Abstract
The TALE homeodomain-containing PBC and MEIS proteins play multiple roles during metazoan development. Mutations in these proteins can cause various disorders, including cancer. In this study, we examined the roles of MEIS proteins in mesoderm development in C. elegans using the postembryonic mesodermal M lineage as a model system. We found that the MEIS protein UNC-62 plays essential roles in regulating cell fate specification and differentiation in the M lineage. Furthermore, UNC-62 appears to function together with the PBC protein CEH-20 in regulating these processes. Both unc-62 and ceh-20 have overlapping expression patterns within and outside of the M lineage, and they share physical and regulatory interactions. In particular, we found that ceh-20 is genetically required for the promoter activity of unc-62, providing evidence for another layer of regulatory interactions between MEIS and PBC proteins.
Keywords: Meis, Hth, unc-62, Pbx, Exd, ceh-20, Hox, C. elegans, mesoderm
Introduction
Hox genes encode homeodomain (HD)-containing proteins that establish body plan and specify cell fates during embryogenesis and organogenesis (McGinnis and Krumlauf, 1992; Krumlauf, 1994; Pearson et al., 2005). As monomers, Hox proteins bind to DNA with poor selectivity and affinity in vitro. However, interactions with other DNA-binding proteins can increase the DNA-binding specificity and affinity of Hox proteins in vitro and therefore facilitate Hox proteins to bind distinct in vivo biological targets. Several members of the TALE (Three-Amino-Acid Loop Extension) class of homeodomain proteins can function as Hox partners to modulate Hox-dependent transactivation in invertebrates and vertebrates (Mann and Affolter, 1998; Moens and Selleri, 2006). These Hox cofactors are further grouped into PBC and MEIS classes based on the additional motifs located N-terminal to the homeodomain (Bürglin, 1997, 1998; Mukherjee and Bürglin, 2007).
The lPBC class of homeodomain proteins is represented by the vertebrate Pbx proteins and the Drosophila Extradenticle (Exd) protein (Bürglin and Ruvkun, 1992; Rauskolb et al., 1993; Bürglin, 1997, 1998; Mukherjee and Bürglin, 2007). In addition to the homeodomain, this class of proteins contains two other conserved domains, PBC-A and PBC-B, and the PBC-A domain mediates the interaction of PBC proteins with the MEIS subfamily of proteins (Chang et al., 1997; Knoepfler et al., 1997). The MEIS class of homeodomain proteins includes the vertebrate Meis and Prep1 proteins and the Drosophila Homothorax (Hth) protein (Bürglin, 1997, 1998; Mukherjee and Bürglin, 2007). Recent studies found that both Hth and Meis1 encode two types of isoforms, one HD-containing and one HD-less (Noro et al., 2006). All MEIS proteins share two domains at their N-termini: HM1 and HM2 (also referred to as MEIS A and MEIS B), which are essential for mediating interactions between MEIS and PBC proteins (Rieckhof et al., 1997; Ryoo et al., 1999, Bürglin, 1997, 1998; Mukherjee and Bürglin, 2007).
PBC and MEIS proteins primarily modulate Hox functions by forming heterodimeric or trimeric complexes with Hox proteins on DNA (For examples, see Maconochie et al., 1997; Kroon et al., 1998; Swift et al., 1998; Ryoo et al., 1999; Shanmugam et al., 1999; Shen et al., 1999; Ferretti et al., 2000; Ebner et al., 2005). In these complexes, the PBC or MEIS proteins either directly bind DNA or serve as non-DNA binding partners to stabilize the complex. PBC and MEIS can also form stable heterodimers with each other in the presence or absence of DNA (Chang et al., 1997; Rieckhof et al., 1997; Abu-Shaar et al., 1999; Berthelsen et al., 1999; Jaw et al., 2000). In addition to their physical interaction, PBC and MEIS proteins also display mutual regulatory interactions. A number of studies have shown that MEIS proteins are required for the nuclear localization and stability of PBC proteins (Pai et al., 1998; Abu-Shaar et al., 1999; Affolter et al., 1999; Berthelsen et al., 1999; Kurant et al., 1998; Stevens and Mann, 2007). Work in Drosophila showed that in exd mutants, HTH protein levels are undetectable in the developing brain and epidermis and greatly reduced in multiple other embryonic tissues, suggesting that the maintenance of HTH protein level is dependent on EXD activity (Kurant et al., 1998; Nagao et al., 2000).
C. elegans contains both PBC and MEIS proteins. CEH-20 and two divergent members, CEH-40 and CEH-60, comprise the PBC class (Burglin, 1997, 1998; Van Auken et al., 2002; Mukherjee and Burglin, 2007). UNC-62 is the HD-containing MEIS1/hth ortholog, while PSA-3 is a HD-less variant of the PREP family that belongs to the ancient MEIS class (Burglin, 1997; Van Auken et al., 2002, Arata et al., 2006; Mukherjee and Burglin, 2007). A number of studies have shown that Hox and CEH-20 function in a complex to directly regulate downstream gene expression during mesodermal and vulval development, and in regulating apoptosis (Liu and Fire, 2000; Koh et al., 2002; Shemer and Podbilewicz, 2002; Liu et al., 2006; Takacs-Vellai et al., 2007; Potts et al., 2009). However, the roles of MEIS proteins and their functional interactions with Hox and PBC proteins in C. elegans are not well characterized. unc-62 has been shown to function during embryogenesis, Q neuroblast migration, vulval formation and VC motor neuron survival (Van Auken et al., 2002; Yang et al., 2005; Potts et al., 2009). While unc-62 interacts genetically with ceh-20 and ceh-40 during embryogenesis, these genes do not appear to function together with any of the Hox genes (Van Auken et al., 2002). Similarly, loss of unc-62 or ceh-20 activity causes Q cell migration and vulval formation defects different than those seen upon loss of Hox activity, suggesting that UNC-62 and CEH-20 may function independently of Hox proteins in regulating these processes (Yang et al., 2005). Finally, the HD-less protein PSA-3 appears to function as a non-DNA binding partner of CEH-20 and the Hox protien NOB-1 in regulating T cell asymmetric cell division (Arata et al., 2006).
We use the C. elegans postembryonic mesodermal lineage, the M lineage, as a model system to investigate the roles of MEIS proteins and their relationships with PBC and Hox proteins during mesodermal development. The C. elegans M lineage originates from a single embryonic pluripotent precursor, the M mesoblast (Sulston and Horvitz, 1977). At the first larval stage (L1) in hermaphrodites, the M lineage undergoes four to five rounds of cell division and produces 18 descendants, including two non-muscle coelomocytes (CCs), 14 striated bodywall muscles (BWMs) and two sex myoblasts (SMs) (Fig. 1A). The two SMs migrate from the posterior to the presumptive vulval region and during the L3 stage divide three times to generate 16 precursor cells that eventually differentiate into four classes of non-striated sex muscles required for egg-laying: type I and II vulval muscles (VM1 and VM2) and type I and II uterine muscles (UM1 and UM2). Vulval muscles form tubular muscles and attach to the vulva, while uterine muscles ingress, form thin sheets and attach to the uterus (Sulston and Horvitz, 1977).
Figure 1. unc-62 is required for proper development of the M lineage.
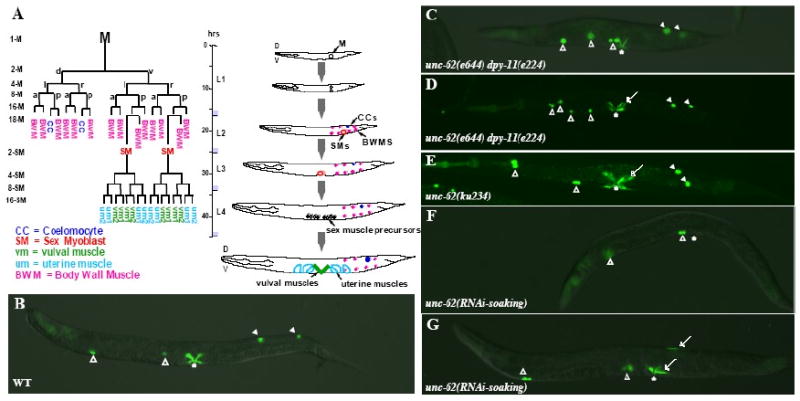
(A) The C. elegans hermaphrodite postembryonic M lineage. Times are indicated post-hatching at 25°C. The M lineage showing all differentiated cell types is shown on the left (modified from Sulston and Horvitz, 1977), while a schematic lateral view of the M lineage through larval development is shown on the right. The different stages of the M lineage are also depicted as 1-M through 16-SM. D, dorsal; V, ventral; L, left; R, right; A, anterior; P, posterior. (B-H) Wild-type (B), unc-62(e644) dpy-11(e224) (C-D), unc-62(ku234), and unc-62(RNAi-soaking) (F-G) adult animals visualized using an intrinsic CC∷gfp and egl-15∷gfp. Open arrowheads point to embryonic CCs, while solid arrowheads point to M-derived CCs. Arrows point to either extra type I vulval muscles (VM1s) in the vulval region (the vulva is denoted by an asterisk) or ectopic egl-15∷gfp-positive cells in the posterior. (C) A unc-62(e644) dpy-11(e224) animal with normal numbers of CCs and VM1s. (D) A unc-62(e644) dpy-11(e224) animal with normal numbers of CCs but extra VM1s. (E) A unc-62(ku234) animal with normal numbers of CCs but extra VM1s. (F) A unc-62(RNAi-soaking) animal with no M-derived CCs and VM1s. (G) A unc-62(RNAi-soaking) animal with no M-derived CCs and extra VM1-like cells located both in the vulval region and on the dorsal side of the animal.
Previous studies have shown that two medial group Hox genes, mab-5 and lin-39, share redundant roles in the M lineage, and that they function together with the Hox cofactor ceh-20 in regulating the specification and diversification of the M lineage (Liu and Fire, 2000). One of the direct target genes of the Hox/CEH-20 complex is the hlh-8 gene, which encodes the C. elegans Twist ortholog and is expressed in all undifferentiated cells in the M lineage (Harfe et al., 1998; Liu and Fire, 2000). The homeobox gene mls-2 is likely another direct target of CEH-20, although the activation of mls-2 by CEH-20 appears to be Hox-independent (Jiang et al., 2005; Jiang et al., 2008).
