Abstract
Retinol-binding protein (RBP4) transports retinol in the circulation from hepatic stores to peripheral tissues. Since little is known regarding the regulation of this gene, we analysed the cis-regulatory sequences of the mouse RBP4 gene. Our data show that transcription of the gene is regulated through a bipartite promoter: a proximal region necessary for basal expression and a distal segment responsible for cAMP-induction. This latter region contains several binding sites for the structural HMGA1 proteins, which are important to promoter regulation. We further demonstrate that HMGA1s play a key role in basal and cAMP-induction of Rbp4 transcription and the RBP4 and HMGA1 genes are coordinately regulated in vitro and in vivo. HMGA1 acts to recruit transcription factors to the RBP4 promoter and we specifically identified p54nrb/NonO and Protein-associated Splicing Factor (PSF) as components that interact with this complex. Steroidogenic factor 1 (SF1) or the related Liver Receptor Homologue 1 (LRH-1) are also associated with this complex upon cAMP-induction. Depletion of SF1/LRH-1 by RNA interference resulted in a dramatic loss of cAMP-induction.
Collectively, our results demonstrate that basal and cAMP-induced Rbp4 transcription is regulated by a multiprotein complex that is similar to ones that modulate expression of genes of steroid hormone biosynthesis. Since genes related to glucose metabolism are regulated in a similar fashion, this suggests that Rbp4 expression may be regulated as part of a network of pathways relevant to the onset of type 2 diabetes.
1. Introduction
Retinoids (vitamin A and its analogs) are needed to maintain normal growth and development, immunity, reproduction, vision and other important physiological processes (Napoli, 1996). Retinol is not biologically active per se, and within tissues is oxidized to retinaldehyde, active in the visual cycle (Saari, 1999), and to retinoic acid, that regulates the transcription of a variety of target genes (Clagett-Dame and Plum, 1997; Gudas et al., 1994) through receptor-mediated events (Mangelsdorf et al., 1995; Glass et al., 1997; Lefebvre et al., 2005).
All retinoids present in the body originate from the diet. They are esterified in the intestine to retinyl esters and incorporated into chylomicrons along with other dietary lipids. Chylomicrons are secreted into lymph and then into the bloodstream where they are rearranged to chylomicron remnants through the release of retinyl esters promoted by lipoprotein lipase on the surface of endothelial cells (Vogel et al., 1999). The majority of chylomicrons are cleared by the hepatocyte upon hydrolysis of retinyl esters into retinol. The newly formed retinol can be either stored in hepatic stellate cells (also called Ito cells) in the form of retinyl esters or secreted into the blood and transported to target tissues exclusively by means of a specific 21 kDa carrier protein, retinol-binding protein (RBP4) (Soprano and Blaner, 1994). Liver is the major, but not the only, site of RBP synthesis. Retinol-RBP circulates in the bloodstream in a 1:1 molar complex with transthyretin (TTR), a 55 kDa protein that is synthesized in and secreted from liver. This ternary complex prevents retinol-RBP excretion by kidney (Monaco et al., 1995).
cyclic AMP (cAMP) is a second messenger involved in the transduction of hormonal or growth signals that regulate many cell functions such as proliferation, differentiation, neuronal signalling and metabolism. Interaction of an extracellular ligand with its cognate G-protein coupled receptor dissociates G-proteins and induces GDP-nucleotide exchange (Choi et al., 1993). The active G-protein, in turn, binds to and stimulates transmembrane adenylyl cyclase. The resultant elevation in cAMP concentration increases Protein Kinase A (PKA) activity, which, among many other proteins, phosphorylates transcription factors known as Cyclic AMP Responsive Elements Binding Proteins (CREB) (Gonzales and Montminy, 1989; Harootunian et al., 1993). Phosphorylated CREB interacts with transcription coactivator CBP/p300 and this multiprotein complex is able to recruit enzymes with histone acetylase activity (HAT) (Servillo et al., 2002). These events lead to the assembly of a functional transcriptional machinery on the promoter regions of target genes (Montminy, 1997). CREBs belong to the bZip (basic–leucine zipper domain) family of transcription factors and at least four members have been identified so far: CREB I and II, Activating Transcription Factor 1 (ATF-1), CRE modulator protein (CREM) which recognize and bind conserved cAMP-responsive elements (CRE) in the regulatory regions of responsive genes (Foulkes and Sassone-Corsi, 1992). CRE usually consists of an eight bp palindromic sequence (TGACGTCA) although sequence variations are possible (Sassone-Corsi, 1995). In addition to the large number of cAMP-target genes modulated by mechanisms involving the binding of CREB (or CREB-like factors) to CRE sequences, there is extensive documentation of genes whose transcription is mediated by novel PKA/cAMP-dependent but CREB-independent pathways (Ying et al., 1997; Zanger et al., 1999; Sewer et al., 2002).
In this paper we investigated the molecular mechanisms of cAMP induction of Rbp4 transcription and showed that in cultured Hepa1 cells stimulation occurs at the mRNA and protein levels, confirming published reports (Jessen and Satre, 1998). We dissected the 5′ flanking region of the gene and identified the sequence motifs responsible for basal and cAMP-induced transcription. Interestingly, we showed that the upstream element required for the induction contains several copies of an AT-rich motif, known to be the binding site for HMGA1 proteins, structural components of chromatin. Upon synergistic binding to multiple sites, HMGA1s uncover chromatin and facilitate the recruitment of multiple transcription factors (Reeves, 2003); in the case of Rbp4, they stimulate cAMP-induction. We identified the proteins involved in this interaction as p54nrb/NonO, polypyrimidine tract-binding protein-Associated Splicing Factor (PSF) and the associated Steroidogenic factor 1 (SF1) or the related Liver Receptor Homologue-1 (LRH-1). These factors have been shown to form diverse multiprotein complexes in different cell contexts to regulate gene expression. By different experimental approaches we verified their role in Rbp4 expression in vitro and in vivo. The data suggest a relationship between transcription regulation of genes involved in retinol metabolism, steroid hormone biosynthesis and glucose homeostasis.
Materials and Methods
2.1 Reagents
8-Bromo-cAMP and luciferin were obtained from Sigma. Deoxyribonuclease I (DNase I) was purchased from Boehringer-Mannheim Biochemicals. Acetyl-CoA was obtained from Roche Molecular Biochemicals. Trypsin, dithiothreitol (DTT) iodoacetamide and α-cyano-4-hydroxycinnamic acid were purchased from Sigma. Trifluoracetic acid (TFA) HPLC grade was from Carlo Erba. All other chemicals and HPLC solvents were from Baker.
2.2 Plasmids
Plasmid constructs were generated by inserting different Rbp4 promoter regions in the polylinker of pCAT3 basic vector. To generate mRBP-CAT1 plasmid an Rbp4 genomic DNA fragment extending from -852 to +300 with respect to the transcription start site was cloned at the SmaI site. From this plasmid a series of 5′ end deletion mutants were generated taking advantage of unique restriction enzyme sites present in the original DNA fragment. mRBP-CAT2, 3, 4 and 5 constructs containing 345, 208, 164 and 81 bp of the promoter region, respectively, were generated by KpnI/StuI, KpnI/SacI, KpnI/ApaI, KpnI/SacII digestions. KpnI is located in the 5′ region of polylinker, while all other sites are included in the Rbp4 promoter fragment. The plasmids RBP-DEL1, 2, and 3, were generated by internal deletions of the -852 to +300 segment contained in mRBP-CAT1. RBP-DEL1 carries a deletion from -345 to -81 and was generated by StuI/SacII digestion of mRBP-CAT1. RBP-DEL2 and 3 carry deletions of the -345 to -164 and -164 to -81 segments, respectively, and were originated by digesting mRBP-CAT1 with StuI/ApaI and StuI/SacII. RBP-DEL4 was generated by cloning the puc18 83 bp-long polylinker in RBP-DEL3.
2.3 Cell culture and transient transfections
Hepa1 murine hepatoma cells were obtained from American Type Culture Collection and were cultured routinely in Dulbecco's modified Eagle's medium (DMEM) (Gibco Laboratories) supplemented with 10% fetal bovine serum (FBS) (Gibco Laboratories) and penicillin-streptomycin (100 IU/ml and 100mg/ml, respectively). Hepa1 cells were seeded onto 60 mm dishes 4-5 h before transfection at 70/80% confluence. Transfections were carried out by the calcium phosphate co-precipitation procedure (Graham and Van der Eb, 1973). Hepa1 were serum-deprived for 24 h and subsequently cotransfected for 16 h with the Chloramphenicol Acetyl Transferase (CAT) reporter construct (4 μg), the Protein Kinase A Inhibitor (PKI) subunit expression vector and pSV-luciferase plasmid (0.4 μg). When indicated, cells were treated for additional 8 h with 0.5 mM 8-bromo cAMP. The pSV-luciferase plasmid was used as internal control to normalize for transfection efficiency. CAT assay was performed as described (Panariello et al., 1996). Cotrasfection experiments of the CAT reporter plasmid and p54nrb/NonO, PSF and SF1 expression vectors alone or in various combinations were performed under similar conditions.
2.4 RNA isolation and Northern blot analysis
Total RNA was extracted from control or cAMP-treated Hepa1 cells with Trizol (Invitrogen), following the manifacturer's conditions. Total RNA, preliminary tested for its concentration and integrity, was analysed by Northern blot. Twenty μg of each sample were separated on denaturing formaldehyde-agarose gel electrophoresis (1%), transferred to a membrane (Amersham Life Sciences) and subsequently hybridized at 65°C with 32P-labeled probes for rat RBP and β-actin as control by random priming (Boehringer-Mannheim Biochemicals). Hybridization was carried out in a 2× sodium citrate-sodium chloride (SSC) buffer containing 50% formamide, 5× Denhardt's solution, 2% SDS, 100μg/ml denatured single strand salmon sperm DNA. Autoradiography was obtained by exposing the hybridized membranes to Kodak AR-2 film at −80°C.
2.5 Radioimmunoassay
A sensitive and accurate radioimmunoassay (RIA) was performed to measure RBP protein concentration in cells and medium of control and cAMP-treated Hepa1 cells, as described (Blaner, 1990). Proteins present in the culture media were concentrated using Centriprep-10 centrifugal concentrators (Amicon Corp.) and after this step RIA was performed on the concentrate.
