Abstract
A variety of analytical techniques are used to probe the mechanisms by which RNA molecules fold to discrete three dimensional structures. Methods such as small angle X-ray scattering (SAXS) report global properties such as the overall size and shape of the RNA. Other methods such as chemical or enzymatic mapping (footprinting) report properties with resolution as fine as single nucleotide. The hydroxyl radical (•OH) is a footprinting probe which cleaves the oligonucleotide backbone independently of sequence and thus is a valuable reporter of backbone solvent accessibility. Combinations of global and local measures of folding reactions are uniquely able to distinguish specific from nonspecific processes. This article highlights the application of •OH footprinting as a complement to SAXS for kinetics analysis of RNA folding. We illustrate this combination of techniques using a study of the role played by the stiffness of a hinge in determining the rate limiting step of a Mg2+-mediated RNA folding reaction.
Keywords: RNA folding, hydroxyl radical, footprinting, SAXS
1. Introduction
In the process of adopting a functional structure, RNA must fold from a highly disordered polymer to a discrete structure. This process can be viewed from different perspectives. Information about global size and shape can be derived from biophysical techniques such as small angle X-ray scattering (SAXS) and analytical ultracentrifugation [1, 2]. The data obtained from such studies can be used to develop coarse grain models describing the global compaction of RNA molecules. However, information about the conformation of individual residues is not available from these techniques. Therefore, time-resolved SAXS studies are productively complemented by time-resolved chemical mapping (footprinting) experiments that report on local properties with as fine as single nucleotide resolution.
Footprinting describes assays which investigate ligand binding or conformational changes by monitoring the accessibility of a nucleic acid backbone to an exogenous probe. Quantitation of the accessibility is achieved by chemical and enzymatic probes which modify or cleave the nucleic acids [3, 4] such that at most each molecule is subject to a single modification or cleavage event [5]. Amongst the available footprinting probes [6] •OH radicals offer a unique combination of properties as they are highly reactive [7] and cleave the RNA backbone without nucleotide or base pairing specificity [8]. Due to the similar size of water molecules and •OH, •OH footprinting reports the solvent accessible surface of oligonucleotides. The •OH reaction products are readily analyzed by electrophoretic methods [8, 9]. RNA cleavage by •OH radicals can be adapted to reveal time-resolved information of folding processes.
Fe2+-EDTA [8, 10], radiolysis [11], UV photolysis [12–14], peroxonitrite [15], and synchrotron X-ray radiolysis [16] have been used to produce •OH for footprinting. The Fenton reaction, Fe2+ + H2O2 → Fe3+ + •OH + OH−, generates •OH from H2O2 by oxidizing Fe2+ to Fe3+, chelation of the Fe2+ by EDTA prevents its interactions with the RNA molecule. In general, •OH cleaves the phosphate backbone of oligonucleotides by abstracting the solvent accessible sugar hydrogens [17]. An implementation of the Fenton reaction that produces •OH radicals sufficient to footprint nucleic acids on a millisecond and shorter time scales is the subject of this article [18].
The latter technique termed Fast Fenton Footprinting is an inexpensive laboratory based method of time-resolved •OH footprinting. When implemented with quench-flow mixing technology this method allows fast RNA processes such as folding events to be studied with a detection limit of several milliseconds. This time scale allows the visualization of early intermediate folding species [19]. We utilize a KinTek® three-syringe mixer and readily available reagents such as H2O2, Fe(NH4)2(SO4)2, EDTA, and ethanol to conduct these studies. The single-hit kinetics conditions necessary for quantitative analysis are achieved with as little as 0.75 mM Fe2+.
RNA folding reactions studied in vitro are often initiated by the addition of cations that neutralize backbone charge and/or participate in specific interactions. In •OH footprinting experiments, radicals that cleave the oligonucleotide backbone are briefly generated after a specific folding time. The cleavage reaction is quenched by a potent radical scavenger (ethanol or thiourea) and the resultant fragments separated by denaturing gel [20] or capillary [9] electrophoresis. Quantitative analysis of the reaction products is accomplished either by SAFA (Semi-Automated Footprinting Analysis) for gel separations [20] or CAFA (Capillary Automated Footprinting Analysis) [20] or ShapeFinder [21] for capillary separations. Data processing and transformation yields time progress curves following the folding transitions with as fine as single nucleotide resolution. Clustering of progress curves to identify comparable curves can provide model independent correlations between folding and structure and is a necessary prelude to structural – kinetic modeling [19, 22]. Detailed Fast Fenton time-resolved footprinting protocols have been recently published [23–25].
