Abstract
Although myosin VI has properties that would allow it to function optimally as a dimer, full-length myosin VI exists as a monomer in isolation. Based on the ability of myosin VI monomers to dimerize when held in close proximity, we postulated that cargo binding normally regulates dimerization of myosin VI. We tested this hypothesis by expressing a known dimeric cargo adaptor protein of myosin VI, optineurin, and the myosin VI-binding segment from a monomeric cargo adaptor protein, Dab2. In the presence of these adaptor proteins, full-length myosin VI has ATPase properties of a dimer, appears as a dimer in electron micrographs, and moves processively on actin filaments. The results support a model in which cargo binding exposes internal dimerization sequences within full-length myosin VI. Because, unexpectedly, a monomeric fragment of Dab2 triggers dimerization, it would appear that myosin VI is designed to function as a dimer in cells.
Keywords: Dab2, directionality, motility, optineurin, unconventional myosin
Within the myosin superfamily there are at least 35 classes of molecular motors that move along actin filaments (1). Myosin VI is the only class of myosin known to move toward the minus-end of actin filaments (2, 3). Myosin VI dimers take large and variable steps on actin (average of 30–36 nm) using a short lever arm and a unique lever arm extension (4–6). Not only is the myosin VI dimer capable of processive movement (i.e., can move as a single molecule) along an actin filament (4–6), it also functions as a load-dependent actin anchoring protein (7). Thus myosin VI can potentially fulfill a number of specialized cell biological functions (8–10). Paradoxically, although these functional features suggest that myosin VI is designed to work in cells as a dimer, myosin VI as isolated from cells is a monomer, and expressed full-length myosin VI is also monomeric (4, 5, 11).
A number of cargo adaptor proteins that recruit myosin VI have been identified (8). For example, it has been demonstrated that optineurin is essential for myosin VI localization to the Golgi complex (12), and binds to a site within the globular tail of myosin VI. Dab2 (13, 14) and Sap97 (15) mediate the recruitment of myosin VI to clathrin-coated pits and vesicles, whereas GipC serves this role on uncoated vesicles (16, 17).
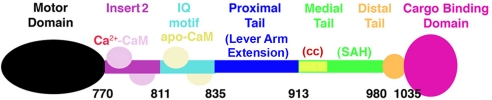
The myosin VI molecule has discrete structural domains, as diagrammed in Fig. 1, using the terminology of Spink et al. (18). We have demonstrated that internal dimerization (probably coiled coil) occurs between residues 913 and 936 (6). However, we noted that this dimerization is weak and forms only if the molecules are held in close proximity (5, 6). We postulated that in vivo dimerization is initiated upon binding of myosin VI to a dimeric cargo, which would then trigger the weak internal dimerization (5, 6).
Fig. 1.
Diagram of myosin VI structural domains. Myosin VI contains a motor domain (N terminus), followed by a short lever arm that is composed of a unique insert (insert 2), containing a calmodulin (CaM)-binding site, and an IQ motif that binds a second CaM (3). This short lever arm is followed by a region that has been referred to as the proximal tail (PT) domain, which exists as a folded three-helix bundle in myosin VI monomers and unfolds to for a lever arm extension in dimers (6). This region is followed by the medial tail (MT), which likely has a short segment of weak coiled coil (cc) followed by a stable single α-helix, or SAH (18). This is followed by the distal tail (DT) and finally by the cargo-binding domain (CBD).
Subsequently, it was shown that a headless myosin VI construct containing the entire tail and cargo-binding domains dimerized with relatively high affinity (18). Thus there may be two separate regions of the myosin VI molecule that can contribute to dimerization. Given our earlier results suggesting that the cargo-binding domain inhibits dimerization in the context of the full-length molecule (5), the collective data suggest that intramolecular interactions within the full-length myosin VI monomer, involving the cargo-binding domain, normally prevent dimerization. We hypothesize that binding to cargo must break these interactions and allow dimerization to occur. To test this hypothesis, we examined the behavior of full-length myosin VI in the presence of full-length optineurin, a cargo adaptor protein predicted to be dimeric based on sequence (19) (Swiss-Prot, accession number Q96CV9), and in the presence of a myosin VI-binding fragment from Dab2 (a monomeric adaptor protein).
Results
Optineurin Functions as a Homodimer.