2.6 Nuclear extracts and Dnase I footprinting assay
Nuclear extracts from Hepa1 cells were prepared according to Shapiro et al. (Shapiro et al., 1988). DNase I footprinting analysis was performed as described (Lichtsteiner et al., 1987). F1 (-852 /-695) and F2 (-694 /-481) DNA fragments were obtained by PCR from mRBP-CAT1 plasmid, using the following oligonucleotides as primers. F1 forward primer (CTAGATCTTTAACGACGGCCAGTGAAATTC) was derived from the upstream region of pCAT3 basic vector; F1 reverse primer TGGCCGAGTGGTTGATCGTCGGATCCTC was from -695/-678. F2 forward primer (-678/-695) GAGGATCCGACGATCAACCACTCGGCCA; F2 reverse primer (-481/-470): TGT TGTCAGAAGCTTTTTGGC. F1 and F2 DNA fragments were end-labeled with [32P]dATP, using the Klenow fragment of DNA polymerase I. Approximately 105cpm of each end-labeled probe were mixed with the indicated amounts of nuclear extracts from Hepa1 cells in a solution containing 25mM HEPES pH 7.6, 50mM NaCl, 1mM DTT, 1mM EDTA, 10μg BSA, 0.01% NP-40 and 1μg poly(dG-dC) for 20 min at room temperature. Samples were digested with DNase I for 2 min, and the reaction was stopped with 30μl of DNase I stop solution containing 50mM EDTA, 0.1% SDS, 150μg/ml yeast tRNA and 200μg/ml Proteinase K. The reaction mixtures were then incubated at 42° C for 30 min, the DNA purified by phenol-chloroform extraction and ethanol precipitated in presence of 0.1 volume of 3M sodium acetate. The pellet was resuspended in 4μl of formamide containing loading buffer, incubated at 95°C and loaded on 8% acrylamide, 7M urea sequencing gel. The gel was dried and exposed at −80°C.
2.7 Electrophoresis Mobility Shift Assay (EMSA)
Double-stranded oligonucleotides were 5′-end-labelled with [32P]γ-dATP and 10U of T4 polynucleotide Kinase. 50.000 cpm of each probe were incubated with 2-6 μg of Hepa1 cells nuclear extracts in 20 μl reaction mixture containing 20mM Tris-HCl, pH 7.5, 75mM KCl, 5μg BSA, 1mM DTT, 13% glycerol and 1μg of poly(dG/dC) (Amersham Pharmacia Biotech.) for 20 min at room temperature. In competition assays, a 100-fold molar excess of unlabeled competitor oligonucleotide was added to the reaction mixture. In supershift assays, nuclear extracts were incubated with the indicated antibodies prior addition of the labelled oligonucleotides as probes. The anti-HMGA1 antibody was a gift from Prof. G. Manfioletti Università degli Studi di Trieste, Italy; the anti-p54nrb/NonO and PSF antibodies were a gift from Dr. Phil Tucker, University of Texas, Austin, Texas; the anti-SF1 was from Santa Cruz Biotechnology. After incubation, the DNA-protein complexes were separated from the free probes on native 6% polyacrylamide gel run in 0.5X TBE, at 15V/cm at 4°C. Gels were dried end exposed to X-ray films.
The sense oligonucleotides used were:
PRDII-5′GGGAAATTCCGTGGGAAATTCCGAGCT3′
Oligo1-5′GCATGAAATAAAAATACG 3′
Oligo1A-5′TCTGGAAATAAATAAATCCTAATATGGCTTAGAAATAAAAATGCA3′
Oligo2- 5′ GGTTTCAGGGGAGTGTGGCAAGAGAAGTGGA 3′
Oligo2A - 5′GAATTCAGGGGAGTGTGGCAAGAGAAGTGGA 3′
Oligo2B - 5′ GGTTTCGAATTCGTGTGGCAAGAGAAGTGGA 3′
Oligo2C -5′ GGTTTCAGGGGAGGAATTCAAGAGAAGTGGA 3′
Oligo2D −5′ GGTTTCAGGGGAGTGTGGCGAATTCAAGTGGA 3′
Oligo2E -5′ 5′ GGTTTCAGGGGAGTGTGGCAAGAGGAATTCA 3′
Oligo2F -5′ GGTTTCAGGGGAGTGGAATTCGAGAAGTGGA 3′
2.8 DNA-binding Protein Purification, SDS-PAGE and in-situ Digestion
Protein purification was performed by DNA affinity chromatography using the purification kit (Roche Molecular Biochemicals) as recommended by the manufacturer. Briefly, concatamers of double-stranded 2B and 2F oligonucleotides were obtained by self-primed PCR technique. The PCR products were coupled to streptavidin-magnetic particles by ligation to the tethered biotinylated oligonucleotides. Nuclear extracts were mixed with magnetic particles and the specific proteins were captured by binding to the multiple sites present on the concatamerized oligonucleotides. The eluted fractions were separated on SDS-PAGE and detected by Coomassie Brilliant Blue G-250.
2.9 In situ hydrolysis and mass spectrometry analysis
Protein bands were excised from the gel and destained by repetitive washings with 50mM NH4HCO3 pH 8 and acetonitrile. Samples were reduced and carboxyamidomethylated with 10 mM DTT and 55 mM iodoacetamide in 50mM NH4HCO3 buffer pH 8. Tryptic digestion of the alkylated samples was performed for 2 h at 4°C and overnight at 37°C using 20 μl of trypsin 10ng/l. The resulting peptide mixtures were directly analyzed on a reflectron Voyager DE PRO MALDITOF mass spectrometer (Applied Biosystems). The mass range was calibrated using the [M+H]+ ions from the standard peptide mixture provided by the manufacturer. About 1.0 μl of sample was applied to the sample plate and mixed with 1.0 μl of a 10 mg/ml α-cyano-4-hydroxycinnamic acid solution in acetonitrile/0.2% TFA (70:30, v/v) before air-drying. Peptide mass values recorded as monoisotopic masses, were then introduced into MASCOT peptide fingerprinting search program (Matrix Science) available on the net and used for protein identification.
2.10 Chromatin immunoprecipitation and RNA interference assays
Chromatin immunoprecipitation assays were performed as indicated by the manufacturer (Epigentek Inc.). Briefly, Hepa1 cells were seeded onto 60 mm dishes at 70-80% confluence and cultured in 10% FBS. Cells were serum-deprived for 48 h and then stimulated for additional 8 h with 0.5mM 8-bromo cAMP. Both proliferating and cAMP-induced cells were tripsinized and harvested in PBS. About 106 cells of each sample were fixed in 1% formaldehyde for 10 min and the reaction blocked with 0.125M glycine. Chromatin was sonicated in lysis buffer (1% SDS, 10 mM EDTA and 50 mM Tris pH 8.0, plus protease inhibitors), centrifugated and purified. Ten μl of the supernatants was used as input; the remaining lysate was subjected to the assay with each of the different antibodies directed against HMGA1, PSF, p54/NonO, SF1, LRH-1, Pol II, respectively, as mentioned above. About 50 μg of immunoprecipitated DNA was used for the PCR reaction using as primers oligonucleotides spanning the Rbp4 promoter fragment from -825 to -635 (forward: TGAGCCAGTTTTCCTCGCT; reverse: CAACCTGGTCAGGACAGGA). Immunoprecipitated DNA was analysed also by quantitative PCR (qPCR) using a Master Mix SYBR (Biorad) and the above reported oligonucleotides.
Gene knockdown experiments were performed by transiently transfecting a pool of siRNAs (Sc-37116) directed against HMGA1 mRNAs, following the protocol suggested by the manufacturer (Santa Cruz). As a control, a pool of unrelated siRNAs (scrambled siRNAs, Sc-37007) was also used. Briefly, Hepa1 cells were transfected with 100 pM of HMGA1 and scrambled siRNAs, respectively. After 24 h, cells were trypsinized, seeded in 60 mm plates and transfected again with 100 pM of the corresponding siRNAs. After additional 24 h, cells were harvested and total protein extracts prepared for Western blot analysis.
Stable knockdown of SF1 gene was achieved by transfecting a retroviral vector pSM2c (Open Biosystem, Huntsville, AL) carrying a short hairpin RNA based on a target sequence (ACCTGGCCTGCAGTGCGTGAAA) coding for the Ligand Binding Domain of the protein. This sequence is homologous to that of the closely related LRH-1 gene, so we would expect to knock down both genes. 48 h after transfection, cells were harvested and seeded in a selection medium containing 1ug/ml of puromycin for the next 6 days. A vector containing a short unrelated hairpin RNA was used as control. Single clones were isolated and characterized for SF1/LRH-1 expression by western blot analysis. The specificity of the silencing was tested assessing the expression of proteins as PSF, p54/NonO, RXRα, cyclin A.
2.11 Glucagon injection and mRNA analysis by qRT-PCR
Wild type C57/black 6 mice were intraperitoneally injected with human glucagon (1 mg/kg body weight) or saline after 3-4 h fasting. At different times from the injection, the mice were killed by cervical dislocation and the liver was rapidly removed, frozen into liquid nitrogen and stored at −80 °C. For qRT-PCR, total cellular RNA was extracted from tissue and subjected to DNaseI treatment. RNA levels were normalized against 18S ribosomal RNA in each sample, and cDNAs were synthesized from 1 μg of total RNA using Omniscript reverse transcriptase (Qiagen). Primers for RBP4 (5′-AGGAGAACTTCGACAAGGCT-3′; 5′-TTCCCAGTTGCTCAGAAGAC-3′) and HMGA1 (5′-GCAGGAAAAGGATGGGACTG-3′ and 5′-AGCAGGGCTTCCAGTCCCAG-3′) cDNAs were designed according to sequences from the GeneBank database and relative quantification was made against GAPDH cDNA used as an internal standard. All PCR reactions were carried out in triplicates.
3. Results
3.1 cAMP stimulates RBP4 mRNA and protein synthesis
We confirmed and extended the observation by Jessen and Satre (Jessen and Satre, 1998) that Rbp4 is induced by cAMP in Hepa1 cells, which are derived from a murine hepatoma (Supplementary Fig.1A). The cells were kept in serum-free medium for 48 h and then treated with increasing concentrations of 8-bromo cAMP (Br-cAMP) for 24 h. Northern blot analysis of total RNA showed a robust increase of RBP4 mRNA over the basal level in cells treated with 0.5 mM Br-cAMP (lane 5) before decreasing at higher Br-cAMP concentrations. This value (0.5 mM) corresponds to the standard concentration reported in the literature for cAMP responsive genes (Jessen and Satre, 1998). Induction kinetics was determined in a time-course experiment (Suppl. Fig.1B). In the presence of 0.5 mM Br-cAMP, RBP4 mRNA levels increased starting at 3 h, peaked at 24 h and then declined, suggesting a transient transcriptional stimulation (lane 6). mRNA induction was paralleled by an equivalent elevation of the protein (Suppl. Fig 1C). A Radio Immuno Assay (RIA) carried out on the medium of cells exposed to 0.5 mM Br-cAMP indicated de novo synthesis in response to Br-cAMP with a peak around 48 h.
3.2 Identification of the cis-regulatory sequences that mediate cAMP-induction of Rbp4 transcription
To identify the cis-regulatory sequences responsible for induction, a DNA fragment from the 5′ flanking region of Rbp4 was subjected to computational analysis to recognize and locate putative binding sites for transcription factors. This fragment was derived from a lambda clone containing the complete gene, originally isolated from a mouse genomic library. Several canonical sites for ubiquitous factors (Ap1 and Sp1) were identified in the more proximal promoter sequences (Fig. 1A). Remarkably, two CREs were identified at position -177/-171 and -63/-57 from the transcription start site, implying that CREB bound to these two sites could mediate Rbp4 induction. To determine if these two sites are involved in Rbp4 induction, we generated a set of chimeric CAT fusion plasmids (Fig. 1B). The fusion carrying the largest segment of Rbp4, mRBP-CAT1, contains a DNA segment extending from -852 to +300, with respect to the Rbp4 transcription start site, fused to the CAT reporter gene.
