Abstract
Circular permutation of Candida antarctica lipase B yields several enzyme variants with substantially increased catalytic activity. To better understand the structural and functional consequences of protein termini reorganization, we have applied protein engineering and x-ray crystallography to cp283, one of the most active hydrolase variants. Our initial investigation has focused on the role of an extended surface loop, created by linking the native N and C-termini, on protein integrity. Incremental truncation of the loop partially compensates for observed losses in secondary structure and the permutants’ temperature of unfolding. Unexpectedly, the improvements are accompanied by quaternary structure changes from monomer to dimer. The crystal structures of one truncated variant (cp283Δ7) in the apo-form determined at 1.49Å resolution and with a bound phosphonate inhibitor at 1.69Å resolution confirmed the formation of a homodimer by swapping of the enzyme’s 35-residue N-terminal region. Separately, the new protein termini at amino acid positions 282/283 convert the narrow access tunnel to the catalytic triad into a broad crevice for accelerated substrate entry and product exit while preserving the native active site topology for optimal catalytic turnover.
INTRODUCTION
Lipase B from Candida antarctica (CALB) is an important biocatalyst for asymmetric synthesis. The enzyme’s broad substrate specificity and high enantioselectivity, as well as its ability to function in aqueous and organic reaction environments makes CALB a popular choice for a wide range of applications including kinetic resolution, transesterification, and polymerization reactions. 1–6 Structurally, CALB is a monomer and belongs to the 〈/® hydrolase fold family, represented by a conserved core structure with a eight-stranded mostly parallel twisted ®-sheet, flanked on both sides by 〈-helices.7,8 The fold presents a stable scaffold for the residues of the catalytic triad (Ser-His-Asp). Separately, helix 7–9 (also known as the lid region) and helix 16/17 in the C-terminal portion of the protein form the cap domain which plays a critical role by defining the narrow substrate binding pocket and controlling access to the active site.
The modularity and functional versatility of CALB has made the enzyme a target for protein engineering, tailoring substrate specificity and improving catalytic performance by rational protein design, random mutagenesis and DNA shuffling.9–14 Complementing these efforts, we recently employed a less conventional technique called circular permutation (CP) to explore the effects of termini relocation on catalytic performance of CALB.15 During the process of CP, the natural N and C termini of a protein are covalently linked by a short peptide and new termini are created elsewhere within the original sequence (Fig. 1). As a consequence, the amino acid composition of the protein remains unchanged, yet its primary sequence is rearranged. We hypothesize that such reorganization could benefit enzyme catalysis through changes in local protein conformation and increased backbone flexibility. Supporting our idea, a recent report by Miki and coworkers on the crystal structure of the polyhydroxybutyrate depolymerase from Penicillium funiculosum concluded that CP of the 〈/® hydrolase fold in nature resulted in topological changes of the substrate binding site, allowing the enzyme to accommodate the polymeric substrate without the need for a distinct binding domain.16 Additional examples of protein reorganization by CP in nature has been reported although its evolutionary benefit remains unclear.17,18 In the laboratory, the technique has traditionally been used for protein folding studies whereas its application in protein engineering led to improvements in stability, specificity and activity.19–26 Upon random circular permutation of CALB, we found that almost 20% of permutated enzymes retained ester hydrolase activity. More importantly, some permutants showed increases in hydrolytic activity of 11-fold and 175-fold for p-nitrophenol butyrate (p-NB) and 6,8-difluoro-4-methylumbelliferyl octanoate (DiFMU octanoate), respectively.27
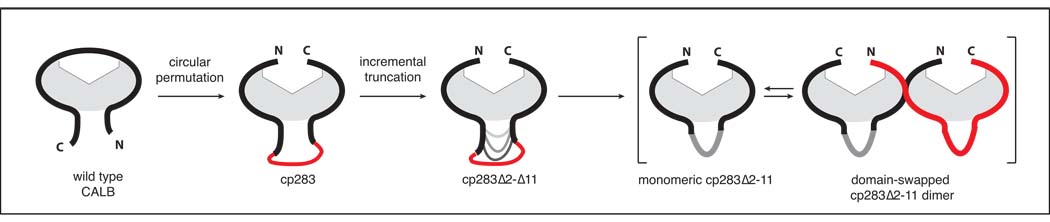
Figure 1.
Schematic overview of CALB engineering by circular permutation and subsequent incremental truncation of the newly created surface loop in cp283, the most active variant among the lipase permutants. Depending on the extent of loop truncation, we observed a change in the enzyme quaternary structure, shifting from a strictly monomeric form to a dimer with a domain-swapped N-terminal segment.
The substantial enhancements in catalytic performance raised questions concerning the consequences of CP on CALB structure. Several earlier crystallographic studies of laboratory-permutated proteins have shown only minor deviations in those proteins’ tertiary structures except in two regions: the original termini and the newly created N and C-termini.28–32 The fusion of the original termini often creates structurally poorly defined loops, in particular in the presence of extra amino acids to link distant residues. Similarly, the cleavage of the peptide backbone at the site of the new protein termini appears to result in a higher degree of flexibility and disorder. To explore whether similar changes occurred upon CP of CALB, we had initially conducted studies using circular dichroism spectroscopy to evaluate the structural integrity and stability of our engineered lipases.27 The results from these early experiments were consistent with the previous literature reports, suggesting a significant destabilization of the overall protein upon CP. We hypothesized that the effect was related to the extended surface loop, created by covalently fusing the native CALB termini with a hexapeptide. Together with the 12 and 30 amino acids of the structurally poorly defined native N and C-termini, respectively, the glycin-rich linker was believed to form a flexible loop region. In model studies with unstructured loops in proteins, such regions were shown to potentially compromise overall stability due to their unfavorable entropy of loop closure.33,34
Reevaluating our original linker design, we have now investigated the importance of this region’s length and sequence composition by incremental truncation. For the current study, we selected cp283, the variant with the most dramatic rate enhancements, whose new N and C-termini are located at position 283 and 282 of the wild type CALB sequence (Fig. 1). Following loop truncation, the characterization of selected candidates indicated that deletion of up to 11 residues has little effect on the catalytic activity, yet can significantly improve enzyme stability. Unexpectedly, the loop truncation also changed the protein’s quaternary structure. The shortening of the loop seems to favor protein dimerization and formation of higher order oligomers. To verify dimer creation and explore the effects of CP on old and new termini regions, we consequently solved the crystal structure of cp283Δ7, a truncated cp283 with a seven-amino acid deletion in the linker region connecting the native termini, which showed homodimer formation via N-terminal domain swapping. Interestingly, protein dimerization creates a new major cleft at the protein interface which supports the idea that circular permutation could play an important role in protein evolution by enabling the fundamental reorganization of existing protein scaffolds at the quaternary structure level. At the tertiary structure level, the crystal structure indicates a substantial redesign of the active site binding pocket of CALB, converting the restricting active site tunnel of the wild type lipase into a wide open crevice.