Optineurin, as expressed and purified from SF9 cells, was not monodisperse and aggregated over time. We postulated that this was due either to misfolded protein or to exposure of sites normally bound to partner proteins. In either case, binding to the target, myosin VI, might select for and/or stabilize the proper protein conformation. To test this, full-length myosin VI (with a Flag tag at its N terminus) was preincubated with anti-Flag resin at low saturation. Optineurin was then flowed through the column. The rationale was that if there were properly folded optineurin dimers in the preparation, then each myosin VI tail would be able to bind to one optineurin dimer. Densitometric gel analysis of the eluted complexes showed that the molar binding ratio of optineurin to myosin VI under these conditions was 2:1 (supporting information (SI) Fig. S1A, Protocol I), demonstrating that the expressed optineurin contained a dimer fraction capable of binding to myosin VI. To see whether two myosin VI molecules could bind to each optineurin dimer, we first mixed optineurin and full-length myosin VI together and then purified the resulting complexes using the Flag affinity tag on the myosin. The observed molar binding ratio of the complexes made in this manner was 1:1, optineurin to myosin VI (Fig. S1A, Protocol II), consistent with a complex of one optineurin dimer and two bound myosin VI monomers.
Optineurin Triggers Myosin VI Dimerization.
The next question was whether binding to the dimeric optineurin triggered internal dimerization of myosin VI that allows gating of the heads and processive walking along actin filaments (5). Gating occurs when the lead head of the dimer is stalled until the rear head detaches (20), so that the ATPase activity per head is approximately half that of monomeric myosin VI.
To demonstrate that optineurin does not directly influence the ATPase activity of myosin VI, we measured the activity under conditions where the optineurin to myosin VI ratio was 2:1 (i.e., one full-length myosin VI bound to each optineurin dimer formed under Protocol I) and compared this with the activity of the full-length myosin VI monomer. Under these conditions, the myosin VI ATPase activity (Vmax) was 4.6 ± 0.13/sec per head, which is nearly identical to full-length monomeric myosin VI (Table 1). When the myosin VI/optineurin complex was formed so that two myosin VI molecules were bound to each optineurin dimer (Protocol II), the actin-activated ATPase showed a kinetic signature of myosin VI working as dimer with gating (Vmax = 2.6 ± 0.3/sec per head, Table 1), similar to the forced (zippered) dimer (Table 1). The fact that the binding of myosin VI to the optineurin dimer allows gating of the myosin VI heads is consistent with internal dimerization of the myosin VI molecule (5, 6).
Table 1.
Actin-activated ATPase activity (Vmax) and rotary shadowing electron microscopy of myosin VI constructs
| Construct (protocol) | Vmax | Dimers, % |
|---|---|---|
| FL-Myosin VI | 4.6 ± 0.1 | 0 |
| FL-Myosin VI + optineurin (Protocol I) | 4.6 ± 0.1 | N.D. |
| FL-Myosin VI + optineurin (Protocol II) | 2.6 ± 0.3 | 16.9 ± 8 |
| FL-Myosin VI + tDab2 | 3.3 ± 0.4 | 1.8 ± 0.3 |
| FL-Myosin VI + 5 μM tDab2 | 2.8 ± 0.4 | N.D. |
| Myosin VI-1050 | 4.7 ± 0.3 | 0 |
| Myosin VI-1050 + tDab2 | 4.4 ± 0.4 | N.D. |
| Myosin VI zippered dimer | 2.4 ± 0.8 | 100 |
Actin-activated ATPase: mean values (±SD) of ATP hydrolyzed head−1sec−1 for three or four independent protein preparations are shown for each construct and condition. Protocol I results in one myosin VI monomer bound to an optineurin dimer, whereas Protocol II results in two myosin VI molecules bound to an optineurin dimer. Rotary shadowing EM (mean ± SD): Data were obtained from a single spray experiment, 2–10 electron micrographs, and a total of 40–100 molecules. The following populations are significantly different (Student's t test with confidence at 95%): FL-MVI/FL-MVI + optineurin (P < 0.003); FL-MVI/FL-MVI + tDab2 (P < 0.001). FL-MVI is the full-length construct (amino acids 1–1,273).
To demonstrate that the dimers are functional, the optimal assay would be to observe the movement of single myosin VI/optineurin complexes on actin filaments. However, myosin VI binding to optineurin has been characterized as weak by using coimmunoprecipitation (12), which would preclude dilution of the complexes to the low or subnanomolar concentrations necessary for single-molecule studies. Thus, we first quantitatively measured myosin VI binding to optineurin with surface plasmon resonance. When histidine tagged optineurin was immobilized on a Ni-NTA sensor chip, myosin VI bound optineurin with a KD = 6.3 nM (Fig. S2). The previous report of weak binding (12) may have resulted from heterogeneity in the optineurin preparation, which would underestimate the affinity as determined by immunoprecipitation but not by our approach.