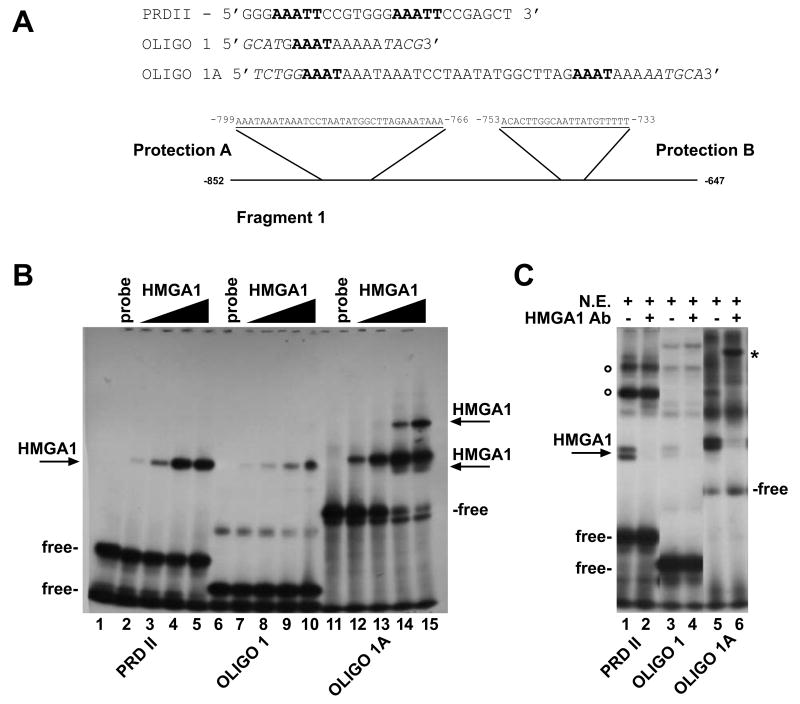
Figure 1. Nucleotide sequence of the Rbp4 promoter and schematic representation of the chimeric plasmids used to identify the cis-regulatory elements that mediate cAMP induction.
(A) Upper panel: 1.1 kb of the 5′ flanking region of Rbp4 cloned in the p-CAT3 basic vector are reported. Circa 800 bp of the flanking region are numbered in negative. Only the 300 bp transcribed sequences carried by the mRBP-CAT1 plasmid are numbered in positive. The two CRE sites, the TATA box and the transcriptional start site are indicated in boldface. Lower panel: enlargement of the more proximal promoter region. The binding sites for transcription factors and their relative positions are indicated. The more upstream regulatory region is also reported with the position of the 5′ and 3′ nucleotide borders. (B) mRBP-CAT chimeric constructs were transfected into Hepa1 cells either treated with 8-Br-cAMP or cotransfected with the PKI (Protein Kinase A Inhibitory Subunit) expression vector as control. The mRBP-CAT plasmids were generated by fusing fragments of various length from the 5′ flanking region of Rbp4 to the CAT reporter gene. Both 5′ and internal deletion mutants are depicted in scale and “breaks” have been included. The results are the mean +/= S.E. of at least five separate experiments carried out in duplicate with different DNA preparations. P < 0.05 was calculated and was statistically significant in induced versus basal level. Data refer to CAT/Luciferase activity ratio to normalize for transfection efficiency and are expressed as percentage of pSV2CAT activity taken as 100%. Fold induction refers to the basal CAT activity of the mRBP-CAT1 construct. A CRE-TK-CAT plasmid was used as positive control in all experiments. (C) mRBP-TK-CAT constructs 1-5 were transfected into Hepa1 cells either treated with 8-Br-cAMP or cotransfected with the PKI (Protein Kinase A Inhibitory Subunit) expression vector as control. The results are the mean +/= S.E. of at least five separate experiments carried out in duplicate with different DNA preparations. Data refer to CAT/Luciferase activity ratio to normalize for transfection efficiency and are expressed as fold induction referred to the basal CAT activity of the RBP-TK-CAT plasmid 3. The P value was calculated and was < 0.05 versus basal levels.
Transfection of the pCAT3 empty expression vector in Hepa1 cells yielded negligible levels of CAT expression either in the presence or absence of cAMP. In contrast, mRBP-CAT1 induced a detectable level of expression that was enhanced about 6-fold in the presence of cAMP (Fig. 1B). Hepa1 cells contain substantial levels of active PKA. To determine true basal expression levels, therefore, each reporter plasmid was cotransfected with a plasmid carrying the cDNA for the PKA inhibitory subunit (PKI) in the absence of exogenous cAMP. Fold induction was thus referred to these values. As a positive transfection control, we used the CRE-TK-CAT construct. This plasmid bears three copies of a CRE-containing oligonucleotide, derived from the Somatostatin gene and fused upstream to the Thymidine Kinase minimal promoter, which in turn drives transcription of the CAT gene.
To identify the minimal promoter fragment that mediates the cAMP response, we generated 5′ deletion mutants of the mRBP-CAT1 construct, yielding plasmids mRBP-CAT 2, 3, 4 and 5 (Fig. 1B, upper part). CAT expression from mRBP-CAT2 and 3, carrying 345 and 208 bp of Rbp4 promoter sequence, respectively, was not stimulated by cAMP, even though both carry the two CRE elements. Note that plasmid mRBP-CAT 3 expressed high basal CAT activity. Plasmids mRBP-CAT 4 and 5, containing 164 and 81 bp of the 5′ flanking region, showed little activity. These results indicate that sequences upstream to -345 contain necessary elements for cAMP stimulation and sequences downstream to -345 are responsible for basal transcription. We conclude that the two CRE sites present in this promoter fragment do not contribute to cAMP enhancement of Rbp4 transcription.
To further analyse the sequence elements of the Rbp4 promoter, we generated plasmids carrying internal deletions (Fig. 1B, lower part). Plasmid DEL-CLONE1, which carries upstream DNA from -852 to -345 and downstream DNA from -81 to + 300, showed no cAMP induction. In contrast, plasmid DEL-CLONE2, which contains additional DNA from -81 up to -164, was induced by cAMP. Both plasmids carry only the CRE2 site, but DEL-CLONE2, in addition, carries three downstream Sp1 sites. Recall that mRBP-CAT4, which lacks the upstream elements, shows no cAMP induction. The importance of the AP1 and Sp1 sites is again shown by the results obtained with plasmids DEL-CLONE3 and 4 in which the AP1 and three Sp1 sites present in the proximal promoter between the two CRE sites (from -164 to -81) were deleted or replaced by a polylinker sequence of the same length, keeping unchanged the DNA phasing of this promoter segment. These plasmids showed little basal transcription and no cAMP induction. Taken together these data demonstrate that cAMP induction is mediated by: 1) an upstream region located between -852 and -345, and 2) a proximal region that contains binding sites for the general transcription factors AP1 and Sp1. These results were corroborated by transfections with plasmids carrying segments of the Rbp4 promoter fused to a heterologous promoter (minimal TK promoter), in cells exposed to cAMP or cotransfected with the PKI expression vector (Fig. 1C). Chimeric mRBPTK-CAT 1 and 2 plasmids, carrying only the upstream (-852 to -345) or the downstream (-242 to -23) promoter elements, respectively, displayed no cAMP induction. In contrast, plasmid mRBPTK-CAT 3, containing both elements, was induced by cAMP to a level similar to that obtained with the mRBP-CAT1 construct carrying the native promoter. The 3′ border of the upstream element was further defined at position -482 because the mRBPTK-CAT 4 plasmid in which an upstream element extending from -835 to -482 fused to the -242 to -23 element was induced by 6-fold, as the mRBPTK-CAT 3 construct. mRBPTK-CAT5 plasmid that carries only the -242 to -23 element fused to the -482 to -345 fragment elicited no induction. Finally, the role of the cAMP-PKA pathway in Rbp4 induction was confirmed by cotransfecting the various reporter constructs with the PKA catalytic subunit instead of exposing the cells to Br-cAMP. Equivalent results were obtained (data not shown).
3.3 HMGA1 proteins specifically bind cis-elements in the more distal promoter
To identify the sequence motifs contained in the -852/-482 fragment that specifically interact with transcription factors to mediate the cAMP response, we performed DNase I footprinting experiments. The original Rbp4 segment was divided in two shorter fragments (F1 from -852 to -697 and F2 from -667 to -482), which were 32P-labelled, mixed with nuclear extracts from Hepa1 cells and subjected to DNAse I digestion (Suppl. Fig. 2A and B). Protected tracts were detected on both strands, indicating that they are binding sites for transcription factors contained in basal or cAMP-induced nuclear extracts. Inspection of the protected sequences revealed several AT-rich motifs homologous to the binding sites of high mobility group proteins (HMGA1), members of HMGAs protein family that is involved in chromatin architecture (Reeves, 2003). No additional AT-rich sequences were present in the remainder of the promoter segment analyzed in this study. This suggests that interactions between these DNA motifs and HMGA1 proteins play a role in Rbp4 transcription, consistent with the results obtained with deletion mutants. To verify that HMGA1 proteins indeed bind these sequences, we performed Electrophoretic Mobility Shift Assay (EMSA) using several oligonucleotides based on the AT-motifs contained in the distal promoter segment and recombinant HMGA1. Retarded complexes of variable intensity were obtained, indicating that HMGA1 binds its cognate sequences with different affinities (data not shown). Oligo1 and 1A, based on the F1 fragment protected sequences (Fig. 2A), formed the more intense complexes; consequently, they were selected and further analyzed. Oligo1 interacted with HMGA1, generating a retarded complex (Fig. 2B, lanes 7-10), whose electrophoretic mobility was similar to that obtained with PRDII, an oligonucleotide based on the HMGA1 binding site of β-interferon promoter (lanes 2-5). Oligo 1A, which carries two HMGA1 binding sites, had higher affinity for HMGA1 than Oligo 1 and generated an additional, slower-moving complex (Fig. 2B, lanes 12-15).
Figure 2. HMGA1 proteins bind the AT-rich motifs in the Rbp4 distal promoter.