This article focuses on the investigation of RNA folding by time resolved •OH footprinting conducted in coordination with time-resolved SAXS experiments. Detailed descriptions of time-resolved small-angle X-ray scattering can be found elsewhere [26–29]. Ideally both experiments should be performed at identical experimental conditions. However, each experiment has its own requirements necessitating some compromises to achieve complementarity. We discuss these compromises along with general guidelines for experimental design and data analysis so that valid comparisons and co-analysis can be made from complementary global and local measures of folding.
2. Description of method
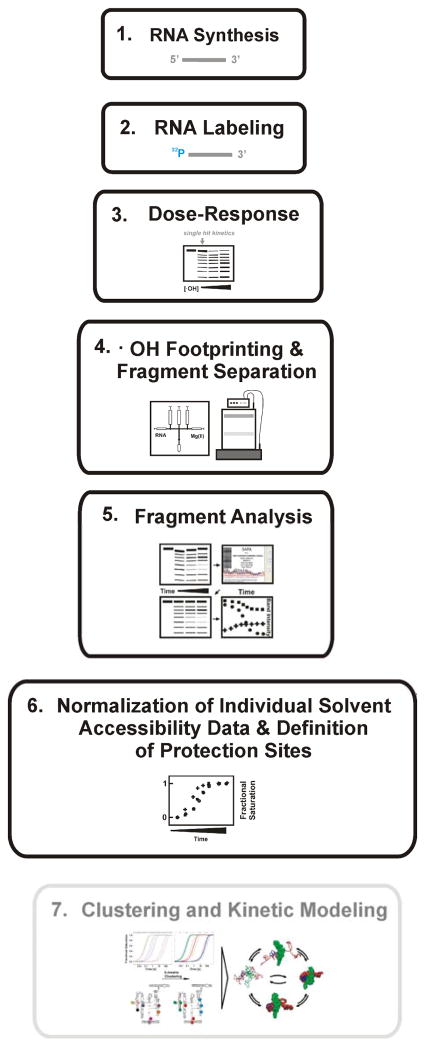
Figure 1 outlines the steps required to conduct a time-resolved •OH footprinting experiment. Each step is separately discussed in the sections that follow.
Figure 1.
A schematic diagram illustrating the general procedure for conducting time –resolved rapid - reaction •OH footprinting studies. In our example steps 1 – 6 were performed. The grey box indicates an additional tool for the development of ‘structural – kinetic’ models of complex folding reactions. The individual steps are explained in detail in the text and in recently published protocols [23, 25].
2.1 RNA samples
Sample integrity is important to both footprinting and SAXS studies. Proper handling of RNA that includes maintaining a high standard of laboratory cleanliness allows the integrity of stored samples to be maintained for many months [30]. Helpful procedures include: 1) assign a dedicated space for work with RNA; 2) always wear gloves; 3) prevent contamination of your RNA sample with ‘oral secrets’ while working with opened test tubes and reagent vessels (i.e., do not talk or cough); 4) clean laboratory benches every day and shelves every week using either detergent or a product such as RNaseZAP (Ambion); 5) exclusively use RNase free reaction tubes; 6) utilize filter pipet tips for measuring small quantities of liquid; 7) every month clean the shafts of pipets outside and the inside with water and ethanol; 8) exclusively utilize ultra pure water; 9) consider using RNase inhibitors such as RNaseOut® (Invitrogen) or SUPERase•In® (Ambion) in reaction solutions although be sure to check these additives are inert with regard to the reaction being studied; 10) insure that water bathes and circulators are free of bacterial growth by using inhibitors. Clean water bathes every month.
Ideally, sufficient RNA can be produced so that both footprinting and SAXS studies experiments can be conducted using a common batch of RNA. RNA molecules are generated by standard in vitro transcription of DNA templates [31] followed by gel purification. SAXS studies require RNA concentrations of ~15 μM. A total of ~ 50 mg of RNA may be required for a complete SAXS study. Much less material is required for •OH footprinting; the nM concentrations typically used result in a complete set of studies using less than 0.01 mg of RNA.
It should be noted that the high RNA concentrations used in time resolved SAXS experiments can lead to oligomerization if the molecules have a propensity to do so. For example, the P4–P6 domain slowly undergoes dimerization following the addition of Mg2+ [27]. Identification of an experimental time window can occur by inspection of the corresponding scattering profiles [27]. However, the low RNA concentrations used in time resolved •OH footprinting studies can minimize if not eliminate this issue, as is the case in the P4–P6 domain. Native gel electrophoresis is an easy method to evaluate oligomerization at low RNA concentrations [27].
