Abstract
We conducted a SNP-based genome-wide association study (GWAS) focused on the high-risk subset of neuroblastoma1. As our previous unbiased GWAS showed strong association of common 6p22 SNP alleles with aggressive neuroblastoma2, we now restricted our analysis to 397 high-risk cases compared to 2,043 controls. We detected new significant association of six SNPs at 2q35 within the BARD1 gene locus (Pallelic = 2.35×10−9 − 2.25×10−8). Each SNP association was confirmed in a second series of 189 high-risk cases and 1,178 controls (Pallelic = 7.90×10−7 − 2.77×10−4). The two most significant SNPs (rs6435862, rs3768716) were also tested in two additional independent high-risk neuroblastoma case series, yielding combined allelic odds-ratios of 1.68 each (P = 8.65×10−18 and 2.74×10−16, respectively). Significant association was also found with known BARD1 nsSNPs. These data show that common variation in BARD1 contributes to the etiology of the aggressive and most clinically relevant subset of human neuroblastoma.
Neuroblastoma, one of the most common solid tumors in childhood, is characterized by diverse clinical phenotypes1. While a substantial proportion of patients may show a favorable outcome and may even have spontaneous regression of a localized, or even disseminated, tumor3,4, approximately 50% of cases show an aggressive clinical course with widespread metastases to bone and bone marrow at diagnosis1. These latter children have survival rates of less than 35% despite aggressive therapy with dose-intensive induction chemotherapy and surgery, followed by myeloablative therapy with stem cell rescue, local radiation therapy and biological response modification using retinoids and/or immunotherapy1,5,6.
Our recent GWAS demonstrated that three common SNP alleles within the predicted genes FLJ22536 and FLJ44180 at chromosome band 6p22 were associated with neuroblastoma2. No other region of the genome contained SNPs that reached genome-wide significance and survived our replication effort. Of particular interest was the finding that not only were the three 6p22 SNPs associated with the likelihood of developing neuroblastoma, but patients who carried the 6p22 risk alleles were more likely to develop the clinically aggressive form of the disease and suffer tumor recurrence (dbGaP: http://www.ncbi.nlm.nih.gov/projects/gap/cgi-bin/study.cgi?study_id=phs000124.v1.p1)2. These data support the hypothesis that the benign and malignant forms of neuroblastoma may represent distinct entities in terms of the genetic events that initiate tumorigenesis.
We therefore performed a second genome-wide analysis, this time limiting the cases to those patients with high-risk neuroblastoma as defined by the Children’s Oncology Group (COG)1. We identified 397 high-risk cases from the 1032 neuroblastoma patients included in the discovery set from our original study, and analyzed them against the same 2043 unaffected children from the discovery set control group. Quality control filters were applied to SNP genotype data as previously described2, resulting in a total of 462,866 autosomal SNPs available for analysis (see Supplementary Information). This analysis confirmed the three previously identified SNPs at chromosome band 6p22 (rs6939340, rs4712653, rs9295536) being significantly associated to high-risk neuroblastoma (Fig. 1; Pallelic = 3×10−11, 8×10−11, 6×10−10). These results in just 397 cases showed more highly significant P-values than those observed in the analysis of all 1032 cases with the identical control group2. In addition, we were able to identify a new association with common intronic SNPs at the BARD1 (BRCA1-associated RING domain-1) gene locus at chromosome band 2q35 (Fig. 1), with a total of six SNPs showing allelic test P-values less than 1×10−7 (Table 1, Fig. 2 and Supplementary Table 1). All BARD1 SNP P-values were minimally affected by correction for population stratification based on principal component analysis (Supplementary Table 1 and Supplementary Figure 1).
Figure 1. Summary of the neuroblastoma GWAS results in the discovery set restricted to the high-risk group of patients.
The y-axis represents the level of significance for each SNP (log transformed P-values) at the relative genomic position (Build 35) on each chromosome along the x-axis from short arm terminus (left) to long arm terminus (right). The horizontal line indicates our threshold for follow-up analysis (P-value < 1×10−7). The previously reported 6p22 signal is more significant in this discovery case series enriched for the subset of 397 high-risk patients compared to the 1032 unselected neuroblastoma cases previously studied,2 and a new chromosome 2q35 signal emerged.
