Abstract
Severe acute respiratory syndrome (SARS) is a newly emerging infectious disease. The potential recurrence of the disease from animal reservoirs highlights the significance of development of safe and efficient vaccines to prevent a future SARS epidemic. In this study, we expressed the recombinant receptor-binding domain (rRBD) in mammalian (293T) cells, insect (Sf9) cells, and E. coli, respectively, and compared their immunogenicity and protection against SARS-CoV infection in an established mouse model. Our results show that all rRBD proteins expressed in the above systems maintained intact conformation, being able to induce highly potent neutralizing antibody responses and complete protective immunity against SARS-CoV challenge in mice, albeit the rRBD expressed in 293T cells elicited stronger humoral immune responses with significantly higher neutralizing activity (P < 0.05) than those expressed in Sf9 and E. coli cells. These results suggest that all three rRBDs are effective in eliciting immune responses and protection against SARS-CoV and any of the above expression systems can be used for production of rRBD-based SARS subunit vaccines. Preference will be given to rRBD expressed in mammalian cells for future evaluation of the vaccine efficacy in a non-human primate model of SARS because of its ability to refold into a native conformation more readily and to induce higher level of neutralizing antibody responses than those expressed in E. coli and insect cells.
Keywords: SARS-CoV, Receptor-binding domain, Neutralizing antibody, Protective immunity, Subunit vaccines
Introduction
Severe acute respiratory syndrome (SARS) is a novel emerging infectious disease that was first identified in late 2002 in Guangdong, China (Zhong et al., 2003). The worldwide outbreak of the disease in 2003 led to hundreds of deaths among thousands of total infected individuals (Skowronski et al., 2005). Although no new cases of SARS have been reported since 2004, the study of SARS and its causative agent, SARS coronavirus (SARS-CoV), is continuing. SARS is still a safety concern because of the presence of possible animal reservoirs, including its natural reservoir bats and intermediate hosts such as palm civets and raccoon dogs (Guan et al., 2003, Kan et al., 2005, Lau et al., 2005, Li et al., 2005). Therefore, it is essential to develop vaccines against SARS for the prevention of future outbreaks.
The genome of SARS-CoV encodes four structural proteins, including spike (S), membrane (M), envelope (E), and nucleocapsid (N), and some non-structural proteins (Marra et al., 2003, Rota et al., 2003). Among all of the SARS-CoV proteins, the S protein plays an essential role in receptor-binding, virus entry and membrane fusion (Liu et al., 2004, Tripet et al., 2004, Wong et al., 2004, Xu et al., 2004, Zhu et al., 2004). SARS-CoV first binds to the host cellular receptor angiotensin-converting enzyme 2 (ACE2) (Dimitrov, 2003, Kuhn et al., 2004, Li et al., 2003, Prabakaran et al., 2004), via its receptor-binding domain (RBD), a 193-amino acid (aa) fragment spanning the residues 318–510 of the S1 region (Babcock et al., 2004, Wong et al., 2004, Xiao et al., 2003). The S protein is also the main domain in inducing neutralizing antibodies against SARS and is thus considered the main component for developing SARS vaccines (Du et al., 2009a).
The S protein-based vaccines may be developed on the full-length of S protein or its fragments (Hu et al., 2007b, Zakhartchouk et al., 2007), or in various vectors encoding S protein, including DNA-based (Martin et al., 2008, Wang et al., 2008, Yang et al., 2004) and viral vector-based vaccines (Gao et al., 2003, Liniger et al., 2008). These S protein-based vaccine candidates may induce humoral immune responses and/or neutralizing antibodies, as well as cellular immune responses, in vaccinated animals (Hu et al., 2007a, Huang et al., 2006, Kobinger et al., 2007). A DNA vaccine encoding the S protein induces neutralizing antibody and cellular immune responses in healthy adults in a phase I clinical trial (Martin et al., 2008). Since the full-length S protein-based vaccines might induce potential harmful immune responses (Czub et al., 2005, Weingartl et al., 2004), vaccines based on the fragments of S protein, such as RBD, have a greater potential for developing into effective vaccines against SARS (Lee et al., 2006, Zhao et al., 2006, Zhou et al., 2006).
We previously showed that a 293T cell-expressing fusion protein RBD-Fc, which contains RBD of SARS-CoV and Fc fragment of human IgG, elicited neutralizing antibody response and protection in the vaccinated animals (Du et al., 2007b, He et al., 2004, He et al., 2005), suggesting a potential of developing RBD-based subunit SARS vaccines. However, the big molecule Fc in the fusion protein may cause some unwanted responses when it is used in humans as a SARS vaccine in the future. In addition, it is unknown whether insect cell and E. coli expression systems, which are suitable for production of recombinant proteins in large quantity, can be used for expression of rRBD with functionality, although it was reported that a truncated antigenic fragment (aa 441–700) of S protein of SARS-CoV expressed in insect Sf9 cells exhibited high specificity and sensitivity in detection of anti-SARS-CoV antibodies in sera of SARS patients and lacked cross-reactivity with sera of patients with infectious bronchitis virus (IBV) and transmissible gastroenteritis virus (TGEV) infection (Manopo et al., 2005). In this study, we expressed the rRBD protein without Fc in three different expression systems, including mammalian 293T cells, insect Sf9 cells and E. coli, and compared their immunogenicity and protection in a mouse model.
Results
All rRBD proteins expressed in mammalian cell, insect cell, and E. coli expression systems maintained intact conformation and authentic antigenicity
The purified rRBD proteins expressed in the above expression systems were detected by Western blot using a panel of monoclonal antibodies (mAbs) that contain different conformational and linear epitopes in RBD (He et al., 2005, He et al., 2006a). As shown in Fig. 1 , all rRBD proteins expressed in mammalian 293T cells (RBD-293T), insect cells (RBD-Sf9) and E. coli (RBD-Ec) reacted with the majority (5 of 6) of the conformational epitope-specific mAbs and one linear epitope-specific mAb, having the strongest reaction with the Conf V mAb 33G4 and the linear mAb 17H9. These results suggest that the rRBD proteins expressed in mammalian cells, insect cells and E. coli maintain intact conformation and authentic antigenicity.
Fig. 1.
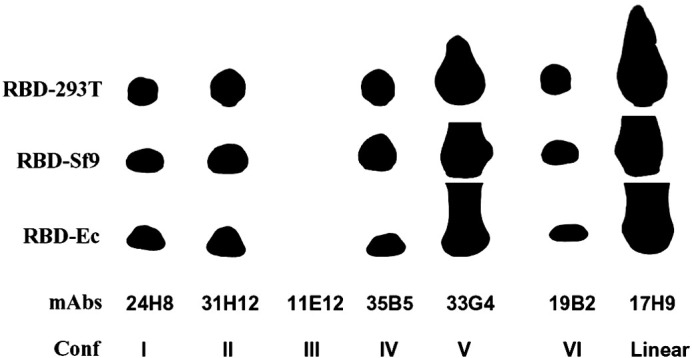
Expressed rRBD proteins (RBD-293T, RBD-Sf9 and RBD-Ec) were detected by Western blot for their reactivity with a panel of mAbs that recognize conformational and linear epitopes in the SARS-CoV RBD region.