For •OH footprinting, RNA must be labeled at one terminus to uniquely visualize the cleavage products and subsequently quantify the backbone solvent accessibility of each nucleotide. The approach most likely easiest for laboratories initiating quantitative footprinting studies is to 32P label the 5′ end of RNA’s by kinasing [32] and conduct fragment separation by denaturing gel electrophoresis. Other labeling methods for footprinting studies, including the use of fluorescence labels for capillary product separation, are discussed elsewhere [21] [9, 25]. Radio-labeled RNA is gel purified, precipitated by ethanol or isopropanol and dried. The RNA is resuspended in a buffer system compatible with ·OH footprinting and also matches, as closely as possible, the conditions used in the corresponding SAXS studies (see Section 2.3). Background RNA cleavage by trace metal ions and 32P induced radiolysis can significantly affect the signal-to-noise ration of footprinting experiments. Best results are achieved by: 1) the use of buffer systems containing EDTA; 2) RNA is resuspended in buffer just before the experiment is executed; 3) The 32P-RNA solution is stored at −70 °C; and 4) The 32P-RNA is used within one week after labeling.
2.2 Chemicals and buffers
Stock solutions for fast Fenton footprinting conducted in conjunction with SAXS include 100 mM Fe(NH4)2(SO4)2 (Sigma- Aldrich) in water (store at −70 °C), aqueous 500 mM Na2-EDTA (Ambion) (store at room temperature) and 30% H2O2 (Fluka) (store at 4 °C). 10X “SAXS buffer” (500 mM potassium [3-(N-morpholino)propanesulfonic acid, pH 7.0) and pure ethanol (Pharmco-AAPER), 1 M MgCl2 (Ambion) solution are stored at room temperature. 10X “Footprinting buffer” (180 mM potassium cacodylate, pH 7.0) is also stored at room temperature.
2.3 Choice of reaction conditions
The radiolysis of water by X-rays produces a variety of reactive species including •OH. While •OH is the boon of footprinting, it is the bane of SAXS. Therefore, SAXS experiments usually employ radical scavengers in the buffer to protect the irradiated macromolecule from degradation. It is often convenient to use a buffer such as Mops to maintain pH as well as scavenge radicals [27, 28]. In contrast, time resolved footprinting protocols seek to minimize •OH scavenging and thus extend their lifetime in solution. Cacodylate has been used to buffer numerous footprinting studies since it is unreactive with •OH [9, 19, 22, 28, 33, 34].
Since polynucleotides both attract cations and repel anions from the surrounding solution [35], achieving the same cation strength in cacodylate and MOPS buffers requires some attention [27, 28]. For example, 50 mL of 10X ‘SAXS buffer’ (500 mM potassium MOPS, pH 7.0) is generated by dissolving 5.23 g 3-(N-morpholino)propanesulfonic acid in 45 mL water, titration of the solution with 0.98 mL 10 M KOH followed by adjustment of the solution volume to 50 mL by addition of ultra pure water. The corresponding 50 mL of 10X ‘Footprinting buffer’ (180 mM potassium cacodylate, pH 7.0) is generated by titrating a solution of 0.98 mL 10 M KOH in 40 mL water with 9 mL 1 M cacodylic acid and subsequent adjustment of the volume to 50 mL by addition of ultra pure water. These resulting Footprinting and SAXS buffers have similar cation strengths.
The presence of •OH scavengers in solutions requires higher concentrations of Fe2+-EDTA and H2O2 and/or longer reaction times to achieve the RNA cleavage necessary for footprinting. Thus, it is not optimal to conduct footprinting experiments in 1X SAXS buffer. Another problem is that strong crosslinking amongst the P4–P6 RNA molecules (see section 2.8) is observed when •OH footprinting is conducted in the MOPS buffer; about 90 % of the modified RNA molecules barely entered the 15% denaturing polyacrylamide gel used to analyze them (unpublished data). The detected cleavage pattern was weak and thus generally unsuitable for quantitative analysis. However, some detectable bands could be transformed into time progress curves for the folding reaction initiated by the addition of 10 mM Mg2+; the folding rates resolved for these were identical within experimental error to those obtained in the Footprinting buffer. These results confirm the utility of using separate buffers and matching the cation concentration of cacodylate and MOPS buffers for concordant footprinting and SAXS studies.