Table 1.
Summary of discovery and replication results for six BARD1 SNPs with allelic P<10−7 in discovery case series.
| SNP (bp position) |
Case- control group |
Alleles * |
Case genotypes |
Control genotypes |
Case MAF |
Control MAF |
Pallelic | Ptrend | PCMH | ORallelic (95% CI) |
ORhet (95% CI) |
OR hom (95% CI) |
ORCMH (95% CI) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| rs6435862 | A | T/G | 149/177/71 | 1015/850/176 | 0.40 | 0.29 | 2.35×10−9 | 3.29×10−9 | 1.61 (1.38–1.88) | 1.42 1.12–1.80) | 2.75 (1.99–3.80) | ||
| (215,498,052) | B | 77/85/27 | 642/458/78 | 0.37 | 0.26 | 1.51×10−5 | 1.58×10−5 | 1.65 (1.31–2.07) | 1.55 (1.11–2.15) | 2.89 (1.75–4.75) | |||
| C | 28/39/19 | 397/329/48 | 0.45 | 0.27 | 2.16×10−6 | 1.41×−10−6 | 2.14 (1.55–2.95) | 1.68 (1.01–2.79) | 5.61 (2.91–10.80) | ||||
| D | 40/39/14 | 75/74/10 | 0.36 | 0.30 | 1.33×10−1 | 1.27×10−1 | 1.34 (0.91–1.97) | 0.99 (0.57–1.70) | 2.62 (1.07–6.44) | ||||
| Combined | 8.65×10−18 | 1.68 (1.49–1.90) | |||||||||||
| rs3768716 | A | A/G | 181/163/51 | 1192/738/112 | 0.34 | 0.24 | 3.19×10−9 | 4.44×10−9 | 1.64 (1.39–1.93) | 1.45 (1.15–1.83) | 2.99 (2.08–4.32) | ||
| (215,461,300) | B | 85/81/21 | 712/424/42 | 0.33 | 0.22 | 1.39×10−6 | 8.28×10−7 | 1.78 (1.41–2.26) | 1.60 (1.15–2.22) | 4.19 (2.37–7.41) | |||
| C | 35/42/7 | 452/302/28 | 0.33 | 0.23 | 2.56×10−3 | 1.55×10−3 | 1.68 (1.20–2.37) | 1.79 (1.12–2.88) | 3.23 (1.32–7.91) | ||||
| D | 35/31/28 | 88/62/9 | 0.46 | 0.25 | 1.05×10−6 | 6.31×10−6 | 2.56 (1.75–3.76) | 1.26 (0.70–2.25) | 7.82 (3.35–18.25) | ||||
| Combined | 2.74×10−16 | 1.68 (1.48–1.91) | |||||||||||
| rs17487792 | A | C/T | 181/163/50 | 1189/737/111 | 0.33 | 0.24 | 5.66×10−9 | 7.48×10−9 | 1.63 (1.38–1.92) | 1.45 (1.15–1.83) | 2.96 (2.05–4.28) | ||
| (215,469,006) | B | 85/82/21 | 715/421/42 | 0.33 | 0.21 | 7.90×10−7 | 4.74×10−7 | 1.80 (1.42–2.28) | 1.64 (1.18–2.27) | 4.21 (2.38–7.44) | |||
| Combined | 3.11×10−14 | 1.68 (1.47–1.92) | |||||||||||
