Abstract
The let-7 family of microRNAs (miRNAs) are important regulators of developmental timing and cell differentiation and are often misexpressed in human cancer. In C. elegans, let-7 controls cell fate transitions from larval stage 4 (L4) to adulthood by post-transcriptionally down-regulating lineage-abnormal 41 (lin-41) and hunchback-like 1 (hbl-1). Primary let-7 (pri-let-7) transcripts are up-regulated in the L3, yet little is known about what controls this transcriptional up-regulation. We sought factors that either turn on let-7 transcription or keep it repressed until the correct time. Here we report that one of let-7's targets, the transcription factor Hunchback-like 1 (HBL-1), is responsible for inhibiting the transcription of let-7 in specific tissues until the L3. hbl-1 is a known developmental timing regulator and inhibits adult development in larval stages. Therefore, one important function of HBL-1 in maintaining larval stage fates is inhibition of let-7. Indeed, our results reveal let-7 as the first known target of the HBL-1 transcription factor in C. elegans and suggest a negative feedback loop mechanism for let-7 and HBL-1 regulation.
Keywords: let-7, hbl-1, hunchback, microRNA, heterochronic, transcription, C. elegans
Introduction
MicroRNAs (miRNAs) are a class of 22 nucleotide (nt), single-stranded RNA molecules that control many biological processes including, development, cellular differentiation, cell signaling, life span, metabolism and apoptosis (Kloosterman and Plasterk, 2006; Stefani and Slack, 2008). MiRNAs post-transcriptionally regulate their messenger RNA (mRNA) targets by binding with imperfect complementarity to sites in their 3’ untranslated regions (UTRs). While miRNAs generally act as negative regulators of their targets’ translation, recent evidence has also documented positive effects (Vasudevan et al., 2007; Vasudevan et al., 2008)
The first two miRNAs identified, lin-4 and let-7, were originally found in C. elegans (Lee et al., 1993; Reinhart et al., 2000). These two genes are important components of the heterochronic pathway in C. elegans. The heterochronic pathway regulates the timing of C. elegans development through four larval stages and into adulthood (Moss, 2007). Mutations in heterochronic genes can lead to accelerated (precocious) or delayed (retarded) development (Moss, 2007). let-7's role in the pathway is to promote the transition from larval stage 4 (L4) to adulthood by down-regulating at least two targets lin-41 and hbl-1 (Abrahante et al., 2003; Lin et al., 2003; Reinhart et al., 2000). Consistently, let-7 loss-of-function (lf) mutants exhibit retarded phenotypes, specifically a well-characterized delay in the formation of adult seam cell fates and alae (Ambros, 1989; Reinhart et al., 2000). Alae are ridges on the cuticle of the animal that are secreted by a population of stem-cell like cells called seam cells as C. elegans transition from the L4 stage to adulthood (Ambros, 1989). In let-7 mutants, the seam cells undergo an additional cycle of cell division before they terminally differentiate and produce alae (Reinhart et al., 2000). let-7 was the first miRNA shown to be highly conserved from C. elegans to humans (Pasquinelli et al., 2000). In humans, let-7 is involved in cellular differentiation and cell cycle regulation. Reduced levels of let-7 have been observed in several types of cancer (Esquela-Kerscher and Slack, 2006; Kloosterman and Plasterk, 2006; Shi et al., 2008; Stefani and Slack, 2008).
Understanding what regulates the expression of miRNAs is important in developmental biology and medicine since they regulate many areas of development in different phyla (Stefani and Slack, 2008) and are implicated as factors in several cancers (Esquela-Kerscher and Slack, 2006). Some aspects of miRNA biogenesis and regulation have been elucidated in recent years. MiRNA genes are found in intergenic and intronic regions of the genome and can be mono- or polycistronic (Baskerville and Bartel, 2005; Rodriguez et al., 2004). The majority of miRNA genes are first transcribed by RNA polymerase II resulting in primary miRNAs transcripts (pri-miRNA). Like protein-coding targets of RNA Pol II, pri-miRNAs are polyadenylated and 5′-methyl G-capped (Bracht et al., 2004; Lee et al., 2004). Pri-miRNAs are processed into pre-miRNAs in the nucleus by the Microprocessor complex (Denli et al., 2004; Gregory et al., 2004). The premiRNAs are then exported to the cytoplasm by Exportin 5 where they are further processed by Dicer into mature miRNAs (Bohnsack et al., 2004; Yi et al., 2003).