2.4 Preparation of the three syringe quench flow apparatus
The quench-flow mixer is made RNase free utilizing the procedures described in section 2.1 and the recommendations summarized below. Our laboratory has a flow-box (Figure 2A) dedicated to RNA experiments. Prior to an experiment the drive syringes of the mixer (Figure 2, item 1) are loaded with a solution of RNaseZAP® (Ambion) and flushed three times by moving the liquid between the 5 mL fill (item 2) and drive syringes twice in sequence. Treat all loading, exit, and sample loops at least three times with RNaseZAP®. Wash all drive syringes, loops, and lines with water at least six times and subsequently with 1X footprinting buffer [23]. Use 0.5 μM non-radioactive RNA in 1X buffer to rinse all loading, sample and exit loops to minimize nonspecific binding of 32P labeled RNA to the plastic tubes and fittings. This last procedure results in the consistent recovery of expelled RNA solution. Wash all loading, sample and exit loops with 1X footprinting buffer and apply vacuum to dry the loading and reaction loops.
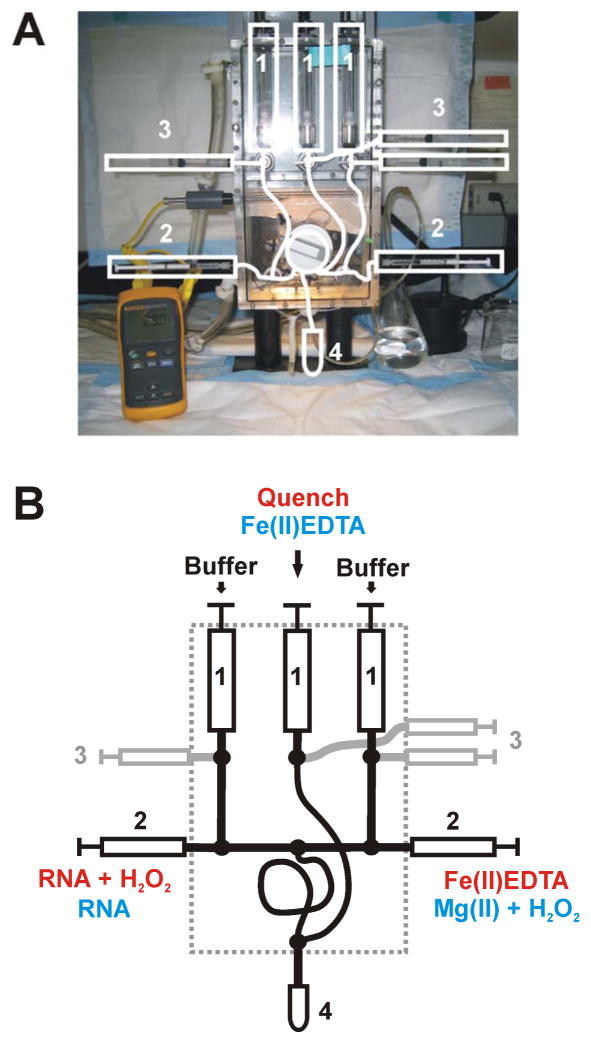
Figure 2.
Representations of a three-syringe quench flow mixer: (A) Photograph of a KinTek® RQF-3 three syringe mixer. The white lines highlight the basic components of the mixer. The numbers 1 – 3 represent the drive syringes, loading syringes, and sample syringes, respectively. The collection tube is depicted as 4. The thermometer in yellow allows continuous temperature monitoring in the box which is connected to a Nexlab RTE 111 water bath and circulator: (B) Schematic representation of the KinTek® RQF-3 three syringe mixer. This cartoon simplifies the syringe and line settings. The loading pattern for dose-response and time - resolved •OH footprinting experiments are shown in red and blue, respectively. The dashed line indicates the mixer box. All numbers refer to items as described for panel A.
2.5 The experiment
Appropriate cleavage is essential for quantitative footprinting experiments as described in the introduction. The presence of radical scavengers requires a higher concentration of Fe2+-EDTA and H2O2 and/or longer reaction times. Therefore, dose-response determinations must be conducted to achieve approximately 10% cleavage of the full length RNA in order to ensure single-hit kinetics [23]. Figure 2 shows the KinTek three-syringe mixer. Highlighted in red in panel B is the solution setup for the dose-response experiments. The sample solution and H2O2 are in the left loading syringe, the right loading syringe is filled with Fe2+-EDTA. The drive syringes are loaded with buffer- (left, right) and the quench (middle) solutions. Detailed descriptions of dose-response experiments have been published recently [23].
The loading setup for the Fast Fenton Footprinting experiment is different (Figure 2B, highlighted in blue). The left and right drive syringes are filled with 1X Footprinting buffer whereas the Fe2+-EDTA solution is loaded into the middle drive syringe. The left loading syringe contains the RNA sample in 1X Footprinting buffer, the freshly prepared initiator containing solution, in this example Footprinting buffer containing Mg2+ at double the final concentration and H2O2 at triple the final concentration.