RESULTS & DISCUSSION
Truncating the extended surface loop region of cp283
Circular permutation of CALB can significantly improve the catalytic performance of the lipase which has raised questions concerning the principal changes in enzyme structure and function. From a structural perspective, previous studies of circular permutated proteins have consistently identified the old and new amino and carboxy termini as the regions with the most substantial changes in protein topology. Hence the initial focus of our present study was the glycine-rich six amino acid linker to connect the native enzyme termini. The fusion of the original N and C-termini of CALB with the hexapeptide linker created a 44-residue surface loop which is believed responsible for the observed thermal destabilization of our engineered lipases.27 To test our hypothesis, we created a library of cp283 with variable loop length by incremental truncation (Fig. 1). The library was constructed by cleaving the gene encoding cp283 at the unique SpeI restriction site in the hexapeptide coding region.15 Next, the two new DNA ends were incrementally truncated, using the ITCHY protocol in combination with 〈-phosphothioate dNTPs.35 Given cp283’s gene size of ~1000 nucleotides, truncated DNA fragments of 750 to 1000 nucleotides in length were selected by agarose gel electrophoresis.36 By restricting loop truncation to ~250 nucleotides, we reduced the library size and improved the efficiency of library analysis as functional members with larger truncations are considered highly unlikely. Following transformation into P. pastoris, the size of the naïve library was estimated at ~450,000 members. The DNA sequence analysis of 44 randomly selected library members showed truncation of up to 80 amino acids with no apparent bias besides the limited extend of truncation due to size selection (Fig. S1).
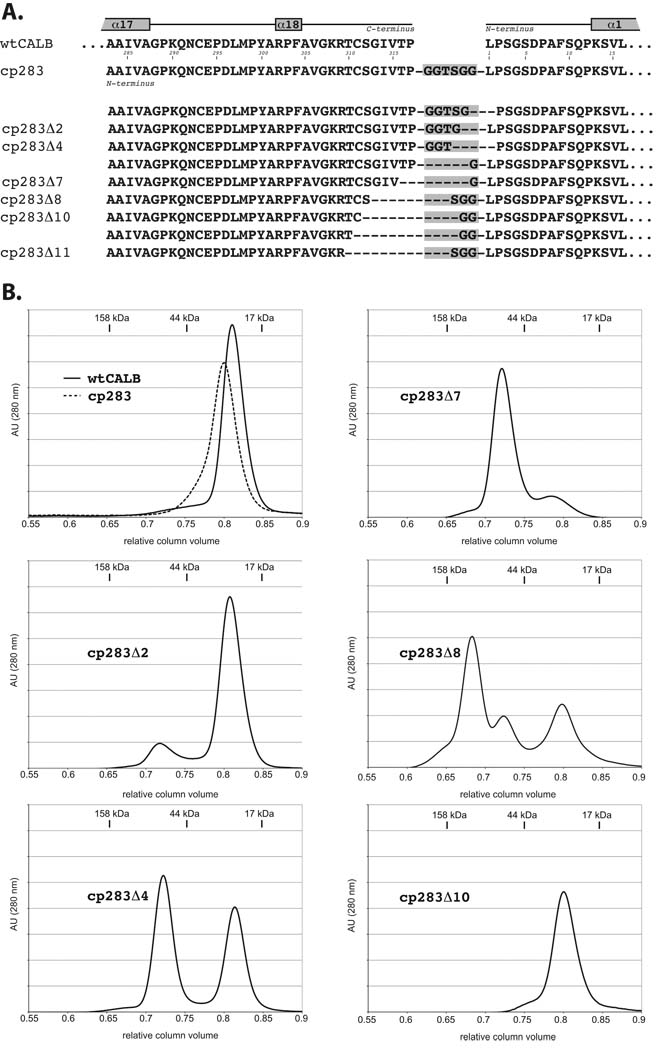
Library members were screened on tributyrin plates and active lipases were identified based on halo formation.15 Subsequent DNA sequence analysis of the calB genes of 100 positive colonies yielded 31 variants with distinct truncation patterns, showing deletion of up to 11 residues (25%) of the loop (Fig. 2A). Even though the naive library showed equal amount of truncation on either terminus, the distribution after functional selection appears to be skewed towards elimination of residues from the native C-terminus. The cause for such a bias is unclear in the absence of known specific interactions in the N-terminal region. The abrupt loss of function upon truncation of more than 11 residues at the C-terminus might also points towards a critical role of Lys308 and Arg309 for catalysis or enzyme stability. Furthermore, our data suggest that length rather than the precise nature of the amino acid composition is important to the linker design as exemplified by the multiple solutions for cp283Δ2, Δ4, and Δ10. Interestingly though, all truncated variants kept at least one glycine residue from our original linker sequence, indicating some benefit from added backbone flexibility. From among the 31 truncated cp283 variants, we chose six members with 2 to 11 amino acid deletions (Δ2 – Δ11) for further characterization (Fig. 2A). All candidates were overexpressed and purified to homogeneity (>95% based on SDS-PAGE analysis).
Figure 2.
Correlation of loop truncation with quaternary structure changes. A) Sequence alignment of N and C-terminal region of CALB. The original variant cp283 carries a six-amino acid linker (grew box) and has its new N-terminus moved to position 283. Subsequent incremental truncation of cp283, followed by screening for functional lipases, yields a series of variants (cp283Δ2–11) with shortened linker regions. Representatives with various linker lengths were selected for overexpression and in vitro characterization (indicated by labels). B) Size exclusion chromatograms for wild type CALB, cp283, and truncated variants. While CALB and cp283 are monomeric (RCV ~0.81, estimated MW: 32 kDa), a second peak at RCV ~0.72, corresponding to dimer, emerges in variants with truncated loops. The third peak (RCV ~0.68) in cp283Δ8 like represents the trimer form. Molecular weights were calculated based on the calibration curve with protein size standards (data not shown).
The final protein purification step for the truncated cp283 variants by size exclusion chromatography (SEC) revealed some fundamental changes in their quaternary structures. Both wild type CALB and cp283 eluted at a relative column volume (RCV) of 0.8 to 0.82 which, based on size standards, corresponds to a monomer with a calculated molecular weight of ~31 kDa (Fig. 2B). In contrast, most cp283 variants with truncated loops showed a second peak at approximately 0.72 RCV. Such a RCV corresponds to a molecular mass of ~64 kDa and suggests formation of a dimeric species. The fraction of dimer grows with increasing length of truncation, shifting from only a shoulder in cp283Δ2 to roughly 50:50 for cp283Δ4 to becoming the dominant product in cp283Δ7. Surprisingly, the trend stops abruptly upon deletion of an additional amino acid in cp283Δ8, creating instead a complex mixture of monomer, dimer and oligomer (RCV ~ 0.68). Further truncation of residues in the loop region (cp283Δ10) eliminated all higher-order quaternary structures and once again yields exclusively monomer.