Recently researchers have begun to address the question of what regulates specific miRNAs. In humans, regulation of let-7 expression has only begun to be examined. It has been shown that one of the transcription factors controlling let-7 expression is MYC (Chang et al., 2008; Chang et al., 2009; Sampson et al., 2007). However, there is growing evidence that the temporal expression of let-7 in humans is regulated post-transcriptionally by LIN28, a gene known to be involved in maintaining stem cell identity (Heo et al., 2008; Newman et al., 2008; Piskounova et al., 2008; Rybak et al., 2008; Viswanathan et al., 2008). Human LIN28 is the homolog of C. elegans lin-28, which is also a component of the heterochronic pathway in C. elegans (Ambros, 1989). The involvement of LIN28 in let-7 maturation and cellular development suggests the possibility that other members of the C. elegans heterochronic pathway could play a role in these same processes.
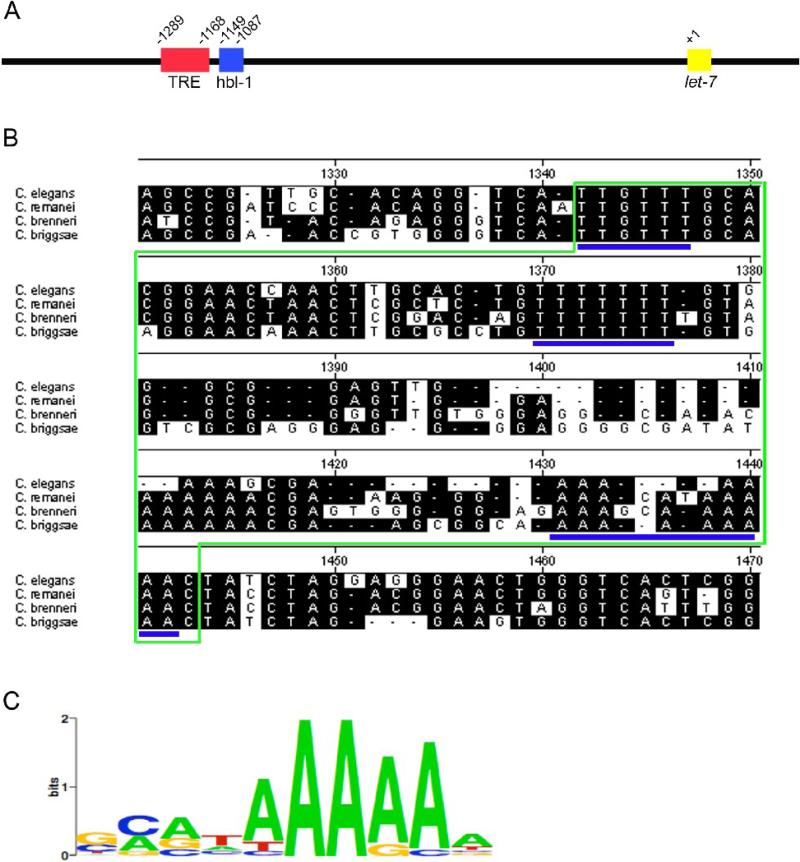
In C. elegans, little is known about the regulation of let-7 (Roush and Slack, 2008). Three pri-miRNAs transcripts have been detected in C. elegans by RT-PCR beginning at larval stage 3 (L3) (Bracht et al., 2004). One of these transcripts has the spliced leader 1 sequence (SL1) at its 5’ end, and it is assumed that this transcript is formed from the trans-splicing of the longer transcripts to the SL1 sequence (Bracht et al., 2004). One cis-acting transcriptional regulatory element, called the temporal regulatory element (TRE), that begins ∼1289 nt upstream of the mature let-7 has also been identified (Fig. 1A). This 116 nt element is needed for the activation of let-7 transcription at the correct developmental time in the seam cells of C. elegans. The TRE is also specifically bound by an unidentified TRE binding protein designated TREB (Johnson et al., 2003).