The time resolved footprinting folding experiment is initiated by mixing the RNA solution with the initiator solution, in this case Mg2+/H2O2. After a defined aging time in the “reaction line”, the distribution of reactants and products is sampled by generating a pulse of •OH radicals by the addition of Fe2+-EDTA from the “quench syringe”. The footprinting reaction takes place within the exit tube until the reaction mixture is expelled into the collection tube containing ~ 0.4 ml absolute ethanol that quenches the footprinting reaction by scavenging the radicals. Details of this protocol have been recently published [23, 25].
The footprinted RNA is ethanol precipitated, washed with 70% ethanol and dried. The RNA pellets are resuspended in ~ 10 μl of Gel Loading Buffer II (Ambion) by shaking and 1 min incubation at 90°C to prepare them for electrophoretic separation. In parallel, a reference ladder is prepared. We recommend using RNase T1 (e.g. Fermentas) to achieve guanosine specific RNA cleavage. Use ~30% of the 32P RNA which was used for a footprinting experiment per reference lane. Incubate 32P RNA in 6.6 M urea, 20 mM sodium citrate, 1 mM EDTA, 0.25 μg/μl tRNA, 0.025 % xylene cyanol, 0.025 % bromphenol blue for 1 min at 90°C. Cool the sample on ice for 2 min and incubate at 50°C for 5 min before adding 5U RNase T1. Incubate the sample a further 25 min. Load the reference ladder immediately on the 8% denaturing polyacrylamide gel immediately followed by the footprinting samples. After electrophoresis and drying of the gel the separated RNA fragments are visualized by phosphor storage plate imaging.
2.6 Fragment analysis
Insight into the formation of specific tertiary interactions during RNA folding can be gained by separately following the change of solvent accessibility of each nucleotide. The first step in this analysis is to individually quantitate each band present on an autoradiogram. Peak fitting software such as SAFA (Semi-Automated Footprinting Analysis) simplifies the procedure tremendously [20] (Fig. 3c). This software achieves accurate gel quantification by the correction of geometric gel distortion, lane and band assignment, and an optimized band deconvolution algorithm. The output of SAFA is a spreadsheet containing columns representing lanes on the gel and rows representing the density of the individual bands corresponding to the RNA fragment. SAFA implements the automatic normalization of footprint transitions that corrects for variation in the 32P-RNA loading among the lanes of an autoradiogram. As depicted in Figure 3 (Panels B & C) some nucleotides show a distinct transition from an unprotected state to and protected state or vice versa.
Figure 3.
Example of an •OH radical footprinting experiment complementing time resolved SAXS studies [27]. RNA folding was initiated by addition of 10 mM Mg2+. Green and orange emphasize the exemplary protection sites 163–164 and 185–187, respectively. (A) Secondary structure of the wild-type P4–P6 domain of the Tetrahymena thermophila ribozyme. The yellow label indicates the 32P marked 5′ terminus of the RNA. (B) Autoradiogram of a 15% denaturing polyacrylamide gel. “T1” indicates a guanosine ladder generated by RNase T1 digest of P4–P6 RNA. The control lane shows a radio-labeled RNA that has not been exposed to •OH. From the left to the right footprinted P4–P6 RNA samples with increasing folding times were loaded. (C) Solvent accessibility of P4–P6 nucleotides after SAFA [20] analysis as false color map. Blue and red refer to decrease and increase in solvent exposure, respectively. Lines connecting panel B with C indicate the corresponding areas of the gel and the thermo-blot. (D) Time progression of the solvent accessibility of the RNA backbone. Nucleotides 163–164 and 185–187 participate in local structure formation with rate constants of 9 ± 0.8 s−1 and 10 ± 0.6 s−1, respectively.
2.7 Time progression curves
Kinetic information can be gained by creating transition curves of band intensities versus time. These transitions are individually scaled to the fractional saturation function by fi = L + (U − L) ·Ȳ where f denotes the integrated density of the band(s) being analyzed, L and U represent the lower and upper limits to the transition and Ȳ is fractional saturation (Figure 3D). In our example, L and U were determined from samples lacking MgCl2 and samples equilibrated in 10 mM MgCl2. Thus, L is the initial state and U is the final state of the reaction. At this point in the analysis, multiple data sets, rapid-mix, hand-mix and replicate experiments, are combined so that the necessary time scale is sampled and the integrity of the data insured. Typically the concurrent bands that define a unique ‘protection’ are combined for analysis [16, 36]. This combination increases the sampling of the solvent accessibility change and hence the precision of the time progress curves.