Structural and functional properties of truncated cp283
The impact of truncation on the structure and function of the circular permutated CALB was next investigated by steady-state kinetics and circular dichroism spectroscopy. The kinetic parameters of purified cp283Δ2 through Δ11 were determined for the reference substrate p-nitrophenol butyrate (pNB) and compared with the catalytic properties of wild type CALB and cp283 (Table S1). At ambient temperature, the kcat and KM values of the truncated variants deviates less than 3-fold from cp283, leaving the catalytic efficiency largely unchanged and retaining an approximately 10-fold higher relative efficiency over wild type CALB. Overall, these data suggest that the shortened loop in our candidates does not directly impact catalysis. To address concerns over using a mixture of oligomeric forms for the kinetic analysis, we separated the monomeric and dimeric form of cp283Δ4 by SEC. Re-injection of the two purified proteins showed no re-equilibration, even after extended sample storage over several days, suggesting a significant thermodynamic barrier between the two structural forms (data not shown). Next, the catalytic performance of purified cp283Δ4 monomer and dimer was tested with pNB (Table S1). The results did not show any significant difference in apparent binding affinity or turnover rate, supporting the earlier conclusion that truncation of up to 11 residues in the loop region does not directly influence catalysis.
The circular dichroism experiments in the far UV range show good correlation with the trend observed in size exclusion chromatography. A gain in signal strength at 195 nm suggests recovery of helical content upon truncation of up to 7 residues (Fig. S2). The observed increase in secondary structure is paralleled by a raise in the temperature of unfolding (TM) from 40°C for cp283 to 45°C for cp283Δ7 (TM for wild type CALB = 53°C). While the spectrum of cp283Δ7 closely resembles that of wild type CALB, the trend stops abruptly upon removal of an additional residue in cp283Δ8, resulting in declining signal intensity and a drop in the temperature of unfolding to 35 °C (Table S1). As such, the findings are consistent with the trend observed for the SEC data. More interestingly, the TM measurement of the cp283Δ4 monomer/dimer mixture shows a temperature of unfolding of 41.7 °C, yet its purified monomer has a transition midpoint of 40.0 °C while the TM for the presumptive dimer shifts to 42.3 °C. These results suggest that loop length per se does not contribute to increased stability as the TM for monomeric cp283Δ4 remains unchanged compared to cp283 (TM = 39.9°C). Instead, the ΔTM of 2.3°C upon cp283Δ4 dimerization points towards minimization of exposed surface area as the driving force behind increased thermostability. The additional gain of 3°C in the temperature of unfolding from cp283Δ4 to Δ7 could be rationalized by improved side chain packing at the dimer interface. Calculations of free energy differences (ΔΔG40), based on the fraction of folded versus unfolded proteins at 40°C (as determined by thermodenaturation CD spectroscopy) indicated a gain of up to 1.1 kcal/mol in cp283Δ7 compared to parental cp283 with the hexapeptide linker (Table S1).
Crystallographic analysis of cp283Δ7
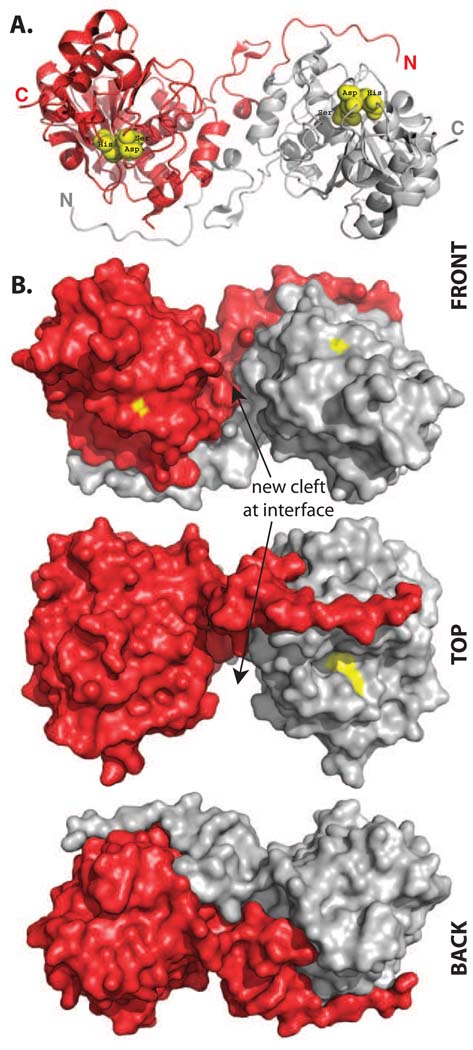
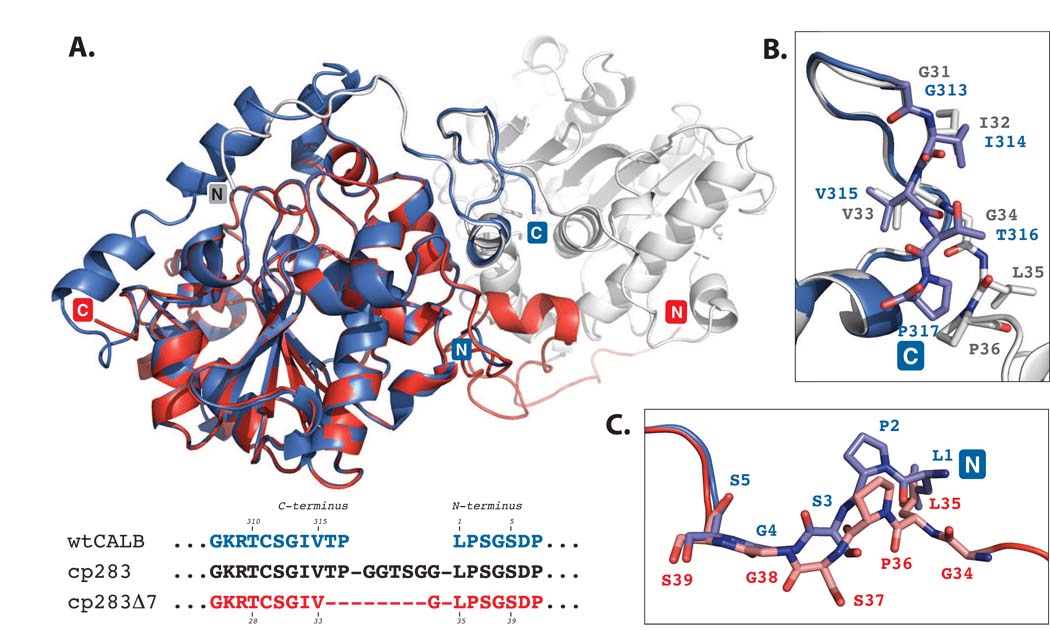
To further investigate the nature of the newly formed oligomeric species of truncated cp283, we crystallized the most stable variant, cp283Δ7. Crystals were obtained for two states, the apo-protein and an inhibitor-bound form of the enzyme, and diffracted to resolutions of 1.49 Å (apo) and 1.69 Å (inhibitor-bound), respectively (Table 1). The structures were solved by molecular replacement using the wild type CALB (PDB: 1LBS)8 as a search model. cp283Δ7 forms a homodimer as predicted from the gel filtration data (Fig. 3). The two subunits exhibit C2-symmetry and are nearly perfect superimposable with the wild type enzyme, showing a root square mean deviation (rmsd) of only 0.5 Å over 263 C〈 atoms (out of 298 residues). Most noticeable, a large portion of the new interface is created by swapping of the two subunits’ N-terminal segments. Circular permutation of CALB in combination with incremental truncation has positioned the 35-residue N-terminus of the lipase in an awkward extended orientation, pointing away from the rest of the protein. The benefit of such a conformational change becomes apparent upon superposition of the dimer structure with wild type CALB (Fig. 4). The engineering of the loop region allows the two subunits to orient themselves in such a way that the extended N-terminal segment of first subunit assumes precisely the orientation of the wild type enzyme’s native C-terminus on the second subunit (Fig 4B). At the same time, the N-terminus connects almost seamlessly with the body of its own subunit (Fig. 4C).
