Abstract
A FRET peptide substrate was synthesized and evaluated for enzymatic cleavage by the BoNT/B light chain protease. The FRET substrate was found to be useful in both a high throughput assay to uncover initial “hits” and a low throughput HPLC assay to determine kinetic parameters and modes of inhibition.
The botulinum neurotoxins (BoNTs), are one of the deadliest substances known to man, and are produced by Clostridum Botulinum, a gram-positive soil dwelling bacteria.1 This toxic substance is listed by the Centers for Disease Control as a category A agent, representing the most deadliest of agents that could pose a serious threat to humanity. The neurotoxin, composed of both a heavy (100 kDa) and light (50 kDa) chain is the etiological agent accountable for a loss of neurotransmitter release. Specifically, the light chain which is a metalloprotease is responsible for the enzymatic damage of the SNARE protein complex.2 Thus, cleavage of the SNARE protein causes termination of neurotransmission resulting in flaccid paralysis and in severe cases death. There are seven different serotypes of the neurotoxin, (A-G), produced by different strains of the C. Botulinum.3 Each serotype is sequence specific for one of the SNARE proteins; and each serotype has a different level of toxicity. Of the seven serotypes BoNT/A, B and E are the most common causes of botulism in humans.4 The most toxic BoNT is the A serotype which has a LD50 of 1 ng/kg of body weight in humans, followed by the B and E serotypes.
The frequency of botulism in the human population is low, occurring mainly from consuming contaminated food due to improper storage of homemade canned foods. However, due to the high toxicity of the neurotoxin and ease at which both the bacteria can be cultured and the neurotoxin isolated, has caused concern among many that this deadly substance can be used as a biological weapon.5, 6 To defend against such an attack research is ongoing for development of small molecule inhibitors of the light chain protease. Indeed, progress has been made on the development of small molecule inhibitors of the BoNT/A light chain7-9, however, there has been little progress in terms of the development of BoNT/B protease inhibitors, which is the second most common agent of human botulism.4 To date there have been only two reports of small molecule inhibitors of the BoNT/B protease10, 11 and a scattering of peptide inhibitors12-16. We believe a lack of potent inhibitors of BoNT/B is due to short comings within current assays of this serotype of the neurotoxin. Herein, we report a robust substrate that allows for both high/low throughput assay and readily determination of basic enzymatic parameters of the BoNT/B protease.
There are reported enzymatic assays for the light chain of BoNT/B.16-24 Many of the assays, however, are rather cumbersome and not readily applicable to determine kinetic parameters and inhibition modes of potential inhibitors.19, 21, 22, 24 Additionally, some of these assays involve expensive equipment such as capillary electrophoresis17, 20, which reduces the amount of material needed, but is not readily available to many laboratories conducting inhibitor screens. Alternative assays employ fluorophores which are incorporated within the peptide substrate so as to monitor the enzymatic reaction via fluorescence.10, 21 Building upon these fluorescence based assays, peptide substrates were synthesized with internal FRET pairs that allowed BoNT/B light chain enzyamtic evaluation.16, 23
To increase research efforts for the development of BoNT/B protease inhibitors we have designed a substrate that can be incorporated into both a high throughput assay, to quickly identify potential inhibitors and a low throughput assay to obtain accurate kinetic parameters. The high throughput assay is based on a fluorescence resonance energy transfer (FRET) substrate. Substrates containing a FRET pair have been used for a variety of high throughput assays to identify protease inhibitors. For example, a commercially available FRET substrate composed of a truncated version of SNAP-25, the native substrate for the BoNT/A LC is known as SNAPtide™. However, a drawback with many FRET substrates is the ability to accurately measure the fluorescence of the fluorophore once the enzyme has cleaved the substrate. This phenomenon known as the inner filter effect; has plagued accurate kinetic determination of inhibition modes.25 Keeping these short comings in mind we designed our FRET-substrate so that it can also be utilized in a HPLC low throughput assay where the products are separated and quantified to determine accurate kinetic parameters such as Km, kcat and Ki. Hence, a substrate that can be used in a variety of assays is beneficial in terms of time, cost and reproducibility.