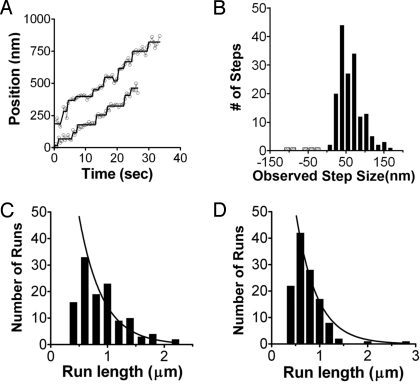
Given that the optineurin affinity for myosin VI was tight enough to allow some dimers to persist at the concentrations of single-molecule assays, we next asked whether the myosin VI/optineurin complexes could move processively on actin filaments. Full-length myosin VI was labeled with Cy3 calmodulin, and its stepping in the presence of optineurin was analyzed by using total internal reflection fluorescence microscopy (TIRF) as described below. We observed myosin VI/optineurin complexes that moved processively on actin tracks with an average (but highly variable) step size of 30.8 ± 15.3 nm (half of the observed average steps in Fig. 2B), which was similar to the previously reported value (27.6 ± 9.8 nm) for the zippered dimer step size (5). The average (from pooled data) run length was 0.90 ± 0.39 μm, and the decay constant (λ) was 0.46 μm (Fig. 2C). The decay constant was determined as previously described (21, 22) by using an exponential fit of the cumulative probability distribution of the data (see Fig. S3A), as indicated in Fig. 2C.
Fig. 2.
Movement of myosin VI/adaptor protein complexes on actin filaments. (A) Raw traces and steps defined by step-fitting for myosin VI/optineurin complexes. (B) Distribution of observed steps of Cy3-labeled full-length myosin VI bound to optineurin. Myosin VI molecules in which 10–20% of the molecules were labeled on the IQ-bound calmodulin with Cy3 were mixed with optineurin. The resulting complexes (two myosin VI monomers per optineurin dimer) displayed the step size distribution shown in the histogram. The average forward step size was 61.58 ± 30.67 nm (n = 163), which represents two steps of the center of mass (≈30 nm per step) of the myosin VI dimer. The average backward step size of full-length myosin VI was −20.4 ± 8.3 nm (n = 8). (C) Histogram of the distribution of the lengths of individuals for myosin VI/optineurin complexes. The average run length from pooled data for the myosin VI/optineurin complex was 0.90 ± 0.39 μm (n = 119). The distribution fit an exponential with a decay constant (λ) of 0.46 μm. (D) Histogram of the distribution of the lengths of individuals for myosin VI/tDab2 complexes. The average run length for the myosin VI/tDab2 complex (pooled data) was 0.75 ± 0.33 μm (n = 121). The distribution fit an exponential with a decay constant (λ) of 0.30 μm. For both optineurin and tDab2, the decay constants were determined by an exponential fit of the data cumulative probability distribution (not to the histogram distributions shown here), as previously described (21).
When full-length myosin VI was introduced into the assay at these low concentrations without optineurin, no single-molecule movement was observed. This contrasts with our previous demonstration that by clustering full-length myosin VI molecules on actin filaments at high density in rigor to increase the effective concentration before adding ATP, dimerization was induced and single-molecule movement observed (5).
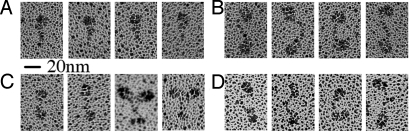
Last, we performed rotary shadowing electron microscopy on the myosin VI/optineurin complex. The electron micrographs demonstrate myosin VI dimerization in the presence of optineurin (Fig. 3B). Although the percentage of dimers was surprisingly low (≈20%) given the actin-activated ATPase results, we discovered that this is at least in part due to the destabilization of complexes by glycerol that must be added to the solutions to permit rotary shadowing. This was verified by showing that the same amount of glycerol added to the ATPase assays resulted in loss of gating. (In the presence of 10% glycerol, the actin-activated ATPase activity of the full-length construct was the same with or without optineurin: ≈4.6/sec per head).
Fig. 3.
