Abstract
Acute toxicity of organophosphorus poisons (OP) is explained by inhibition of acetylcholinesterase in nerve synapses. Low dose effects are hypothesized to result from modification of other proteins, whose identity is not yet established. The goal of the present work was to obtain information that would make it possible to identify tubulin as a target of OP exposure. Tubulin was selected for study because live mice injected with a nontoxic dose of a biotinylated organophosphorus agent appeared to have OP-labeled tubulin in brain as determined by binding to avidin beads and mass spectrometry. The experiments with live mice were not conclusive because binding to avidin beads could be nonspecific. To be convincing, it is necessary to find and characterize the OP-labeled tubulin peptide. The search for OP-labeled tubulin peptides was begun by identifying residues capable of making a covalent bond with OP. Pure bovine tubulin (0.012 mM) was treated with 0.01–0.5 mM chlorpyrifos oxon for 24 h at 37 °C in pH 8.3 buffer. The identity of labeled amino acids and percent labeling was determined by mass spectrometry. Chlorpyrifos oxon bound covalently to tyrosines 83, 103, 108, 161, 224, 262, 272, 357, and 399 in bovine alpha tubulin, and to tyrosines 50, 51, 59, 106, 159, 281, 310, and 340 in bovine beta tubulin. The most reactive were tyrosine 83 in alpha and tyrosine 281 in beta tubulin. In the presence of 1 mM GTP, percent labeling increased 2-fold. Based on the crystal structure of the tubulin heterodimer (PDB 1jff) tyrosines 83 and 281 are well exposed to solvent. In conclusion seventeen tyrosines in tubulin have the potential to covalently bind chlorpyrifos oxon. These results will be useful when searching for OP-labeled tubulin in live animals.
Keywords: mass spectrometry, tubulin, chlorpyrifos oxon, tyrosine
1. Introduction
Although some organophosphorus compounds (OP) have been banned or severely restricted (www.epa.gov) they still comprise more than 50% of all pesticides used worldwide, causing much of the human population to be exposed. An especially high percentage of exposure is reported for farmers and workers dealing with pesticides in El Salvador, Peru and Southern Spain (Azaroff, 1999; Roldan-Tapia et al., 2006).
The acute toxicity of OP is due to inhibition of acetylcholinesterase (EC 3.1.1.7, AChE). Inhibition of greater than 50% of an individual’s AChE produces an acute “cholinergic syndrome” initiated by accumulation of the neurotransmitter acetylcholine at cholinergic synapses (McDonough and Shih, 1997; Brown and Brix, 1998; Pope, 1999). Respiratory failure due to inhibition of AChE can cause death (Brown and Brix, 1998; Eddleston et al., 2006). In addition to the acute toxicity that occurs with high doses of OP, low-dose exposure has been implicated in insomnia, fatigue, inability to concentrate, memory deficits, depression, and generalized weakness (Stephens et al., 1995; Salvi et al., 2003; Roldan-Tapia et al., 2006; Kamel et al., 2007). Low-dose exposure is defined as a dosage that causes minimal inhibition of AChE and no obvious cholinergic symptoms.
The idea that AChE is the only physiologically important target for OP exposure was undisputed for many decades. However, toxicologists demonstrated that signs of toxicity in animals depended on the identity of the OP (Moser, 1995; Pope, 1999). For example, a low dose of fenthion decreased motor activity in rats by 86% but did not alter the tail-pinch response, whereas a low dose of parathion did not lower motor activity but did decrease the tail-pinch response. This led to the conclusion that OP have other biological actions in addition to their cholinesterase-inhibitory properties.
A variety of enzymes have been shown to react with OP, not all of which contain the serine hydrolase active site that is a hallmark of OP targets (Casida and Quistad, 2004). These findings re-enforce the proposal that non-cholinesterase targets may contribute to the toxicity of OP.
Possible non-cholinesterase targets
Cytoskeletal proteins including tubulin have been implicated in the neurotoxicity of OP (Abou-Donia, 2003). Rats treated with subthreshold doses of chlorpyrifos oxon had impaired axonal transport in the nervous system, indicative of structural damage to the kinesin motor protein whose task is to transport cell components along microtubules (Gearhart et al., 2007; Terry et al., 2007). Studies on organotypic slice cultures of rat hippocampus suggested that chlorpyrifos oxon produces a progressive decrease in neuronal viability that may be associated with impaired microtubule synthesis and/or function (Prendergast et al., 2007). Furthermore, the polymerization of pure bovine brain tubulin was inhibited by low doses of chlorpyrifos oxon (0.1–10 µM) (Prendergast et al., 2007). These studies suggest that the function of cytoskeletal proteins, including tubulin, may be adversely affected by OP.
Tubulin was selected for study because tubulin appeared to be labeled by the organophosphorus ester, FP-biotin, when mice were treated with a low dose that did not inhibit acetylcholinesterase. The evidence for labeling of tubulin was indirect. In efforts to provide direct evidence of labeling in vivo, we need to know the identity of the potentially modified residues. For this purpose we undertook the mass spectrometry study of purified bovine tubulin presented here.
Materials and methods
Materials
Bovine tubulin (TL238) > 99% pure, isolated from bovine brain, was from Cytoskeleton, Inc (Denver, CO). This tubulin preparation contains both alpha and beta-tubulin (MW is approximately 50 kDa for each). Chlorpyrifos oxon (MET-674B from ChemService Inc, West Chester, PA) was dissolved in dimethyl sulfoxide and stored at −80°C. 10-Fluoroethoxyphosphinyl-N-biotinamidopentyldecanamide (FP-biotin) was custom synthesized in the laboratory of Dr. Charles M. Thompson at the University of Montana, Missoula, MT (Schopfer et al., 2005a). Sequencing grade modified porcine trypsin (V5113) was from Promega (Madison, WI). Slide-A-Lyzer 7K dialysis cassettes (No. 66370) were from Pierce Biotechnology Inc. (Rockford, IL), PVDF (polyvinylidene difluoride) membrane (Immun-Blot #162–0177), Coomassie blue G250 (Bio-Safe), and broad range biotinylated SDS PAGE molecular weight markers (#161–0319) were from BioRad Laboratories (Hercules, CA). Performa Spin Columns (#73328) with 0.8 ml capacity, packed with a gel-filtration matrix were from Edge BioSystems (Gaithersburg, MD). Avidin-Agarose (A-9207) was from Sigma (St. Louis, MO), Streptavidin Alexa-680 (S-21378) was from Molecular Probes (Eugene, OR). All other chemicals were of analytical grade.
Detection of FP-biotinylated proteins from mouse brain supernatant
In vitro studies were performed as follows. Mouse brain was homogenized in 10 volumes of 50 mM potassium phosphate buffer pH 7.0 and centrifuged at 100,000×g. The clear supernatant (1.4 mg protein/ml) was reacted with 10 µM FP-biotin at 25 °C for 5 hours in 50 mM TrisCl buffer, pH 8.0, containing 5 mM EDTA. The reaction was stopped by passage over a G-25 gel filtration column to separate the excess FP-biotin from the protein, followed by boiling in 0.5% SDS for 3 minutes. The FP-biotinylated proteins were enriched by binding to avidin beads, and analyzed by mass spectrometry as described (Peeples et al., 2005).
Mice treated with FP-biotin
Animal work was carried out in accordance with the Guide for the Care and Use of Laboratory Animals as adopted by the National Institutes of Health. Two mice in strain 129Sv were injected intraperitoneally with 5.5 mg/kg FP-biotin (9.3 µM) dissolved in ethanol. Mice were euthanized 120 min later by inhalation of carbon dioxide. Blood was washed out by intracardial perfusion with 0.1M phosphate buffered saline. Brains were homogenized in 10 volumes of 50 mM TrisCl pH 8.0 containing 5 mM EDTA, and centrifuged for 10 min to partially clarify the suspension. FP-biotinylated proteins were isolated by adsorption onto Avidin-Agarose beads (Peeples et al., 2005). The boiled beads were loaded on an SDS PAGE gel where proteins were visualized with Coomassie blue. Coomassie stained bands were excised, digested with trypsin, and the peptides identified by tandem quadrupole LC/MS/MS mass spectrometry. Proteins from a separate SDS PAGE gel were transferred to a PVDF membrane and hybridized with the fluorescent probe, Streptavidin Alexa-680.
Chlorpyrifos oxon-labeled tryptic peptides from bovine tubulin
Bovine tubulin (0.6 mg/ml or 12 µM in terms of tubulin monomer) dissolved in 15 mM ammonium bicarbonate (pH 8.3) was treated with a 40-fold molar excess (0.5 mM) of chlorpyrifos oxon. The reaction mixture was incubated at 37 °C for 24 hours. The protein was denatured by incubating in a boiling water bath for 10 min. Excess chlorpyrifos oxon was removed by dialysis against 10 mM ammonium bicarbonate at 4°C for 12 h. Sixty µg of dialyzed tubulin were digested with 1.5 µg of Promega trypsin at 37 °C for 16 hours.
Tubulin labeling by different concentrations of chlorpyrifos oxon
A 0.05 ml aliquot of 0.6 mg/ml bovine tubulin, in 15 mM ammonium bicarbonate pH 8.3, was treated with 0.01; 0.05; 0.1; 0.25 and 0.5 mM chlorpyrifos oxon. The reaction mixtures were incubated at 37°C for 24 hours. Tubulin was denatured in a boiling water bath for 10 min. Excess chlorpyrifos oxon was removed by dialysis against 10 mM ammonium bicarbonate at 4°C for 12 h. Thirty µg of dialyzed tubulin were digested with 0.75 µg of Promega trypsin at 37°C overnight.
Time course for labeling of bovine tubulin by chlorpyrifos oxon
Bovine tubulin, 0.3 mg dissolved in 0.5 ml of 15 mM ammonium bicarbonate pH 8.3, was treated with a 40-fold molar excess (0.5 mM) of chlorpyrifos oxon at 37 °C for 1, 3, 6.5, 10, and 24 h. At each time point 0.1 ml of reaction mixture was removed for analysis of chlorpyrifos oxon-labeled peptides. Samples were immediately centrifuged (Sorvall MC 12V benchtop microcentrifuge) through a Performa Spin Column to remove excess chlorpyrifos oxon (2 minutes at 3000 rpm) and the pass-thru was frozen at −70 °C. After all samples were collected, each was digested with 1.5 µg of Promega trypsin at 37 °C for 16 hours.
GTP effect on OP binding to tubulin
One tenth ml of 0.6 mg/ml tubulin, dissolved in 15 mM ammonium bicarbonate (pH 8.3) containing 1 mM GTP, was treated with a 40-fold molar excess of chlorpyrifos oxon or 8-fold molar excess of FP-biotin dissolved in dimethylsulfoxide. The reaction mixtures were incubated at 37 °C for 24 hours. The tubulin was denatured by boiling in a water bath for 10 min. Excess OP was removed by dialysis against 10 mM ammonium bicarbonate at 4 °C overnight. The 60 µg of dialyzed tubulin were digested with 1.5 µg of Promega trypsin at 37°C overnight.
MALDI-TOF/TOF mass spectrometry
The MALDI-TOF/TOF 4800 mass spectrometer (Applied Biosystems, Foster City, CA) was used for analysis of tryptic peptides from the tubulin heterodimer. One µl of a tryptic digest was spotted on a 384 well Opti-TOF plate (P/N 1016491, Applied Biosystems) and air dried. The spot was overlaid with 1 µl of 10 mg/ml alpha-cyano-4-hydroxycinnamic acid matrix dissolved in 50% acetonitrile, 0.1% trifluoroacetic acid and allowed to air dry. Mass spectra were collected in positive ion reflector mode. The final spectrum was the average of 500 laser shots. Masses were calibrated using CalMix 5 (Applied Biosystems).