The zinc endopeptidase of the light chain of BoNT/B cleaves human VAMP-2 between Gln76 and Phe77.26 As such our substrate incorporates the native substrate binding sites of human VAMP-2. Thus, a 40 amino acid peptide similar to the conserved central region of human VAMP-2 (residues 55-94) was synthesized. The 40 amino acid sequence was determined to be the smallest peptide fragment to produce cleavage rates similar to the native substrate by the BoNT/B protease.27 In addition to the length of the peptide, the position of the FRET pair is important as this can have a substantial effect on the binding and catalytic efficiency of the protease. In order to incorporate the FRET pair without impacting enzymatic catalysis, two substitutions were made at positions 74 and 77. Position 74 was mutated to a lysine in order to couple 7-methoxycoumarin-4-acetic acid (MCA) to the amine via an amide bond. The selection of lysine was made in part to aid in the flexibility of the MCA to allow free rotation away from the active site and not affect binding and/or cleavage of the peptide. At position 77 the native phenylalanine was replaced with the unnatural amino acid diaminopropionic acid to facilitate coupling of 2, 4-dinitrophenyl (DNP) to the peptide. We did not anticipate the addition of DNP to this residue adjacent to the cleavage site would have a large effect on the catalytic efficiency; as mutations made at position 77 with other aromatic residues where shown to not have a major effect on the catalytic rates of BoNT/B protease.28
In addition to the FRET pair, Thr79 was mutated to a serine. This mutation has been shown to drastically increase the kcat by two fold, without affecting the Km.28 In sum the addition of the FRET pair within our substrate should increase Km and decrease kcat, thus, by mutating Thr79 to Ser79 we hoped to retrieve some of the lost catalysis.
The synthesis of the substrate was performed using Fmoc chemistry on tentagel NH2 resin involving orthogonal protecting group strategies to incorporate the FRET pair. The N-terminus was acylated and the peptide cleaved from the resin by treatment with 95 % TFA and purified by prep HPLC to give 1 (Figure 1) as an orange solid (7.8 % overall yield). To determine if our FRET peptide was cleaved by the BoNT/B light chain protease the substrate (50 μM) was incubated with the light chain (50 nM) in 40 mM HEPES buffer pH 7.4 for 15 min at room temperature. The mixture was separated by analytical HPLC, monitoring at 280 nm provided three peaks representing the two products and the parent substrate (Suppl. Mater.). To further quantify the kinetic parameters of this new substrate we synthesized the C-terminus product containing DNP to obtain a calibration curve and to determine the lower limit of detection (Figure 2). The lower limit for the DNP product was determined to be 36 pmole by analytical HPLC. Resolving the products by HPLC combined with the low level of detection, we could efficiently determine kinetic parameters of the BoNT/B light chain with our substrate. Using this assay we determined the Km of our substrate to be 70.9 ± 17 μM with a kcat of 3.81 sec−1, compared to the values of the native substrate of 1.6 μM and 1.0 sec−1 for the Km and kcat respectively.26
Figure 1.
Synthetic FRET substrate for the BoNT/B light chain protease. K = lysine with 7-methoxycoumarin-4-acetic acid coupled; X = diaminopropionic acid coupled to 2,4-dinitrophenyl.
Figure 2.
Calibration curve for the C-terminus DNP product of 1 as determined by analytic HPLC; monitoring was accomplished at 280 nm.
With our low throughput HPLC assay in place and suitable kinetic values for our substrate we focused upon the high throughput assay with 1. Here, the assay is dependent on cleavage of 1 to produce an increase in is fluorescence that can be measured by a fluorescence plate reader. Upon addition of BoNT/LC to a buffered solution of 1 in HEPES buffer we observed an increase in the fluorescence signal over time at excitation and emission wavelengths of 325 and 400 nm, respectively. Measuring the initial rates of reaction at various enzyme concentrations gave a linear response (Figure 3).
Figure 3.