| rs6712055 | A | T/C | 131/189/69 | 960/859/205 | 0.42 | 0.31 | 6.63×10−9 | 9.13×10−9 | 1.59 (1.36–1.86) | 1.61 (1.27–2.05) | 2.47 (1.77–3.42) | ||
| (215,492,410) | B | 71/88/29 | 574/510/92 | 0.39 | 0.30 | 2.77×10−4 | 2.25×10−4 | 1.52 (1.21–1.90) | 1.39 (0.99–1.95) | 2.55 (1.57–4.14) | |||
| Combined | 8.04×10−12 | 1.56 (1.37–1.78) | |||||||||||
| rs7587476 | A | C/T | 168/169/57 | 1106/795/129 | 0.36 | 0.26 | 9.66×10−9 | 1.03×10−8 | 1.60 (1.36–1.88) | 1.40 (1.11–1.76) | 2.91 (2.05–4.13) | ||
| (215,479,393) | B | 82/81/25 | 659/458/61 | 0.35 | 0.25 | 2.73×10−5 | 2.16×10−5 | 1.64 (1.30–2.06) | 1.42 (1.02–1.98) | 3.29 (1.96–5.53) | |||
| Combined | 1.22×10−12 | 1.61 (1.41–1.84) | |||||||||||
| rs6715570 | A | C/T | 141/180/76 | 948/899/196 | 0.42 | 0.32 | 2.25×10−8 | 2.39×10−8 | 1.56 (1.33–1.82) | 1.35 (1.06–1.71) | 2.61 (1.89–3.58) | ||
| (215,498,946) | B | 73/85/31 | 609/476/93 | 0.39 | 0.28 | 1.99×10−5 | 2.29×10−5 | 1.63 (1.30–2.04) | 1.49 (1.07–2.08) | 2.78 (1.73–4.46) | |||
| Combined | 2.19×10−12 | 1.58 (1.39–1.79) |
A total of 23 SNPs are located at the BARD1 locus on the HH550 SNP chip (Supplemental Table 1), and shown here are the six SNPs showing allelic P<10−7 in the discovery series, ranked according to their P-values. A. Discovery case series; B. CHOP replication case series; C. UK replication case series D. CCG replication case series. Breslow-Day tests for heterogeneity of the allelic odds-ratios among the case-control groups were not significant for all SNPs. All P-values for deviation from Hardy Weinberg equilibrium > 0.01 except SNP rs3768716 in control series C (P=0.01) and case series D (P=0.001).
Major/minor alleles. Pallelic: Fisher’s exact test on allele frequencies; Ptrend: Armitage-Cochran trend test on genotype frequencies; PCMH: Cochran-Mantel-Haenszel test for stratified tables on allele frequencies.
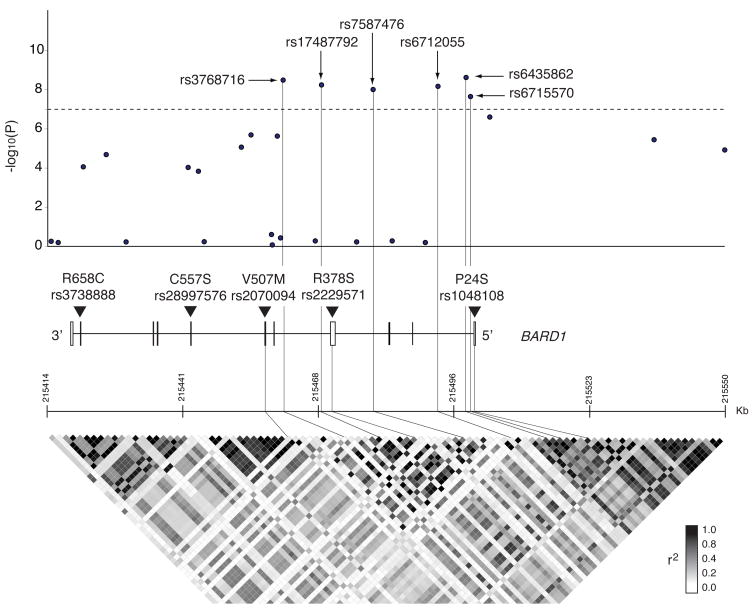
Figure 2. Regional plot of the BARD1 locus associated with high-risk neuroblastoma.