Percent labeled peptide estimated from MALDI-TOF peak areas
MALDI-TOF spectra are saved as DATA EXPLORER files. The DATA EXPLORER file has an output window that lists the area for each peak in an MS spectrum. Percentage of chlorpyrifos oxon labeling was calculated from the isotope cluster areas of peaks corresponding to unlabeled and labeled peptides. Cluster areas for both labeled and unlabeled masses were taken from the same MALDI spectrum where the unlabeled peptide served as the internal control. The percentage labeling was calculated by dividing the cluster area for the labeled mass by the sum of the cluster areas for the labeled and the unlabeled masses. This method assumes that addition of the organophosphate does not significantly alter the intrinsic properties of the peptide (i.e. charge state, peptide size, peptide hydrophobicity, or secondary structure) which are considered to be responsible for the signal intensity in MALDI TOF mass spectrometry (Pashkova et al., 2004). This ratio method has proven to be valid in kinetic studies of modified peptides and in the quantitative determination of other peptide modifications (Tang et al., 1996; Jennings et al., 2003; Li et al., 2007b; Sun and Lynn, 2007; Lockridge et al., 2008).
LC/MS/MS mass spectrometry
Tryptic peptides of chlorpyrifos oxon treated tubulin were dried in a vacuum centrifuge (SpeedVac from Jouan, RC10-10) and dissolved in 5% acetonitrile, 0.1% formic acid. A 10 µl aliquot was subjected to HPLC using a nanocolumn (#218MS3.07515 Vydac C18 polymeric revphase, 75 µm I.D. ×150 mm long; P.J. Cobert Assoc, St. Louis, MO). Peptides were separated with a 90 min linear gradient from 0 % to 60 % acetonitrile at a flow rate of 0.3 µl/min and electrosprayed through a fused silica emitter (360 µm O.D., 75 µm I.D., 15 µm taper, New Objective) directly into the QTRAP 2000 (Applied Biosystems, Foster City, CA), a hybrid quadrupole linear ion trap mass spectrometer. An ion-spray voltage of 1900 volts was maintained between the emitter and the mass spectrometer. Information-dependent acquisition was used to collect MS, high resolution MS and MS/MS spectra for the 3 most intense peaks in each cycle, having a charge of +1 to + 4, a mass between 200 and 1700 m/z, and an intensity >10,000 cps. All spectra were collected in the enhanced mode, using the trap function. Precursor ions were excluded for 30 seconds after one MS/MS spectrum had been collected. The collision cell was pressurized to 40 µTorr with pure nitrogen and collision energies between 20 and 40 eV were determined automatically by the software based on the mass and charge of the precursor ion. The mass spectrometer was calibrated on selected fragments from the MS/MS spectrum of Glu-Fibrinopeptide B. The MS/MS data were processed using Analyst 1.4.1 software.
Initial identification of peptide sequences was obtained using the Mascot search engine licensed to the University Nebraska Medical Center (http://www.matrixscience.com (Perkins et al., 1999). Mascot used the NCBI database version Dec 29, 2005. To make it possible to search for chlorpyrifos oxon labeled and FP-biotin labeled Ser, Thr, Tyr, and Lys we introduced these modifications into the UNIMOD database according to the instructions on the web site http://www.unimod.org. Access to the modifications is freely available to all Mascot users in the Variable Modifications menu under the names O-diethylphosphate (for chlorpyrifos oxon), and FP-biotin. The search parameters included a variable modification for oxidized methionine, a fixed modification for carbamidomethylated cysteine, and charge states +1, +2, and +3. One missed cleavage was allowed for digestion by trypsin. The peptide tolerance was ±1.2 Da. The MS/MS tolerance was ±0.6 Da. The sequences of labeled peptides that were identified by Mascot were further checked manually with the aid of MS/MS Fragment Ion Calculator from Systems Biology (http://db.systemsbiology.net).
Results
Preliminary detection of FP-biotinylated tubulin
Preliminary screening for OP-targets other than AChE was conducted with the biotinylated organophosphorus agent FP-biotin, whose structure is shown in Figure 1. FP-biotin was employed because the biotin moiety provided a convenient way for enriching and extracting OP-reactive proteins from crude mixtures (Kidd et al., 2001; Nomura et al., 2005; Peeples et al., 2005; Schopfer et al., 2005a; Schopfer et al., 2005b; Ding et al., 2008; Grigoryan et al., 2008; Nomura et al., 2008; Tuin et al., 2009).
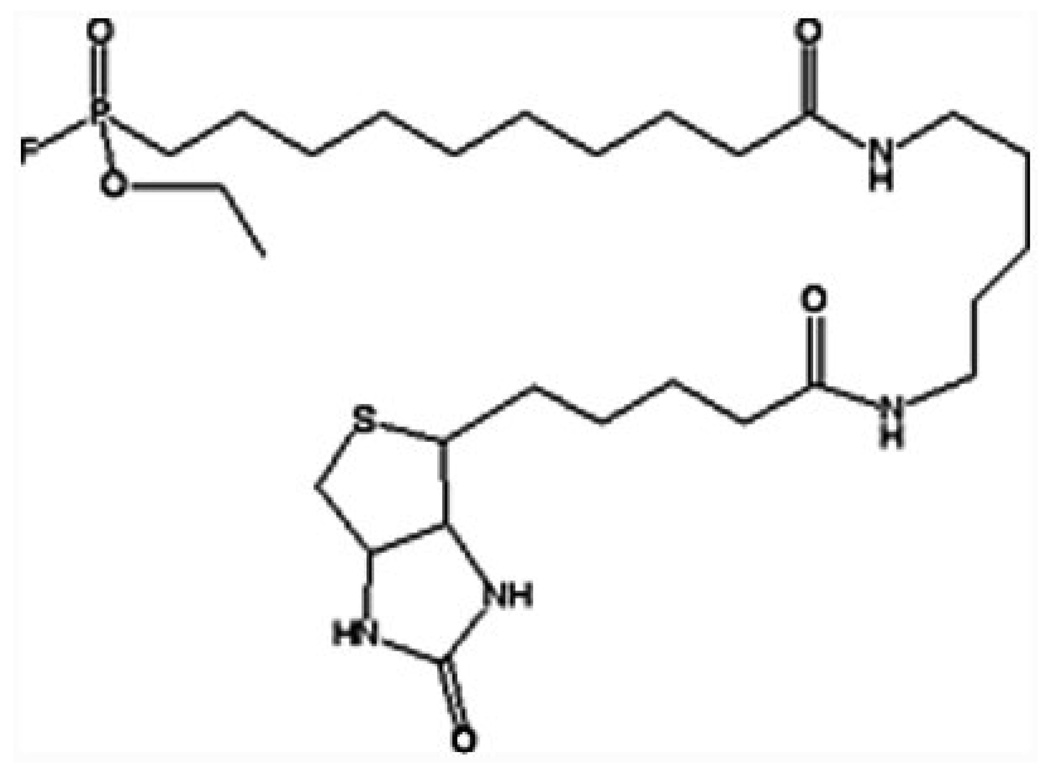
Fig. 1.
Structure of FP-biotin. The second order rate constant for the reaction of FP-biotin with human butyrylcholinesterase is 1.6 × 108 M−1 min−1 and for the reaction with human acetylcholinesterase is 1.8 × 107 M−1 min−1. These rates are comparable to the rates of reaction with chlorpyrifos oxon which are 1.7×109 M−1 min−1 for human butyrylcholinesterase and 1.0 × 107 M−1 min−1 for human acetylcholinesterase (Schopfer et al., 2005b).
In vitro labeling of mouse brain supernatant (100,000 × g) with 10 µM FP-biotin, was followed by binding to avidin beads, SDS gel electrophoresis, transfer of proteins to PVDF membrane, and staining with Streptavidin Alexa-680. This sensitive, fluorescent dye revealed as many as 55 separate bands from a single SDS PAGE gel lane for FP-biotinylated mouse brain supernatant (Figure 2, lane B). In the absence of FP-biotin, no more than four bands were normally detected.
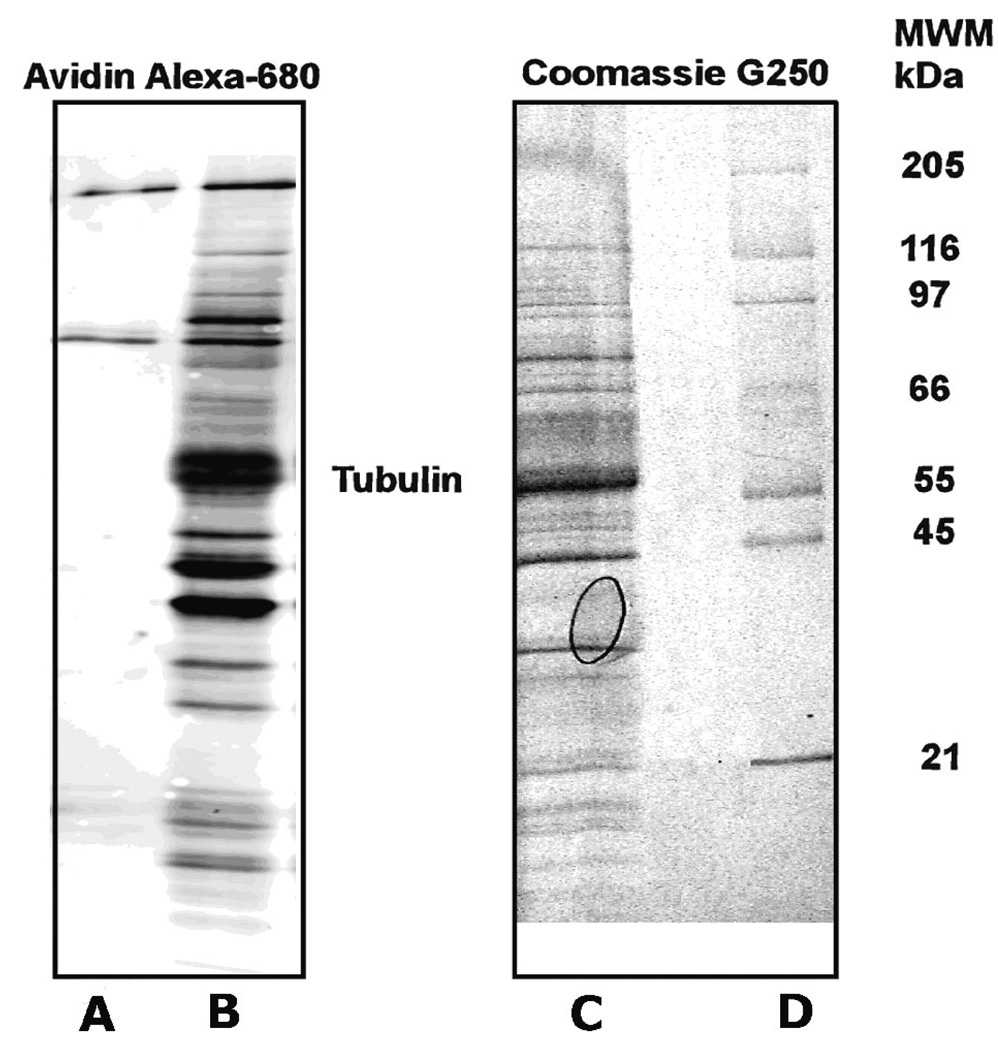
Fig. 2.
