Abstract
Sum frequency generation (SFG) vibrational spectroscopy has been demonstrated to be a powerful technique to study the molecular structures of surfaces and interfaces in different chemical environments. This review summarizes recent SFG studies on hybrid bilayer membranes and substrate-supported lipid monolayers and bilayers, the interaction between peptides/proteins and lipid monolayers/bilayers, and bilayer perturbation induced by peptides/proteins. To demonstrate the ability of SFG to determine the orientations of various secondary structures, studies on the interaction between different peptides/proteins (melittin, G proteins, almethicin, and tachyplesin I) and lipid bilayers are discussed. Molecular level details revealed by SFG in these studies show that SFG can provide a unique understanding on the interactions between a lipid monolayer/bilayer and peptides/proteins in real time, in situ and without any exogenous labeling.
Keywords: Sum frequency generation, SFG, cell membrane, antimicrobial peptide, membrane protein, lipid bilayer, melittin, G Proteins, alamethicin, tachyplesin I
1. Introduction
Cell membranes play a crucial role in many biological functions of cells. They govern all interactions between cells and their environments, such as the exchange of information and ions/molecules between the inside and outside of the cells. The cell membrane can be quite complex and dynamic, with a thickness of several nanometers. A cell membrane mainly consists of a lipid bilayer, which is an arrangement of lipids in which the acyl chains of each layer interact through hydrophobic interactions and the hydrophilic head groups face the inside and outside of the cell. A variety of peptides and proteins are also embedded inside or associated with the cell membranes, helping to fulfill various cellular functions (Katsaras et al., 2001; Mateo et al., 2006; Yeagle, 2005). It is important to study the structures and kinetics of membrane embedded/associated peptides/proteins in order to understand their functions. Results from these types of studies can help in the rational design of molecules that can more effectively mediate or interfere with various cellular events in the desired manner.
Different experimental tools have been used to study peptides/proteins in the membrane environment, and excellent results have been obtained. For example, surface plasmon resonance spectroscopy measurements allow for the determination of the peptide/protein coverage on membranes (Beseničar et al., 2006; Devanathan et al., 2006; Salamon et al., 1997). Neutron reflection studies have also permitted the determination of the amount of adsorbed peptides/proteins (Fragneto-Cusani, 2001; Haas et al., 2007; Kučerka et al., 2007). Atomic force microscopy techniques, on the other hand, are very useful in studying the packing and surface ordering of membrane-bound proteins (Alessandrini et al., 2005; Brasseur et al., 2008; Engel et al., 2008; Johnston, 2007; Laflamme et al., 2008; Richter et al., 2006). Ellipsometry, in contrast, can be utilized to follow proteins’ adsorption kinetics onto membranes and determine the adsorbed protein film thickness (Faiss et al., 2008). Excellent review articles have been published that summarize the applications of these techniques (and others) to the study of the interactions between peptides/proteins and lipid membranes (Cabiaux, 2004; Lee, 2005; McIntosh et al., 2006). Even though some molecular level information can be acquired by probing the interactions between cell membranes and proteins/peptides using these analytical tools, further details regarding such molecular interactions need to be elucidated.
Several spectroscopic techniques that can probe detailed structural information have been applied to study membrane-related peptides and proteins, leading to in-depth understanding of such molecular interactions. For example, Attenuated Total Reflectance Fourier Transform Infrared (ATR-FTIR) measurements can provide vibrational spectra (or fingerprints) of membrane proteins and peptides. However, the technique may suffer from a lack of intrinsic surface sensitivity and therefore the signals from the molecules in the cell membrane environment might be confused with those in the bulk environment, e.g., in the bulk solution. Also, ATR-FTIR only gives one vibrational measurable of the tilt angle <cos2θ> for orientation determination of a membrane protein or peptide (Tamm et al., 1997). Nuclear magnetic resonance (NMR) spectroscopy is another powerful spectroscopic technique that is widely used to characterize peptide/protein-lipid interactions (Bader et al., 2003; Bechinger, 1999; Bechinger et al., 2004; Lindblom et al., 2006; Naito et al., 2007; Wang, 2008). Solution and solid-state NMR have been successfully used to determine the site-specific secondary structures and dynamics of membrane-bound proteins (Andronesi et al., 2005; Dürr et al., 2007; Fernández et al., 2001). Both solution and solid-state NMR can also be used to study several aspects of the lipid-protein interactions, such as looking at the proteins’ effects on the lipid dynamics and determining which parts of the protein are interacting with the headgroups or tail region of the lipids (Lee et al., 2008; Dvinskikh et al., 2007). However, due to NMR’s low sensitivity, large amounts of protein/peptides are required (milligram quantities), sometimes isotope-labeled proteins are needed, and experimental time can be very long. Difficulties can also be encountered in sample preparation (i.e. high concentrations of sample can cause membrane proteins to aggregate). The different model membranes used in NMR also make it complicated to create an asymmetric lipid bilayer (i.e. controlling the composition of each leaflet).
Over the last two decades, sum frequency generation (SFG) vibrational spectroscopy has been developed into a very powerful and highly versatile spectroscopic tool for surface and interfacial studies (Anglin, 2008; Bain et al., 1995; Baldelli et al., 2008; Belkin et al., 2005; Chen, 2007; Dreesen et al., 2004; Fourkas et al., 2007; Gautam et al., 2002; Gopalakrishnan et al., 2006; Gracias et al., 1999; Holman et al., 2004; Hopkins et al., 2005; Iwahashi et al., 2008; Kim et al., 2008; Koffas et al., 2004; Li et al., 2008; Ma et al., 2006; Moore et al., 2008; Opdahl et al., 2004; Richmond, 2002; Rupprechter et al., 2008; Shen et al., 2006; Shultz et al., 2002; Stiopkin et al., 2008; Voges et al., 2007; Yang et al.,2002; Ye et al., 2008). SFG is a vibrational technique that is intrinsically surface-sensitive, requires small amounts of sample, and with which the experiments can be done in situ and in real-time (Allen et al., 2000; Bain, 1995; Belkin, et al., 2005; Buck et al., 2001; Chen, 2007; Chen and Chen, 2006; Chen and Clarke, 2005; Chen and Shen, 2002; Eisenthal, 1992; Gopalakrishnan et al., 2006; Gracias et al., 1999; Hopkins, et al., 2005; Koffas et al., 2004; Lambert et al., 2005; Miranda et al., 1999; Moore et al., 2008; Opdahl et al., 2004; Perry, et al., 2006; Richmond, 2001; Richmond, 2002; Shen, 1989; Shen et al., 2006; Shultz et al., 2000; Tadjeddine et al., 1996; Wang and Gan, 2005; Williams et al., 2002; Zhuang et al., 1996). SFG permits the identification of interfacial molecular species (or chemical groups), and also provides information about the interfacial structure, such as the orientation and the orientation distribution of functional groups on the surface. SFG has been applied to study the structure and orientation of biomolecules, such as lipids (Anderson et al., 2006; Anglin et al., 2007; Anglin et al., 2008; Chen and Wang, 2007a; Doyle et al., 2004; Harper et al., 2007; Kim and Kim, 2001; Levy et al., 2007; Liu et al., 2004a; Liu et al., 2004b; Liu et al., 2005a; Liu et al., 2005b; Liu et al., 2007; Lobau et al., 1999; Ma et al., 2006; Ma et al., 2007; Nickolov et al., 2006; Ohe et al., 2004; Petralli-Mallow et al., 1999; Sovago et al., 2007; Watry et al., 2003; White et al., 2006), and peptides/proteins (including membrane-related proteins/peptides) (Chen and Chen; 2006; Chen and Clarke, 2005; Chen and Wang, 2005; Clarke et al., 2005; Dreesen et al., 2004; Evans-Nguyen et al., 2006; Humbert et al. 2006; Kim and Cremer, 2001; Kim and Gurau, 2002; Kim and Gurau, 2003; Kim and Somorjai, 2003; Knoesen et al., 2004; Mermut et al., 2006; Rocha-Mendoza et al., 2007; Sartenaer et al., 2007; Wang and Buck, 2003; Wang and Chen, 2005; Wang and Chen, 2006; Wang and Clarke, 2003; Wang and Even, 2003; York et al., 2008).
Planar substrate-supported lipid bilayers have been widely used as a model to mimic cell membranes. Their suitability for biological studies has been extensively tested. These lipid bilayers are readily prepared by directly depositing lipid monolayers or bilayers onto the substrates such as glass, mica, quartz, and silicon surfaces using Langmuir-Blodgett method or vesicle fusion method (Kalb et al., 1992; Steinem et al., 2000; Tamm et al., 1985; Tamm, 1988; Tamm et al., 1997; Thompson et al., 1988). In addition, many research groups are also employing different strategies to improve the properties of supported lipid bilayers (i.e. using ultrathin polymer to support lipid bilayers) (Sackmann, 1996; Tanaka et al., 2005; Zhao et al., 2003). Previous studies have indicated that planar substrate-supported lipid bilayers can offer several advantages over other model membranes, such as free-standing lipid bilayers, solvent-free lipid bilayers, or phospholipid vesicles. Planar substrate-supported lipid bilayers are unilamellar and geometrically well defined. They can maintain excellent mechanical stability without losing their fluid nature. These advantages of substrate-supported bilayers make it possible to carry out experiments that probe structural and dynamic properties of membranes and protein-lipid interactions, using the surface analytical techniques mentioned above (Castellana et al., 2006; Kalb et al., 1992; McConnell et al., 1986; Sackmann, 1996; Tamm et al., 1985; Tamm, 1988; Tamm et al., 1997; Tanaka et al., 2005).
In this paper, we will first present a brief introduction of the theoretical background needed to understand SFG, and then summarize recent studies on the interactions between lipid membranes (monolayers and bilayers, focusing especially on substrate-supported lipid bilayers) and biomolecules monitored by SFG in real time and in situ. The names of lipids mentioned in this paper and their respective abbreviations are listed in Table 1.
Table 1.
The full names of lipids and their abbreviations that appear in this paper.
| Abbr. | Full Name |
|---|---|
| POPC | 1-Palmitoyl-2-Oleoyl-sn-Glycero-3-Phosphocholine |
| POPG | 1-Palmitoyl-2-Oleoyl-sn-Glycero-3-[Phospho-rac-(1-glycerol)] (Sodium Salt) |
| DMPC | 1,2-Dimyristoyl-sn-Glycero-3-Phosphocholine |
| d-DMPC | 1,2-Dimyristoyl-D54-sn-Glycero-3-Phosphocholine-1,1,2,2-D4-N,N,N-trimethyl-D9 |
| DMPC-d54 | 1,2-Dimyristoyl-D54-sn-Glycero-3-Phosphocholine |
| DPPC | 1,2-Dipalmitoyl-sn-Glycero-3-Phosphocholine |
| d-DPPC | 1,2-Dipalmitoyl-D62-sn-Glycero-3-Phosphocholine-1,1,2,2-D4-N,N,N-trimethyl-D9 |
| DPPG | 1,2-Dipalmitoyl-sn-Glycero-3-[Phospho-rac-(1-glycerol)] (Sodium Salt) |
| d-DPPG | 1,2-Dipalmitoyl-D62-sn-Glycero-3-[Phospho-rac-(1-glycerol)] (Sodium Salt) |
| DSPC | 1,2-Distearoyl-sn-Glycero-3-Phosphocholine |
| d-DSPC or DSPC-d83 | 1,2-Distearoyl-D70-sn-Glycero-3-Phosphocholine-1,1,2,2-D4-N,N,N-trimethyl-D9 |
| DSPC-d70 | 1,2-Distearoyl-D70-sn-Glycero-3-Phosphocholine |
| DSPG | 1,2-Distearoyl-sn-Glycero-3-[Phospho-rac-(1-glycerol)] (Sodium Salt) |
2. Theoretical Background of SFG
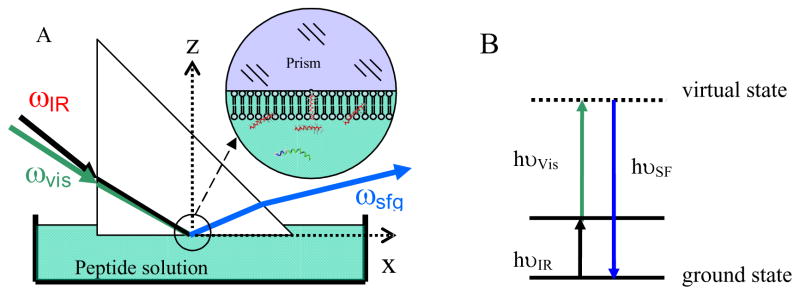
Although it has only been about twenty years since the first SFG spectra were recorded by Shen (Guyotsionnest et al., 1987; Hunt et al., 1987; Zhu et al., 1987), SFG is now being used by a growing number of research groups for a variety of applications, including polymer and biological interface studies. Many excellent review papers have summarized details about SFG theory and research (Allen et al., 2000; Bain, 1995; Belkin, et al., 2005; Buck et al., 2001; Chen, 2007; Chen and Chen, 2006; Chen and Clarke, 2005; Chen and Shen, 2002; Eisenthal, 1992; Gopalakrishnan et al., 2006; Gracias et al., 1999; Hopkins, et al., 2005; Koffas et al., 2004; Lambert et al., 2005; Miranda et al., 1999; Moore et al., 2008; Opdahl et al., 2004; Perry, et al., 2006; Richmond, 2001; Richmond, 2002; Shen, 1989; Shen et al., 2006; Shultz et al., 2000; Tadjeddine et al., 1996; Wang and Gan, 2005; Williams et al., 2002; Zhuang et al., 1996), which will not be repeated here. In a typical SFG experiment, two pulsed laser beams, one with a fixed frequency in the visible frequency range (ωVis) and one with a tunable frequency in the infrared frequency range (ωIR), are overlapped spatially and temporally on the sample (Fig. 1A). The SFG signal is generated at the sum frequency of the two input beams by the nonlinear process, ωSF = ωVis + ωIR. Therefore, the SFG process can be simply viewed as a combination of infrared (IR) absorbance and Raman scattering, as shown in the energy diagram in Fig. 1B. The intensity of the sum frequency signal is proportional to the square of the vibration’s second-order nonlinear susceptibility (Bain, 1995; Buck et al., 2001; Chen and Chen, 2006; Chen and Clarke, 2005; Chen and Shen, 2002; Eisenthal, 1992; Lambert et al., 2005; Miranda and Shen, 1999; Richmond, 2001; Shen, 1984; Shen, 1989; Tadjeddine et al., 1996; Wang and Gan, 2005; Williams et al., 2002; Zhuang et al., 1999). Different components of can be probed using different polarization combinations of the input and output laser beams. From such measurements, orientation information of surface molecules and functional groups can be deduced (Hirose and Akamatsu, 1992a; Hirose and Akamatsu, 1992b; Hirose and Yamatoto, 1993; Gautam et al., 2001; Kim and Somorjai, 2003; Shen, 1984; Wang and Chen, 2001). More details about SFG theory and data analysis can be found in the appendix.
Fig. 1.
SFG experimental geometry and energy diagram. (A) Total reflection geometry employed for the experiments described in sections 3.3 and 3.4. A bilayer is immersed in a small reservoir with an approximate volume of 1.6–2.0 mL; (B) Simplified energy level diagram of vibrational sum frequency generation process.
The selection rules of SFG make the technique surface sensitive. As we stated above, the SFG signal intensity is proportional to the square of the vibration’s second-order nonlinear susceptibility vanishes when a material has inversion symmetry under the dipole approximation (Bain, 1995; Buck et al., 2001; Chen and Chen, 2006; Chen and Clarke, 2005; Chen and Shen, 2002; Eisenthal, 1992; Hirose and Akamatsu, 1992a; Hirose and Akamatsu, 1992b; Hirose and Yamatoto, 1993; Gautam et al., 2001; Lambert et al., 2005; Miranda et al., 1999; Richmond, 2001; Shen, 1984, Shen, 1989; Tadjeddine et al., 1996; Wang and Chen, 2001; Wang and Gan, 2005; Williams et al., 2002; Zhuang et al., 1999). The majority of bulk materials exhibits inversion symmetry, thus they do not generate SFG signals. However, the presence of an interface causes a break in the symmetry, and therefore molecules on surfaces or at interfaces can generate SFG signal. Therefore SFG is an intrinsically surface/interface specific technique, different from linear vibrational spectroscopic techniques such as ATR-FTIR and Raman scattering.
