Abstract
The stepwise process of Ag receptor gene assembly, termed V(D)J recombination, is coordinated during lymphocyte development by sweeping changes in chromatin that permit or deny access to a single recombinase enzyme. We now show that switching/sucrose nonfermenting (SWI/SNF) chromatin remodeling complexes are recruited to the Igh locus by an enhancer-dependent process and that these complexes are essential for generating recombinase accessibility throughout the locus. Depletion of SWI/SNF in pro-B cells also inhibits antisense transcription through all clusters of Igh gene segments, a pioneering process that has been implicated in the initial opening of chromatin. We conclude that SWI/SNF complexes play multiple roles in Igh gene assembly, ranging from initial locus activation to the spreading and maintenance of chromatin accessibility over large VH, DH, and JH domains.
Antigen receptor gene assembly occurs via a genetic reorganization, termed V(D)J recombination, which generates the enormous diversity of the Ig and Tcr variable regions required for adaptive immunity (1). V(D)J recombination is mediated at all Ag receptor loci by a single enzymatic complex consisting of the recombinases RAG1 and RAG2, which recognize recombination signal sequences flanking all variable (V), diversity (D), and joining (J) gene segments. Prior studies have shown that RAG complexes are targeted to specific regions within Ag receptor loci by changes in chromatin accessibility that either enhance or diminish the availability of recombination signal sequences to recombinase (1, 2). Chromatin accessibility at Ig and Tcr loci is regulated by cis-acting elements (promoters and enhancers) that communicate to drive the transcription of intervening gene segments in sense and antisense directions (3, 4).
These cis elements also sculpt regional patterns of histone modifications on chromatin associated with each cluster of gene segments. For example, deletion of Tcr enhancer elements alters histone modifications from active (e.g., histone 3 lysine 9 (H3K9)4 acetylation) to inaccessible patterns (e.g., H3K9 methylation) over neighboring chromatin (5, 6). Histone modification patterns constitute regional codes that are recognized by other nuclear factors, including nucleosome remodeling complexes (7). These multisubunit enzymes use ATP to render neighboring chromatin either accessible or inaccessible for transcription, recombination, and DNA repair (1). We recently showed that recruitment of switching/sucrose nonfermenting (SWI/SNF) remodeling complexes to a promoter in the Tcrb locus is required to activate transcription and recombination of neighboring DβJβ gene segments (8). However, the generality of SWI/SNF requirements in the mechanisms that regulate Ag receptor gene assembly remains to be established.
For this purpose, we explored SWI/SNF-dependent activation of Igh loci in pro-B cells. Prior studies have shown that the intronic enhancer Eμ directs sense and antisense transcription (αST) of the DHJH cluster in pro-B cells (9, 10). Eμ-directed transcription may be crucial for opening chromatin throughout the DHJH region (3), making it accessible to recombinase and recruiting RAG complexes via transcription-coupled histone modifications (2). Indeed, DHJH rearrangement is significantly impaired in pro-B cells harboring a targeted deletion of Eμ (9, 11). In contrast, the large VH cluster becomes accessible via Eμ-independent mechanisms and is opened in a regional manner, with DH-proximal gene segments opening before distal VH segments (12). Emerging evidence is consistent with a role for αST in converting inaccessible chromatin to a more accessible state over the 2-Mb VH cluster (3). We now show that SWI/SNF complexes are essential for initiation of αSTs throughout the Igh locus. Consequently, pro-B cells lacking SWI/SNF fail to activate sense Igh transcription and assembly of VH, DH, and JH gene segments. These findings underscore a central role for SWI/SNF complexes in the genetic-epigenetic crosstalk that directs locus-wide opening of Ag receptor genes for tissue- and stage-specific assembly during lymphocyte development.