Figure 1.
The let-7 promoter contains a 62 bp region of high conservation wherein lie three putative HBL-1 binding sites. (A) Schematic of the C. elegans let-7 promoter showing the positions of TRE and putative HBL-1 binding sites. Red box: Temporal Regulatory Region (TRE); blue box: 62 bp region containing putative HBL-1 binding sites (HBL-1); yellow box: mature let-7. (B) Sequence alignment of region containing putative HBL-1 binding sites in the let-7 promoter from 4 nematode species. Green outlines the 62 bp element. Potential HBL-1 binding sites are underlined in blue. These sites do show variation with the Drosophila melanogaster Hunchback consensus binding site. (C) Drosophila melanogaster Hunchback consensus binding site from the TESS database (Schug, 1997).
We show in the present study that another heterochronic gene, Hunchback-like 1 (HBL-1), acts to transcriptionally inhibit the temporal expression of its regulator, let-7. C. elegans HBL-1 is a Cys2-His2, zinc-finger transcription factor homologous to the segmentation gene Hunchback (Hb) of Drosophila melanogaster , where it regulates the development of the syncytial embryo (Fay et al., 1999; Lehmann and Nusslein-Volhard, 1987; Struhl et al., 1992) and the temporal identity of neuroblasts (Isshiki et al., 2001; Pearson and Doe, 2003). It has been proposed that at low concentrations Hb acts as an activator, while at high concentrations it acts as an inhibitor of the transcription of its regulatory targets (Schulz and Tautz, 1994). In C. elegans, hbl-1 acts to inhibit the precocious expression of later fates during the L2 to L3 transition and also the L4 to adult transition. At both of these developmental transitions, hbl-1 must be down-regulated by let-7 or other let-7 miRNA family members for the animal to continue developing normally (Abbott et al., 2005; Abrahante et al., 2003; Lin et al., 2003). From the Drosophila research, several Hb targets have been identified and a consensus binding site has been elucidated (Hoch et al., 1991; Qian et al., 1991; Small et al., 1991; Stanojevic et al., 1989; Stanojevic et al., 1991; Treisman and Desplan, 1989; Zuo et al., 1991). However, no direct transcriptional target has as yet been identified for this critical developmental regulator in C. elegans. Here we provide evidence that let-7 is a transcriptional target of HBL-1 and that together they function in a negative feedback loop to control timing of cell fate during development.
Materials and Methods
Promoter alignments and binding site identification
let-7 promoter regions starting 2 kb upstream to the mature let-7 sequence were obtained from for C. elegans, C. remanei, C. briggsae and C. brenneri (Han, 2008) (www.wormbase.org). The sequences were aligned using MegAlign from DNASTAR Lasergene. The promoter region was run through the TESS program database (Schug, 1997) (www.cbil.upenn.edu/cgi-bin/tess/tess) from the University of Pennsylvania for identification of potential transcription factor binding sites.
Strain Construction and Analysis
Δ3hbl-1::gfp was amplified using overlap PCR from the let-7::gfp plasmid (pSJ11) (Johnson et al., 2003) using LET7SMJ2 (5′ GGTACCCTCCCTCTTTTAAGCCTG 3′) and HBLD1 (5′ CCCAGTTCCCTCCTAGATATGACCTGTGCAACGGCTAG 3′) to amplify the first half of the construct and HBLD2 (5′ CTAGCCGTTGCACAGGTCATATCTAGGAGGGAACTGGG 3′) and GFP-2C (5′ GGAAACAGTTATGTTTGGTATATTGGG 3′) to amplify the second half. The two PCR products were then used as the templates for the final PCR that deleted a 62 bp region. The primers used for the final PCR were LET7SMJ2 and GFP-2C. The purified PCR product was injected at 50 ng/μl with 10 ng/μl myo-2::DsRED2 as an injection marker.
The let-7 Rescue Fragment was amplified from pSK+let-7 plasmid (Reinhart et al., 2000) using LET7SMJ2 and PSKDOWN (5′ CCGCGGTGGCGGCCGCTCTAG 3′). The purified PCR product was injected at 4 ng/μl with 15 ng/μl myo-2::gfp as an injection marker.
