Abstract
Inducible nitric oxide synthase (iNOS) and cyclooxygenase 2 (COX-2) have been known to be involved in various pathophysiological processes such as inflammation. This study was performed to determine the regulatory function of superoxide dismutase (SOD) on the LPS-induced expression of iNOS, and COX-2 in RAW 264.7 cells. When a cell-permeable SOD, Tat-SOD, was added to the culture medium of RAW 264.7 cells, it rapidly entered the cells in a dose-dependent manner. Treatment of RAW 264.7 cells with Tat-SOD led to decrease in LPS-induced ROS generation. Pretreatment with Tat-SOD significantly inhibited LPS-induced expression of iNOS and NO production but had no effect on the expression of COX-2 and PGE2 production in RAW 264.7 cells. Tat-SOD inhibited LPS-induced NF-κB DNA binding activity, IκBα degradation and activation of MAP kinases. These data suggest that SOD differentially regulate expression of iNOS and COX-2 in LPS-stimulated RAW 264.7 cells.
Keywords: cyclooxygenase 2, lipopolysaccharides, NF-κB, nitric oxide synthase type II, reactive oxygen species, superoxide dismutase
Introduction
Macrophages become activated by a variety of stimuli such as LPS and participate in the process of immune responses. Activated macrophages produce various inflammatory mediators such as cytokines/chemokines, nitric oxide (NO), and prostaglandin E2 (PGE2) (Reviewed in Guha and Mackman, 2001). NO is produced from L-arginine by iNOS and PGE2 is derived form arachidonic acid metabolism by cyclooxygenase-2 (COX-2) during the inflammatory response.
Reactive oxygen species (ROS) is considered to be a causal factor in the inflammation responses induced by a variety of stimuli including LPS (Keller et al., 1999; Wang and Joseph, 1999). Cells maintain a balanced cellular redox state by using a complex ROS regulating network which is composed of antioxidant enzymes such as superoxide dismutase (SOD), catalase and glutathione peroxidase, and low molecular mass antioxidants such ascorbic acid, glutathione and tocopherols (Mates, 2000). SOD catalyzes the decomposition of superoxide to generate hydrogen peroxide that is subsequently converted to water and oxygen by glutathione peroxidase and catalase (Halliwell and Gutteridge, 1990).
Although excessive generation of ROS is cytotoxic, sub-lethal levels of ROS act as second messengers in signaling cascades involved in gene expression. ROS such as superoxide anion (O2-) and hydrogen peroxide (H2O2) directly and indirectly activates redox-sensitive transcriptional factors. Depending on the amount of ROS generated, different redox-sensitive transcription factors are activated (Reviewed in Gloire et al., 2006). Low level of ROS has been shown to induce Nrf2 transcription factor involved in expression of antioxidant enzymes (Jaiswal, 2004). An intermediate amount of ROS triggers an inflammatory response through the activation of NF-κB and AP-1 (Halliwell and Gutteridge, 1990), which are involved in expression of many pro-inflammatory genes (Schulze-Osthoff et al., 1997).
Several antioxidants were reported to exert the anti-inflammatory effects by suppressing the activity of NF-κB and the expression of proinflammatory genes (Zafarullah et al., 2003; Surh et al., 2005). However, due to broad effects of antioxidants on cellular physiology, it would be difficult to study the exact role of ROS in NF-κB activation and subsequent inflammatory gene expression. Furthermore, it is not clear whether supplementation of SOD can suppress NF-κB-dependent proinflammatory processes. To elucidate a role of superoxide dismutase in regulating inflammation processes induced by ROS, we generated a cell permeable SOD by use of HIV-1 Tat protein transduction domain (PTD) that enhances the uptake of extracellular proteins into cells (reviewed in Schwarze et al., 1999; Schwarze and Dowdy, 2000). Tat-SOD has been shown to inhibit ROS production in various types of cells under the oxidative stress (Kwon et al., 2000; Eum et al., 2004; Song et al., 2008).
In this study, we investigated the possible immuno-modulatory roles of SOD by determining its effects on LPS-induced expression of proinflammatory cytokines by RAW 264.7 cells. We observed that pretreatment of RAW 264.7 cells with Tat-SOD significantly suppressed LPS-induced iNOS expression and NO production. Unexpectedly, Tat-SOD was not capable of inhibiting LPS-induced COX-2 expression and PGE2 production under the same experimental conditions. Tat-SOD inhibited LPS-induced NF-κB DNA binding activity, IκBα degradation and activation of MAP kinases in RAW 264.7 cells. These results show that SOD selectively suppresses iNOS expression and subsequent production of NO in RAW cells. These results are the first report on the differential effect of SOD on expressions of iNOS and COX-2 in LPS stimulated macrophages.