In this study, we set out to determine the functions of MEIS proteins in M lineage development. We showed that while psa-3 does not have an apparent role in the M lineage, unc-62 has multiple functions throughout M lineage development. Furthermore, unc-62 genetically interacts with ceh-20 and mab-5 to regulate the expression of downstream target genes such as hlh-8. Finally, we provide evidence that unc-62 and ceh-20 can bind to each other in the yeast two-hybrid system and that they exhibit mutual regulatory interactions: while unc-62 is required for the nuclear accumulation of CEH-20, ceh-20 is required for the promoter activity of unc-62.
Materials and Methods
C. elegans strains
Strains were manipulated under standard conditions as described by Brenner (1974). Analyses were performed at 20°C, unless otherwise noted. The wild-type reference strain was LW0081 [ccIs4438 (intrinsic CC∷gfp) III; ayIs2 (egl-15∷gfp) IV; ayIs6 (hlh-8∷gfp) X] (Jiang et al., 2005). Other M lineage-specific reporters, including Nde-box∷gfp, arg-1∷gfp, rgs-2∷gfp and mls-2∷lacZ, were as described in Kostas and Fire (2002). The mutant alleles used in this work are: LGIII, ceh-20(ay9, ay42, n2513, ok541, os39, os114), unc-36(e251) and unc-119(ed3); LGV, unc-62(e917, e644, ku234), dpy-11(e224) and him-5(e1467); LGX, psa-3(os8).
RNA interference
Plasmids to make dsRNA are: p15.121A (Wang et al., 1993) for lin-39, pJKL422.1 (Liu and Fire, 2000) for ceh-20, and yk414a1 (gift from Yuji Kohara, National Institute of Genetics, Japan) for unc-62. For mab-5 RNAi, primers JKL-601 and JKL-602 were used to amplify the insert in the mab-5 RNAi clone III-4I09 from the Ahringer library (Kamath et al., 2003). dsRNA synthesis was following the protocol of Fire and colleagues (Fire et al., 1998). For ceh-20, mab-5 or lin-39 RNAi, dsRNA was injected into gravid adults of different genotypes. Progeny of injected animals were scored for M lineage and vulval defects. Water-injected animals were used as controls. RNAi soaking was used for unc-62(RNAi-soaking) by modifying the soaking protocol described in Ahringer (2006). Embryos and the subsequent newly hatched L1s were soaked in dsRNA against unc-62 or in soaking buffer alone for 24-48 hours at 20°C. The soaked L1 worms were transferred to NGM plates with OP50 and followed through development for M lineage phenotypes or gfp expression.
Transgenic animals and mls-1∷lacZ expression
2.9 kb of the unc-62 promoter sequence (-2883 to −1 exon 1a) were amplified using iProof DNA polymerase (Bio-Rad Laboratories) using genomic DNA as template and YJ-178 and YJ-179 as primers. The PCR products were used to generate a reporter construct, pYJ167 (-2.9 kb unc-62p∷gfp∷let-858 3′ UTR). pJKL790 and pJKL791 are unc-62 rescuing constructs that contain the unc-62 promoter driving unc-62 1a-7b and unc-62 1b-7b cDNAs respectively.
pHK110 (a gift from Thomas Burglin, Karolinska Institutet, Sweden) and pJKL775 were used for analyzing ceh-20 expression pattern. pHK110 contains 2.4 kb of the ceh-20 promoter sequence (-2385 to −1), the entire ceh-20 coding region fused with gfp sequence at the C-terminus, and the unc-54 3′ UTR. pJKL775 is a transcriptional reporter of ceh-20 with nls∷gfp under the control of the ceh-20 promoter (-2385 to −1).
Reporters were microinjected with the plasmid pRF4 (Mello et al., 1991) or the pha-1 rescuing plasmid pC1 (Granato et al., 1994) as markers using standard techniques. LW1175 was a spontaneous integrant of pHK110. pSAK534 (mls-1∷lacZ) (Kostas and Fire, 2002) was microinjected with pRF4 and pJKL449.1(myo-2∷gfp) into N2 worms using standard techniques (Mello et al., 1991). Animals transgenic for jjEx[pSAK534.1(mls-1∷lacZ)+rol-6(su1006)+ pJKL449.1(myo-2∷gfp)] were crossed to animals of unc-62(ku234)V; ayIs6 (hlh-8∷gfp) X to generate unc-62(ku234)V; ayIs6 (hlh-8∷gfp) X; jjEx[pSAK534.1(mls-1∷lacZ)+rol-6(su1006)+ pJKL449.1(myo-2∷gfp)] animals.. L3 larvae were fixed following the protocol of Harfe and colleagues (Harfe et al., 1998) and used for immunostaining. The following antibodies were used: goat anti-GFP antibodies (Rockland Immunochemicals) (1:5000), mouse anti-β-galactosidase antibodies (Promega) (1:50), Cy3-conjugated donkey anti-mouse antibodies (Jackson Immunoresearch Laboratories) (1:100) and FITC-conjugated donkey anti-goat antibodies (Jackson Immunoresearch Laboratories) (1:100).
Yeast two-hybrid assay
Two-hybrid analysis was performed using the protocol described by James and colleagues (James et al., 1996). The following plasmids were used in the two-hybrid assay:
pYJ188: ceh-20 aa1-196 in pGAD-C2
pJKL841: ceh-20 aa1-196 in pGBD-C2
pJKL842: unc-62 isoform 1a7b aa1-287 in pGBD-C2
pJKL843: unc-62 isoform 1a7b aa1-287 in pGAD-C2
pJKL844: unc-62 isoform 1b7b aa1-250 in pGAD-C1
pJKL845: unc-62 isoform 1b7b aa1-250 in pGBD-C1
Plasmids were transformed into the yeast strain PJ69-4A and selected for on -Trp (pYJ188, pJKL843 and pJKL844) or -Leu (pJKL841, pJKL842 and pJKL845) plates. Transformed strains were tested for auto-activation (growth) on -Ade plates. Transformed yeast colonies were mated in liquid culture and plated in serial dilutions on control (-Leu -Trp) and selection (-Leu -Trp -Ade) plates and grown at 30°C for 2-3 days before being scored for growth.
Results
The Meis protein UNC-62 plays multiple roles in the postembryonic M lineage
C. elegans contains two Meis-related genes, unc-62 and psa-3 (Van Auken et al., 2002 and Arata et al., 2006). We first examined the role of psa-3 in the M lineage. Neither psa-3(RNAi) animals nor mutant animals homozygous for a loss-of-function psa-3(os8) allele (Arata et al., 2006) exhibited any M lineage defects, suggesting that psa-3 is not likely to play a role in M lineage development. In contrast, animals with reduced levels of unc-62 displayed multiple M lineage defects.
There are multiple unc-62 alleles available, and the majority of them cause either zygotic or maternal-effect embryonic lethality (Van Auken et al., 2002). However, two weak zygotic alleles, ku234 and e644, can produce viable progeny. ku234 has a point mutation specifically affecting the start codon of the 1b isoforms, changing the first amino acid from Met to Ile, while e644 has an early stop codon in the homeodomain region encoded by exon 7b (Van Auken et al., 2002). We introduced the M lineage specific GFP markers into ku234 and e644 mutants and examined their M lineage phenotypes.
The early M lineage appeared completely normal in ku234 mutants, producing two SMs, two CCs and fourteen BWMs (Table 1, Figure 1E). Just like in wild-type animals, the two SMs in ku234 mutants migrated to the presumptive vulval region and underwent three rounds of division to produce 16 hlh-8∷gfp-positive precursor cells. However, unlike wild-type animals in which the 16 sex muscle precursors differentiate into 8 vulval muscles (4 VM1s and 4 VM2s) and 8 uterine muscles (4 UM1s and 4 UM2s), in 100% (n=200) of ku234 mutants examined, the majority (ranging between 9 and 14) of the 16 sex muscle precursor cells differentiated into VM1-like cells that expressed egl-15∷gfp (Fig. 1E, compare Figure 2A-B). These extra VM1-like cells appeared to be a result of fate transformation from UMs and VM2s to VM1s based on the following observations. First, in wild-type animals, NdE-box∷gfp labels all 16 differentiated sex muscles while highlighting the distinct cell morphologies of VMs and UMs (Harfe et al., 1998; Fig. 2C). ku234 mutants had 16 Nde-box∷gfp-expressing cells; however, there was an increase of VM-like cells and a concomitant decrease of UM-like cells (Fig. 2D). Second, rgs-2∷gfp, which preferentially labels UMs but also faintly labels VMs in wild-type animals (Kostas and Fire, 2002; Fig. 2E), showed an increase of the faint VM-like signal and a decrease of the bright UM-like signal in ku234 mutants (Fig. 2F). Finally, all the eight vulval muscles (both VM1s and VM2s) in wild-type animals express arg-1∷gfp (Kostas and Fire, 2002; Fig. 2G). ku234 mutants had 10 to 14 arg-1∷gfp-positive cells that all exhibited the morphology of VM1-like cells (Fig. 3H). Thus, most of the UMs and VM2s are transformed to VM1s in ku234 mutants.