Materials and Methods
Cell lines and mice
The RAG2-deficient cell lines P5424 (pro-T) and 63−12 (pro-B) as well as their derivative clones were described previously (4, 6). Mice lacking Eμ or both PDQ52 and Eμ were reported previously (9). All experiments were conducted according to guidelines of the Animal Care and Use Committee at Vanderbilt University (Nashville, TN). Data shown in the manuscript are all representative of at least two independent experiments (three for Fig. 2A and Fig. 4).
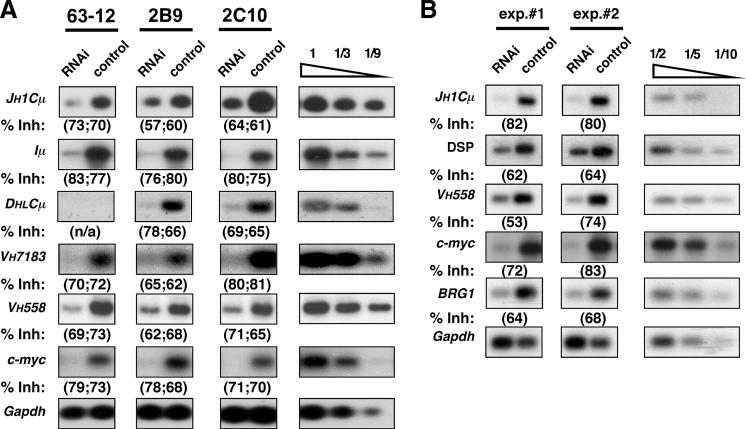
FIGURE 2.
SWI/SNF depletion in pro-B cell lines inhibits Igh germline transcription. A, RAG2-deficient pro-B cell lines (63−12, 2B9, and 2C10) were transfected transiently with an RNAi vector specific for BRG1 and BRM transcripts or with an empty control vector. 63−12 was transiently transfected with a Rag2 expression vector to generate subclones 2B9 and 2C10, which harbor stable Igh rearrangements (see Results and Discussion). Total RNA from transfected cells (GFP+) was analyzed by RT-PCR for germline Igh transcripts, specific upstream DHJH transcripts (DHLCμ), and mRNAs corresponding to SWI/SNF-dependent (c-myc) or -independent genes (Gapdh). Assay linearity was confirmed using sample titrations (far right). B, RT-PCR assays described in A were performed on pro-B cell cultures from RAG-deficient bone marrows supplemented with IL-7. Cells were infected with either control or RNAi retroviral vectors specific for BRG1/BRM. Quantified data were normalized for GAPDH and the relative inhibition (% Inh) resulting from SWI/SNF depletion is shown below each panel for two independent experiments.
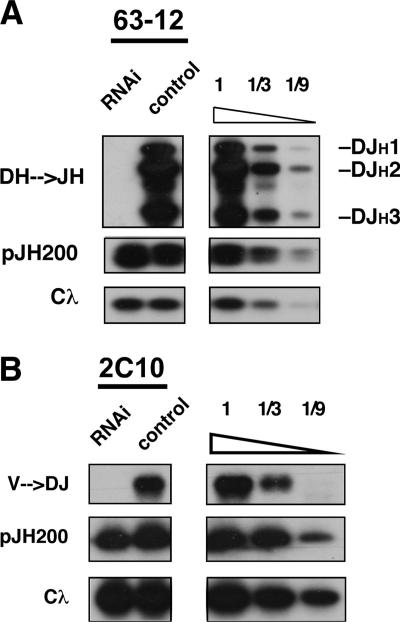
FIGURE 4.
Igh rearrangement requires SWI/SNF. A, DHJH coding joins in RAG2-deficient 63−12 cells cotransfected with a RAG2 expression vector and the episomal recombination substrate pJH200. Transfected cells were sorted on the basis of GFP expression and harvested for DNA. PCR assays were used to measure levels of DHJH rearrangements (DFL16 and all DSP elements), recombinase activity (pJH200), and total DNA (Cλ). B, The 2C10 pro-B cell line harboring a DFL16.1JH1 rearrangement was transfected and analyzed as in A except that PCR assays were used to detect VH→DHJH joins (VH558 family).