Preliminary detection of FP-biotinylated tubulin in mouse brain. Mouse brain supernatant was treated with 10 µM FP-biotin. Excess FP-biotin was separated by gel filtration. FP-biotinylated proteins were extracted by binding to avidin-agarose beads. The avidin-agarose beads were washed with 0.2% SDS, boiled in SDS gel loading buffer, and the extract and beads loaded on an SDS PAGE gradient gel (10–20%). Protein in Lanes A and B were transferred to PVDF membrane and stained with streptavidin Alexa-680. Lane A shows endogenously biotinylated proteins that appear in the absence of FP-biotinylation. Lane B shows 55 FP-biotinylated bands including an intense band for FP-biotinylated tubulin. Lanes C and D were stained with Coomassie blue. Lane C is the same sample as in lane B at a 20-fold higher concentration. Lane D contains molecular weight marker proteins. The molecular weight values for the marker proteins are shown to the right of the panel (MWM/kDa). The oval in lane C is an air bubble.
Coomassie staining is less sensitive than staining with Streptavidin Alexa-680; twenty-two bands were detected in the Coomassie-stained lane shown in Figure 2C. Proteins identified by mass spectrometry from tryptic, in-gel digests of these bands included serine hydrolases, dehydrogenases, a phosphatase, a monooxygenase, peroxiredoxin, heat shock proteins, albumin, and tubulin. See Table 1. In addition, two endogenously biotinylated enzymes (pyruvate carboxylase and propionyl CoA-carboxylase) were identified.
Table 1.
Proteins in mouse brain homogenates that bind FP-biotin.
| Protein name | Accession # | MOWSE | kDa | GXSXG |
|---|---|---|---|---|
| Fatty acid synthase | gi: 28461372 | 49 | 270 | yes |
| Puromycin-sensitive aminopeptidase | gi 6679491 | 174 | 103 | no |
| Formyltetrahydrofolate dehydrogenase | gi 27532959 | 426 | 97 | no |
| Heat shock protein 90-Beta | gi 123681 | 352 | 84 | no |
| Prolyl endopeptidase | gi 6755152 | 827 | 81 | yes |
| acylpeptide hydrolase | gi 22122789 | 254 | 80 | yes |
| RIKEN D030028o16 | gi 22122431 | 372 | 73 | no |
| Serum albumin precursor | gi 5915682 | 270 | 67 | no |
| RIKEN 4931406N15 | gi 28076969 | 173 | 70 | no |
| Heat shock protein 8 | gi 13242237 | 154 | 71 | yes |
| Acetylcholinesterase | gi: 28279461 | 64 | 68 | yes |
| Protein phosphatase 2 | gi 8394027 | 70 | 65 | no |
| Fatty acid amide hydrolase | gi: 30354073 | 72 | 64 | yes |
| Dihydropyrimidinase-like 2 | gi 6753676 | 62 | 61 | no |
| Dihydropyrimidinase-like 3 | gi 6681219 | 59 | 62 | no |
| Dihydropyrimidinase-like 5 | gi 12746424 | 43 | 61 | yes |
| Esterase 1 | gi: 6679689 | 91 | 61 | yes |
| D-3-phosphoglycerate dehydrogenase | gi 3122875 | 195 | 51 | no |
| Tubulin | gi 20455323 | 5320 | 50 | no |
| Tubulin alpha-1 | gi 6755901 | 576 | 50 | no |
| Tubulin alpha 6 | gi: 74186501 | 327 | 50 | no |
| Tubulin beta | gi: 21746161 | 453 | 50 | no |
| Tubulin beta 2 | gi: 13542680 | 106 | 50 | no |
| Tubulin beta 5 | gi 7106439 | 423 | 50 | no |
| Acyl-CoA hydrolase | gi 19923052 | 296 | 38 | no |
| Glyceraldehyde-3-phosphate dehydrogenase | gi 6679937 | 275 | 36 | no |
| Lactate dehydrogenase 1 | gi 6754524 | 337 | 36 | no |
| Esterase 10 | gi 12846304 | 261 | 35 | yes |
| Platelet-activating factor acetylhydrolase alpha-2 | gi 6679199 | 65 | 25 | no |
| Tyrosine 3-monooxygenase/tryptophan 5- monooxygenase activation protein |
gi 6756039 | 354 | 28 | no |
| Platelet-activating factor acetylhydrolase alpha-1 | gi 6679201 | 273 | 26 | no |
| Platelet activating factor alpha1 | gi: 44890813 | 110 | 26 | no |
| Platelet activating factor 1b2 | gi: 1373363 | 158 | 25 | no |
| Lysophospholipase 1 | gi 6678760 | 206 | 25 | yes |
| Lysophospholipase 2 | gi 7242156 | 160 | 25 | yes |
| Peroxiredoxin 1 | gi 6754976 | 245 | 22 | no |
The accession number is unique to each protein in the NCBI database in PubMed. The accession number typed into the Protein subsection of PubMed or into Google brings up the protein sequence, the species,and references. The MOWSE score is the statistical probability that the identification is correct. A score greater than 34 is significant. A score greater than 69 is significant with p<0.05. kDa indicates the protein molecular weight in kilodaltons. GXSXG is the consensus sequence for the active site serine.Yes indicates proteins that have this motif.
An intensely stained band at 55 kDa from the SDS PAGE gel contained tubulin as the major component. Both the alpha and beta subunits of tubulin were found through probability-based searching of the NCBI database using the Mascot algorithm (Perkins et al., 1999). The MOWSE scores showed high confidence in the identification. The band at 55 kDa did not appear in brain preparations that had not been treated with FP-biotin (Figure 2A).
Of the 36 proteins in Table 1, only 11 have an active site serine as indicated by Yes in the column headed by GXSXG. Proteins that have an active site serine are expected to covalently bind organophosphorus agents on serine. However, the 25 proteins without the consensus sequence GXSXG bind organophosphorus agent at unknown sites.
Mice treated with FP-biotin
Similar analyses were conducted on brain supernatant from mice that had been injected intraperitoneally with 5.5 mg/kg FP-biotin. These animals showed no signs of cholinergic toxicity and no AChE inhibition (Peeples et al., 2005). Proteins that bound to avidin beads were separated by SDS gel electrophoresis, digested with trypsin, and analyzed by mass spectrometry. A gel band at about 55 kDa contained tubulin as the primary component. Alpha tubulin gave a MOWSE score of 322, which was based on 9 peptides, 6 of which scored in the identity range and 3 in the homology range. Beta tubulin gave a MOWSE score of 217, which was based on 4 peptides, 3 of which scored in the identity range and 1 in the homology range.
Mascot clearly identified tubulin as the major component in the avidin-bound material extracted from the brain of a mouse that had been treated with FP-biotin. However, the FP-biotin-labeled peptides for neither alpha nor beta tubulin were found. Identification of the labeled peptide is essential to confirm that tubulin truly reacts with OP. This question is particularly important in the case of tubulin, because this protein does not carry the serine hydrolase active site, which is the traditionally accepted target for OP.
Chlorpyrifos oxon labeled tryptic peptides from bovine tubulin identified by mass spectrometry
In an effort to find an OP-labeled peptide from tubulin, the reaction of pure bovine tubulin with a different OP was studied. The OP chosen for these more focused studies was chlorpyrifos oxon. Chlorpyrifos oxon was chosen because it is the active metabolite of chlorpyrifos, a commercially used pesticide that is more relevant than FP-biotin in terms of human exposure. The chlorpyrifos oxon studies complement those from the more exotic FP-biotin and address the question of whether the FP-biotin results are relevant to real-world exposure situations. Though FP-biotin is very convenient for finding unknown proteins that react with OP, it is subject to the criticism that it is an atypical OP and that its reactions may not reflect the selectivity that can be expected for commonly used OP.
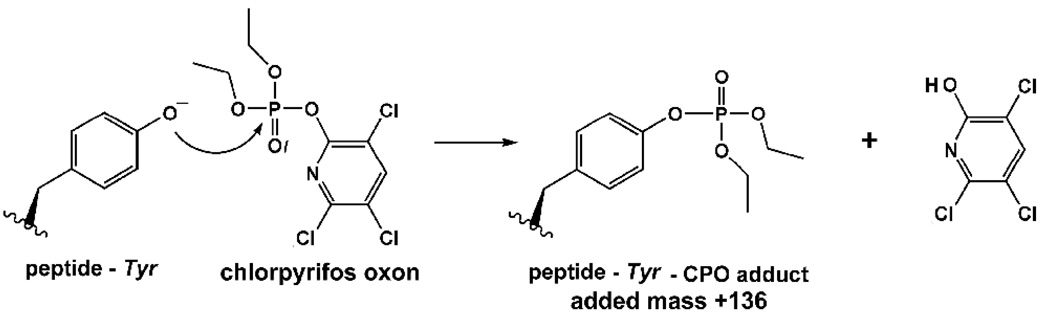
Initial analysis of the tryptic digest of chlorpyrifos oxon treated bovine tubulin was made on the MALDI-TOF/TOF 4800 mass spectrometer. The MS peaks from the labeled preparation were compared with theoretical mass lists of tryptic peptides from unlabeled tubulin (alpha1, gi 73586894 and beta2 gi 75773583 NCBI database). Alpha1 and beta2 are the most abundant tubulin isotypes in the brain. Therefore we took these isotypes for generation of the theoretical mass lists by Protein Prospector. The list of theoretical masses was generated with Protein Prospector software (UCSF, www.prospector.ucsf.edu). Peaks from the labeled preparation whose mass was equal to that of an unlabeled peptide plus the added mass from chlorpyrifos oxon (136 amu) were considered to be candidates for chlorpyrifos oxon modified peptides.
Labeling did not reach 100 % under the experimental conditions employed. Therefore, masses for both labeled and unlabeled peptides appeared in the same mass spectrum. A MALDI-TOF MS spectrum showing some unlabeled peptides and their corresponding chlorpyrifos oxon-labeled candidates is shown in Figure 3.
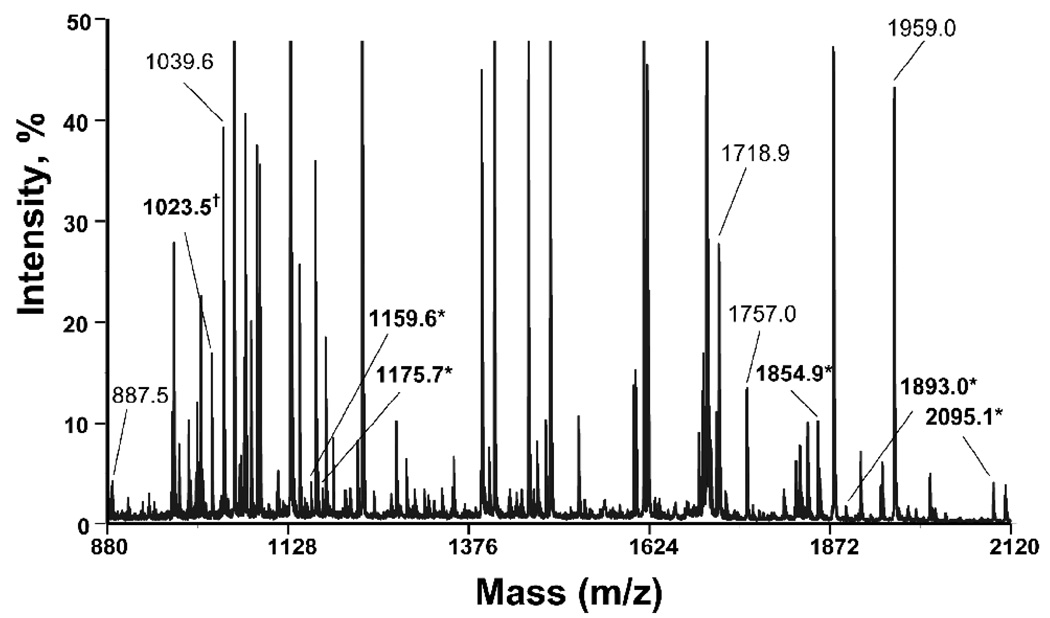
Fig. 3.
