Abstract
Nucleostemin was first identified in neural stem cells and has become a focus of research in cell cycle control, tumorigenesis and cellular senescence. As the biology of nucleostemin begins to be unveiled in multiple species, an ensuing task is to resolve the apparent differences between the functions of mammalian and invertebrate nucleostemin and its homologues, an issue of pressing interest given the role of nucleostemin in stem cell self-renewal and tissue regeneration. A genome-wide search reveals that nucleostemin and its closest homologue, GNL3L, only emerge as separate genes in vertebrates and possess conserved protein sequences as evolution proceeded to the Mammalia. The invertebrate orthologue of nucleostemin and GNL3L resembles GNL3L more than it does nucleostemin in function, raising the idea that nucleostemin acquires new properties while GNL3L inherits an evolutionarily fixed role, and that the birth of nucleostemin may signify the appearance of new functional features in the vertebrate lineage.
Keywords: nucleostemin, GNL3L, GNL3, stem cells, regeneration
1. Introduction
Mammalian nucleostemin was originally identified as a gene enriched in neural stem cells (NSCs) and human cancer cells (Tsai and McKay, 2002), and later found abundantly expressed by several other types of stem cells (Baddoo et al., 2003, Tsai and McKay, 2002). It is functionally required for in vitro propagation of embryonic NSCs and human cancer cells (Tsai and McKay, 2002), as well as for early embryogenesis (Beekman et al., 2006, Zhu et al., 2006). Since its initial report in 2002, growing numbers of studies have begun to explore the biology of nucleostemin and nucleostemin-related genes in mammal (Baddoo et al., 2003, Beekman et al., 2006, Dai et al., 2008, Ma and Pederson, 2007, Meng et al., 2008, Ohmura et al., 2008, Zhu et al., 2006), frog (Beekman et al., 2006), newt (Maki et al., 2007), nematode (Kudron and Reinke, 2008), fruitfly (Kaplan et al., 2008), and yeast (Du et al., 2006). While an obligatory role of nucleostemin and nucleostemin-related genes in early embryogenesis was noted in all species, a critical question has yet to be resolved, i.e. how much does the mammalian nucleostemin abide by the rule set by its invertebrate counterpart?
2. Structure
2.1 Nucleostemin Family Proteins
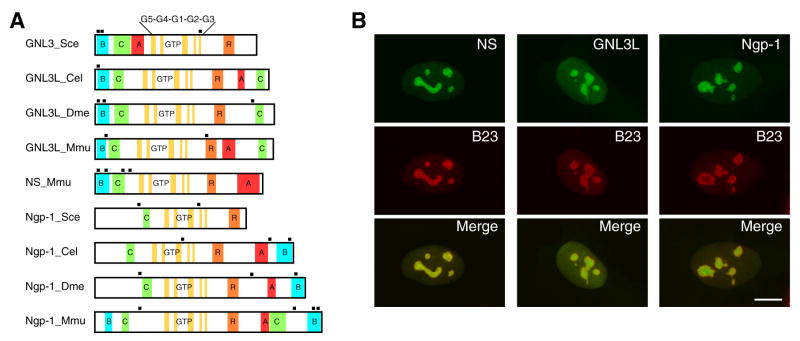
Based on protein structure, nucleostemin is categorized in the YlqF/YawG GTPase family present from prokaryotes to eukaryotes (Leipe et al., 2002), which is uniquely characterized by a MMR1_HSR1 GTP-binding domain of five GTP-binding motifs arranged in a circularly permuted order. Among the eukaryotic members of the YlqF/YawG family, nucleostemin, GNL3L (guanine nucleotide binding protein-like 3), and Ngp-1 constitute a distinct subgroup of proteins that are most closely related to each other (Fig. 1A) and localized predominantly in the nucleolus (Fig. 1B). Many YlqF/YawG proteins were shown to bind GTP, including nucleostemin, GNL3L, and Ngp-1 (Meng et al., 2006, Tsai and McKay, 2005), but few were experimentally confirmed to possess intrinsic GTPase activity.