Single marker results (−log10P-values) for association testing of all SNPs at the BARD1 locus with six SNPs showing P-values <1×10−7 indicated in relation to the BARD1 genomic structure (exons indicated by vertical boxes) and in relation to the r2 linkage disequilibrium structure at all BARD1 SNPs with MAF>0.05 from HapMap CEU data, generated with the use of Haploview software. The position of five additional nsSNPs subsequently genotyped is also indicated (rs7585356 located 3′ downstream of BARD1 was also genotyped but is not shown).
Associations for all SNPs with P-values < 1×10−7 in the discovery set from both the 2q35 and 6p22 regions showed replication in a second independent group of 189 high-risk cases and 1178 unaffected controls genotyped genome-wide (Table 1 for BARD1 SNPs and Pallelic = 0.010, 0.008, 0.005 for chromosome 6p22 SNPs rs6939340, rs4712653, rs9295536, respectively). In contrast, association with the BARD1 SNPs was not significant in 575 low- and intermediate-risk patients from the discovery series (P-values > 0.05), and just three of them reached nominal significance in the total 756 low- and intermediate-risk patients from the discovery and replication sets combined, the smallest allelic P-value being 0.003 for rs3768716 (Supplementary Table 2). We then tested the two most significantly associated BARD1 SNPs (rs6435862 and rs3768716) for association with high-risk neuroblastoma, utilizing the cases within our previous two separate independent case series from the United Kingdom (UK) and the US-based legacy Children’s Cancer Group (CCG) with a high-risk phenotype.2 Both BARD1 SNPs were found significantly associated to high-risk neuroblastoma in the UK case series, whereas only rs3768716 showed a significant allelic P-value in the smaller CCG case series (Table 1). Combining data from all four groups of cases and controls showed allelic odds-ratios for these two SNPs of 1.68 each, with P-values of 9×10−18 for rs6435862 and of 3×10−16 for rs3768716 (Table 1).
The six genome-wide significant SNPs in the discovery phase are located in introns 1, 3 and 4 of BARD1 (Fig. 2). Pairwise linkage disequilibrium (LD) analysis showed that these SNPs are in relatively strong LD with each other (r2=0.47–0.96), but are not in LD with the non-significant SNPs elsewhere within BARD1 (Supplementary Fig. 2). Haplotype analysis on the six most significant SNPs in cases (586) and controls (3221) from CHOP discovery and replication sets combined revealed only seven haplotypes with frequency greater than 1% in both cases and controls, and only four with frequency greater than 2% (Supplementary Table 3). The most frequent haplotype was composed of all major alleles, and the second most frequent of all minor alleles, in both cases and controls. These were the only haplotypes with different frequencies in cases and controls (0.50 in cases and 0.60 in controls the first; 0.31 in cases and 0.21 in controls the second). These results are consistent with the high correlation observed among all SNPs in this region. Logistic regression analysis performed in the same cases and controls showed that a model including only rs3768716 explained the observed association as well as a model including any additional SNPs. It is therefore possible that a single variant in high LD with these SNPs explains all of the observed association, but regional resequencing will likely be required to address this definitively.
BARD1 contains several known coding variants (http://www.ncbi.nlm.nih.gov/SNP/snp_ref.cgi?locusId=580)7–10, and it was possible that one or more of these may be disease causal. We therefore selected the three common non-synonymous SNPs (minor allele frequency [MAF] > 0.10) for genotyping; rs1048108 (P24S) in exon 1, rs2229571 (R378S) in exon 4, and rs2070094 (V507M) in exon 6 (Fig. 2). According to HapMap CEU data, these three coding SNPs are in strong LD with the neuroblastoma-associated intronic SNPs rs6744811 (r2=0.86), rs7557557 (r2=0.76), and rs7584646 (r2=0.75). In addition, we also selected two rare non-synonymous SNPs for additional genotyping because of previously reported associations with breast cancer;7,9,10 rs28997576 (C557S) in exon 7 and rs3738888 (R658C) in exon 10 (Fig. 2). Finally, we also genotyped the common SNP rs7585356 located 3′ downstream of BARD1 because of it being in LD with a SNP rs16852600 (r2=0.89) that was of borderline significance, and because of it being located in a highly conserved region of the genome (SNPseek; http://snp.wustl.edu/cgi-bin/SNPseek/index.cgi). These six SNPs were assayed by TaqMan®-based genotyping in a case series consisting of 540 high-risk neuroblastoma cases from the discovery and replication set, and 1142 controls from the replication set.