MALDI-TOF MS spectrum of tryptic peptides from bovine tubulin treated with a 40 fold molar excess of chlorpyrifos oxon (0.5 mM). Singly-charged, unlabeled peptides with masses of 887.5, 1023.5, 1039.5, 1718.9, 1757.0, and 1959.0 amu are shown in regular font. Corresponding candidates for chlorpyrifos oxon labeled peptides (plus 136 amu) with masses of 1023.5, 1159.5, 1175.7, 1854.9, 1893.0, and 2095.1 amu are shown in bold and marked with an asterisk. The mass at 1023.5 m/z coincides with both the unlabeled peptide EDAANNYAR and the chlorpyrifos oxon labeled peptide FDLMYAK.
Not all labeled peptides could be detected in the MALDI mass spectrometer and MS/MS fragmentation in the MALDI mass spectrometer was not always sufficient to fully characterize the labeled peptides. Complementary MS/MS spectra for the candidate peptides were obtained via LC/MS/MS in the tandem, quadrupole, linear-ion trap, QTRAP 2000 mass spectrometer, using electrospray ionization to introduce the sample.
MS/MS data from the QTRAP 2000 were submitted to the Mascot search engine for identification of chlorpyrifos oxon labeled peptides. The Mascot results were checked manually to confirm the sequences of putative, chlorpyrifos oxon-labeled peptides and to identify the covalently modified amino acid residue. In total, 16 labeled peptides and 17 amino acid residues were identified (one peptide carried two labels). Each labeled peptide was covalently attached to an O-diethyl phosphate fragment. Nine tyrosines were from alpha tubulin and 8 tyrosines were from beta tubulin. Four of the tyrosines (59, 83, 159 and 281) had been identified and reported previously (Grigoryan et al., 2008). Representative MS/MS spectra of labeled peptides are shown in Figure 4.
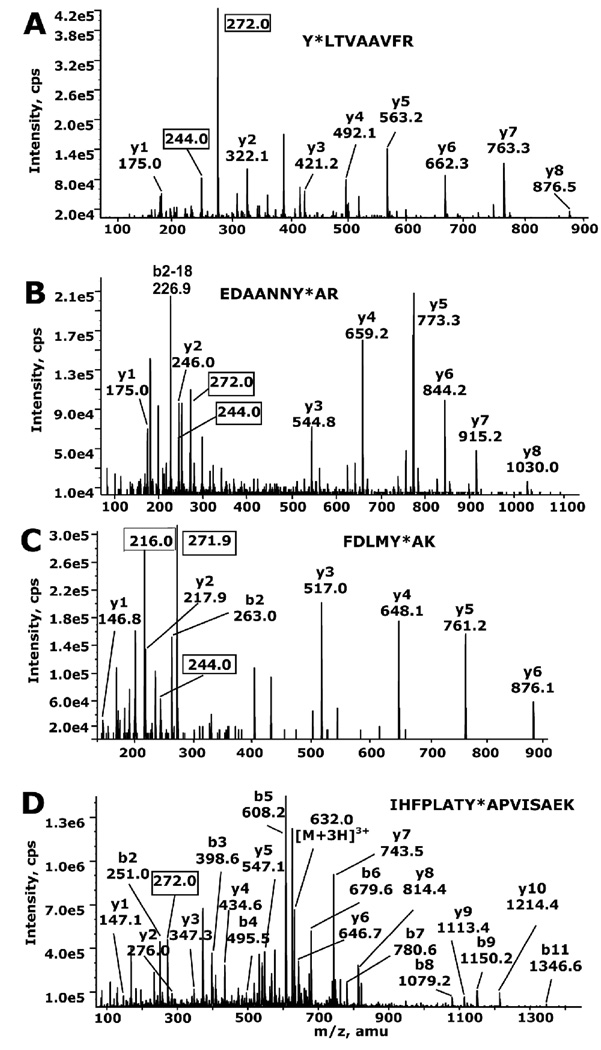
Figure 4.
MS/MS spectra of chlorpyrifos oxon-labeled peptides from bovine tubulin obtained by LC/MS/MS on the QTRAP 2000 mass spectrometer. Panel (A) peptide Y310LTVAAVFR from beta tubulin; (B) peptide EDAANNY103AR from alpha tubulin; (C) peptide, FDLMY399AK from alpha tubulin; (D) peptide IHFPLATY272APVISAEK from alpha tubulin. In panels A-C, singlycharged y ion series derived from doubly-charged parent ions at 588.43 (A), 580.23 (B) and 512.32 (C) m/z are indicated. For IHFPLATY272APVISAEK, in panel D, singly-charged y and b series ions derived from a triply-charged parent ion at 632.01 m/z are marked. In all cases the masses of the y and b ions are consistent with O-diethylphosphate attached to tyrosine (marked by an asterisk in the peptide sequence). Masses enclosed in a box are the immonium ion of phosphotyrosine (216), the immonium ion of O-monoethylphosphotyrosine (244), and the immonium ion of O-diethylphosphotyrosine (272). Peaks that are not labeled include internal fragments and masses that have lost a molecule of water or ammonia.
Theoretical fragment ion masses for each labeled peptide were calculated using the MS/MS Fragment Ion Calculator from Systems Biology (http://db.systemsbiology.net) and compared to the observed data. Extensive y-ion series were identified for each peptide. A delta mass corresponding to the labeled tyrosine fit into the sequence ions for the y-series of each peptide.
Characteristic, non-sequence fragments at 216, 244, and/or 272 m/z were found for each peptide. The mass at 216 m/z is consistent with the immonium ion of phosphotyrosine, the mass at 244 m/z is consistent with the immonium ion of O-monoethylphosphotyrosine, and the mass at 272 m/z is consistent with the immonium ion of O-diethylphosphotyrosine. These characteristicions provide additional evidence that the chlorpyrifos oxon is bound to tyrosine in these peptides. In Figure 4, the characteristic ion masses are enclosed in boxes (Schopfer et al., 2009).
The scheme for the reaction of chlorpyrifos oxon with tyrosine is presented in Figure 5.
Figure 5.
Reaction of chlorpyrifos oxon with tyrosine. Covalent binding of chlorpyrifos oxon to tyrosine increases the mass of tyrosine by 136 amu due to the addition of diethoxyphosphate.
No aging
Aging is a secondary, enzymatically catalyzed reaction that occurs with OP-labeled acetyl- and butyrylcholinesterase. It results in the loss of one alkoxy side-chain from the phosphorus atom. For example, aging of chlorpyrifos oxon modified acetylcholinesterase would result in loss of ethylene from one ethoxy group (28 amu). Chlorpyrifos oxon-labeled tyrosine did not lose an ethylene group from the phosphate. If aging had occurred the added mass from chlorpyrifos oxon modification would have been +108 rather than +136. No peptides with an added mass of +108 were found.
OP-modified and phosphate-modified tyrosines
The complete list of organophosphorylated peptides from bovine alpha and beta tubulin is presented in Table 2. Four of the peptides (TGTYR, YVPR, GSQQYR and EEYPDR) were previously reported to have been labeled with a variety of OP (Grigoryan et al., 2008).
Table 2.
Chlorpyrifos oxon-labeled tryptic peptides from bovine alpha1 and beta2 tubulin identified by LC/MS/MS.
| Tubuli n chain |
Peptide | chlorpyrifos oxon labeled tyrosine a |
Unlabeled m/z |
Mass+136 m/z |
Phosphor ylation b |
Referencec |
|---|---|---|---|---|---|---|
| alpha | TGTY*R | 83 | 597.4 | 733.4 | no | |
| EDAANNY*AR | 103 | 1023.5 | 1159.5 | yes | (Rikova et al., 2007) | |
| GHY*TIGK | 108 | 775.4 | 911.4 | no | ||
| LSVDY*GK | 161 | 781.4 | 917.4 | no | ||
| NLDIERPTY*TNLNR | 224 | 1718.9 | 1854.9 | yes | (Rikova et al., 2007) | |
| FDGALNVDLTEFQTNLVPY*PR | 262 | 2409.2 | 2545.2 | no | ||
| IHFPLATY*APVISAEK | 272 | 1757 | 1893 | yes | (Rush et al., 2005; Rikova et al., 2007) |
|
| VGINY*QPPTVVPGGDLAK | 357 | 1825 | 1961 | yes | (Rush et al., 2005; Rikova et al., 2007; Guo et al., 2008) |
|
| FDLMY*AK | 399 | 887.5 | 1023.5 | no | ||
| beta | INVY*YNEATGGK | 50 | 1328.6 | 1464.6 | yes | (Rikova et al., 2007) |
| INVYY*NEATGGK | 51 | 1328.6 | 1464.6 | yes | (Rikova et al., 2007) | |
| Y*VPR | 59 | 534.4 | 670.4 | no | ||
| GHY*TEGAELVDSVLDVVR | 106 | 1959 | 2095.1 | no | ||
| EEY*PDR | 159 | 808.5 | 944.5 | no | ||
| GSQQY*R | 281 | 738.5 | 874.5 | no | ||
| Y*LTVAAVFR | 310 | 1039.6 | 1175.6 | no | ||
| NSSY*FVEWIPNNVK | 340 | 1696.8 | 1832.8 | yes | (Guo et al., 2008) |
Numbering of the organophosphorylated residues corresponds to accession numbers gi 73586894 for alpha and gi 75773583 for bovine beta tubulin in the NCBI database. The chlorpyrifos oxon-modified tyrosine (Y) is marked by an asterisk in the peptide sequence.
Entries in this column indicate whether or not phosphorylation of the tyrosine has been reported. Phosphorylation is by GTP.
Reference(s) where the phosphorylation information was reported. Data on phosphorylation are from http://www.phosphosite.org.
No serine in tubulin was found to be organophosphorylated by chlorpyrifos oxon.
Phosphoproteomic data for a variety of proteins from different sources are available from www.phosphosite.org. In vivo and in vitro experiments showed that in cancer cells alpha and beta tubulin can be phosphorylated on tyrosine using GTP and different kinases (Rush et al., 2005; Zheng et al., 2005; Rikova et al., 2007; Guo et al., 2008). Some of the tyrosines that have been reported to be phosphorylated are organophosphorylated by chlorpyrifos oxon as well. Both phosphorylation and organophosphorylation occur at positions 103, 224, 272 and 357 on alpha and 50, 51, and 340 on beta tubulin (Table 2). Hyper phosphorylation can interfere with the polymerization of tubulin (Wandosell et al., 1987; Ley et al., 1994). It follows that multiple organophosphorylation of tubulin might also interfere with tubulin polymerization and consequently with its functions.