(A) The nucleotide sequences of the probes used in EMSA are depicted. The AT-rich motifs are shown in boldface. PRDII was based on the HMGA1 binding site of β-interferon promoter. Oligo 1 and Oligo 1A were based on the F1 fragment of Rbp4 distal promoter. The coordinates of Oligo 1 and 1A in the context of the F1 fragment-protected regions are indicated. The nucleotides not included in the protections are indicated in italics. (B) EMSAs were performed with labelled PRDII (lanes 1-5), Oligo1 (lanes 6-10) or Oligo1A (lanes 11-15), as probes treated with increasing amounts (5-10-25 and 50 ng) of purified HMGA1. The DNA-HMGA1 complexes are indicated by arrows. Free indicates the unbound probes; (C) EMSAs were performed with 3 μg of nuclear extracts from Hepa1 cells (lanes 1-6), and radiolabeled PRDII (lanes 1 and 2), Oligo 1 (lanes 3 and 4) and Oligo 1A (lanes 5 and 6). Nuclear extracts were incubated with 1 μl of a specific anti-HMGA1 antibody (200 mg/ml) (lanes 2, 4 and 6) prior addition of the probes. The position of the DNA-HMGA1 complexes is indicated by the arrow, unbound probes are indicated as free at the bottom of the figure. The supershifted band is indicated by an asterisk (lane 6). Empty circles indicate the position of the DNA-NFkB complex on its site contained in PRDII.
We then demonstrated that these oligonucleotides bind HMGA1 also in Hepa1 nuclear extracts as shown by EMSA. Retarded bands with similar mobility as seen with the recombinant protein were detected (Fig. 2C lanes 1, 3, 5). The retarded doublet that migrates at the level of the purified protein when the HMGA1 specific oligonucleotide is challenged with nuclear extracts, is representative of two HMGA1 isoforms produced by alternative splicing (Reeves, 2003). Addition of an anti-HMGA1 antibody to the reaction mixtures reduced the intensity of the HMGA1 containing-retarded bands (lanes 2, 4, 6). In the case of oligonucleotide 1A, a supershifted band was detected (lane 6). Recall that oligo 1A bears two binding sites for HMGA1 and thus is more efficient in protein binding so that the supershifted band observed can be explained. Recall that oligo 1A bears two HMGA1 binding sites. We assume that the increased amount of bound HMGA1 binding accounts for the supershifted band. Moreover, DNA-protein complexes of different migration and intensity were generated, suggesting that the oligonucleotides carry binding sites for other transcription factors, as previously reported for PRDII (Mantovani et al., 1998). These results demonstrated that HMGA1 proteins bind upstream sequences of the Rbp4 promoter.
3.4 HMGA1 proteins plays a pivotal role in basal and cAMP-induced Rbp4 expression
We then performed a series of experiments designed to test whether HMGA1 binding to the Rbp4 promoter modulates its transcription. Nuclear and cytosolic extracts were prepared from Hepa1 cells kept in the presence or absence of 0.5 mM Br-cAMP for 24 h and analysed by Western blot. A 21 kDa band was detected in the cytosolic extracts by a specific anti-RBP4 antibody, whose intensity increased in the presence of cAMP. Interestingly, an antibody to HMGA1 detected a specific band in the nuclear extracts whose intensity also increased after cAMP. In contrast, β-actin and RXRα, representative proteins of the cytosolic and nuclear compartments, respectively, did not vary after cAMP exposure (Fig. 3A, left panel). Densitometric analysis of the bands obtained in three separate experiments and normalization to β-actin and RXRα are reported in the histograms in the right panel. RBP4 increased about 7-fold, consistent with the experiments reported above, while HMGA1 increased by 2-fold, indicating modest induction of this protein by cAMP.
Figure 3. HMGA1 proteins play a pivotal role in basal and cAMP-induced Rbp4 transcription.
(A, left panel) Hepa1 cells were serum starved for 48 h and kept in the presence or absence of cAMP for additional 24 h. Nuclear and cytosolic extracts were then prepared and analyzed by western blot with antibodies against HMGA1, RBP4, RXRα and β-actin. (A, right panel) Densitometric analysis of the bands of at least three independent experiments and quantitation to RXRα and β-actin, respectively, is reported. The results are the mean +/= S.E. of at least five separate experiments. RBP4 and HMGA1 basal levels are indicated as 1 on the relative density arbitrary scale. Fold-induction over basal level is also reported. The P value was calculated and was < 0.05 versus basal levels. (B) Hepa1 cells were transfected with siRNA specific to HMGA1 (lane 1) or unrelated, scrambled siRNA (lane 2) for 48 h. HMGA1 and RBP4 protein levels were detected by immunoblotting with anti-HMGA1 and anti-RBP4 antibodies, respectively. Antibodies against β-actin were used for proteins unaffected by either siRNA treatment. The results reported in the histograms on the right side of the figure are the mean +/= S. E. of at least three separate experiments. The HMGA1 and RBP4 basal expression is reported as 1 on the arbitrary scale along with the extent of the reduction following silencing. The P value was calculated and was < 0.05 versus basal levels. (C) HMGA1 and RBP4 mRNA liver expression in vivo following glucagon-injection in mice. Total RNA was isolated from liver of fasted mice, before and after i. p. glucagon injection. HMGA1 and RBP4 mRNA levels were measured at the indicated time intervals by quantitative RT-PCR as described in the Material and Methods section. Results are the mean values ± S.E. obtained from four to six animals per group. Black bars, HMGA1 mRNA; dashed bars, RBP4 mRNA. P < 0.05 versus control group (time 0).
We then asked if HMGA1 regulates transcription from an RBP-CAT chimeric plasmid. To this goal, we first cotransfected mRBP-CAT1 reporter construct and the pKI expression vector into Hepa 1 cells to determine the basal level of expression. CAT activity was induced by ∼6 fold in the presence of cAMP, confirming the results reported above (Suppl. Figure 3). Overexpression of exogenous HMGA1 did not affect basal CAT activity. This probably reflects the abundant levels of HMGA1 already present in Hepa1 cells. However, in the presence of cAMP a dose-dependent stimulation of CAT activity was observed with increasing amounts of the transfected HMGA1 expression vector. Induction declined at doses higher than 300 ng of transfected plasmid. It has been reported that HMGA1 at high concentration forms inactive multiprotein complexes that no longer bind DNA (Reeves, 2003). The effect of overexpressed exogenous HMGA1 was tested also on the endogenous RBP4 gene. Basal transcription was not affected, probably due to the abundant levels of HMGA1 present in the cells. Upon cAMP exposure, RBP4 gene was induced as already reported above (data not shown).
To further assess the role that HMGA1 plays in Rbp4 transcription, we transiently transfected Hepa1 cells with a pool of siRNAs specific to HMGA1 or with control scrambled siRNA, and prepared protein extracts 72 h later. HMGA1 diminished by almost 50% in cells transfected with specific siRNAs as compared to control siRNAs (Fig. 3B). Interestingly, RBP4 was similarly reduced in cells transfected with HMGA1 siRNAs. β-actin, in contrast, was not affected by either pool of siRNAs.
To test whether RBP4 and HMGA1 genes respond to similar stimuli in vivo, we administered the hormone glucagon, which stimulates cAMP synthesis, to wild type mice. HMGA1 and RBP4 mRNA levels were determined in liver extracts: HMGA1 mRNA began to increase at 3 h, peaked at 6 h and remained elevated up to 9 h after injection, suggesting an early response to the hormone treatment (Fig. 3C). RBP4 mRNA showed a later induction that started at 6 h and reached a plateau at 9 h. These results indicate that Hmga1 and Rbp4 are both responsive to glucagon and thus to cAMP pathway stimulation in vivo.
Taken together the experiments reported in this section demonstrate that the HMGA1 binding to the RBP4 promoter has a key role in Rbp4 basal and cAMP induced transcription. siRNA and in vivo experiments support this notion suggesting that the HMGA1 and RBP4 genes respond to similar stimuli, and implying that they are part of a novel biochemical pathway induced by cAMP.
3.5 HMGA1 binding facilitates DNA-protein interactions at the Rbp4 promoter
HMGA1s regulate gene expression by recruiting transcription factors to specific DNA sites. To establish whether such a mechanism underlies cAMP induction of Rbp4 transcription, we carried out DNase I footprinting assays with the F1 fragment as probe, purified HMGA1 protein and Hepa1 nuclear extracts (Suppl. Fig. 4, lanes 5-7). When the F1 probe was mixed with nuclear extracts, we observed protection of the AT-rich elements, as well as protection of DNA between positions -725 and -699 (indicated as Oligo 2 sequence, Suppl. Fig 4, lane 4). This suggests that other proteins, distinct from and recruited by HMGA1, interact with the sequences from -725 to -699. To verify that HMGA1 indeed facilitates binding of proteins to the nearby site, the F1 probe was mixed with Hepa1 nuclear extracts and increasing concentrations of HMGA1 protein. The intensity of the protection from nucleotide -725 to -699 enhanced proportionally to the amount of the recombinant protein added to the reaction (lanes 8-10).
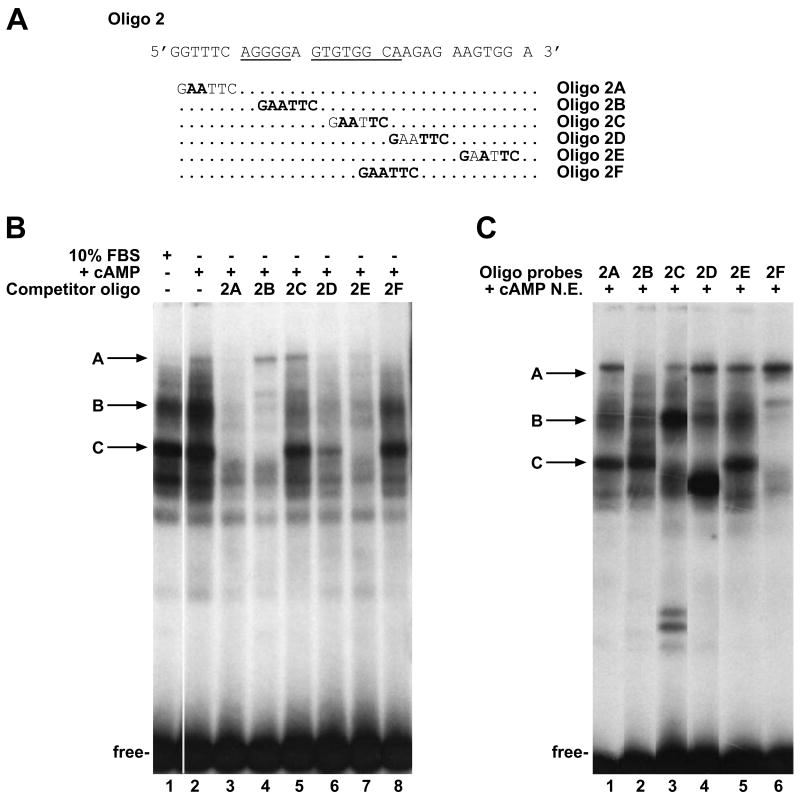
To define the protein binding sites contained in the -725 to -699 DNA segment, an oligonucleotide (Oligo 2, Fig. 4A) was synthesized, labelled and used as a probe in EMSA with nuclear extracts from proliferating Hepa1 cells (10% FBS) or Hepa1 cells induced with Br-cAMP (Fig. 4B, lanes 1 and 2, respectively). Two major bands, B and C, were seen with proliferating cells and three, A, B and C, after cAMP induction. To identify the protein binding sites of each complex, we used Oligo 2 mutants as unlabelled competitors (see the scheme in Fig. 4A). Oligo 2A and 2E competed for bands A, B and C as efficiently as unlabelled Oligo 2 (lanes 3, 7 and Suppl. Fig. 7). The mutations in these oligos, therefore, do not affect complex formation. Oligo 2B competed for the B and C complexes, without affecting the A complex (lane 4). Oligo 2C competed for the B complex, but not for the A and C complexes (lane 5), whereas Oligo 2F prevented the formation of the A complex without affecting the B and C complexes (lane 8). Finally, Oligo 2D competed for the A and B complexes and partially abrogated the C complex (lane 6). These results indicate that the complexes are generated by interaction of proteins with separate and distinct DNA motifs.