Four of these SNPs showed statistically significant association with high-risk neuroblastoma and that these were in strong LD as there were only 6 observed haplotypes with an observed frequency > 2% (Pallelic ranging from 1×10−8 for rs2070094 to 3×10−6 for rs2229571; Table 2; Supplementary Table 4). However, none of the nsSNPs was more strongly associated with high-risk neuroblastoma than the GWAS intronic SNPs rs3768716 and rs6435862, which yielded allelic ORs of 1.82 (rs3768716: 95% CI: 1.56–2.13, P = 5×10−14; rs6435862: 95% CI: 1.57–2.11, P = 2×10−15), when analyzed in a comparable group of 586 cases (genome-wide discovery and replication cases combined) and 1178 controls (genome-wide replication controls). SNP rs28997576 was not polymorphic in our dataset, and association with rs3738888, which had a very low minor allele frequency, was not significant.
Table 2.
Results for BARD1 regulatory and coding SNPs genotyped in CHOP cases and controls
| SNP (bp position) | Role | AA change | Alleles* | Case genotypes | Control genotypes | Case MAF | Control MAF | Pallelic | Ptrend | ORallelic (95% CI) | ORhet (95% CI) | ORhom (95% CI) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| rs7585356 (215,417,812) | 3′ regulatory region | - | G/A | 333/188/22 | 527/423/98 | 0.21 | 0.30 | 7.95×10−7 | 1.02×10−6 | 0.65 (0.55–0.77) | 0.70 (0.57–0.88) | 0.36 (0.22–0.58) |
| rs3738888 (215,420,670) | Coding exon 10 | R658C | C/T | 547/12/1 | 1059/20/0 | 0.01 | 0.01 | 3.86×10−1 | 4.00×10−1 | 1.35 (0.68–2.69) | 1.16 (0.56–2.39) | 5.81 (0.24–142.75) |
| rs28997576 (215,442,684) | Coding exon 7 | C557S | G/C | 567/−/− | 1109/−/− | 0.0 | 0.0 | - | - | - | - | - |
| rs2070094 (215,457,761) | Coding exon 6 | V507M | G/A | 277/161/10 1 | 398/326/28 2 | 0.34 | 0.44 | 1.21×10−8 | 9.03×10−7 | 0.64 (0.55–0.75) | 0.71 (0.56–0.91) | 0.52 (0.39–0.68) |
| rs2229571 (215,470,970) | Coding exon 4 | R378S | C/G | 139/278/14 5 | 363/552/18 8 | 0.51 | 0.42 | 3.41×10−6 | 3.13×10−6 | 1.41 (1.22–1.63) | 1.32 (1.03–1.68) | 2.01 (1.50–2.70) |
| rs1048108 (215,499,730) | Coding exon 1 | P24S | C/T | 298/218/45 | 448/488/15 5 | 0.27 | 0.37 | 1.43×10−7 | 2.50×10−7 | 0.66 (0.56–0.77) | 0.67 (0.54–0.83) | 0.44 (0.30–0.63) |
Major/minor alleles. Pallelic: Fisher’s exact test on allele frequencies; Ptrend: Armitage-Cochran trend test on genotype frequencies
To assess the joint impact on disease risk of the genetic variants at chromosome regions 2q35 and 6p22, we estimated the two-locus genotype odds-ratios for the two most significant SNPs from each region (rs6435862 and rs9295536) in the CHOP discovery and replication sets combined (Table 3). Each locus independently contributed to disease risk with odds-ratios for carriers of 1 or 2 risk alleles relatively to non-carriers of 1.52 (95% CI: 1.03–2.24, P = 0.037) for rs6435862, of 1.75 (95% CI: 1.26–2.45, P = 0.001) for rs9295536, and of 2.99 (95% CI: 2.17–4.11, P = 3×10−13) for the two SNPs together. No significant interaction between the two SNPs was detected (P = 0.6).