Table 1.
Statistics of X-ray Data Reduction and Phasing, as well as Structure Refinement
| Crystal | Native | Inhibitor Soak | |
|---|---|---|---|
| Space group | P3221 | ||
| Cell constants | a (Å) = | 111.680 | 111.746 |
| b (Å) = | 111.680 | 111.746 | |
| c (Å) = | 54.997 | 54.806 | |
| Beamline | SER-CAT, APS | ||
| Wavelength (Å) | 1.00000 | 1.00000 | |
| Resolution Range, Å (Highest Resolution Shell) |
27.8–1.49 (1.56–1.49) |
27.8–1.69 (1.75–1.69) |
|
| Measured Reflections | 675,200 | 572,878 | |
| Unique Reflections | 64,040 | 44,608 | |
| <I/σ> | 23.3 | 14.4 | |
| Completeness (%) | 99.1 (98.5) | 99.9 (99.8) | |
| Rlinear=Σ|I-<I>|/Σ<I> | 0.077 (0.345) | 0.121 (0.473) | |
| R factor | 0.177 (0.220) | 0.180 (0.238) | |
| R-free (5% data) | 0.193 (0.225) | 0.199 (0.259) | |
| Non-hydrogen atoms | |||
| Protein | 2057 | 2078 | |
| Hetrogen (NAG,BTB) | 44 | ||
| Hetrogen (NAG, PO4 3−, methylhexylphosphonate) |
45 | ||
| Water | 413 | 338 | |
| r.m.s. deviation from ideality | |||
| Bond Lengths (Å) | 0.008 | 0.005 | |
| Bond Angles (°) | 1.6 | 1.5 | |
| Dihedral (°) | 24.1 | 23.9 | |
| Improper (°) | 1.08 | 0.96 | |
| Estimated coordinate error | |||
| From Luzzati plot (Å) | 0.15 | 0.18 | |
Figure 3.
Structure of the homodimeric cp283Δ7 (PDB: 3ICW). A) Ribbon representation of the dimer, showing the domain swapping of the N-terminal segments between the two subunits. The position of the catalytic triad is marked with yellow spheres. B) Space-filling model of the dimer. Yellow patches mark the location of residues in the catalytic triad. The two exchanged N-termini fit snug to the body of the adjacent subunit. The domain swapping creates a new major cleft at the protein subunit interface, measuring approximately 11 Å × 20 Å × 10 Å (width/length/depth). Figures were created using the program PyMol (Delano Scientific).
Figure 4.
Superposition of wild type CALB (PDB: 1LBS)8 and cp283Δ7 (PDB: 3ICW). CALB is colored in blue while the two subunits of cp283Δ7 are shown in red and light grey. The structure overlay is calculated based on minimal root square mean deviation (rsmd), using PyMol software. A) Overview of superimposed proteins. The core regions of the 〈/® hydrolase fold for CALB and cp283Δ7 align closely (rsmd: 0.5 Å over 263 C〈 atoms out of 298 residues). Deviations can be seen near the new termini of cp283Δ7 where the 5 N-terminal residues are slightly reoriented. The 17 amino acids at the C-terminus are invisible due to a lack of electron density, suggesting increased flexibility. B) Close-up view of the region near the C-terminus of CALB. The backbone structure and side chain orientation of the five amino acids at CALB’s carboxy end and the corresponding residues in cp283Δ7 are shown. The overlay illustrates the similar positioning of all residues up to Val315 (which corresponds to Val33 in cp283Δ7). Incremental truncation of cp283 eliminated Thr316 and Pro317, replacing them with the more flexible Gly34 from the linker sequence. Gly34 facilitates the rotation of the backbone required to properly align the domain-swapped N-terminal segment with the protein body. C) Close-up of the region at the N-terminus of CALB. A 180°-rotation at the peptide bond connecting Ser37-Gly38 (Ser3-Gly4 in CALB) results in the reorganization of Leu35-Ser37 (Leu1-Ser3 in CALB), forming a bridge to Gly34 which itself ties in with the swapped N-terminal segment of cp283Δ7.
Dimerization creates a new, extended protein-protein interface that includes 51 residues with a buried surface area of approximately 1850 Å2.37 Closer evaluation of the protein interface does not indicate formation of any covalent or disulfide bonds between the two subunits. Furthermore, no electrostatic interactions are detected and all nine hydrogen bonds between the N-terminal segment and the complementary subunit in the domain-swapped dimer are also present in the wild type structure. One weak hydrogen bond between Gln9 and Ser184 is slightly decreased from 3.8 Å (monomer) to 3.1 Å (dimer). It has been estimated that an interfacial H-bond could contribute as much as 1.8 kcal/mol.38
The driving force for dimerization could be the reduction in overall exposed surface area. While an accurate analysis of the differences between the monomeric cp283 and the dimeric cp283Δ7 are currently not possible due to no available structural information for the former, the comparison of the accessible surface area of wild-type CALB (11,672 Å2) with cp283Δ7 (10,712 Å2 per subunit) indicates that the quaternary structure formation does result in a noticeable reduction (~8%) in the surface area which could contribute towards the overall increase in protein stability. Separately, the ratio of total surface to interface area of cp283Δ7 falls well within the limits, characteristic for native dimers.39
The analysis of the apo-protein diffraction pattern indicated a lack of electron density for five residues at the new N-terminus, as well as 17 amino acids that make up the new C-terminus. Furthermore, nine residues of 〈-helix 8 (Gly142 – Val149), the enzyme’s active site lid, are missing. Speculating that these regions might become structured in the presence of a substrate analog in the active site, we incubated the enzyme with the suicide inhibitor 4-methylumbelliferyl hexylphosphonate and crystallized the enzyme adduct. Despite good electron density for the inhibitor and the N-terminus, which rotates by ~45° away from the active site and assumes an extended conformation, the lid and C-terminal region remained invisible (Fig. 5B). The presence of the substrate analog has no significant effect on the geometry of the active site binding pocket (Fig. S3) or the overall protein structure as reflected in the low rmsd of 0.2 Å over 294 residues between the apo protein and the inhibitor-bound enzyme. The substrate analog assumes an orientation resembling the transition state (Fig. S3). While the phosphonate oxygen is pointing towards the oxyanion hole, the hexyl moiety is placed into the acyl binding pocket. The methoxy group occupies the alcohol binding site, positioning the oxygen of the alkoxy group within hydrogen-bonding distance (3.2 Å) of Nε of His224 in the active site which is consistent with the mechanistic model for productive ester hydrolysis.40 These findings also complement earlier reports of wild type CALB, co-crystallized with a similar hexylphosphonate suicide substrate (PDB: 1LBS), which place the inhibitor in the opposite orientation, representing the less-favored enantiomer.8
Figure 5.