Initial velocities of the BoNT/B LC enzymatic reaction versus concentration of the enzyme, as measured on a fluorescence plate reader
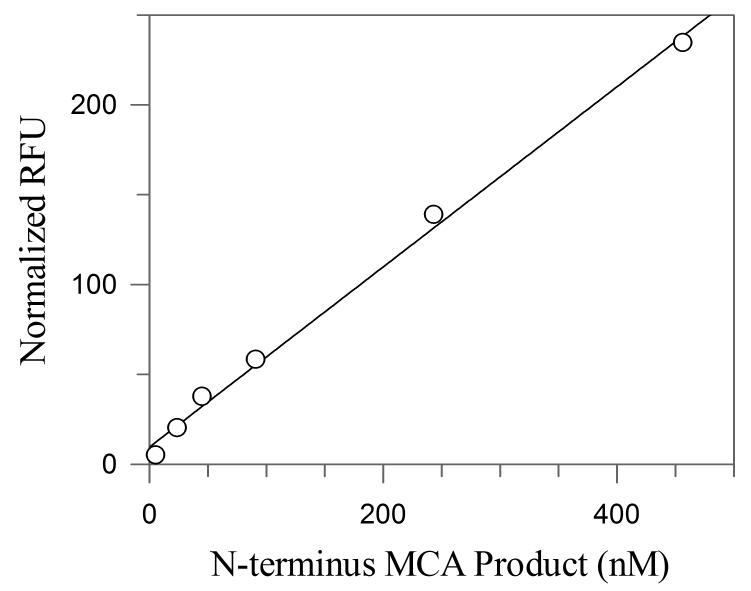
Importantly, rates can be effectively measured on a fluorescence plate reader at enzyme concentrations as low as 2 nM. We also determined the lower limit of detection for the MCA product to be 15 nM (Figure 4.). The ability to measure low concentrations of the MCA product means less time needed to monitor the enzymatic reaction, which in turn increases the number of compounds that can be screened.
Figure 4.
Normalized RFU of the N-terminus MCA product at various concentrations, as measured by a fluorescence plate reader.
In summary we report the synthesis of 1 in conjunction with two assays that will be useful in the search for inhibitors of the BoNT/B light chain. The usefulness of 1 is that it is amenable to both a high throughput assay to uncover potential inhibitors and a low throughput assay to determine accurate kinetic parameters and inhibition modes. With these two assays we envision the rapid screening of chemical libraries to uncover inhibitors of the BoNT/B light chain protease. Inhibitors uncovered from these screens will be published in due course.
Supplementary Material
Acknowledgments
This work is supported by National Institutes of Health (NO-AI-30050) and the Skaggs Institute for Chemical Biology.
Footnotes
Supplementary data Supplementary data associated with this article can be found, in the online version
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Shukla HD, Sharma SK. Crit Rev Microbiol. 2005;31:11. doi: 10.1080/10408410590912952. [DOI] [PubMed] [Google Scholar]
- 2.Schiavo G, Rossetto O, Santucci A, DasGupta BR, Montecucco C. J Biol Chem. 1992;267:23479. [PubMed] [Google Scholar]
- 3.Sebaihia M, Peck MW, Minton NP, Thomson NR, Holden MT, Mitchell WJ, Carter AT, Bentley SD, Mason DR, Crossman L, Paul CJ, Ivens A, Wells-Bennik MH, Davis IJ, Cerdeno-Tarraga AM, Churcher C, Quail MA, Chillingworth T, Feltwell T, Fraser A, Goodhead I, Hance Z, Jagels K, Larke N, Maddison M, Moule S, Mungall K, Norbertczak H, Rabbinowitsch E, Sanders M, Simmonds M, White B, Whithead S, Parkhill J. Genome Res. 2007;17:1082. doi: 10.1101/gr.6282807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Woodruff BA, Griffin PM, McCroskey LM, Smart JF, Wainwright RB, Bryant RG, Hutwagner LC, Hatheway CL. J Infect Dis. 1992;166:1281. doi: 10.1093/infdis/166.6.1281. [DOI] [PubMed] [Google Scholar]
- 5.Villar RG, Elliott SP, Davenport KM. Infect Dis Clin North Am. 2006;20:313. doi: 10.1016/j.idc.2006.02.003. [DOI] [PubMed] [Google Scholar]
- 6.Bossi P, Garin D, Guihot A, Gay F, Crance JM, Debord T, Autran B, Bricaire F. Cell Mol Life Sci. 2006;63:2196. doi: 10.1007/s00018-006-6308-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boldt GE, Kennedy JP, Hixon MS, McAllister LA, Barbieri JT, Tzipori S, Janda KD. J Comb Chem. 2006;8:513. doi: 10.1021/cc060010h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Capkova K, Hixon MS, McAllister LA, Janda KD. Chem Commun (Camb) 2008:3525. doi: 10.1039/b808305c. [DOI] [PubMed] [Google Scholar]