Figure 1.
Protein structures and subcellular distributions of nucleostemin family proteins.
(A) Schematic diagrams of nucleostemin (NS), GNL3L, and Ngp-1 proteins from different species. Abbreviations: Sce, Saccharomyces cerevisiae; Cel, Caenorhabditis elegans; Dme, Drosophila melanogaster; Mmu, Mus musculus; B, basic; C, coiled-coil; GTP, GTP-binding motifs (G5*, G4, G1, G2, and G3); R, RNA-binding; A, acidic. The protein structures are the same in human as in mouse. GNL3 and Ngp-1 in S. cerevisiae are also named as Nug1p and Nog2p, respectively. (B) Subcellular distributions of NS, GNL3L and Ngp-1 in U2OS cells were shown by a C-terminally fused green fluorescent protein (green) and counterstained with anti-B23 immunofluorescence (red). Scale bar: 10 um.
2.2. Arrival of nucleostemin in vertebrate
The existence and transition of the nucleostemin subfamily genes in phylogeny is revealed by a genome-wide search of all available databases, which identifies nucleostemin and GNL3L Ngp-1 as separate gene entities in all vertebrate species. Interestingly, only one homologue (thereafter named GNL3) was found for both nucleostemin and GNL3L in the completely sequenced genomes of D. melanogaster (CG3983), C. elegans (K01C8.9), S. cerevisiae (Nug1), and S. pombi (Grn1). For the invertebrate species whose genomes are incomplete sequenced, such as A. gambiae and A. thaliana, still only one GNL3 and one Ngp-1 were identified. The protein products of GNL3 bear similar homology to human nucleostemin and GNL3L, whereas Ngp-1 clearly exists as a single gene and is highly conserved from yeast to human.
3. Expression, interaction and localization
Then comes the question of whether the gene duplication event of nucleostemin and GNL3L was accompanied by creation of distinct expression patterns and functions. In adult mammalian tissues, nucleostemin is highly expressed only in the testis (Tsai and McKay, 2002), whereas GNL3L is preferentially expressed in the cerebellum and forebrain (Yasumoto et al., 2007). During neural development, GNL3L is expressed lower in the undifferentiated NSCs than in their differentiated progeny (Tsai, unpublished data), while nucleostemin is mainly expressed by the undifferentiated NSCs and early neuroepithelial precursors (Tsai and McKay, 2002).
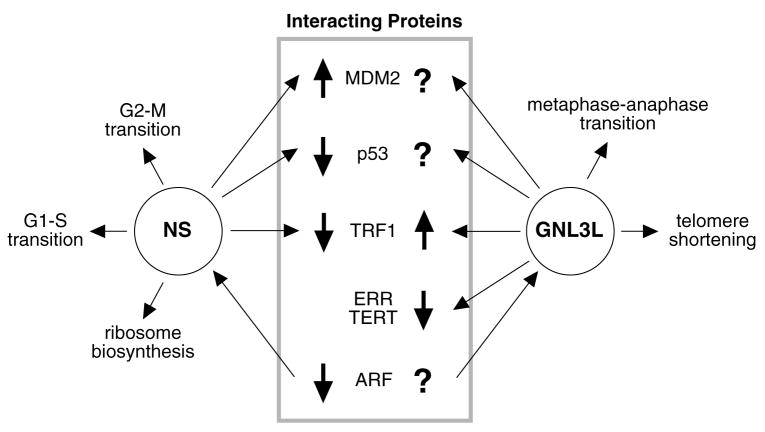
A multitude of proteins have been shown to interact with nucleostemin in mammal. Functional outcomes have been reported in p53 (Ma and Pederson, 2007, Tsai and McKay, 2002), MDM2 (mouse double minute 2) (Dai et al., 2008, Meng et al., 2008), TRF1 (telomeric repeat binding factor 1) (Zhu et al., 2006), ARF (alternative reading frame) (Ma and Pederson, 2007), and RSL1D1 (ribosomal L1 domain containing 1) (Meng et al., 2006) as a result of their interaction with nucleostemin (Fig. 2). Most nucleostemin-interacting proteins show comparable binding affinities to GNL3L. The rare exceptions are RSL1D1, which interacts only with nucleostemin (Meng et al., 2006), and the estrogen-related receptor (ERR) family proteins and TERT (telomerase reverse transcriptase), which bind only GNL3L (Fu and Collins, 2007, Yasumoto et al., 2007). RSL1D1 binding plays a role in the nucleolar accumulation of nucleostemin, and therefore the lack of interaction between GNL3L and RSL1D1 may explain the short nucleolar residence time of GNL3L (Meng et al., 2006).