Dose dependent organophosphorylation of tyrosines in tubulin by chlorpyrifos oxon
With so many chlorpyrifos oxon binding sites on tubulin, it was of interest to determine which was more reactive and to what extent. To characterize the dynamics of chlorpyrifos oxon-binding, both the reactivity (i.e. the fraction of each tyrosine that became organophosphorylated) and the kinetics of organophosphorylation were determined.
Eight peptides were studied: TGTY83R, NLDIERPTY224TNLNR, IHFPLATY272APVISAEK, Y59VPR, GHY106TEGAELVDSVLDVVR, EEY159PDR, GSQQY281R and Y310LTVAAVFR. The reactivity of the remaining peptides could not be determined for one of the following reasons: i) the mass of the unlabeled peptide coincided with the m/z value for another peptide labeled with chlorpyrifos oxon. For example, 1023.5 amu is the mass of both unlabeled EDAANNYAR and chlorpyrifos oxon-labeled FDLMY*AK, ii) some peptides were labeled on more than one tyrosine. For example, the peptide INVYYNEATGGK was labeled on either tyrosine, iii) the isotope clusters for some labeled peptides overlapped isotope clusters of other peptides. For example, the isotope cluster for VGINY*QPPTVVPGGDLAK at 1961.0 m/z overlapped the isotope cluster of a peak at 1959.0 m/z, and the isotope cluster for FDGALNVDLTEFQTNLVPY*PR at 2545.2 m/z overlapped the isotope cluster of a peak at 2544.2 m/z and iv) labeled peptides were not detectable by MALDI -TOF/TOF though they could be identified in the QTRAP. For example, masses 911.4, 917.4, 1832.8 amu for labeled-peptides GHY*TIGK, LSVDY*GK and NSSY*FVEWIPNNVK could not be detected in the MALDI.
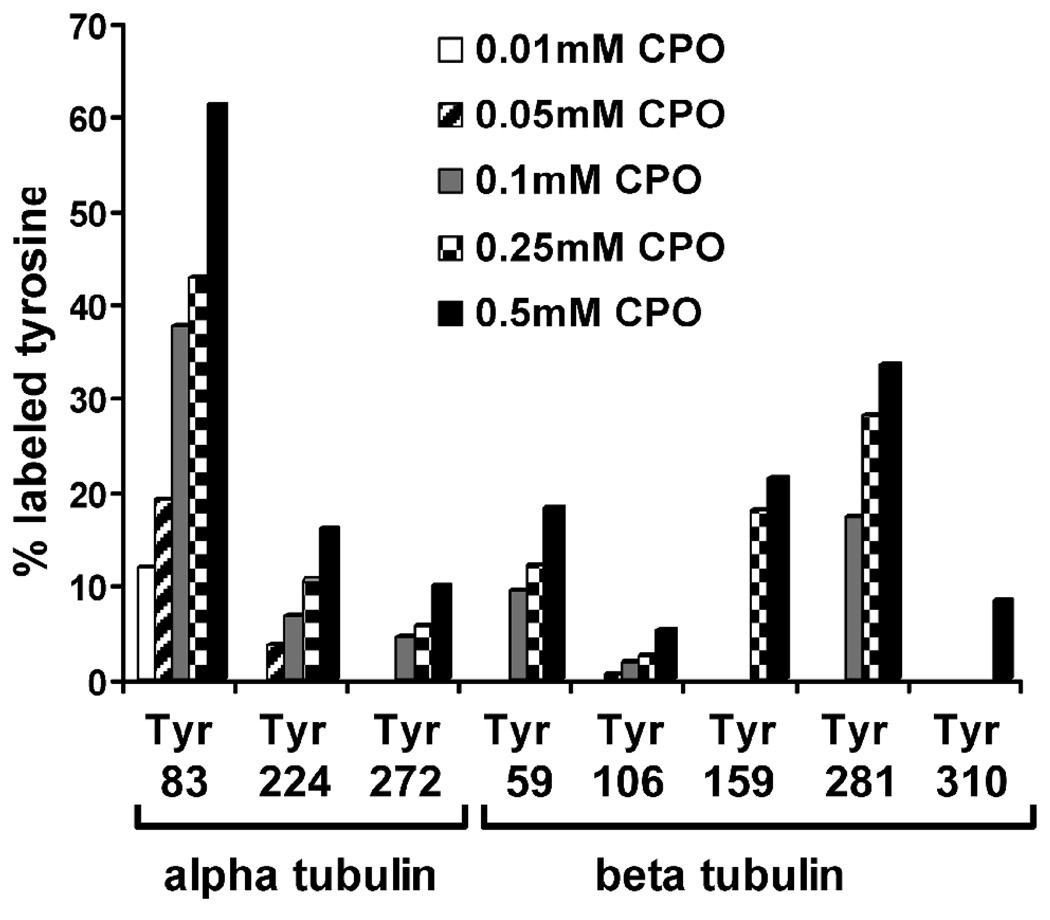
For those peptides that were studied, the percentage of each tyrosine that was labeled is presented in Figure 6.
Figure 6.
Reactivity of chlorpyrifos oxon with various tyrosines from tubulin. Bars represent the percentage of chlorpyrifos oxon-modified tyrosine in 3 peptides from alpha tubulin and 5 peptides from beta tubulin. Twelve μM tubulin was incubated with various concentrations of chlorpyrifos oxon at 37 °C for 24 h (pH 8.3).
As shown in Figure 6, Tyr 83 in alpha tubulin was the most reactive tyrosine. It was the only tyrosine labeled (12%) by an equimolar concentration of chlorpyrifos oxon. Tyr 83 was the most extensively labeled residue (61 %) in the presence of 0.5 mM chlorpyrifos oxon. For beta tubulin, tyrosine 281 exhibited the largest amount of labeling, with 34% labeling at 0.5 mM chlorpyrifos oxon. Residues 224, 272, 59, 106, 159 and 310 were substantially less reactive than 83 and 281. Reaction with 0.5 mM chlorpyrifos oxon resulted in 17%, 10%, 19%, 5%, 22% and 8.3% labeling of these tyrosines, respectively. Tyr 83 showed 3 – 12-fold higher reactivity than all other tyrosines. Tyrosines 106 and 310 from beta tubulin were the least reactive. The difference in the reactivity between tyrosines can be rationalized by their location in the crystal structure of the tubulin heterodimer and by the effect of neighboring amino acids.
Kinetics of the chlorpyrifos oxon reaction with tubulin tyrosines
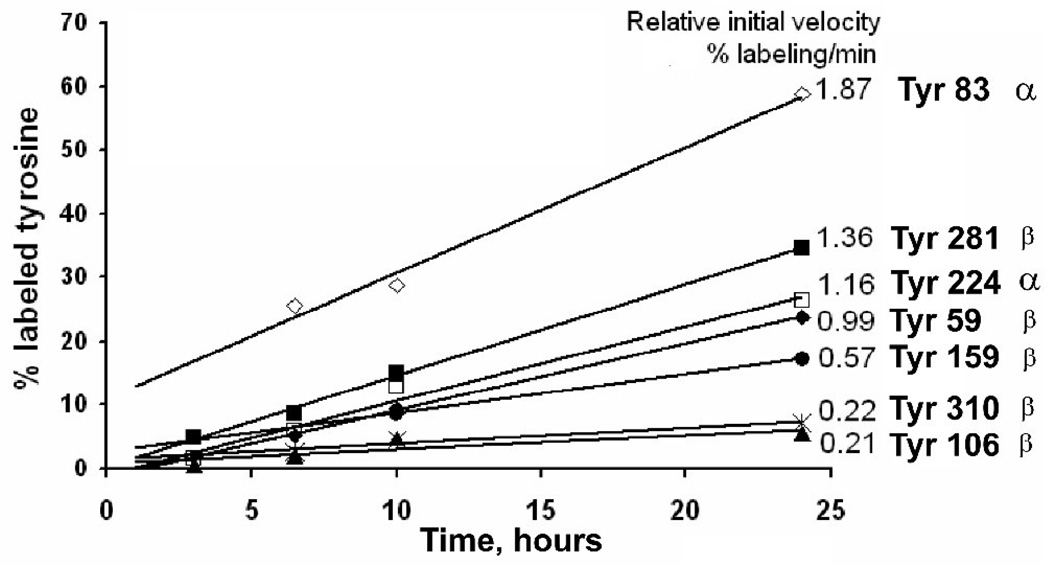
Progress curves for chlorpyrifos oxon binding to tyrosine were obtained by plotting the percentage of labeled peptide against the corresponding incubation time. Figure 7 shows the progress curves are linear (R2 = 0.7 – 1).
Figure 7.
Progress curves for chlorpyrifos oxon binding to tyrosines from bovine tubulin. The percentage of labeled peptides was calculated after 3, 6.5, 10 and 24 hr of incubation at 37 °C. Experimental data for alpha tubulin (tyrosines 83 and 224) are presented with empty symbols for TGTY83R and NLDIERPTY224TNLNR. Data for beta tubulin (tyrosines 281, 59, 159, 310 and 106) are shown with filled symbols for GSQQY281R, Y59VPR, EY159PDR, Y310LTVAAVFR and GHY106TEGAELVDSVLDVVR. The values of the initial velocities are presented next to each curve. Each point is the average of 3 trials, with an error of approximately 10%.
The initial velocity for each tyrosine was taken from the slope of the plot. No significant labeling was observed for any peptide during the first hour of incubation. Tyr 272 is not depicted in Figure 7 because it showed labeling only after 24 hr of incubation. Tyr 83 yielded the fastest rate at 1.87 % labeling/min, but no labeling was detected until the 6.5 h time point. At that time it was found to be 25 % labeled. Thereafter, the rate of labeling was rapid. This observation suggests that there might have been a reorientation of tyrosine 83 within the structure of tubulin. Tyr 83 reacted with chlorpyrifos oxon 1.5 – 9 times faster than other tyrosines. Tyrosine 281 showed the second fastest rate at 1.36 % labeling/min. These findings are consistent with the reactivity measurements described above.
Bovine alpha tubulin (accession # 73586894) has 19 tyrosines, of which 9 can be labeled by chlorpyrifos oxon. Bovine beta tubulin (accession # 75773583) has 16 tyrosines of which 8 can be labeled. About half of the tyrosines are reactive and half are unreactive.
In summary, Tyr 83 from alpha and Tyr 281 from beta bovine tubulin were more reactive toward chlorpyrifos oxon than the other tyrosines that became labeled. They both showed a higher percentage of labeling: Tyr 83 and Tyr 281 were labeled 61 % and 34 %, respectively, after 24 h of incubation at 37°C (Figure 6). And, they reacted with chlorpyrifos oxon more rapidly than the other tyrosines, being labeled at a rate of 1.87 and 1.36 % labeled peptide/min, respectively (Figure 7).