Results
Delivery efficiency of SOD fusion proteins into RAW 264.7 cells and its effect on ROS generation by LPS
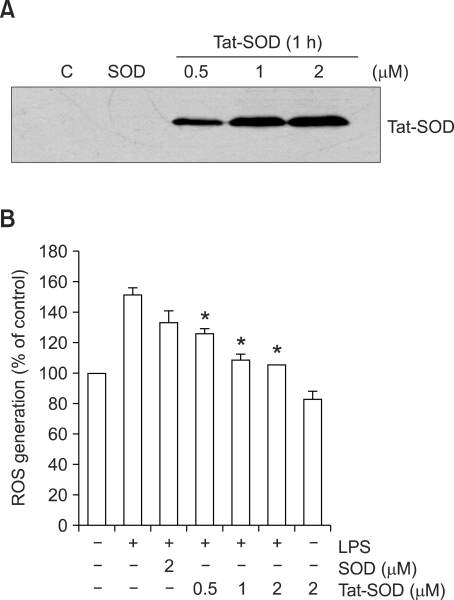
We have demonstrated that the genetic in-frame Tat PTD-superoxide dismutase (Tat-SOD) was efficiently delivered in vitro and in vivo and exerted its biological effects (Eum et al., 2004; Song et al., 2008). To evaluate the cellular uptake of SOD fusion proteins into the RAW 264.7 cells, SOD fusion proteins were incubated with cells at varying concentrations for 1 h, and the amounts of protein taken up were analyzed by immunoblot analysis. As shown in Figure 1A, Tat-SOD efficiently entered the cells in a dose-dependent manner whereas the control SOD did not. LPS, as a main component of endotoxin, can induce ROS generation in macrophage/monocyte and activate them to produce proinflammatory factors (Sanlioglu et al., 2001; Park et al., 2004). To examine the effect of SOD on ROS generation induced by LPS, RAW 264.7 cells were pretreated with SOD fusion proteins for 1 h and then exposed to LPS (100 ng/ml) for 15 min and the levels of intracellular ROS was evaluated by DCFH2-DA staining (Figure 1B). Pretreatment with Tat-SOD inhibited LPS-induced increase of intracellular ROS levels in RAW 264.7 cells whereas the control SOD had minimal effect (Figure 1B). These results indicate that Tat-SOD can efficiently remove ROS generation by LPS stimulation in RAW 264.7 cells.
Figure 1.
Delivery of SOD fusion proteins into RAW 264.7 cells and its effect on ROS generation by LPS. (A) Cellular uptake of SOD fusion proteins. RAW 264.7 cells were incubated with 0.5, 1 and 2 µM Tat-SOD or 2 µM SOD fusion proteins for 1 h. Cellular lysates were prepared for immunoblot analysis to determine the cellular uptake of SOD fusion proteins. C, untreated control cell lysates. (B) Effects of SOD fusion proteins on ROS generation in the LPS-stimulated RAW 264.7 cells. RAW 264.7 cells were treated with various concentrations of Tat-SOD or SOD for 1 h, and then stimulated with LPS (100 ng/ml) for 15 min. Intracellular ROS levels were measured after staining with a fluorescent dye, DCFH2-DA. The data are the mean ± SEM of three separate experiments. *P < 0.001 compared with cell treated with LPS alone.
Effects of SOD on LPS-induced expression of iNOS and COX-2 in RAW 264.7 cells
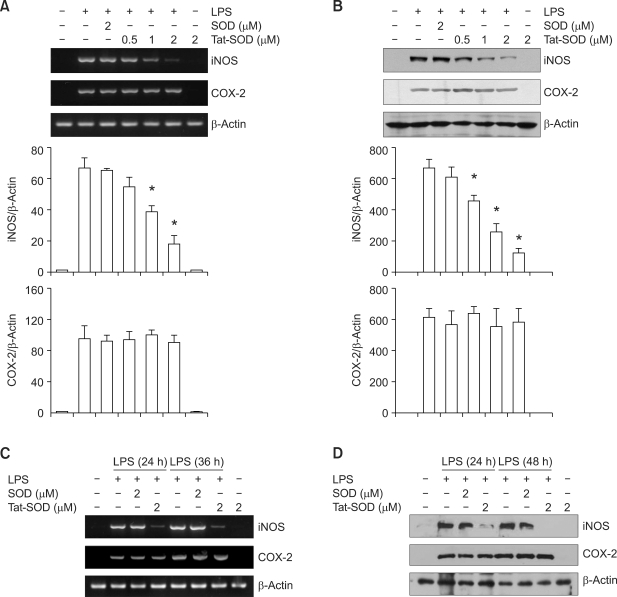
Previous studies reported that ROS was involved in LPS-induced expression of proinflammatory genes such as iNOS and COX-2 by macrophage (Kabe et al., 2005). We further used these SOD fusion proteins to analyze the effects of SOD on LPS-induced up-regulation of iNOS and COX-2 expression in RAW 264.7 cells. Cells were incubated in the absence or presence of SOD fusion proteins for 1 h, treated with LPS (100 ng/ml), and then iNOS and COX-2 expression were analyzed by RT-PCR. The level of iNOS mRNA was markedly inhibited by pretreatment with Tat-SOD, while COX-2 mRNA was not (Figure 2A). In addition, immmunoblot analysis showed that pretreatment with Tat-SOD inhibited LPS-induced expression of iNOS protein but had no effect on the expression of COX-2 protein (Figure 2B). We next performed an experiment to determine the effect of Tat-SOD on lately stage inflammatory response in time-dependent manner. As shown in Figure 2C, Tat-SOD fusion protein significantly inhibited LPS-induced mRNA expression of iNOS but not COX-2 at the 24 h and 36 h. In addition, Tat-SOD fusion protein significantly inhibited LPS-induced protein expression of iNOS but not COX-2 at the 24 h and 48 h (Figure 2D).