3. Recent SFG studies on membrane-related peptides/proteins
In this section, we will summarize recent SFG studies on membrane-related peptides and proteins. Such studies have demonstrated that SFG can provide a unique understanding of the interactions between a lipid monolayer/bilayer and peptides/proteins without any exogenous labeling, in real time and in situ.
3.1. SFG studies on lipid monolayers, substrate-supported lipid bilayers and hybrid bilayer membranes
As mentioned above, it has been extensively shown that substrate-supported lipid bilayers are valid models for cell membranes (Castellana et al., 2006; Kalb et al., 1992; McConnell et al., 1986; Sackmann, 1996; Steinem et al., 2000; Tamm et al., 1985; Tamm, 1988; Tamm et al., 1997; Tanaka et al., 2005; Thompson et al., 1988; Zhao et al., 2003). SFG has been applied to investigate the structures of these lipid bilayer systems (Anglin et al., 2007; Anglin et al., 2008; Chen and Wang, 2007a; Doyle et al., 2004; Levy et al., 2007; Liu et al., 2004a; Liu et al., 2004b; Liu et al., 2005a; Liu et al., 2005b; Liu et al., 2007; Petralli-Mallow et al., 1999), with a recent review article (Chen and Chen, 2006) summarizing these SFG studies. Here, we do not attempt to reiterate the details, but instead will give some brief discussions on how some fundamental questions regarding the interactions between peptide/proteins and lipid bilayers can be answered using SFG. For example, knowledge of the conformations of lipids in the bilayer, both before and after their interactions with antimicrobial peptides, aids in understanding the peptide’s mode of interaction. These modes of interaction can be generally separated into the carpet, barrel-stave and toroidal pore modes (Yang et al., 2001; Chen and Chen, 2006). Theoretically, each of these peptide/protein–lipid bilayer interaction modes should induce different conformational changes on the lipid bilayers, resulting in distinct SFG spectral changes. Therefore, using these kinds of SFG lipid studies, details regarding the interactions of antimicrobial peptides, and other peptides/proteins, with lipid systems may be elucidated.
Before we go further to discuss the interactions between peptides/proteins and lipid bilayers, knowledge regarding the membrane model system, the lipid bilayer, is needed. When studying such interactions with SFG, substrate-supported lipid bilayers are used as the typical model for a cell membrane. Optical techniques using fluorescence labeling methods have previously been used to study the physical properties of such bilayers. Although these techniques offer important insights (e.g., about the flip-flop of lipids in a membrane, the transition temperature of a lipid system, and lipid domain segregation) (Bagatolli, 2006; Heberle et al., 2005; Lee, 2005), the incorporation of a fluorescent label on the lipids, or of a fluorescent probe inside the bilayer, may affect the behavior of the system and generate some artifacts in the experiments. An advantage of using SFG to study these types of supported bilayers is that it is not necessary to introduce any bulky labels. The only form of labeling required to study the bilayers is the introduction of isotope labeling. In order to monitor each leaflet of a lipid bilayer individually, it is necessary to have one leaflet deuterated, in order to break the inversion symmetry of the bilayer and obtain SFG signal.
One important aspect of the behavior of lipids in cell membranes is their ability to translocate both laterally and across the membrane (flip-flop). With this in mind, Conboy and his colleagues have investigated some important physical properties of substrate-supported lipid bilayers, such as the kinetics of the flip-flop and the transition temperatures of various lipid bilayers using SFG (Liu et al., 2004a; Liu et al., 2004b; Liu et al., 2005a; Liu et al., 2005b). More recently, they investigated the asymmetric distribution of domains in lipid bilayers by carrying out SFG spectroscopic measurements of symmetric C-H stretching modes of the fatty acid methyl groups (Liu et al., 2007). In this research, they correlated the intensity of the C-H symmetric stretch of the fatty acid methyl groups with the symmetry of the lipid bilayer, with the stronger intensity obtained when the bilayer became more asymmetric. The breakage in symmetry was caused by the dislocation of the gel and liquid-crystalline phase domains at the transition temperature. They have also done SFG studies on the lateral pressure dependence of the phospholipid transmembrane diffusion rate in supported lipid bilayers (Anglin et al., 2008). Their results indicated that the kinetics of lipid flip-flop in these membranes show a strong lateral pressure dependence. Based on these data, they successfully determined the activation area for phospholipid flip-flop (Anglin et al., 2008).
Briggman and coworkers have employed hybrid bilayer membranes (HBM) as an alternative for supported lipid bilayers in their research (Anderson et al., 2004; Anderson et al., 2006; Anderson et al., 2007; Levy et al., 2007; Petralli-Mallow, 1999). The HBM system studied has one single lipid layer deposited on top of a hydrophobic self-assembled monolayer (SAM). Briggman’s group has extensively studied the properties of HBMs and found that the transition temperature of the lipid layer depends greatly on the packing density and the crystallinity of the SAM layer underneath it (Anderson et al., 2007). They also carried out SFG studies on the effect of cholesterol on phospholipids (Levy et al., 2007). Their research has shown that HBMs could be used as excellent model membranes for biological studies on peripherally-bound proteins. However, it is challenging to use HBMs as cell membrane models to study lipid flip-flop and some transmembrane proteins that extensively interact with the inner-leaflet.
Many research groups also model cell membranes by using lipid monolayers. The fundamental properties such as transition temperatures and effects of cholesterol on monolayers have been investigated using SFG (Bonn, et al., 2004; Ohe and Sasaki, 2007; Ohe and Goto, 2007; Roke, et al., 2003).
3.2. SFG studies on the interactions between peptides/proteins and lipid monolayers/bilayers: C-H stretching frequency region
Early SFG studies on the interactions between proteins/peptides and lipid monolayers, HBMs and supported lipid bilayers focused on the C-H stretching frequency region. Table 2 contains a list of some of those studies. One of the very first SFG studies on the interaction between proteins and lipid monolayers was done by Cremer and his colleagues (Kim and Gurau, 2003). They investigated the orientation of gramicidin A in a DMPC monolayer using the C-H SFG stretching signals generated from the side chains of gramicidin A. Their results indicated that the orientation of gramicidin A was concentration dependent (Kim and Gurau, 2003).
Table 2.
SFG studies on the interactions between proteins/peptides and lipid monolayers, HBMs and supported lipid bilayers
| Peptides or Proteins | Membrane | Studied region | Reference |
|---|---|---|---|
| Gramicidin A | DMPC-d54 monolayer | CH | Kim and Gurau, 2003 |
| Polymyxin B | DPPC monolayer DPPG monolayer |
CH | Ohe et al., 2004 |
| FGF-1 | DSPG hybrid bilayer membranes | CH | Doyle et al., 2004 |
| Antimicrobial Oligomers | DPPG/d-DPPG bilayer | CH, CD, Amide I | Chen and Tang, 2006 |
| Antimicrobial peptides | DPPG/d-DPPG bilayer | CH, CD, amide I | Chen and Chen, 2006 |
| Gramicidin A | DSPC/DSPC-d70 DSPC-d83/DSPC-d83 |
CH, CD | Anglin et al., 2007 |
| Melittin | dDPPG/dDPPG dDPPG/DPPG |
CH, CD | Chen and Wang, 2007a |
| Melittin | DPPG/DPPG | Amide I | Chen and Wang, 2007b |
| Heterotrimeric | POPC/POPC bilayer | Amide I | Chen and Boughton, 2007 |
| G Protein βγ Subunit | POPC/POPG bilayer |
Conboy and coworkers studied the effect of gramicidin A on the flip-flop of DSPC lipids in a substrate-supported phospholipid bilayer (Anglin et al., 2007). Using SFG and ATR-FTIR, they showed that gramicidin A induced rapid flip-flop of the DSPC lipids. In this study, the C-H stretching signals of the lipid’s hydrophobic chain were studied by SFG and the amide signal of gramicidin A was observed by ATR-FTIR. The detection of amide signal from the peptide using ATR-FTIR indicated that gramidicin A was bound to the bilayer. SFG signal of the lipid C-H stretches could be observed because an asymmetric bilayer (with one leaflet deuterated) was used in the study. The time-dependence of the SFG C-H signals can be used to monitor the flip-flop rate, because as the system becomes more symmetric (from interchange between outer- and inner-leaflet lipids), the signal should decrease. By studying the time-dependent C-H signals both with and without gramicidin A bound, they showed that gramicidin A induced a faster flip-flop rate than when no gramicidin A was added to the bilayer.
Neivandt and coworkers examined a protein in the fibroblast growth factor (FGF) family, FGF 1, and its interaction with an HBM (Doyle et al., 2004). Using SFG, they showed that this protein caused deformation of the DSPG lipid layer even at very low concentrations. They also found that this process was reversible to a certain extent when the protein was washed off the HBM. Itoh and his colleagues investigated the interactions between antibiotic polymyxin B (PMB) and monolayers composed of DPPG, as well as DPPC (Ohe et al., 2004). By monitoring the SFG C-H stretching signals of the lipids’ side chains and the O-H stretching signals of water molecules, along with the measurements of the pressure-area isotherms, they found that this antibiotic peptide bound to the negatively charged lipids (DPPG), but not to the neutral lipids (DPPC). They also showed that the binding of PMB to the lipid monolayer affected the phase transition of the lipid monolayer
Because SFG can provide structural information such as functional group composition, orientation, and ordering at a surface or interface with a submonolayer sensitivity, it has also been applied to determine the average minimal inhibitory concentration (MIC) of antimicrobial peptides (or analogue oligomers) in membranes. Chen et al. used SFG to investigate the molecular interactions between a small antimicrobial oligomer and a single substrate-supported lipid bilayer with a hydrogenated leaflet (DPPG) and a deuterated leaflet (d-DPPG) (Chen and Tang, 2006). It was observed from the C-H and C-D stretching signals that the distal leaflet was disrupted at a very low peptide/lipid ratio, while the proximal leaflet remained intact below a threshold concentration very close to the MIC (0.8 mg/mL) value. The orientation of this oligomer was deduced by SFG C-H signals and the results indicated that the small antimicrobial oligomer acted as a “molecular knife” by disrupting primarily the outer leaflet of the bilayer at lower concentrations and further inserting into the entire bilayer at a certain critical (MIC) concentration.
3.3. SFG studies on the interaction between peptides/proteins and lipid bilayers: amide I frequency region
3.3.1. Detection of SFG amide I signal
The SFG results presented in the previous section are obtained from SFG signals acquired mainly in the C-H stretching frequency region, which are comprised of signals from the lipids and protein side chains. The feasibility of detecting SFG amide I signals from interfacial proteins/peptides was demonstrated by Chen and his coworkers in 2003 (Wang and Even, 2003). Because the water bending mode does not contribute noticeable SFG signals, it is not necessary to perform a background subtraction to obtain the SFG amide I signals from interfacial proteins and peptides; instead, they can be detected directly (Clarke and Wang, 2005; Wang and Clarke, 2005; Wang and Even, 2003). In addition, proteins in the bulk solution do not generate SFG signals, and thus SFG can selectively probe interfacial proteins/peptides. Moreover, SFG is able to provide more measurements than ATR-FTIR in studying the orientation of interfacial proteins/peptides. In SFG studies, our group has also adopted a near total reflection experimental geometry (Fig. 1A) that enables us to obtain very strong SFG amide I signals of interfacial proteins, which makes the data analysis easier and more accurate (Wang and Even, 2003).
SFG amide I signals of proteins can be affected by the surface coverage, orientation, and secondary structures of the adsorbed proteins (Clarke and Wang, 2005; Wang and Clarke, 2005; Wang and Even, 2003). The amide I mode contains predominately the peptide C=O stretching bands. These C=O groups are held together by hydrogen bonds within the secondary structures and the frequency of the C=O stretch depends heavily on its hydrogen-bonded environment. The peak center of the amide I band therefore depends on the secondary structure adopted by the peptide/protein. The Chen group has successfully demonstrated that SFG amide I signals can be used to distinguish α-helical and β-sheet structures of peptides and proteins on polymers and lipid bilayers (Chen and Boughton, 2007; Chen and Chen, 2006; Chen and Wang, 2005; Chen and Wang, 2007b; Clarke and Wang, 2005; Wang and Chen, 2005; Wang and Clarke, 2005; Wang and Even, 2003; Wang and Lee, 2008; Wang and Paszti, 2007). Recently, Somorjai’s group developed a new optical parametric amplifier (OPA) in the SFG spectrometer that can also create a tunable infrared light between 1500 cm−1 and 2000 cm−1; this IR beam can be used to study the amide I signals of interfacial peptides (York et al., 2008).
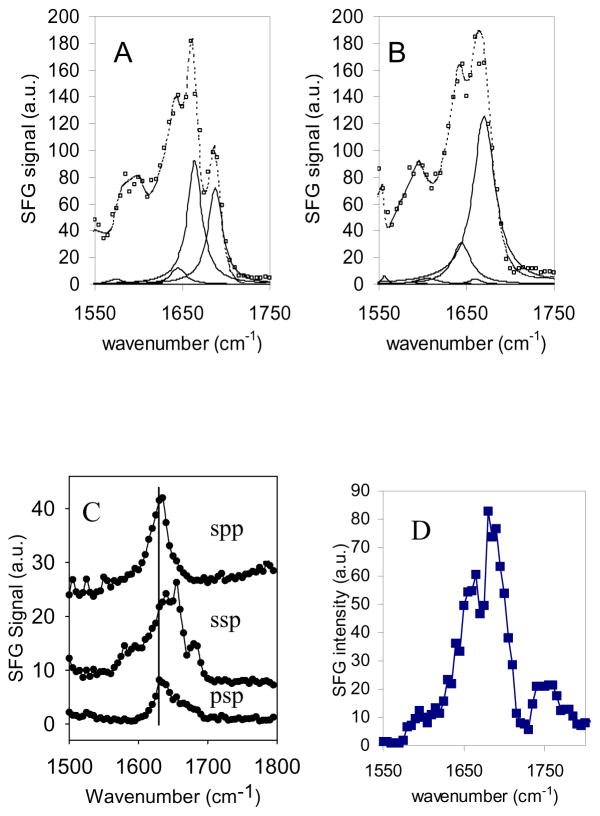
Based on previous FTIR and Raman studies, SFG amide signals of different secondary structures can be assigned. Using antimicrobial peptides that are known to adopt an α-helical structure in lipid bilayers, Chen et al. were able to detect SFG amide I signals of α-helical structures centered at around 1650 cm−1 (Chen and Wang, 2007b). Using model β-sheet peptide tachyplesin I, Chen et al. also showed that SFG amide I signals from a β-sheet structure has characteristic peaks at 1635 and 1685 cm−1, corresponding to the B2 mode and B1/B3 modes, respectively (Chen and Chen, 2006). Recent results in our laboratory on alamethicin indicate that SFG amide I signals of 310 helical structures have peaks at around 1635 cm−1 and 1670 cm−1. These results are indeed in good agreement with previous FTIR and Raman studies. More details regarding the interfacial structures of different secondary domains of proteins/peptides can be obtained by the careful data analysis of SFG amide I signals collected using different polarization combinations, which will be presented below.