The rate constants defined by the appropriate grouping of the time progress curves are determined by global non-linear, least-squares to
| (1) |
where ai and ki are the amplitude and rate constant, respectively, of the ith kinetic phase. The reaction is the sum of the delay time plus half of the Fenton reaction time since the macromolecular transition is ongoing during Fenton footprinting. It is these time progress curves that are compared with time progress curves derived from the companion SAXS studies (Figure 3D).
2.8 Conclusions drawn from concordant footprinting and SAXS studies
The shape and size evolution of biopolymers during a folding process can be determined by SAXS experiments. The global compaction of the molecule is assessed by calculating the time-dependent radius of gyration (Rg) from the angular dependence of the X-ray scattered to the lowest angle. For example, Mg2+-mediated folding of the Tetrahymena ribozyme showed that the transition from the unfolded to the folded RNA occurs in two distinct phases [28]. The first transition is reflected by a change in Rg from 75 Å to 55 Å. During the second phase Rg decreases to 45 Å.
A question posed by these observations is whether these compaction transitions result from the formation of specific tertiary contacts or from nonspecific neutralization of backbone charge by the cation that initiates folding. It is this question that the coordinate conduct of time resolved SAXS and footprinting studies can answer. We illustrate this approach with the Tetrahymena ribozyme. Comparison of the compaction time progress curves with those following the formation of individual tertiary contacts unequivocally demonstrated that the second but not the first compaction transition is due to tertiary contact formation. Furthermore, the mapping of each footprinting time progress curve to specific nucleotides allowed structuring of the ribozyme’s P4–P6 domain to be correlated with the second transition [28].
We sought further insight into the physical nature of this process by embarking upon comparable studies of the isolated P4–P6 domain [27] which is by itself capable of folding to its native structure [37]. We asked the question: What is the rate limiting step of the folding reaction. However, before answering these questions we needed to address a consequence of the different RNA concentration requirements of the two techniques noted in Section 2.3. At the high RNA concentrations required for the acquisition of high quality SAXS time progress curves, the P4–P6 domain slowly undergoes dimerization following the addition of Mg2+ that is dependent upon the concentration of the cation. In contrast, dimerization of this RNA is undetectable at the nM concentrations utilized in •OH footprinting. We therefore needed to identify a window during which folding of the P4–P6 domain in the SAXS experiment could be followed without the complication of oligomerization. Such windows were identified and the acquisition of •OH footprinting data was concentrated within these time frames.
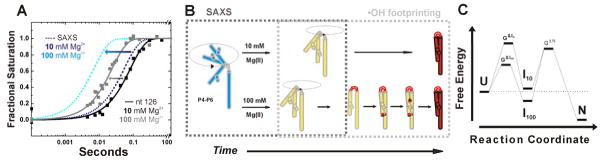
The comparison of the global and local time evolutions resolved for the P4–P6 domain is illustrated in Figure 4a. The Mg2+-dependent molecular compactions measured by SAXS data are shown as dashed lines. The solid lines refer to the local structure formation of nucleotide 126 at 10 mM (black) and 100 mM (grey) Mg2+. This reporter is located in the hinge of the domain and exhibits the overall largest folding rate within the RNA. The inspection of the curves shows that at low ionic strength global compaction is slightly faster than local structure formation at the RNA hinge. At 100 mM Mg2+ the rate of compaction far exceeds the assembly of the hinge measured by hydroxyl radicals. The analysis of rate constants and time progression curves leads to the determination of folding hierarchies at different ionic conditions and demonstrates the ionic strength dependent hinge stiffness as depicted in Figure 4b. Furthermore, SAXS and •OH footprinting of P4–P6 allows to propose the physical barriers to RNA folding as depicted in the tri-state- free-energy diagram in Figure 4c.
Figure 4.
Folding of P4-P6 RNA at 10 and 100 mM Mg2+ condensed from time resolved SAXS and complementary •OH radical footprinting studies [27]. The schemes are adapted from Schlatterer et al. [27]. (A) Assessment of the time constants for RNA compaction measured by SAXS (dashed lines) with time constant for structuring of nucleotide 126 (solid squares reflect data points with their global fit as solid lines). The SAXS data are depicted as simulated curves at 10 mM and 100 mM Mg2+ in dark blue and light blue, respectively. Nucleotide 126 is located in the P4–P6 hinge. Footprinting data of nucleotide 126 report the time dependent structuring of the RNA hinge at 10 mM and 100 mM Mg2+ in black and grey, respectively. The arrows indicate the increased compaction and hinge structuring rate at high Mg2+ concentration. (B) Schematic representation of the ionic strength dependent folding pathway. The dashed boxes in grey show the individual contributions of the complementary techniques. The initial position is flexible and extended (blue). Upon addition of 10 mM Mg2+ (top route) hinge bending results in simultaneous formation of tertiary contacts (red). In contrast, addition of 100 mM Mg2+ (bottom route) results in the sequential formation of the local structures such as hinge, tetraloop/tetraloop receptor, and A-bulge/P4 helix. (B) Findings derived from SAXS and fast Fenton footprinting experiments can suggest free energy diagrams of RNA folding. Here, the tri-state free energy diagram shows that at high ionic strength the intermediate (I100) resides at a lower free energy level than the unfolded molecule (U) and I10.