Figure 2.
Effects of SOD fusion proteins on LPS-induced expression of iNOS and COX-2 in RAW 264.7 cells. (A) RAW 264.7 cells were treated with 0.5, 1 and 2 µM Tat-SOD or 2 µM SOD fusion proteins for 1 h, and then exposed to LPS (100 ng/ml) for 6 h. Total RNA was extracted. iNOS, COX-2 and β-actin mRNA were analyzed by RT-PCR using specific primers. The level of mRNA detected by RT-PCR was quantified by densitometer. The data are the mean ± SEM of three separate experiments. (B) RAW 264.7 cells were treated with LPS (100 ng/ml) for 12 h with or without pretreatment with Tat-SOD or SOD fusion protein for 1 h. Cell lysates were prepared and analyzed for iNOS and COX-2 protein expression by immunoblotting. The level of proteins detected by immunoblot was quantified by densitometer. The data are the mean ± SEM of three separate experiments. *P < 0.001 compared with cell treated with LPS alone. (C) RAW 264.7 cells were treated with 0.5, 1 and 2 µM Tat-SOD or 2 µM SOD fusion proteins for 1 h, and then exposed to LPS (100 ng/ml) for 24 and 36 h. Total RNA was extracted. iNOS, COX-2 and β-actin mRNA were analyzed by RT-PCR. (D) RAW 264.7 cells pretreated with Tat-SOD or SOD fusion protein for 1 h were exposed to LPS (100 ng/ml) for 24 h and 48 h. Cell lysates were analyzed for the levels of iNOS and COX-2 protein by immunoblotting.
Effects of SOD on LPS-induced production of NO and PGE2 in RAW 264.7 cells
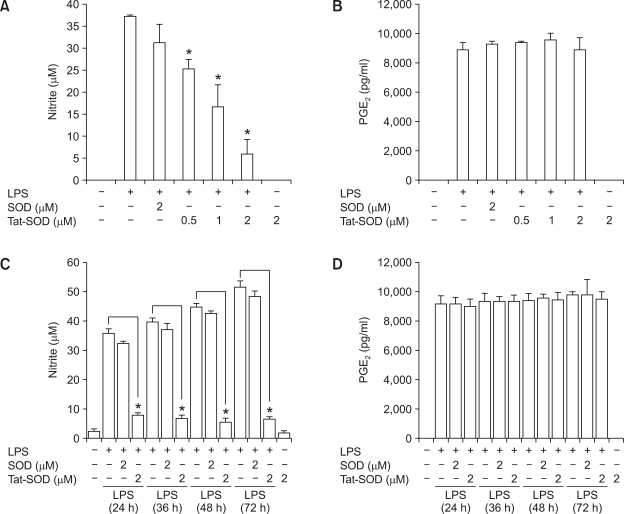
Since NO and PGE2 are products of iNOS and COX-2 enzymes, respectively, we next analyzed the effect of SOD on the production of NO and PGE2 in LPS-activated RAW 264.7 cells. Cells were pretreated with SOD fusion proteins for 1 h and then exposed to LPS for 18 h. The levels of NO and PGE2 in the culture medium were measured by using the Griess reagent system and an ELISA kit, respectively. As shown in Figure 3A, Tat-SOD significantly inhibited LPS-induced NO production in RAW 264.7 cells. However, pretreatment of Tat-SOD was not able to abrogate PGE2 production in LPS stimulated RAW 264.7 cells (Figure 3B). Tat-SOD fusion protein efficiently inhibited production of nitrate but not PGE2 in LPS stimulated RAW 264.7 cells until 72 h (Figure 3C and 3D). These results reflect the same effects of Tat-SOD on iNOS and COX-2 expression.
Figure 3.
Effects of SOD fusion proteins on production of NO and PGE2 in LPS-stimulated RAW 264.7 cells. RAW 264.7 cells were stimulated with LPS (100 ng/ml) for 18 h (A, B), 24 h, 36 h, 48 h and 72 h (C, D) with or without pretreatment with Tat-SOD or SOD fusion proteins for 1 h. Culture medium of RAW 264.7 cells was harvested. The levels of NO (A, C) and PGE2 (B, D) in the culture medium were determined using a spectrophotometric method based on the Griess reactions and an ELISA kit, respectively. The data are the mean ± SEM of three separate experiments. *P < 0.001 compared with cell treated with LPS alone.