Figure 2.
Distinct modes of operation for NS and GNL3L
Mammalian NS and GNL3L share similar but non-identical interacting proteins and distinct functions. Up and down thick arrows show promoting and inhibitory activities, respectively. Thin arrows point the direction of action. Question marks indicate interactions with unknown functional effect.
4. Biological function
These interactions result in functional consequences in cell cycle control and telomere maintenance (Fig. 2). Nucleostemin depletion causes defects in the G2-M and G1-S transition. GNL3L binding suppresses the transcriptional activities of ERR family proteins by competing for their coactivator binding and represses telomere elongation, suggesting that GNL3L exerts a broader activity than nucleostemin. The only evidence supporting that nucleostemin and GNL3L may exert opposite effects on a single pathway comes from their interaction with TRF1. Overexpression of nucleostemin promotes the degradation of TRF1 (Zhu et al., 2006), whereas GNL3L shows the opposite effect on TRF1 protein stability by preventing its ubiquitylation (Zhu et al., 2009).
The best proof for functional conservation and divergence of nucleostemin and GNL3L is provided by the rescue approach. So far, functional complementation between nucleostemin and GNL3L in mammalian cells has yet to be done. Rescue experiments of yeast GNL3 by human nucleostemin and GNL3L have been tried in S. pombe (Du et al., 2006). Deletion of Grn1 in S. pombe results in a slow-growth phenotype and defects in 35S rRNA processing and nucleolar export of 60S-complex. What is important is that the Grn1-null phenotype can be rescued by human GNL3L but not by nucleostemin. A similar experiment was performed in C. elegans (Kudron and Reinke, 2008), which showed that murine nucleostemin fails to rescue the GNL3/nst-1-deficient growth phenotype in C. elegans. The nematode GNL3/nst-1 (K01C8.9) is expressed by both proliferating and differentiated cells, and required for both larval growth and germline stem cell division. Another notable feature of nematode GNL3 is its relatively high nucleoplasmic distribution, which is reminiscent of the localization of mammalian GNL3L rather than that of nucleostemin.
One common feature of all invertebrate GNL3 proteins is their functional connection with the 60S ribosome complex. In S. cerevisiae, GNL3 (or Nug1p) is required for its viability and 60S ribosome export, but has little effect on the pre-rRNA processing (Bassler et al., 2001). In S. pombi, GNL3 is associated with both 35S rRNA processing and 60S-complex export (Du et al., 2006). In the nst-1 mutant C. elegans, rRNA analyses revealed a decrease of 18S and 26S rRNAs. A recent study indicates a role of nucleostemin in pre-rRNA processing (Romanova et al., 2008). A direct relationship between mammalian nucleostemin and the ribosome biosynthetic pathway awaits further clarification and needs to be reconciled with its localization in a subnucleolar compartment deficient of nascent rRNAs and 28S RNA-containing ribosomes (Politz et al., 2005). Considering that invertebrate GNL3 functionally resembles mammalian GNL3L more than it does nucleostemin, one theory would be that during the gene divergence of nucleostemin and GNL3L in the vertebrate lineage, the primary ribosomal activity of invertebrate GNL3 may have been passed on to GNL3L, thereby freeing nucleostemin for other cell type-specific functions.