Table 3.
Estimates of high-risk neuroblastoma risk (odds-ratios) by genotype (number of risk alleles) at BARD1 (rs6435862) and 6p22 (rs9295536)
| rs9295536 | |||||||
|---|---|---|---|---|---|---|---|
| 0 | 1 or 2 | ||||||
| Cases | Controls | OR (95% CI) | Cases | Controls | OR (95% CI) | ||
| rs6435862 | 0 | 49 | 541 | 1 | 177 | 1114 | 1.75 (1.26, 2.45) |
| 1 or 2 | 66 | 481 | 1.52 (1.03, 2.24) | 292 | 1080 | 2.99 (2.17, 4.11) | |
No significant interaction was detected between the two loci (P-value = 0.6).
Taken together, these data strongly support BARD1 as the second identified susceptibility locus to sporadically occurring neuroblastoma. Coupled with the recent discovery of highly penetrant mutations in the ALK oncogene as the major cause of hereditary neuroblastoma,11 the genetic basis of human neuroblastoma is now coming into focus. This report further confirms that common genomic variants are highly associated with this childhood cancer, and unlike our 6p22 discovery within putative transcripts of unknown function,2 these data clearly implicate a known and well characterized gene. BARD1 heterodimerizes with the familial breast cancer gene product BRCA1,12 and is considered to be essential for the latter genes known tumor suppressive function. Since pathogenic BRCA1 mutations interfere with heterodimerization to BARD1, it has been postulated that BARD1 may also function as a breast cancer susceptibility gene. However, while many studies have investigated the potential role of BARD1 in breast cancer susceptibility, there is currently no compelling evidence that DNA sequence alterations influence breast cancer pathogenesis and the locus has not emerged in breast cancer GWAS efforts.13–15 We now report the first definitive evidence that this gene is involved in cancer susceptibility. Ongoing studies are now focused on understanding the biological consequences of these SNP variations at the BARD1 locus in the developing sympathetic neuroblast, and how these influence malignant transformation.
It is also important to emphasize that this work clearly demonstrates the power of having robust phenotypic information available in GWAS approaches to human disease. Because the 6p22 association was enriched in the more aggressive subset of neuroblastoma cases, we were able to focus a new discovery case series on this clinically important group of patients. It is clear that at a somatic level, a single cancer histology may represent multiple different genomic subsets. Our data suggest that genetic initiating events may predispose not only to cancer, but to a particular subphenotype of the disease, and thus to patient outcome. This may have implications for both screening and identifying critical pathways for targeted therapeutics.
METHODS
Subjects and neuroblastoma risk grouping
For genome-wide genotyping, cases were defined as a child diagnosed with neuroblastoma or ganglioneuroblastoma and registered through the Children’s Oncology Group (COG). The blood samples from the neuroblastoma cases were identified through the COG Neuroblastoma bio-repository for specimen collection at the time of diagnosis. The majority of specimens were annotated with clinical and genomic information that included: age at diagnosis, site of origin, disease stage by the International Neuroblastoma Staging System, INPC International Neuroblastoma Pathology Classification, MYCN oncogene copy number, and DNA index, and assignment to the low-, intermediate- or high-risk subsets based on these data as described.1 The COG definition of high-risk disease included patients greater than 12 months of age at diagnosis with metastatic (Stage 4) disease or Stage 3 disease and unfavorable histologic features and any patient with a tumor showing MYCN amplification EXCEPT for a completely resected (Stage 1) tumor, All cases were determined to be of European ancestry as determined by self-report or parental report. Of the 1032 cases used in our previously reported discovery case series,2 397 were identified as high-risk (575 low- or intermediate-risk, 60 unknown) and were used for the discovery case series in this report. Of the 409 cases in our previously reported initial replication effort,2 189 were identified as high-risk (182 low- or intermediate-risk, 30 unknown) and used for the initial replication effort here.