Table 1. unc-62 genetically interacts with ceh-20 and mab-5 to regulate M lineage development.
| Genotype1 | hlh-8∷gfp expression | Early M lineage phenotypes2 | SM lineage phenotypes3 |
|---|---|---|---|
| Wild-type | bright (100%) n=200 | 2 CCs, 2 SMs, 14 BWMs (100%) n=200 | 4 VM1s (100%) n=200 |
| ceh-20(ay9) | bright (80%) faint4 (20%) n=200 | 2 CCs, 2 SMs, 14 BWMs (91%) 1-3 CCs, 1 SM & 13-14 BWMs (9%) n=2005 | 4 VM1s (44%) >4 VM1s (56%) n=75 |
| unc-62(e644) dpy-11(e224) | bright (100%) n=200 | 2 CCs, 2 SMs, 14 BWMs (78%) 0-1 CCs, 3-4 SMs, 12-14 BWMs (22%) n=2006 | 4 VM1s (44%) >4 VM1s (56%) n=69 |
| unc-62(ku234) | bright (100%) n=200 | 2 CCs, 2 SMs, 14 BWMs (100%) n=2007 | >4 VM1s (100%) n=200 |
| mab-5(RNAi) | no early expression bright SM lineage expression (100%) n=156 | 2 CCs, 2 SMs, 14 BWMs (6%) 0 CCs, 3-5 SMs, 13-14 BWMs (94%) n=156 | 4 VM1s (6%) >4 VM1s (94%) |
| ceh-20(ay9)/+; unc-62(e644) dpy-11(e224) | bright (100%) n=123 | 2 CCs, 2 SMs, 14 BWMs (55%) 0-1 CCs, 3-4 SMs (45%) n=60 | 4 VM1s (7%) >4 VM1s (93%) n=123 |
| ceh-20(ay9); unc-62(e644) dpy-11(e224) | faint (3%) no expression (97%) N=200 | 0-1 CCs8 (100%) n=83 | >4 VM1s (100%) n=45 |
| ceh-20(ay9)/+; unc-62(ku234)/+ | bright (100%) n=23 | 2 CCs, 2 SMs, 14 BWMs (100%) n=23 | 4 VM1s (100%) n=23 |
| ceh-20(ay9)/+; unc-62(ku234) | bright (100%) n=163 | 2 CCs, 2 SMs, 14 BWMs (72%) 0-1 CCs, 2 SMs (28%) n=163 | >4 VM1s (100%) n=163 |
| ceh-20(ay9); unc-62(ku234) | faint (7%) no expression (93%) N=200 | 2 CCs, 2 SMs, 14 BWMs (13%) 0-1 CCs, 9-14 BWMs9 (87%) n=200 | >4 VM1s (100%) n=200 |
| mab-5(RNAi); unc-62(e644) dpy-11(e224) | no expression (41%) bright in SM lineage10 (59%) n=200 | 2 CCs (1%) 0-1 CCs8 (99%) n=200 | 0 VM1s11 (58%) 4 VM1s (1%) >4 VM1s (41%) n=200 |
| mab-5(RNAi) ceh-20(ay9) | no expression (52%) bright in SM lineage10 (48%) n=200 | 2 CCs (9%) 0-1 CCs8 (91%) n=200 | 0 VM1s11 (60%) 4 VM1s (9%) >4 VM1s (31%) n=200 |
All animals examined carried the following GFP markers for the M lineage: intrinsic CC∷gfp(ccIs4438) III, ayIs2(egl-15∷gfp) IV; hlh-8∷gfp(ayIs6) X.
Phenotypes focus on specification of the 18 M lineage descendants (14 BWMs, 2 CCs and 2 SMs) produced at the L1 stage.
The SM lineage phenotypes shown here focused on the number of egl-15∷gfp expressing VM1-like cells.
hlh-8∷gfp signal was faint in both the early M lineage and the SM lineage.
The number of BWMs was obtained via examining 6 animals.
The number of BWMs was obtained via examining 5 animals.
The number of BWMs was obtained via examining 3 animals.
The number of BWMs and SMs was not determined in this mutant strain.
The number of BWMs was obtained via examining 5 animals.
There was no hlh-8∷gfp expression in the early M lineage.
Some of the animals had 1-3 elongated egl-15∷gfp positive cells randomly located in the posterior of the worm.
Figure 2. Uterine muscles (UMs) and type II vulval muscles (VM2s) are transformed to type I vulval muscles (VM1s) in unc-62(ku234) mutants.
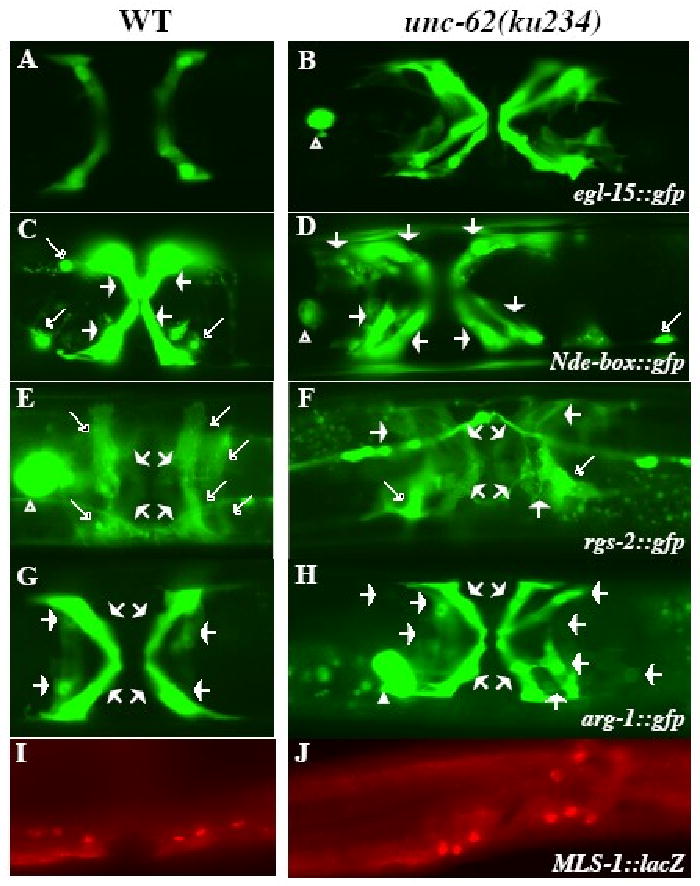
Ventral views of wild-type (A, C, E, G) and unc-62(ku234) (B, D, F, H) animals showing the differentiated sex muscles. Four VM1s in wild-type (A) and 12 VM1s in unc-62(ku234) (B) animals visualized by egl-15∷gfp. UMs and VMs visualized by Nde-box∷gfp (C, D) and rgs-2∷gfp (E, F) in wild-type (C, E) and unc-62(ku234) (D, F) animals. The morphology of UMs (marked by longer open arrows) and VMs (marked by short solid arrows) are distinct. unc-62(ku234) animals (D, F) have reduced numbers of cells with the UM morphology and extra numbers of cells with the VM morphology. arg-1∷gfp in wild-type (G) and unc-62(ku234) (H) animals showing both VM1s and VM2s. The unc-62(ku234) animal in panel H has 12 VM1s. (I, J) mls-1∷LacZ expression pattern in wild-type (I) and unc-62(ku234) (J) animals. Six out of the 12 (I) and 8 out of the 12 (J) mls-1∷lacZ-expressing cells are shown in panels I and J, respectively. The other mls-1∷lacZ-expressing cells are on a different focal plane. Both wild-type and unc-62(ku234) animals show similar mls-1∷lacZ expression patterns.
Figure 3. ceh-20 genetically interacts with unc-62 to regulate M lineage development.
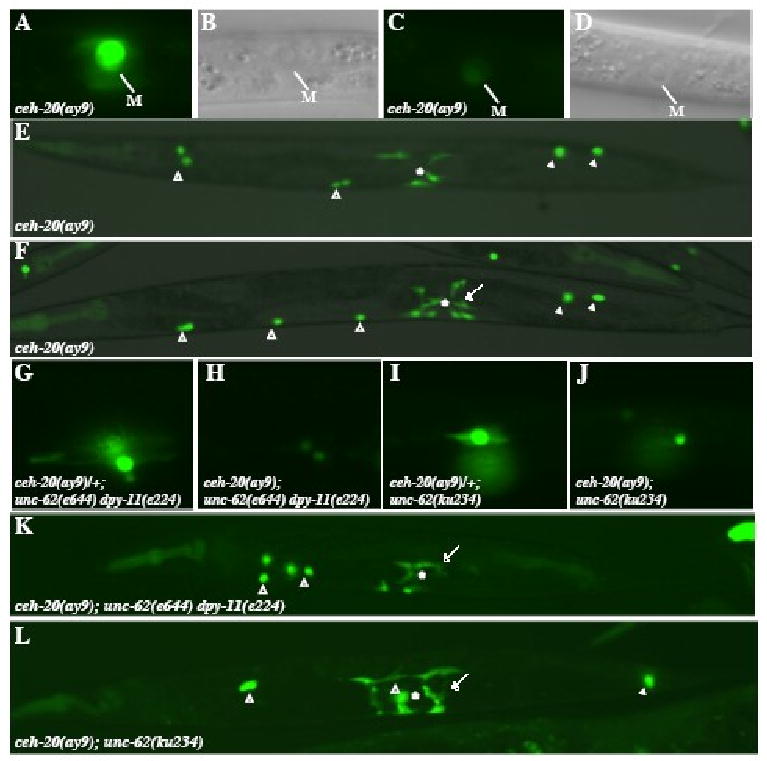
(A-F) various M lineage phenotypes of ceh-20(ay9) animals. (A-D) ceh-20(ay9) animals at the 1-M stage with hlh-8∷gfp (A, C) and the corresponding DIC images (B, D). 80% (n=200) of ceh-20(ay9) animals exhibit strong hlh-8∷gfp expression (A), while 20% showing much reduced hlh-8∷gfp expression (C). Panels A and C are under the same exposure. (E-F) Adult ceh-20(ay9) animals visualized using an intrinsic CC∷gfp and egl-15∷gfp. (E) An ay9 animal with the normal number of M-derived CCs and VM1s that are not properly attached. (F) An ay9 animal with the normal number of M-derived CCs but a total number of 10 VM1s. (G-J) hlh-8∷gfp expression in the 2 SMs of ceh-20(ay9)/+; unc-62(e644) dpy-11(e224) (G), ceh-20(ay9); unc-62(e644) dpy-11(e224) (H), ceh-20(ay9)/+; unc-62(ku234) (I) and ceh-20(ay9); unc-62(ku234) (J) animals. Notice the reduced level of hlh-8∷gfp expression in ceh-20(ay9); unc-62(e644) dpy-11(e224) (H) and ceh-20(ay9); unc-62(ku234) (J) animals compared to that in ceh-20(ay9)/+; unc-62(e644) dpy-11(e224) (G) and ceh-20(ay9)/+; unc-62(ku234) (I) animals. (K-L) Adult ceh-20(ay9); unc-62(e644) dpy-11(e224) (K) and ceh-20(ay9); unc-62(ku234) (L) animals visualized using an intrinsic CC∷gfp and egl-15∷gfp. (K) A ceh-20(ay9); unc-62(e644) dpy-11(e224) animal with no M-derived CCs and extra VM1s. (L) A ceh-20(ay9); unc-62(ku234) animal with one M-derived CCs and extra VM1s. Open arrowheads: embryonically-derived CCs. Solid arrowheads: M-derived CCs. Arrows: ectopic egl-15∷gfp-expressing VM1-like cells. *: location of the vulva.