Effects of SOD on LPS-induced activation of NF-κB
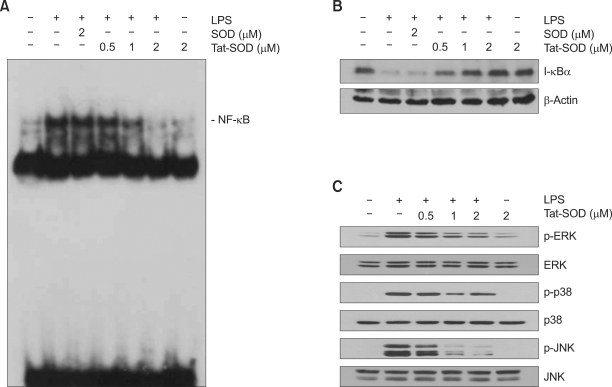
Expression of iNOS and COX-2 is regulated by different transcription factors including NF-κB in macrophage (Reviewed in Guha and Mackman, 2001). NF-κB is one of redox-dependent transcription factors (Sen and Packer, 1996; Flohe et al., 1997), and ROS was involved in LPS-induced activation of NF-κB in RAW 264.7 cells (Kabe et al., 2005). Next, we examined the effect of SOD on LPS-induced DNA binding ability of NF-κB in RAW 264.7 cells. RAW 264.7 cells were incubated in the presence of SOD fusion proteins for 1 h, followed by exposure to LPS (100 ng/ml) for 30 min. Pretreatment with Tat-SOD resulted in a decrease in LPS-induced DNA binding activity of NF-κB p65 in a dose-dependent manner (Figure 4A). Next, we examined the regulatory effect of Tat-SOD on LPS-induced signal cascade of NF-κB activation, such as IκBα degradation by immunoblot analysis using an antibody against IκBα. As shown in Figure 4B, Tat-SOD significantly inhibited LPS-induced IκBα degradation in RAW 264.7 cells, while control SOD had the minimal effect.
Figure 4.
Inhibitory effects of Tat-SOD on NF-κB activation, IκBα degradation and MAPK activation in LPS-stimulated RAW 264.7 cells. RAW 264.7 cells were treated with Tat-SOD or SOD fusion proteins for 1 h and then stimulated with LPS (100 ng/ml) for 30 min. (A) DNA-binding activity of NF-κB in the nuclear extracts of the RAW 264.7 cells was measured by EMSA. (B) Cell lysates were prepared from the RAW 264.7 cells treated with LPS and the levels of IκBα were determined by immunoblot analysis. (C) RAW 264.7 cells were incubated with Tat-SOD or SOD fusion protein for 1 h and then stimulated with LPS (100 ng/ml) for 30 min. Cell lysates were prepared and analyzed for MAP kinase protein activation by immunoblot using phospho-specific antibodies against MAPK proteins.
Inhibition of LPS-induced MAP kinase activation by Tat-SOD in RAW 264.7 cells
The MAPK signaling cascades have been shown to be involved in activation of NF-κB upon LPS stimulation (Guha and Mackman, 2001). Several lines of evidence have indicated that ROS has a regulatory role in the activity of MAP kinases such as p38, JNK and ERK protein kinase (Kyaw et al., 2004). To examine the effect of SOD on LPS induced MAPK activation, cells were incubated in the absence or presence of Tat-SOD or SOD fusion protein for 1 h, followed by exposure to LPS (100 ng/ml) for 30 min, and then MAP kinase activation was analyzed by immunoblot analysis (Figure 4C). Tat-SOD suppressed LPS-induced MAP kinase activation in a dose-dependent manner in RAW 264.7 cells.
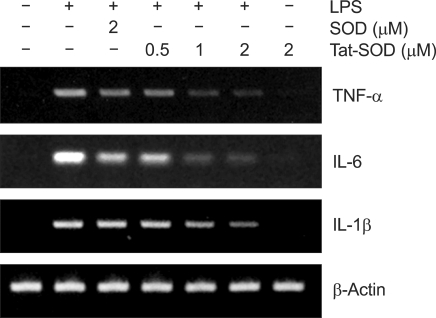
Effects of Tat-SOD on LPS-induced production of pro-inflammatory cytokines in RAW 264.7 cells
Since it has been reported that the pro-inflammatory cytokines such as TNF-α, IL-1β and IL-6 are involved in LPS-induced inflammation (Reviewed in Guha and Mackman, 2001), we further examined the effects of Tat-SOD on the expression of pro-inflammatory genes such as TNF-α, IL-1β and IL-6 in LPS-stimulated RAW 264.7 cells by RT-PCR. Pretreatment with Tat-SOD inhibited markedly LPS-induced mRNA expression of theses cytokines in a dose-dependent manner, while control SOD had a minimal effect (Figure 5). These results suggest that Tat-SOD is capable of modulating LPS-induced expression of pro-inflammatory genes through the transcriptional downregulation in LPS-stimulated RAW 264.7 cells.
Figure 5.