Rotary shadowing EM of myosin VI constructs. Representative images for each myosin VI complex are shown. (A) Monomeric myosin VI appears as a single globular motor domain and a narrow lever arm. With the addition of adaptor proteins (either optineurin or truncated Dab-2) dimers are found. (B) Dimeric myosin VI/optineurin molecules are mainly found in a linear configuration with the two globular motor domains separated by ≈180° and the lever arms connected at the tail tips. The distances between heads along the dimer are slightly variable, probably due to different positions of the molecule on the mica surface. The optineurin molecule is not resolved. (C) Myosin VI dimers formed by addition of truncated Dab-2 are in two different configurations: V-shaped and linear with higher prevalence of the latter. The two differ in the position of the heads, which are located at a smaller angle in the V configuration. Note that occasionally myosin VI dimerized in the presence of tDab-2 shows two globular structures at the tail end (C, column 3), likely corresponding to tDab-2 attached to the cargo-binding region. (D) As a comparison, the myosin VI zippered dimer constructs also show two distinct globular heads connected by a common tail region and present either a linear or a V-shaped configuration (5).
Truncated Dab2 Triggers Dimerization of Myosin VI.
To avoid potential aggregation problems with full-length Dab2, we took advantage of the fact that the myosin VI-binding region of Dab2 has previously been mapped to amino acids 440–770, and the expressed fragment did not aggregate (13). Thus, we studied the interaction of this portion of Dab2 (denoted tDab2) with myosin VI. First, we repeated the mixing studies that were done for optineurin, wherein tDab2 was mixed with myosin VI, and the complex then was isolated by using an affinity tag on the myosin, or the myosin was bound to an affinity resin at low density, and the tDab2 then flowed through the column. In either case, the ratio of tDab2 to myosin VI was 1:1, consistent with monomeric tDab2 binding to a single full-length myosin VI (Fig. S1B).
Surprisingly, the tDab2/myosin VI complexes displayed gated actin-activated ATPase activity (reduction to 3.3 ± 0.4/sec per head), consistent with dimerization of myosin VI (Table 1). However, the ATPase activity was not reduced to the level of the optineurin-induced dimers or the zippered dimer, perhaps because of the previously documented weak binding affinity of the truncated Dab2 (13). An extrapolation from the monomeric (full-length) ATPase values and those of the zippered dimer would suggest that ≈60% of the heads are in dimers in the presence of tDab2. Thus, we repeated the ATPase measurements in the presence of 5 μM tDab2 to saturate the myosin-binding sites with tDab2. Under those conditions, the Vmax fell to 2.8 ± 0.3/sec per head (Table 1), similar to the value obtained with optineurin.
There was no reduction in actin-activated ATPase activity when a truncated myosin VI construct, MVI-1050 (truncated at residue 1,050 and thus lacking the cargo-binding domain), was incubated with tDab2 (Table 1). This ruled out the possibility that the myosin ATPase activity was inhibited by direct binding of tDab2 to the myosin motor domain.
To provide further evidence for dimerization of myosin VI by tDab2 binding, we performed rotary shadowing. As we had seen for optineurin, the percentage of dimers was unexpectedly low based on the gating in the ATPase activity. Again, we confirmed that glycerol abolished gating in the ATPase assays (4.6 ± 0.8/sec per head with tDab2 and 10% glycerol), explaining the low percentage of dimers in electron micrographs. Nevertheless, some dimers (≈2% of the population) were observed as shown in Fig. 3C. No dimers were observed in the case of full-length myosin VI alone, as we have previously reported (5).
We next investigated whether the individual tDab2:myosin VI complexes (1:1 ratio) could move processively on actin. We could not detect any movement, likely because of the weak binding affinity of the truncated Dab2 (13). To overcome this weak affinity, we performed single-molecule assays in the presence of a vast excess (1 μM final concentration) of tDab2. Under these conditions, myosin VI dimers were observed moving on actin tracks with an average (from pooled data) run length of 0.75 ± 0.33 μm and a decay constant (λ) of 0.30 μm (Fig. 2D).
Discussion
The results demonstrate that, upon binding to a dimeric cargo adaptor protein, optineurin, myosin VI is induced to internally dimerize, as evidenced by the gating of its actin-activated ATPase activity (Table 1) and processive movement on actin (Fig. 2 A–C). Somewhat surprisingly, binding to a monomeric cargo adaptor protein (truncated Dab2) is also able to trigger internal dimerization of myosin VI, allowing gating of the actin-activated ATPase activity (Table 1) and processive movement on actin (Fig. 2D). These results imply that in the full-length molecule, the cargo-binding domain must block dimerization. Cargo binding removes this inhibition and allows the close proximity between cargo domains from two unfolded monomers to initiate dimer formation.