Previous work by Kostas and Fire (2002) showed that the T-box gene mls-1 is a cell fate determinant of uterine muscles (UMs): mls-1 is specifically expressed in the UM and VM2 precursor cells (Fig. 2I) and loss of mls-1 activity results in the doubling of the numbers of both VM1s and VM2s due to a fate transformation of UMs to VMs. The ku234 mutant phenotype is similar, but not identical, to that of mls-1 mutants. We therefore tested if mls-1 expression is affected in unc-62(ku234) mutants. We detected normal expression of a functional mls-1∷lacZ reporter in the SM lineage of ku234 mutants (Fig. 2J), suggesting that the UM to VM fate transformation in ku234 mutants is not due to the loss of or altered mls-1 expression.
Unlike ku234 mutants, e644 mutants exhibited variable M lineage defects. We used hlh-8∷gfp to follow the M lineage in e644 mutants as hlh-8∷gfp expression appeared unaffected in e644 mutants. Among 200 e644 worms examined, 22% of them had fewer (0-1) M-derived CCs and extra (3-4) SMs (Table 1). Direct observation M lineage cell divisions in three e644 animals showed that while M lineage patterning and proliferation were normal, both M-derived CCs and 1 or 2 M-derived BWMs were transformed to SMs (data not shown). The remaining 78% of e644 animals contained normal numbers of SMs; however, 69% of these mutant animals with 2 SMs (n=69) gave rise to extra VM1s, ranging from 5 to 10 (Fig. 1D, Table 1). Using the cell type specific GFP markers that were used for analyzing the ku234 mutants, we found that these extra VM1s were produced at the expense of UMs and VM2s (data not shown), similar to the phenotypes observed in the ku234 mutants. The multiple M lineage defects in e644 mutants suggest that unc-62 is not only involved in UM and VM decisions in the SM lineage, but also involved in CC, SM and BWM cell fate decisions in the early M lineage.
Because both ku234 and e644 are partial loss-of-function alleles of unc-62 and unc-62 is essential for embryogenesis (Van Auken et al., 2002), we also soaked synchronized L1 wild-type worms with dsRNA against unc-62 (referred to as unc-62(RNAi-soaking), see Materials and Methods) and examined the M lineage phenotypes in the soaked animals. Soaking buffer alone was used as a negative control. Unlike the control animals, unc-62(RNAi-soaking) worms exhibited defects in the vulva and the somatic gonad (data not shown), as well as in the M lineage. The M lineage phenotypes can be grouped into two categories. The majority of unc-62(RNAi-soaking) worms (87%, n=137) had no differentiated M-derived CCs, VM1s, or BWMs (Fig. 1F), or occasionally no M-derived CCs and BWMs, but with 1-4 elongated egl-15∷gfp-positive cells (data not shown). These phenotypes resemble phenotypes observed in ceh-20(RNAi) or lin-39(0) mab-5(0) mutants (Liu and Fire, 2000). Unlike ceh-20(RNAi) or lin-39(0) mab-5(0) mutants in which hlh-8∷gfp expression is undetectable throughout postembryonic development (Liu and Fire, 2000), unc-62(RNAi-soaking) worms showed normal hlh-8∷gfp expression in the early M lineage during the L1 stage when CC, SM and BWM fates are specified. The early expression of hlh-8∷gfp in the unc-62(RNAi-soaking) worms is likely due to the timing of soaking being too late to affect the early M lineage. The remaining 13% of unc-62(RNAi-soaking) animals had hlh-8∷gfp expression throughout the M lineage, but exhibited fate transformation from one or both M-derived CCs to SMs (Fig. 1G) and randomized division orientation in the SM lineage (data not shown).
The range of M lineage defects observed in unc-62(RNAi-soaking) animals and in ku234 and e644 mutants suggest that unc-62 plays multiple roles at multiple stages in the M lineage, including regulating CC/SM/BWM fate specification and differentiation, and VM/UM fate specification.
Analysis of a weak ceh-20 allele, ay9, revealed multiple roles of the PBC protein CEH-20 in the M lineage
We have previously shown that CEH-20, but not CEH-40, is the PBC protein that functions in the M lineage (Liu and Fire, 2000; Jiang et al., 2008). Furthermore, CEH-20 is required for the diversification of the M lineage (Liu and Fire, 2000). These previous studies are based on analyses of ceh-20(RNAi) and a strong loss-of-function allele n2513. We extended our analysis to additional alleles of ceh-20, including os114 and os39 (Arata et al., 2006), and ay42 and ay9 (Takacs-Vellai et al., 2007). Among these alleles, os114 and os39 appear to be the strongest loss-of-function, and possibly null, alleles of ceh-20, as 99% of os114 and os39 mutant animals (n=87 for os114 and n=56 for os39) failed to generate any M lineage descendants, consistent with previous studies of ceh-20(RNAi or n2513) (Liu and Fire, 2000). ay42 is likely a partial loss-of-function allele as 62% of ay42 worms (n=224) failed to produce any M lineage products (data not shown).
ay9 contains a missense mutation, changing a highly conserved Met78 residue to Ile (Takacs-Vellai K. et al., 2006), and exhibits multiple M lineage phenotypes. In ay9 mutant animals, hlh-8∷gfp expression was detectable throughout the M lineage; however, compared to wild-type worms, the intensity of hlh-8∷gfp signal was moderately decreased throughout the M lineage in 20% of ay9 mutants (n=200) (Fig. 3A-D). All ay9 mutants exhibited normal M lineage patterning and proliferation; however, 9% of ay9 animals examined (n=200) exhibited earlier defects in CC/SM/BWM fate decisions, resulting in variable numbers of M-derived CCs, SMs, and BWMs in ay9 mutants (Table 1). In addition to these earlier M lineage defects, ay9 mutants also exhibited defects in the SM lineage. 56% of the ay9 animals (n=75) that contained the normal number of SMs exhibited fate transformations from UMs and VM2s to VM1s, resulting in an increase of the number of egl-15∷gfp-positive VM1s (5-10 VM1s) in ay9 mutants (Table 1, Fig. 3F). VM1s often did not display proper orientation and attachment (Fig. 3E), likely due to the vulval defects in ay9 mutants.
The broad range of M lineage defects seen in the various ceh-20 mutant animals suggest that like unc-62, ceh-20 plays multiple roles at multiple stages in the M lineage, including regulating CC/SM/BWM fate specification, VM/UM fate specification and hlh-8 expression.
unc-62 genetically interacts with ceh-20 and mab-5 to regulate hlh-8/CeTwist expression and cell fate specification in the M lineage
The similar M lineage defects caused by mutations or RNAi of unc-62 and ceh-20 suggest that unc-62 and ceh-20 may function together to regulate proper M lineage development. To test this hypothesis, we generated double mutants between ceh-20(ay9) and unc-62(e644) or unc-62(ku234).
Compared to unc-62(e644) single mutants, ceh-20(ay9)/+; unc-62(e644)/unc-62(e644) animals showed a roughly 20% increase (from 22% to 45%) in penetrance of fate transformations from M-derived CCs to SMs (Table 1). ceh-20(ay9); unc-62(e644) double homozygous animals exhibited significantly more penetrant and severe M lineage defects compared to ceh-20(ay9) and unc-62(e644) single mutants (Table 1). First, unlike in each single mutant, hlh-8∷gfp was undetectable (97%) or very faint (3%, n=200) throughout the M lineage in ceh-20(ay9); unc-62(e644) double mutants (Fig. 3G-H). Second, 100% of ceh-20(ay9); unc-62(e644) double mutants (n=83) had no M-derived CCs, compared to 9% (n=200) of ceh-20(ay9) animals and 22% (n=200) of unc-62(e644) animals that had a CC to SM fate transformation (Table 1, Fig. 3K). Thus there appeared to be a dose-dependent effect of ceh-20 on unc-62(e644) mutants regarding hlh-8 expression and early M lineage fate specification. Similar synergistic effect on M lineage development was also observed between ceh-20(ay9) and unc-62(ku234) (Table 1, Fig. 3I, J, L). These observations are consistent with ceh-20 and unc-62 functioning together in regulating fate specification and hlh-8∷gfp expression during M lineage development. In addition to the M lineage defects, we also observed enhanced penetrance of embryonic lethality and vulval defects in ceh-20; unc-62 double mutants (data not shown), suggesting that these two genes function together in multiple tissues at multiple developmental stages.