Δhbl-1 Rescue Fragment was amplified using overlap PCR from pSK+let-7 plasmid using LET7SMJ2 and HBLD1 to amplify the first half of the construct and HBLD2 and PSKDOWN to amplify the second half. The two PCR products were then used as the templates for the final PCR that deleted a 62 bp region. The primers used for the final PCR were LET7SMJ2 and PSKDOWN The purified PCR product was injected at 4 ng/μl with 15 ng/μl myo-2::gfp injection marker. For each strain, at least 30 animals of each stage were scored.
RNAi Experiments
Larval stage 4 let-7::gfp (zaEx5) C. elegans were placed on plates containing HT115 E. coli bacteria expressing empty vector (pL4440) RNAi (Kamath et al., 2003; Timmons et al., 2001) or a mix of 1:1 empty vector to hbl-1(RNAi) from the Ahringer RNAi library. The mixed RNAi was called 50% hbl-1(RNAi). These animals were allowed to grow overnight and to begin laying eggs on the RNAi plates. The gravid C. elegans were transferred to new RNAi plates leaving their progeny behind every two hours to obtain staged progeny. At least 30 animals were scored at each larval stage for both pL4440 and 50% hbl-1 (RNAi).
Wild-type (N2) C. elegans grown on RNAi for protein extraction were first placed on the RNAi as synchronized L4 animals and grown overnight. Eggs from these gravid animals were isolated the next day and immediately plated on the RNAi bacteria.
Mixed staged C. elegans whole cell extracts
Packed, mixed staged N2, hbl-1(mg285), or 50% hbl-1(RNAi) C. elegans were snap frozen in T+120 buffer (Tris-HCl, pH 7.5, 120mM NaCl, 1% glycerol, 1mM DTT, 0.01% Igepal, 1× Roche Complete protease inhibitors, 0.5mM EDTA). Two times the packed C. elegans volume of T+120 was added to thawed C. elegans pellet, which was then incubated for 20 minutes on ice with intermittent vortexing. C. elegans were homogenized using a motorized mortar and pestle for 30 seconds to 1 minute on ice. Extract was spun at 16,000 g for 20 minutes at 4°C. The supernatant was transferred to a polycarbonate centrifuge tube and spun at 100,000 g at 4°C for 1 hour. The resulting supernatant from this spin was snap frozen and stored at −80°C.
Electrophoretic Mobility Shift Assays
For wild-type, mutant and RNAi extracts: In 10 μl reactions, 0.5μg of whole cell extract was pre-incubated for 10 minutes at room temperature in Tris-HCl binding buffer (10mM Tris-HCl pH 7.5, 4% glycerol, 1mM MgCl2, 0.5mM EDTA, 0.5mM DTT, 50mM NaCl) with 0.3 μg poly dI•dC as non-specific competitor. Two fmol of radiolabeled probe were added and the reaction mixture was incubated for 60 minutes at room temperature. Reactions were loaded, without loading buffer, into a 7.5% TBE gel. The gel was run at 150 volts/10mA at 4°C until the bromophenol blue in marker lanes was near the bottom of the gel. A phosphoimager plate was exposed to dried gels overnight and visualized using a Storm Phosphoimager.
Cold competition assays were performed as above but with a pre-incubation of 5 minutes followed by the inclusion of unlabeled wild-type or mutant probes at 20, 100, or 200 fmol (10×, 50×, or 100×) and incubation for another 5 minutes before adding radiolabeled probes.