- 9.Capkova K, Yoneda Y, Dickerson TJ, Janda KD. Bioorg Med Chem Lett. 2007;17:6463. doi: 10.1016/j.bmcl.2007.09.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Adler M, Nicholson JD, Hackley BE., Jr FEBS Lett. 1998;429:234. doi: 10.1016/s0014-5793(98)00492-x. [DOI] [PubMed] [Google Scholar]
- 11.Hanson MA, Oost TK, Sukonpan C, Rich DH, Stevens RC. Journal of the American Chemical Society. 2000;122:11268. [Google Scholar]
- 12.Anne C, Blommaert A, Turcaud S, Martin AS, Meudal H, Roques BP. Bioorg Med Chem. 2003;11:4655. doi: 10.1016/s0968-0896(03)00450-4. [DOI] [PubMed] [Google Scholar]
- 13.Anne C, Turcaud S, Blommaert AG, Darchen F, Johnson EA, Roques BP. Chembiochem. 2005;6:1375. doi: 10.1002/cbic.200400398. [DOI] [PubMed] [Google Scholar]
- 14.Anne C, Turcaud S, Quancard J, Teffo F, Meudal H, Fournie-Zaluski MC, Roques BP. J Med Chem. 2003;46:4648. doi: 10.1021/jm0300680. [DOI] [PubMed] [Google Scholar]
- 15.Blommaert A, Turcaud S, Anne C, Roques BP. Bioorg Med Chem. 2004;12:3055. doi: 10.1016/j.bmc.2004.03.006. [DOI] [PubMed] [Google Scholar]
- 16.Schmidt JJ, Stafford RG. Appl. Environ. Microbiol. 2003;69:297. doi: 10.1128/AEM.69.1.297-303.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Adler M, Shafer HF, Manley HA, Hackley BE, Jr., Nicholson JD, Keller JE, Goodnough MC. J Protein Chem. 2003;22:441. doi: 10.1023/b:jopc.0000005459.00492.60. [DOI] [PubMed] [Google Scholar]
- 18.Barr JR, Moura H, Boyer AE, Woolfitt AR, Kalb SR, Pavlopoulos A, McWilliams LG, Schmidt JG, Martinez RA, Ashley DL. Emerg Infect Dis. 2005;11:1578. doi: 10.3201/eid1110.041279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hallis B, James BA, Shone CC. J Clin Microbiol. 1996;34:1934. doi: 10.1128/jcm.34.8.1934-1938.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Purcell AL, Hoard-Fruchey HM. Anal Biochem. 2007;366:207. doi: 10.1016/j.ab.2007.04.048. [DOI] [PubMed] [Google Scholar]
- 21.Schmidt JJ, Stafford RG, Millard CB. Anal Biochem. 2001;296:130. doi: 10.1006/abio.2001.5236. [DOI] [PubMed] [Google Scholar]
- 22.Keller JE, Nowakowski JL, Filbert MG, Adler M. J Appl Toxicol. 1999;19(Suppl 1):S13. doi: 10.1002/(sici)1099-1263(199912)19:1+<s13::aid-jat607>3.0.co;2-3. [DOI] [PubMed] [Google Scholar]
- 23.Anne C, Cornille F, Lenoir C, Roques BP. Anal Biochem. 2001;291:253. doi: 10.1006/abio.2001.5028. [DOI] [PubMed] [Google Scholar]
- 24.Hines HB, Kim AD, Stafford RG, Badie SS, Brueggeman EE, Newman DJ, Schmidt JJ. Appl Environ Microbiol. 2008;74:653. doi: 10.1128/AEM.01690-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu Y, Kati W, Chen CM, Tripathi R, Molla A, Kohlbrenner W. Anal Biochem. 1999;267:331. doi: 10.1006/abio.1998.3014. [DOI] [PubMed] [Google Scholar]
- 26.Chen S, Hall C, Barbieri JT. J Biol Chem. 2008;283:21153. doi: 10.1074/jbc.M800611200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shone CC, Quinn CP, Wait R, Hallis B, Fooks SG, Hambleton P. Eur J Biochem. 1993;217:965. doi: 10.1111/j.1432-1033.1993.tb18327.x. [DOI] [PubMed] [Google Scholar]
- 28.Shone CC, Roberts AK. Eur J Biochem. 1994;225:263. doi: 10.1111/j.1432-1033.1994.00263.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.