The rRBD proteins expressed in 293T and Sf9 cells induced higher level of RBD-specific antibodies in vaccinated mice than that expressed in E. coli
To evaluate humoral immune responses induced by the rRBD expressed in three different systems, mice were vaccinated with RBD-293T, RBD-Sf9 and RBD-Ec, respectively, and sera were tested by ELISA for RBD-specific IgG. As shown in Fig. 2A, all three rRBD proteins induced RBD-specific IgG antibody responses with increased antibody binding after each boost, reaching the highest level after the last boost. The mean end-point titer of antibodies in the sera, collected 10 days post-last boost, of the mice immunized with RBD-293T and RBD-Sf9 (1:3.3 × 106) were significantly higher than that in the sera of the mice vaccinated with RBD-Ec (1:4.1 × 105) (Fig. 2B). There were no detectable RBD-specific antibodies in the pre-immune sera of the vaccinated mice or those from mice injected with PBS control (Fig. 2). The above results suggest that rRBD proteins expressed in mammalian cells (293T) and insect cells (Sf9) are able to elicit stronger SARS-CoV RBD-specific antibody responses than that expressed in E. coli.
Fig. 2.
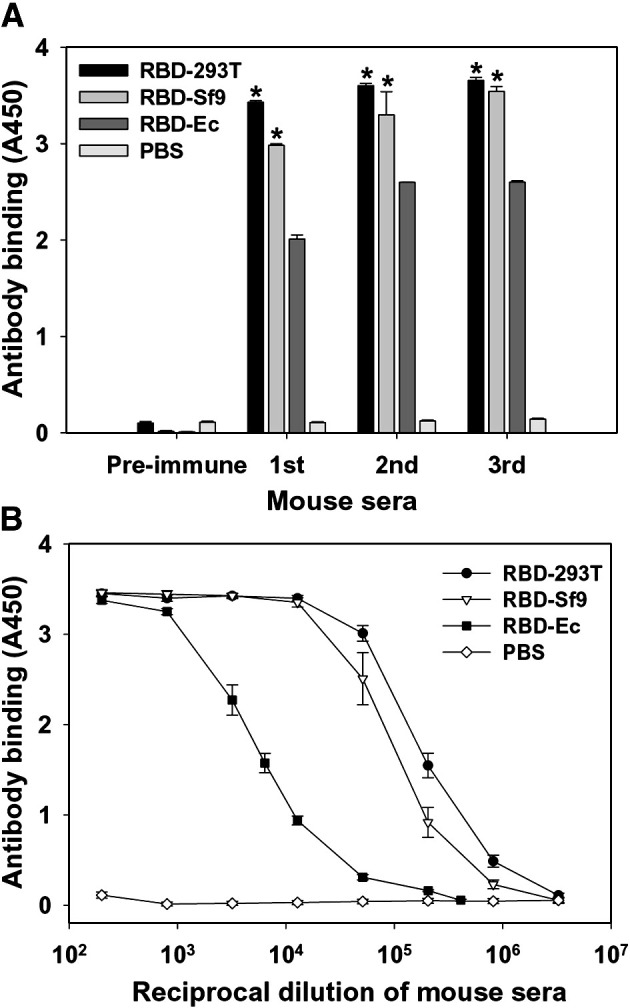
Humoral immune responses were detected using sera of mice vaccinated with rRBD proteins. PBS was used as the negative control. (A) Reactivity of serum antibodies with rRBDs. RBD-specific IgG was detected by ELISA using sera (1:3000 dilution) from mice before (pre-immune) and 10 days after each vaccination. The data are presented as mean A450 ± standard error (SE) of five mice per group. ⁎ indicates significant difference (P < 0.05) between RBD-293T or RBD-Sf9, respectively, and RBD-Ec group. (B) The ability of antibody binding to rRBD was detected by ELISA using serial dilutions of sera collected from mice 10 days post-last vaccination. The data are presented as mean A450 ± SE of five mice per group at various dilution points. RBD-293T, RBD-Sf9 and RBD-Ec indicate rRBD proteins expressed in 293T, Sf9 and E. coli cells, respectively.
All rRBD induced highly potent neutralizing antibody responses, albeit the neutralizing antibody titer elicited by RBD-293T was significantly higher than that induced by rRBD expressed in Sf9 and E. coli cells
To assess whether the induced IgG antibody could neutralize infection of SARS-CoV in cell cultures in vitro, sera from 10 days post-last boost were detected by neutralization assays using pseudotyped and live SARS-CoV. As shown in Fig. 3A, sera from all three vaccination groups effectively neutralized infection of SARS pseudovirus in 293T cells expressing the receptor ACE2 (ACE2/293T). Particularly, RBD-293T protein induced a significantly higher titer of neutralizing antibody than the other two groups, with the mean 50% neutralizing antibody titer (NT50) about 6.9 × 105. The neutralization titer against live SARS-CoV infection in Vero E6 cells is also significantly higher in the RBD-293T vaccination group (mean NT50 = 1.6 × 103) than in the RBD-Sf9 and RBD-Ec vaccination groups (Fig. 3B), while no neutralizing activity was detected in the PBS control group (Fig. 3). The above results suggest that the rRBD proteins expressed in mammalian, insect and E. coli cells are able to induce potent neutralizing antibody responses in the vaccinated mice.
Fig. 3.

Neutralizing antibody activity was detected using sera of mice vaccinated with rRBD proteins. PBS was used as the negative control. Sera collected at 10 days post-last vaccination were used for the detection. (A) neutralizing antibody titers against SARS pseudovirus infection. The data are presented as mean ± SE of 50% neutralizing antibody titers (NT50) from five mice per group. ⁎ indicates significant difference (P < 0.05) between RBD-293T and other groups, or RBD-Sf9 and RBD-Ec groups. (B) neutralizing antibody titers against live SARS-CoV infection. The titers were determined as the highest dilutions of sera that could completely prevent CPE in at least 50% of the wells (NT50) and are presented as mean ± SE of five mice per group. ⁎ indicates significant difference (P < 0.05) between the RBD-293T vaccination group and other groups.