Retrovirus production
BRG1/BRM-specific short hairpin RNA sequences (13) were cloned into the pGIPZ lentiviral vector (OpenBiosystems) modified as described (14). For viral production, the EcoPack2 packaging cell line (Clontech) was transfected with retroviral plasmid using Fugene6 (Roche) according to manufacturer's protocols. Viral supernatant was collected 48h later and filtered before use.
Cell line transfections
The pro-B clones 63−12, 2B9, and 2C10 were transfected with a vector expressing short hairpin RNA for BRG1/BRM (RNA interference (RNAi)) or a control GFP-only vector as described (8). Live GFP+ cells were sorted 72 h post-transfection for molecular analysis (BD FACSAria). For recombination assays, cells were cotransfected with PGK-Rag2 and pJH200 plasmids in addition to the RNAi vector (8).
Primary pro-B cell cultures and retroviral infections
Bone marrow cells from RAG2-deficient mice (6−8 wk) were cultured in RPMI plus IL-7 as described (14). Cultures were infected with a high-titer viral supernatant corresponding to RNAi or GFP control vectors, as described for transfections, containing Polybrene and IL-7 in 12-well plates. Fresh viral supernatant was substituted after 24 h of incubation and cultures maintained an additional 24 h. Cells were harvested for RNA 48 h postinfection.
PCR assays
Total RNA was treated with RNase-free DNase I and reverse transcribed using random hexanucleotides or strand-specific primers. PCR assays to measure μ0, Iμ, upstream DHJH, VH, and antisense transcripts were described previously (4, 9, 10). Total input cDNA in each reaction was measured with an RT-PCR assay specific for Gapdh transcripts (8). To measure Igh rearrangement, DNA extracts were prepared from cells and analyzed by PCR for DHJH and VHDHJH joins as described (9).
Chromatin immunoprecipitation (ChIP) assay
IL-7-cultured pro-B cells were crosslinked with formaldehyde and chromatin was sheared using a Bioruptor sonicator (Diagenode) according to manufacturer's protocol. Chromatin from 106 cells (average size 200−700 bp) was immunoprecipitated with Abs specific for BRG1 (07−478; Upstate Biotechnology) or with rabbit IgG (AB-105-C; R&D Systems). Immunocomplexes were washed and crosslinks were reversed as described (6). DNA from bound and input samples was analyzed by PCR using primers I plus K (Fig. 1A), which are specific for the Jh4-Eμ region of Igh (10). Dilutions of each sample confirmed assay linearity (input, 0.08 and 0.02% of total collected chromatin; bound, 5 and 1.25%).
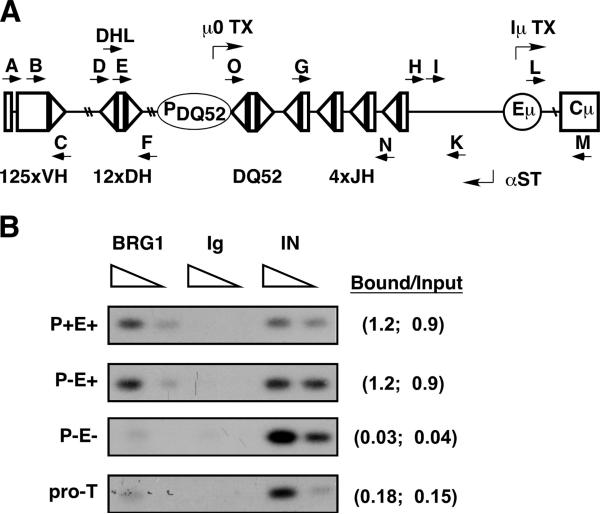
FIGURE 1.