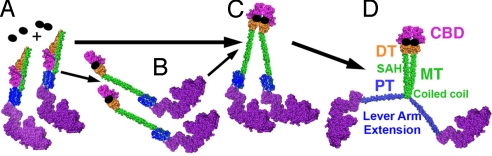
Spink et al. (18) concluded that dimerization of the full-length molecule involves only dimerization of the cargo-binding domains. However the construct that dimerized in their study contained residues 835–1,285, and the cargo-binding domain is from ≈1,035 to 1,285 (Fig. 1). Their logic was that a construct containing 835–1,035 did not dimerize and that the region of myosin VI that was originally thought to form a coiled coil has been proposed to be a stable single α-helix (SAH) (18, 23, 24). Thus, Spink et al. (18) postulated that the SAH domain forms an extension of the short myosin VI lever arm, enabling the dimer to take large steps on actin. However, we subsequently demonstrated that a lever arm extension is formed upon dimerization by unfolding of a three-helix bundle (6), formerly known as the proximal tail (Fig. 1). This unfolded bundle is immediately followed by a region that participates in dimerization (6), as illustrated in Fig. 4D. This region was present in the construct that was shown to be a dimer in the Spink et al. (18) study.
Fig. 4.
Model for cargo-mediated dimerization of myosin VI. Our working model for myosin VI function in a cell is that the full-length myosin VI molecule primarily exists as a monomer folded in such a manner as to form intramolecular interactions involving the cargo-binding domain (magenta) that block potential dimerization sites, as shown in A. This is consistent with small-angle X-ray scattering data obtained by Spink et al. (18). (B) Binding to monomeric cargo adaptors (represented by single black ovals) leads to an unfolding of the monomers, exposing potential dimerization sites. (C) The unfolded monomers can then be held in close proximity, either via tethering by the adaptor protein (25) and/or via as yet unidentified cargo-binding domain interactions. Alternatively, binding to a dimeric cargo adaptor protein (represented by black oval doublet) leads to simultaneous unfolding and close opposition of the cargo-binding domains. (D) This distal tethering of two cargo-binding domains allows internal dimerization (likely coiled coil) to occur at the proximal end of the medial tail (MT), and may include part of the last helix of the three-helix bundle (not depicted). This internal dimerization causes the three-helix bundle, formerly known as the proximal tail (PT), to unfold, forming an extension of the myosin VI lever arm (6).
While this study was under review, a publication appeared that presented the structure of a Dab2 peptide bound to a fragment of the cargo-binding domain (CBD) of myosin VI (25). The CBD-Dab2 peptide complexes dimerized, consistent with our observations. However the surprising finding was that the dimerization was not due to interactions between the cargo-binding domains themselves, but due to tethering provided by an extended Dab2 peptide binding to two different myosin VI CBDs. Although this is consistent with our current observations, it is difficult to reconcile with the observations of Spink et al. (18) that suggested that there was dimerization between the cargo-binding domains. Dimerization might occur between regions of the distal tail and the proximal portion of the CBD that were not present in the CBD fragment that was crystallized (25). Alternatively, internal dimerization within myosin VI may be limited to the region adjacent to the lever arm extension as diagrammed in Fig. 4.
It is also noteworthy that the short (39-aa) Dab2 peptide used in the work of Yu et al. (25) has a high affinity for the myosin VI CBD (≈50 nM), in contrast to our much larger tDab2 fragment. Our tDab2 contains the 39-aa peptide, but it must not be readily accessible. Within the context of full-length Dab2 bound to cargo, the peptide must be made accessible (perhaps in a regulated manner) and thus allow full-length Dab2 to bind two myosin VI monomers with high affinity (similar to what we measure for full-length optineurin).
We propose that the only model consistent with all published data are as follows (Fig. 4). First, there is at least one region within myosin VI capable of dimerizing. However, in the monomer, there are intramolecular interactions involving parts of the cargo-binding domain that inhibit this dimerization. These inhibitory interactions are destabilized by the binding of either dimeric or monomeric adaptor proteins, allowing the cargo-binding domains of two different monomers to closely associate with each other. This distal dimerization, whether it occurs simply by adaptor protein tethering, or whether it also involves an internal dimerization of a portion of the cargo-binding domain of myosin VI, allows a more proximal region to dimerize. This proximal dimerization (likely coiled-coil formation) is responsible for the dimerization seen in truncated constructs (5, 6). It is this proximal dimerization that triggers the unfolding of the three-helix bundle, creating an extension of the myosin VI lever arm (6). In this model the SAH domain serves as a spacer (Fig. 4D) and may provide interactions for folding the monomer (Fig. 4B).