Inhibitory effect of Tat-SOD on LPS-induced expression of other pro-inflammatory cytokines in RAW 264.7 cells. Inhibitory effect of Tat-SOD on LPS-induced pro-inflammatory cytokines, TNF-α, IL-1β and IL-6 in the RAW 264.7 cells. Cells were stimulated with LPS (100 ng/ml) for 12 h with or without pretreatment with Tat-SOD or SOD fusion protein for 1 h. Total RNA was harvested and analyzed for TNF-α, IL-1β and IL-6 by RT-PCR.
Discussion
Since ROS play a crucial role in the inflammation processes, in this study, we examined the potential regulatory effects of SOD on the redox-regulated NF-κB signaling pathway and the expression of proinflammatory genes such as iNOS and COX-2 in the LPS-stimulated RAW 264.7 cells. Pretreatment with Tat-SOD significantly decreased protein and mRNA levels of iNOS upregulated by LPS. However, Tat-SOD had no effect on LPS-induced expression of COX-2 mRNA and protein. NO production induced by LPS was significantly suppressed by pretreatment with Tat-SOD while PGE2 production was not affected. Activation of NF-κB and MAP kinases was inhibited by Tat-SOD in LPS-stimulated RAW 264.7 cells. These experiments demonstrate differential action of SOD on iNOS and COX-2 expression in LPS stimulated RAW 264.7 cells.
Macrophages stimulated with LPS can produce ROS which can act as a second messenger to activate NF-κB signaling cascades leading to production of a wide spectrum of proinflammatory mediators, including TNF-α, IL-1β, NO and PGE2 (Schulze-Osthoff et al., 1997). Although it has been suggested that ROS is involved in LPS-induced NF-κB activation by using antioxidants, broad effects of antioxidants has limited their use in further study. Antioxidant enzymes such as SOD play a major role in controlling the intracellular redox balance (Mates, 2000). To elucidate a regulatory role of SOD in the inflammation processes induced by oxidative stress, we generated a cell-permeable SOD by exploiting HIV-1 Tat PTD peptide in the form of fusion with enzyme. Tat-SOD has been demonstrated to protect against oxidative stress in various cell types and in vivo disease models (Eum et al., 2004; Song et al., 2008). Tat-SOD significantly enhanced cellular uptake of SOD fusion protein and resulted in the efficient removal of ROS generated by LPS stimulation in RAW 264.7 cells (Figure 1).
ROS generated via the activation of NADPH oxidase (Park et al., 2004) is capable of activating NF-κB in LPS-stimulated macrophages (Gloire et al., 2006). The NF-κB signaling cascades involve the activation of the IκB-kinase (IKK) complex consisted of two kinase subunits (IKKα and IKKβ) and a regulatory subunit IKKγ/NEMO. In the presence of low or physiological levels of ROS, the IKK complex phosphorylates IκBα on Ser32 and Ser36, resulting in its ubiquitination and subsequent proteasomal degradation (Gloire et al., 2006). This allows NF-κB to translocate into the nucleus, where it activates the transcription of target genes such as proinflammatory genes. As shown in Figure 4A and 4B, pretreatment of Tat-SOD significantly inhibited IκBα degradation as well as NF-κB activation in LPS-stimulated RAW 264.7 cells, suggesting that SOD exert its effect on the NF-κB signaling cascades by removing intracellular ROS. Growing evidences support that MAPK pathway is involved in the expression of these genes by LPS (Reviewed in Guha and Mackman, 2001). Major MAPK subfamily members include ERK, JNK and p38 MAPK. Previous studies have demonstrated that ROS can activate the MAPK pathway as well as the NF-κB pathway in proinflammatory responses. Pretreatment with Tat-SOD significantly inhibited activation of MAPKs such as ERK, JNK and p38 in LPS-stimulated RAW 264.7 cells.
Although Tat-SOD significantly inhibited LPS-induced NF-κB activation, IκBα degradation and MAPK activation, it differently affected expression of iNOS and COX-2 in LPS-stimulated RAW 264.7 cells. As shown in Figure 2, expression of iNOS and NO production were significantly suppressed by Tat-SOD in LPS-stimulated RAW 264.7 cells while expression of COX-2 and PGE2 production were not. Although the signaling pathways for expression of COX-2 and iNOS are complex, these differential effects may due to the degree of dependency of iNOS and COX-2 promoters on the various transcription factors. iNOS promoter contains several cis-acting elements such as NF-κB, AP-1, C/EBPβ, and Stat, while COX-2 promoter has NF-κB, C/EBPβ, and CRE cis-acting elements (Chun and Surh, 2004; Kleinert et al., 2004). Activity of these promoters may vary according to the cell type and the stimulus applied. Our results suggest that COX-2 promoter show less dependency on NF-κB transcription factor comparing to iNOS promoter. In supporting this notion, expression of other pro-inflammatory cytokines such as TNF-α, IL-1β and IL-6, which was known to be critically dependent on the activation of NF-κB, were significantly inhibited by Tat-SOD at the transcriptional levels (Figure 5). These results suggest that coordinate activation of several transcription factors including NF-κB or activation of other transcription factors rather than NF-κB is responsible for COX-2 expression in LPS-stimulated RAW 264.7 cells. It remains to be determined whether Tat-SOD modulates other transcription factors than NF-κB in our experimental conditions.