3.3.2. SFG studies on α-helical proteins/peptides in lipid bilayers
(a). Data analysis for orientation determination using amide I band
As mentioned above, it has been shown experimentally that amide I peak centers are different for each type of secondary structure. Therefore, analyses of amide I signals provide insights into the protein backbone secondary structures. This section will focus on how to deduce the orientation of α-helical structures. Conventional polarized ATR-FTIR has been widely applied to study the orientation of α-helical structures of membrane peptides/proteins (Tamm et al 1997). In ATR-FTIR studies, the tilt angle of the α-helices can be calculated from the order parameter (Sθ), which is defined as:
| (1) |
with θ being the tilt angle between the helix’s principal axis and the surface normal. The bracket denotes the time and ensemble average. Theoretically, <cos2θ> can be determined from the measured intensity ratio in ATR-FTIR using p- and s-polarized IR light (Tamm et al., 1997). If we assume θ to have the simplest delta distribution, the orientation of the helix can be determined from this intensity ratio. The orientation in reality can be more complicated and such a simple distribution may not be adequate enough to describe the orientation of the peptide/protein. For example, a helix may adopt two different orientations or have a broad orientation distribution. To characterize such a more complicated orientation distribution, more measured parameters are needed. For example, when the parameter Sθ approaches zero, <cos2θ> is equal to 1/3, and thus there is always ambiguity in whether all helices have the same tilt angle around 54.7°, or a completely random orientation, or other orientations/distributions in between. In order to determine which of the three cases is correct, more measurements using different spectroscopic methods would be required (Chen and Wang, 2007b).
It has been shown that it is possible to deduce the orientation of functional groups, such as methyl, methylene, phenyl, and carbonyl, by measuring different tensor elements using SFG spectra collected with different polarization combinations (Briggman et al., 2001; Gautam et al., 2000; Hirose and Akamatsu, 1992a; Hirose and Akamatsu, 1992b; Hirose and Yamamoto, 1993; Oh-e et al., 2002; Tyrode et al., 2005). By analyzing the SFG amide I signals using group theory and projection operators, the Chen group has been able to investigate interfacial protein structures and deduce the orientation of α-helical peptides (Lee et al., 2006; Wang and Lee, 2008; Wang and Paszti, 2007). They showed that both the amide I A mode and amide I E1 mode of an α-helix can contribute to SFG signals (Wang and Lee, 2008). Using the near total internal reflection geometry (Wang and Even, 2003), ssp and ppp amide I signals can be collected. These signals are mainly due to contributions from the χyyz and χzzz susceptibility components, respectively. The dependence of χyyz and χzzz susceptibility components on the molecular hyperpolarizability is described by the following equations:
For the A mode:
| (2) |
| (3) |
For the E1 mode:
| (4) |
| (5) |
where βaca and βccc are the molecular hyperpolarizability elements. The hyperpolarizability elements of an α-helix can be obtained from the product of the components of the Raman polarizability and IR transition dipole moment. Chen et al. (Chen and Wang, 2007b) deduced the relations among different hyperpolarizability tensor elements to be r = βaac/βccc≈0.54 and βaca≈0.32βccc (Lee et al.,1998a; Lee et al., 1998b; Marsh et al., 2000; Rintoul et al., 2000). Ns is the number density of ideal α-helix units composed of 18 amino acid residues. Due to the limited resolution of many SFG spectrometers (~5 cm−1 or more), the A mode and E1 mode cannot be readily resolved in the frequency domain, and therefore, the total susceptibility is often assumed to be the sum of the susceptibilities from these two modes (Chen and Wang, 2007b):
| (6) |
| (7) |
As Eqs. (2–5) indicate, only two measurables related to the orientation angle are independent: <cosθ> and <cos3θ>. Using different polarization combinations of the input and output laser beams, <cosθ> and <cos3θ> can be deduced. If all α-helical structures on the surface/interface adopt the same orientation, <cosθ> and <cos3θ> can be replaced by cosθ and cos3θ. Subsequently, the relationship between an SFG measurable and the orientation angle of the α-helix can be depicted. However, for most cases, it may not be correct to assume that all the α-helical structures on a surface/interface have the same orientation. For example, one protein may have two α-helical segments pointing two different directions. The following section discusses the orientation analysis of α-helical structures in some of these complex cases.
(b). Example: α-helical melittin in membrane
Recently, Chen et al. used melittin as a model peptide to study the orientation of α-helical peptides in substrate-supported lipid DPPG bilayers (Chen and Wang, 2007b). Chen et al. found that the SFG measurements were not compatible to those of a δ-distribution or a Gaussian distribution. These two distributions were therefore not adequate to describe the melittin orientation distribution inside a DPPG bilayer and the orientation distribution had to be more complex. Chen et al. assumed two δ-distributions as the orientation function, meaning that melittin was assumed to adopt two distinct orientations in the lipid bilayer. A fraction (N) of melittin may orient with an angle of θ1, and a fraction (1-N) of melittin can orient with another angle of θ2. By combined ATR-FTIR and SFG studies, all of these parameters, θ1, θ2 and N, were successfully deduced. The obtained results from SFG and ATR-FTIR experiments indicated that melittin helices existed in two main populations in the lipid bilayer. About three-fourths of melittin molecules oriented parallel to the bilayer surface with a slight tilt, while the rest oriented more or less parallel to the surface normal, as shown in Fig. 2A and Fig. 2B.
Fig. 2.
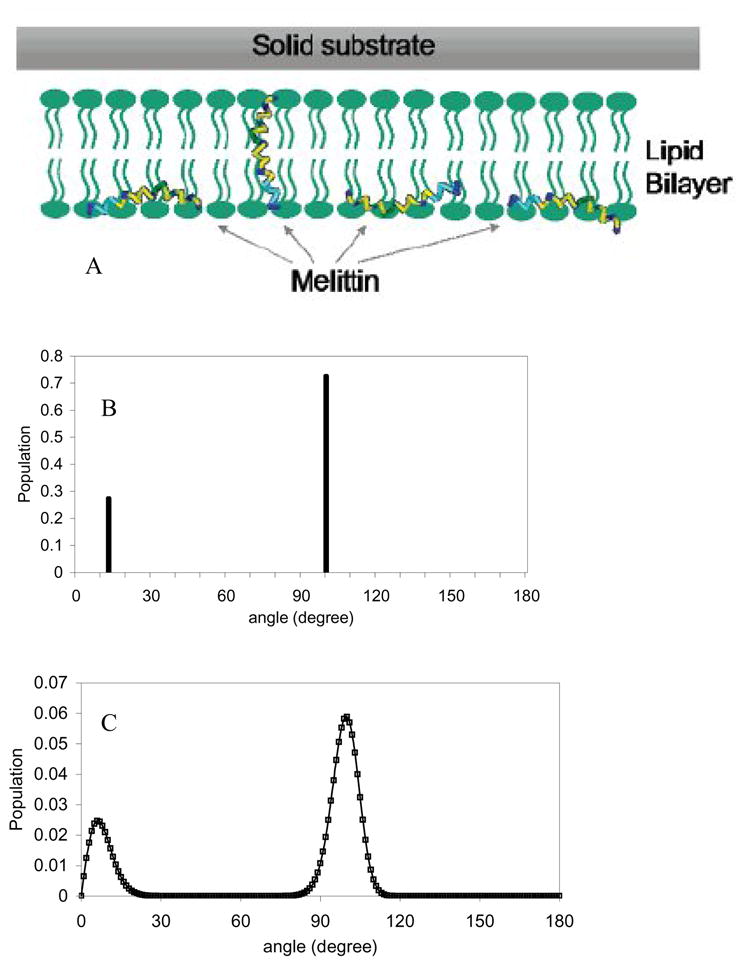
A. Schematic of melittin’s two orientations in the lipid bilayer. B. Orientation distribution function derived based on a dual δ–distribution. C. Orientation distribution function derived based on the maximum entropy theory. Reproduced with permission from J. Am. Chem. Soc. 2007, 129, 1420–1427. Copyright 2007, American Chemical Society.
In addition, Chen et al. also introduced the maximum entropy function to deduce the orientation distribution of melittin in a single lipid bilayer based on the ATR-FTIR and SFG measurements. Such a treatment does not have any assumptions regarding the orientation distribution function, e.g., assuming two δ-distributions. The deduced orientation distribution using the maximum entropy function was very similar to that obtained from the two δ-distributions, as shown in Fig. 2C. This research demonstrated the power of combining ATR-FTIR measurements, SFG data and the maximum entropy function analysis for deducing complicated orientations of membrane-bound peptides. These kinds of orientation determination results can be correlated to different modes of action of peptides’ interactions with bilayers, and ultimately lead to an understanding of the mechanism of antimicrobial activity, for example. Such an analysis can also be applied to study interfacial proteins with two (or even three) α-helical segments that adopt different orientations. A similar method has also been used to study the orientation distribution of two α-helical coiled-coils of fibrinogen at the polystyrene/fibrinogen solution (phosphate buffered solution with a total ionic strength of 0.14 M and a pH value of 7.4) interface (Wang and Lee, 2008).
(c). Example: α-helical structure in G protein in lipid bilayer
Heterotrimeric guanine nucleotide-binding proteins (G proteins) are a family of peripheral membrane proteins that transduce extracellular signals (e.g. hormones and neurotransmitters), as sensed by G protein-coupled-receptors (GPCRs), to intracellular effector systems (e.g. ion channels and cell transcription machinery) (Cabrera-Vera et al., 2003; Neves et al., 2002). Each G protein is comprised of Gα, Gβ, and Gγ subunits, with Gβ and Gγ forming a tightly associated dimer. In the resting state, a G protein exists in the Gαβθ form. Upon GPCR activation, Gαβθ releases the GDP originally bound to the Gα subunit and the binding of GTP allows dissociation of Gαβθ into Gα·GTP and Gβγ (Gaudet et al., 1999; Lodowski et al., 2003). Gα and Gβγ can then associate with their own effectors and trigger downstream signaling cascades. The cycle returns to the resting state when Gα hydrolyzes GTP back to GDP (Neves et al., 2002; Oldham et al., 2006; Koch, 2004; Pitcher et al., 1992).
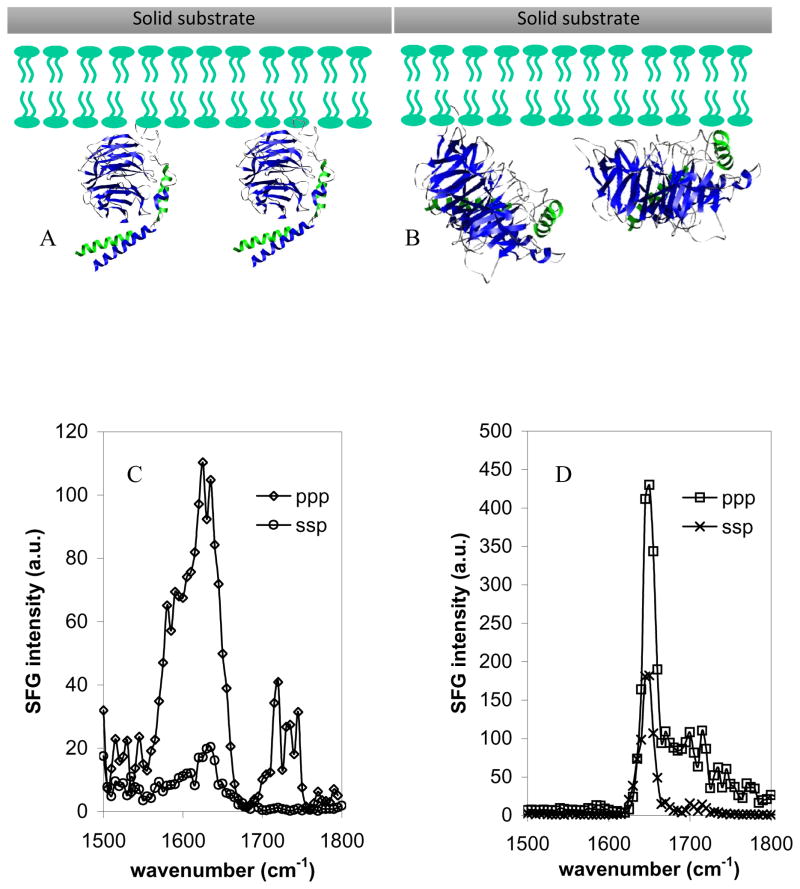
Recently, Chen et al. (Chen and Boughton, 2007) investigated how the Gβ1γ2 subunit binds to and orients on a substrate-supported lipid bilayer using SFG. Both wild-type Gβ1γ2 subunits, which contain a geranylgeranyl anchor group, and only the soluble domain of the Gβ1γ2 subunits were used in this research. SFG spectra were collected from both types of G protein subunits in a hydrated POPG/POPG bilayer. These two Gβ1γ2 subunits showed very different SFG spectral properties (Figs. 3C and 3D). Even at relatively high concentrations (125 μg/mL), soluble Gβ1γ2 generated weaker signals than geranylgeranylated Gβ1γ2, with a peak centered at 1630 cm−1, indicative of β-sheet secondary structure. The presence of the geranylgeranyl anchor group resulted in a significant enhancement of SFG amide I signal that was dominated by a peak at around 1650 cm−1, which is characteristic of contributions from an α-helical structure. Chen et al. (Chen and Boughton, 2007) suggested that without the geranylgeranyl group, Gβ1γ2 adsorbs onto the surface with the β-propeller domain facing the membrane surface and the helical domains orienting more or less parallel to the surface (Fig. 3B, Chen and Boughton, 2007; Wang and Even, 2005). On the other hand, for the wild-type Gβ1γ2 subunit, the β-propeller more or less orients perpendicular to the bilayer surface and the helical domains are ordered and no longer parallel to the surface. This orientation allows the β-propeller to keep its native semi-centrosymmetry, resulting in very weak β-sheet signal and causing the amide I signal to be dominated by a peak at 1650 cm−1 (originating from the ordered helical domains), as shown in Fig. 3A. From the measured SFG ppp and ssp intensity ratio, Chen et al. deduced the orientation angle of the wild-type Gβ1γ2 to be −35° from a reference orientation in which the β-sheets within the β-propeller are parallel to the membrane surface (Tesmer et al., 2005; Wall et al., 1995). This research demonstrates that SFG can be used to deduce the membrane protein orientation in situ by studying the orientation of α-helical components of a protein.
Fig. 3.
Schematics of Gβ1γ2 adsorbed onto a POPG/POPG bilayer deduced from SFG spectra: A. wild-type Gβ1γ2 with geranylgeranyl group, B. soluble Gβ1γ2 without geranylgeranyl group. SFG amide I spectra of the interfacial Gβ1γ2 adsorbed onto a POPG/POPG bilayer. C. 250 μg of the soluble form of Gβ1γ2 was first injected into the subphase (~ 2 mL) of the bilayer. Only relatively weak signals indicative of β-sheet were observed. D. After the spectra in panel C were collected, 50 μg wild type Gβ1γ2 was injected into the subphase and stronger SFG signals indicated of α-helix structure were observed. Reproduced with permission from J. Am. Chem. Soc., 2007, 129, 12658 -12659. Copyright 2007, American Chemical Society.
3.3.3. SFG studies on 310 helical peptides in membrane bilayer: alamethicin
Alamethicin is a 20-residue hydrophobic antibiotic peptide that can form voltage-gated ion channels in membranes. It has been used frequently as a model for larger channel proteins (Tamm et al., 1997). In addition to the regular amino acids, the peptide contains eight aminoisobutyric acid units. Its crystal structure contains an α-helical domain and a 310 helical domain (Fox et al., 1982).
An extensive amount of research has been performed to examine the alamethicin action mechanism on membranes (Cafiso, 1994; Duclohier et al., 2001; Hall et al., 1984; Leitgeb et al., 2007; Mathew et al., 1983a; Mathew et al., 1983b; Nagaraj et al., 1981; Sansom, 1993a; Sansom, 1993b; Woolley et al., 1992). It is currently believed that alamethicin interacts with cell membranes through the barrel stave mode (Duclohier, 2004; Fox et al., 1982; Laver, 1994; Mathew et al., 1983; Sansom, 1993a; Sansom, 1993b) with the resulting conducting pores in the membrane formed by parallel bundles of three to twelve helical alamethicin monomers surrounding a central, water-filled pore. However, further details on the structural origin of some important properties of alamethicin channels in the membrane, such as the strong dependence of their conductivity on the transmembrane potential (Stella, et al., 2007), are not known. In addition, contradicting orientations of alamethicin in the membrane in the absence of voltage have been reported. Alamethicin has been suggested to adopt a transmembrane orientation (Bak et al., 2001; Kessel et al., 2000; Marsh et al., 2007a; North et al., 1995), lie on the membrane surface (Banerjee et al., 1985; Ionov et al., 2000; Mottamal et al., 2006), or both (depending on the experimental conditions) (Chen and Lee, 2002; Huang et al., 1991). A continuous distribution of orientations has also been proposed (Spaar et al., 2004). Because of this lack of agreement in the literature, our lab has also been studying the molecular interactions between alamethicin and lipid bilayers in situ and in real time using SFG.
