hien TT, Relman DA, Farrar J. Patterns of host genome-wide gene transcript abundance in the peripheral blood of patients with acute dengue hemorrhagic fever. J Infect Dis. 2007;195:1097–107. doi: 10.1086/512162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Jacobsen M, Repsilber D, Gutschmidt A, Neher A, Feldmann K, Mollenkopf HJ, Ziegler A, Kaufmann SH. Candidate biomarkers for discrimination between infection and disease caused by Mycobacterium tuberculosis. J Mol Med. 2007;85:613–21. doi: 10.1007/s00109-007-0157-6. [DOI] [PubMed] [Google Scholar]
- 164.Ockenhouse CF, Hu WC, Kester KE, Cummings JF, Stewart A, Heppner DG, Jedlicka AE, Scott AL, Wolfe ND, Vahey M, Burke DS. Common and divergent immune response signaling pathways discovered in peripheral blood mononuclear cell gene expression patterns in presymptomatic and clinically apparent malaria. Infect Immun. 2006;74:5561–73. doi: 10.1128/IAI.00408-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Tang BM, McLean AS, Dawes IW, Huang SJ, Lin RC. The Use of Gene-Expression Profiling to Identify Candidate Genes in Human Sepsis. Am J Respir Crit Care Med. 2007 doi: 10.1164/rccm.200612-1819OC. [DOI] [PubMed] [Google Scholar]
- 166.Flechner SM, Kurian SM, Head SR, Sharp SM, Whisenant TC, Zhang J, Chismar JD, Horvath S, Mondala T, Gilmartin T, Cook DJ, Kay SA, Walker JR, Salomon DR. Kidney transplant rejection and tissue injury by gene profiling of biopsies and peripheral blood lymphocytes. Am J Transplant. 2004;4:1475–89. doi: 10.1111/j.1600-6143.2004.00526.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Martinez-Llordella M, Puig-Pey I, Orlando G, Ramoni M, Tisone G, Rimola A, Lerut J, Latinne D, Margarit C, Bilbao I, Brouard S, Hernandez-Fuentes M, Soulillou JP, Sanchez-Fueyo A. Multiparameter immune profiling of operational tolerance in liver transplantation. Am J Transplant. 2007;7:309–19. doi: 10.1111/j.1600-6143.2006.01621.x. [DOI] [PubMed] [Google Scholar]
- 168.Deng MC, Eisen HJ, Mehra MR, Billingham M, Marboe CC, Berry G, Kobashigawa J, Johnson FL, Starling RC, Murali S, Pauly DF, Baron H, Wohlgemuth JG, Woodward RN, Klingler TM, Walther D, Lal PG, Rosenberg S, Hunt S. Noninvasive discrimination of rejection in cardiac allograft recipients using gene expression profiling. Am J Transplant. 2006;6:150–60. doi: 10.1111/j.1600-6143.2005.01175.x. [DOI] [PubMed] [Google Scholar]
- 169.Baron C, Somogyi R, Greller LD, Rineau V, Wilkinson P, Cho CR, Cameron MJ, Kelvin DJ, Chagnon P, Roy DC, Busque L, Sekaly RP, Perreault C. Prediction of graft-versus-host disease in humans by donor gene-expression profiling. PLoS Med. 2007;4:e23. doi: 10.1371/journal.pmed.0040023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Takamura T, Honda M, Sakai Y, Ando H, Shimizu A, Ota T, Sakurai M, Misu H, Kurita S, Matsuzawa-Nagata N, Uchikata M, Nakamura S, Matoba R, Tanino M, Matsubara K, Kaneko S. Gene expression profiles in peripheral blood mononuclear cells reflect the pathophysiology of type 2 diabetes. Biochem Biophys Res Commun. 2007;361:379–84. doi: 10.1016/j.bbrc.2007.07.006. [DOI] [PubMed] [Google Scholar]
- 171.Tang Y, Xu H, Du X, Lit L, Walker W, Lu A, Ran R, Gregg JP, Reilly M, Pancioli A, Khoury JC, Sauerbeck LR, Carrozzella JA, Spilker J, Clark J, Wagner KR, Jauch EC, Chang DJ, Verro P, Broderick JP, Sharp FR. Gene expression in blood changes rapidly in neutrophils and monocytes after ischemic stroke in humans: a microarray study. J Cereb Blood Flow Metab. 2006;26:1089–102. doi: 10.1038/sj.jcbfm.9600264. [DOI] [PubMed] [Google Scholar]
- 172.Solmi R, Ugolini G, Rosati G, Zanotti S, Lauriola M, Montroni I, del Governatore M, Caira A, Taffurelli M, Santini D, Coppola D, Guidotti L, Carinci P, Strippoli P. Microarray-based identification and RT-PCR test screening for epithelial-specific mRNAs in peripheral blood of patients with colon cancer. BMC Cancer. 2006;6:250. doi: 10.1186/1471-2407-6-250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Osman I, Bajorin DF, Sun TT, Zhong H, Douglas D, Scattergood J, Zheng R, Han M, Marshall KW, Liew CC. Novel blood biomarkers of human urinary bladder cancer. Clin Cancer Res. 2006;12:3374–80. doi: 10.1158/1078-0432.CCR-05-2081. [DOI] [PubMed] [Google Scholar]