In conclusion, this study describes the molecular effect of SOD on the redox-regulated NF-κB signaling pathway and the expression of proinflammatory genes, suggesting that SOD can modulate ROS-dependent signaling pathways during the inflammatory responses. Availability of cell-permeable SOD allowed us to unravel a putative role of SOD in the inflammation processes. Since the redoxsensitive NF-κB signaling cascade can be interrupted by regulating the levels of ROS, a cell-permeable SOD can be useful for modulating inflammation processes caused by oxidative stress.
Methods
Cell culture
Murine macrophage cell line RAW 264.7 was cultured in DMEM including 2 mM L-glutamine, 100 U/ml penicillin, 100 µg/ml streptomycin and 10% FBS 37℃ in a humidified atmosphere containing 5% CO2 and 95% air.
Reagents
2',7'-dichlobrofluorescin diacetate (DCF-DA) was purchased from Sigma (St. Louis, MO). Primary antibodies against iNOS, (BD Biosciences PharMingen, San Diego, CA), COX-2 and actin (Santa Cruz, CA), and phosphor-IκBα, phosphor-p38, phosphor-ERK, or phosphor-JNK, total-IκBα, total-p38 MAPK, total-ERK, and total-JNK (Cell Signaling Technology, Beverly, MA) were obtained commercially.
Expression and purification of SOD fusion proteins
Expression and purification of SOD fusion proteins were carried out as described previously (Song et al., 2008). SOD fusion proteins were expressed by inducing E. coli BL21 cells transformed with plasmids encoding the SOD or Tat-SOD fusion proteins with IPTG. SOD fusion proteins were purified under denaturing conditions that use a Ni++-IDA column, followed by desalting with a PD10 column (Amersham). The SOD preparation eluted from column was subjected to a Detoxi-Gel™ Endotoxin Removing Gel (Pierce, Rockford, IL) to remove endotoxin. Endotoxin levels for the SOD preparation were below the detection limit (< 0.1 EU/ml) as tested by Limulus Amoebocyte Lysate assay (BioWhitaker, Walkersville, MD). The purified SOD fusion proteins dissolved in PBS containing 20% glycerol were then aliquoted and stored at -80℃.
Immunoblot analysis
Cell lysates were prepared by incubating cells in lysis buffer (125 mM Tris-HCl pH 6.8, 2% SDS, 10% v/v glycerol) at 4℃ for 30 min. Samples of fifty µg protein were separated on a 10% SDS-polyacrylamide gel and the proteins electrotransferred to a nitrocellulose membrane, which was blocked with 10% dry milk in PBS. The membrane was probed with the indicated antibodies, and the immunoreactive bands were visualized by enhanced chemiluminescence (ECL; Amersham) as recommended by the manufacturer.
RT-PCR analysis
Total RNA was isolated from RAW 264.7 cells using a Trizol reagent kit (Invitrogen, Gaithersburg, MD). The RNA (2 µg) was converted to cDNA by treatment with 10,000 units of reverse transcriptase and 0.5 µg/µl oligo-(dT)15 primer (Promega, Madison, WI). PCR amplification of cDNA aliquots was performed with the following sense and antisense primers (5'→3'): iNOS sense, CCCTTCCGAAGTTTCTGGCAGCAGC; iNOS antisense, GGCTGTCAGAGCCTCGTGGCTTTGG; COX-2 sense, ACTCACTCAGTTTGTTGAGTCATTC; COX-2 antisense, TTTGATTAGTACTGTAGGGTTAATG; TNF-α sense, ACAAGCCTGTAGCCCACG; TNF-α antisense, TCCAAAGTAGACCTGCCC; IL-1β sense, TGCAGAGTTCCCCAACTGGTACATC; IL-1β antisense, GTGCTGCCTAATGTCCCCTTGAATC; IL-6 sense, CAAGAAAGACAAAGCCAGAGTCCTT; IL-6 antisense, TGGATGGTCTTGGTCCTTAGCC; beta-actin sense, GCGGGAAATCGTGCGTGACATT; and beta-actin antisense, GATGGAGTTGAAGGTAGTTTCGTG. PCR was performed in 50 µl of 10 mM Tris-HCl (pH 8.3), 25 mM MgCl2, 10 mM dNTP, 100 U of Taq DNA polymerase, and 0.1 µM of each primer and was terminated by heating at 70℃ for 15 min. PCR products were resolved on a 1% agarose gel and visualized with UV light after ethidium bromide.
Measurement of intracellular ROS
Intracellular ROS was measured using a ROS-sensitive fluorescent dye, DCFH2-DA. RAW 264.7 cells (1 × 105 cells) were incubated in the absence or presence of Tat-SOD or SOD fusion proteins for 1 h, and then treated with LPS (100 ng/ml) for 15 min. Cells were treated with DCFH2-DA (10 µM) for 30 min and then washed twice with PBS. The level of DCF fluorescence, reflecting the concentration of ROS, was measured at 485 nm excitation and 538 nm emission by a Fluoroskan ELISA plate reader (Labsystems Oy, Helsinki, Finland) (Song et al., 2008).