Control subjects were recruited from the Philadelphia region through the Children’s Hospital of Philadelphia (CHOP) Health Care Network, including four primary care clinics and several group practices and outpatient practices that included well child visits. Eligibility criteria for control subjects were European ancestry as determined by self-report or parental report and no serious underlying medical disorder including cancer. We utilized the same 3414 control subjects as our prior report,2 with the same 2236 controls used in the discovery phase, and 1178 in the initial replication effort.
Two additional case series were used for the purpose of replication, both subsets from our prior GWAS replication efforts.2 Of the 252 Caucasian neuroblastoma cases from the United Kingdom (UK), 86 met criteria for high-risk disease and were compared to 782 controls. Likewise, of the 96 unrelated Caucasian neuroblastoma studied previously from the US-based Children’s Cancer Group (CCG), all were high-risk and compared to 159 controls.
Written informed consent was obtained for all participants and this study was approved by each participating center’s Institutional Review Board as well as the COG Scientific Council, COG Neuroblastoma Disease Committee and Cancer Therapy Evaluation Program (CTEP) at the NCI.
Genotyping and statistical methods
Since this analysis was a subset of a previously reported GWAS, we utilized identical methods for genome-wide genotyping, controlling for population substructure, and data analysis.2 Description of these methods are included in the Supplementary Information materials.
The initial replication effort was with samples genotyped genome-wide on the same platform as the discovery case series. The subsequent two replication efforts focused on the two most significantly associated SNPs with genotyping by conventional TaqMan®assays. Finally, we performed additional PCR-based allelic discrimination assays of all nsSNPs and potential regulatory SNPs in LD with significant SNPs from the discovery series. To maximize power, we utilized all of the high-risk cases available at CHOP (N=586) by combining the discovery and initial replication sets, but utilized only the replication controls (N=1178) to contain costs while providing sufficient statistical power.
The primary statistical tests for association in the discovery case series were carried out using the software package PLINK.16 We set 1×10−7 as the threshold for follow-up analysis, and single marker analyses for the genome-wide data were carried out using the χ2 test based on allele count differences and the Cochran-Armitage test for trends on genotype frequencies. Allelic odds ratios and the corresponding 95% confidence intervals were calculated for the association analyses. Association analyses were also performed after correction for substructure based on principal components analysis as implemented in Eigenstrat and as previously described2,17,18. Combined odds-ratios and p-values over the different datasets were calculated using the Cochran-Mantel-Haenszel test, and heterogeneity of the allelic odds-ratio was tested using the Breslow-Day test. Haplotypes were estimated from the unphased genotypes by means of the EM algorithm implemented in Haploview19. Interactions between variants at the 6p and 2q loci were tested by logistic regression analysis.
Supplementary Material
Acknowledgments
The authors acknowledge the Children’s Oncology Group for providing neuroblastoma specimens. Dr. Mario Capasso is a fellow of Fondazione Italiana per la Ricerca sul Cancro, FIRC, and Kristopher Bosse is a Howard Hughes Medical Institute Research Training Fellow. This work was supported in part by R01-CA124709 (JMM), the Giulio D’Angio Endowed Chair (JMM), the Alex’s Lemonade Stand Foundation (JMM), Andrew’s Army Foundation (JMM), the Rally Foundation (JMM), the Evan Dunbar Foundation (JMM), the Abramson Family Cancer Research Institute (JMM) and the Center for Applied Genomics (HH) at the Joseph Stokes Research Institute.
Footnotes
The authors declare no competing financial interests.
AUTHOR CONTRIBUTIONS
M.C., M.D, H.H. and J.M.M. designed the experiment and drafted the manuscript. C.H. E.F.A., Y.P.M., K.A.C., M.L., C.W. M.G., K.B., M.D. and C.K. performed sample collection and quality control, and the genome-wide genotyping. W.B.L. identified high-risk neuroblastoma cases and assisted in sample selection. M.C., M.D, J.P.B., S.J.D., J.J., J.T.G., S.G. and H.L. analyzed SNP data and performed association analyses. J.B., H.H., S.G. and H.L. performed the corrections for population stratification. S.A., R.H.S., R.C.S., C.M. and N.R. performed the BARD1 replication genotyping and analyses. C.H. and E.R. performed the genotyping of potential regulatory SNPs. All authors commented on or contributed to the manuscript.