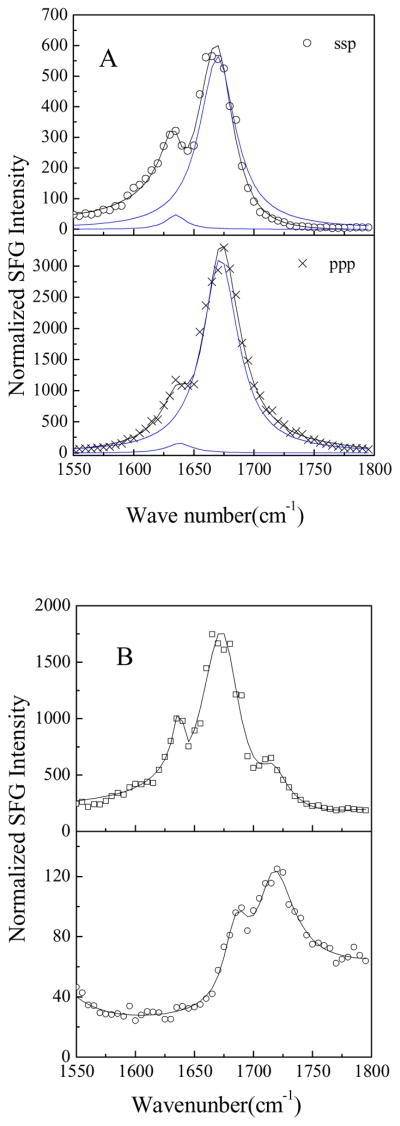
SFG ssp and ppp spectra of alamethicin in a d-DMPC/DMPC bilayer are shown in Fig. 4A. The spectra were collected after 37.5 μg of alamethicin (dissolved in water) was injected into the water subphase (~1.6 mL) of d-DMPC/DMPC bilayer for 71 min at pH 6.7. The SFG spectra were dominated by two peaks at 1635 and 1670 cm−1. Peak assignments in the literature indicate that the 1635 cm−1 peak is due to the 310-helix, while the peak at 1670 cm−1 has contributions by both the 310 helix and the α-helix (Haris et al., 1988; Haris et al., 2004; Vogel, 1987, Kennedy et al. 1991, Dwivedi et al., 1984). The orientation analysis method for a 310-helix in the membrane, using the SFG amide I band, has been developed in our lab, similar to the method for an α-helix discussed in section 3.3.2 (a). This method is now being applied to deduce the orientation of alamethicin in a d-DMPC/DMPC bilayer using the spectra shown in Fig. 4A. Detailed results will be reported in the future.
Fig. 4.
A. The SFG spectra of the alamethicin after 37.5μg alamethicin was injected into the subphase (~ 1.6 mL) of d-DMPC/DMPC bilayer for 71 min at pH 6.7. B. The ppp SFG spectra of the alamethicin after 37.5 μg alamethicin was injected into the subphase (~ 1.6 mL) of the bilayers for 60–80 min at pH 6.7. Top: POPC/POPC; Bottom d-DPPC/DPPC.
It has been shown that the membrane lipid chain length affects the interaction between alamethicin and the cell membrane (Archer et al, 1991; Barranger-Mathys, 1994; Hall et al., 1984; Marsh et al., 2007a; Marsh et al., 2007b; Marsh, 2008). We observed markedly different SFG signal intensities from alamethicin in lipid bilayers with lipids of different chain lengths (Table 3). The length of the lipid chain is one of the factors that determine the physical phase in which the bilayer will exist at room temperature: longer chain lipids tend to exist in the gel phase, whereas shorter chain lipids tend to exist in the fluid phase. Fig. 4B shows the ppp SFG spectra collected after 37.5 μg of alamethicin was injected into the subphase (~ 1.6 mL) of the bilayer for 60–80 min at pH 6.7. In the fluid phase bilayers (Fig. 4A and top spectrum in Fig. 4B), the strong SFG signal of alamethicin is dominated by two peaks at 1635 and 1670 cm−1 contributed by 310-helices and α-helices, respectively. When alamethicin was present in gel-phase lipid bilayers (the lower spectrum in Fig. 4B), only two weak peaks at 1685 and 1720 cm−1 were observed. The peak at 1685 cm−1 was assigned to antiparallel β-sheet or aggregated strands of peptides (Tamm et al., 1997, see more details in the next section), and the 1720 cm−1 signal originates from the bilayer. These results suggest that alamethicin is able to insert into fluid-phase membranes, but that it lies or aggregates on the gel-phase membrane surface and does not have significant insertion into these membranes. These results align well with the results obtained using other analytical tools in the literature (Banerjee et al., 1985; Ionov et al., 2000; Mottamal et al., 2006).
Table 3.
The interaction between alamethicin and different lipid bilayers.
| Inner layer | Outer layer | Transition temperature of outer layer lipid (°C)* | Phase of outer layer lipid at experimental condition | SFG signal |
|---|---|---|---|---|
| POPC | POPC | −2 | Fluid | Very strong |
| POPC | POPG | −2 | Fluid | Very strong |
| d-DMPC | d-DMPC | 23 | Fluid | Very strong |
| d-DMPC | DMPC | 23 | Fluid | Very strong |
| d-DPPC | DPPC | 41 | Gel | Weak |
| d-DPPG | DPPG | 41 | Gel | Weak |
| d-DSPC | DSPC | 55 | Gel | Weak |
We also studied the pH effects on alamethicin in the lipid bilayer using a POPC/POPC bilayer as a model. According to the ppp SFG spectra (not shown) of alamethicin in POPC/POPC bilayer at pH 6.7 and pH 11.9 (the pH was adjusted by adding K3PO4), after adjusting the pH to 11.9, the SFG amide I intensity from alamethicin increased by ~10 fold and the peak at ~1720 cm−1 disappeared. These results suggested that higher pH values, which may affect membrane potential, can induce significant insertion or incorporation of alamethicin into membranes. These results may provide molecular information on the voltage dependence of the alamethicin channels formed in membranes.
3.3.4. SFG studies on β-sheet peptides in membrane bilayers: tachyplesin I
After having discussed the α-helices and 310 helices studied by SFG, we will now present SFG results on another important secondary structure, the β-sheet. Tachyplesin I has been widely used as a model antimicrobial peptide (AMP) that has a β-sheet structure (Katsu et al., 1993; Matsuzaki et al., 1993; Miziguchi et al., 2003; Nakamura et al., 1988). The antiparallel β-sheet structure is held rigidly together by two intra-strand disulfide bonds. The role of the disulfide bonds has been the focus of several research articles. Decreased antimicrobial activity has been reported when the four cysteines are protected by acetamidomethyl groups (Matsuzaki et al., 1993), but another study, which used linear analogs with cysteines mutated to other residues, indicated that the rigidly held disulfide-bonded β-sheet structure may not be absolutely required for antimicrobial activity (Miziguchi et al., 2003). The exact mode of action for tachyplesin I is therefore still controversial.
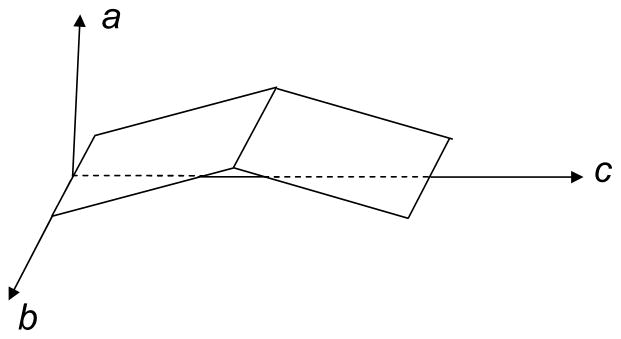
Recently, the Chen group discovered that tachyplesin I induced bilayer structural changes that also exhibited concentration dependence (Chen and Chen, 2006). The SFG results indicated that tachyplesin I was very active in disrupting DPPG bilayers (more details in section 3.4). To carry out the SFG data analysis on β-sheet structures, Chen et al. first studied tachyplesin I at a polystyrene/peptide solution interface (Chen and Wang, 2005; Wang and Chen, 2005). Due to the D2 symmetry of β-sheets (Fig. 5), which differs from that of helical structures, one angle θ is not enough to determine β-sheet interfacial orientation because the twist angle φ cannot be random. For D2 symmetry, SFG measured parameters (nonlinear susceptibility tensor components) can be relate to molecular properties of β-sheets (hyperpolarizability tensor components) through orientation parameters (Wang and Chen, 2005):
Fig. 5.
Molecular coordinate for an antiparallel β-sheet. Reproduced with permission from Proc. Natl. Acad. Sci. USA. 2005, 102, 4978–4983. Copyright 2005, The National Academy of Sciences of the USA.
B1 mode:
| (8) |
| (9) |
| (10) |
B2 mode:
| (11) |
B3 mode:
| (12) |
where Ns is the surface number density of the repeating units of the β-sheet. The standard (or achiral) susceptibility components χxxz, χyyz, χxzx, χyzy, χzxx, χzyy and χzzz can be obtained by fitting achiral SFG spectra, and chiral tensors χzxy, χzyx, χyzx and χxzy can be deduced from chiral SFG spectra. The achiral susceptibility tensor elements for the B2 and B3 modes have the same form as the B1 mode, except that βabc should be replaced by βacb and βbca, respectively. The molecular hyperpolarizability components of β-sheets can be calculated from literature values in IR and Raman measurements and confirmed by ab-initio calculations. Therefore, for SFG experiments, orientation parameters such as <cos2θ>, <cosθsinφcosφ> and <sin2θcos2φ> of β-sheets can be measured.
The Chen group successfully collected SFG amide I signals from tachyplesin I at the polystyrene/solution interface (Fig. 6A) (Chen and Wang, 2005). The fitting results showed three major peaks at 1645, 1664 and 1688 cm−1, respectively. A large number of reports have stated that amide I signals at 1688 and 1633 cm−1 can be ascribed to the B1/B3 and B2 modes of antiparallel β-sheets, respectively (Barth et al., 2002; Hilario et al., 2003; Krimm et al., 1986; Vass et al., 2003). The 1664 and 1645 cm−1 peaks in the tachyplesin I SFG spectra are due to turns, random structures or a combination thereof (Barth et al., 2002; Krimm et al., 1986; Vass et al., 2003). Following the addition of dithiothreitol (DTT), the 1688 cm−1 peak disappeared, confirming that this peak was due to β-sheet structure of tachyplesin I at the interface (Fig. 6B). The addition of DTT broke the two disulfide bonds in tachyplesin I, which are essential for it to maintain its β-sheet structure (Li, et al., 1998; Matsuzaki et al., 1993). Furthermore, Chen and colleagues also detected very strong SFG chiral spp and psp spectra directly from the polystyrene/tachyplesin I solution interface (Fig. 6C) (Wang and Chen, 2005). Their intensities were comparable to those in the ssp spectrum, but with distinct spectral features. For the spp and psp spectra, only two peaks at 1635 and 1685 cm−1 were observed from the spectral fitting results, showing that SFG chiral signals were dominated by contributions from β-sheet structures. Such signals can provide more measurements to determine β-sheet orientation.
Fig. 6.
SFG Spectra and fitting results for 0.1 mg/mL tachyplesin I adsorbed at solution/polystyrene interface with squares representing the actual spectrum, dotted line the fitted spectrum and solid lines the component peaks used to fit the spectra. A. SFG spectra before contacting with 10 mM DTT; B. SFG spectra after contacting with 10 mM DTT. Reproduced with permission from Langmuir 2005, 21, 2662–2664. Copyright 2005, American Chemical Society. C. SFG spectra of adsorbed tachyplesin I at the polystyrene/solution interface with different polarization combinations. Reproduced with permission from Proc. Natl. Acad. Sci. USA. 2005, 102, 4978–4983. Copyright 2005, The National Academy of Sciences of the USA. D. Amide I SFG spectrum of tachyplesin I (5.6 μg/mL) adsorbed onto a DPPG/DPPG bilayer collected using ssp polarization combination. Reproduced with permission from Biochim. Biophys. Acta. 2006, 1758, 1257–1273. Copyright 2006, Elsevier B.V.
Chen and colleagues have also collected SFG amide I signals from tachyplesin I in a DPPG/DPPG bilayer. These spectra indicated the presence of antiparallel β-sheet structure, with a dominant band around 1685 cm−1 (Fig. 6D) (Chen and Chen, 2006). Some differences in the spectral features in these spectra compared to the spectra in Fig. 6A and Fig. 6C indicated that tachyplesin I adopted a different conformation at the bilayer interface compared to that adsorbed onto a polystyrene surface (Chen and Wang, 2005; Wang and Chen, 2005). Chen and colleagues observed that the addition of DTT to the solution caused no changes to the spectrum of tachyplesin I already adsorbed onto a bilayer, as opposed to the changes observed when tachyplesin I was adsorbed onto a polystyrene surface, where addition of DTT led to the disappearance of the 1685 cm−1 peak of tachyplesin I (Chen and Chen, 2006; Chen and Wang, 2005; Wang and Chen, 2005). It appeared that the membrane, in contrast to the polystyrene surface, offered protection to the β-sheet structure, either by shielding the interfacial tachyplesin I from the reducing agent or by inducing the β-sheet structure even without the presence of the disulfide bonds. Currently we are investigating the detailed orientation of tachyplesin I in the lipid bilayer using the methods we presented above and the results will be reported in the future.
3.4. Real time monitoring of bilayer perturbation induced by peptides/proteins
In addition to the structural studies of membrane proteins and peptides in lipid bilayers, time-dependent SFG studies have also been applied to monitor the kinetics of changes in bilayers themselves, or as they are interacting with membrane proteins/peptides (Liu et al., 2004a; Chen and Chen, 2006; Chen and Wang, 2007a). The Conboy group deduced the flip-flop rate for a planar supported lipid bilayer (DSPC/DSPC-d83) by measuring the time-dependent SFG intensity of terminal CH3 symmetric modes at various temperatures (Liu et al., 2004a). Chen et al. applied SFG to monitor the time-dependent interactions between antimicrobial peptides and lipid bilayers (Chen and Chen, 2006; Chen and Wang, 2007a). Different modes of actions were observed between lipid bilayers and different molecules such as melittin, tachyplesin I, D-magainin 2, MSI-843, and a synthetic antibacterial oligomer (Chen and Chen, 2006).