BRG1 recruitment requires Eμ. A, Schematic of mouse Igh showing relative positions of gene segment clusters and cis-elements. Initiation regions for major sense (μ0, Iμ) and antisense transcripts (αST) within the DHJH cluster are shown. Lettered arrows indicate primers used in various PCR assays (μ0: G plus M; upstream DHJH transcripts: DHL plus M; Iμ: L plus M; DQ52 rearrangement: O plus N; VH558 rearrangement: B plus N; αST-JH:H (RT) and I plus K (PCR); αST-DH: D (RT) and E plus F (PCR); αST-VH:A (RT) and B plus C (PCR). B, BRG1-specific ChIP assays detect SWI/SNF association within the JH cluster. Chromatin was prepared from IL-7 cultures of bone marrow cells derived from the indicated mouse genotypes and analyzed by PCR using primers I plus K. Signals were quantified using the Fuji Phosphor-Imager MultiGauge program and presented on the right as bound to input ratios for two independent experiments (separated by semicolons).
Results and Discussion
Eμ-dependent recruitment of SWI/SNF to the JH cluster
Prior studies have shown that Eμ is essential for germline transcription and recombination throughout the DHJH cluster (9, 11). In what may be a related finding, accessible Igh loci in transformed pro-B cells are hyperacetylated at H3K9 and associate with BRG1, an essential ATPase subunit of many SWI/SNF complexes (12, 15). These findings suggest that Eμ may recruit SWI/SNF remodeling complexes to generate chromatin accessibility at some or all DHJH gene segments. As an initial test of this hypothesis, we analyzed whether Igh cis-acting elements are required for localized recruitment of BRG1 to the JH cluster (Fig. 1A). Accordingly, ChIP assays were performed on primary pro-B cells from Rag-deficient mice harboring targeted deletions of the DQ52 germline promoter (P−E+) or a dual deletion of the promoter and Eμ (P−E−) (9). Consistent with reported effects on DH→JH recombination, tissue-specific recruitment of BRG1 to JH segments was impaired at the P−E− allele when compared with either wild-type (P+E+) or promoter-deficient Igh loci (Fig. 1B). These findings indicate that Eμ is required for efficient recruitment and/or continued occupancy of BRG1-containing complexes at the JH cluster.
Classic germline Igh transcription requires SWI/SNF
To establish a causal relationship between Eμ-dependent recruitment of remodeling complexes and accessibility control at Igh, we used RNAi to simultaneously deplete BRG1 and BRM, the two known catalytic subunits of SWI/SNF. Depletion of SWI/SNF activity in a RAG2-deficient pro-B cell line (63−12) inhibits both μ0 and Iμ transcription (see Fig. 1A) through the germline DQ52-JH-Cμ region (Fig. 2A, left column). In contrast to DQ52, other germline DH elements are embedded in semirepressive chromatin but switch to a highly transcribed state upon rearrangement with a JH segment, presumably because they are brought into the Eμ regulatory domain (4). To determine whether upstream DH promoters also require SWI/SNF, we examined RAG-deficient cell lines harboring an upstream DHJH rearrangement on one allele (2B9: DSP2.2-JH1; 2C10: DFL16-JH1) and a VHDHJH rearrangement on the second Igh allele (2B9: VH7183DJH2; 2C10: VH7183DJH1) (Ref. 4) and our unpublished data). As shown in Fig. 2A (top two panels), transcription of the JHCμ region on both Igh alleles is inhibited upon depletion of BRG/BRM1. To specifically detect transcripts originating from the rearranged DH promoters, we used a PCR primer (DHL) that recognizes a region within all upstream DH segments but not in the JH-proximal DQ52 gene segment (Fig. 1A and Ref. 9). Consistent with this primer specificity, the new PCR assay fails to detect transcripts originating from the germline DQ52JH region in the 63−12 cell line (Fig. 2A, third panel from top). However, BRG1/BRM depletion again led to a significant decrease in the level of rearranged DHJH transcripts in 2B9 and 2C10 cells. These data suggest that Eμ-directed transcription from any of the DH promoters requires continual SWI/SNF activity.