References
- 1.Maris JM, Hogarty MD, Bagatell R, Cohn SL. Neuroblastoma. Lancet. 2007;369:2106–20. doi: 10.1016/S0140-6736(07)60983-0. [DOI] [PubMed] [Google Scholar]
- 2.Maris JM, et al. Chromosome 6p22 locus associated with clinically aggressive neuroblastoma. N Engl J Med. 2008;358:2585–93. doi: 10.1056/NEJMoa0708698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hero B, et al. Localized infant neuroblastomas often show spontaneous regression: results of the prospective trials NB95-S and NB97. J Clin Oncol. 2008;26:1504–10. doi: 10.1200/JCO.2007.12.3349. [DOI] [PubMed] [Google Scholar]
- 4.Nickerson HJ, et al. Favorable biology and outcome of stage IV-S neuroblastoma with supportive care or minimal therapy: a Children’s Cancer Group study. J Clin Oncol. 2000;18:477–86. doi: 10.1200/JCO.2000.18.3.477. [DOI] [PubMed] [Google Scholar]
- 5.Matthay KK, et al. Treatment of high-risk neuroblastoma with intensive chemotherapy, radiotherapy, autologous bone marrow transplantation, and 13-cis-retinoic acid. Children’s Cancer Group. New England Journal of Medicine. 1999;341:1165–73. doi: 10.1056/NEJM199910143411601. [DOI] [PubMed] [Google Scholar]
- 6.Berthold F, et al. Myeloablative megatherapy with autologous stem-cell rescue versus oral maintenance chemotherapy as consolidation treatment in patients with high-risk neuroblastoma: a randomised controlled trial. Lancet Oncol. 2005;6:649–58. doi: 10.1016/S1470-2045(05)70291-6. [DOI] [PubMed] [Google Scholar]
- 7.Stacey SN, et al. The BARD1 Cys557Ser variant and breast cancer risk in Iceland. PLoS Med. 2006;3:e217. doi: 10.1371/journal.pmed.0030217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Onay VU, et al. SNP-SNP interactions in breast cancer susceptibility. BMC Cancer. 2006;6:114. doi: 10.1186/1471-2407-6-114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Johnatty SE, et al. The BARD1 Cys557Ser polymorphism and breast cancer risk: an Australian case-control and family analysis. Breast Cancer Res Treat. 2008 doi: 10.1007/s10549-008-0045-y. [DOI] [PubMed] [Google Scholar]
- 10.Vahteristo P, et al. BARD1 variants Cys557Ser and Val507Met in breast cancer predisposition. Eur J Hum Genet. 2006;14:167–72. doi: 10.1038/sj.ejhg.5201542. [DOI] [PubMed] [Google Scholar]
- 11.Mosse YP, et al. Identification of ALK as a major familial neuroblastoma predisposition gene. Nature. 2008;455:930–5. doi: 10.1038/nature07261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wu LC, et al. Identification of a RING protein that can interact in vivo with the BRCA1 gene product. Nat Genet. 1996;14:430–40. doi: 10.1038/ng1296-430. [DOI] [PubMed] [Google Scholar]
- 13.Irminger-Finger I, Jefford CE. Is there more to BARD1 than BRCA1? Nat Rev Cancer. 2006;6:382–91. doi: 10.1038/nrc1878. [DOI] [PubMed] [Google Scholar]
- 14.Easton DF, et al. Genome-wide association study identifies novel breast cancer susceptibility loci. Nature. 2007 doi: 10.1038/nature05887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hunter DJ, et al. A genome-wide association study identifies alleles in FGFR2 associated with risk of sporadic postmenopausal breast cancer. Nat Genet. 2007 doi: 10.1038/ng2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Purcell S, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–75. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Price AL, et al. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006;38:904–9. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 18.Patterson N, Price AL, Reich D. Population Structure and Eigenanalysis. PLoS Genet. 2006;2:e190. doi: 10.1371/journal.pgen.0020190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–5. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.