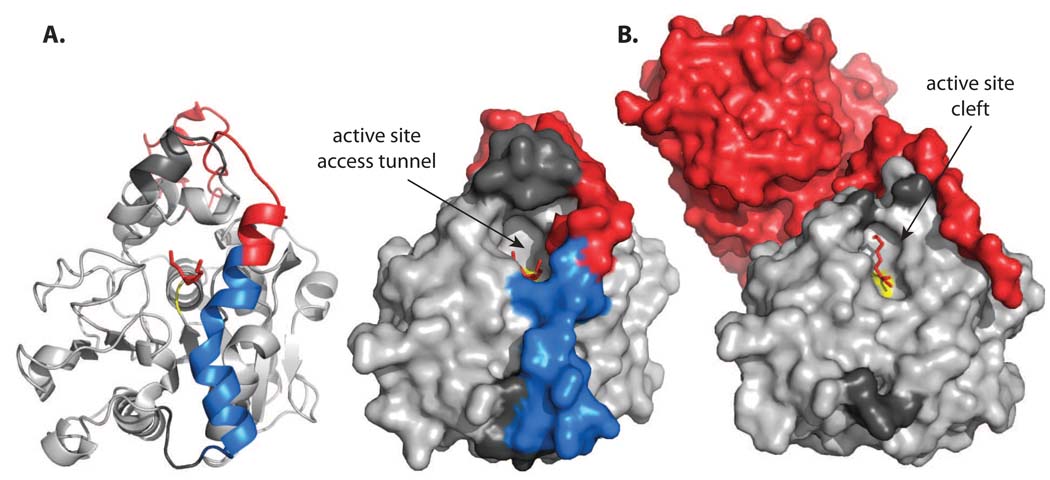
Comparison of the active site accessibility in wild type CALB (A) and cp283Δ7 (B). The structure of the parental CALB is shown as ribbon and surface representation. In all structures, the active site is marked in yellow and the bound phosphonate inhibitor is shown in red sticks. The dark-grey area above the active site marks the enzyme’s lid region which could not be assigned in cp283Δ7 due to a lack of electron density. Comparing the structures of the wild type and engineered lipases, the cleavage at Ala283 (position at red-blue crossover in helix 17) during circular permutation results in widening of the active site pocket. A 45°-rotation of the tip of the new N-terminus (highlighted in red), away from the active site, combined with a loss in helical conformation at the new C-terminus (colored in blue in wild type CALB, invisible in cp283Δ7 due to greater flexibility), eliminates most elements of the original binding pocket, converting the narrow access tunnel in wild type CALB into a open cleft-like structure.
Changes in cp283Δ7 ternary structure
At the ternary structure level, the x-ray diffraction data for the cp283Δ7 core structure shows little deviation from wild type enzyme, yet significant changes in the topology of the active site binding pocket can be detected (Fig. 5). Instead of the narrow tunnel leading to the catalytic triad in wild type CALB, the cleavage of the peptide bond between residues Ala282 and Ala283 in helix 17 causes a disruption of the local secondary structures, opening up the tunnel and converting it into a cleft-like feature. The five amino acids of the new N-terminus loose their helicity and instead assume an extended conformation (Fig. 5B). In combination with a ~120° rotation of the peptide bond linking Ala287 and Gly288, the segment undergoes a 45° right-turn and positions itself in a small groove paralleling the active site cleft. At the new carboxy terminus, electron density for the 17 amino acids of helix 16 and portions of helix 17 is missing in both cp283Δ7 structures (Fig. 5, region highlighted in blue). Beyond indicating an increase in flexibility for this segment, the data is inconclusive as to the conformational state of these residues which could range from an orderly helical arrangement to an entirely unstructured tether. Studies are ongoing in our laboratory to resolve the structure and function of this portion of the enzyme. In preliminary experiments, the deletion of the 17-residue C-terminal segment resulted in a substantial drop in catalytic activity (Yu & Lutz, unpublished results). In the absence of declining protein stability, these findings point towards a direct functional contribution of this portion of the enzyme. These observations are also consistent with the idea that the entire region represents a structural element which undergoes a functionally relevant conformational change, maybe even performing a lid-like function as part of the reaction mechanism.7 Additional support for significant flexibility in the helix 16/17 region and its potential correlation with catalytic performance was found in recent computational models of CALB in aqueous environment and organic solvents.41 Finally, we predict that the ternary structure changes observed in cp283Δ7 are not exclusive but apply to all permutants with new termini in this helical region. As such, our crystal structure serves as a good model for structure changes in the active site, rationalizing not just the improved performance of cp283Δ7 but all other permutants and truncated cp283 variants.
From the perspective of catalysis, the crystal structure also provides at least in part a rational for the observed 10 to 100-fold rate enhancements and the preservation of the enzyme’s enantioselectivity. As seen in Figure 5B, the greater accessibility of the active site in cp283Δ7 compared to the wild type CALB likely translates into faster substrate binding and product release. Although we have not conducted pre-steady state measurements, we assume the latter to be the rate-determining step of the catalytic cycle. In contrast, the topology of the active site residues itself appears unperturbed, preserving the catalytic triad and the substrate binding pocket. The latter is a significant factor in controlling the enzyme’s enantioselectivity and its conservation is consistent with the high degree of stereoselectivity previously reported for our variants.27 Separately, the possibility for allosteric regulation of the catalytic activity in the newly created dimer structure of cp283Δ7 was raised. Allostery has been observed for naturally occurring domain-swapped dimers such as RNase. However, our steady state kinetics did not indicate cooperativity between the two active sites as the data clearly fit the Michaelis-Menten model.
Changes in cp283Δ7 quaternary structure
Besides significant changes at the ternary structure level, equally dramatic changes in quaternary structure are observed for truncated cp283 variants (Fig. 3). The crystal structure of cp283Δ7 confirmed our working hypothesis that dimerization takes place via domain swapping. The structure overlay of cp283Δ7 and wild type CALB in Figure 4 strongly suggests that shortening the linker sequence induces conformational constraints which leads to an alternate, energetically favorable orientations of the N-terminal segment. Instead of assuming its normal position in the monomer, considered the closed conformation, the N-terminus adjusts by acquiring an open conformation which allows for its incorporation into the equivalent position on the adjoining monomer. This conformational change is accommodated by rotations of the peptide bonds after Gly34 and Ser37 (Fig. 4B/C) and is thermodynamically driven by an overall reduction in solvent accessible surface area.