RNA Extraction and qRT-PCR
L2 and L3 staged N2, hbl-1(ve18) and hbl-1(mg285) C. elegans were obtained by isolating eggs from gravid adults and then immediately plating the eggs onto bacteria. The C. elegans were collected and washed 3 times in excess M9 buffer and then were snap frozen and stored at −80°C. RNA was extracted from thawed C. elegans pellets using Ambion's PARIS kit. The RNA was DNAse treated following the manufacturer's protocol using Ambion's DNA-free kit. cDNA was produced from 2 μg of RNA using random hexamer primers and Invitrogen's Superscript III reverse transcriptase, following the manufacturer's protocol. 1 μl of RNAse H treated cDNA was used to amplify and quantitate SL1-spliced pri-let-7 in a 10 μl qPCR reaction using Roche Diagnostic's LightCycler 480 DNA SYBR Green I Master kit. eft-2 was used as a control gene. Primers used in the SL1-spliced pri-let-7 qPCR were A92 (5′ GGTTTAATTACCCAAGTTTGAGgcaa 3′) and A102 (5′ CTTCGAAGAGTTCTGTCTCC 3′). Primers used in the eft-2 qPCR were 158 (5′ TGCCCAGAAGCCGCCGTTGGAG 3′) and 159 (5′ CGGTGTTGGAGCGGAGATCGGCGG 3′). The ratio of the fold change was calculated using an equation from Pfaffl (Pfaffl, 2001).
The same RNA samples used to measure pri-let-7 were used to measure mature let-7 levels. From 10 ng of total RNA, let-7 and an internal control, U18, were measured using Applied Biosystems’ TaqMan MicroRNA Assay following the manufacturer's protocol. Applied Biosystems’ hsa-let-7a and U18 probes were used in the assay. The ratio of the fold change was calculated using an equation from Pfaffl (Pfaffl, 2001).
Results
Deletion of putative hbl-1 sites in the let-7 promoter leads to precocious let-7::gfp expression
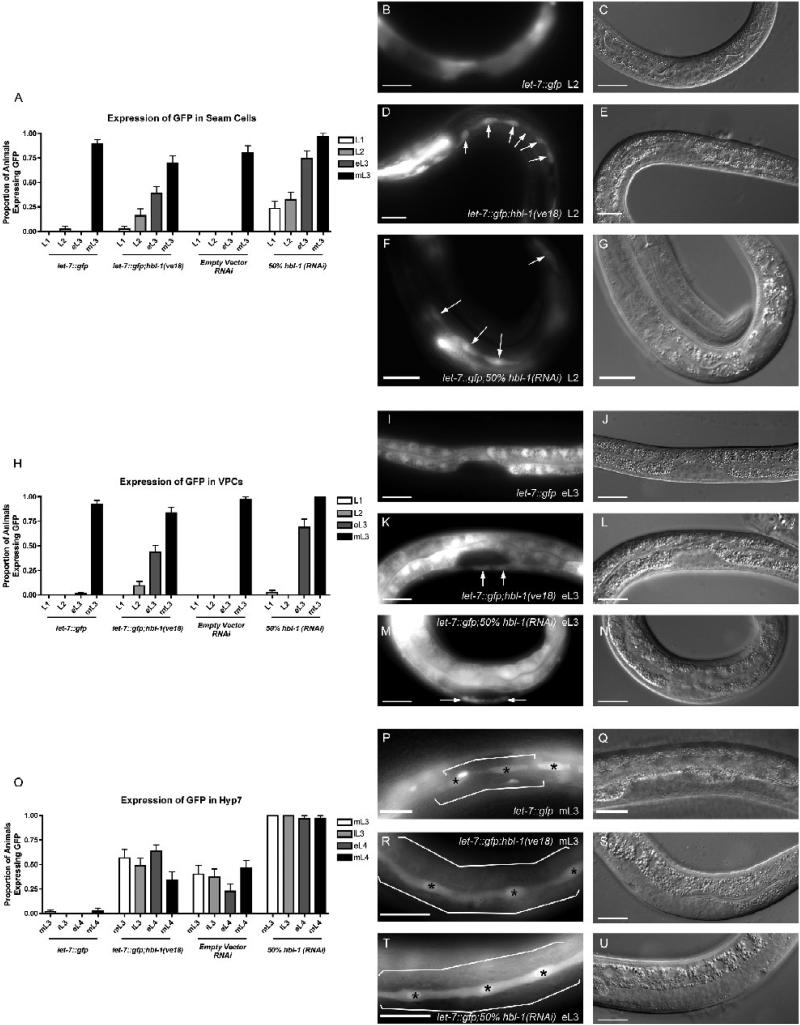
To look for potential transcription factor binding sites in the let-7 gene, we aligned the region ∼1.8 kb upstream of the mature let-7 sequence from four Caenorhabditis species. The alignment revealed a region of high conservation ∼1149 nt upstream of the mature let-7 sequence (Fig 1A and Fig. 1B). When this sequence was computationally analyzed using the TESS transcription factor database (Schug, 1997) (www.cbil.upenn.edu/cgi-bin/tess/tess), a 62 bp region containing 3 highly conserved, putative hunchback binding sites was identified (Fig. 1B and 1C) (Hoch et al., 1991; Qian et al., 1991; Small et al., 1991; Stanojevic et al., 1989; Stanojevic et al., 1991; Treisman and Desplan, 1989; Zuo et al., 1991). These sites begin ∼18 nt downstream of the previously identified TRE element in the promoter, which was responsible for the activation of GFP expression in the seam cells in a let-7 promoter driven reporter strain, let-7::gfp(zaEx5) (Fig. 1A) (Johnson et al., 2003). To examine if this potential Hb/HBL-1 consensus region was involved in regulating let-7 transcription, the putative HBL-1 sites were