Effect of GTP on the reaction of chlorpyrifos oxon with tubulin
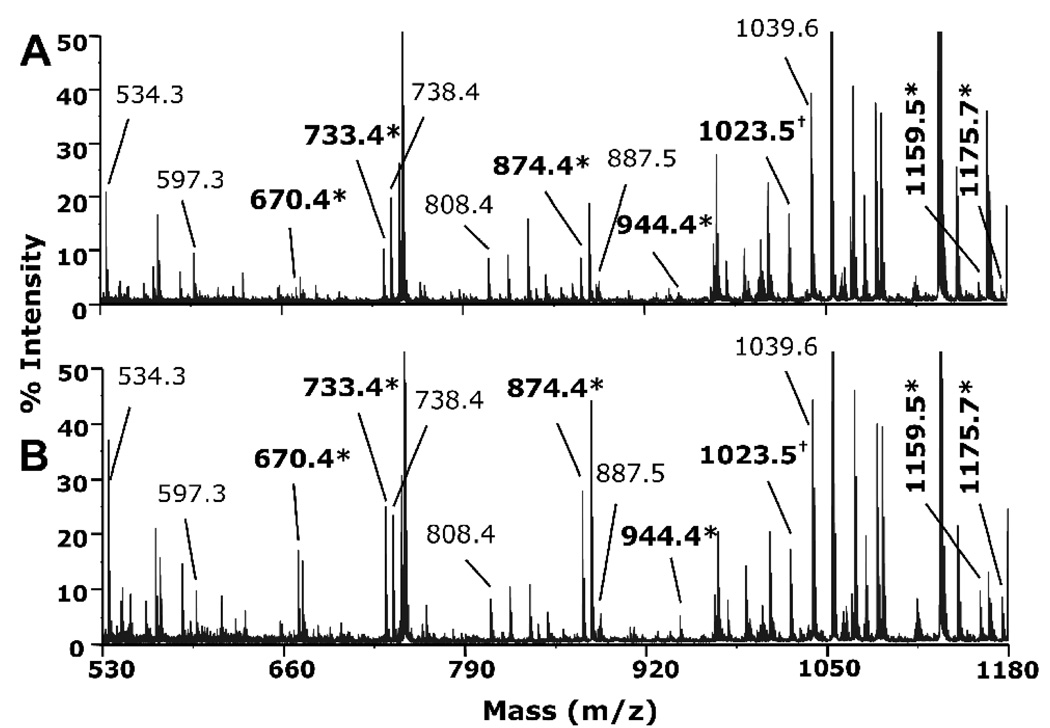
Tubulin dimers are incorporated into microtubules only after they have bound GTP to the exchangeable GTP binding site on beta tubulin. The binding of GTP alters the structure of the tubulin dimer. It was of interest to determine whether the structural change induced by GTP binding was reflected in the amount of chlorpyrifos oxon covalently bound to tubulin. The effect of GTP on chlorpyrifos oxon binding to tubulin was determined by incubation of 0.012 mM bovine tubulin heterodimer with 0.5 mM chlorpyrifos oxon at 37 °C for 24 hr in the presence and absence of 1 mM GTP. Tryptic peptides were analyzed in the MALDI-TOF/TOF 4800 mass spectrometer. Figure 8 presents the MS spectra of pairs of unlabeled and chlorpyrifos oxon-labeled tryptic peptides without (panel A) and with GTP (panel B). The presence of 1 mM GTP increased the extent of tubulin organophosphorylation. The peak intensities of chlorpyrifos oxon-labeled peptides were increased about 2 fold in the presence of GTP relative to the intensities of the unlabeled peaks.
Figure 8.
Effect of GTP on extent of chlorpyrifos oxon labeling. MS spectra were acquired on the MALDI TOF mass spectrometer. (A) labeled in the absence of GTP; (B) labeled in the presence of 1 mM GTP. Unlabeled peptides are indicated by regular font and labeled peptides (+136 amu added mass from chlorpyrifos oxon) are indicated in bold and marked by asterisks. Seven pairs of peptides are represented. Their masses and amino acid sequences are listed in Table 2. The peak at 1023.5 m/z corresponds to the unlabeled EDAANNYAR and the chlorpyrifos oxon labeled FDLMY*AK peptides. The peak height of each labeled peptide compared to the unlabeled peptide is higher in panel B (with GTP) than in panel A (without GTP).
Location of chlorpyrifos oxon labeled tyrosines in the crystal structure of the tubulin heterodimer
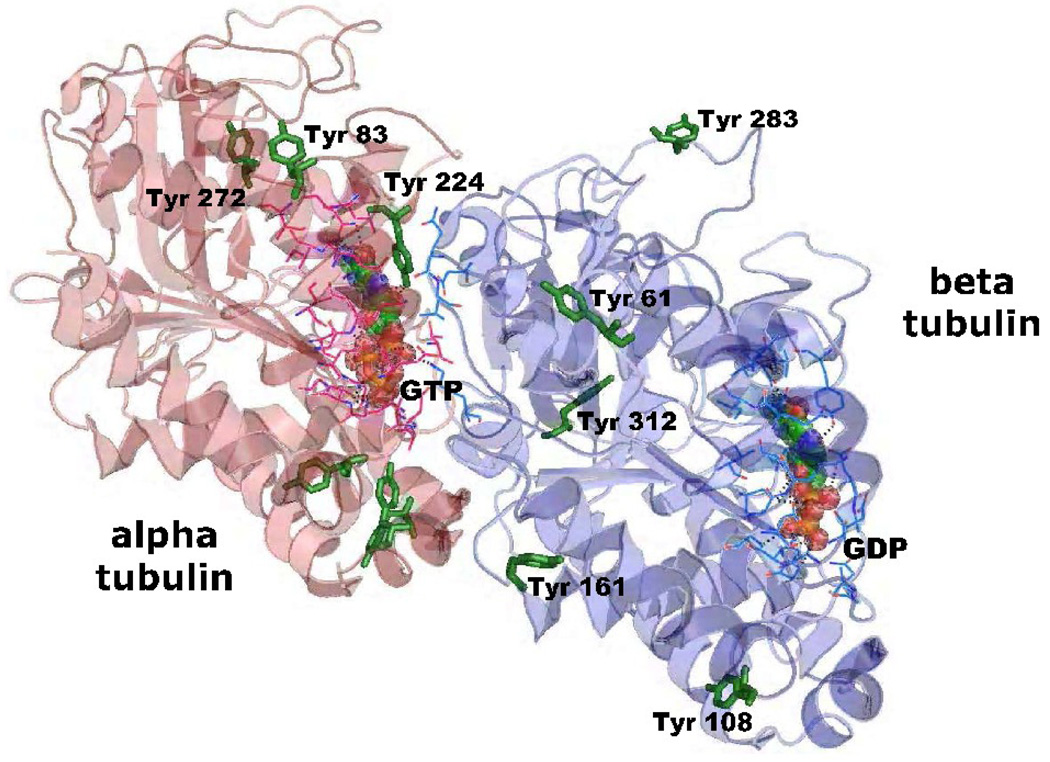
Tyrosines of bovine tubulin differed in their sensitivity toward chlorpyrifos oxon. To explain these differences, the location of modified tyrosines was of interest. The crystal structure of bovine brain tubulin heterodimer, reported by Nogales et al (Protein Data Bank number, 1jff) is shown in Figure 9 (Nogales et al., 1999). The 8 tyrosines for which reactivity data were obtained are indicated. Nogales et al introduced gaps into the numbers of the beta tubulin amino acid sequence, which explains why the numbers in the crystal structure are higher than the numbers in the protein database.
Fig. 9.
Crystal structure of the bovine brain tubulin heterodimer with subunit alpha in red and beta in blue (Protein Data Bank number, 1jff). Eight chlorpyrifos oxon labeled tyrosines (3 in alpha and 5 in beta tubulin) are shown as solid green sticks. GTP in the GTP non-exchangeable binding site in alpha tubulin and GDP in the exchangeable binding site in beta tubulin are shown as balls surrounded by amino acid residues that directly interact with these nucleotides. Amino acid numbers correspond to the numbers in pdb 1jff. These numbers are high compared to the numbers in the protein sequence because a gap of two residues was introduced in the numbers of the beta tubulin crystal structure (Nogales et al., 1999). The structure was drawn with PyMol software www.pymol.sourceforge.net
The phenolic oxygen of Tyr 272 in alpha tubulin and Tyr 108 (Y106) and Tyr 312 (Y310) in beta tubulin face the inside of the protein. As a consequence, it can be assumed that access of chlorpyrifos oxon to these tyrosines was limited, which is consistent with the relatively low percentage of chlorpyrifos oxon-labeling observed for these tyrosines (see Figure 6). Tyrosines 61 (59), 83, 161 (159), 224 and 283 (281) are exposed on the surface of tubulin. Tyr 224, in addition to being exposed to solvent, makes a π-πinteraction with the guanine moiety of the GTP molecule. Tyrosine 83 and Tyr 283 (281) are exposed on the surface of the tubulin dimer making them readily available for reaction with chlorpyrifos oxon, which would contribute to their high reactivity.
Discussion
Tubulin labeled in living mice
Living mice treated with a nontoxic dose of FP-biotin appeared to have FP-biotin labeled tubulin in their brains. The evidence for OP-labeled tubulin in mice is not complete because the OP-labeled peptide was not identified. The following observations led to the idea that tubulin was labeled by FP-biotin. 1) Avidin beads bound tubulin from brains of FP-biotin treated mice, but not from brains of untreated mice. 2) A protein of 55 kDa contained covalently bound FP-biotin as visualized on a blot hybridized with Streptavidin Alexa-680. Mass spectrometry identified the major component of this 55 kDa SDS gel band as tubulin. The data presented in this report are intended to aid the search for OP-labeled tubulin peptides, to provide the proof that is currently missing.
OP-labeled tyrosines in bovine tubulin
In preparation for renewed attempts to find the OP-labeled tubulin peptide in mouse brain, we set out to identify the potentially labeled sites. For this purpose we studied the labeling of highly purified bovine tubulin by chlorpyrifos oxon. We identified tyrosine as the site of covalent binding by OP and identified the specific tyrosines that are most reactive with OP, i.e. Tyr83 from alpha tubulin and Tyr281 from beta tubulin.
OP target tyrosines are conserved in bovine, mouse, and human tubulin
The NCBI database shows up to 8 alpha and 8 beta tubulin sequences in bovine, mouse, and human species. Their amino acid sequences are highly conserved. For example, alignment of accession numbers 73586894 (bovine alpha), 55977479 (mouse alpha) and 17986283 (human alpha) shows 98% sequence identity with all tyrosines conserved. Alignment of accession numbers 75773583 (bovine beta), 33859488 (mouse beta), and 4507729 (human beta) shows 97% identity with all tyrosines conserved. It can therefore be proposed that the tyrosines modified by OP in bovine tubulin are likely to be modified in mouse and human tubulin.
Tyrosine as a motif for OP binding to proteins
The presence of the consensus sequence GXSXG defines a protein as a member of the serine hydrolase family. These proteins make a covalent bond with OP on the active site serine. However, the majority of proteins that bound FP-biotin covalently in Table 1 do not have the consensus sequence GXSXG. Except for albumin and tubulin, where OP make a covalent bond with tyrosine (Li et al., 2007a; Grigoryan et al., 2008), the site of covalent attachment of FP-biotin is unknown. The most extensive data are for OP labeling of albumin. Several laboratories agree that albumin is covalently modified by OP both in vitro and in vivo (Adams et al., 2004; Peeples et al., 2005; Carter et al., 2007; Williams et al., 2007; Ding et al., 2008; Tarhoni et al., 2008).
Non-acetylcholinesterase mechanisms of OP toxicity
Tubulin dimers polymerize to form microtubules. Microtubules are part of the cytoskeletal system that maintains the morphology of neurons including axonal and dendritic processes (Gendron and Petrucelli, 2009). Microtubules serve as tracks for transport of mitochondria, synaptic vesicles, components of ion channels, receptors, and scaffolding proteins to and from synaptic sites. Synapses are highly vulnerable to impairments in transport. Perturbations to microtubule transport could cause malfunctions in neurotransmission and signal propagation and lead to synaptic degradation (Gendron and Petrucelli, 2009). Drugs that inhibit tubulin polymerization, for example colchicine, trigger apoptosis (Yeste-Velasco et al., 2008). Thus, the ability of tubulin to polymerize is very important for the life of a cell.