We previously suggested that the cargo-binding domain inhibits dimerization based on the fact that when myosin VI molecules were clustered on actin to create a high local concentration, constructs that were truncated after residue 1,050 dimerized much more readily than full-length myosin VI (5). Therefore, we postulated that two monomeric myosin VI molecules would be induced to dimerize when they bind to a dimeric cargo. The data in this article demonstrate that internal dimerization of myosin VI can be triggered by binding to either a dimeric or a monomeric cargo adaptor protein. Given all that is known about the design of myosin VI (3–7), it clearly has been engineered to function as a dimer. With the observation that internal dimerization of myosin VI is triggered by binding to either a monomeric or dimeric cargo adaptor protein, it appears increasingly likely that myosin VI always functions as a dimer in cells.
Materials and Methods
Protein Constructs, Expression, Purification, and Actin-Activated ATPase Assays.
Full-length porcine myosin VI (amino acids 1–1,273) was constructed with an N-terminal Flag tag. Truncated porcine myosin VI lacking the cargo-binding domain (amino acids 1–1,050) was constructed with a Flag tag appended at the C terminus. The zippered dimer myosin VI construct was created by adding a GCN4 motif after amino acid 991, as previously described (4, 5). Human full-length optineurin (clone ID 3457195; Invitrogen) was expressed with a hexahistidine tag at the C terminus, allowing its purification with Ni-NTA fast-flow resin (Qiagen). These constructs were all expressed in SF9 cells (26). Truncated human Dab2, residues 440–770, was cloned into pET28b vector (Novagen) with a hexahistidine tag fused to the C terminus, and expressed in T7 ExpressIq Escherichia coli cells (New England BioLabs). Actin-activated ATPase assays were performed as previously described (27).
Electron Microscopy.
Myosins and adaptor protein specimens were prepared, and electron microscopy was performed essentially as in our previous reports (5, 6). Regions of the specimen were selected for imaging and the frequency of dimers calculated as previously reported (6). Student's t tests were performed to verify the significance of the data, by using the number of micrographs as the sample size, n. The individual profiles within each micrograph were classified into categories based on the known structure of the full-length myosin VI, which is monomeric. A single globular structure, with a visible small tail, was considered monomeric myosin VI; two globular structures within a short distance of each other (30 nm or less, center to center) were classified as a single dimeric myosin.
Surface Plasmon Resonance (SPR) Analysis.
SPR analysis was performed with a BIACORE 3000 and a Sensor Chip NTA (Biacore GE). Optineurin with a C-terminal hexahistidine tag was immobilized on the NTA chip. The KD values were determined by injecting 40 μL of wild-type myosin VI solution ranging from 0.1 to 0.5 μM at a flow rate of 10 μL/min. Curve fitting used BIAEVALUATION 3.0.1 software (Biacore GE), based on a Langmuir model.
Determination of Binding of Myosin VI to Adaptor Proteins.
Two preincubation conditions were used for binding experiments (Fig. S1). In the first condition (Protocol I), full-length myosin VI was preincubated with anti-Flag peptide resin (Sigma), and optineurin or tDab2 was added subsequently. In the second condition (Protocol II), full-length myosin VI was preincubated with optineurin or tDab2 before binding to anti-Flag peptide resin. For both conditions, complexes were eluted from resin in KMg50 (50 mM KCl, 20 mM imidazole, 1 mM EGTA, 5 mM MgCl2 at pH 7.0) with 200 μg/mL FLAG peptide and analyzed either by SDS/PAGE and densitometry or by actin-activated ATPase assays.
Single Complex Motility Assays.
A FIONA type of assay (28) was used for single step-size measurements. Both step-size and run-length assays were conducted by using the protocols described by Park et al. (5). In brief, a mixture of rabbit muscle G-actin and biotin-labeled G-actin at 10:1 ratio was polymerized, diluted 250 times in assay buffer, and introduced into a flow chamber where filaments bound to the film surface through streptavidin–biotin–BSA complexes. Unbound filaments were washed out with assay buffer. The assays were initiated by flowing in the dimerization mixture of myosin/Cy3-labeled calmodulin construct and optineurin (final dilution 7 nM) into a motility assay buffer supplemented with an oxygen scavenging system (5). Importantly, this procedure does not induce dimerization by clustering monomers in rigor on actin filaments, unlike in our previous study with full-length myosin VI alone (5). The motility was imaged by using a MultiColor Leica AM TIRF MC system. A high-sensitivity and high-speed EMCCD camera (ImagEM-CCD Camera C9100–13; Hamamatsu Corporation) was used with the system for image acquisition. The assay was carried out at 30 °C.