Oligomerization by domain swapping has been observed in a number of native, as well as engineered proteins.42 The arrangement can involve the exchange of entire domains or protein termini and has been considered a key step in the evolution of multifunctional enzyme complexes, allosteric regulation, as well as increased protein stability. In the context of our engineered CALBs, results from single-chain antibody fragments (sFv) and staphylococcal nuclease are particularly interesting.43,44 In both cases, truncation in hinge regions led to conformational changes favoring dimerization. Nevertheless, oligomerization by domain swapping has not been observed in native CALB and, more generally, /® hydrolase-fold family members. Higher-order assemblies have been reported for family members, carboxyesterases and polyhydroxyalkanoate synthases, yet those trimer and hexamer structures differ fundamentally from our engineered variant in that their protein-protein interactions involving simple surface contacts without the exchange of structure elements.45,46
Beyond the novelty of domain swapping in an 〈/® hydrolase-fold family member, we are intrigued the formation of the major cleft at the dimer interface. As clearly visible in the top view of the dimer structure (Fig. 3B), the exchange of the N-terminal portions of both subunits creates a groove about 10 Å deep and 11 Å wide. Although this new groove does not show any particular organization of functional groups that would indicate an immediate functional relevance, it could be viewed as a primordial binding site, a template for evolution to create new function. In analogy to our two-step laboratory procedure of circular permutation and incremental truncation, such gene rearrangements could be accomplished in nature via a permutation-by-duplication mechanism as recently demonstrated on DNA methyltransferases.47 Rather than generating a fusion of the precisely duplicated gene sequence, it seems feasible for recombination to result in small insertions and deletions, creating what accounts for the truncated linker region and allowing for optimization of the connector in the fusion protein. Our results further demonstrate that CP can have more serious consequences on protein quaternary structure than the previously reported fold changes.47,48
Finally, our findings might offer one possible answer to the question of the driving force and potential benefits of circular permutation in nature. Besides the observed increases in catalytic activity, our results show that circular permutation can also give rise to new protein topology at the ternary and quaternary structure-level. While cp283Δ7 appears to represent the most stable dimer, a range of deletions in the linker region are tolerated and result in oligomerization of the protein. Furthermore, oligomerization does not seem to interfere with the original lipase activity, consistent with the hypothesis that evolving enzymes maintain their original function, at least in the early stages of gene duplication and functional divergence.
MATERIAL AND METHODS
Materials
Reagents were purchased from Sigma-Aldrich (St. Louis, MO) unless otherwise indicated. Pfu Turbo DNA polymerase from Stratagene (La Jolla, CA) was used for all cloning. Restriction enzymes were acquired from New England Biolabs (Ipswich, MA). Oligonucleotides were ordered from Integrated DNA Technologies (Coralville, IA). Plasmid DNA was isolated using the QiaPrep Spin MiniPrep kit and PCR products were purified with the QiaQuick PCR purification kit (Qiagen, Valencia, CA). All gene constructs were confirmed by DNA sequencing.
Strains and media
E. coli strain DH5〈–E (Invitrogen, Carlsbad, CA) was used in gene manipulations. Bacteria were grown under standard conditions in Luria-Bertani (LB) liquid media or on LB agar plates supplemented with the appropriate antibiotics. Lipases were overexpressed in Pichia pastoris GS115 (his4) (Invitrogen, Carlsbad, CA). P. pastoris was cultured in BMGY medium and transferred to BMMY medium for protein overexpression.15
Incremental truncation
Incremental truncation libraries were generated as reported.51 Briefly, the plasmid was amplified with Taq DNA polymerase in the presence of spiked dNTPs (dNTP :〈S-dNTP = 7 : 1). Following exonuclease III treatment, 5’-overhangs were removed with mung bean nuclease and the truncated DNA was repaired with Klenow-fragment DNA polymerase. The DNA underwent intramolecular ligation and was then subjected to Not and Xho restriction digestion. The reaction mixture was loaded on an agarose gel and DNA fragments, 750 to 1000 nucleotides in size, were recovered by gel excision and DNA extraction. The recovered DNA was ligated into pPIC9 (Invitrogen), and transferred into P. pastoris for expression. When grown on media containing tributyrin, library members with lipase activity were detected based on halo formation.15
Protein expression, purification and kinetics
The overexpression and purification of wild type CALB and the variants was performed as previously described.15,49 Lipase activity was measured by spectrophotometric assay with the reference substrate p-nitrophenol butyrate.15 All experiments were performed in triplicate. Kinetic constants were calculated by fitting the initial rates to the Michaelis-Menten equation using Origin7 (OriginLab, Northhampton, MA).
Circular dichroism and gel filtration
CD spectra were determined using a J-810 spectropolarimeter (Jasco, Easton, NJ). Spectra of proteins (1 mg/ml) in potassium phosphate buffer (50 mM, pH 7) were recorded at 10°C from 260–190 nm (0.5 nm increments) using a 0.1 mm pathlength cell, 20 nm/min scan rate, 4 s response time, and 2 nm bandwidth (5 scans per data set). Thermal denaturation was monitored by following the ellipticity at 222 nm from 10 to 80°C (1°C/min). Gel filtration was performed on a Superdex-200 10/300 GL column (AmershamBiosciences, Piscataway, NJ), using potassium phosphate buffer (50 mM, pH 7.0) containing 150 mM NaCl.
Crystallization
Using the hanging drop method, single crystals appeared with 1.4 M sodium/potassium phosphate (pH 6.9), as well as with 0.1 M Bis-Tris (pH 6.5) and 25% PEG3350. Some crystals were soaked in inhibitor solution overnight. Crystals, acquired with a nylon loop (Hampton Research), were transferred quickly to mother liquor containing either 25% (v/v) glycerol or ethylene glycol before being flash-frozen directly in liquid nitrogen or in a stream of nitrogen gas at 100 K. Using data from a well-diffracting crystal grown in phosphate buffer, structural determination of the protein proceeded by molecular replacement with the program REPLACE50 using protein coordinates of the previously reported wild type CALB structure (PDB: 1LBT).8 The 1LBT model was modified on the basis of evident backbone connectivity in the electron density using the program ‘O’.51 Refinement of the model with and without inhibitor proceeded computationally with the program CNS and manually using ‘O’.52
Coordinates
Coordinates for cp283Δ7 in its apo-form (1.49 Å) and inhibitor-bound form (1.69 Å) were deposited in the Protein Data Bank (accession codes 3ICV and 3ICW, respectively).
Supplementary Material
ACKNOWLEDGEMENTS
This work was supported in part by grants from the US National Science Foundation to SL (CRC-040677 & CBET-0730312). JH and XC were supported by grants from the US National Institutes of Health (GM068680 & GM049245). We thank Dr. Da Jia for his technical assistance with the crystallization experiments. Data were collected at Southeast Regional Collaborative Access Team (SER-CAT) beamline at the Advanced Photon Source, Argonne National Laboratory. Use of the Advanced Photon Source was supported by the U. S. Department of Energy, Office of Science, Office of Basic Energy Sciences, under Contract No. W-31-109-Eng-38. Financial support for the beamline operation of Emory's shares comes from the Department of Biochemistry, Emory University School of Medicine.