All rRBD proteins expressed in the three expression systems induced equally effective protection in mice against virus challenge
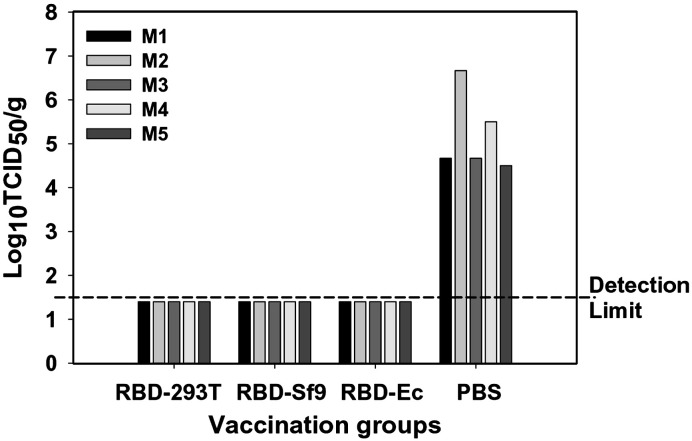
To evaluate the efficacy of protective immunity induced by rRBD proteins, vaccinated mice were challenged with SARS-CoV GZ50, and virus replication was detected by titration of the infectious virus in mouse lung tissues. As shown in Fig. 4 , all mice from three individual rRBD vaccination groups could be completely protected from SARS-CoV challenge, with the virus titer below the detection limit of 1.5 Log10 50% tissue-culture infectious dose (TCID50)/g of lung tissues. However, all five mice from the PBS control group maintained high titer of virus replication in the lung tissues post SARS-CoV challenge, with the virus titer highly above the detection limit. These results suggest that all rRBD proteins elicit protective immunity with the sufficiency required to prevent vaccinated mice from subsequent virus challenge.
Fig. 4.
Protective immunity was detected in mice vaccinated with rRBD proteins. Mice respectively vaccinated with RBD-293T, RBD-Sf9 and RBD-Ec were challenged with SARS-CoV GZ50. The SARS-CoV replication in the challenged mouse lung tissues was detected and expressed as Log10TCID50/g of tissues. M1–M5 indicates five mice per group. The limit of detection was 1.5 Log10TCID50/g of lung tissues.
Suppression of SARS-CoV replication may be associated with the neutralizing antibodies induced by the rRBD
The rRBD protein-vaccinated mice were sacrificed 5 days post-challenge with SARS-CoV, and mouse lung tissues were collected to further investigate the protection. The viral load in the lung tissues was determined by quantitative reverse-transcriptase polymerase chain reaction (Q-RT-PCR) and indicated as viral RNA copies/μg of lung tissues. As shown in Table 1 , no RNA copies were detected in the mice vaccinated with RBD-293T, RBD-Sf9, and RBD-Ec, respectively, while a high level of RNA copies was detected in the lung tissues of control mice administrated with PBS. Suppression of virus replication in the lung tissues may be associated with the high titer of virus-neutralizing antibodies induced by all three rRBD proteins. The high level of RNA copies in the lung tissues of the PBS control mice may be attributed to the undetectable neutralizing antibody responses in these mice (Table 1).
Table 1.
Measurement of SARS-CoV RNA copies in the lung tissues by Q-RT-PCR and the potential correlation between RNA copies and neutralizing antibody titer.
| Immunogen | Neutralizing titer (NT50) |
RNA copies/μg of lung tissues |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| M1 | M2 | M3 | M4 | M5 | Mean ± SE | M1 | M2 | M3 | M4 | M5 | Mean ± SE | |
| RBD-293T | 1810 | 1613 | 905 | 1810 | 1613 | 1550 ± 167 | UD⁎ | UD | UD | UD | UD | UD |
| RBD-Sf9 | 1280 | 1280 | 640 | 320 | 320 | 768 ± 217 | UD | UD | UD | UD | UD | UD |
| RBD-Ec | 905 | 905 | 905 | 905 | 905 | 905 ± 0 | UD | UD | UD | UD | UD | UD |
| PBS | < 20 | < 20 | < 20 | < 20 | < 20 | < 20 | 1305 | 1543 | 1626 | 2055 | 3124 | 1931 ± 322 |
Each group consists of five mice (M1–M5).
UD indicates undetectable.
Discussion
The main purpose of this study was to determine whether the rRBD without human IgG Fc could induce effective neutralizing antibody responses and protective immunity, and if the insect cell and E. coli expression systems, like the mammalian cell expression system, can also be used for production of functional RBD. As shown in Fig. 1, rRBD without Fc expressed in mammalian cell 293T (RBD-293T), insect cell Sf9 (RBD-Sf9) and E. coli (RBD-Ec) were able to react with majority of a panel of mAbs that recognize different conformational and linear epitopes in RBD of SARS-CoV S protein, except the mAb 11E12. This Conf III mAb was generated by immunization with recombinant RBD-Fc fusion protein and characterized to recognize a specific conformational epitope presented on RBD when fused with the Fc fragment of human IgG (He et al., 2005), suggesting that this epitope may not be presented when rRBDs were independently expressed. The currently expressed three rRBD proteins without Fc could be obtained at > 90% purity (data not shown). In addition, all three rRBD proteins elicited strong RBD-specific antibody responses in the vaccinated mice, albeit the titers of antibodies induced by RBD-293T and RBD-Sf9 were significantly higher than that by RBD-Ec (Fig. 2). These results suggest that while all the three RBD proteins retain proper immunogenicity, the RBD expressed in E. coli is less immunogenic than those expressed in mammalian and insect cells. Similarly, all three rRBD proteins elicited high titers of neutralizing antibody that neutralized both SARS pseudovirus and live SARS-CoV infection in cell cultures in vitro although the titer of neutralizing antibodies induced by RBD-293T was higher than those by RBD-Sf9 and RBD-Ec (Fig. 3).
Previous study has demonstrated that protection of immunized animals from virus challenge is mediated by a humoral but not a T-cell-dependent immune mechanism (Yang et al., 2004). Here we also evaluated the protective immunity in the vaccinated mice against SARS-CoV challenge. We found that all rRBD proteins expressed in mammalian, insect and E. coli cells could induce equally protective immunity that protected all vaccinated mice from SARS-CoV challenge, since no RNA copies were detected in the challenged mouse lung tissues, and the viral replication was significantly below the detection limit in the vaccination groups (Fig. 4, Table 1). Consistent with the report by Yang et al. (2004), the protection is mainly associated with the neutralizing antibodies. These results are also in agreement with our previous study on RBD-Fc vaccine (Du et al., 2007b).
The present study provides answers for our first question about the role of human IgG Fc in the induction of neutralizing antibody response and protective immunity. Although the Fc fragment in fusion proteins containing HIV-1 antigen indeed significantly enhances the immunogenicity of the HIV-1 antigen and increases the production of neutralizing antibodies through different mechanisms, e.g., assisting the antigen binding to the FcR (Chen et al., 2007) or prolonging the in vivo half-life of the antigen (Zhang et al., 2009), Fc seems to be playing no critical role in enhancing the immunogenicity of rRBD of SARS-CoV. In the absence of Fc, the rRBD expressed in 293T cells elicited a similar level of neutralizing antibody responses and protective immunity in the vaccinated animals as the RBD-Fc expressed in 293T cells (He et al., 2004, He et al., 2005).
