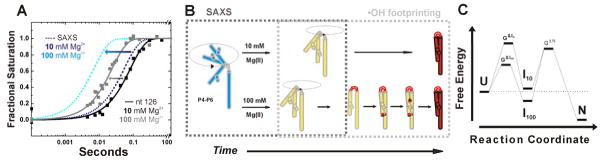
Figure 4.
Folding of P4-P6 RNA at 10 and 100 mM Mg2+ condensed from time resolved SAXS and complementary •OH radical footprinting studies [27]. The schemes are adapted from Schlatterer et al. [27]. (A) Assessment of the time constants for RNA compaction measured by SAXS (dashed lines) with time constant for structuring of nucleotide 126 (solid squares reflect data points with their global fit as solid lines). The SAXS data are depicted as simulated curves at 10 mM and 100 mM Mg2+ in dark blue and light blue, respectively. Nucleotide 126 is located in the P4–P6 hinge. Footprinting data of nucleotide 126 report the time dependent structuring of the RNA hinge at 10 mM and 100 mM Mg2+ in black and grey, respectively. The arrows indicate the increased compaction and hinge structuring rate at high Mg2+ concentration. (B) Schematic representation of the ionic strength dependent folding pathway. The dashed boxes in grey show the individual contributions of the complementary techniques. The initial position is flexible and extended (blue). Upon addition of 10 mM Mg2+ (top route) hinge bending results in simultaneous formation of tertiary contacts (red). In contrast, addition of 100 mM Mg2+ (bottom route) results in the sequential formation of the local structures such as hinge, tetraloop/tetraloop receptor, and A-bulge/P4 helix. (B) Findings derived from SAXS and fast Fenton footprinting experiments can suggest free energy diagrams of RNA folding. Here, the tri-state free energy diagram shows that at high ionic strength the intermediate (I100) resides at a lower free energy level than the unfolded molecule (U) and I10.