Measurement of nitrite and PGE2
RAW 264.7 cells were plated at a density of 5 × 105 cells in a 24-well cell culture plate with 500 µl of culture medium and incubated for 12 h. Cells were treated with Tat-SOD or SOD for 1 h and then stimulated with LPS (100 ng/ml) for 18 h. The amount of nitrite, the oxidized product of NO, was measured in cell culture media using the Griess reagent system (Promega). The amount of PGE2 produced was measured using an ELISA kit (R&D, Minneapolis, MI) according to the manufacturer's instructions.
Electrophoretic mobility shift assay (EMSA)
RAW 264.7 cells were treated with LPS (100 ng/ml) for 30 min, then nuclear extracts of cells were prepared and analyzed for NF-κB binding activity, as described previously (Song et al., 2007). Five micrograms of nuclear extracts were equilibrated for 15 min in binding buffer (10 mM Tris-HCl, pH 8.0, 75 mM KCl, 2.5 mM MgCl2, 0.1 mM EDTA, 10% glycerol, 0.25 mM DTT) and 1 µg of poly dI/dC. 32P-labeled oligonucleotide probe (20,000 cpm) was then added and the reaction was incubated on ice for an additional 20 min. Samples were then resolved by electrophoresis on a 6% native polyacrylamide gel in TBE buffer (89 mM Tris-HCl, 89 mM boric acid, and 2 mM EDTA). The gel was then dried and exposed to X-ray films.
Statistical analysis
Data are presented as means ± SEM. Values were evaluated by one-way ANOVA, followed by Duncan's multiple range tests using the GraphPad Prism 4.0 software (GraphPad Software, Inc., San Diego, CA). Differences were considered significant at P < 0.05.
Acknowledgments
This work was supported in part by a Next Generation Growth Engine Program Grant from the Korean Science and Engineering Foundation, and in part by a Regional Innovation Center Grant from the Korean Ministry of Commerce, Industry and Energy.
Abbreviations
- COX-2
cyclooxygenase 2
- ROS
reactive oxygen species
- SOD
superoxide dismutase
References
- 1.Chun KS, Surh YJ. Signal transduction pathways regulating cyclooxygenase-2 expression: potential molecular targets for chemoprevention. Biochem Pharmacol. 2004;68:1089–1100. doi: 10.1016/j.bcp.2004.05.031. [DOI] [PubMed] [Google Scholar]
- 2.Eum WS, Choung IS, Li MZ, Kang JH, Kim DW, Park J, Kwon HY, Choi SY. HIV-1 Tat-mediated protein transduction of Cu,Zn-superoxide dismutase into pancreatic beta cells in vitro and in vivo. Free Radic Biol Med. 2004;37:339–349. doi: 10.1016/j.freeradbiomed.2004.04.036. [DOI] [PubMed] [Google Scholar]
- 3.Flohe L, Brigelius-Flohe R, Saliou C, Traber MG, Packer L. Redox regulation of NF-kappa B activation. Free Radic Biol Med. 1997;22:1115–1126. doi: 10.1016/s0891-5849(96)00501-1. [DOI] [PubMed] [Google Scholar]
- 4.Gloire G, Legrand-Poels S, Piette J. NF-kappaB activation by reactive oxygen species: fifteen years later. Biochem Pharmacol. 2006;72:1493–1505. doi: 10.1016/j.bcp.2006.04.011. [DOI] [PubMed] [Google Scholar]
- 5.Guha M, Mackman N. LPS induction of gene expression in human monocytes. Cell Signal. 2001;13:85–94. doi: 10.1016/s0898-6568(00)00149-2. [DOI] [PubMed] [Google Scholar]
- 6.Halliwell B, Gutteridge JM. Role of free radicals and catalytic metal ions in human disease: an overview. Methods Enzymol. 1990;186:1–85. doi: 10.1016/0076-6879(90)86093-b. [DOI] [PubMed] [Google Scholar]
- 7.Jaiswal AK. Nrf2 signaling in coordinated activation of antioxidant gene expression. Free Radic Biol Med. 2004;36:1199–1207. doi: 10.1016/j.freeradbiomed.2004.02.074. [DOI] [PubMed] [Google Scholar]
- 8.Kabe Y, Ando K, Hirao S, Yoshida M, Handa H. Redox regulation of NF-κB activation: distinct redox regulation between the cytoplasm and the nucleus. Antioxidants and Redox Signaling. 2005;7:395–403. doi: 10.1089/ars.2005.7.395. [DOI] [PubMed] [Google Scholar]
- 9.Keller JN, Hanni KB, Gabbita SP, Friebe V, Mattson MP, Kindy MS. Oxidized lipoproteins increase reactive oxygen species formation in microglia and astrocyte cell lines. Brain Res. 1999;830:10–15. doi: 10.1016/s0006-8993(99)01272-x. [DOI] [PubMed] [Google Scholar]