In addition to the genetic interactions between unc-62 and ceh-20, we also observed genetic interactions between unc-62 and the Hox gene mab-5. mab-5(0) or mab-5(RNAi) mutants lack hlh-8∷gfp expression in the early M lineage, but show normal hlh-8∷gfp expression in the SM lineage. Furthermore, the M-derived CCs and 1-3 M-derived BWMs are transformed to SMs in mab-5(0) or mab-5(RNAi) mutants (Harfe et al., 1998; Liu and Fire, 2000; Amin et al., 2007, Table 1). 41% (n=85) of mab-5(RNAi); unc-62(e644) mutant animals showed no hlh-8∷gfp expression throughout the M lineage, lacked all M-derived CCs, and had zero or 1-5 elongated egl-15∷gfp positive cells. These phenotypes resemble those observed in lin-39(0) mab-5(0) and ceh-20(RNAi) animals (Liu and Fire, 2000). These observations suggest that in addition to ceh-20, unc-62 also functions together with the Hox gene mab-5 to regulate hlh-8∷gfp expression and M lineage fate specification. Similar synergistic interactions were also observed in mab-5(RNAi) ceh-20(ay9) animals (Table 1), consistent with previous findings on the cooperative actions of mab-5 and ceh-20 in regulating hlh-8 expression and M lineage fate specification.
unc-62 and ceh-20 are co-expressed throughout the M lineage and in other cell types
Because ceh-20 and unc-62 appear to function together to regulate multiple processes of M lineage development at multiple developmental stages, we next determined when and where these two genes are expressed in the M lineage.
unc-62 uses two alternative start codons, exon 1a or 1b (Van Auken et al., 2002). To determine the temporal and spatial expression pattern of unc-62, we generated a transcriptional gfp reporter (unc-62p∷gfp) using 2.9kb sequences upstream of the start codon of exon 1a (see Materials and Methods). We first found that unc-62 cDNAs under the control of this 2.9kb promoter element can rescue the M lineage defects of unc-62(e644) and unc-62(ku234) mutants. We then examined the expression pattern of unc-62p∷gfp in transgenic animals carrying the reporter construct. Similar GFP expression patterns were observed in three independent transgenic lines. unc-62p∷gfp expression was first detectable during mid-embryogenesis in a subset of cells that include AB lineage derivatives (data not shown). In newly hatched L1 larvae, unc-62p∷gfp was observed in multiple cell types including the M mesoblast, as well as a few neurons in the head and tail, and dorsal and ventral hypodermis (Fig. 4A-B). In the L1 larva, unc-62p∷gfp expression continued throughout the M lineage in all dividing M descendants (data not shown). The GFP signal was transiently detectable in M-derived CCs and BWMs before they terminally differentiate, but remained detectable in the SMs (Fig. 4C-D) and throughout the SM lineage (data not shown), including the differentiated vulval muscles (Fig. 4E-F). Outside of the M lineage, the neuronal expression in the head and tail was retained throughout post-embryonic development, while hypodermal expression appeared more variable. unc-62p∷gfp expression was also observed in a subset of ventral nerve cord (VNC) cells after the L2 stage (data not shown) and in the P lineage with initial faint GFP expression in P3.p, P4.p and P5.p in L1 animals, which expanded to P6.p, P7.p and P8.p cells at the L2 stage (Fig. 4C-D). Upon vulval induction, L3 and L4 larvae displayed strong GFP expression in P5.p, P6.p and P7.p and their descendants, but expression in P3.p, P4.p and P8.p and their descendants became weaker and eventually undetectable by mid-L4 stage. unc-62p∷gfp expression persisted in all differentiated vulval cells after vulval morphogenesis in adults (data not shown).
Figure 4. Both unc-62 and ceh-20 are expressed in the M lineage.
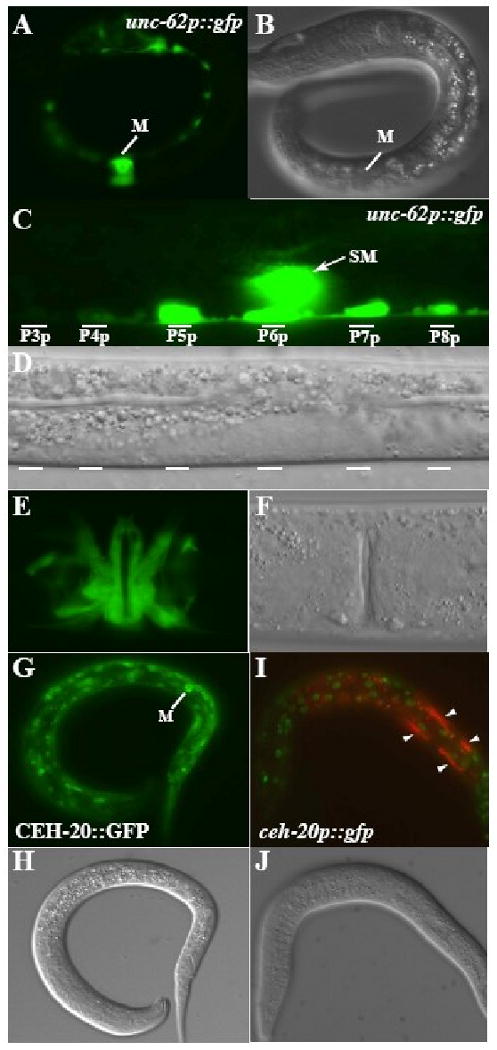
(A-F) unc-62p∷gfp (A, C, E) and corresponding DIC images (B, D, F) of 1-M stage L1 (A-B), 2-SM stage L2 (C-D) and adult stage animals. unc-62p∷gfp is expressed throughout the M lineage, including mature vulval muscles (E) and in the P lineages (C). (G-J) GFP (G, I) and corresponding DIC images showing the expression of a functional translational CEH-20∷GFP in the M mesoblast (G-H) and the lack of M lineage expression of the transcriptional ceh-20p∷gfp in the M lineage (I-J). Green in I: ceh-20p∷gfp. Red in I: hlh-8p∷rfp. Notice that ceh-20p∷gfp is expressed in the other cell types that express CEH-20∷GFP, but not in cells of the M lineage (marked by arrowheads).
ceh-20 shared an overlapping, but not identical, expression pattern with unc-62 both within and outside of the M lineage. Using a functional ceh-20∷gfp construct (see Materials and Methods), we detected nuclear GFP signal in a wide array of cells outside of the M lineage, including a subset of embryonic cells, hypodermal cells, gut cells, bodywall muscles, VNC neurons and VPCs (data not shown), consistent with previous reports on the expression pattern of ceh-20 (Yang et al., 2005; Liu et al., 2006; Takacs-Vellai et al., 2006). Within the M lineage, CEH-20∷GFP was detected in the M cell (Fig. 4G-H) and throughout the early M lineage. It persisted in the differentiated BWMs as well as in the SMs and all the SM descendants (data not shown). In addition to the translational gfp reporter, we also generated a transcriptional ceh-20p∷gfp reporter (see Materials and Methods). Transgenic animals carrying ceh-20p∷gfp showed similar expression pattern to the translational ceh-20∷gfp reporter outside of the M lineage, but failed to show expression in the M lineage (Fig. 4I-J). Because the only difference between the translational ceh-20∷gfp and the transcriptional ceh-20p∷gfp constructs is the presence or absence of the introns, the above observations suggest that the introns of ceh-20 may contain enhancer elements(s) required for ceh-20 expression in the M lineage.
The similar expression patterns of unc-62 and ceh-20 throughout the M lineage are consistent with their multiple and cooperative roles in the M lineage, as indicated by the genetic interactions described earlier.
unc-62 and ceh-20 display physical and regulatory interactions
Because both unc-62 and ceh-20 share similar expression patterns in the M lineage, the synergistic interactions between them could be due to direct physical interactions between these two factors, or regulatory interactions, or both. To test these possibilities, we first used the yeast two-hybrid system and tested whether UNC-62 and CEH-20 can interact with each other. When fused with the GAL4 DNA binding domain, full length UNC-62 and CEH-20 each alone activated reporter expression (data not shown). We thus made truncations of each protein and found that the region containing the PBC domain of CEH-20 (aa1-196) interacts with the N-terminal 287aa (isoform 1a) or 250aa (isoform 1b) of UNC-62, which contains the HM domain (Fig. 5A). These results suggest that CEH-20 and UNC-62 can physically interact with each other.
Figure 5. UNC-62 and CEH-20 exhibit physical and regulatory interactions.
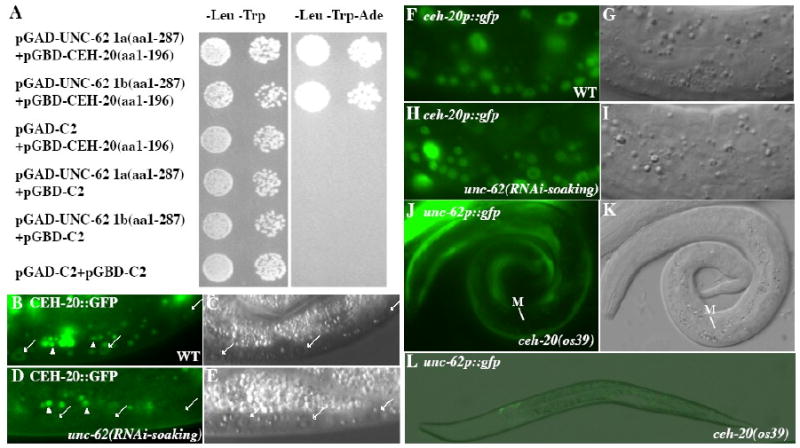
(A) The N-termini of both the 1a and 1b splicing isoforms of unc-62 that contain the HM domain can bind to the PBC domain (aa 1-196) of CEH-20 in the yeast two-hybrid assay. Interaction was detected in one direction using the PBC domain (aa 1-196) of CEH-20 fused to the GAL4 DNA binding domain (GBD). The GBD-UNC-62 fusions autoactivate and were not included in the tests. (B-E) CEH-20∷GFP (B, D) and the corresponding DIC images (C, E) in wild-type (B-C) and unc-62(RNAi-soaking) (D-E) animals. CEH-20∷GFP is present in the nucleus, but not the nucleolus, of multiple cell types in wild-type animals (B-C). Notice the significant reduction or loss of nuclear CEH-20∷GFP in unc-62(RNAi-soaking) animals (arrows in panel D) compared to wild-type animals (arrows in panel B). Additional bright gut autofluorescence signals in panels B and D are marked by arrowheads. (F-I) ceh-20p∷gfp (F, H) and the corresponding DIC images (G, I) in wild-type (F-G) and unc-62(RNAi-soaking) (H-I) animals. The expression of the transcriptional ceh-20p∷nls∷gfp is not affected by unc-62(RNAi-soaking). (J-L) unc-62p∷gfp in ceh-20(os39) animals. Expression of unc-62p∷gfp is lost in the M cell in an L1 animal (J-K) and most of the cells in an L2 animal (L).