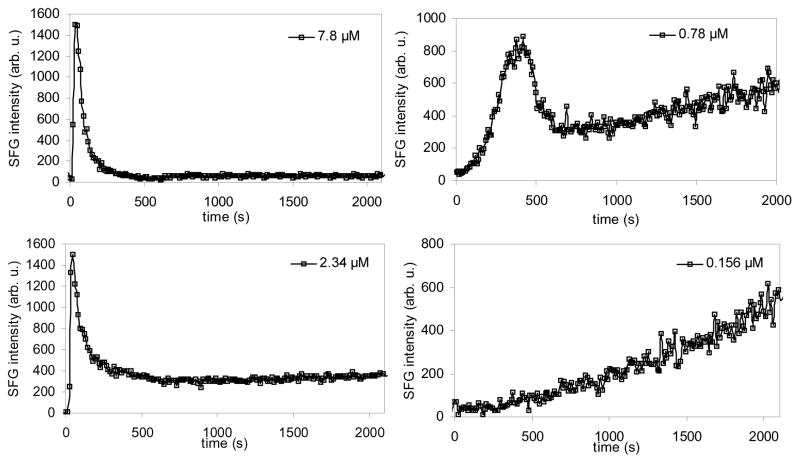
For example, Chen et al. (Chen and Wang 2007a) monitored the time-dependent and concentration-dependent disruption of a substrate supported lipid bilayer by melittin. In this research, both a symmetric d-DPPG/d-DPPG bilayer and an asymmetric d-DPPG/DPPG bilayer were used. It was found that the extent and kinetics of the bilayer disruption induced by melittin were greatly affected by the peptide concentration. For a symmetric d-DPPG/d-DPPG bilayer, Chen et al. monitored the intensity of the CD3 symmetric stretching peak at 2070 cm−1 as a function of time after injecting melittin at time 0 seconds (Fig. 7). At high melittin solution concentrations (2.34 μM to 7.8 μM), the peak intensity reached a maximum immediately after the injection of melittin, and then decreased. The initial signal increase was due to the fact that the first leaflet was disrupted more substantially initially and the inversion symmetry of the bilayer was broken. Because the second leaflet was disrupted shortly after, a new inversion symmetry was created causing the SFG signal to decrease. For lower melittin concentrations, the disruption process was much slower. For melittin concentrations at 0.78 μM, the CD3 peak intensity underwent a gradual increase after the initial increase/decrease pattern. For concentrations even lower (e.g., 0.156 μM), the initial intensity increase process took a much longer time to complete, showing that the disruption process that creates a break in the inversion symmetry was much slower than at higher concentrations of melittin.
Fig. 7.
Signal intensity change at 2070 cm−1 monitoring melittin’s interaction with a dDPPG/dDPPG bilayer on a CaF2 prism. Melittin stock solution was injected into the subphase of the bilayer at time 0 seconds. Four solution concentrations were used and dramatically different kinetics were observed. Reproduced with permission from Biophys. J. 2007, 93, 866–875. Copyright 2007, the Biophysical Society.
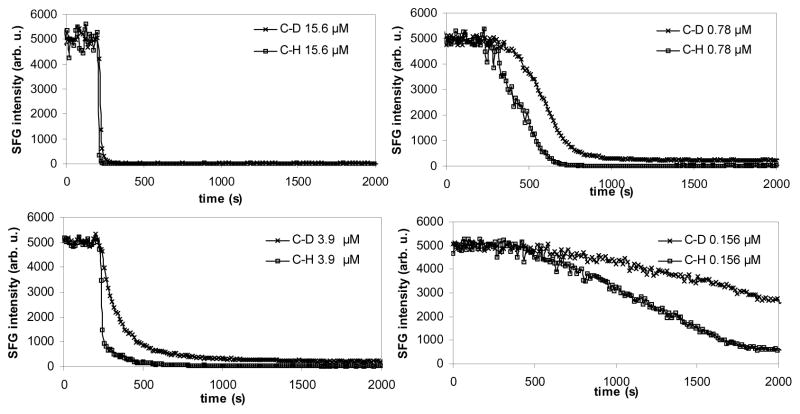
For asymmetric d-DPPG/DPPG bilayers (Fig. 8), Chen et al. monitored the SFG intensity of CD3 and CH3 symmetric stretching peaks at 2070 and 2875 cm−1, respectively. It was found that the d-DPPG/DPPG bilayer had a similar disruption trend to that observed in the d-DPPG/d-DPPG bilayer. The spectral changes occurred very quickly and then the signal became stable rapidly at higher concentrations, while a much longer time was required for the intensity to become stable for lower melittin concentrations. When 0.78 μM or higher melittin solution concentrations were used, very weak final signals from both leaflets were observed after the peptide-lipid interactions. At a solution concentration of 0.156 μM or lower, strong SFG signals from the proximal d-DPPG leaflet could still be observed after a prolonged period of time. This experiment also indicated that the lipid bilayer was disrupted at a much faster rate at a higher peptide concentration. In addition, Fig. 8 shows that melittin disrupted the two leaflets at different rates: the distal leaflet was disrupted first, before the disruption of the proximal leaflet.
Fig. 8.
Signal intensity changes at 2070 cm−1 and 2875 cm−1 monitoring melittin’s interaction with dDPPG/DPPG bilayers on a CaF2 prism. Melittin stock solution was injected into the subphase of bilayer at 200 seconds. Four solution concentrations were used and dramatically different kinetics were observed. Reproduced with permission from Biophys. J. 2007, 93, 866–875. Copyright 2007, the Biophysical Society.
The Chen group also detected time-dependent SFG signals from the asymmetric d-DPPG-DPPG bilayer/tachyplesin I solution interface with a solution concentration of 5.6 μg/mL (Chen and Chen, 2006). However, unlike the time delay between the onset of SFG C-H and C-D signal decrease observed for melittin, the signal decreases for both leaflets, were coincident with each other. This indicated that there were possibly different interaction patterns among various antimicrobial peptides. With tachyplesin I, the insertion probably occurred more readily and both leaflets were disrupted immediately and at the same rate. Time-dependent interactions between DTT-treated tachyplesin I and a dDPPG/DPPG bilayer were different from those observed for the untreated tachyplesin I. The DTT-treated tachyplesin I induced a decrease in C-D/C-H stretching mode similar to that observed for 0.44 μg/mL of melittin. This shows that the DTT-treated tachyplesin I was actually more potent in causing the signal intensity decrease during the initial period of interaction, but the untreated tachyplesin I could induce a more thorough destabilizing effect, which may be the critical factor in killing microbes.
4. Summary
In this paper, we summarized the recent applications of SFG studies on lipid monolayers, HBMs and substrate-supported lipid bilayers, as well as the interactions between peptides/proteins and lipid monolayers/bilayers, and bilayer perturbation induced by peptides/proteins. We introduced early SFG studies that focused on the C-H stretching frequency region and the more recent SFG studies in the amide I region investigated by our group. As discussed above, the peptide/protein secondary structures (α-helical, 310-helical and β-sheet) can be determined and their orientation information can be deduced. We discussed examples of studies on the interaction of peptides/proteins (melittin, G proteins, almethicin, and tachyplesin I) and lipid bilayers using SFG. These studies showed that SFG is a powerful technique to study the membrane-related peptides and proteins and that SFG can provide a unique understanding of the interactions between a lipid monolayer/bilayer and peptides/proteins without any exogenous labeling in real time and in situ.
Acknowledgments
This research is supported by National Institute of Health (1R01GM081655-01A2) and Office of Naval Research (N00014-02-1-0832 and N00014-08-1-1211). SVLC acknowledges the Molecular Biophysics Training Grant from the University of Michigan.
Appendix
The intensity of the sum frequency signal is proportional to the square of the vibration’s second-order nonlinear susceptibility :
| (A1) |
where ni(ω) is the refractive index of medium i at frequency ω, β is the reflection angle of the sum-frequency field, I1(ωVis) and I2(ωIR) are the intensities of the two input fields. The effective nonlinear susceptibility is:
| (A2) |
with ê(ω) being the unit polarization vector and L(ω) the Fresnel factor at frequency ω. Details about Fresnel factors have been discussed extensively and will not be repeated here (Zhuang et al., 1999).
For Visible-IR SFG, if the IR frequency (ωIR) is near vibrational resonances, the effective surface nonlinear susceptibility can be enhanced. The frequency dependence of can be written as:
| (A3) |
where Aq, ωq, and Γq are the strength, resonant frequency, and damping coefficient of the vibrational mode q respectively, is the nonresonant background, and is the resonant contribution (Hirose and Akamatsu, 1992a; Hirose and Akamatsu, 1992b; Hirose and Yamatoto, 1993; Gautam et al., 2001; Kim and Somorjai, 2003; Shen, 1984; Wang and Chen, 2001).
The components of can be probed by using different polarization combinations of the input and output laser beams, such as ssp (s-polarized SFG signal, s-polarized visible beam, p-polarized IR beam), sps, pss and ppp. Each measured polarization combination is related to the different components of χ(2) defined in the lab coordinate system. For an azimuthally isotropic interface only four independent components of χ(2) are nonzero from Eq. (A2). If the lab coordinate system is defined as the z-axis being along the surface normal and the x-axis being in the incident plane (Fig. 1A), these four components will have the following properties: χxxz=χyyz, χxzx= χyzy, χzxx= χzyy, and χzzz. The expressions under ssp, sps, pss and ppp polarization combinations are then (Miranda et al., 1999; Shen, 1984; Wang and Chen, 2001; Zhuang et al., 1999):
| (A4) |
| (A5) |
| (A6) |
| (A7) |
where βSF, βVis and βIR are the angles between the surface normal and the sum frequency beam, the input visible beam, and the input IR beam, respectively; and Lii’s (i = x, y or z) are the Fresnel coefficients.
The tensor components of χ(2) defined in the lab coordinate system are macroscopic properties of a surface or interface. They can be directly related to the microscopic hyperpolarizability tensor components βλμν of molecules on the surface or interface. In certain papers, microscopic hyperpolarizability tensor components have also been referred to as . In short, each is a transformation of individual microscopic hyperpolarizability tensor components βλμν in the molecular coordinate system to its macroscopic quantity of the ensemble average over all possible orientations in the lab frame (Miranda and Shen, 1999; Shen, 1984; Wang and Chen, 2001; Zhuang et al., 1999):
| (A8) |
where N is the number density of molecules and the brackets denote averaging over the molecular orientation distribution. Both χ(2) and βλμν are third rank tensors with 27 elements whose values are functions of the frequencies of the three beams involved in the SFG process. For the SFG technique we are discussing here, βλμν is a product of the IR transition dipole moment component and Raman polarizability component, which can be measured experimentally or calculated. Therefore, by measuring in SFG experiments, the orientation and the number density of interfacial molecules in a sample may be deduced.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alessandrini A, Facci P. AFM: a versatile tool in biophysics. Meas Sci Technol. 2005;16:R65–R92. [Google Scholar]
- Allen HC, Raymond EA, Richmond GL. Non-linear vibrational sum frequency spectroscopy of atmospherically relevant molecules at aqueous solution surfaces. Curr Opin Colloid Interface Sci. 2000;5:74–80. [Google Scholar]
- Anderson NA, Richter LJ, Stephenson JC, Briggman KA. Protein deformation of lipid hybrid bilayer membranes studied by sum frequency generation vibrational spectroscopy. Langmuir. 2004;20:8961–8965. doi: 10.1021/la0484220. [DOI] [PubMed] [Google Scholar]
- Anderson NA, Richter LJ, Stephenson JC, Briggman KA. Determination of lipid phase transition temperatures in hybrid bilayer membranes. Langmuir. 2006;22:8333–8336. doi: 10.1021/la061819e. [DOI] [PubMed] [Google Scholar]
- Anderson NA, Richter LJ, Stephenson JC, Briggman KA. Characterization and control of lipid layer fluidity in hybrid bilayer membranes. J Am Chem Soc. 2007;129:2094–2100. doi: 10.1021/ja066588c. [DOI] [PubMed] [Google Scholar]
- Andronesi OC, Becker S, Seidel K, Heise H, Young HS, Baldus M. Determination of membrane protein structure and dynamics by magic-angle-spinning solid-state NMR spectroscopy. J Am Chem Soc. 2005;127:12965–12974. doi: 10.1021/ja0530164. [DOI] [PubMed] [Google Scholar]
- Anglin TC, Liu J, Conboy JC. Facile lipid flip-flop in a phospholipid bilayer induced by gramicidin A measured by sum-frequency vibrational spectroscopy. Biophys J. 2007;92:L1–L3. doi: 10.1529/biophysj.106.096057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anglin TC, Conboy JC. Lateral pressure dependence of the phospholipid transmembrane diffusion rate in planar-supported lipid bilayers. Biophys J. 2008;95:186–193. doi: 10.1529/biophysj.107.118976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Archer SJ, Ellena JF, Cafiso DS. Dynamics and aggregation of the peptide ion channel alamethicin. Biophys J. 1991;60:389–398. doi: 10.1016/S0006-3495(91)82064-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bader R, Lerch M, Zerbe O. Methods and Principles in Medicinal Chemistry: BioNMR in Drug Research. Vol. 16. Wiley; Weinheim: 2003. NMR of membrane-associated peptides and proteins; pp. 95–120. [Google Scholar]
- Bagatolli LA. To see or not to see: Lateral organization of biological membranes and fluorescence microscopy. Biochim Biophys Acta. 2006;1758:1541–1556. doi: 10.1016/j.bbamem.2006.05.019. [DOI] [PubMed] [Google Scholar]
- Bain CD. Sum-frequency vibrational spectroscopy of the solid–liquid interface. J Chem Soc, Faraday Trans. 1995;91:1281–1296. [Google Scholar]
- Bak M, Baywater RP, Hohwy M, Thomsen JK, Adelhorst K, Jakobsen HJ, Sorensen OW, Nielsen NC. Conformation of alamethicin in oriented phospholipid bilayers determined by N-15 solid-state nuclear magnetic resonance. Biophys J. 2001;81:1684–1698. doi: 10.1016/S0006-3495(01)75822-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldelli S. Surface structure at the ionic liquid-electrified metal interface. Acc Chem Res. 2008;41:421–431. doi: 10.1021/ar700185h. [DOI] [PubMed] [Google Scholar]
- Banerjee U, Zidovetzki R, Birge RR, Chan SI. Interaction of alamethicin with lecithin bilayers - a P-31 and H-2 NMR-study. Biochemistry. 1985;24:7621–7627. doi: 10.1021/bi00347a019. [DOI] [PubMed] [Google Scholar]
- Barranger-Mathys M, Cafiso DS. Collisions between helical peptides in membranes monitored using electron paramagnetic resonance: evidence that alamethicin is monomeric in the absence of a membrane potential. Biophys J. 1994;67:172–176. doi: 10.1016/S0006-3495(94)80466-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barth A, Zscherp C. What vibrations tell us about proteins. Quart Rev Biophys. 2002;35:369–430. doi: 10.1017/s0033583502003815. [DOI] [PubMed] [Google Scholar]
- Bechinger B. The structure, dynamics and orientation of antimicrobial peptides in membranes by multidimensional solid-state NMR spectroscopy. Biochim Biophys Acta. 1999;1462:157–183. doi: 10.1016/s0005-2736(99)00205-9. [DOI] [PubMed] [Google Scholar]
- Bechinger B, Aisenbrey C, Bertani P. The alignment, structure and dynamics of membrane-associated polypeptides by solid-state NMR spectroscopy. Biochim Biophys Acta. 2004;1666:190–204. doi: 10.1016/j.bbamem.2004.08.008. [DOI] [PubMed] [Google Scholar]