Microtubule function is also disrupted by hyperphosphorylation. It has been reported that aberrant phosphorylation of cytoskeletal proteins results in destabilization of microtubules and neurofilaments, leading to their aggregation. Consequently, normal function of the axons of central and peripheral nervous systems is disrupted (Abou-Donia, 1995; Abou-Donia, 2003). Organophosphorylation occurs on many of the same tyrosines that are phosphorylated (Table 2). It is reasonable to suggest that, like hyperphosphorylation, organophosphorylation of tubulin on multiple sites could impede tubulin polymerization and normal functioning of microtubules
It has also been reported that dichlorvos, tri-o-cresyl phosphate, and diisopropyl phosphofluoridate induce hyperphosphorylation of cytoskeletal proteins (Suwita et al., 1986; Gupta and Abou-Donia, 1994; Choudhary et al., 2001). Phosphorylation is mediated by the microtubule associated endogenous cAMP dependent protein kinases and Ca2+/Calmodulin kinase. This could provide an alternate means by which organophosphates could disrupt the function of microtubules.
These observations make disruption of tubulin function a very likely source of toxicity related to low dose exposure to OP. However, disruption of other systems is also possible.
Hyperphosphorylation of CREB, induced by OP has been proposed as a mechanism of OP toxicity (Schuh et al., 2002). CREB is the nuclear transcription factor that controls cell differentiation. CREB is critical to synaptic plasticity and transcription-dependent forms of memory. Exposure to OP has been correlated with increased phosphorylation of CREB and it has been suggested that the increased phosphorylation alters the function of CREB. Alteration in the function of CREB could explain the memory impairment following OP exposure. It is not known whether the hyperphosphorylated CREB is a result of the direct action of OP on CREB or an indirect effect from OP-modification of proteins involved in phosphorylation or dephosphorylation of CREB.
The developmental neurotoxicity of chlorpyrifos may be due to disruption of the adenylyl cyclase cell signaling cascade (Song et al., 1997). Perturbation of the synthesis of the second messenger cyclic AMP affects cell replication and differentiation. The serotonergic signal transduction system has also been implicated in chlorpyrifos neurotoxicity. Abnormal levels of serotonin may alter the differentiation and architectural organization of the developing brain (Aldridge et al., 2003). Either of these effects could be the consequence of direct reaction of chlorpyrifos oxon with tyrosine in critical proteins of these systems.
Direct action of chlorpyrifos on muscarinic receptors causes M2 receptor dysfunction and bronchoconstriction in the absence of AChE inhibition. This mechanism is proposed to explain symptoms of asthma including airway hyperreactivity and wheezing following OP exposure (Lein and Fryer, 2005). Covalent binding of 3H-chlorpyrifos oxon to M2 muscarinic acetylcholine receptors in rat heart has been demonstrated, though the modified amino acid has not been identified (Bomser and Casida, 2001).
Some OP inhibit neuronal α4β2 nicotinic acetylcholine receptors at low concentrations that have no effect on AChE activity (Smulders et al., 2004). The desensitized receptors may contribute to neurological and neurobehavioral deficits.
Though we have demonstrated a clear reaction of OP with tubulin, it is likely that disruption of some or all of these other systems is involved in low dose OP neurotoxicity. The dominant mechanism would depend on the identity of the OP, the developmental age of the exposed individual, the health of the organs that metabolize and dispose of OP, and genetic factors, for example the genotype of enzymes involved in OP metabolism.
Acknowledgements
Mass spectra were obtained with the support of the Mass Spectrometry and Proteomics core facility at the University of Nebraska Medical Center. Supported by the U.S. Army Medical Research and Materiel Command W81XWH-07-2-0034, the National Institutes of Health U01 NS058056, U01 ES016102, and P30CA36727, and DGA/PEA grants 08co501 and ANR-06-BLAN-0163.
Abbreviations
- AChE
acetylcholinesterase
- CPO
chlorpyrifos oxon
- CREB
calcium/cyclic AMP response element binding protein
- FP-biotin
10-Fluoroethoxyphosphinyl-N-biotinamidopentyldecanamide
- GTP
guanosine-5′-triphosphate
- PVDF
polyvinylidene difluoride
- MS
mass spectrometry
- MS/MS
tandem mass spectrometry
- LC/MS/MS
liquid chromatography tandem mass spectrometry
- MALDI-TOF/TOF
matrix-assisted laser desorption/ionization time of flight mass spectrometry
- OP
organophosphorus compounds
- QTRAP
a tandem quadrupole linear ion trap mass spectrometer
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abou-Donia MB. Involvement of cytoskeletal proteins in the mechanisms of organophosphorus ester-induced delayed neurotoxicity. Clin Exp Pharmacol Physiol. 1995;22:358–359. doi: 10.1111/j.1440-1681.1995.tb02015.x. [DOI] [PubMed] [Google Scholar]
- Abou-Donia MB. Organophosphorus ester-induced chronic neurotoxicity. Arch Environ Health. 2003;58:484–497. doi: 10.3200/AEOH.58.8.484-497. [DOI] [PubMed] [Google Scholar]
- Adams TK, Capacio BR, Smith JR, Whalley CE, Korte WD. The application of the fluoride reactivation process to the detection of sarin and soman nerve agent exposures in biological samples. Drug Chem. Toxicol. 2004;27:77–91. doi: 10.1081/dct-120027901. [DOI] [PubMed] [Google Scholar]
- Aldridge JE, Seidler FJ, Meyer A, Thillai I, Slotkin TA. Serotonergic systems targeted by developmental exposure to chlorpyrifos: effects during different critical periods. Environ Health Perspect. 2003;111:1736–1743. doi: 10.1289/ehp.6489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azaroff LS. Biomarkers of exposure to organophosphorous insecticides among farmers' families in rural El Salvador: factors associated with exposure. Environ Res. 1999;80:138–147. doi: 10.1006/enrs.1998.3877. [DOI] [PubMed] [Google Scholar]
- Bomser JA, Casida JE. Diethylphosphorylation of rat cardiac M2 muscarinic receptor by chlorpyrifos oxon in vitro. Toxicol Lett. 2001;119:21–26. doi: 10.1016/s0378-4274(00)00294-0. [DOI] [PubMed] [Google Scholar]
- Brown MA, Brix KA. Review of health consequences from high-, intermediate- and low-level exposure to organophosphorus nerve agents. J Appl Toxicol. 1998;18:393–408. doi: 10.1002/(sici)1099-1263(199811/12)18:6<393::aid-jat528>3.0.co;2-0. [DOI] [PubMed] [Google Scholar]
- Carter WG, Tarhoni M, Rathbone AJ, Ray DE. Differential protein adduction by seven organophosphorus pesticides in both brain and thymus. Hum. Exp. Toxicol. 2007;26:347–353. doi: 10.1177/0960327107074617. [DOI] [PubMed] [Google Scholar]
- Casida JE, Quistad GB. Organophosphate toxicology: safety aspects of nonacetylcholinesterase secondary targets. Chem Res Toxicol. 2004;17:983–998. doi: 10.1021/tx0499259. [DOI] [PubMed] [Google Scholar]
- Choudhary S, Joshi K, Gill KD. Possible role of enhanced microtubule phosphorylation in dichlorvos induced delayed neurotoxicity in rat. Brain Res. 2001;897:60–70. doi: 10.1016/s0006-8993(00)03222-4. [DOI] [PubMed] [Google Scholar]
- Ding SJ, Carr J, Carlson JE, Tong L, Xue W, Li Y, Schopfer LM, Li B, Nachon F, Asojo O, Thompson CM, Hinrichs SH, Masson P, Lockridge O. Five tyrosines and two serines in human albumin are labeled by the organophosphorus agent FP-biotin. Chem Res Toxicol. 2008;21:1787–1794. doi: 10.1021/tx800144z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eddleston M, Mohamed F, Davies JO, Eyer P, Worek F, Sheriff MH, Buckley NA. Respiratory failure in acute organophosphorus pesticide self-poisoning. Qjm. 2006;99:513–522. doi: 10.1093/qjmed/hcl065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gearhart DA, Sickles DW, Buccafusco JJ, Prendergast MA, Terry AV., Jr Chlorpyrifos, chlorpyrifos-oxon, and diisopropylfluorophosphate inhibit kinesin-dependent microtubule motility. Toxicol Appl Pharmacol. 2007;218:20–29. doi: 10.1016/j.taap.2006.10.008. [DOI] [PubMed] [Google Scholar]
- Gendron TF, Petrucelli L. The role of tau in neurodegeneration. Mol Neurodegener. 2009;4:13. doi: 10.1186/1750-1326-4-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grigoryan H, Schopfer LM, Thompson CM, Terry AV, Masson P, Lockridge O. Mass spectrometry identifies covalent binding of soman, sarin, chlorpyrifos oxon,diisopropyl fluorophosphate, and FP-biotin to tyrosines on tubulin: A potential mechanism of long term toxicity by organophosphorus agents. Chem Biol Interact. 2008;175:180–186. doi: 10.1016/j.cbi.2008.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo A, Villen J, Kornhauser J, Lee KA, Stokes MP, Rikova K, Possemato A, Nardone J, Innocenti G, Wetzel R, Wang Y, MacNeill J, Mitchell J, Gygi SP, Rush J, Polakiewicz RD, Comb MJ. Signaling networks assembled by oncogenic EGFR and c-Met. Proc Natl Acad Sci U S A. 2008;105:692–697. doi: 10.1073/pnas.0707270105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta RP, Abou-Donia MB. In vivo and in vitro effects of diisopropyl phosphorofluoridate (DFP) on the rate of hen brain tubulin polymerization. Neurochem Res. 1994;19:435–444. doi: 10.1007/BF00967321. [DOI] [PubMed] [Google Scholar]
- Jennings LL, Malecki M, Komives EA, Taylor P. Direct analysis of the kinetic profiles of organophosphate-acetylcholinesterase adducts by MALDI-TOF mass spectrometry. Biochemistry. 2003;42:11083–11091. doi: 10.1021/bi034756x. [DOI] [PubMed] [Google Scholar]
- Kamel F, Engel LS, Gladen BC, Hoppin JA, Alavanja MC, Sandler DP. Neurologic symptoms in licensed pesticide applicators in the Agricultural Health Study. Hum Exp Toxicol. 2007;26:243–250. doi: 10.1177/0960327107070582. [DOI] [PubMed] [Google Scholar]
- Kidd D, Liu Y, Cravatt BF. Profiling serine hydrolase activities in complex proteomes. Biochemistry. 2001;40:4005–4015. doi: 10.1021/bi002579j. [DOI] [PubMed] [Google Scholar]