Previous studies have shown that unc-62 is required for the nuclear localization of CEH-20 (Van Auken et al., 2002; Potts et al., 2009). Consistent with these findings, we found that unc-62(RNAi) led to the reduction of the translational CEH-20∷GFP signal (Fig. 5B-E), but not the transcriptional ceh-20p∷gfp signal (Fig. 5F-I). To determine whether CEH-20 plays a role in regulating the expression of unc-62, we assayed the expression of the unc-62p∷gfp∷let-858 3′UTR reporter in the strong loss-of-function, and possibly null, alleles of ceh-20, os39. We found that 96 of 97 ceh-20(os39) mutant animals lacked unc-62p∷gfp signal in the M mesoblast and the P lineages, but did express unc-62p∷gfp in neuronal or hypodermal cells (Fig. 5J-L). Similar observations were made using the strong loss-of-function ceh-20(n2513) mutants (data not shown). Since the M mesoblast is still present in the os39 mutant animals and the unc-62p∷gfp reporter uses the well characterized let-858 3′UTR (Kelly et al., 1997), the above results suggest that ceh-20 is required to positively regulate the promoter activity of unc-62 in the M lineage and P lineage, but not in neuronal or hypodermal cells.
Discussion
Meis/Hth/unc-62 functions in mesodermal development
MEIS family proteins have been previously shown to play important roles in multiple aspects of development in both vertebrates and invertebrates (for example, see Abu-Shaar and Mann, 1998; Henderson and Andrew, 2000; Choe et al., 2002; Choe and Sagerstrom, 2004; Mercader et al., 2005). Similarly, the C. elegans MEIS-related proteins psa-3 and unc-62 have been shown to function in regulating T cell asymmetric cell division (psa-3, Arata et al., 2006), regulate embryogenesis, Q neuroblast migration, vulval formation and VC motor neuron survival (unc-62, Van Auken et al., 2002; Yang et al., 2005; Potts et al., 2009). In this study, we have found that a loss-of-function mutation psa-3(os8) or psa-3(RNAi) did not affect the postembryonic mesodermal lineage, the M lineage. However, reducing unc-62 activity led to pleiotropic M lineage phenotypes including defects in cell fate specification and differentiation, indicating that unc-62 is a crucial regulator of mesodermal development in C. elegans.
In Drosophila, the MEIS protein Hth functions in the visceral mesoderm to regulate dpp expression (Stultz et al., 2006). Murine meis-2 exhibits a dynamic expression pattern in differentiating somitic mesodermal cells (Cecconi et al., 1997). Similar temporal expression of meis-2 has also been observed in the lateral mesoderm in zebrafish (Biemar et al., 2001; Zerucha and Prince, 2001). Thus MEIS family proteins may share a conserved role in mesodermal development.
UNC-62 functionally cooperates with CEH-20 to regulate mesoderm development
Previous work has shown that ceh-20 is required for the specification and diversification of the M lineage, as well as the proper specification of CC and BWM fates (Liu and Fire, 2000). Our analysis of the phenotypes of a weak ceh-20(ay9) allele revealed another role of ceh-20 in the proper specification of uterine muscles (UMs) and vulval muscles (VMs). ceh-20 and unc-62 appear to function together in this process, as two partial loss-of-function unc-62 alleles, e644 and ku234, both caused fate transformations from UMs and type II vulval muscles (VM2s) to type I vulval muscles (VM1s). In particular, the ku234 mutants exhibited 100% penetrance of the UM and VM2 to VM1 fate transformation without affecting the early M lineage (Table 1, Fig, 2). Because ku234 is a point mutation located in the first intron of the 1a isoform, but mutating the translation initiation codon of the 1b isoform, either the ku234 mutation disrupts a VM/UM precursor-specific enhancer element located in the first intron, or the 1b isoform of unc-62 is specifically expressed in the VM/UM precursor cells. unc-62 is also required for proper specification and differentiation of M-derived CCs and BWMs. In addition to the similar M lineage phenotypes shared by ceh-20 and unc-62 mutants, we observed synergistic genetic interactions between ceh-20 and unc-62. As shown in Table 1, both ceh-20(ay9); unc-62(e644) and ceh-20(ay9); unc-62(ku234) double mutants exhibited more severe M lineage defects compared to each of the corresponding single mutants. For example, even though unc-62(ku234) single mutants did not exhibit any early M lineage defects, the ceh-20(ay9); unc-62(ku234) double mutant animals showed severe early M lineage defects in CC/BWM/SM fate specification.
The genetic interactions between unc-62 and ceh-20 could be due to 1) a physical interaction, 2) regulatory interactions, or 3) both physical and regulatory interactions between the two factors. We have found that unc-62 and ceh-20 share similar expression patterns within and outside of the M lineage, and that they can physically interact in the yeast two-hybrid system. We further showed that the two genes also share regulatory interactions: while unc-62 is required for the nuclear localization of the CEH-20 protein, ceh-20 is required for the expression of unc-62 in the M lineage. Because the M cell is present in ceh-20 mutants and the unc-62p∷gfp reporter uses the well characterized let-858 3′UTR (Kelly et al., 1997), the lack of expression of the unc-62p∷gfp reporter in the M and P lineages in ceh-20 mutants suggests that ceh-20 may positively regulate the transcription of unc-62. However, at this point, we do not know whether this regulation is direct or indirect. The regulation of the nuclear localization of PBC proteins by MEIS proteins have been previously reported in multiple systems (Pai et al., 1998; Abu-Shaar et al., 1999; Affolter et al., 1999; Berthelsen et al., 1999; Jaw et al., 2000; Rieckhof et al., 1997; Potts et al., 2009). However, transcriptional regulation of MEIS by PBC proteins has not been previously reported. In Drosophila, hth transcription appears not to be affected by the loss of exd (Abu-Shaar and Mann, 1998) even though Hth protein level is greatly reduced in embryos lacking both maternal and zygotic EXD (Kurant et al., 1998) or in exd mutant clones (Abu-Shaar and Mann, 1998). Thus our findings add one more level to the complex interactions between MEIS and PBC family of proteins: possible transcriptional regulation of meis genes by PBC proteins.
The MEIS and PBC proteins have been implicated as cofactors of Hox proteins that cooperatively bind to target DNA sequences with Hox proteins (Maconochie et al., 1997; Kroon et al., 1998; Swift et al., 1998; Ryoo et al., 1999; Shanmugam et al., 1999; Shen et al., 1999; Ferretti et al., 2000; Ebner et al., 2005). The early M lineage defects of unc-62 (e664), unc-62(RNAi-soaking) and ceh-20(ay9) mutants regarding CC/BWM/SM fate specification and differentiation are very similar to the previously described phenotypes of mab-5(0) mutants (Harfe et al., 1998). Furthermore, the ceh-20(ay9); unc-62(e644), ceh-20(ay9); unc-62(ku234), mab-5(RNAi); unc-62(e644) and mab-5(RNAi) ceh-20(ay9) double mutants exhibited more severe M lineage phenotypes that resemble the lin-39(0) mab-5(0) double mutant phenotypes. These observations suggest that UNC-62 may interact with CEH-20 and Hox to activate downstream targets required for the diversification and proper BWM/CC/SM fate specification of the M lineage. At this point, it is not clear whether the cooperative role of UNC-62 and CEH-20 in UM/VM fate specification is dependent on the Hox factors. lin-39(0) mutants have no defects in the M lineage, while lin-39(0) mab-5(0) mutants and mab-5(0) mutants both have earlier defects in the M lineage, which could potentially mask the role of mab-5 and lin-39 in the late SM lineage.
It has been shown that LIN-39 or MAB-5 and CEH-20 can form heterodimers and bind to the corresponding Hox-PBC binding sites in the promoter region of hlh-8 to regulate its expression (Liu and Fire, 2000). Since hlh-8∷gfp expression is absent or barely detectable in ceh-20(ay9); unc-62(e644), ceh-20(ay9); unc-62(ku234), mab-5(RNAi); unc-62(e644) and mab-5(RNAi) ceh-20(ay9) mutants, it is possible that UNC-62 may participate in the DNA-bound Hox-CEH-20 complex in regulating hlh-8 expression. However, we did not find any potential MEIS binding site (TGACAG, Chang et al., 1997; Knoepfler et al., 1997; Shen et al., 1997; Berthelsen et al., 1998) adjacent to the Hox-CEH-20 consensus sequence in the characterized hlh-8 promoter region (Liu and Fire, 2000). Furthermore, attempts using chromatin immunoprecipitation (ChIP) on the hlh-8 protomter failed, possibly due to the short time window of early M lineage development and the small number of M-derived cells (1-18) relative to the whole animal (∼900 somatic cells). It is possible that UNC-62 binds to some novel sequences in the hlh-8 promoter that are different from the canonical MEIS binding site, or UNC-62 acts as a non-DNA-binding partner in the Hox-CEH-20-UNC-62 trimeric complex. The physical interaction that we observed between CEH-20 and UNC-62 is consistent with the second hypothesis.
In addition to regulating hlh-8 expression, unc-62 and ceh-20 may have additional targets in the M lineage. We have previously found that the HMX factor MLS-2 is likely a direct target of CEH-20 in the M lineage (Jiang et al., 2008). We observed normal mls-2 expression in the early M lineage of unc-62(RNAi-soaking) animals (data not shown). However, since unc-62(RNAi-soaking) may not completely knock out unc-62 activity in the early M lineage, we cannot rule out the possibility that mls-2 is regulated by unc-62.
In the SM lineage, a T-box transcription factor MLS-1 is both necessary and sufficient for UM fate specification (Kostas and Fire, 2002). mls-1 is expressed in both UM and VM2 precursor cells and is regulated by hlh-8 (Kostas and Fire, 2002). While unc-62(ku234) mutants exhibited fate transformations from UMs and VM2s to VM1s (Fig. 2A-H), mls-1 expression appeared normal (Fig. 2I-J). Thus, unc-62 must be involved in regulating mls-1 activity through post-transcriptional processes. For instance, unc-62 could be required to activate MLS-1 cofactor(s) in the SM lineage, which then function(s) together with MLS-1 to promoter UM fate.