- Belkin MA, Shen YR. Non-linear optical spectroscopy as a novel probe for molecular chirality. Int Rev Phys Chem. 2005;24:257–299. [Google Scholar]
- Beseničar M, Maček P, Lakey JH, Anderluh G. Surface plasmon resonance in protein–membrane interactions. Chem Phys Lipids. 2006;141:169–178. doi: 10.1016/j.chemphyslip.2006.02.010. [DOI] [PubMed] [Google Scholar]
- Bonn M, Roke S, Berg O, Juurlink LBF, Stamouli A, Muller M. A molecular view of cholesterol-induced condensation in a lipid monolayer. J Phys Chem B. 2004;108:19083–19085. [Google Scholar]
- Brasseur R, Deleu M, Mingeot-Leclercq MP, Francius G, Dufréne YF. Probing peptide– membrane interactions using AFM. Surf Interface Anal. 2008;40:151–156. [Google Scholar]
- Briggman KA, Stephenson JC, Wallace WE, Richter LJ. Absolute molecular orientational distribution of the polystyrene surface. J Phys Chem B. 2001;105:2785–2791. [Google Scholar]
- Buck M, Himmelhaus M. Vibrational spectroscopy of interfaces by infrared-visible sum frequency generation. J Vac Sci Technol, A. 2001;19:2717–2736. [Google Scholar]
- Cabiaux V. PH-sensitive toxins: interactions with membrane bilayers and application to drug delivery. Adv Drug Del Rev. 2004;56:987– 997. doi: 10.1016/j.addr.2003.10.044. [DOI] [PubMed] [Google Scholar]
- Cabrera-Vera TM, Vanhauwe J, Thomas TO, Medkova M, Preininger A, Mazzoni MR, Hamm HE. Insights into G protein structure, function, and regulation. Endocr Rev. 2003;24:765–781. doi: 10.1210/er.2000-0026. [DOI] [PubMed] [Google Scholar]
- Cafiso DS. Alamethicin - a peptide model for voltage gating and protein membrane interactions. Annu Rev Biophys Biomol Struct. 1994;23:141–165. doi: 10.1146/annurev.bb.23.060194.001041. [DOI] [PubMed] [Google Scholar]
- Castellana ET, Cremer PS. Solid supported lipid bilayers: From biophysical studies to sensor design. Surf Sci Rep. 2006;61:429–444. doi: 10.1016/j.surfrep.2006.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen FY, Lee MT, Huang HW. Sigmoidal concentration dependence of antimicrobial peptide activities: A case study on alamethicin. Biophys J. 2002;82:908–914. doi: 10.1016/S0006-3495(02)75452-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Clarke ML, Wang J, Chen Z. Sum frequency generation vibrational spectroscopy studies on molecular conformation and orientation of biological molecules at interfaces. Int J Mod Phys B. 2005;19:691–713. [Google Scholar]
- Chen X, Wang J, Sniadecki J, Even MA, Chen Z. Probing α-helical and β-sheet structures of peptides at solid/liquid interfaces with SFG. Langmuir. 2005;21:2662–2664. doi: 10.1021/la050048w. [DOI] [PubMed] [Google Scholar]
- Chen X, Chen Z. SFG studies on interactions between antimicrobial peptides and supported lipid bilayers. Biochim Biophys Acta. 2006;1758:1257–1273. doi: 10.1016/j.bbamem.2006.01.017. [DOI] [PubMed] [Google Scholar]
- Chen X, Tang H, Even MA, Wang J, Tew GN, Chen Z. Observing a molecular knife at work. J Am Chem Soc. 2006;128:2711–2714. doi: 10.1021/ja057029t. [DOI] [PubMed] [Google Scholar]
- Chen X, Boughton AP, Tesmer JJG, Chen Z. In situ investigation of heterotrimeric G protein βγ subunit binding and orientation on membrane bilayers. J Am Chem Soc. 2007;129:12658–12659. doi: 10.1021/ja075542w. [DOI] [PubMed] [Google Scholar]
- Chen X, Wang J, Kristalyn CB, Chen Z. Real-time structural investigation of a lipid bilayer during its interaction with mlittin using sum frequency generation vibrational spectroscopy. Biophys J. 2007a;93:866–875. doi: 10.1529/biophysj.106.099739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Wang J, Boughton AP, Kristalyn CB, Chen Z. Multiple orientation of melittin inside a single lipid bilayer determined by combined vibrational spectroscopic studies. J Am Chem Soc. 2007b;129:1420–1427. doi: 10.1021/ja067446l. [DOI] [PubMed] [Google Scholar]
- Chen Z. Understanding surfaces and buried interfaces of polymer materials at the molecular level using sum frequency generation vibrational spectroscopy. Polymer Int. 2007;56:577–587. [Google Scholar]
- Chen Z, Shen YR, Somorjai GA. Studies of polymer surfaces by sum frequency generation vibrational spectroscopy. Annu Rev Phys Chem. 2002;53:437–465. doi: 10.1146/annurev.physchem.53.091801.115126. [DOI] [PubMed] [Google Scholar]
- Clarke ML, Wang J, Chen Z. Conformational changes of fibrinogen after adsorption. J Phys Chem B. 2005;109:22027–22035. doi: 10.1021/jp054456k. [DOI] [PubMed] [Google Scholar]
- Devanathan S, Salamon Z, Lindblom G, Grobner G, Tollin G. Effects of sphingomyelin, cholesterol and zinc ions on the binding, insertion and aggregation of the amyloid Aβ1–40 peptide in solid-supported lipid bilayers. FEBS J. 2006;273:1389–1402. doi: 10.1111/j.1742-4658.2006.05162.x. [DOI] [PubMed] [Google Scholar]
- Doyle AW, Fick J, Himmelhaus M, Eck W, Graziani I, Prudovsky I, Grunze M, Maciag T, Neivandt DJ. Protein deformation of lipid hybrid bilayer membranes studied by sum frequency generation vibrational spectroscopy. Langmuir. 2004;20:8961–8965. doi: 10.1021/la0484220. [DOI] [PubMed] [Google Scholar]
- Dreesen L, Humbert C, Sartenaer Y, Caudano Y, Volcke C, Mani AA, Peremans A, Thiry PA, Hanique S, Frere JM. Electronic and molecular properties of an adsorbed protein monolayer probed by two-color sum-frequency generation spectroscopy. Langmuir. 2004;20:7201–7207. doi: 10.1021/la0488001. [DOI] [PubMed] [Google Scholar]
- Dreesen L, Sartenaer Y, Humbert C, Mani AA, Methivier C, Pradier CM, Thiry PA, Peremans A. Probing ligand-protein recognition with sum-frequency generation spectroscopy: The avidin-biocytin case. ChemPhysChem. 2004;5:1719–1725. doi: 10.1002/cphc.200400213. [DOI] [PubMed] [Google Scholar]
- Duclohier H. Helical kink and channel behaviour: a comparative study with the peptaibols alamethicin, trichotoxin and antiamoebin. Eur Biophys J. 2004;33:169–174. doi: 10.1007/s00249-003-0383-y. [DOI] [PubMed] [Google Scholar]
- Duclohier H, Wroblewski H. Voltage-dependent pore formation and antimicrobial activity by alamethicin and analogues. J Membr Biol. 2001;184:1–12. doi: 10.1007/s00232-001-0077-2. [DOI] [PubMed] [Google Scholar]
- Dürr UHN, Yamamoto K, Im SC, Waskell L, Ramamoorthy A. Solid-state NMR reveals structural and dynamical properties of a membrane-anchored electron-carrier protein, cytochrome b5. J Am Chem Soc. 2007;129:6670–6671. doi: 10.1021/ja069028m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dvinskikh SV, Dürr UHN, Yamamoto K, Ramamoorthy A. High-resolution 2D NMR spectroscopy of bicelles to measure the membrane interaction of ligands. J Am Chem Soc. 2007;129:794–802. doi: 10.1021/ja065536k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dwivedi AM, Krimm S. Vibrational analysis of peptides, polypeptides, and proteins. 24 conformation of poly(alpha-aminoisobutyric acid) Biopolymers. 1984;23:2025–2065. [Google Scholar]
- Eisenthal KB. Equilibrium and dynamic processes at interfaces by second harmonic and sum frequency generation. Annu Rev Phys Chem. 1992;43:627–661. [Google Scholar]
- Engel A, Gaub HE. Structure and mechanics of membrane proteins. Annu Rev Biochem. 2008;77:127–48. doi: 10.1146/annurev.biochem.77.062706.154450. [DOI] [PubMed] [Google Scholar]
- Evans-Nguyen KM, Fuierer RR, Fitchett BD, Tolles LR, Conboy JC, Schoenfisch MH. Changes in adsorbed fibrinogen upon conversion to fibrin. Langmuir. 2006;22:5115–5121. doi: 10.1021/la053070y. [DOI] [PubMed] [Google Scholar]
- Faiss S, Kastl K, Janshoff A, Steinem C. Formation of irreversibly bound annexin A1 protein domains on POPC/POPS solid supported membranes. Biochim Biophys Acta. 2008;1778:1601–1610. doi: 10.1016/j.bbamem.2008.01.003. [DOI] [PubMed] [Google Scholar]
- Fernández C, Adeishvili K, Wuthrich K. Transverse relaxation-optimized NMR spectroscopy with the outer membrane protein OmpX dihexanoyl phosphatidylcholine micelles. Proc Natl Acad Sci. 2001;98:2358–2363. doi: 10.1073/pnas.051629298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fourkas JT, Walker RA, Can SZ, Gershgoren E. Effects of reorientation in vibrational sum-frequency spectroscopy. J Phys Chem C. 2007;111:8902–8915. [Google Scholar]
- Fox RO, Richards FM. A voltage-gated ion channel model inferred from the crystal-structure of alamethicin at 1.5-Å resolution. Nature. 1982;300:325–330. doi: 10.1038/300325a0. [DOI] [PubMed] [Google Scholar]
- Fragneto-Cusani G. Neutron reflectivity at the solid/liquid interface: examples of applications in biophysics. J Phys: Condens Matter. 2001;13:4973–4989. [Google Scholar]
- Gaudet R, Savage JR, McLaughlin JN, Willardson BM, Sigler PB. A molecular mechanism for the phosphorylation-dependent regulation of heterotrimeric G proteins by phosducin. Mol Cell. 1999;3:649–660. doi: 10.1016/s1097-2765(00)80358-5. [DOI] [PubMed] [Google Scholar]
- Gautam KS, Schwab AD, Dhinojwala A, Zhang D, Dougal SM, Yeganeh MS. Molecular structure of polystyrene at air/polymer and solid/polymer interfaces. Phys Rev Lett. 2000;85:3854–3857. doi: 10.1103/PhysRevLett.85.3854. [DOI] [PubMed] [Google Scholar]
- Gautam KS, Dhinojwala A. Molecular structure of hydrophobic alkyl side chains at comb polymer-air interface. Macromolecules. 2001;34:1137–1139. [Google Scholar]
- Gautam KS, Dhinojwala A. Melting at alkyl side chain comb polymer interfaces. Phys Rev Lett. 2002;88:145501. doi: 10.1103/PhysRevLett.88.145501. [DOI] [PubMed] [Google Scholar]
- Gopalakrishnan S, Liu DF, Allen HC, Kuo M, Shultz MJ. Vibrational spectroscopic studies of aqueous interfaces: Salts, acids, bases, and nanodrops. Chem Rev. 2006;106:1155–1175. doi: 10.1021/cr040361n. [DOI] [PubMed] [Google Scholar]
- Gracias DH, Chen Z, Shen YR, Somorjai GA. Molecular characterization of polymer and polymer blend surfaces. Combined sum frequency generation surface vibrational spectroscopy and scanning force microscopy studies. Acc Chem Res. 1999;32:930–940. [Google Scholar]
- Guyotsionnest P, Hunt JH, Shen YR. sum-frequency vibrational spectroscopy of a Langmuir film - study of molecular-orientation of a two-dimensional system. Phys Rev Lett. 1987;59:1597–1600. doi: 10.1103/PhysRevLett.59.1597. [DOI] [PubMed] [Google Scholar]
- Haas H, Steitz R, Fasano A, Liuzzi GM, Polverini E, Cavatorta P, Riccio P. Laminar order within Langmuir-Blodgett multilayers from phospholipid and Myelin basic protein: A neutron reflectivity study. Langmuir. 2007;23:8491–8496. doi: 10.1021/la700733y. [DOI] [PubMed] [Google Scholar]
- Hall JE, Vodyanoy I, Balasubramanian TM, Marshall GR. Alamethicin - A rich model for channel behavior. Biophys J. 1984;45:233–247. doi: 10.1016/S0006-3495(84)84151-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haris PI, Chapman D. Fourier-transform infrared-spectra of the polypeptide alamethicin and a possible structural similarity with bacteriorhodopsin. Biochim Biophys Acta. 1988;943:375–380. doi: 10.1016/0005-2736(88)90571-8. [DOI] [PubMed] [Google Scholar]
- Haris PI, Molle G, Duclohier H. Conformational changes in alamethicin associated with substitution of its alpha-methylalanines with leucines: A FTIR spectroscopic analysis and correlation with channel kinetics. Biophys J. 2004;86:248–253. doi: 10.1016/S0006-3495(04)74100-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harper KL, Allen HC. Competition between DPPC and SDS at the air-aqueous interface. Langmuir. 2007;23:8925–8931. doi: 10.1021/la7006974. [DOI] [PubMed] [Google Scholar]
- Heberle FA, Buboltz JT, Stringer D, Feigenson GW. Fluorescence methods to detect phase boundaries in lipid bilayer mixtures. Biochim Biophys Acta. 2005;1746:186–92. doi: 10.1016/j.bbamcr.2005.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilario J, Kubelka J, Keiderling TA. Optical spectroscopic investigations of model beta-sheet hairpins in aqueous solution. J Am Chem Soc. 2003;125:7562–7574. doi: 10.1021/ja030039e. [DOI] [PubMed] [Google Scholar]
- Hirose C, Akamatsu N, Domen K. Formulas for the analysis of the surface SFG spectrum and transformation coefficients of cartesian SFG tensor components. Appl Spectrosc. 1992a;46:1051–1072. [Google Scholar]
- Hirose C, Akamatsu N, Domen K. Formulas for the analysis of surface sum-frequency generation spectrum by CH stretching modes of methyl and methylene groups. J Chem Phys. 1992b;96:997–1004. [Google Scholar]
- Hirose C, Yamamoto H, Akamatsu N, Domen K. Orientation analysis by simulation of vibrational sum-frequency generation spectrum - CH stretching bands of the methyl-group. J Phys Chem. 1993;97:10064–10069. [Google Scholar]
- Holman J, Ye S, Neivandt DJ, Davies PB. Studying nanoparticle-induced structural changes within fatty acid multilayer films using sum frequency generation vibrational spectroscopy. J Am Chem Soc. 2004;126:14322–14323. doi: 10.1021/ja046954x. [DOI] [PubMed] [Google Scholar]
- Hopkins AJ, McFearin CL, Richmond GL. Investigations of the solid-aqueous interface with vibrational sum-frequency spectroscopy. Curr Opin Solid State Mater Sci. 2005;9:19–27. [Google Scholar]
- Huang HW, Wu Y. Lipid-alamethicin interactions influence alamethicin orientation. Biophys J. 1991;60:1079–1087. doi: 10.1016/S0006-3495(91)82144-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humbert C, Dreesen L, Sartenaer Y, Peremans A, Thiry PA, Volcke C. On the protoporphyrin monolayers conformation. Chem Phys Chem. 2006;7:569–571. doi: 10.1002/cphc.200500475. [DOI] [PubMed] [Google Scholar]
- Hunt JH, Guyot-Sionnest P, Shen YR. Observation of C–H stretching vibrations of monolayers of molecules optical sum frequency generation. Chem Phys Lett. 1987;133:189–192. [Google Scholar]
- Ionov R, El-Abed A, Angelova A, Goldmann M, Peretti P. Asymmetrical ion-channel model inferred from two-dimensional crystallization of a peptide antibiotic. Biophys J. 2000;78:3026–3035. doi: 10.1016/S0006-3495(00)76841-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwahashi T, Miyamae T, Kanai K, Seki K, Kim D, Ouchi Y. Anion configuration at the air/liquid interface of ionic liquid [bmim]OTf studied by sum-frequency generation spectroscopy. J Phys Chem B. 2008;112:11936–11941. doi: 10.1021/jp8021908. [DOI] [PubMed] [Google Scholar]