To analyze the data, individual trajectories were extracted from image 2sequences by means of ImageJ (National Institutes of Health, Bethesda, MD) and ImageJ plugins: ParticleTracker (29) and SpotTracker (30). A step-fitting algorithm (31) implemented in MatLab (The MathWorks) was used for step-finding and step-size measurements. The mean run length was determined by averaging pooled data. The run length distribution was also analyzed by nonlinear least-squares fitting of cumulative distribution from X0 to infinity to 1 − exp[(X0 − X/λ] as described by Thorn et al. (21). GraphPad Prism 4 (GraphPad Software, Inc.) was used for statistical analysis and data plotting.
Acknowledgments.
We thank C.F.-A. for providing facilities for electron microscopy and for sponsoring Mirko Travaglia. This work was supported by grants from the National Institutes of Health to H.L.S. and C.F.-.A.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0909748106/DCSupplemental.
References
- 1.Odronitz F, Kollmar M. Drawing the tree of eukaryotic life based on the analysis of 2,269 manually annotated myosins from 328 species. Genome Biol. 2007;8:R196. doi: 10.1186/gb-2007-8-9-r196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wells AL, Lin AW, Chen L-Q, Safer D, Cain SM, Hasson T, Carragher BO, Milligan RA, Sweeney HL. Myosin VI is an actin-based motor that moves backwards. Nature. 1999;401:505–508. doi: 10.1038/46835. [DOI] [PubMed] [Google Scholar]
- 3.Ménétrey J, et al. The structure of the myosin VI motor reveals the mechanism of directionality reversal. Nature. 2005;435:779–785. doi: 10.1038/nature03592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rock RS, et al. A flexible domain is essential for the large step size and processivity of myosin VI. Mol Cell. 2005;17:603–609. doi: 10.1016/j.molcel.2005.01.015. [DOI] [PubMed] [Google Scholar]
- 5.Park H, et al. Full-length myosin VI dimerizes and moves processively along actin filaments upon monomer clustering. Mol Cell. 2006;21:331–336. doi: 10.1016/j.molcel.2005.12.015. [DOI] [PubMed] [Google Scholar]
- 6.Mukherjea M, et al. Myosin VI dimerization triggers an unfolding of a three-helix bundle in order to extend its reach. Mol Cell. 2009;35:305–315. doi: 10.1016/j.molcel.2009.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Altman D, Sweeney HL, Spudich JA. The mechanism of myosin VI translocation and its load-induced anchoring. Cell. 2004;116:737–749. doi: 10.1016/s0092-8674(04)00211-9. [DOI] [PubMed] [Google Scholar]
- 8.Buss F, Spudich G, Kendrick-Jones J. Myosin VI: Cellular functions and motor properties. Annu Rev Cell Dev Biol. 2004;20:649–676. doi: 10.1146/annurev.cellbio.20.012103.094243. [DOI] [PubMed] [Google Scholar]
- 9.Frank DJ, Noguchi T, Miller KG. Myosin VI: Cellular functions and motor properties. Curr Opin Cell Biol. 2004;16:189–194. doi: 10.1016/j.ceb.2004.02.001. [DOI] [PubMed] [Google Scholar]
- 10.Sweeney HL, Houdusse A. What can myosin VI do in cells? Curr Opin Cell Biol. 2007;19:57–66. doi: 10.1016/j.ceb.2006.12.005. [DOI] [PubMed] [Google Scholar]
- 11.Lister I, et al. A monomeric myosin VI with a large working stroke. EMBO J. 2004;23:1729–1738. doi: 10.1038/sj.emboj.7600180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sahlender DA, et al. Optineurin links myosin VI to the Golgi complex and is involved in Golgi organization and exocytosis. J Cell Biol. 2005;169:285–295. doi: 10.1083/jcb.200501162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Inoue A, Sato O, Homma K, Ikebe M. DOC-2/DAB2 is the binding partner of myosin VI. Biochem Biophys Res Commun. 2002;292:300–307. doi: 10.1006/bbrc.2002.6636. [DOI] [PubMed] [Google Scholar]
- 14.Morris SM, et al. Myosin VI binds to and localizes with Dab2, potentially linking receptor-mediated endocytosis and the actin cytoskeleton. Traffic. 2002;3:331–341. doi: 10.1034/j.1600-0854.2002.30503.x. [DOI] [PubMed] [Google Scholar]