Abbreviations
- CALB
Candida antarctica lipase B
- CP
circular permutation
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Berglund P. Controlling lipase enantioselectivity for organic synthesis. Biomol. Eng. 2001;18:13–22. doi: 10.1016/s1389-0344(01)00081-8. [DOI] [PubMed] [Google Scholar]
- 2.Bornscheuer UT, Bessler C, Srinivas R, Krishna SH. Optimizing lipases and related enzymes for efficient application. Trends Biotechnol. 2002;20:433–437. doi: 10.1016/s0167-7799(02)02046-2. [DOI] [PubMed] [Google Scholar]
- 3.Bornscheuer UT, Kazlauskas RJ. Hydrolases in organic synthesis - regio- and stereoselective biotransformations. Weinheim: Wiley-VCH; 1999. [Google Scholar]
- 4.Freemantle M. Biocatalysis in polymer science. Chem. Eng. News. 2004;82:25–27. [Google Scholar]
- 5.Jaeger KE, Eggert T. Lipases for biotechnology. Curr. Opin. Biotechnol. 2002;13:390–397. doi: 10.1016/s0958-1669(02)00341-5. [DOI] [PubMed] [Google Scholar]
- 6.Fjerbaek L, Christensen KV, Norddahl B. A review of the current state of biodiesel production using enzymatic transesterification. Biotechnol. Bioeng. 2009;102:1298–1315. doi: 10.1002/bit.22256. [DOI] [PubMed] [Google Scholar]
- 7.Uppenberg J, Hansen MT, Patkar S, Jones TA. The sequence, crystal structure determination and refinement of two crystal forms of lipase B from Candida antarctica. Structure. 1994;2:293–308. doi: 10.1016/s0969-2126(00)00031-9. [DOI] [PubMed] [Google Scholar]
- 8.Uppenberg J, Ohrner N, Norin M, Hult K, Kleywegt GJ, Patkar S, Waagen V, Anthonsen T, Jones TA. Crystallographic and molecular-modeling studies of lipase B from Candida antarctica reveal a stereospecificity pocket for secondary alcohols. Biochemistry. 1995;34:16838–16851. doi: 10.1021/bi00051a035. [DOI] [PubMed] [Google Scholar]
- 9.Suen WC, Zhang N, Xiao L, Madison V, Zaks A. Improved activity and thermostability of Candida antarctica lipase B by DNA family shuffling. Protein Eng. Des. Sel. 2004;17:133–140. doi: 10.1093/protein/gzh017. [DOI] [PubMed] [Google Scholar]
- 10.Zhang N, Suen WC, Windsor W, Xiao L, Madison V, Zaks A. Improving tolerance of Candida antarctica lipase B towards irreversible thermal inactivation through directed evolution. Protein Eng. 2003;16:599–605. doi: 10.1093/protein/gzg074. [DOI] [PubMed] [Google Scholar]
- 11.Rotticci D, Rotticci-Mulder JC, Denman S, Norin T, Hult K. Improved enantioselectivity of a lipase by rational protein engineering. ChemBioChem. 2001;2:766–770. doi: 10.1002/1439-7633(20011001)2:10<766::AID-CBIC766>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- 12.Magnusson AO, Rotticci-Mulder JC, Santagostino A, Hult K. Creating space for large secondary alcohols by rational redesign of Candida antarctica lipase B. ChemBioChem. 2005;6:1051–1056. doi: 10.1002/cbic.200400410. [DOI] [PubMed] [Google Scholar]
- 13.Skjot M, De Maria L, Chatterjee R, Svendsen A, Patkar SA, Ostergaard PR, Brask J. Understanding the plasticity of the alpha/beta hydrolase fold: lid swapping on the Candida antarctica lipase B results in chimeras with interesting biocatalytic properties. ChemBioChem. 2009;10:520–527. doi: 10.1002/cbic.200800668. [DOI] [PubMed] [Google Scholar]
- 14.Kim SY, Sohn JH, Pyun YR, Yang IS, Kim KH, Choi ES. In vitro evolution of lipase B from Candida antarctica using surface display in hansenula polymorpha. J. Microbiol. Biotechnol. 2007;17:1308–1315. [PubMed] [Google Scholar]
- 15.Qian Z, Lutz S. Improving the catalytic activity of Candida antarctica lipase B by circular permutation. J. Am. Chem. Soc. 2005;127:13466–13467. doi: 10.1021/ja053932h. [DOI] [PubMed] [Google Scholar]
- 16.Hisano T, Kasuya KI, Tezuka Y, Ishii N, Kobayashi T, Shiraki M, Oroudjev E, Hansma H, Iwata T, Doi Y, Saito T, Miki K. The crystal structure of polyhydroxybutyrate depolymerase from Penicillum funiculosum provides insights into the recognition and degradation of biopolyesters. J. Mol. Biol. 2006;356:993–1004. doi: 10.1016/j.jmb.2005.12.028. [DOI] [PubMed] [Google Scholar]
- 17.Weiner J, 3rd, Bornberg-Bauer E. Evolution of circular permutations in multidomain proteins. Mol. Biol. Evol. 2006;23:734–743. doi: 10.1093/molbev/msj091. [DOI] [PubMed] [Google Scholar]
- 18.Jeltsch A. Circular permutations in the molecular evolution of DNA methyltransferases. J. Mol. Evol. 1999;49:161–164. doi: 10.1007/pl00006529. [DOI] [PubMed] [Google Scholar]
- 19.Zhang T, Bertelsen E, Benvegnu D, Alber T. Circular permutation of T4 lysozyme. Biochemistry. 1993;32:12311–12318. doi: 10.1021/bi00097a006. [DOI] [PubMed] [Google Scholar]
- 20.Hennecke J, Sebbel P, Glockshuber R. Random circular permutation of DsbA reveals segments that are essential for protein folding and stability. J. Mol. Biol. 1999;286:1197–1215. doi: 10.1006/jmbi.1998.2531. [DOI] [PubMed] [Google Scholar]
- 21.Viguera AR, Blanco FJ, Serrano L. The order of secondary structure elements does not determine the structure of a protein but does affect its folding kinetics. J. Mol. Biol. 1995;247:670–681. doi: 10.1006/jmbi.1994.0171. [DOI] [PubMed] [Google Scholar]
- 22.Zhang P, Schachman HK. In vivo formation of allosteric aspartate transcarbamoylase containing circularly permuted catalytic polypeptide chains: implications for protein folding and assembly. Protein Sci. 1996;5:1290–1300. doi: 10.1002/pro.5560050708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Iwakura M, Nakamura T, Yamane C, Maki K. Systematic circular permutation of an entire protein reveals essential folding elements. Nat. Struct. Biol. 2000;7:580–585. doi: 10.1038/76811. [DOI] [PubMed] [Google Scholar]
- 24.Lindberg M, Tangrot J, Oliveberg M. Complete change of the protein folding transition state upon circular permutation. Nat. Struct. Biol. 2002;9:818–822. doi: 10.1038/nsb847. [DOI] [PubMed] [Google Scholar]