The second question that needs to be answered is whether the insect cell and E. coli expression systems are suitable for producing functional rRBD as vaccines. For large quantity production of a recombinant protein-based vaccine, the insect cell and E. coli expression systems are superior to the mammalian cell expression system. Both insect Sf9 cells and E. coli can grow at high density at room temperature without the supply of CO2, and express recombinant proteins in a fast, easy and reliable way, resulting in lower production cost and higher protein productivity than the mammalian expression system (Boosani and Sudhakar, 2006, Farinha-Arcieri et al., 2008, Marblestone et al., 2006, Saitoh et al., 2009). Although the rRBD expressed in mammalian 293T cells induces stronger RBD-specific antibody responses and higher titers of neutralizing antibodies than those expressed in insect cell and E. coli expression systems, RBD-Sf9 and RBD-Ec also maintained intact conformation and elicited neutralizing antibody responses strong enough to protect vaccinated hosts against SARS-CoV challenge (Fig. 1, Fig. 2, Fig. 3, Fig. 4 and Table 1). Our previous study also demonstrated that rRBD-immunized aged mice with serum neutralizing antibody titers ranging from 1:189 to 1:505 had no detectable SARS-CoV in their lungs after virus challenge, but a marginal level of SARS-CoV replication was detected in one of the mice with serum neutralizing antibody titer of 1:57 (Du et al., 2007b). Other groups reported that mice immunized with attenuated virus or DNA vaccines encoding the SARS-CoV S protein, or with inactivated virus had serum neutralizing antibody titers ranging from 1:50 to 1:640 and most of these mice were protected from SARS-CoV challenge (Bisht et al., 2004, Stadler et al., 2005, Yang et al., 2004). Three patients who failed to respond to the available treatment recovered from serious SARS after transfusion of SARS convalescent plasma with neutralizing titer > 1:640 (Yeh et al., 2005). Based on these data, we suggest that any vaccine that induces in immunized mice the serum neutralizing antibody titers of > 1:500 would be considered as highly effective. According to these criteria, we believe that either insect cell or E. coli expression system can be used for production of RBD-based SARS subunit vaccines on a large scale because RBD-Sf9 and RBD-Ec could induce high titer (mean value > 1:700) of neutralizing antibodies and strong protective immunity that protect all the vaccinated mice from SARS-CoV challenge.
Compared with the insect cell and E. coli expression systems, the mammalian cell system has relatively higher production cost and lower productivity. The main advantage of the mammalian cell system over the other two expression systems is that there is a far higher chance of getting correctly folded soluble proteins with proper glycosylation, making the expressed protein maintain native conformation and keep sufficient bioactivity (Du et al., 2009b, Yin et al., 2007). This advantage may partially explain why RBD-293T expressed in mammalian cells induced stronger immune responses, particularly the neutralizing antibody response, than the rRBD proteins expressed in insect cells and E. coli (Fig. 3).
To summarize, this comprehensive study compares the immune response and protective immunity of three SARS subunit vaccines based on the rRBD proteins respectively expressed in mammalian 293T cells, insect Sf9 cells and E. coli. In the absence of human IgG Fc, the rRBDs expressed in all three expression systems are effective in eliciting potent neutralizing antibody responses and protective immunity. Considering that the rRBD protein expressed in mammalian 293T cells (RBD-293T) induced significantly higher level of neutralizing antibody responses than those expressed in insect Sf9 cells and E. coli, we will select mammalian cell-expressing rRBD protein for further evaluation of its in vivo efficacy in a non-human primate model of SARS before going to clinical trials.
Materials and methods
Construction and expression of rRBD proteins
Genes encoding the fragments containing 193 residues (aa 318–510) of SARS-CoV S protein RBD with a 6× His tag were amplified by PCR using the plasmid containing the full-length S protein (Tor2 strain) as the template (Du et al., 2007a, He et al., 2006b) and were then constructed into the recombinant plasmids named as rRBD-SUMO, rRBD-Bacul, and rRBD-pcDNA6, respectively.
The expression of the rRBD protein in the pET SUMO E. coli system (RBD-Ec) was done following the manufacturer's protocols (Invitrogen, Carlsbad, CA). Briefly, rRBD-SUMO E. coli was cultured in LB medium at 37 °C overnight, which was diluted at 1:100 the next day and cultured with vigorous shaking until the OD600 reached 0.6–1.0. Isopropyl-β-D-thiogalactopyranoside (IPTG, Sigma, St. Louis, MO) was then added at the final concentration of 0.25 mM, followed by a culture at 28 °C for an additional 5 h and a centrifugation at 6000 rpm for 15 min at 4 °C. Sonicated E. coli supernatant containing the expressed rRBD protein was collected and purified using His columns (Promega, Madison, WI) according to the manufacturers' protocols.
The expression of the rRBD protein in the Baculovirus system (RBD-Sf9) was conducted according to the manufacturer's protocols (BD Biosciences, San Jose, CA). In brief, 2 μg rRBD-Bacul plasmid was mixed with 0.5 μg BD BaculoGold linearized Baculovirus DNA which sat for 5 min at room temperature before adding 1 ml BD BaculoGold Transfection Buffer B. The mixture was added dropwisely to Sf9 insect cells (ATCC, Manassas, VA) pre-covered with 1 ml Transfection Buffer A, followed by incubation of the cells at 28 °C for 4 h, and further culture of the transfected cells in fresh SF-900 II SFM (Invitrogen) for an additional 4 days. Supernatant was collected 4 days later, which will be used to infect cells with three more 3-day culture cycles for virus amplification. Cell culture supernatant from the 4th cycle was collected for purification of rRBD protein.
The transient expression of the rRBD protein in the pcDNA6/His A vector (RBD-293T) was performed according to the manufacturer's (Invitrogen) and our previous protocols (Du et al., 2009b). Briefly, the rRBD-pcDNA6 plasmid was transfected using calcium phosphate method into HEK293T cells (ATCC). Culture medium was replaced by fresh OPTI-MEM I Reduced-Serum Medium (Invitrogen) 10 h later. Supernatant containing expressed rRBD protein was collected 72 h later and purified for rRBD protein.
Western blot
The purified rRBD proteins were detected by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and Western blot as described before (Du et al., 2006). Briefly, 2 μg purified protein was separated by 10–20% Tricine gels (Invitrogen) and transferred to nitrocellulose membranes (Bio-Rad Laboratories, Hercules, CA). After blocking overnight at 4 °C, the blots were incubated with a panel of RBD-specific mAbs previously produced in our laboratory (He et al., 2006b) at the final concentration of 0.5 μg/ml for 1 h at room temperature. The blots were then incubated with horseradish peroxidase (HRP)-conjugated goat anti-mouse IgG (1:5,000, Zymed, Carlsbad, CA) for 1 h at room temperature. Signals were visualized with ECL Western blot substrate reagents and Amersham Hyperfilm (GE Healthcare, Piscataway, NJ).