5. Medical implications: Link between nucleostemin and tissue regeneration
The ability to regenerate and repair injured tissues is crucial for the viability of multicellular organisms and can be achieved by two mechanisms. One recruits a reserve population of cells, i.e. tissue-resident stem cells, capable of self-renewal and differentiation into multiple mature cell types. The other one involves cell cycle reentry of the already differentiated cells in response to injurys. On one hand, nucleostemin is linked to tissue regeneration via its association with the multipotent stem/progenitor cells. On the other hand, a recent report has shown that the expression of nucleostemin can also be reactivated in the adult cardiomyocytes in mice in response to injury signals before the cells reenter the cell cycle (Siddiqi et al., 2008). A similar observation has been made in the regenerating lens and limbs in newt (Cynops pyrrhogaster) (Maki et al., 2007). Unlike those seen in high vertebrates, regeneration in newt involves reversal (dedifferentiation) and plasticity (transdifferentiation) of the differentiated state of cells at the site of injury. Although not entirely clear, a number of genes have been shown to turn up during the dedifferentiation process. One of them, none other, is nucleostemin. In this study, it is found that nucleostemin expression is switched on in the originally differentiated pigmented epithelial cells after lentectomy, as well as in the muscle cells after limb amputation. Up-regulation of nucleostemin in those cells occurs before their S-phase reentry or expression of FGF2, Pax6, Sox2, and MafB (Maki et al., 2007). Therefore, a more fitting and inclusive view of the nucleostemin pathway is one used by either tissue-resident stem cells or differentiated cells to stay in or reenter the cell cycle, which may or may not require dedifferentiation.
The role of GNL3 in invertebrate regeneration is unclear. Of the two invertebrate species whose genome information is complete and certain to contain only one orthologue of nucleostemin and GNL3L, adult nematodes show cell constancy, and therefore are not capable of regeneration or contain stem cells outside their reproductive organ. In adult D. melanogaster, the only non-reproductive tissue-resident stem cell is found in the gut. This example by itself may argue against the appearance of nucleostemin gene as the sole determinant for the existence of tissue-resident stem cells, but further clarification is required to determine whether the invertebrate GNL3 takes part in the regenerative process of the fruitfly gut stem cells and how its activity differs from that of vertebrate nucleostemin and GNL3L, as opposed to the yeast GNL3 protein. It is possible that a regenerative role of GNL3 may have already evolved in fruitfly and higher invertebrates as a prelude to its divergence into nucleostemin and GNL3L.
The appearance of nucleostemin as a distinct gene entity represents a recent evolutionary event found only in the vertebrate lineage. Mammalian nucleostemin and GNL3L possess different biological roles. As the story of nucleostemin continues to unfold, it is our hope that one might follow the footprint it left behind and predict the future face of tissue regeneration.