- Johnston LJ. Nanoscale Imaging of Domains in Supported Lipid Membranes. Langmuir. 2007;23:5886–5895. doi: 10.1021/la070108t. [DOI] [PubMed] [Google Scholar]
- Kalb E, Frey S, Tamm LK. Formation of supported planar bilayers by fusion of vesicles to supported phospholipid monolayers. Biochim Biophys Acta. 1992;1103:307–316. doi: 10.1016/0005-2736(92)90101-q. [DOI] [PubMed] [Google Scholar]
- Katsaras J, Gutberlet T. Lipid bilayers: structure and interactions. New York: Springer; 2001. [Google Scholar]
- Katsu T, Nakao S, Iwanaga S. Mode of action of an antimicrobial peptide, tachyplesin I, on biomembranes. Biol Pharm Bull. 1993;16:178–181. doi: 10.1248/bpb.16.178. [DOI] [PubMed] [Google Scholar]
- Kennedy DF, Crisma M, Toniolo C, Chapman D. Studies of peptides forming 310-and a-helices and b-bend ribbon structures in organic solution and in model biomembranes by Fourier transform infrared spectroscopy. Biochemistry. 1991;30:6541–6548. doi: 10.1021/bi00240a026. [DOI] [PubMed] [Google Scholar]
- Kessel A, Cafiso DS, Ben-Tal N. Continuum solvent model calculations of alamethicin-membrane interactions: Thermodynamic aspects. Biophys J. 2000;78:571–583. doi: 10.1016/S0006-3495(00)76617-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Cremer PS. Elucidating changes in interfacial water structure upon protein adsorption. ChemPhysChem. 2001;2:543–546. doi: 10.1002/1439-7641(20010917)2:8/9<543::AID-CPHC543>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- Kim J, Kim G, Cremer PS. Investigations of water structure at the solid/liquid interface in the presence of supported lipid bilayers by vibrational sum frequency spectroscopy. Langmuir. 2001;17:7255–7260. [Google Scholar]
- Kim J, Somorjai GA. Molecular packing of lysozyme, fibrinogen, and bovine serum albumin on hydrophilic and hydrophobic surfaces studied by infrared-visible sum frequency generation and fluorescence microscopy. J Am Chem Soc. 2003;125:3150–3158. doi: 10.1021/ja028987n. [DOI] [PubMed] [Google Scholar]
- Kim G, Gurau MC, Kim J, Cremer PS. Investigations of lysozyme adsorption at the air/water and quartz/water interfaces by vibrational sum frequency spectroscopy. Langmuir. 2002;18:2807–2811. [Google Scholar]
- Kim G, Gurau MC, Lim SM, Cremer PS. Investigations of the orientation of a membrane peptide by sum frequency spectroscopy. J Phys Chem B. 2003;107:1403–1409. [Google Scholar]
- Kim C, Gurau MC, Cremer PS, Yu H. Chain conformation of poly(dimethyl siloxane) at the air/water interface by sum frequency generation. Langmuir. 2008;24:10155–10160. doi: 10.1021/la800349q. [DOI] [PubMed] [Google Scholar]
- Knoesen A, Pakalnis S, Wang M, Wise WD, Lee N, Frank CW. Sum-frequency spectroscopy and imaging of aligned helical polypeptides. IEEE J Sel Top Quantum Electron. 2004;10:1154–1163. [Google Scholar]
- Koch WJ. Genetic and phenotypic targeting of beta-adrenergic signaling in heart failure. Mol Cell Biochem. 2004;263:5–9. doi: 10.1023/B:MCBI.0000041843.64809.48. [DOI] [PubMed] [Google Scholar]
- Koffas TS, Amitay-Sadovsky E, Kim J, Somorjai GA. Molecular composition and mechanical properties of biopolymer interfaces studied by sum frequency generation vibrational spectroscopy and atomic force microscopy. J Biomater Sci Polymer Ed. 2004;4:475–509. doi: 10.1163/156856204323005325. [DOI] [PubMed] [Google Scholar]
- Krimm S, Bandekar J. Vibrational spectroscopy and conformation of peptides, polypeptides, and proteins. Adv Protein Chem. 1986;38:181–364. doi: 10.1016/s0065-3233(08)60528-8. [DOI] [PubMed] [Google Scholar]
- Kučerka N, Nieh MP, Pencer J, Harroun T, Katsaras J. The study of liposomes, lamellae and membranes using neutrons and X-rays. Curr Opin Coll Int Sci. 2007;12:17–22. [Google Scholar]
- Laflamme E, Badia A, Lafleur M, Schwartz JL, Laprade R. Atomic Force Microscopy Imaging of Bacillus thuringiensis Cry1 Toxins Interacting with Insect Midgut Apical Membranes. J Membrane Biol. 2008;222:127–139. doi: 10.1007/s00232-008-9106-8. [DOI] [PubMed] [Google Scholar]
- Lambert AG, Davies PB, Neivandt DJ. Implementing the theory of sum frequency generation vibrational spectroscopy: A tutorial review. Appl Spectrosc Rev. 2005;40:103–145. [Google Scholar]
- Laver DR. The barrel-stave model as applied to alamethicin and its analogs reevaluated. Biophys J. 1994;66:355–359. doi: 10.1016/s0006-3495(94)80784-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee AG. How lipids and proteins interact in a membrane: a molecular approach. Mol BioSyst. 2005;1:203–212. doi: 10.1039/b504527d. [DOI] [PubMed] [Google Scholar]
- Lee D, Walter KFA, Brückner AK, Hilty C, Becker S, Griesinger C. Bilayer in small bicelles revealed by lipid-protein interactions using NMR spectroscopy. J Am Chem Soc. 2008;130:13822–13823. doi: 10.1021/ja803686p. [DOI] [PubMed] [Google Scholar]
- Lee S, Krimm S. Ab initio-based vibrational analysis of alpha-poly(L-alanine) Biopolymers. 1998a;46:283–317. [Google Scholar]
- Lee S, Krimm S. Polarized Raman spectra of oriented films of alpha-helical poly(L-alanine) and its N-deuterated analogue. J Raman Spectrosc. 1998b;29:73–80. [Google Scholar]
- Lee SH, Wang J, Krimm S, Chen Z. Irreducible representation and projection operator application to understanding nonlinear optical phenomena: Hyper-raman, sum frequency generation, and four-wave mixing spectroscopy. J Phys Chem A. 2006;110:7035–7044. doi: 10.1021/jp057200n. [DOI] [PubMed] [Google Scholar]
- Leitgeb B, Szekeres A, Manczinger L, Vagvolgyi C, Kredics L. The history of alamethicin: A review of the most extensively studied peptaibol. Chem BioDivers. 2007;4:1027–1051. doi: 10.1002/cbdv.200790095. [DOI] [PubMed] [Google Scholar]
- Levy D, Briggman KA. Cholesterol/phospholipid interactions in hybrid bilayer membranes. Langmuir. 2007;23:7155–7161. doi: 10.1021/la070204u. [DOI] [PubMed] [Google Scholar]
- Li Q, Hua R, Chou KC. Electronic and conformational properties of the conjugated polymer MEH-PPV at a buried film/solid interface investigated by two-dimensional IR-visible sum frequency generation. J Phys Chem B. 2008;112:2315–2318. doi: 10.1021/jp0745135. [DOI] [PubMed] [Google Scholar]
- Li YJ, Rothwarf DM, Scheraga HA. An unusual adduct of dithiothreitol with a pair of cysteine residues of a protein as a stable folding intermediate. J Am Chem Soc. 1998;120:2668–2669. [Google Scholar]
- Lindblom G, Grobner G. NMR on lipid membranes and their proteins. Curr Opin Colloid Interface Sci. 2006;11:24–29. [Google Scholar]
- Liu J, Conboy JC. Direct measurement of the transbilayer movement of phospholipids by sum-frequency vibrational spectroscopy. J Am Chem Soc. 2004a;126:8376–8377. doi: 10.1021/ja048245p. [DOI] [PubMed] [Google Scholar]
- Liu J, Conboy JC. Phase transition of a single lipid bilayer measured by sum-frequency vibrational spectroscopy. J Am Chem Soc. 2004b;126:8894–8895. doi: 10.1021/ja031570c. [DOI] [PubMed] [Google Scholar]
- Liu J, Conboy JC. Structure of a gel phase lipid bilayer prepared by the Langmuir-Blodgett/Langmuir-Schaefer method characterized by sum-frequency vibrational spectroscopy. Langmuir. 2005a;21:9091–9097. doi: 10.1021/la051500e. [DOI] [PubMed] [Google Scholar]
- Liu J, Conboy JC. 1, 2-diacyl-phosphatidylcholine flip-flop measured directly by sum-frequency vibrational spectroscopy. Biophys J. 2005b;89:2522–2532. doi: 10.1529/biophysj.105.065672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Conboy JC. Asymmetric distribution of lipids in a phase segregated phospholipid bilayer observed by sum-frequency vibrational spectroscopy. J Phys Chem C. 2007;111:8988–8999. [Google Scholar]
- Lobau J, Sass M, Pohle W, Selle C, Koch MHJ, Wolfrum K. Chain fluidity and phase behaviour of phospholipids as revealed by FTIR and sum-frequency spectroscopy. J Mol Struct. 1999;481:407–411. [Google Scholar]
- Lodowski DT, Pitcher JA, Capel WD, Lefkowitz RJ, Tesmer JJG. Keeping G proteins at bay: A complex between G protein-coupled receptor kinase 2 and G beta gamma. Science. 2003;300:1256–1262. doi: 10.1126/science.1082348. [DOI] [PubMed] [Google Scholar]
- Ma G, Allen HC. DPPC Langmuir monolayer at the air-water interface: probing the tail and head groups by vibrational sum frequency generation spectroscopy. Langmuir. 2006;22:5341–5349. doi: 10.1021/la0535227. [DOI] [PubMed] [Google Scholar]
- Ma G, Allen HC. Condensing effect of palmitic acid on DPPC in mixed Langmuir monolayers. Langmuir. 2007;23:589–597. doi: 10.1021/la061870i. [DOI] [PubMed] [Google Scholar]
- Marsh D, Muller M, Schmitt FJ. Orientation of the infrared transition moments for an alpha-helix. Biophys J. 2000;78:2499–2510. doi: 10.1016/S0006-3495(00)76795-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsh D, Jost M, Peggion C, Toniolo C. TOAC spin labels in the backbone of alamethicin: EPR studies in lipid membranes. Biophys J. 2007a;92:473–481. doi: 10.1529/biophysj.106.092775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsh D, Jost M, Peggion C, Toniolo C. Lipid chainlength dependence for incorporation of alamethicin in membranes: EPR studies on TOAC-spin labelled analogues. Biophys J. 2007b;92:4002–4011. doi: 10.1529/biophysj.107.104026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsh D. Energetics of hydrophobic matching in lipid-protein interactions. Biophys J. 2008;94:3996–4013. doi: 10.1529/biophysj.107.121475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mateo CR, Gómez J, Villalaín J, González Ros JM. Protein-lipid interaction: new approaches and emerging concepts. Berlin: Springer; 2006. [Google Scholar]
- Mathew MK, Balaram P. Alamethicin and related membrane channel forming polypeptides. Mol Cell Biochem. 1983a;50:47–64. doi: 10.1007/BF00225279. [DOI] [PubMed] [Google Scholar]
- Mathew MK, Balaram P. A helix dipole model for alamethicin and related transmembrane channels. FEBS Lett. 1983b;157:1–5. [Google Scholar]
- Matsuzaki K, Nakayama M, Fukui M, Otaka A, Funakoshi S, Fujii N, Bessho K, Miyajima K. Role of disulfide linkages in tachyplesin-lipid interactions. Biochemistry. 1993;32:11704–11710. doi: 10.1021/bi00094a029. [DOI] [PubMed] [Google Scholar]
- McConnell HM, Watts TH, Weis RM, Brian AA. Supported planar membranes in studies of cell-cell recognition in the immune system. Biochim Biophys Acta. 1986;864:95–106. doi: 10.1016/0304-4157(86)90016-x. [DOI] [PubMed] [Google Scholar]
- McIntosh TJ, Simon SA. Roles of bilayer material properties in function and distribution of membrane proteins. Annu Rev Biophys Biomol Struct. 2006;35:177–198. doi: 10.1146/annurev.biophys.35.040405.102022. [DOI] [PubMed] [Google Scholar]
- Mermut O, Phillips DC, York RL, McCrea KR, Ward RS, Somorjai GA. In situ adsorption studies of a 14-amino acid leucine-lysine peptide onto hydrophobic polystyrene and hydrophilic silica surfaces using quartz crystal microbalance, atomic force microscopy, and sum frequency generation vibrational spectroscopy. J Am Chem Soc. 2006;128:3598–3607. doi: 10.1021/ja056031h. [DOI] [PubMed] [Google Scholar]
- Miranda PB, Shen YR. Liquid interfaces: a study by sum-frequency vibrational spectroscopy. J Phys Chem B. 1999;103:3292–3307. [Google Scholar]
- Mizuguchi M, Kamata S, Kawabat S, Kawan K. Structure of horseshoe crab antimicrobial peptide tachyplesin I in dodecylphosphocholine micelles. Peptide Science. 2003;39:281–282. [Google Scholar]
- Moore FG, Richmond GL. Integration or segregation: How do molecules behave at oil/water interfaces? Acc Chem Res. 2008;41:739–748. doi: 10.1021/ar7002732. [DOI] [PubMed] [Google Scholar]
- Mottamal M, Lazaridis T. Voltage-dependent energetics of alamethicin monomers in the membrane. Biophys Chem. 2006;122:50–57. doi: 10.1016/j.bpc.2006.02.005. [DOI] [PubMed] [Google Scholar]
- Nagaraj R, Balaram P. Alamethicin, A transmembrane channel. Acc Chem Res. 1981;14:356–362. [Google Scholar]
- Naito A, Kawamura I. Solid-state NMR as a method to reveal structure and membrane-interaction of amyloidogenic proteins and peptides. Biochim Biophys Acta. 2007;1768:1900–1912. doi: 10.1016/j.bbamem.2007.03.025. [DOI] [PubMed] [Google Scholar]
- Nakamura T, Furunaka H, Miyata T, Tokunaga F, Muta T, Iwanaga S, Niwa M, Takao T, Shimonishi Y. Tachyplesin, a class of antimicrobial peptide from the hemocytes of the horseshoe crab (tachypleus tridentatus). isolation and chemical structure. J Biol Chem. 1988;263:16709–16713. [PubMed] [Google Scholar]
- Neves SR, Ram PT, Iyengar R. G protein pathways. Science. 2002;296:1636–1639. doi: 10.1126/science.1071550. [DOI] [PubMed] [Google Scholar]
- Nickolov ZS, Britt DW, Miller JD. Sum-frequency spectroscopy analysis of two-component Langmuir monolayers and the associated interfacial water structure. J Phys Chem. 2006;110:15506–15513. doi: 10.1021/jp0631578. [DOI] [PubMed] [Google Scholar]
- North CL, Barranger-Mathys M, Cafiso DS. Membrane orientation of the N-terminal segment of alamethicin determined by solid-state N-15 NMR. Biophys J. 1995;69:2392–2397. doi: 10.1016/S0006-3495(95)80108-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohe C, Ida Y, Matsumoto S, Sasaki T, Goto Y, Noi A, Tsurumaru T, Itoh K. Investigations of polymyxin B-phospholipid interactions by vibrational sum frequency generation spectroscopy. J Phys Chem B. 2004;108:18081–18087. [Google Scholar]
- Ohe C, Sasaki T, Noi M, Goto Y, Itoh K. Sum frequency generation spectroscopic study of the condensation effect of cholesterol on a lipid monolayer. Anal Bioanal Chem. 2007;388:73–79. doi: 10.1007/s00216-006-1030-0. [DOI] [PubMed] [Google Scholar]
- Ohe C, Goto Y, Noi M, Arai M, Kamijo H, Itoh K. Sum frequency generation spectroscopic studies on phase transitions of phospholipid monolayers containing poly(ethylene oxide) lipids at the air-water interface. J Phys Chem B. 2007;111:1693–1700. doi: 10.1021/jp064179x. [DOI] [PubMed] [Google Scholar]