deleted from the let-7::gfp reporter construct to create Δhbl-1::gfp. This new reporter was injected into C. elegans and three independent transgenic lines were generated. In the wild-type reporter strain, three tissues exhibited temporally expressed let-7::gfp: seam cells, vulval precursor cells (VPCs), and the hypodermal syncytium 7 (hyp7) (Johnson et al., 2003). Therefore, these tissues were examined for changes in temporal GFP expression in Δhbl-1::gfp lines (Fig. 2). Deletion of the HBL-1 sites consistently led to precocious GFP expression in the seam cells and VPCs starting in larval stage 1 (L1) compared to starting in the mid-larval stage 3 (mL3) as with the wild-type promoter (Fig. 2A-2J). In the hyp7, GFP expression was abnormally high in larval stages, contrary to the repression normally observed in the wild-type reporter strain (Fig. 2K-2O). These trends were observed in all three independent lines. Precocious and sustained let-7::gfp expression upon deletion of this region suggests that one or more inhibitory regulatory element(s) exists within this 62 bp region.
Figure 2.
Deletion of the 62 bp region from the let-7::gfp transcriptional reporter causes precocious and sustained GFP expression in transgenic lines. (A,F,K) N ≥ 30 animals for each stage. (A) GFP is expressed precociously in the seam cells of animals where the 62 bp element has been deleted from the reporter construct (Δhbl-1::gfp). (B) GFP image of an L1 let-7::gfp animal showing that there is no GFP expression in the seam cells. (C) Nomarski DIC image showing the stage of the animal in (B). (D) GFP image of an L1 Δhbl-1::gfp animal showing expression in the seam cells, as indicated by arrows. (E) Nomarski DIC image showing the stage of the animal in (D). (F) GFP is expressed precociously in the VPCs of Δhbl-1::gfp animals. (G) GFP image of an L2 let-7::gfp animal showing that there is no GFP expression in the VPCs. (H) Nomarski DIC image showing the stage of the animal in (G). (I) GFP image of an L2 Δhbl-1::gfp animal showing expression in the VPCs, as indicated by arrows. (J) Nomarski DIC image showing the stage of the animal in (I). (K) GFP expression persists in later larval stages in Δhbl-1::gfp animals. (L) GFP image of an mL3 let-7::gfp animal showing no GFP expression in the hyp7, indicated by brackets, while there is expression in the seam cells, indicated by asterisks. (M) Nomarski DIC image showing the stage of the animal in (L). (N) GFP and Nomarski DIC images of an mL3 Δhbl-1::gfp animal showing expression in the hyp7, as indicated by brackets. Asterisks indicate seam cells. Lack of GFP expression in all of the seam cells may be due to mosaicism in the transgenic line. (O) Nomarski DIC image showing the stage of the animal in (N). Scale bars all represent 20 microns. Anterior is left and dorsal is top on all images.