Mouse vaccination and sample collection
A group of five female BALB/c mice aged 4–6 weeks were vaccinated subcutaneously (s.c.) with 20 μg/mouse of purified rRBD proteins re-suspended in PBS in the presence of Sigma Adjuvant System (SAS, Sigma) and boosted twice with 10 μg/mouse of immunogen containing SAS at 3-week intervals. Serum samples were collected at 10 days post-each vaccination and used for detection of IgG and neutralizing antibody responses. Vaccinated mice were challenged with live SARS-CoV 10 days post-last vaccination, and viral replication was detected in challenged mouse lungs 5 days post-challenge. All mouse study was maintained in accordance with the national animal care protocols.
ELISA
IgG specific for RBD of SARS-CoV S protein in mouse sera was tested by ELISA using the protocol described previously (Du et al., 2007b) with some modifications. Briefly, 96-well microtiter plates were precoated respectively with the recombinant rRBD proteins overnight at 4 °C and blocked with 2% non-fat milk at 37 °C for 2 h. Serially diluted mouse sera were added to the plates and incubated at 37 °C for 1 h, followed by four washes. Bound antibodies were then reacted with HRP-conjugated goat anti-mouse IgG for 1 h at 37 °C. After four washes, the substrate 3,3′,5,5′-tetramethylbenzidine (TMB) (Zymed) was added to the plates, and the reaction was stopped by adding 1 N H2SO4. The absorbance at 450 nm (A450) was measured by an ELISA plate reader (Tecan, San Jose, CA).
Neutralization assay for SARS pseudovirus infection
The neutralization assay against SARS pseudovirus infection was done as previously described (He et al., 2005). Briefly, 293T cells were cotransfected with a plasmid encoding SARS-CoV S protein and a plasmid encoding Env-defective, luciferase-expressing HIV-1 genome (pNL4-3.luc.RE) using FuGENE 6 transfection reagents (Roche Applied Science). Supernatant was harvested 72 h post-transfection and used for single-cycle infection of ACE2/293T cells. The pseudovirus-containing supernatant was preincubated with serially diluted mouse sera at 37 °C for 1 h before adding to the cells. Fresh medium was added 24 h later, followed by lysing cells using cell lysis buffer (Promega). After addition of luciferase substrate (Promega), relative luciferase activity was determined in Ultra 384 luminometer (Tecan). SARS pseudovirus neutralization was calculated and expressed as 50% neutralizing antibody titer, NT50 (Chou, 2006).
Neutralization assay for live SARS-CoV infection
The neutralization assay against infection by live SARS-CoV (GZ50 strain, GenBank accession no. AY304495) was performed, and titers of neutralizing antibody in vaccinated mouse sera were detected in Vero E6 cells (ATCC) as previously described (Du et al., 2008a, Du et al., 2008b). Briefly, serial 2-fold dilutions of sera were mixed with 100 TCID50 of SARS-CoV, incubated at 37 °C for 1 h, and added to the monolayer of Vero E6 cells in triplicate. Cytopathic effect (CPE) in each well was observed daily and recorded on day 3 post-infection. The neutralizing titers of mouse anti-sera that completely prevented CPE in 50% of the wells (NT50) were calculated as before (Du et al., 2008a, Du et al., 2008b).
Live SARS-CoV challenge
The rRBD protein-vaccinated mice were anesthetized with isoflurane and intranasally inoculated with 50 μl of SARS-CoV GZ50 (100 TCID50) according to the National Animal Care and Use Guidelines in an approved animal BSL-3 laboratory. The mice were sacrificed 5 days after virus challenge, and the lungs were removed for virological tests.
Q-RT-PCR
The viral RNA copies in lung tissues were determined by Q-RT-PCR according to our previous protocols (Du et al., 2007b). Briefly, total RNA was extracted from 20 mg of lung tissues using RNeasy Mini kit (Qiagen, Valencia, CA). SARS-CoV RNA was quantified in a 30 μl mixture containing 10 μl RNA, 15 μl 2× TaqMan® one-step RT-PCR Master Mix (ABI, Foster, CA), 0.75 μl 40× multiscribe, 0.25 μM each forward primer Af (Taq-772F 5′-AAG CCT CGC CAA AAA CGT AC-3′), reverse primer Ar (Taq-1000R 5′-AAG TCA GCC ATG TTC CCG AA-3′) and probe (Taq-955T 5′-FAM-TCA CGC ATT GGC ATG GAA GTC ACA CT-TAMRA) (TIB Molbiol, Berlin, Germany) using a fluorometric PCR instrument (ABI 7300).
Determination of viral titer and total virus
The viral replication was determined by titration of the inoculated virus in the lung tissues as previously described (Du et al., 2007b). Briefly, the lung tissues were homogenized to a final concentration of 10% (w/v) suspension in DMEM. Tissue homogenates were centrifuged, filtrated and inoculated into the monolayer of Vero E6 cells. Results were evaluated 3 days later under phase-contrast microscopy, and viral titer using a CPE-based TCID50 test was calculated by the Reed–Muench method. The total amount of virus was calculated by multiplying the weight of the lung tissues and the viral titer measured in 10% tissue homogenates.
Statistical analysis
Values were presented as mean with standard error, SE. Statistical significance among different vaccination groups was calculated by Student's t test using Stata statistical software. P values less than 0.05 were considered significant.
Acknowledgments
This study was supported by the National Institutes of Health (NIH) of the United States (RO1 AI68002), by the Research Fund for the Control of Infectious Diseases, the Health, Welfare and Food Bureau of the Hong Kong SAR Government, and by the National 973 Basic Research Program of China (2005CB523001).