Figure 4. Identification of the binding sites for transcription factors.
(A) The sequences of the wild-type (Oligo2) and mutated Oligos (2A-2F) used in EMSA are illustrated. (B) EMSAs were performed with 3 μg of nuclear extracts from proliferating (lane 1) and cAMP induced Hepa1 cells (lanes 2-8), in the presence of labelled Oligo2 as probe (lanes 1-8). Incubation with 100-fold molar excess of the unlabelled mutated oligos as competitors (lanes 3-8) was carried out before adding the probe. The positions of the A, B and C complexes are indicated by the arrows; the unbound (free) probe is indicated at the bottom of the figure. (C) EMSAs were performed with 3 μg of nuclear extracts from cAMP-induced Hepa1 cells in the presence of the radiolabelled Oligo2 mutants as indicated (lanes 1-6). The positions of the A, B and C complexes are indicated by the arrows; the unbound (free) probe is indicated at the bottom of the figure.
To confirm these data, the above Oligo 2 mutants were used as labelled probes with extracts from induced cells (Fig. 4C). As expected, Oligo 2A and 2E yielded complexes A, B and C (lanes 1 and 5). Oligo 2B formed complexes B and C but not A (lane 2). With Oligo 2C complex C was eliminated, complex A was reduced and complex B was enhanced (lane 3). In addition, two fast moving complexes were observed. Oligo 2D eliminated complex C and formed a faster moving complex (lane 4). Oligo 2F formed only the A complex (lane 6). These results demonstrate that the B and C complexes are generated by the interaction of protein(s) with the GTGTGGCA sequence motif located in the central part of Oligo 2 (Fig. 4A). The A complex, which is formed in response to cAMP (Fig. 4B), involves an AGGGG motif (Fig. 4A).
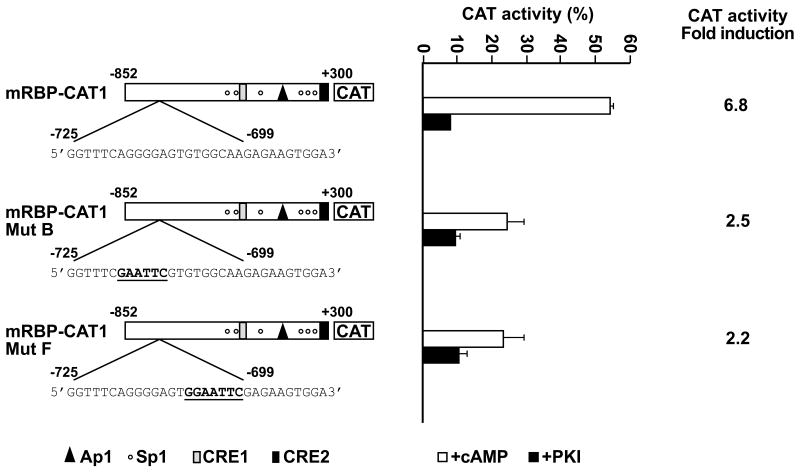
To correlate these biochemical results with functional data, the binding sites for the A and B complexes were site-directed mutated in the context of the mRBP-CAT1 plasmid (Fig. 5). The resulting mRBP-CAT1mutB and mRBP-CAT1mutF plasmids were transfected into proliferating Hepa1 cells in the presence of pKI, or into Hepa1 cells induced with Br-cAMP. For comparison, cells were transfected with the mRBP-CAT1 control plasmid. In the presence of Br-cAMP, mRBP-CAT1mutB and mRBP-CATmutF plasmids resulted into CAT activity that was only 30% that obtained with the mRBP-CAT1 plasmid. Thus, the DNA segment from -725 to -699 carries binding sites for factors directly involved in cAMP-dependent Rbp4 transcription, although a role in basal transcription cannot be ruled out.
Figure 5. Site-directed mutation analysis of Rbp4 promoter region.
Site-directed mutations, underlined and in bold face, were inserted, between nucleotides -725 and -699, in the context of the mRBP-CAT1 plasmid. The resulting RBP-CAT mutB and RBP-CATmutF plasmids were transiently transfected along with the wild type mRBP-CAT1 construct into Hepa1 cells, as reported (see Fig. 1). The results are the mean +/= S.E. of at least five independent experiments carried out in duplicate with different DNA preparations. Data refer to CAT/Luciferase activity ratio to normalize for transfection efficiency and are expressed as fold induction to basal CAT activity of the mRBP-CAT1 construct taken arbitrarly as 1. The P value was calculated and was < 0.05 versus basal levels.
3.6 Purification and identification of proteins that bind the distal promoter fragment
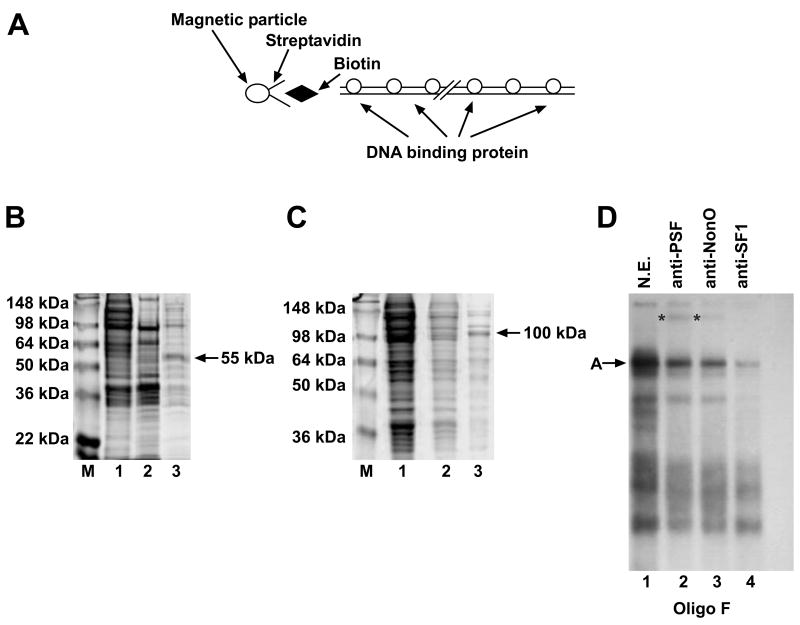
To identify the protein species contained in retarded bands A, B and C, we established a DNA affinity chromatography purification step (Fig. 6A). Mutant Oligo 2B and 2F were multimerized and used as bait to isolate cognate binding proteins, as described in Materials and Methods. Oligo 2B enriched a 55 kDa molecular mass protein from nuclear extracts of proliferating or cAMP induced cells. Coomassie blue staining of an SDS-PAGE gel loaded with the input (1), the excluded (2) and retained proteins (3) of proliferating Hepa1 cells is illustrated in Fig. 6B. Oligo 2F enriched a protein of approximately 100 kDa molecular mass from nuclear extracts of proliferating or cAMP induced Hepa1 cells (Fig. 6C). The 55 kDa and 100 kDa bands were isolated, trypsin-digested and analysed by MALDI-TOF. The 55 kDa protein was identified as p54nrb, the human homologue of murine NonO, a non-Pou domain-containing octamer– binding protein. The 100-kDa band was identified as PSF, the polypyrimidine tract-binding protein-associated splicing factor.
Figure 6. Different DNA binding proteins constitute the A and B complexes that regulate basal and cAMP-dependent Rbp4 transcription.
(A) Scheme of the affinity chromatography strategy used to purify the DNA binding proteins present in the A and B complexes. (B) Comassie blue staining of a 10% SDS-PAGE loaded with 30 μg of the input crude extract (lane 1), 30 μg of the flow-through (lane 2) and 3 μg of the retained proteins (lane 3) from the multimerized Oligo 2B used as bait. The 55 kDa molecular mass of the enriched protein is indicated by an arrow. M indicates the molecular weight markers. (C) Comassie blue staining of a 10% SDS-PAGE loaded with 30 μg of the input crude extract (lane 1), 30 μg of the flow-through (lane 2) and 3 μg of the retained proteins (lane 3) from the multimerized Oligo 2F used as bait. The 100 kDa molecular mass of the enriched protein is indicated by an arrow. M indicates the molecular weight markers. (D) EMSA was performed with the labeled Oligo 2F as probe and 3 μg of nuclear extracts from cAMP induced Hepa1 cells (lanes 1-4). Incubation with 1 μg of anti-PSF (lane 2), anti-p54nrb/NonO (lane 3) or anti-SF1 (lane 4) antibodies, respectively, was carried out before adding the probe. The position of the A complex is indicated by the arrow, supershifted bands are indicated by asterisks. The pattern of retarded bands obtained with Hepa1 nuclear extracts and the same probe is shown in lane 1.
3.7 p54nrb/NonO, PSF and SF1 constitute a functional complex that mediates cAMP induction of Rbp4 transcription in vitro and in vivo
To determine the components of band A, which is specifically formed in cAMP-induced cells, we carried out EMSA with the labelled oligonucleotide 2F, induced extracts and anti-p54nrb/NonO and PSF antibodies. A reduction in the recovery of band A as well as a supershift was observed with both antibodies (Fig. 6D). Because p54nrb/NonO and PSF (Sewer et al., 2002) are reported to form a ternary complex with Steroidogenic Factor 1 (SF1), we tested for the presence of this factor in complex A. The anti-SF1 antibodies employed in these experiments were specifically directed against the DNA binding domain of the protein, thus significantly reducting the retarded complex (Fig. 6D, lane 4). All three antibodies reduced the intensity of other retarded bands, that are unrelated to the complex A band, as already shown in Fig. 4 and as proved also by an unrelated antibody or a preimmune serum (data not shown). SF1 is structurally and functionally related to LRH-1 and is recruited by p54nrb/NonO and PSF to activate gene expression in different cell types (Fayard et al., 2004). To demonstrate that LRH-1 is present in the multiprotein complex described, we performed EMSA in the presence of an anti-LRH-1 antibody. The intensity of the retarded complex was reduced similarly to the anti-SF1 antibody (data not shown). We conclude that band A contains at least these proteins.