In contrast to the DHJH cluster, transcription of unrearranged VH gene segments is independent of Eμ function (9, 11). As shown in Fig. 2A, depletion of SWI/SNF inhibits germline VH transcription at both the 5′ (VH558) and 3′ ends (VH7183) of this large cluster. Thus, SWI/SNF controls Eμ-dependent and -independent expression of germline gene segments positioned throughout the Igh locus.
To confirm and extend these findings from transformed cells, we knocked down BRG1/BRM in primary pro-B cell cultures using retroviral infection of the requisite RNAi vector (14). Bone marrow-derived pro-B cells from RAG2-deficient animals were expanded in medium containing IL-7, and cycling cells were infected with either RNAi or control retroviral vectors (>85% infection). As expected, RNAi-mediated depletion of Brg1 and Brm transcripts led to a dramatic reduction in the level of SWI/SNF-dependent c-myc mRNAs (Fig. 2B). Consistent with results from transformed cell lines, inhibition of SWI/SNF activity significantly impaired locus-wide expression of Igh as measured by steady-state levels of VH, DH, and JH germline transcripts. We conclude that SWI/SNF remodeling complexes are essential for establishing the accessible chromatin at Igh promoters that drive classical germline VH, DH, and JH transcription.
SWI/SNF is required for antisense Igh transcription
In addition to conventional germline transcripts that traverse Igh gene segments in a 5′ to 3′ (sense) orientation, recent studies showed that primary activation of Igh in pro-B cells is accompanied by locus-wide αST (3, 4). This type of intergenic αST has been implicated as a pioneering process in the opening of chromatin at several genetic loci that serves as a prelude to gene expression (16). In the JH region, αST and DH→JH recombination are both Eμ dependent, with αST initiating within or near this enhancer (10). The cis-elements and initiation sites for αST of VH segments remain unknown. These observations are consistent with the hypothesis that αST plays an important role in establishing chromatin accessibility at the DHJH cluster and perhaps throughout the entire Igh locus.
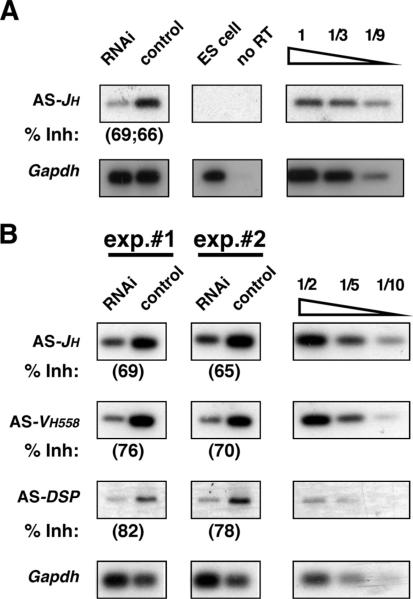
To determine whether αST within distinct Igh domains requires SWI/SNF, we primed total RNA from pro-B cells (SWI/SNF depleted or control) with strand-specific oligonucleotides to generate cDNAs corresponding to αSTs (10). Consistent with prior studies, PCR analysis of the resultant cDNAs showed that αST of Igh is specific to pro-B cells (Fig. 3A and data not shown). Importantly, αST throughout the VH, DH, and JH clusters is significantly impaired in primary pro-B cell cultures upon depletion of BRG1/BRM (Fig. 3B). These data indicate that pioneering αST throughout the entire Igh locus is dependent upon the nucleosome remodeling activity of SWI/SNF complexes.
FIGURE 3.
Pioneering αST is SWI/SNF-dependent. A, Pro-B cells (63−12) transfected with RNAi or control vectors were analyzed using a strand-specific RT-PCR assay for αST (AS) of the JH region. RNA from mouse embryonic stem (ES) cells or pro-B cell mRNA untreated with reverse transcriptase (no RT) are shown as negative controls. B, Strand-specific RT-PCR assays were performed as in A on RNA from primary pro-B cell cultures using primers for αST (AS) of the JH, VH, and DH regions indicated. An assay for Gapdh transcripts is shown as a control for RNA levels.