In summary, UNC-62/MEIS/HTH, CEH-20/PBX/EXD and Hox factors (MAB-5 and LIN-39) play multiple roles in the C. elegans postembryonic mesoderm. Previous work have found roles of CEH-20 and Hox factors MAB-5 and LIN-39 in cell proliferation in the M lineage as well as in the specification and differentiation of M lineage-derived striated body-wall muscles (BWMs) and non-muscle coelomocytes (CCs) (Harfe et al., 1998; Liu and Fire, 2000; Jiang et al. (2008). We showed in this work that UNC-62 also participates in these functions. Furthermore, we demonstrated that UNC-62 and CEH-20 are also required for the proper specification of non-striated vulval and uterine muscles (VM and UMs). While all three factors are required for the M lineage expression of CeTwist, HLH-8 (Liu and Fire, 2000; this work), CEH-20 appears to function in a Hox-independent manner in regulating the HMX gene mls-2 (Jiang et al., 2005; 2008). The mode in which these three factors function together or independently will be elucidated upon the identification and characterization of additional M lineage specific targets of each.
Acknowledgments
We thank the C. elegans Genetics Center (which is funded by the NIH), Thomas Bürglin, Yuji Kohara, Hitoshi Sawa, Lois Edgar and Bill Wood for strains and plasmids; Nirav Amin and Chenxi Tian for helpful discussions and valuable comments on the manuscript. This work was supported by NIH R01 GM066953 (to J.L.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abu-Shaar M, Ryoo HD, Mann RS. Control of the Nuclear Localization of Extradenticle by Competing Nuclear Import and Export Signals. Genes Dev. 1999;13:935–945. doi: 10.1101/gad.13.8.935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Affolter M, Marty T, Vigano MA. Balancing Import and Export in Development. Genes Dev. 1999;13:913–915. doi: 10.1101/gad.13.8.913. [DOI] [PubMed] [Google Scholar]
- Amin NM, Hu K, Pruyne D, Terzic D, Bretscher A, Liu J. A Zn-finger/FH2-Domain Containing Protein, FOZI-1, Acts Redundantly with CeMyoD to Specify Striated Body Wall Muscle Fates in the Caenorhabditis Elegans Postembryonic Mesoderm. Development. 2007;134:19–29. doi: 10.1242/dev.02709. [DOI] [PubMed] [Google Scholar]
- Arata Y, Kouike H, Zhang Y, Herman MA, Okano H, Sawa H. Wnt Signaling and a Hox Protein Cooperatively Regulate Psa-3/Meis to Determine Daughter Cell Fate After Asymmetric Cell Division in C. Elegans. Dev Cell. 2006;11:105–115. doi: 10.1016/j.devcel.2006.04.020. [DOI] [PubMed] [Google Scholar]
- Biemar F, Devos N, Martial JA, Driever W, Peers B. Cloning and Expression of the TALE Superclass Homeobox Meis2 Gene during Zebrafish Embryonic Development. Mech Dev. 2001;109:427–431. doi: 10.1016/s0925-4773(01)00554-8. [DOI] [PubMed] [Google Scholar]
- Bürglin TR, Ruvkun G. New motif in PBX genes. Nature Genet. 1992;1:319–320. doi: 10.1038/ng0892-319. [DOI] [PubMed] [Google Scholar]
- Burglin TR. Analysis of TALE Superclass Homeobox Genes (MEIS, PBC, KNOX, Iroquois, TGIF) Reveals a Novel Domain Conserved between Plants and Animals. Nucleic Acids Res. 1997;25:4173–4180. doi: 10.1093/nar/25.21.4173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burglin TR. The PBC Domain Contains a MEINOX Domain: Coevolution of Hox and TALE Homeobox Genes? Dev Genes Evol. 1998;208:113–116. doi: 10.1007/s004270050161. [DOI] [PubMed] [Google Scholar]
- Cecconi F, Proetzel G, Alvarez-Bolado G, Jay D, Gruss P. Expression of Meis2, a Knotted-Related Murine Homeobox Gene, Indicates a Role in the Differentiation of the Forebrain and the Somitic Mesoderm. Dev Dyn. 1997;210:184–190. doi: 10.1002/(SICI)1097-0177(199710)210:2<184::AID-AJA10>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- Chan SK, Ryoo HD, Gould A, Krumlauf R, Mann RS. Switching the in Vivo Specificity of a Minimal Hox-Responsive Element. Development. 1997;124:2007–2014. doi: 10.1242/dev.124.10.2007. [DOI] [PubMed] [Google Scholar]
- Chan SK, Jaffe L, Capovilla M, Botas J, Mann RS. The DNA Binding Specificity of Ultrabithorax is Modulated by Cooperative Interactions with Extradenticle, another Homeoprotein. Cell. 1994;78:603–615. doi: 10.1016/0092-8674(94)90525-8. [DOI] [PubMed] [Google Scholar]
- Chang CP, Jacobs Y, Nakamura T, Jenkins NA, Copeland NG, Cleary ML. Meis Proteins are Major in Vivo DNA Binding Partners for Wild-Type but Not Chimeric Pbx Proteins. Mol Cell Biol. 1997;17:5679–5687. doi: 10.1128/mcb.17.10.5679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choe SK, Sagerstrom CG. Paralog Group 1 Hox Genes Regulate Rhombomere 5/6 Expression of vhnf1, a Repressor of Rostral Hindbrain Fates, in a Meis-Dependent Manner. Dev Biol. 2004;271:350–361. doi: 10.1016/j.ydbio.2004.04.007. [DOI] [PubMed] [Google Scholar]
- Choe SK, Vlachakis N, Sagerstrom CG. Meis Family Proteins are Required for Hindbrain Development in the Zebrafish. Development. 2002;129:585–595. doi: 10.1242/dev.129.3.585. [DOI] [PubMed] [Google Scholar]
- Ebner A, Cabernard C, Affolter M, Merabet S. Recognition of Distinct Target Sites by a Unique Labial/Extradenticle/Homothorax Complex. Development. 2005;132:1591–1600. doi: 10.1242/dev.01721. [DOI] [PubMed] [Google Scholar]
- Ferretti E, Marshall H, Popperl H, Maconochie M, Krumlauf R, Blasi F. Segmental Expression of Hoxb2 in r4 Requires Two Separate Sites that Integrate Cooperative Interactions between Prep1, Pbx and Hox Proteins. Development. 2000;127:155–166. doi: 10.1242/dev.127.1.155. [DOI] [PubMed] [Google Scholar]
- Ferretti E, Schulz H, Talarico D, Blasi F, Berthelsen J. The PBX-Regulating Protein PREP1 is Present in Different PBX-Complexed Forms in Mouse. Mech Dev. 1999;83:53–64. doi: 10.1016/s0925-4773(99)00031-3. [DOI] [PubMed] [Google Scholar]
- Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC. Potent and Specific Genetic Interference by Double-Stranded RNA in Caenorhabditis Elegans. Nature. 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- Granato M, Schnabel H, Schnabel R. Pha-1, a Selectable Marker for Gene Transfer in C. Elegans. Nucleic Acids Res. 1994;22:1762–1763. doi: 10.1093/nar/22.9.1762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harfe BD, Vaz Gomes A, Kenyon C, Liu J, Krause M, Fire A. Analysis of a Caenorhabditis Elegans Twist Homolog Identifies Conserved and Divergent Aspects of Mesodermal Patterning. Genes Dev. 1998;12:2623–2635. doi: 10.1101/gad.12.16.2623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson KD, Andrew DJ. Regulation and Function of Scr, Exd, and Hth in the Drosophila Salivary Gland. Dev Biol. 2000;217:362–374. doi: 10.1006/dbio.1999.9560. [DOI] [PubMed] [Google Scholar]
- James P, Halladay J, Craig EA. Genomic Libraries and a Host Strain Designed for Highly Efficient Two-Hybrid Selection in Yeast. Genetics. 1996;144:1425–1436. doi: 10.1093/genetics/144.4.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaw TJ, You LR, Knoepfler PS, Yao LC, Pai CY, Tang CY, Chang LP, Berthelsen J, Blasi F, Kamps MP, Sun YH. Direct Interaction of Two Homeoproteins, Homothorax and Extradenticle, is Essential for EXD Nuclear Localization and Function. Mech Dev. 2000;91:279–291. doi: 10.1016/s0925-4773(99)00316-0. [DOI] [PubMed] [Google Scholar]
- Jiang Y, Shi H, Amin NM, Sultan I, Liu J. Mesodermal Expression of the C. Elegans HMX Homolog Mls-2 Requires the PBC Homolog CEH-20. Mech Dev. 2008;125:451–461. doi: 10.1016/j.mod.2008.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Y, Horner V, Liu J. The HMX Homeodomain Protein MLS-2 Regulates Cleavage Orientation, Cell Proliferation and Cell Fate Specification in the C. Elegans Postembryonic Mesoderm. Development. 2005;132:4119–4130. doi: 10.1242/dev.01967. [DOI] [PubMed] [Google Scholar]
- Kamath RS, Fraser AG, Dong Y, Poulin G, Durbin R, Gotta M, Kanapin A, Le Bot N, Moreno S, Sohrmann M, Welchman DP, Zipperlen P, Ahringer J. Systematic Functional Analysis of the Caenorhabditis Elegans Genome using RNAi. Nature. 2003;421:231–237. doi: 10.1038/nature01278. [DOI] [PubMed] [Google Scholar]