3. Concluding Remarks
Biological reactions rely on the proper folding of the biopolymers that participate in the reactions. Thus, a complete understanding of biological systems is linked to tracking the biopolymer assembly from earliest folding events such as compaction to the proper formation of intra-molecular elements. Global structure evolution of RNA molecules can be examined by techniques such as SAXS and analytical ultracentrifugation. Here, information about the local RNA confirmation is not accessible.
Time resolved hydroxyl radical footprinting directly probes the structural dynamics and folding of RNA molecules at the individual nucleotide level. Therefore, time resolved •OH footprinting has been used to complement biophysical studies such as time resolved SAXS to illuminate RNA assembly reactions from both the global and local perspectives. Such complementary studies often require flexibility in the experimental conditions. This report showcases the flexibility of the system by a study linking molecular envelop transition to local contact formation within the P4–P6 domain of the Tetrahymena ribozyme. More in depth analysis of time resolved •OH footprinting studies can yield structural - kinetic models describing the detailed routes through the folding landscape of RNA [19, 22]. A future goal for the concordant global and local analysis described in this article is to amalgamate global measures such as time – resolved SAXS into kinetic modeling in order to provide constraints on the global conformation of the intermediate species.
Acknowledgments
The writing of this article was supported by the Institute of General Medical Sciences of the National Institutes of Health grants PO1-GM066275.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Lipfert J, Doniach S. Annu Rev Biophys Biomol Struct. 2007;36:307–327. doi: 10.1146/annurev.biophys.36.040306.132655. [DOI] [PubMed] [Google Scholar]
- 2.Howlett GJ, Minton AP, Rivas G. Curr Opin Chem Biol. 2006;10:430–436. doi: 10.1016/j.cbpa.2006.08.017. [DOI] [PubMed] [Google Scholar]
- 3.Galas DJ, Schmitz A. Nucleic Acids Res. 1978;5:3157–3170. doi: 10.1093/nar/5.9.3157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Latham JA, Cech TR. Science. 1989;245:276–282. doi: 10.1126/science.2501870. [DOI] [PubMed] [Google Scholar]
- 5.Petri V, Brenowitz M. Curr Opin Biotechnol. 1997;8:36–44. doi: 10.1016/s0958-1669(97)80155-3. [DOI] [PubMed] [Google Scholar]
- 6.Giege R, Helm M, Florents C. In: Comprehensive Natural Product Chemistry. Soell D, Nishimura S, editors. Vol. 6. Pergamon Press; Oxford: 1999. pp. 63–80. [Google Scholar]
- 7.Buxton GV, Greenstock CL, Helman WP, Ross AB. J Phys Chem Ref Data. 1988;17:513–886. [Google Scholar]
- 8.Tullius TD, Dombroski BA. Proc Natl Acad Sci U S A. 1986;83:5469–5473. doi: 10.1073/pnas.83.15.5469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mitra S, Shcherbakova IV, Altman RB, Brenowitz M, Laederach A. Nucleic Acids Res. 2008;36:e63. doi: 10.1093/nar/gkn267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tullius TD, Dombroski BA. Science. 1985;230:679–681. doi: 10.1126/science.2996145. [DOI] [PubMed] [Google Scholar]
- 11.Hayes JJ, Kam L, Tullius TD. Methods Enzymol. 1990;186:545–549. doi: 10.1016/0076-6879(90)86148-o. [DOI] [PubMed] [Google Scholar]
- 12.Sharp JS, Becker JM, Hettich RL. Anal Chem. 2004;76:672–683. doi: 10.1021/ac0302004. [DOI] [PubMed] [Google Scholar]
- 13.Aye TT, Low TY, Sze SK. Anal Chem. 2005;77:5814–5822. doi: 10.1021/ac050353m. [DOI] [PubMed] [Google Scholar]
- 14.Hambly DM, Gross ML. J Am Soc Mass Spectrom. 2005;16:2057–2063. doi: 10.1016/j.jasms.2005.09.008. [DOI] [PubMed] [Google Scholar]