Recombinational accessibility of Igh requires SWI/SNF activity
The Igh locus is assembled in a stepwise fashion, with the opening of DHJH chromatin and its recombination preceding the final VH to DHJH joining events. Although the transcription of gene segments can often reflect their accessibility to V(D)J recombinase, these two properties do not always correlate (1). To directly test whether SWI/SNF is required for the recombination of DHJH clusters, we transiently transfected RAG2-null pro-B cells with a RAG2 expression construct, creating a readout for recombinase accessibility at Igh. As shown in Fig. 4A, germline Igh alleles were readily converted to DHJH rearrangements in pro-B cells that received both RAG2 and control vectors. In contrast, RAG2 expression failed to induce DHJH recombination in cells that received a RNAi vector targeting BRG1/BRM (the observed inhibition of rearrangement is >9-fold). However, this was not due to an absence of recombinase activity, because an episomal substrate (pJH200) was rearranged at similar levels in all samples (8).
For probing recombinase accessibility within the VH cluster, we performed similar experiments with RAG2-deficient pro-B cells that harbored a preformed DHJH junction on one allele (2C10; DFL16.1JH1) (4). SWI/SNF depletion also inhibited VH to DHJH recombination in this pro-B cell clone (Fig. 4B; consistent >3-fold inhibition in three independent experiments). Similar results were obtained with other clones harboring distinct DHJH joins (data not shown).
Together, our results demonstrate that SWI/SNF remodeling complexes have a global function in Igh locus activation rather than a localized effect restricted to the Eμ-proximal DQ52JH cluster. SWI/SNF remodeling activity is required for αST as well as conventional germline transcription, the latter of which initiates from well-defined Igh promoters. Our studies support the following stepwise model for accessibility control at Igh. First, Eμ recruits SWI/SNF to activate αST through inaccessible chromatin associated with upstream DHJH gene segments, resulting in a partial opening of this chromatin domain (3, 10). Second, Eμ interacts with promoters in partially accessible DHJH chromatin, activating conventional (sense) germline transcription in a SWI/SNF-dependent manner. Both sense and antisense transcription may be required to generate full chromatin accessibility at DHJH gene segments or to recruit recombinase via transcription-coupled histone modifications (2, 9, 11). Ultimately, SWI/SNF action at Igh must culminate in a recombinase-accessible configuration of chromatin throughout the locus that triggers complete VDJ exon assembly and B cell development. Important future studies will focus on the mechanisms of SWI/SNF recruitment to Eμ and to novel cis-elements that regulate the accessibility of the large VH cluster.
Footnotes
This work is supported by National Institutes of Health Grants AI 079732 and P30 CA68485 and by the Intramural Research Program of the National Institute of Aging (Baltimore, MD).
Abbreviations used in this paper: H3K9, histone 3 lysine 9; αST, antisense transcription; ChIP, chromatin immunoprecipitation; SWI/SNF, switching/sucrose nonfermenting.
Disclosures
The authors have no financial conflict of interest.