References
- Babcock G.J., Esshaki D.J., Thomas W.D., Jr., Ambrosino D.M. Amino acids 270 to 510 of the severe acute respiratory syndrome coronavirus spike protein are required for interaction with receptor. J. Virol. 2004;78:4552–4560. doi: 10.1128/JVI.78.9.4552-4560.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bisht H., Roberts A., Vogel L., Bukreyev A., Collins P.L., Murphy B.R., Subbarao K., Moss B. Severe acute respiratory syndrome coronavirus spike protein expressed by attenuated vaccinia virus protectively immunizes mice. Proc. Natl. Acad. Sci. U. S. A. 2004;101:6641–6646. doi: 10.1073/pnas.0401939101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boosani C.S., Sudhakar A. Cloning, purification, and characterization of a non-collagenous anti-angiogenic protein domain from human alpha1 type IV collagen expressed in Sf9 cells. Protein Expr. Purif. 2006;49:211–218. doi: 10.1016/j.pep.2006.03.007. [DOI] [PubMed] [Google Scholar]
- Chen H., Xu X., Jones I.M. Immunogenicity of the outer domain of a HIV-1 clade C gp120. Retrovirology. 2007;4:33. doi: 10.1186/1742-4690-4-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou T.C. Theoretical basis, experimental design, and computerized simulation of synergism and antagonism in drug combination studies. Pharmacol. Rev. 2006;58:621–681. doi: 10.1124/pr.58.3.10. [DOI] [PubMed] [Google Scholar]
- Czub M., Weingartl H., Czub S., He R., Cao J. Evaluation of modified vaccinia virus Ankara based recombinant SARS vaccine in ferrets. Vaccine. 2005;23:2273–2279. doi: 10.1016/j.vaccine.2005.01.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimitrov D.S. The secret life of ACE2 as a receptor for the SARS virus. Cell. 2003;115:652–653. doi: 10.1016/S0092-8674(03)00976-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du L., He Y., Wang Y., Zhang H., Ma S., Wong C.K., Wu S.H., Ng F., Huang J.D., Yuen K.Y., Jiang S., Zhou Y., Zheng B.J. Recombinant adeno-associated virus expressing the receptor-binding domain of severe acute respiratory syndrome coronavirus S protein elicits neutralizing antibodies: implication for developing SARS vaccines. Virology. 2006;353:6–16. doi: 10.1016/j.virol.2006.03.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du L., Kao R.Y., Zhou Y., He Y., Zhao G., Wong C., Jiang S., Yuen K.Y., Jin D.Y., Zheng B.J. Cleavage of spike protein of SARS coronavirus by protease factor Xa is associated with viral infectivity. Biochem. Biophys. Res. Commun. 2007;359:174–179. doi: 10.1016/j.bbrc.2007.05.092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du L., Zhao G., He Y., Guo Y., Zheng B.J., Jiang S., Zhou Y. Receptor-binding domain of SARS-CoV spike protein induces long-term protective immunity in an animal model. Vaccine. 2007;25:2832–2838. doi: 10.1016/j.vaccine.2006.10.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du L., Zhao G., Lin Y., Chan C., He Y., Jiang S., Wu C., Jin D.Y., Yuen K.Y., Zhou Y., Zheng B.J. Priming with rAAV encoding RBD of SARS-CoV S protein and boosting with RBD-specific peptides for T cell epitopes elevated humoral and cellular immune responses against SARS-CoV infection. Vaccine. 2008;26:1644–1651. doi: 10.1016/j.vaccine.2008.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du L., Zhao G., Lin Y., Sui H., Chan C., Ma S., He Y., Jiang S., Wu C., Yuen K.Y., Jin D.Y., Zhou Y., Zheng B.J. Intranasal vaccination of recombinant adeno-associated virus encoding receptor-binding domain of severe acute respiratory syndrome coronavirus (SARS-CoV) spike protein induces strong mucosal immune responses and provides long-term protection against SARS-CoV infection. J. Immunol. 2008;180:948–956. doi: 10.4049/jimmunol.180.2.948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du L., He Y., Zhou Y., Liu S., Zheng B.J., Jiang S. The spike protein of SARS-CoV—a target for vaccine and therapeutic development. Nat. Rev., Microbiol. 2009;7:226–236. doi: 10.1038/nrmicro2090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du L., Zhao G., Li L., He Y., Zhou Y., Zheng B.J., Jiang S. Antigenicity and immunogenicity of SARS-CoV S protein receptor-binding domain stably expressed in CHO cells. Biochem. Biophys. Res. Commun. 2009;384:486–490. doi: 10.1016/j.bbrc.2009.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farinha-Arcieri L.E., Porchia B.M., Carromeu C., Simabuco F.M., Tamura R.E., Ferreira L.C., Zerbini L.F., Ventura A.M. Expression and purification of a recombinant adenovirus fiber knob in a baculovirus system. Intervirology. 2008;51:189–195. doi: 10.1159/000151532. [DOI] [PubMed] [Google Scholar]
- Gao W., Tamin A., Soloff A., D Aiuto L., Nwanegbo E., Robbins P.D., Bellini W.J., Barratt-Boyes S., Gambotto A. Effects of a SARS-associated coronavirus vaccine in monkeys. Lancet. 2003;362:1895–1896. doi: 10.1016/S0140-6736(03)14962-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan Y., Zheng B.J., He Y.Q., Liu X.L., Zhuang Z.X., Cheung C.L., Luo S.W., Li P.H., Zhang L.J., Guan Y.J., Butt K.M., Wong K.L., Chan K.W., Lim W., Shortridge K.F., Yuen K.Y., Peiris J.S., Poon L.L. Isolation and characterization of viruses related to the SARS coronavirus from animals in southern China. Science. 2003;302:276–278. doi: 10.1126/science.1087139. [DOI] [PubMed] [Google Scholar]
- He Y., Zhou Y., Liu S., Kou Z., Li W., Farzan M., Jiang S. Receptor-binding domain of SARS-CoV spike protein induces highly potent neutralizing antibodies: implication for developing subunit vaccine. Biochem. Biophys. Res. Commun. 2004;324:773–781. doi: 10.1016/j.bbrc.2004.09.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y., Lu H., Siddiqui P., Zhou Y., Jiang S. Receptor-binding domain of severe acute respiratory syndrome coronavirus spike protein contains multiple conformation-dependent epitopes that induce highly potent neutralizing antibodies. J. Immunol. 2005;174:4908–4915. doi: 10.4049/jimmunol.174.8.4908. [DOI] [PubMed] [Google Scholar]
- He Y., Li J., Du L., Yan X., Hu G., Zhou Y., Jiang S. Identification and characterization of novel neutralizing epitopes in the receptor-binding domain of SARS-CoV spike protein: revealing the critical antigenic determinants in inactivated SARS-CoV vaccine. Vaccine. 2006;24:5498–5508. doi: 10.1016/j.vaccine.2006.04.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y., Li J., Li W., Lustigman S., Farzan M., Jiang S. Cross-neutralization of human and palm civet severe acute respiratory syndrome coronaviruses by antibodies targeting the receptor-binding domain of spike protein. J. Immunol. 2006;176:6085–6092. doi: 10.4049/jimmunol.176.10.6085. [DOI] [PubMed] [Google Scholar]