- Lein PJ, Fryer AD. Organophosphorus insecticides induce airway hyperreactivity by decreasing neuronal M2 muscarinic receptor function independent of acetylcholinesterase inhibition. Toxicol Sci. 2005;83:166–176. doi: 10.1093/toxsci/kfi001. [DOI] [PubMed] [Google Scholar]
- Ley SC, Verbi W, Pappin DJ, Druker B, Davies AA, Crumpton MJ. Tyrosine phosphorylation of alpha tubulin in human T lymphocytes. Eur J Immunol. 1994;24:99–106. doi: 10.1002/eji.1830240116. [DOI] [PubMed] [Google Scholar]
- Li B, Schopfer LM, Hinrichs SH, Masson P, Lockridge O. Matrix-assisted laser desorption/ionization time-of-flight mass spectrometry assay for organophosphorus toxicants bound to human albumin at Tyr411. Anal. Biochem. 2007a;361:263–272. doi: 10.1016/j.ab.2006.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Schopfer LM, Nachon F, Froment MT, Masson P, Lockridge O. Aging pathways for organophosphate-inhibited human butyrylcholinesterase, including novel pathways for isomalathion, resolved by mass spectrometry. Toxicol Sci. 2007b;100:136–145. doi: 10.1093/toxsci/kfm215. [DOI] [PubMed] [Google Scholar]
- Lockridge O, Xue W, Gaydess A, Grigoryan H, Ding SJ, Schopfer LM, Hinrichs SH, Masson P. Pseudo-esterase activity of human albumin: slow turnover on tyrosine 411 and stable acetylation of 82 residues including 59 lysines. J Biol Chem. 2008;283:22582–22590. doi: 10.1074/jbc.M802555200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonough JH, Jr, Shih TM. Neuropharmacological mechanisms of nerve agent-induced seizure and neuropathology. Neurosci Biobehav Rev. 1997;21:559–579. doi: 10.1016/s0149-7634(96)00050-4. [DOI] [PubMed] [Google Scholar]
- Moser VC. Comparisons of the acute effects of cholinesterase inhibitors using a neurobehavioral screening battery in rats. Neurotoxicol Teratol. 1995;17:617–625. doi: 10.1016/0892-0362(95)02002-0. [DOI] [PubMed] [Google Scholar]
- Nogales E, Whittaker M, Milligan RA, Downing KH. High-resolution model of the microtubule. Cell. 1999;96:79–88. doi: 10.1016/s0092-8674(00)80961-7. [DOI] [PubMed] [Google Scholar]
- Nomura DK, Blankman JL, Simon GM, Fujioka K, Issa RS, Ward AM, Cravatt BF, Casida JE. Activation of the endocannabinoid system by organophosphorus nerve agents. Nat Chem Biol. 2008;4:373–378. doi: 10.1038/nchembio.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nomura DK, Leung D, Chiang KP, Quistad GB, Cravatt BF, Casida JE. A brain detoxifying enzyme for organophosphorus nerve poisons. Proc Natl Acad Sci U S A. 2005;102:6195–6200. doi: 10.1073/pnas.0501915102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pashkova A, Moskovets E, Karger BL. Coumarin tags for improved analysis of peptides by MALDI-TOF MS and MS/MS. 1. Enhancement in MALDI MS signal intensities. Anal Chem. 2004;76:4550–4557. doi: 10.1021/ac049638+. [DOI] [PubMed] [Google Scholar]
- Peeples ES, Schopfer LM, Duysen EG, Spaulding R, Voelker T, Thompson CM, Lockridge O. Albumin, a new biomarker of organophosphorus toxicant exposure, identified by mass spectrometry. Toxicol. Sci. 2005;83:303–312. doi: 10.1093/toxsci/kfi023. [DOI] [PubMed] [Google Scholar]
- Perkins DN, Pappin DJ, Creasy DM, Cottrell JS. Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis. 1999;20:3551–3567. doi: 10.1002/(SICI)1522-2683(19991201)20:18<3551::AID-ELPS3551>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- Pope CN. Organophosphorus pesticides: do they all have the same mechanism of toxicity? J Toxicol Environ Health B Crit Rev. 1999;2:161–181. doi: 10.1080/109374099281205. [DOI] [PubMed] [Google Scholar]
- Prendergast MA, Self RL, Smith KJ, Ghayoumi L, Mullins MM, Butler TR, Buccafusco JJ, Gearhart DA, Terry AV., Jr Microtubule-associated targets in chlorpyrifos oxon hippocampal neurotoxicity. Neuroscience. 2007;146:330–339. doi: 10.1016/j.neuroscience.2007.01.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rikova K, Guo A, Zeng Q, Possemato A, Yu J, Haack H, Nardone J, Lee K, Reeves C, Li Y, Hu Y, Tan Z, Stokes M, Sullivan L, Mitchell J, Wetzel R, Macneill J, Ren JM, Yuan J, Bakalarski CE, Villen J, Kornhauser JM, Smith B, Li D, Zhou X, Gygi SP, Gu TL, Polakiewicz RD, Rush J, Comb MJ. Global survey of phosphotyrosine signaling identifies oncogenic kinases in lung cancer. Cell. 2007;131:1190–1203. doi: 10.1016/j.cell.2007.11.025. [DOI] [PubMed] [Google Scholar]
- Roldan-Tapia L, Nieto-Escamez FA, del Aguila EM, Laynez F, Parron T, Sanchez-Santed F. Neuropsychological sequelae from acute poisoning and long-term exposure to carbamate and organophosphate pesticides. Neurotoxicol Teratol. 2006;28:694–703. doi: 10.1016/j.ntt.2006.07.004. [DOI] [PubMed] [Google Scholar]
- Rush J, Moritz A, Lee KA, Guo A, Goss VL, Spek EJ, Zhang H, Zha XM, Polakiewicz RD, Comb MJ. Immunoaffinity profiling of tyrosine phosphorylation in cancer cells. Nat Biotechnol. 2005;23:94–101. doi: 10.1038/nbt1046. [DOI] [PubMed] [Google Scholar]
- Salvi RM, Lara DR, Ghisolfi ES, Portela LV, Dias RD, ouza DO. Neuropsychiatric evaluation in subjects chronically exposed to organophosphate pesticides. Toxicol Sci. 2003;72:267–271. doi: 10.1093/toxsci/kfg034. [DOI] [PubMed] [Google Scholar]
- Schopfer LM, Champion MM, Smith B, Tamblyn N, Thompson CM, Lockridge O. Characteristic mass spectral fragments of the organophosphorus agent FP-biotin and FPbiotinylated peptides from trypsin and bovine albumin (Tyr410) Anal. Biochem. 2005a;345:122–132. doi: 10.1016/j.ab.2005.07.016. [DOI] [PubMed] [Google Scholar]
- Schopfer LM, Grigoryan H, Li B, Nachon F, Masson P, Lockridge O. Mass Spectral Characterization of Organophosphate-Labeled, Tyrosine-Containing Peptides: Characteristic Mass Fragments and A New Binding Motif for Organophosphates. J Chromatogr B. 2009 doi: 10.1016/j.jchromb.2009.07.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schopfer LM, Voelker T, Bartels CF, Thompson CM, Lockridge O. Reaction kinetics of biotinylated organophosphorus toxicant, FP-biotin, with human acetylcholinesterase and human butyrylcholinesterase. Chem. Res. Toxicol. 2005b;18:747–754. doi: 10.1021/tx049672j. [DOI] [PubMed] [Google Scholar]
- Schuh RA, Lein PJ, Beckles RA, Jett DA. Noncholinesterase mechanisms of chlorpyrifos neurotoxicity: altered phosphorylation of Ca2+/cAMP response element binding protein in cultured neurons. Toxicol Appl Pharmacol. 2002;182:176–185. doi: 10.1006/taap.2002.9445. [DOI] [PubMed] [Google Scholar]
- Smulders CJ, Bueters TJ, Vailati S, van Kleef RG, Vijverberg HP. Block of neuronal nicotinic acetylcholine receptors by organophosphate insecticides. Toxicol Sci. 2004;82:545–554. doi: 10.1093/toxsci/kfh269. [DOI] [PubMed] [Google Scholar]
- Song X, Seidler FJ, Saleh JL, Zhang J, Padilla S, Slotkin TA. Cellular mechanisms for developmental toxicity of chlorpyrifos: targeting the adenylyl cyclase signaling cascade. Toxicol Appl Pharmacol. 1997;145:158–174. doi: 10.1006/taap.1997.8171. [DOI] [PubMed] [Google Scholar]
- Stephens R, Spurgeon A, Calvert IA, Beach J, Levy LS, Berry H, Harrington JM. Neuropsychological effects of long-term exposure to organophosphates in sheep dip. Lancet. 1995;345:1135–1139. doi: 10.1016/s0140-6736(95)90976-1. [DOI] [PubMed] [Google Scholar]
- Sun J, Lynn BC. Development of a MALDI-TOF-MS method to identify and quantify butyrylcholinesterase inhibition resulting from exposure to organophosphate and carbamate pesticides. J Am Soc Mass Spectrom. 2007;18:698–706. doi: 10.1016/j.jasms.2006.11.009. [DOI] [PubMed] [Google Scholar]
- Suwita E, Lapadula DM, Abou-Donia MB. Calcium and calmodulin stimulated in vitro phosphorylation of rooster brain tubulin and MAP-2 following a single oral dose of tri-o-cresyl phosphate. Brain Res. 1986;374:199–203. doi: 10.1016/0006-8993(86)90412-9. [DOI] [PubMed] [Google Scholar]
- Tang X, Sadeghi M, Olumee Z, Vertes A, Braatz v, McIlwain LK, Dreifuss PA. Detection and quantitation of beta-2-microglobulin glycosylated end products in human serum by matrix-assisted laser desorption/ionization mass spectrometry. Anal Chem. 1996;68:3740–3745. doi: 10.1021/ac960516u. [DOI] [PubMed] [Google Scholar]
- Tarhoni MH, Lister T, Ray DE, Carter WG. Albumin binding as a potential biomarker of exposure to moderately low levels of organophosphorus pesticides. Biomarkers. 2008;13:343–363. doi: 10.1080/13547500801973563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terry AV, Jr, Gearhart DA, Beck WD, Jr, Truan JN, Middlemore ML, Williamson LN, Bartlett MG, Prendergast MA, Sickles DW, Buccafusco JJ. Chronic, intermittent exposure to chlorpyrifos in rats: protracted effects on axonaltransport, neurotrophin receptors, cholinergic markers, and information processing. J Pharmacol Exp Ther. 2007;322:1117–1128. doi: 10.1124/jpet.107.125625. [DOI] [PubMed] [Google Scholar]
- Tuin AW, Mol MA, van den Berg RM, Fidder A, van der Marel GA, Overkleeft HS, Noort D. Activity-Based Protein Profiling Reveals Broad Reactivity of the Nerve Agent Sarin. Chem Res Toxicol. 2009 doi: 10.1021/tx8004218. [DOI] [PubMed] [Google Scholar]
- Wandosell F, Serrano L, Avila J. Phosphorylation of alpha-tubulin carboxylterminal tyrosine prevents its incorporation into microtubules. J Biol Chem. 1987;262:8268–8273. [PubMed] [Google Scholar]
- Williams NH, Harrison JM, Read RW, Black RM. Phosphylated tyrosine in albumin as a biomarker of exposure to organophosphorus nerve agents. Arch. Toxicol. 2007;81:627–639. doi: 10.1007/s00204-007-0191-8. [DOI] [PubMed] [Google Scholar]
- Yeste-Velasco M, Alvira D, Sureda FX, Rimbau V, Mullins MM, Pallas M, Camins A, Folch J. DNA low-density array analysis of colchicine neurotoxicity in rat cerebellar granular neurons. Neurotoxicology. 2008;29:309–317. doi: 10.1016/j.neuro.2007.11.007. [DOI] [PubMed] [Google Scholar]
- Zheng H, Hu P, Quinn DF, Wang YK. Phosphotyrosine proteomic study of interferon alpha signaling pathway using a combination of immunoprecipitation and immobilized metal affinity chromatography. Mol Cell Proteomics. 2005;4:721–730. doi: 10.1074/mcp.M400077-MCP200. [DOI] [PubMed] [Google Scholar]