References
- 1.Cobb RM, Oestreich KJ, Osipovich OA, Oltz EM. Accessibility control of V(D)J recombination. Adv. Immunol. 2006;91:45–109. doi: 10.1016/S0065-2776(06)91002-5. [DOI] [PubMed] [Google Scholar]
- 2.Matthews AG, Kuo AJ, Ramon-Maiques S, Han S, Champagne KS, Ivanov D, Gallardo M, Carney D, Cheung P, Ciccone DN, et al. RAG2 PHD finger couples histone H3 lysine 4 trimethylation with V(D)J recombination. Nature. 2007;450:1106–1110. doi: 10.1038/nature06431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bolland DJ, Wood AL, Johnston CM, Bunting SF, Morgan G, Chakalova L, Fraser PJ, Corcoran AE. Antisense intergenic transcription in V(D)J recombination. Nat. Immunol. 2004;5:630–637. doi: 10.1038/ni1068. [DOI] [PubMed] [Google Scholar]
- 4.Chakraborty T, Chowdhury D, Keyes A, Jani A, Subrahmanyam R, Ivanova I, Sen R. Repeat organization and epigenetic regulation of the DH-Cμ domain of the immunoglobulin heavy-chain gene locus. Mol. Cell. 2007;27:842–850. doi: 10.1016/j.molcel.2007.07.010. [DOI] [PubMed] [Google Scholar]
- 5.McMurry MT, Krangel MS. A role for histone acetylation in the developmental regulation of VDJ recombination. Science. 2000;287:495–498. doi: 10.1126/science.287.5452.495. [DOI] [PubMed] [Google Scholar]
- 6.Oestreich KJ, Cobb RM, Pierce S, Chen J, Ferrier P, Oltz EM. Regulation of Tcrb gene assembly by a promoter/enhancer holocomplex. Immunity. 2006;24:381–391. doi: 10.1016/j.immuni.2006.02.009. [DOI] [PubMed] [Google Scholar]
- 7.Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293:1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- 8.Osipovich O, Milley Cobb R, Oestreich KJ, Pierce S, Ferrier P, Oltz EM. Essential function for SWI-SNF chromatin-remodeling complexes in the promoter-directed assembly of Tcrb genes. Nat. Immunol. 2007;8:809–816. doi: 10.1038/ni1481. [DOI] [PubMed] [Google Scholar]
- 9.Afshar R, Pierce S, Bolland DJ, Corcoran A, Oltz EM. Regulation of IgH gene assembly: role of the intronic enhancer and 5′DQ52 region in targeting DHJH recombination. J. Immunol. 2006;176:2439–2447. doi: 10.4049/jimmunol.176.4.2439. [DOI] [PubMed] [Google Scholar]
- 10.Bolland DJ, Wood AL, Afshar R, Featherstone K, Oltz EM, Corcoran AE. Antisense intergenic transcription precedes Igh D-to-J recombination and is controlled by the intronic enhancer Eμ. Mol. Cell. Biol. 2007;27:5523–5533. doi: 10.1128/MCB.02407-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Perlot T, Alt FW, Bassing CH, Suh H, Pinaud E. Elucidation of IgH intronic enhancer functions via germ-line deletion. Proc. Natl. Acad. Sci. USA. 2005;102:14362–14367. doi: 10.1073/pnas.0507090102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chowdhury D, Sen R. Stepwise activation of the immunoglobulin μ heavy chain gene locus. EMBO J. 2001;20:6394–6403. doi: 10.1093/emboj/20.22.6394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ramirez-Carrozzi VR, Nazarian AA, Li CC, Gore SL, Sridharan R, Imbalzano AN, Smale ST. Selective and antagonistic functions of SWI/SNF and Mi-2β nucleosome remodeling complexes during an inflammatory response. Genes Dev. 2006;20:282–296. doi: 10.1101/gad.1383206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Amin RH, Schlissel MS. Foxo1 directly regulates the transcription of recombination-activating genes during B cell development. Nat. Immunol. 2008;9:613–622. doi: 10.1038/ni.1612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Morshead KB, Ciccone DN, Taverna SD, Allis CD, Oettinger MA. Antigen receptor loci poised for V(D)J rearrangement are broadly associated with BRG1 and flanked by peaks of histone H3 dimethylated at lysine 4. Proc. Natl. Acad. Sci. USA. 2003;100:11577–11582. doi: 10.1073/pnas.1932643100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gribnau J, Diderich K, Pruzina S, Calzolari R, Fraser P. Intergenic transcription and developmental remodeling of chromatin subdomains in the human β globin locus. Mol. Cell. 2000;5:377–386. doi: 10.1016/s1097-2765(00)80432-3. [DOI] [PubMed] [Google Scholar]