- Hu H., Lu X., Tao L., Bai B., Zhang Z., Chen Y., Zheng F., Chen J., Chen Z., Wang H. Induction of specific immune responses by severe acute respiratory syndrome coronavirus spike DNA vaccine with or without interleukin-2 immunization using different vaccination routes in mice. Clin. Vaccine Immunol. 2007;14:894–901. doi: 10.1128/CVI.00019-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu M.C., Jones T., Kenney R.T., Barnard D.L., Burt D.S., Lowell G.H. Intranasal protollin-formulated recombinant SARS S-protein elicits respiratory and serum neutralizing antibodies and protection in mice. Vaccine. 2007;25:6334–6340. doi: 10.1016/j.vaccine.2007.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J., Ma R., Wu C.Y. Immunization with SARS-CoV S DNA vaccine generates memory CD4+ and CD8+ T cell immune responses. Vaccine. 2006;24:4905–4913. doi: 10.1016/j.vaccine.2006.03.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kan B., Wang M., Jing H., Xu H., Jiang X., Yan M., Liang W., Zheng H., Wan K., Liu Q., Cui B., Xu Y., Zhang E., Wang H., Ye J., Li G., Li M., Cui Z., Qi X., Chen K., Du L., Gao K., Zhao Y.T., Zou X.Z., Feng Y.J., Gao Y.F., Hai R., Yu D., Guan Y., Xu J. Molecular evolution analysis and geographic investigation of severe acute respiratory syndrome coronavirus-like virus in palm civets at an animal market and on farms. J. Virol. 2005;79:11892–11900. doi: 10.1128/JVI.79.18.11892-11900.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobinger G.P., Figueredo J.M., Rowe T., Zhi Y., Gao G., Sanmiguel J.C., Bell P., Wivel N.A., Zitzow L.A., Flieder D.B., Hogan R.J., Wilson J.M. Adenovirus-based vaccine prevents pneumonia in ferrets challenged with the SARS coronavirus and stimulates robust immune responses in macaques. Vaccine. 2007;25:5220–5231. doi: 10.1016/j.vaccine.2007.04.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn J.H., Li W., Choe H., Farzan M. Angiotensin-converting enzyme 2: a functional receptor for SARS coronavirus. Cell. Mol. Life Sci. 2004;61:2738–2743. doi: 10.1007/s00018-004-4242-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau S.K., Woo P.C., Li K.S., Huang Y., Tsoi H.W., Wong B.H., Wong S.S., Leung S.Y., Chan K.H., Yuen K.Y. Severe acute respiratory syndrome coronavirus-like virus in Chinese horseshoe bats. Proc. Natl. Acad. Sci. U. S. A. 2005;102:14040–14045. doi: 10.1073/pnas.0506735102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J.S., Poo H., Han D.P., Hong S.P., Kim K., Cho M.W., Kim E., Sung M.H., Kim C.J. Mucosal immunization with surface-displayed severe acute respiratory syndrome coronavirus spike protein on Lactobacillus casei induces neutralizing antibodies in mice. J. Virol. 2006;80:4079–4087. doi: 10.1128/JVI.80.8.4079-4087.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liniger M., Zuniga A., Tamin A., Azzouz-Morin T.N., Knuchel M., Marty R.R., Wiegand M., Weibel S., Kelvin D., Rota P.A., Naim H.Y. Induction of neutralising antibodies and cellular immune responses against SARS coronavirus by recombinant measles viruses. Vaccine. 2008;26:2164–2174. doi: 10.1016/j.vaccine.2008.01.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., Moore M.J., Vasilieva N., Sui J., Wong S.K., Berne M.A., Somasundaran M., Sullivan J.L., Luzuriaga K., Greenough T.C., Choe H., Farzan M. Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature. 2003;426:450–454. doi: 10.1038/nature02145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., Shi Z., Yu M., Ren W., Smith C., Epstein J.H., Wang H., Crameri G., Hu Z., Zhang H., Zhang J., McEachern J., Field H., Daszak P., Eaton B.T., Zhang S., Wang L.F. Bats are natural reservoirs of SARS-like coronaviruses. Science. 2005;310:676–679. doi: 10.1126/science.1118391. [DOI] [PubMed] [Google Scholar]
- Liu S., Xiao G., Chen Y., He Y., Niu J., Escalante C.R., Xiong H., Farmar J., Debnath A.K., Tien P., Jiang S. Interaction between heptad repeat 1 and 2 regions in spike protein of SARS-associated coronavirus: implications for virus fusogenic mechanism and identification of fusion inhibitors. Lancet. 2004;363:938–947. doi: 10.1016/S0140-6736(04)15788-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manopo I., Lu L., He Q., Chee L.L., Chan S.W., Kwang J. Evaluation of a safe and sensitive spike protein-based immunofluorescence assay for the detection of antibody responses to SARS-CoV. J. Immunol. Methods. 2005;296:37–44. doi: 10.1016/j.jim.2004.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marblestone J.G., Edavettal S.C., Lim Y., Lim P., Zuo X., Butt T.R. Comparison of SUMO fusion technology with traditional gene fusion systems: enhanced expression and solubility with SUMO. Protein Sci. 2006;15:182–189. doi: 10.1110/ps.051812706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marra M.A., Jones S.J., Astell C.R., Holt R.A., Brooks-Wilson A., Butterfield Y.S., Khattra J., Asano J.K., Barber S.A., Chan S.Y., Cloutier A., Coughlin S.M., Freeman D., Girn N., Griffith O.L., Leach S.R., Mayo M., McDonald H., Montgomery S.B., Pandoh P.K., Petrescu A.S., Robertson A.G., Schein J.E., Siddiqui A., Smailus D.E., Stott J.M., Yang G.S., Plummer F., Andonov A., Artsob H., Bastien N., Bernard K., Booth T.F., Bowness D., Czub M., Drebot M., Fernando L., Flick R., Garbutt M., Gray M., Grolla A., Jones S., Feldmann H., Meyers A., Kabani A., Li Y., Normand S., Stroher U., Tipples G.A., Tyler S., Vogrig R., Ward D., Watson B., Brunham R.C., Krajden M., Petric M., Skowronski D.M., Upton C., Roper R.L. The genome sequence of the SARS-associated coronavirus. Science. 2003;300:1399–1404. doi: 10.1126/science.1085953. [DOI] [PubMed] [Google Scholar]
- Martin J.E., Louder M.K., Holman L.A., Gordon I.J., Enama M.E., Larkin B.D., Andrews C.A., Vogel L., Koup R.A., Roederer M., Bailer R.T., Gomez P.L., Nason M., Mascola J.R., Nabel G.J., Graham B.S. A SARS DNA vaccine induces neutralizing antibody and cellular immune responses in healthy adults in a phase I clinical trial. Vaccine. 2008;26:6338–6343. doi: 10.1016/j.vaccine.2008.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prabakaran P., Xiao X., Dimitrov D.S. A model of the ACE2 structure and function as a SARS-CoV receptor. Biochem. Biophys. Res. Commun. 2004;314:235–241. doi: 10.1016/j.bbrc.2003.12.081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rota P.A., Oberste M.S., Monroe S.S., Nix W.A., Campagnoli R., Icenogle J.P., Penaranda S., Bankamp B., Maher K., Chen M.H., Tong S., Tamin A., Lowe L., Frace M., DeRisi J.L., Chen Q., Wang D., Erdman D.D., Peret T.C., Burns C., Ksiazek T.G., Rollin P.E., Sanchez A., Liffick S., Holloway B., Limor J., McCaustland K., Olsen-Rasmussen M., Fouchier R., Gunther S., Osterhaus A.D., Drosten C., Pallansch M.A., Anderson L.J., Bellini W.J. Characterization of a novel coronavirus associated with severe acute respiratory syndrome. Science. 2003;300:1394–1399. doi: 10.1126/science.1085952. [DOI] [PubMed] [Google Scholar]