- 25.Baird GS, Zacharias DA, Tsien RY. Circular permutation and receptor insertion within green fluorescent proteins. Proc. Natl. Acad. Sci. USA. 1999;96:11241–11246. doi: 10.1073/pnas.96.20.11241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ostermeier M. Engineering allosteric protein switches by domain insertion. Protein Eng. Des. Sel. 2005;18:359–364. doi: 10.1093/protein/gzi048. [DOI] [PubMed] [Google Scholar]
- 27.Qian Z, Fields CJ, Lutz S. Investigating the structural and functional consequences of circular permutation on lipase B from Candida antarctica. ChemBioChem. 2007;8:1989–1996. doi: 10.1002/cbic.200700373. [DOI] [PubMed] [Google Scholar]
- 28.Ay J, Hahn M, Decanniere K, Piotukh K, Borriss R, Heinemann U. Crystal structures and properties of de novo circularly permuted 1,3-1,4-beta-glucanases. Proteins. 1998;30:155–167. doi: 10.1002/(sici)1097-0134(19980201)30:2<155::aid-prot5>3.0.co;2-m. [DOI] [PubMed] [Google Scholar]
- 29.Chu V, Freitag S, Le Trong I, Stenkamp RE, Stayton PS. Thermodynamic and structural consequences of flexible loop deletion by circular permutation in the streptavidin-biotin system. Protein Sci. 1998;7:848–859. doi: 10.1002/pro.5560070403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Manjasetty BA, Hennecke J, Glockshuber R, Heinemann U. Structure of circularly permuted DsbA(Q100T99): preserved global fold and local structural adjustments. Acta Crystallogr. D Biol. Crystallogr. 2004;60:304–309. doi: 10.1107/S0907444903028695. [DOI] [PubMed] [Google Scholar]
- 31.Tougard P, Bizebard T, Ritco-Vonsovici M, Minard P, Desmadril M. Structure of a circularly permuted phosphoglycerate kinase. Acta Crystallogr. D Biol. Crystallogr. 2002;58:2018–2023. doi: 10.1107/s0907444902015548. [DOI] [PubMed] [Google Scholar]
- 32.Pieper U, Hayakawa K, Li Z, Herzberg O. Circularly permuted beta-lactamase from Staphylococcus aureus PC1. Biochemistry. 1997;36:8767–8774. doi: 10.1021/bi9705117. [DOI] [PubMed] [Google Scholar]
- 33.Nagi AD, Regan L. An inverse correlation between loop length and stability in a four-helix-bundle protein. Fold Des. 1997;2:67–75. doi: 10.1016/S1359-0278(97)00007-2. [DOI] [PubMed] [Google Scholar]
- 34.Wang L, Rivera EV, Benavides-Garcia MG, Nall BT. Loop entropy and cytochrome c stability. J. Mol. Biol. 2005;353:719–729. doi: 10.1016/j.jmb.2005.08.038. [DOI] [PubMed] [Google Scholar]
- 35.Lutz S, Ostermeier M, Benkovic SJ. Rapid generation of incremental truncation libraries for protein engineering using alpha-phosphothioate nucleotides. Nucleic Acids Res. 2001;29:E16. doi: 10.1093/nar/29.4.e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lutz S, Ostermeier M. Preparation of SCRATCHY hybrid protein libraries: size-and in-frame selection of nucleic acid sequences. Methods Mol. Biol. 2003;231:143–151. doi: 10.1385/1-59259-395-X:143. [DOI] [PubMed] [Google Scholar]
- 37.Krissinel E, Henrick K. Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 2007;372:774–797. doi: 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
- 38.Fersht AR. The hydrogen-bond in molecular recognition. Trends Biochem. Sci. 1987;12:301–304. [Google Scholar]
- 39.Janin J, Miller S, Chothia C. Surface, subunit interfaces and interior of oligomeric proteins. J. Mol. Biol. 1988;204:155–164. doi: 10.1016/0022-2836(88)90606-7. [DOI] [PubMed] [Google Scholar]
- 40.Lutz S. Engineering lipase B from Candida antarctica. Tetrahedron: Asymmetry. 2004;15:2743–2748. [Google Scholar]
- 41.Trodler P, Pleiss J. Modeling structure and flexibility of Candida antarctica lipase B in organic solvents. BMC Struct. Biol. 2008;8:9. doi: 10.1186/1472-6807-8-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schlunegger MP, Bennett MJ, Eisenberg D. Oligomer formation by 3D domain swapping: a model for protein assembly and misassembly. Adv. Protein Chem. 1997;50:61–122. doi: 10.1016/s0065-3233(08)60319-8. [DOI] [PubMed] [Google Scholar]
- 43.Raag R, Whitlow M. Single-chain Fvs. FASEB J. 1995;9:73–80. doi: 10.1096/fasebj.9.1.7821762. [DOI] [PubMed] [Google Scholar]
- 44.Green SM, Gittis AG, Meeker AK, Lattman EE. One-step evolution of a dimer from a monomeric protein. Nat. Struct. Biol. 1995;2:746–751. doi: 10.1038/nsb0995-746. [DOI] [PubMed] [Google Scholar]
- 45.Bencharit S, Morton CL, Xue Y, Potter PM, Redinbo MR. Structural basis of heroin and cocaine metabolism by a promiscuous human drug-processing enzyme. Nat. Struct. Biol. 2003;10:349–356. doi: 10.1038/nsb919. [DOI] [PubMed] [Google Scholar]
- 46.Stubbe J, Tian J, He A, Sinskey AJ, Lawrence AG, Liu P. Nontemplate-dependent polymerization processes: polyhydroxyalkanoate synthases as a paradigm. Annu. Rev. Biochem. 2005;74:433–480. doi: 10.1146/annurev.biochem.74.082803.133013. [DOI] [PubMed] [Google Scholar]
- 47.Peisajovich SG, Rockah L, Tawfik DS. Evolution of new protein topologies through multistep gene rearrangements. Nat. Genet. 2006;38:168–174. doi: 10.1038/ng1717. [DOI] [PubMed] [Google Scholar]
- 48.Grishin NV. Fold change in evolution of protein structures. J. Struct. Biol. 2001;134:167–185. doi: 10.1006/jsbi.2001.4335. [DOI] [PubMed] [Google Scholar]
- 49.Ottosson J, Rotticci-Mulder JC, Rotticci D, Hult K. Rational design of enantio selective enzymes requires considerations of entropy. Protein Sci. 2001;10:1769–1774. doi: 10.1110/ps.13501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tong L, Rossmann MG. Rotation function calculations with GLRF program. Methods Enzymol. 1997;276:594–611. [PubMed] [Google Scholar]
- 51.Jones TA, Zou JY, Cowan SW, Kjeldgaard M. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A. 1991;47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 52.Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, Read RJ, Rice LM, Simonson T, Warren GL. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr. D Biol. Crystallogr. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.