- 15.Osterweil E, Wells DG, Mooseker MS. A role for myosin VI in postsynaptic structure and glutamate receptor endocytosis. J Cell Biol. 2005;168:329–338. doi: 10.1083/jcb.200410091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hasson T. Myosin VI: Two distinct roles in endocytosis. J Cell Sci. 2003;116:3453–3461. doi: 10.1242/jcs.00669. [DOI] [PubMed] [Google Scholar]
- 17.Aschenbrenner L, Naccache SN, Hasson T. Uncoated endocytic vesicles require the unconventional myosin, Myo6, for rapid transport through actin barriers. Mol Biol Cell. 2004;15:2253–2263. doi: 10.1091/mbc.E04-01-0002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Spink BJ, Sivaramakrishnan S, Lipfert J, Doniach S, Spudich JA. Long single alpha-helical tail domains bridge the gap between structure and function of myosin VI. Nat Struct Mol Biol. 2008;6:591–597. doi: 10.1038/nsmb.1429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.De Marco N, Buono M, Troise F, Diez-Roux G. Optineurin increases cell survival and translocates to the nucleus in a Rab8-dependent manner upon an apoptotic stimulus. J Biol Chem. 2006;281:16147–16156. doi: 10.1074/jbc.M601467200. [DOI] [PubMed] [Google Scholar]
- 20.Sweeney HL, Park H, Zong AB, Yang Z, Selvin PR, Rosenfeld SS. How myosin VI coordinates its heads during processive movement. EMBO J. 2007;26:2682–2692. doi: 10.1038/sj.emboj.7601720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Thorn KS, Ubersax JA, Vale RA. Engineering the processive run length of the kinesin motor. J Cell Biol. 2000;151:1093–1100. doi: 10.1083/jcb.151.5.1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hodges AR, Krementsova EB, Trybus KM. Engineering the processive run length of Myosin V. J Biol Chem. 2007;282(37):27192–27197. doi: 10.1074/jbc.M703968200. [DOI] [PubMed] [Google Scholar]
- 23.Knight PJ, et al. The predicted coiled-coil domain of myosin 10 forms a novel elongated domain that lengthens the head. J Biol Chem. 2005;280:34702–34708. doi: 10.1074/jbc.M504887200. [DOI] [PubMed] [Google Scholar]
- 24.Sivaramakrishnan S, Spink BJ, Sim AY, Doniach S, Spudich JA. Dynamic charge interactions create surprising rigidity in the ER/K alpha-helical protein motif. Proc Natl Acad Sci USA. 2008;105:13356–13361. doi: 10.1073/pnas.0806256105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yu C, et al. Myosin VI undergoes cargo-mediated dimerization. Cell. 2009;138:537–548. doi: 10.1016/j.cell.2009.05.030. [DOI] [PubMed] [Google Scholar]
- 26.Sweeney HL, et al. Kinetic tuning of myosin via a flexible loop adjacent to the nucleotide-binding pocket. J Biol Chem. 1998;273:6262–6270. doi: 10.1074/jbc.273.11.6262. [DOI] [PubMed] [Google Scholar]
- 27.De La Cruz EM, Ostap EM, Sweeney HL. Kinetic mechanism and regulation of myosin VI. J Biol Chem. 2001;276:32373–32381. doi: 10.1074/jbc.M104136200. [DOI] [PubMed] [Google Scholar]
- 28.Yildiz A, Selvin P. Fluorescence imaging with one nanometer accuracy: application to molecular motors. Acc Chem Res. 2005;38:574–582. doi: 10.1021/ar040136s. [DOI] [PubMed] [Google Scholar]
- 29.Sbalzarini F, Koumoutsakos P. Feature point tracking and trajectory analysis for video imaging in cell biology. J Struct Biol. 2005;151:182–195. doi: 10.1016/j.jsb.2005.06.002. [DOI] [PubMed] [Google Scholar]
- 30.Sage D, Neumann F-R, Hediger F, Gasser S-M, Unser M. Automatic tracking of individual fluorescence particles: Application to the study of chromosome dynamics. IEEE Trans Image Process. 2005;14:1372–1383. doi: 10.1109/tip.2005.852787. [DOI] [PubMed] [Google Scholar]
- 31.Kerssemakers J-W, et al. Assembly dynamics of microtubules at molecular resolution. Nature. 2006;442:709–712. doi: 10.1038/nature04928. [DOI] [PubMed] [Google Scholar]