- Saitoh H., Uwada J., Azusa K. Strategies for the expression of SUMO-modified target proteins in Escherichia coli. Methods Mol. Biol. 2009;497:211–221. doi: 10.1007/978-1-59745-566-4_14. [DOI] [PubMed] [Google Scholar]
- Skowronski D.M., Astell C., Brunham R.C., Low D.E., Petric M., Roper R.L., Talbot P.J., Tam T., Babiuk L. Severe acute respiratory syndrome (SARS): a year in review. Annu. Rev. Med. 2005;56:357–381. doi: 10.1146/annurev.med.56.091103.134135. [DOI] [PubMed] [Google Scholar]
- Stadler K., Roberts A., Becker S., Vogel L., Eickmann M., Kolesnikova L., Klenk H.D., Murphy B., Rappuoli R., Abrignani S., Subbarao K. SARS vaccine protective in mice. Emerg. Infect. Dis. 2005;11:1312–1314. doi: 10.3201/eid1108.041003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tripet B., Howard M.W., Jobling M., Holmes R.K., Holmes K.V., Hodges R.S. Structural characterization of the SARS-coronavirus spike S fusion protein core. J. Biol. Chem. 2004;279:20836–20849. doi: 10.1074/jbc.M400759200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Xu W., Tong D., Ni J., Gao H., Wang Y., Chu Y., Li P., Yang X., Xiong S. A chimeric multi-epitope DNA vaccine elicited specific antibody response against severe acute respiratory syndrome-associated coronavirus which attenuated the virulence of SARS-CoV in vitro. Immunol. Lett. 2008;119:71–77. doi: 10.1016/j.imlet.2008.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weingartl H., Czub M., Czub S., Neufeld J., Marszal P., Gren J., Smith G., Jones S., Proulx R., Deschambault Y., Grudeski E., Andonov A., He R., Li Y., Copps J., Grolla A., Dick D., Berry J., Ganske S., Manning L., Cao J. Immunization with modified vaccinia virus Ankara-based recombinant vaccine against severe acute respiratory syndrome is associated with enhanced hepatitis in ferrets. J. Virol. 2004;78(22):12672–12676. doi: 10.1128/JVI.78.22.12672-12676.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong S.K., Li W., Moore M.J., Choe H., Farzan M. A 193-amino acid fragment of the SARS coronavirus S protein efficiently binds angiotensin-converting enzyme 2. J. Biol. Chem. 2004;279:3197–3201. doi: 10.1074/jbc.C300520200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao X., Chakraborti S., Dimitrov A.S., Gramatikoff K., Dimitrov D.S. The SARS-CoV S glycoprotein: expression and functional characterization. Biochem. Biophys. Res. Commun. 2003;312:1159–1164. doi: 10.1016/j.bbrc.2003.11.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y., Lou Z., Liu Y., Pang H., Tien P., Gao G.F., Rao Z. Crystal structure of severe acute respiratory syndrome coronavirus spike protein fusion core. J. Biol. Chem. 2004;279:49414–49419. doi: 10.1074/jbc.M408782200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z.Y., Kong W.P., Huang Y., Roberts A., Murphy B.R., Subbarao K., Nabel G.J. A DNA vaccine induces SARS coronavirus neutralization and protective immunity in mice. Nature. 2004;428:561–564. doi: 10.1038/nature02463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh K.M., Chiueh T.S., Siu L.K., Lin J.C., Chan P.K., Peng M.Y., Wan H.L., Chen J.H., Hu B.S., Perng C.L., Lu J.J., Chang F.Y. Experience of using convalescent plasma for severe acute respiratory syndrome among healthcare workers in a Taiwan hospital. J. Antimicrob. Chemother. 2005;56:919–922. doi: 10.1093/jac/dki346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin J., Li G., Ren X., Herrler G. Select what you need: a comparative evaluation of the advantages and limitations of frequently used expression systems for foreign genes. J. Biotechnol. 2007;127:335–347. doi: 10.1016/j.jbiotec.2006.07.012. [DOI] [PubMed] [Google Scholar]
- Zakhartchouk A.N., Sharon C., Satkunarajah M., Auperin T., Viswanathan S., Mutwiri G., Petric M., See R.H., Brunham R.C., Finlay B.B., Cameron C., Kelvin D.J., Cochrane A., Rini J.M., Babiuk L.A. Immunogenicity of a receptor-binding domain of SARS coronavirus spike protein in mice: implications for a subunit vaccine. Vaccine. 2007;25:136–143. doi: 10.1016/j.vaccine.2006.06.084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang M.Y., Wang Y., Mankowski M.K., Ptak R.G., Dimitrov D.S. Cross-reactive HIV-1-neutralizing activity of serum IgG from a rabbit immunized with gp41 fused to IgG1 Fc: possible role of the prolonged half-life of the immunogen. Vaccine. 2009;27:857–863. doi: 10.1016/j.vaccine.2008.11.083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao B., Jin N.Y., Wang R.L., Zhang L.S., Zhang Y.J. Immunization of mice with a DNA vaccine based on severe acute respiratory syndrome coronavirus spike protein fragment 1. Viral Immunol. 2006;19:518–524. doi: 10.1089/vim.2006.19.518. [DOI] [PubMed] [Google Scholar]
- Zhong N.S., Zheng B.J., Li Y.M., Poon, Xie Z.H., Chan K.H., Li P.H., Tan S.Y., Chang Q., Xie J.P., Liu X.Q., Xu J., Li D.X., Yuen K.Y., Peiris, Guan Y. Epidemiology and cause of severe acute respiratory syndrome (SARS) in Guangdong, People's Republic of China, in February, 2003. Lancet. 2003;362:1353–1358. doi: 10.1016/S0140-6736(03)14630-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Z., Post P., Chubet R., Holtz K., McPherson C., Petric M., Cox M. A recombinant baculovirus-expressed S glycoprotein vaccine elicits high titers of SARS-associated coronavirus (SARS-CoV) neutralizing antibodies in mice. Vaccine. 2006;24:3624–3631. doi: 10.1016/j.vaccine.2006.01.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J., Xiao G., Xu Y., Yuan F., Zheng C., Liu Y., Yan H., Cole D.K., Bell J.I., Rao Z., Tien P., Gao G.F. Following the rule: formation of the 6-helix bundle of the fusion core from severe acute respiratory syndrome coronavirus spike protein and identification of potent peptide inhibitors. Biochem. Biophys. Res. Commun. 2004;319:283–288. doi: 10.1016/j.bbrc.2004.04.141. [DOI] [PMC free article] [PubMed] [Google Scholar]