Acknowledgments
We thank Thoru Pederson for his insightful comments and support from NIH R01CA113750 to R.Y.T.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Baddoo M, et al. Characterization of mesenchymal stem cells isolated from murine bone marrow by negative selection. J Cell Biochem. 2003;89:1235–49. doi: 10.1002/jcb.10594. [DOI] [PubMed] [Google Scholar]
- Bassler J, et al. Identification of a 60S preribosomal particle that is closely linked to nuclear export. Mol Cell. 2001;8:517–29. doi: 10.1016/s1097-2765(01)00342-2. [DOI] [PubMed] [Google Scholar]
- Beekman C, et al. Evolutionarily Conserved Role of Nucleostemin: Controlling Proliferation of Stem/Progenitor Cells during Early Vertebrate Development. Mol Cell Biol. 2006;26:9291–301. doi: 10.1128/MCB.01183-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai MS, Sun XX, Lu H. Aberrant expression of nucleostemin activates p53 and induces cell cycle arrest via inhibition of MDM2. Mol Cell Biol. 2008;28:4365–76. doi: 10.1128/MCB.01662-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du X, et al. The homologous putative GTPases Grn1p from fission yeast and the human GNL3L are required for growth and play a role in processing of nucleolar pre-rRNA. Mol Biol Cell. 2006;17:460–74. doi: 10.1091/mbc.E05-09-0848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu D, Collins K. Purification of human telomerase complexes identifies factors involved in telomerase biogenesis and telomere length regulation. Mol Cell. 2007;28:773–85. doi: 10.1016/j.molcel.2007.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan DD, et al. A nucleostemin family GTPase, NS3, acts in serotonergic neurons to regulate insulin signaling and control body size. Genes Dev. 2008;22:1877–93. doi: 10.1101/gad.1670508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kudron MM, Reinke V. C. elegans nucleostemin is required for larval growth and germline stem cell division. PLoS Genet. 2008;4:e1000181. doi: 10.1371/journal.pgen.1000181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leipe DD, et al. Classification and evolution of P-loop GTPases and related ATPases. J Mol Biol. 2002;317:41–72. doi: 10.1006/jmbi.2001.5378. [DOI] [PubMed] [Google Scholar]
- Ma H, Pederson T. Depletion of the nucleolar protein nucleostemin causes G1 cell cycle arrest via the p53 pathway. Mol Biol Cell. 2007;18:2630–5. doi: 10.1091/mbc.E07-03-0244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maki N, et al. Rapid accumulation of nucleostemin in nucleolus during newt regeneration. Dev Dyn. 2007;236:941–50. doi: 10.1002/dvdy.21027. [DOI] [PubMed] [Google Scholar]
- Meng L, Lin T, Tsai RY. Nucloplasmic mobilization of nucleostemin stabilizes MDM2 and promotes G2-M progression and cell survival. J Cell Sci. 2008;121:4037–46. doi: 10.1242/jcs.037952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng L, Yasumoto H, Tsai RY. Multiple controls regulate nucleostemin partitioning between nucleolus and nucleoplasm. J Cell Sci. 2006;119:5124–36. doi: 10.1242/jcs.03292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohmura M, et al. Identification of Stem Cells During Prepubertal Spermatogenesis Via Monitoring of Nucleostemin Promoter Activity. Stem Cells. 2008;26:3237–46. doi: 10.1634/stemcells.2008-0506. [DOI] [PubMed] [Google Scholar]
- Politz JC, et al. A nonribosomal landscape in the nucleolus revealed by the stem cell protein nucleostemin. Mol Biol Cell. 2005;16:3401–10. doi: 10.1091/mbc.E05-02-0106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siddiqi S, et al. Myocardial induction of nucleostemin in response to postnatal growth and pathological challenge. Circ Res. 2008;103:89–97. doi: 10.1161/CIRCRESAHA.107.169334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai RY, McKay RD. A nucleolar mechanism controlling cell proliferation in stem cells and cancer cells. Genes Dev. 2002;16:2991–3003. doi: 10.1101/gad.55671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai RY, McKay RD. A multistep, GTP-driven mechanism controlling the dynamic cycling of nucleostemin. J Cell Biol. 2005;168:179–84. doi: 10.1083/jcb.200409053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasumoto H, et al. GNL3L inhibits activity of estrogen-related receptor {gamma} by competing for coactivator binding. J Cell Sci. 2007;120:2532–43. doi: 10.1242/jcs.009878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Q, et al. GNL3L stabilizes the TRF1 complex and promotes mitotic transition. J Cell Biol. 2009;185 doi: 10.1083/jcb.200812121. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Q, Yasumoto H, Tsai RY. Nucleostemin delays cellular senescence and negatively regulates TRF1 protein stability. Mol Cell Biol. 2006;26:9279–90. doi: 10.1128/MCB.00724-06. [DOI] [PMC free article] [PubMed] [Google Scholar]