- 15.King PA, Anderson VE, Edwards JO, Gustafson G, Plumb RC, Suggs JW. J Am Chem Soc. 1992;114:5430–5432. [Google Scholar]
- 16.Sclavi B, Woodson S, Sullivan M, Chance MR, Brenowitz M. J Mol Biol. 1997;266:144–159. doi: 10.1006/jmbi.1996.0775. [DOI] [PubMed] [Google Scholar]
- 17.Balasubramanian B, Pogozelski WK, Tullius TD. Proc Natl Acad Sci U S A. 1998;95:9738–9743. doi: 10.1073/pnas.95.17.9738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shcherbakova I, Mitra S, Beer RH, Brenowitz M. Nucleic Acids Res. 2006;34:e48. doi: 10.1093/nar/gkl055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Laederach A, Shcherbakova I, Jonikas MA, Altman RB, Brenowitz M. Proc Natl Acad Sci U S A. 2007;104:7045–7050. doi: 10.1073/pnas.0608765104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Das R, Laederach A, Pearlman SM, Herschlag D, Altman RB. RNA. 2005;11:344–354. doi: 10.1261/rna.7214405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vasa SM, Guex N, Wilkinson KA, Weeks KM, Giddings MC. RNA. 2008;14:1979–1990. doi: 10.1261/rna.1166808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Laederach A, Shcherbakova I, Liang MP, Brenowitz M, Altman RB. J Mol Biol. 2006;358:1179–1190. doi: 10.1016/j.jmb.2006.02.075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shcherbakova I, Mitra S, Beer RH, Brenowitz M. Methods Cell Biol. 2008;84:589–615. doi: 10.1016/S0091-679X(07)84019-2. [DOI] [PubMed] [Google Scholar]
- 24.Shcherbakova I, Mitra S, Laederach A, Brenowitz M. Curr Opin Chem Biol. 2008 doi: 10.1016/j.cbpa.2008.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shcherbakova I, Brenowitz M. Nat Protoc. 2008;3:288–302. doi: 10.1038/nprot.2007.533. [DOI] [PubMed] [Google Scholar]
- 26.Lipfert J, Das R, Chu VB, Kudaravalli M, Boyd N, Herschlag D, Doniach S. J Mol Biol. 2007;365:1393–1406. doi: 10.1016/j.jmb.2006.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schlatterer JC, Kwok LW, Lamb JS, Park HY, Andresen K, Brenowitz M, Pollack L. J Mol Biol. 2008:859–870. doi: 10.1016/j.jmb.2008.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kwok LW, Shcherbakova I, Lamb JS, Park HY, Andresen K, Smith H, Brenowitz M, Pollack L. J Mol Biol. 2006;355:282–293. doi: 10.1016/j.jmb.2005.10.070. [DOI] [PubMed] [Google Scholar]
- 29.Pollack L, Doniach S. Methods Enzymol. doi: 10.1016/S0076-6879(09)69012-1. (in preparation) [DOI] [PubMed] [Google Scholar]
- 30.Schlatterer JC, Stuhlmann F, Jaschke A. Chembiochem. 2003;4:1089–1092. doi: 10.1002/cbic.200300676. [DOI] [PubMed] [Google Scholar]
- 31.Milligan JF, Groebe DR, Witherell GW, Uhlenbeck OC. Nucleic Acids Res. 1987;15:8783–8798. doi: 10.1093/nar/15.21.8783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zaug AJ, Grosshans CA, Cech TR. Biochemistry. 1988;27:8924–8931. doi: 10.1021/bi00425a008. [DOI] [PubMed] [Google Scholar]
- 33.Shcherbakova I, Gupta S, Chance MR, Brenowitz M. J Mol Biol. 2004;342:1431–1442. doi: 10.1016/j.jmb.2004.07.092. [DOI] [PubMed] [Google Scholar]
- 34.Sclavi B, Sullivan M, Chance MR, Brenowitz M, Woodson SA. Science. 1998;279:1940–1943. doi: 10.1126/science.279.5358.1940. [DOI] [PubMed] [Google Scholar]
- 35.Draper DE, Grilley D, Soto AM. Annu Rev Biophys Biomol Struct. 2005;34:221–243. doi: 10.1146/annurev.biophys.34.040204.144511. [DOI] [PubMed] [Google Scholar]
- 36.Brenowitz M, Senear DF, Shea MA, Ackers GK. Methods Enzymol. 1986;130:132–181. doi: 10.1016/0076-6879(86)30011-9. [DOI] [PubMed] [Google Scholar]
- 37.Cate JH, Gooding AR, Podell E, Zhou K, Golden BL, Kundrot CE, Cech TR, Doudna JA. Science. 1996;273:1678–1685. doi: 10.1126/science.273.5282.1678. [DOI] [PubMed] [Google Scholar]