Moreover, we cotransfected the expression vectors for the three factors together with the mRBP-CAT1 reporter plasmid. The combined overexpression resulted in levels of CAT activity equivalent to those obtained in the presence of cAMP. The CRE-TK-CAT construct in the same experimental conditions showed a completely different response. These data suggest that the multiprotein complex specifically modulates cAMP-dependent Rbp4 transcription (see Suppl. Fig. 5 and 6).
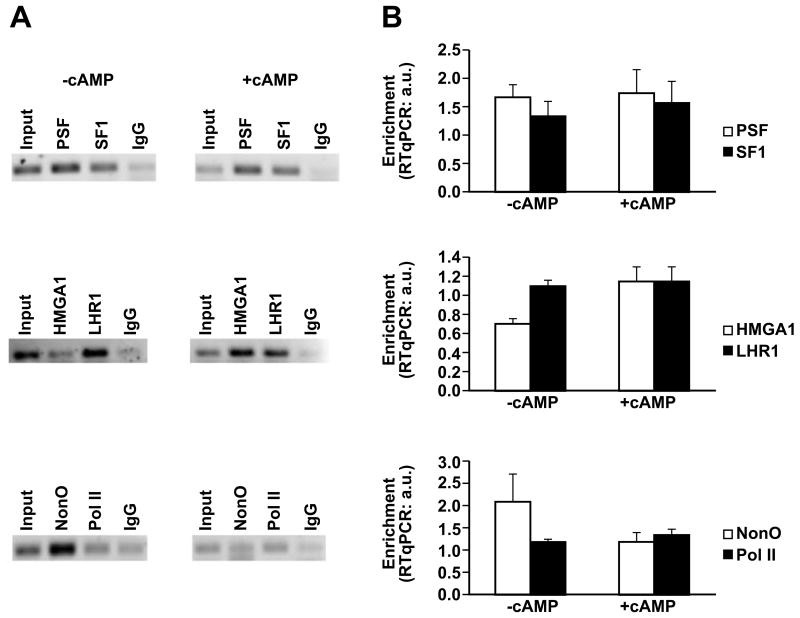
Finally, we carried out Chip assays to demonstrate that these factors regulate Rbp4 expression in vivo (Fig. 7). Chromatin from untreated and cAMP-treated cells was precipitated with antibodies directed against each of these transcription factors. Subsequently, the DNA promoter fragment containing the binding sites for these proteins was PCR-amplified. In proliferating cells, p54nrb/NonO, PSF and SF1/LRH-1 bind the cognate DNA along with HMGA1 (Fig. 7A). Quantification (Fig. 7B) indicates that PSF and SF1 binding does not vary with cAMP exposure. Upon cAMP induction, in contrast, HMGA1 binding is enhanced whereas p54nrb/NonO is reduced. An anti-Polymerase II antibody was used as positive control of transcriptional activity and, in fact, the intensity of the corresponding band does not change; an anti-IgG antibody was used as negative control. Taken together, these results confirm that these factors contribute to Rbp4 transcription both in vivo and in vitro. In addition, they demonstrate that in vivo HMGA1, p54nrb/NonO, PSF, SF1/LRH-1 are all recruited on the promoter in basal conditions whereas, upon cAMP induction, NonO and HMGA1binding are inversely modified. These data suggest that the assembly of the ternary complex required for Rbp4 transcription undergoes higher order rearrangements likely due to posttranslational modifications of one of these factors (SF1/LRH-1) and to a stronger binding of HMGA1. These may also change the relative positioning on the promoter and, in turn, explain the results obtained.
Figure 7. HMGA1, p54nrb/NonO, PSF and SF1/LRH-1 are recruited on the Rbp4 promoter in vivo.
(A) Hepa 1 cells were either treated with 8-Br-cAMP or cotransfected with the PKI expression vector as control. Cross-linked chromatin was immunoprecipitated with the indicated antibodies. The immunoprecipitated DNA was amplified using specific primers for the Rbp4 promoter region located between −825 and − 635. (B) Quantitative analysis by Real Time PCR of the HMGA1, p54nrb/NonO, PSF, SF1, LRH-1 and Pol II binding to the Rbp4 promoter in cAMP-induced and control Hepa1 cells. The results are the mean +/= S.E. of three independent experiments carried out with different chromatin preparations. The P value was calculated and was < 0.05 in induced versus control cells.
3.8 SFI/LRH-1 mediate cAMP stimulation of Rbp4 transcription
To demonstrate that SFI/LRH-1 mediate cAMP induction, we sought to stably knockdown the corresponding genes using siRNA. Hepa1 cells were transfected with a retroviral vector carrying a short hairpin RNA directed against SF1 mRNA. The shRNA corresponded to mRNA sequences encoding the ligand binding domain of the protein. Since this segment is homologous to LRH-1, both genes are expected to be downregulated. Cells were grown for a week in a selection medium containing 1 μg/ml of puromycin and single clones were isolated, expanded and individually analysed for SF1 and LRH-1 protein levels. Among the diverse cell clones showing reduced SF1 levels, we further analysed clone D2, the most effectively silenced. In Hepa1 nuclear extracts, SF1 and LRH-1 were detected, as expected. In D2 nuclear extracts, LRH-1 levels were reduced by about 50%, and SF1 was almost undetectable (Fig. 8A). The levels of PSF, p54nrb/NonO and RXRα were equivalent in both cells, confirming the specificity of the knockdown. No variations were observed in the levels of SF1/LRH-1 and the other proteins investigated in cells transfected with unrelated, control shRNA (data not shown).
Figure 8. SFI/LRH-1 mediate cAMP stimulation of Rbp4 transcription.
(A) SF1 gene was stably depleted by shRNA technology, following transfection of a viral vector as described under Materials and Methods. Nuclear and cytosolic extracts from parental Hepa1 and derived clone D2 cells were tested by Western blot for LRH-1 and SF1 levels. PSF, p54/NonO and RXRα were detected as unrelated, unaffected proteins. Densitometric analysis of the bands and quantitation to RXRα was carried out; the results illustrated in the histograms are the mean +/= S.E obtained in three independent experiments. Black bars refer to Hepa1 and dashed bars to D2 cells. (B) Hepa1 and D2 cells were serum starved for 48 h and exposed to cAMP stimulation for 48 h. Nuclear and cytosolic extracts were then prepared and analyzed by Western blot with antibodies against RBP4, β-actin, HMGA1 and RXRα. Densitometric analysis of the bands and quantitation to β-actin or to RXRα was carried out; the results reported in the histograms on the right are the mean +/= S.E obtained in three independent experiments. Fold-induction is reported as relative density on an arbitrary scale with basal level as 1. (C) Schematic drawing that illustrates the interactions between transcription factors and specific DNA sequences of the Rbp4 promoter in basal and cAMP-induced conditions.
Parental Hepa1 and D2 cells were then serum-starved for 48 h and treated with 1 μM Br-cAMP for an additional 48 h. Cytosolic and nuclear fractions were separated, run on SDS PAGE and analysed by western blot (Fig. 8B). In Hepa1 cells, RBP4 was induced by cAMP; in D2 cells the low basal level was slightly modified by the treatment, as determined by densitometric analysis of the bands and referred to β-actin as control. HMGA1, in contrast, was induced by about 2-fold in both cells, as determined after densitometry and referred to RXRα as control. These results clearly indicate that cAMP-induced Rbp4 transcription depends upon a functional SF1/LRH-1 factor. Since cAMP induces also HMGA1 both in Hepa1 and its derived cell clone, the residual Rbp4 cAMP induction observed in D2 cells could be due to the presence of a still functional and cAMP-responsive HMGA1.
4. Discussion
Mobilization of retinol from liver stores and its delivery to peripheral tissues is the physiological role of the retinoid carrier protein RBP4, especially during fasting or in periods of insufficient Vitamin A dietary intake (Quadro et al., 1999). Despite the central role of RBP4, the transcriptional regulation of the gene in vertebrates is poorly understood. In Xenopus laevis hepatocytes, the RBP4 gene is transcriptionally regulated by estrogens (McKearin et al., 1987). In human HepG2 cells, we demonstrated that retinoic acid up-regulates RBP4 transcription via two retinoic acid response elements (RARE) in the proximal promoter region (Panariello et al., 1996). These findings indicate that differences in the transcriptional control of RBP4 genes occur not only in a tissue-specific, but also in a species-specific fashion, regulated by diverse gene expression programs.
Our present studies have identified that the murine RBP4 gene is regulated via the cAMP-PKA pathway as proposed by others (Jessen and Satre, 1998). We also extended this earlier work by defining the cis-regulatory sequences and the trans-acting factors responsible for this regulation. We demonstrate that full cAMP induction is sustained by a bipartite promoter: a proximal segment is required for basal expression and contains binding sites for general transcription factors such as AP1 and Sp1; a distal fragment is required for cAMP-induction and contains multiple AT-rich DNA motifs, known to be the binding site for HMGA1 proteins (Fig. 1 and Suppl. Fig. 2). These proteins are structural components of chromatin, which bind to the minor groove of the DNA double helix causing a bending of the molecule; thus enhancing the binding affinity of transcription factors to their cognate sites; i. e. they stimulate “enhanceosome” assembly (Merika and Thanos, 2001). Functional cooperation between HMGA1 proteins and Sp1 and/or C/EBPβ transcription factors has been shown to be required to activate transcription of the insulin receptor and leptin genes (Brunetti et al., 2001; Foti et al., 2003; Melillo et al., 2001). These genes show delayed induction by cAMP, like Rbp4, as reported here.
We demonstrate for the first time that HMGA1 proteins play a key role in basal and cAMP-dependent Rbp4 transcription. By several experimental approaches we show that HMGA1 potentiates transcription from both the endogenous Rbp4 gene and an exogenous RBP-CAT reporter gene; consistent with this, HMGA1 silencing reduces Rbp4 transcription (Fig. 3). In addition, we demonstrate that HMGA1 and RBP4 genes respond to glucagon-cAMP stimulation in vivo (Fig. 3C), providing strong support to the hypothesis that both genes are regulated in a coordinated fashion along this pathway. The regulation conferred by HMGA1 proteins is likely due to the structural changes imposed on the DNA and to the recruitment and binding of transcription factors to sequence motifs close to HMGA1 binding sites. We identified such binding motifs in the Rbp4 promoter segment between -725 and -699 and show that three DNA-protein complexes are generated, especially in response to cAMP. The B and C complexes, which are detected under all culture conditions, result from the binding to the TGTGGCAA sequence of p54nrb, the human homologue of murine NonO. The A complex forms on the AGGGGA motif especially after cAMP treatment (Fig. 5) and contains a 100-kDa protein that corresponds to PSF, the polypyrimidine tract-binding protein-associated splicing factor.
p54nrb/NonO belongs to the family of octamer-binding transcription factors, although it does not harbour the POU domain, common to this subset of nuclear proteins. It is involved in RNA processing (Yang et al., 1993) and in transcriptional activation via its DNA binding domain at its C-terminus. Specific interactions between p54nrb/NonO, PSF and the activation function 1 domain (AF-1) of the human androgen receptor have been reported but there are conflicting results regarding the effect that this has on the transcription of the corresponding gene (Ishitani et al., 2003; Dong et al., 2007).