- Kelly WG, Xu S, Montgomery MK, Fire A. Distinct requirements for somatic and germline expression of a generally expressed Caernorhabditis elegans gene. Genetics. 1997;146:227–38. doi: 10.1093/genetics/146.1.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knoepfler PS, Calvo KR, Chen H, Antonarakis SE, Kamps MP. Meis1 and pKnox1 Bind DNA Cooperatively with Pbx1 Utilizing an Interaction Surface Disrupted in Oncoprotein E2a-Pbx1. Proc Natl Acad Sci U S A. 1997;94:14553–14558. doi: 10.1073/pnas.94.26.14553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koh K, Peyrot SM, Wood CG, Wagmaister JA, Maduro MF, Eisenmann DM, Rothman JH. Cell Fates and Fusion in the C. Elegans Vulval Primordium are Regulated by the EGL-18 and ELT-6 GATA Factors -- Apparent Direct Targets of the LIN-39 Hox Protein. Development. 2002;129:5171–5180. doi: 10.1242/dev.129.22.5171. [DOI] [PubMed] [Google Scholar]
- Kostas SA, Fire A. The T-Box Factor MLS-1 Acts as a Molecular Switch during Specification of Nonstriated Muscle in C. Elegans. Genes Dev. 2002;16:257–269. doi: 10.1101/gad.923102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroon E, Krosl J, Thorsteinsdottir U, Baban S, Buchberg AM, Sauvageau G. Hoxa9 Transforms Primary Bone Marrow Cells through Specific Collaboration with Meis1a but Not Pbx1b. EMBO J. 1998;17:3714–3725. doi: 10.1093/emboj/17.13.3714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krumlauf R. Hox Genes in Vertebrate Development. Cell. 1994;78:191–201. doi: 10.1016/0092-8674(94)90290-9. [DOI] [PubMed] [Google Scholar]
- Kurant E, Pai CY, Sharf R, Halachmi N, Sun YH, Salzberg A. Dorsotonals/homothorax, the Drosophila Homologue of meis1, Interacts with Extradenticle in Patterning of the Embryonic PNS. Development. 1998;125:1037–1048. doi: 10.1242/dev.125.6.1037. [DOI] [PubMed] [Google Scholar]
- Liu H, Strauss TJ, Potts MB, Cameron S. Direct Regulation of Egl-1 and of Programmed Cell Death by the Hox Protein MAB-5 and by CEH-20, a C. Elegans Homolog of Pbx1. Development. 2006;133:641–650. doi: 10.1242/dev.02234. [DOI] [PubMed] [Google Scholar]
- Liu J, Fire A. Overlapping Roles of Two Hox Genes and the Exd Ortholog Ceh-20 in Diversification of the C. Elegans Postembryonic Mesoderm. Development. 2000;127:5179–5190. doi: 10.1242/dev.127.23.5179. [DOI] [PubMed] [Google Scholar]
- Maconochie MK, Nonchev S, Studer M, Chan SK, Popperl H, Sham MH, Mann RS, Krumlauf R. Cross-Regulation in the Mouse HoxB Complex: The Expression of Hoxb2 in Rhombomere 4 is Regulated by Hoxb1. Genes Dev. 1997;11:1885–1895. doi: 10.1101/gad.11.14.1885. [DOI] [PubMed] [Google Scholar]
- Mann RS, Affolter M. Hox Proteins Meet More Partners. Curr Opin Genet Dev. 1998;8:423–429. doi: 10.1016/s0959-437x(98)80113-5. [DOI] [PubMed] [Google Scholar]
- McGinnis W, Krumlauf R. Homeobox Genes and Axial Patterning. Cell. 1992;68:283–302. doi: 10.1016/0092-8674(92)90471-n. [DOI] [PubMed] [Google Scholar]
- Mello CC, Kramer JM, Stinchcomb D, Ambros V. Efficient Gene Transfer in C.Elegans: Extrachromosomal Maintenance and Integration of Transforming Sequences. EMBO J. 1991;10:3959–3970. doi: 10.1002/j.1460-2075.1991.tb04966.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercader N, Tanaka EM, Torres M. Proximodistal Identity during Vertebrate Limb Regeneration is Regulated by Meis Homeodomain Proteins. Development. 2005;132:4131–4142. doi: 10.1242/dev.01976. [DOI] [PubMed] [Google Scholar]
- Moens CB, Selleri L. Hox Cofactors in Vertebrate Development. Dev Biol. 2006;291:193–206. doi: 10.1016/j.ydbio.2005.10.032. [DOI] [PubMed] [Google Scholar]
- Mukherjee K, Burglin TR. Comprehensive Analysis of Animal TALE Homeobox Genes: New Conserved Motifs and Cases of Accelerated Evolution. J Mol Evol. 2007;65:137–153. doi: 10.1007/s00239-006-0023-0. [DOI] [PubMed] [Google Scholar]
- Nagao T, Endo K, Kawauchi H, Walldorf U, Furukubo-Tokunaga K. Patterning Defects in the Primary Axonal Scaffolds Caused by the Mutations of the Extradenticle and Homothorax Genes in the Embryonic Drosophila Brain. Dev Genes Evol. 2000;210:289–299. doi: 10.1007/s004270050316. [DOI] [PubMed] [Google Scholar]
- Noro B, Culi J, McKay DJ, Zhang W, Mann RS. Distinct Functions of Homeodomain-Containing and Homeodomain-Less Isoforms Encoded by Homothorax. Genes Dev. 2006;20:1636–1650. doi: 10.1101/gad.1412606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pai CY, Kuo TS, Jaw TJ, Kurant E, Chen CT, Bessarab DA, Salzberg A, Sun YH. The Homothorax Homeoprotein Activates the Nuclear Localization of another Homeoprotein, Extradenticle, and Suppresses Eye Development in Drosophila. Genes Dev. 1998;12:435–446. doi: 10.1101/gad.12.3.435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson JC, Lemons D, McGinnis W. Modulating Hox Gene Functions during Animal Body Patterning. Nat Rev Genet. 2005;6:893–904. doi: 10.1038/nrg1726. [DOI] [PubMed] [Google Scholar]
- Potts MB, Wang DP, Cameron S. Trithorax, Hox, and TALE-Class Homeodomain Proteins Ensure Cell Survival through Repression of the BH3-Only Gene Egl-1. Dev Biol. 2009;329:374–385. doi: 10.1016/j.ydbio.2009.02.022. [DOI] [PubMed] [Google Scholar]
- Rauskolb C, Peifer M, Wieschaus E. extradenticle, a regulator of homeotic gene activity, is a homolog of the homeobox-containing human proto-oncogene pbx1. Cell. 1993;74:1101–1112. doi: 10.1016/0092-8674(93)90731-5. [DOI] [PubMed] [Google Scholar]
- Rieckhof GE, Casares F, Ryoo HD, Abu-Shaar M, Mann RS. Nuclear Translocation of Extradenticle Requires Homothorax, which Encodes an Extradenticle-Related Homeodomain Protein. Cell. 1997;91:171–183. doi: 10.1016/s0092-8674(00)80400-6. [DOI] [PubMed] [Google Scholar]
- Ryoo HD, Marty T, Casares F, Affolter M, Mann RS. Regulation of Hox Target Genes by a DNA Bound Homothorax/Hox/Extradenticle Complex. Development. 1999;126:5137–5148. doi: 10.1242/dev.126.22.5137. [DOI] [PubMed] [Google Scholar]
- Shanmugam K, Green NC, Rambaldi I, Saragovi HU, Featherstone MS. PBX and MEIS as Non-DNA-Binding Partners in Trimeric Complexes with HOX Proteins. Mol Cell Biol. 1999;19:7577–7588. doi: 10.1128/mcb.19.11.7577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shemer G, Podbilewicz B. LIN-39/Hox Triggers Cell Division and Represses EFF-1/fusogen-Dependent Vulval Cell Fusion. Genes Dev. 2002;16:3136–3141. doi: 10.1101/gad.251202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen WF, Rozenfeld S, Kwong A, Kom ves LG, Lawrence HJ, Largman C. HOXA9 Forms Triple Complexes with PBX2 and MEIS1 in Myeloid Cells. Mol Cell Biol. 1999;19:3051–3061. doi: 10.1128/mcb.19.4.3051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens KE, Mann RS. A Balance between Two Nuclear Localization Sequences and a Nuclear Export Sequence Governs Extradenticle Subcellular Localization. Genetics. 2007;175:1625–1636. doi: 10.1534/genetics.106.066449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stultz BG, Jackson DG, Mortin MA, Yang X, Beachy PA, Hursh DA. Transcriptional Activation by Extradenticle in the Drosophila Visceral Mesoderm. Dev Biol. 2006;290:482–494. doi: 10.1016/j.ydbio.2005.11.041. [DOI] [PubMed] [Google Scholar]
- Sulston JE, Horvitz HR. Post-Embryonic Cell Lineages of the Nematode, Caenorhabditis Elegans. Dev Biol. 1977;56:110–156. doi: 10.1016/0012-1606(77)90158-0. [DOI] [PubMed] [Google Scholar]
- Swift GH, Liu Y, Rose SD, Bischof LJ, Steelman S, Buchberg AM, Wright CV, MacDonald RJ. An Endocrine-Exocrine Switch in the Activity of the Pancreatic Homeodomain Protein PDX1 through Formation of a Trimeric Complex with PBX1b and MRG1 (MEIS2) Mol Cell Biol. 1998;18:5109–5120. doi: 10.1128/mcb.18.9.5109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takacs-Vellai K, Vellai T, Chen EB, Zhang Y, Guerry F, Stern MJ, Muller F. Transcriptional Control of Notch Signaling by a HOX and a PBX/EXD Protein during Vulval Development in C. Elegans. Dev Biol. 2007;302:661–669. doi: 10.1016/j.ydbio.2006.09.049. [DOI] [PubMed] [Google Scholar]
- Van Auken K, Weaver D, Robertson B, Sundaram M, Saldi T, Edgar L, Elling U, Lee M, Boese Q, Wood WB. Roles of the Homothorax/Meis/Prep Homolog UNC-62 and the Exd/Pbx Homologs CEH-20 and CEH-40 in C. Elegans Embryogenesis. Development. 2002;129:5255–5268. doi: 10.1242/dev.129.22.5255. [DOI] [PubMed] [Google Scholar]
- Yang L, Sym M, Kenyon C. The Roles of Two C. Elegans HOX Co-Factor Orthologs in Cell Migration and Vulva Development. Development. 2005;132:1413–1428. doi: 10.1242/dev.01569. [DOI] [PubMed] [Google Scholar]
- Zerucha T, Prince VE. Cloning and Developmental Expression of a Zebrafish meis2 Homeobox Gene. Mech Dev. 2001;102:247–250. doi: 10.1016/s0925-4773(01)00299-4. [DOI] [PubMed] [Google Scholar]