- Oh-e M, Hong SC, Shen YR. Orientations of phenyl sidegroups and liquid crystal molecules on a rubbed polystyrene surface. Appl Phys Lett. 2002;80:784–786. [Google Scholar]
- Oldham WM, Hamm HE. Structural basis of function in heterotrimeric G proteins. Quart Rev Biophys. 2006;39:117–166. doi: 10.1017/S0033583506004306. [DOI] [PubMed] [Google Scholar]
- Opdahl A, Koffas TS, Amitay-Sadovsky E, Kim J, Somorjai GA. Characterization of polymer surface structure and surface mechanical behaviour by sum frequency generation surface vibrational spectroscopy and atomic force microscopy. J Phys: Condens Matter. 2004;16:R659–R677. [Google Scholar]
- Perry A, Neipert C, Space B, Moore PB. Theoretical modeling of interface specific vibrational spectroscopy: Methods and applications to aqueous interfaces. Chem Rev. 2006;106:1234–1258. doi: 10.1021/cr040379y. [DOI] [PubMed] [Google Scholar]
- Petralli-Mallow TP, Briggman KA, Richter LJ, Stephenson JC, Plant AL. Nonlinear optics as a detection scheme for biomimetic sensors: SFG spectroscopy of hybrid bilayer membrane formation. Proc SPIE. 1999;3858:25–31. [Google Scholar]
- Pitcher JA, Inglese J, Higgins JB, Arriza JL, Casey PJ, Kim C, Benovic JL, Kwatra MM, Caron MG, Lefkowitz RJ. Role of beta-gamma-subunits of g-proteins in targeting the beta-adrenergic-receptor kinase to membrane-bound receptors. Science. 1992;257:1264–1267. doi: 10.1126/science.1325672. [DOI] [PubMed] [Google Scholar]
- Richmond GL. Structure and bonding of molecules at aqueous surfaces. Annu Rev Phys Chem. 2001;52:357–389. doi: 10.1146/annurev.physchem.52.1.357. [DOI] [PubMed] [Google Scholar]
- Richmond GL. Molecular bonding and interactions at aqueous surfaces as probed by vibrational sum frequency spectroscopy. Chem Rev. 2002;102:2693–2724. doi: 10.1021/cr0006876. [DOI] [PubMed] [Google Scholar]
- Richter RP, Bérat R, Brisson AR. Formation of solid-supported lipid bilayers: An Integrated View. Langmuir. 2006;22:3497–3505. doi: 10.1021/la052687c. [DOI] [PubMed] [Google Scholar]
- Rintoul L, Carter EA, Stewart SD, Fredericks PM. Keratin orientation in wool and feathers by polarized Raman spectroscopy. Biopolymers. 2000;57:19–28. doi: 10.1002/(SICI)1097-0282(2000)57:1<19::AID-BIP4>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- Rocha-Mendoza I, Yankelevich DR, Wang M, Reiser KM, Frank CW, Knoesen A. Sum frequency vibrational spectroscopy: The molecular origins of the optical second-order nonlinearity of collagen. Biophys J. 2007;93:4433–4444. doi: 10.1529/biophysj.107.111047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roke S, Schins J, Muller M, Bonn M. Vibrational spectroscopic investigation of the phase diagram of a biomimetic lipid monolayer. Phys Rev Lett. 2003;90:128101. doi: 10.1103/PhysRevLett.90.128101. [DOI] [PubMed] [Google Scholar]
- Rupprechter G, Unterhalt H, Morkel M, Galletto P, Hu LJ, Freund HJ. Sum frequency generation vibrational spectroscopy at solid-gas interfaces: CO adsorption on Pd model catalysts at ambient pressure. Surf Sci. 2002;502:109–122. [Google Scholar]
- Rupprechter G, Weilach C. Spectroscopic studies of surface-gas interactions and catalyst restructuring at ambient pressure: mind the gap! . J Phys : Condens Matter. 2008;20:184019. [Google Scholar]
- Sackmann E. Supported membranes: Scientific and practical applications. Science. 1996;271:43–48. doi: 10.1126/science.271.5245.43. [DOI] [PubMed] [Google Scholar]
- Salamon Z, Macleod HA, Tollin G. Surface plasmon resonance spectroscopy as a tool for investigating the biochemical and biophysical properties of membrane protein systems. II: Applications to biological systems. Biochim Biophys Acta. 1997;1331:131–152. doi: 10.1016/s0304-4157(97)00003-8. [DOI] [PubMed] [Google Scholar]
- Sansom MSP. Alamethicin and related peptaibols - model ion channels. Eur Biophys J. 1993;22:105–124. doi: 10.1007/BF00196915. [DOI] [PubMed] [Google Scholar]
- Sansom MSP. Structure and function of channel-forming peptaibols. Quart Rev Biophys. 1993;26:365–421. doi: 10.1017/s0033583500002833. [DOI] [PubMed] [Google Scholar]
- Sartenaer Y, Tourillon G, Dreesen L, Lis D, Mani AA, Thiry PA, Peremans A. Sum-frequency generation spectroscopy of DNA monolayers. Biosens Bioelectron. 2007;22:2179–2183. doi: 10.1016/j.bios.2006.10.005. [DOI] [PubMed] [Google Scholar]
- Shen YR. The Principles of Nonlinear Optics. John Wiley& Sons; New York: 1984. [Google Scholar]
- Shen YR. Surface-properties probed by 2nd-harmonic and sum-frequency generation. Nature. 1989;337:519–525. [Google Scholar]
- Shen YR, Ostroverkhov V. Sum-frequency vibrational spectroscopy on water interfaces: Polar orientation of water molecules at interfaces. Chem Rev. 2006;106:1140–1154. doi: 10.1021/cr040377d. [DOI] [PubMed] [Google Scholar]
- Shultz MJ, Schnitzer C, Simonelli D, Baldelli S. Sum frequency generation spectroscopy of the aqueous interface: ionic and soluble molecular solutions. Inter Rev Phys Chem. 2000;19:123–153. [Google Scholar]
- Shultz MJ, Baldelli S, Schnitzer C, Simonelli D. Aqueous solution/air interfaces probed with sum frequency generation spectroscopy. J Phys Chem B. 2002;106:5313–5324. doi: 10.1021/jp014466v. [DOI] [PubMed] [Google Scholar]
- Sovago M, Wurpel GWH, Smits M, Muller M, Bonn M. Calcium-induced phospholipid ordering depends on surface pressure. J Am Chem Soc. 2007;129:11079–11084. doi: 10.1021/ja071189i. [DOI] [PubMed] [Google Scholar]
- Spaar A, Munster C, Salditt T. Conformation of peptides in lipid membranes studied by X-ray grazing incidence scattering. Biophys J. 2004;87:396–407. doi: 10.1529/biophysj.104.040667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinem C, Galla H, Janshoff A. Interaction of melittin with solid supported membranes. Phys Chem Chem Phys. 2000;2:4580–4585. [Google Scholar]
- Stella L, Burattini M, Mazzuca C, Palleschi A, Venanzi M, Coin I, Peggion C, Toniolo C, Pispisa B. Alamethicin interaction with lipid membranes: A spectroscopic study on synthetic analogues. Chem Biodivers. 2007;4:1299–1312. doi: 10.1002/cbdv.200790111. [DOI] [PubMed] [Google Scholar]
- Stiopkin IV, Jayathilake HD, Bordenyuk AN, Benderskii AV. Heterodyne-detected vibrational sum frequency generation spectroscopy. J Am Chem Soc. 2008;130:2271–2275. doi: 10.1021/ja076708w. [DOI] [PubMed] [Google Scholar]
- Tadjeddine A, Peremans A. Vibrational spectroscopy of the electrochemical interface by visible infrared sum-frequency generation. Surf Sci. 1996;368:377–383. [Google Scholar]
- Tamm LK, McConnell HM. Supported phospholipid-bilayers. Biophys J. 1985;47:105–113. doi: 10.1016/S0006-3495(85)83882-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamm LK. Lateral diffusion and fluorescence microscope studies on a monoclonal-antibody specifically bound to supported phospholipidbilayers. Biochemistry. 1988;27:1450–1457. doi: 10.1021/bi00405a009. [DOI] [PubMed] [Google Scholar]
- Tamm LK, Tatulian UA. Infrared spectroscopy of proteins and peptides in lipid bilayers. Quart Rev Biophys. 1997;30:365–429. doi: 10.1017/s0033583597003375. [DOI] [PubMed] [Google Scholar]
- Tanaka M, Sackmann E. Polymer-supported membranes as models of the cell surface. Nature. 2005;437:656–663. doi: 10.1038/nature04164. [DOI] [PubMed] [Google Scholar]
- Tesmer VM, Kawano T, Shankaranarayanan A, Kozasa T, Tesmer JJG. Snapshot of Activated G Proteins at the Membrane: The Gq-GRK2-G Complex. Science. 2005;310:1686–1690. doi: 10.1126/science.1118890. [DOI] [PubMed] [Google Scholar]
- Thompson NL, Palmer AG., III Model cell membranes on planar substrates. Comments Mol Cell Biophys. 1988;5:39–56. [Google Scholar]
- Tyrode E, Johnson CM, Baldelli S, Leygraf C, Rutland MW. A vibrational sum frequency spectroscopy study of the liquid-gas interface of acetic acid-water mixtures: 2. Orientation analysis. J Phys Chem B. 2005;109:329–341. doi: 10.1021/jp047337y. [DOI] [PubMed] [Google Scholar]
- Vass E, Hollosi M, Besson F, Buchet R. Vibrational spectroscopic detection of beta-and gamma-turns in synthetic and natural peptides and proteins. Chem Rev. 2003;103:1917–1954. doi: 10.1021/cr000100n. [DOI] [PubMed] [Google Scholar]
- Vogel H. Comparison of the conformation and orientation of alamethicin and melittin in lipid-membranes. Biochemistry. 1987;26:4562–4572. doi: 10.1021/bi00388a060. [DOI] [PubMed] [Google Scholar]
- Voges AB, Stokes GY, Gibbs-Davis JM, Lettan RB, Bertin PA, Pike RC, Nguyen ST, Scheidt KA, Geiger FM. Insights into heterogeneous atmospheric oxidation chemistry: Development of a tailor-made synthetic model for studying tropospheric surface chemistry. J Phys Chem C. 2007;111:1567–1578. [Google Scholar]
- Wall MA, Coleman DE, Lee E, Iniguez-Lluhi JA, Posner BA, Gilman AG, Sprang SR. The structure of the G protein heterotrimer Gi alpha 1 beta 1 gamma 2. Cell. 1995;83:1047–1058. doi: 10.1016/0092-8674(95)90220-1. [DOI] [PubMed] [Google Scholar]
- Wang GS. NMR of membrane-associated peptides and proteins. Curr Protein Pept Sci. 2008;9:50–69. doi: 10.2174/138920308783565714. [DOI] [PubMed] [Google Scholar]
- Wang HF, Gan W, Lu R, Rao Y, Wu BH. Quantitative spectral and orientational analysis in surface sum frequency generation vibrational spectroscopy (SFG-VS) Int Rev Phys Chem. 2005;24:191–256. [Google Scholar]
- Wang J, Chen CY, Buck SM, Chen Z. Molecular chemical structure on poly(methyl methacrylate) (PMMA) surface studied by sum frequency generation (SFG) vibrational spectroscopy. J Phys Chem B. 2001;105:12118–12125. [Google Scholar]
- Wang J, Buck SM, Chen Z. The effect of surface coverage on conformation changes of bovine serum albumin molecules at the air-solution interface detected by sum frequency generation vibrational spectroscopy. Analyst. 2003;128:773–778. doi: 10.1039/b212551j. [DOI] [PubMed] [Google Scholar]
- Wang J, Even MA, Chen X, Schmaier AH, Waite JH, Chen Z. Detection of amide I signals of interfacial proteins in situ using SFG. J Am Chem Soc. 2003;125:9914–9915. doi: 10.1021/ja036373s. [DOI] [PubMed] [Google Scholar]
- Wang J, Clarke ML, Zhang YB, Chen X, Chen Z. Using isotope-labeled proteins and sum frequency generation vibrational spectroscopy to study protein adsorption. Langmuir. 2003;19:7862–7866. [Google Scholar]
- Wang J, Chen X, Clarke ML, Chen Z. Detection of chiral sum frequency generation vibrational spectra of proteins and peptides at interfaces in situ. Proc Natl Acad Sci USA. 2005;102:4978–4983. doi: 10.1073/pnas.0501206102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Clarke ML, Chen XY, Even MA, Johnson WC, Chen Z. Molecular studies on protein conformations at polymer/liquid interfaces using sum frequency generation vibrational spectroscopy. Surf Sci. 2005;587:1–11. [Google Scholar]
- Wang J, Chen X, Clarke ML, Chen Z. Vibrational spectroscopic studies on fibrinogen adsorption at polystyrene/protein solution interfaces: hydrophobic side chain and secondary structure changes. J Phys Chem B. 2006;110:5017–5024. doi: 10.1021/jp0534683. [DOI] [PubMed] [Google Scholar]
- Wang J, Paszti Z, Clarke ML, Chen XY, Chen Z. Deduction of structural information of interfacial proteins by combined vibrational spectroscopic methods. J Phys Chem B. 2007;111:6088–6095. doi: 10.1021/jp070383o. [DOI] [PubMed] [Google Scholar]
- Wang J, Lee SH, Chen Z. Quantifying the ordering of adsorbed proteins in situ. J Phys Chem B. 2008;112:2281–2290. doi: 10.1021/jp077556u. [DOI] [PubMed] [Google Scholar]
- Watry MR, Tarbuck TL, Richmond GI. Vibrational sum-frequency studies of a series of phospholipid monolayers and the associated water structure at the vapor/water interface. J Phys Chem. 2003;107:512–518. [Google Scholar]
- White RJ, Zhang B, Daniel S, Tang JM, Ervin EN, Cremer PS, White HS. Ionic conductivity of the aqueous layer separating a lipid bilayer membrane and a glass support. Langmuir. 2006;22:10777–10783. doi: 10.1021/la061457a. [DOI] [PubMed] [Google Scholar]
- Williams CT, Beattie DA. Probing buried interfaces with non-linear optical spectroscopy. Surf Sci. 2002;500:545–576. [Google Scholar]
- Woolley GA, Wallace BA. Model ion channels - gramicidin and alamethicin. J Membr Biol. 1992;129:109–136. doi: 10.1007/BF00219508. [DOI] [PubMed] [Google Scholar]
- Yang CSC, Richter LJ, Stephenson JC, Briggman KA. In situ, vibrationally resonant sum frequency Spectroscopy study of the self-assembly of dioctadecyl disulfide on gold. Langmuir. 2002;18:7549–7556. [Google Scholar]
- Yang L, Harroun TA, Weiss TM, Ding L, Huang HW. Barrel-stave model or toroidal model? A case study on melittin pores. Biophys J. 2001;81:1475–1485. doi: 10.1016/S0006-3495(01)75802-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye SJ, McClelland A, Majumdar P, Stafslien SJ, Daniels J, Chisholm B, Chen Z. Detection of tethered biocide moiety segregation to silicone surface using sum frequency generation vibrational spectroscopy. Langmuir. 2008;24:9686–9694. doi: 10.1021/la800769z. [DOI] [PubMed] [Google Scholar]
- Yeagle PL. The structure of biological membranes. Boca Raton: Fla. CRC Press; 2005. [Google Scholar]
- York RL, Holinga GJ, Guyer DR, McCrea KR, Ward RS, Somorjai GA. A new optical parametric amplifier based on lithium thioindate used for sum frequency generation vibrational spectroscopic studies of the amide I mode of an interfacial model peptide. Appl Spectrosc. 2008;62:937–1047. doi: 10.1366/000370208785793227. [DOI] [PubMed] [Google Scholar]
- Zhao J, Tamm LK. FTIR and fluorescence studies of interactions of synaptic fusion proteins in polymer-supported bilayers. Langmuir. 2003;19:1838–1846. [Google Scholar]
- Zhu XD, Suhr H, Shen YR. Surface vibrational spectroscopy by infrared-visible sum frequency generation. Phys Rev B. 1987;35:3047–3050. doi: 10.1103/physrevb.35.3047. [DOI] [PubMed] [Google Scholar]
- Zhuang X, Miranda PB, Kim D, Shen YR. Mapping molecular orientation and conformation at interfaces by surface nonlinear optics. Phys Rev B. 1999;59:12632–12640. [Google Scholar]
- Zhuang X, Shen YR. The application of nonlinear optics to the study of polymers at interfaces. Trends Polym Sci. 1996;4:258–264. [Google Scholar]