PSF forms a complex with PTB (Polypyrimidine Tract-Binding protein) and is required for the correct identification of the 3′-splice site (Patton et al., 1993). PSF contains a proline/glutamine rich N-terminus involved in protein-protein interactions (Dye and Patton, 2001). Recently, PSF has been shown to bind the insulin–like growth factor response elements and silence transcription of the porcine P450 scc gene which encodes a steroidogenic enzyme. In this promoter, PSF recognizes a palindromic CTGAGTC sequence located 5′ to a GC-rich Sp1 binding site (Urban et al., 2000). A homologous AGGGGAGTGT motif is contained in oligonucleotide 2F used as bait to isolate PSF from Hepa1 extracts. PSF interacts with the DNA-binding domain of the thyroid hormone and retinoid X receptors and represses transcription of the corresponding target genes by recruiting Sin3A and class I histone deacetylases (Mathur et al., 2001). Recently, PSF and p54nrb/NonO have been found associated with a silencer motif in the promoter region of the phosphate carrier gene (PiC) (Iacobazzi et al., 2005). p54nrb/NonO and PSF not only interact with each other but also cooperate with other factors, specifically with SF1 to regulate cAMP induction of CYP17 gene transcription (Sewer et al., 2002). A PKA-dependent phosphatase modifies SF1 to form an active trimeric complex with p54nrb/NonO and PSF that in turn upregulates the gene (Sewer and Waterman, 2002, 2003). This model has been described in adrenal cells where SF1 is specifically expressed and where this ternary complex regulates transcription of genes coding for steroid and sex hormone biosynthetic enzymes. It is known that other posttranslational modifications can affect SF1 ability to be recruited to several multiprotein complexes involved in gene regulation (Sewer and Jagarlapudi, 2009). In a different cell context, the hepatocyte, a factor closely related to SF1, LRH-1, is expressed and participates in multifactorial complexes to regulate gene transcription (Fayard et al., 2004). Our data suggest that Rbp4 is modulated by a similar, albeit more complicated, mechanism. This is because the Rbp4 promoter complex includes more partners (HMGA1) and structural constraints (the bipartite structure of the promoter). Moreover, our data indicate that SF1 and LRH-1 can functionally substitute each other in the formation of multiprotein complexes to mediate gene expression. The different outcomes may depend on the amount and molecular ratios of these factors that are present in a given cell and also on their binding affinities to promoter sequences.
In the case of Rbp4, all these proteins contribute to gene transcription and in vivo are all recruited on the promoter. cAMP induces HMGA1 to interact more avidly with its cognate sequence motifs and recruits more factors to the complex. The cAMP pathway, in addition, triggers a series of posttranslational modifications that affect specifically SF1/LRH-1 and presumably PSF, influencing the way they are assembled. The new structure may interact differently with the coactivator/corepressor complexes, the histone modification system and the transcription machinery, resulting in enhanced transcription of the Rbp4 gene. A schematic model illustrating the interactions between the multifactorial complex and the Rbp4 promoter before and after cAMP induction is proposed in Fig. 8C.
Our study has two striking physiological implications. First, our data suggest that genes of the steroid hormone biosynthetic pathway and those encoding for the retinoid transport protein RBP4 are regulated in part by the same nuclear factors and structural motifs. This implies similarities in how these genes must respond to physiological signals. It is well known that retinoids and steroids share some metabolic enzymes, specifically some members of the short chain dehydrogenase/reductase family of enzymes (Persson et al., 2003; Napoli, 2001). Our data suggest that exogenous stimuli transmitted via this complex of activators may induce transcription of genes involved in facilitating both retinoid and steroid action. CYP17 catalyses the 17-hydroxylation of pregnenolone and progesterone which is required for cortisol biosynthesis and the 17,20-lyase activity required to produce androgens from 17-hydroxylated steroids. The genes for CYP17 and RBP4 share identical regulatory mechanisms in the mouse and consequently must respond similarly to the physiological status of the animal.
A second striking feature of our data is its demonstration that Rbp4 expression is regulated by cAMP. Since cAMP is an important second intracellular messenger for hormones that regulate glucose homeostasis, this raises the possibility that Rbp4 expression is responsive to hormones and other factors that regulate glucose homeostasis in the liver. Several early studies suggested linkages between blood RBP4 levels and impaired glucose processing (Abahusain et al., 1999; Lu et al., 2000; Shen et al., 2004). Elevations of RBP4 blood levels, due to increased RBP4 synthesized by adipocytes, has been suggested to serve as a signal which results in the loss of insulin responsiveness in muscle and liver (Yang et al, 2005). These data directly implicate RBP4 as having a causal role in the development of type 2 diabetes. Our findings establish that Rbp4 is regulated through cAMP and it is tempting to speculate that these regulatory elements are important for increasing RBP4 levels in blood and ultimately for the reported RBP4 induced loss of insulin responsiveness (Yang et al, 2005). Consistent with these data, we demonstrate that Hmga1 itself is induced by cAMP and the resulting increased protein level plays a crucial role in basal and cAMP-stimulation of Rbp4 expression. As further support, we report that both Hmga1 and Rbp4 are induced by glucagon via-cAMP signalling in mice in vivo, resulting in increased levels of the corresponding liver mRNAs. This provides compelling evidence for a new biochemical pathway for the maintenance of glucose homeostasis induced by cAMP that involves a coordinated regulation of HMGA1 and RBP4 genes. Although much more research in this area needs to be carried out, our data will be important for understanding not only the molecular regulation of retinoid release from tissues but also the development and progression of insulin resistance and type 2 diabetes.
Supplementary Material
Acknowledgments
We wish to thank Prof. G. Manfioletti, University of Trieste, Italy and Dr. Monica Fedele, University of Naples, Italy, for kindly providing HMGA1 recombinant protein and antibodies. We are indebted with Dr. Phil Tucker, University of Texas, Austin, USA, for the PSF and SF1 expression vectors and antibodies and with Dr. James Patton, Vanderbilt University, Nashville, USA, for the p54/NonO expression vector. This work is in part supported by NIH grants R01 DK079221 and R01 DK06843 (WSB) and by MIUR (PRIN 2004 to VC).
The abbreviations used are
- RBP4
retinol-binding protein
- PKA
protein kinase A
- cAMP
cyclic-Adenosyl Mono Posphate
- HMGA1
high mobility group protein A1
- p54nrb/NonO
a non-Pou domain-containing octamer–binding protein
- PSF
the polypyrimidine tract-binding protein-associated splicing factor
- SF1
Steroidogenic Factor 1
- PKI
Protein Kinase A Inhibitor
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abahusain MA, Wright J, Dickerson JW, de Vol EB. Retinol, alpha-tocopherol and carotenoids in diabetes. Eur J Clin Nutr. 1999;53:630–635. doi: 10.1038/sj.ejcn.1600825. [DOI] [PubMed] [Google Scholar]
- Blaner WS. Radioimmunoassays for retinol-binding protein, cellular retinol-binding protein and cellular retinoic acid-binding protein. Methods Enzymol. 1990;189:270–281. doi: 10.1016/0076-6879(90)89298-v. [DOI] [PubMed] [Google Scholar]
- Brunetti A, Manfioletti G, Chiefari E, Goldfine ID, Foti D. Transcriptional regulation of human insulin receptor gene by the high-mobility group protein HMGI (Y) FASEB J. 2001;15:492–500. doi: 10.1096/fj.00-0190com. [DOI] [PubMed] [Google Scholar]
- Choi EJ, Xia Z, Villacres EC, Storm DR. The regulatory diversity of the mammalian adenylyl cyclase. Curr Opin Cell Biol. 1993;5:269–273. doi: 10.1016/0955-0674(93)90115-7. [DOI] [PubMed] [Google Scholar]
- Clagett-Dame M, Plum LA. Retinoid-regulated gene expression in neural development Crit. Rev Eukaryot Gene Expr. 1997;7:299–342. doi: 10.1615/critreveukargeneexpr.v7.i4.20. [DOI] [PubMed] [Google Scholar]
- Dong X, Sweet J, Challis JR, Brown T, Lye SJ. Transcriptional activity of androgen receptor is modulated by two RNA splicing factors, PSF and p54nrb. Mol Cell Biol. 2007;13:4863–4875. doi: 10.1128/MCB.02144-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dye BT, Patton JG. An RNA recognition motif (RRM) is required for the localization of PTB-associated splicing factor (PSF) to subnuclear speckles. Exp Cell Res. 2001;263:131–144. doi: 10.1006/excr.2000.5097. [DOI] [PubMed] [Google Scholar]
- Fayard E, Auwerx J, Schoonjans K. LRH1:an orphan nuclear receptor involved in development, metabolism and steroidogenesis. Trends Cell Biol. 2004;14:250–260. doi: 10.1016/j.tcb.2004.03.008. [DOI] [PubMed] [Google Scholar]
- Foti D, Iuliano R, Chiefari E, Brunetti A. A nucleoprotein complex containing Sp1, C/EBP beta, andHMGI-Y controls human insulin receptor gene transcription. Mol Cell Biol. 2003;23:2720–2732. doi: 10.1128/MCB.23.8.2720-2732.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foulkes NS, Sassone-Corsi P. More is better: activators and repressors from the same gene. Cell. 1992;68:411–414. doi: 10.1016/0092-8674(92)90178-f. [DOI] [PubMed] [Google Scholar]
- Glass CK, Rosenfeld MG, Rose DW, Kurokawa R, Kamei Y, Xu L, Torchia J, Ogliastro MH, Westin S. Mechanisms of transcriptional activation by retinoic acid receptors. Biochem Soc Trans. 1997;25:602–605. doi: 10.1042/bst0250602. [DOI] [PubMed] [Google Scholar]
- Gonzales GA, Montminy M. Cyclic AMP stimulates somatostatin gene transcription by phosphorylation of CREB at serine 133. Cell. 1989;59:675–680. doi: 10.1016/0092-8674(89)90013-5. [DOI] [PubMed] [Google Scholar]