- 10.Kleinert H, Pautz A, Linker K, Schwarz PM. Regulation of the expression of inducible nitric oxide synthase. Eur J Pharmacol. 2004;500:255–266. doi: 10.1016/j.ejphar.2004.07.030. [DOI] [PubMed] [Google Scholar]
- 11.Kwon HY, Eum WS, Jang HW, Kang JH, Ryu J, Lee BR, Ryoung LB, Jin LH, Park J, Choi SY. Transduction of Cu,Zn-superoxide dismutase mediated by an HIV-1 Tat protein basic domain into mammalian cells. FEBS Lett. 2000;485:163–167. doi: 10.1016/s0014-5793(00)02215-8. [DOI] [PubMed] [Google Scholar]
- 12.Kyaw M, Yoshizumi M, Tsuchiya K, Izawa Y, Kanematsu Y, Tamaki T. Atheroprotective effects of antioxidants through inhibition of mitogen-activated protein kinases. Acta Pharmacol Sin. 2004;25:977–985. [PubMed] [Google Scholar]
- 13.Mates JM. Effects of antioxidant enzymes in the molecular control of reactive oxygen species toxicology. Toxicology. 2000;153:83–104. doi: 10.1016/s0300-483x(00)00306-1. [DOI] [PubMed] [Google Scholar]
- 14.Park HS, Jung HY, Park EY, Kim J, Lee WJ, Bae YS. Cutting edge: direct interaction of TLR4 with NAD(P)H oxidase 4 isozyme is essential for lipopolysaccharide-induced production of reactive oxygen species and activation of NF-kappa B. J Immunol. 2004;173:3589–3593. doi: 10.4049/jimmunol.173.6.3589. [DOI] [PubMed] [Google Scholar]
- 15.Sanlioglu S, Williams CM, Samavati L, Butler NS, Wang G, McCray PB, Jr, Ritchie TC, Hunninghake GW, Zandi E, Engelhardt JF. Lipopolysaccharide induces Rac1-dependent reactive oxygen species formation and coordinates tumor necrosis factor-alpha secretion through IKK regulation of NF-kappa B. J Biol Chem. 2001;276:30188–30198. doi: 10.1074/jbc.M102061200. [DOI] [PubMed] [Google Scholar]
- 16.Schulze-Osthoff K, Bauer MK, Vogt M, Wesselborg S. Oxidative stress and signal transduction. Int J Vitam Nutr Res. 1997;67:336–342. [PubMed] [Google Scholar]
- 17.Schwarze SR, Ho A, Vocero-Akbani A, Dowdy SF. In vivo protein transduction: delivery of a biologically active protein into the mouse. Science. 1999;285:1569–1572. doi: 10.1126/science.285.5433.1569. [DOI] [PubMed] [Google Scholar]
- 18.Schwarze SR, Dowdy SF. In vivo protein transduction: intracellular delivery of biologically active proteins, compounds and DNA. Trends Pharmacol Sci. 2000;21:45–48. doi: 10.1016/s0165-6147(99)01429-7. [DOI] [PubMed] [Google Scholar]
- 19.Sen CK, Packer L. Antioxidant and redox regulation of gene transcription. FASEB J. 1996;10:709–720. doi: 10.1096/fasebj.10.7.8635688. [DOI] [PubMed] [Google Scholar]
- 20.Song HY, Ju SM, Lee JA, Kwon HJ, Eum WS, Jang SH, Choi SY, Park J. Suppression of HIV-1 Tat-induced monocyte adhesiveness by a cell-permeable superoxide dismutase in astrocytes. Exp Mol Med. 2007;39:778–786. doi: 10.1038/emm.2007.84. [DOI] [PubMed] [Google Scholar]
- 21.Song HY, Lee JA, Ju SM, Yoo KY, Won MH, Kwon HJ, Eum WS, Jang SH, Choi SY, Park J. Topical transduction of superoxide dismutase mediated by HIV-1 Tat proteintransduction domain ameliorates 12-O-tetradecanoylphorbol-13-acetate (TPA)-induced inflammation in mice. Biochem Pharmacol. 2008;75:1348–1357. doi: 10.1016/j.bcp.2007.11.015. [DOI] [PubMed] [Google Scholar]
- 22.Surh YJ, Kundu JK, Na HK, Lee JS. Redox-sensitive transcription factors as prime targets for chemoprevention with anti-inflammatory and antioxidative phytochemicals. J Nutr. 2005;135(12) Suppl:2993S–3001S. doi: 10.1093/jn/135.12.2993S. [DOI] [PubMed] [Google Scholar]
- 23.Wang H, Joseph JA. Quantifying cellular oxidative stress by dichlorofluorescein assay using microplate reader. Free Radic Biol Med. 1999;27:612–616. doi: 10.1016/s0891-5849(99)00107-0. [DOI] [PubMed] [Google Scholar]
- 24.Zafarullah M, Li WQ, Sylvester J, Ahmad M. Molecular mechanisms of N-acetylcysteine actions. Cell Mol Life Sci. 2003;60:6–20. doi: 10.1007/s000180300001. [DOI] [PMC free article] [PubMed] [Google Scholar]