- Graham FL, Van der Eb AJ. A new technique for the assay of infectivity of human adenovirus 5 DNA. Virology. 1973;52:456–467. doi: 10.1016/0042-6822(73)90341-3. [DOI] [PubMed] [Google Scholar]
- Gudas LJ, Sporn MB, Roberts AB. Cellular biology and biochemistry of the retinoids. In: Sporn MB, Roberts AB, Goodman DS, editors. The Retinoids, Biology, Chemistry and Medicine. Raven Press; New York, NY: 1994. pp. 443–520. [Google Scholar]
- Harootunian AT, Adams SR, Wen W, Meinkoth JL, Taylor SS, Tsien RY. Movement of the free catalytic subunit of cAMP-dependent protein kinase into and out of the nucleus can be explained by diffusion. Mol Biol Cell. 1993;4:993–1002. doi: 10.1091/mbc.4.10.993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iacobazzi V, Infantino V, Costanzo P, Izzo P, Palmieri F. Functional analysis of the promoter of the mitochondrial phosphate carrier human gene: identification of activator and repressor elements and their transcription factors. Biochem J. 2005;391:613–621. doi: 10.1042/BJ20050776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishitani K, Yoshida T, Kitagawa H, Ohat H, Nozawa S, Kato S. p54nrb acts as a transcriptional coactivator for activation function 1 of the human androgen receptor. Biochem Biophys Res Commun. 2003;306:660–665. doi: 10.1016/s0006-291x(03)01021-0. [DOI] [PubMed] [Google Scholar]
- Jessen KA, Satre MA. Induction of mouse retinol-binding protein gene expression by cyclic AMP in Hepa 1-6 cells. Arch Biochem Biophys. 1998;357:126–130. doi: 10.1006/abbi.1998.0821. [DOI] [PubMed] [Google Scholar]
- Lefebvre P, Martin PJ, Flajollet S, Dedieu S, Billaut X, Lefebvre B. Transcriptional activities of retinoic acid receptors Vitam. Horm. 2005;70:199–264. doi: 10.1016/S0083-6729(05)70007-8. [DOI] [PubMed] [Google Scholar]
- Lichtsteiner S, Wuarin J, Shibler U. The interplay of DNA-binding proteins on the promoter of the mouse albumin gene. Cell. 1987;51:963–973. doi: 10.1016/0092-8674(87)90583-6. [DOI] [PubMed] [Google Scholar]
- Lu J, Dixon WT, Tsin AT, Basu TK. The metabolic availability of vitamin A is decreased at the onset of diabetes in BB rats. J Nutr. 2000;130:1958–1962. doi: 10.1093/jn/130.8.1958. [DOI] [PubMed] [Google Scholar]
- Mangelsdorf DJ, Thummel C, Beato M, Herrlich P, Shutz G, Umesono K, Blumberg B, Kastner P, Mark M, Chambon P, et al. The nuclear receptor superfamily: the second decade. Cell. 1995;83:835–839. doi: 10.1016/0092-8674(95)90199-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mantovani F, Covaceuszach S, Rustighi A, Sgarra R, Heath C, Goodwin GH, Manfioletti G. NF-kappa B mediated transcriptional activation is enhanced by the architectural factor HMGI-C. Nucleic Acid Research. 1998;26:1433–1439. doi: 10.1093/nar/26.6.1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathur M, Tucker PW, Samuels HH. PSF is a novel corepressor that mediated its effect through Sin3A and the DNA binding domain of nuclear hormone receptors. Mol Cell Biol. 2001;21:2298–2311. doi: 10.1128/MCB.21.7.2298-2311.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKearin DM, Barton MC, Keller MJ, Shapiro DJ. Estrogen induces transcription of the Xenopus laevis serum retinol-binding protein gene. J Biol Chem. 1987;262:4939–4942. [PubMed] [Google Scholar]
- Melillo RM, Pierantoni GM, Scala S, Battista S, Fedele M, Stella A, De Blasio MC, Chiappetta G, Fidanza V, Condorelli G, Santoro M, Croce CM, Viglietto G, Fusco A. Critical role of the HMGI(Y) proteins in adipocytic cell growth and differentiation. Mol Cell Biol. 2001;21:2485–2495. doi: 10.1128/MCB.21.7.2485-2495.2001. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Merika M, Thanos D. Enhanceosomes. Curr Opin Genet Dev. 2001;11:205–208. doi: 10.1016/s0959-437x(00)00180-5. [DOI] [PubMed] [Google Scholar]
- Monaco HL, Rizzi M, Coda A. Structure of a complex of two plasma proteins: transthyretin and retinol binding protein. Science. 1995;268:1039–1041. doi: 10.1126/science.7754382. [DOI] [PubMed] [Google Scholar]
- Montminy M. Transcriptional regulation by cyclic AMP. Annu Rev Biochem. 1997;66:807–822. doi: 10.1146/annurev.biochem.66.1.807. [DOI] [PubMed] [Google Scholar]
- Napoli JL. Biochemical pathways of retinoid transport, metabolism and signal transduction. Clin Immunol Immunopathol. 1996;80:S52–62. doi: 10.1006/clin.1996.0142. [DOI] [PubMed] [Google Scholar]
- Napoli JL. 17beta-hydroxysteroid dehydrogenase type 9 and short-chain dehydrogenase /reductases that catalyze retinoid, 17beta- and 3alpha-hydroxysteroid metabolism. Mol Cell Endocrinology. 2001;171:103–109. doi: 10.1016/s0303-7207(00)00392-0. [DOI] [PubMed] [Google Scholar]
- Panariello L, Quadro L, Trematerra S, Colantuoni V. Identification of a novel retinoic acid response element in the promoter region of the retinol-binding protein gene. J Biol Chem. 1996;271:25524–25532. doi: 10.1074/jbc.271.41.25524. [DOI] [PubMed] [Google Scholar]
- Patton JG, Porro EB, Galceran J, Tempst P, Nadal-Ginard B. Cloning and characterization of PSF, a novel pre-mRNA splicing factor. Genes Dev. 1993;7:393–406. doi: 10.1101/gad.7.3.393. [DOI] [PubMed] [Google Scholar]
- Persson B, Kallberg Y, Oppermann U, Jörnvall H. Coenzyme-based functional assignments of short-chain dehydrogenase/reductase (SDRs) Chem Biol Interact. 2003:143–144. 271–278. doi: 10.1016/s0009-2797(02)00223-5. [DOI] [PubMed] [Google Scholar]
- Quadro L, Blaner WS, Salchow DJ, Vogel S, Piantedosi R, Gouras P, Freeman S, Cosma MP, Colantuoni V, Gottesman ME. Impaired retinal function and vitamin A availability in mice lacking retinol-binding protein. EMBO J. 1999;18:4633–4644. doi: 10.1093/emboj/18.17.4633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reeves R. HMGA proteins: flexibility finds a nuclear niche? Biochem Cell Biol. 2003;81:185–195. doi: 10.1139/o03-044. [DOI] [PubMed] [Google Scholar]
- Saari JC. Retinoids in mammalian vision. In: Nau Retinoids. H, Blaner W., editors. The Handbook of experimental pharmacology. Springer Verlag Publishing; Heidelberg, Germany: 1999. pp. 563–588. [Google Scholar]
- Sassone-Corsi P. Transcription factors responsive to cAMP. Annu Rev Cell Dev Biol. 1995;11:355–377. doi: 10.1146/annurev.cb.11.110195.002035. [DOI] [PubMed] [Google Scholar]
- Servillo G, Della Fazia MA, Sassone-Corsi P. Coupling cAMP signaling to transcription in the liver: pivotal role of CREB and CREM. Exp Cell Res. 2002;275:143–154. doi: 10.1006/excr.2002.5491. [DOI] [PubMed] [Google Scholar]
- Sewer MB, Nguyen VQ, Huang C, Tucker PW, Kagawa N, Waterman MR. Transcriptional activation of human CYP17 in H295R adrenocortical cells depends on complex formation among p54 (nrb)/NonO, protein-associated splicing factor, and SF1, a complex that also participates in repression of transcription. Endocrinology. 2002;143:1280–1290. doi: 10.1210/endo.143.4.8748. [DOI] [PubMed] [Google Scholar]
- Sewer MB, Waterman MR. Adrenocorticotropin/cyclic adenosine 3′, 5′-monophosphate-mediated transcription of the human CYP17 gene in the adrenal cortex is dependent on phosphatase activity. Endocrinology. 2002;143:1769–1777. doi: 10.1210/endo.143.5.8820. [DOI] [PubMed] [Google Scholar]
- Sewer MB, Waterman MR. ACTH modulation of transcription factors responsible for steroid hydroxylase gene expression in the adrenal cortex. Microsc Res Tech. 2003;61:300–307. doi: 10.1002/jemt.10339. [DOI] [PubMed] [Google Scholar]
- Sewer MB, Jagarlapudi S. Complex assembly on the human CYP17 promoter. Mol Cell End. 2009;300:109–114. doi: 10.1016/j.mce.2008.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapiro DJ, Sharp PA, Wahli WW, Keller MJ. A high-efficiently HeLa cell nuclear transcription extract. DNA. 1988;7:47–55. doi: 10.1089/dna.1988.7.47. [DOI] [PubMed] [Google Scholar]
- Shen Q, Cline GW, Shulman GI, Leibowitz MD, Davies PJA. Effects of rexinoid on glucose transport and insulin-mediated signaling muscles of diabetic (db/db) mice. J Biol Chem. 2004;279:19721–19731. doi: 10.1074/jbc.M311729200. [DOI] [PubMed] [Google Scholar]
- Soprano DR, Blaner WS. Retinol and retinoic acid metabolism. In: Sporn MB, Roberts AB, Goodman DS, editors. The Retinoids, Biology, Chemistry and Medicine. Raven Press; New York, NY: 1994. pp. 257–282. [Google Scholar]
- Urban RJ, Bodenburg Y, Kurosky A, Wood TG, Gasic S. Polypyrimidine tract-binding protein-associated splicing factor is a negative regulator of transcriptional activity of the porcine p450scc insulin-like growth factor response element. Mol Endocrinol. 2000;14:774–782. doi: 10.1210/mend.14.6.0485. [DOI] [PubMed] [Google Scholar]
- Vogel S, Gamble MV, Blaner WS. Biosynthesis, adsorption, metabolism and transport of retinoids, Chapter 2. In: Nau H, Blaner WS, editors. The Handbook of Experimental Pharmacology. The Retinoids, Heidelberg: Springer-Verlag; 1999. pp. 31–96. [Google Scholar]
- Yang YS, Hanke JH, Carayannopoulos L, Craft CM, Capra JD, Tucker PW. NonO, a non-Pou-domain-containing octamer-binding protein, is the mammalian homolog of Drosophila nonAdiss. Mol Cell Biol. 1993;13:5593–5603. doi: 10.1128/mcb.13.9.5593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Q, Graham TE, Mody N, Preitner F, Peroni OD, Zabolotny JM, Totani K, Quadro L, Kahn BB. Serum retinol binding protein 4 contributes to insulin resistance in obesity and type 2 diabetes. Nature. 2005;436:356–362. doi: 10.1038/nature03711. [DOI] [PubMed] [Google Scholar]
- Ying L, Morris BJ, Sigmund CD. Transactivation of the human renin promoter by cyclic AMP/protein kinase A pathway is mediated by both cAMP-responsive element binding protein −1 (CREB) –dependent and CREB-independent mechanisms in Calu-6 cells. J Biol Chem. 1997;272:2412–2420. doi: 10.1074/jbc.272.4.2412. [DOI] [PubMed] [Google Scholar]
- Zanger K, Cohen LE, Hashimoto K, Radovick S, Wondisford FE. A novel mechanism for cyclic adenosine 3′, 5′-monophosphate regulation of gene expression by CREB-binding protein. Mol Endocrinol. 1999;13:268–275. doi: 10.1210/mend.13.2.0245. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.