Deletion of putative hbl-1 sites from the let-7 rescue fragment causes precocious alae formation
To test whether this region was biologically important, the 62 bp region was deleted from the let-7 construct that rescued the let-7 retarded alae and lethal mutant phenotypes (Reinhart et al., 2000). Alae are cuticular structures that form along the sides of the animal as it develops from the late L4 stage (lL4) into adulthood. In let-7(lf) mutants, alae formation is delayed until after the animal has passed into adulthood. Injection of the rescue fragment into wild-type C. elegans resulted in some precocious alae developing before the L4 to adult transition (Abrahante et al., 2003; Hayes and Ruvkun, 2006; Reinhart et al., 2000), and a construct that had the lin-4 promoter driving early let-7 expression also showed a high level of precocious alae (Hayes and Ruvkun, 2006). When the rescuing DNA containing the deletion and wild-type sequence were each injected at the same concentration into wild-type animals, there was a significantly higher formation of precocious alae in the early L4 (eL4) in lines containing the deletion construct (Fig. 3A-3E). As observed in previous studies (Abrahante et al., 2003; Lin et al., 2003), depletion of hbl-1 function led to precocious alae formation in eL4 (Fig. 3F and 3G). This increase in precocious alae expression indicates that this region contains biologically relevant, cis-acting regulatory elements and we propose that they correspond to HBL-1 binding sites.
Figure 3.
Deletion of the 62 bp region from the let-7 rescue fragment causes an increase in precocious alae expression. (A) At the eL4 stage, wild-type (N2) animals do not express alae, while wild-type animals injected with the wild-type rescue fragment express precocious alae at a low level. Significantly more wild-type animals injected with a rescue fragment that has the 62 bp region deleted show precocious alae. n ≥ 30 for each strain. Three independent lines were analyzed for each construct. (B) Nomarski DIC images of wild-type eL4 C. elegans showing that no alae are expressed. (C) Nomarski DIC image showing the stage of the animal in (B). (D) Nomarski DIC images of wild-type eL4 C. elegans injected with the let-7 rescue fragment which has the 62 bp region deleted showing precociously expressed alae. Arrows indicate alae. (E) Nomarski DIC image showing the stage of the animal in (D). (F) Wild-type eL4 animal fed 50% hbl-1(RNAi) which shows precocious alae formation similar to precocious alae seen in C. elegans injected with the 62 bp deleted rescue fragment. Arrows indicate alae. Precocious alae in hbl-1(lf) has previously been identified in other studies (Abrahante et al., 2003; Lin et al., 2003). (G) Nomarski DIC image showing the stage of the animal in (F). Scale bars all represent 20 microns. Anterior is left and dorsal is top on all images.
Reduced HBL-1 activity causes precocious let-7::gfp expression
In order to test the requirement of HBL-1 activity for the establishment of the proper temporal pattern of let-7 expression, the let-7::gfp reporter line was crossed into the hbl-1 loss-of-function mutant, hbl-1(ve18), or subjected to hbl-1(RNAi). The RNAi was a mixture of 50% hbl-1(RNAi) vector and 50% empty RNAi vector, which diluted the effect of the RNAi allowing for a higher percentage of animals to survive and easier staging of the animals for analysis. We predicted that if HBL-1 is needed for inhibition of let-7 expression, then reduced HBL-1 function should lead to precocious let-7::gfp expression in transgenic animals.
Similar to the reporter with the putative HBL-1 binding sites removed, reduction of HBL-1 by either the mutant allele or 50% hbl-1(RNAi) led to precocious expression of the let-7::gfp in the seam cells and the VPCs and persistence of GFP expression in the hyp7 (Fig. 4). In the seam cells of let-7::gfp; hbl-1(ve18) animals, increased precocious expression was observed at all larval stages, including L1 (Fig. 4A, 4D and 4E) compared to wild-type controls. A greater proportion of animals with precocious GFP expression in the L1 stage is seen in the seam cells when the reporter strain was grown on 50% hbl-1(RNAi) compared to the cross with the hbl-1 mutant (Fig. 4A, 4F and 4G). In the VPCs of let-7::gfp; hbl-1(ve18) and let-7::gfp; 50% hbl-1(RNAi) animals, there was a small proportion of animals with precocious GFP expression in the L2 and L1, respectively (Fig. 4H). However, most of the precocious expression is observed in the eL3 stage (Fig. 4H-4N). Both let-7::gfp; hbl-1(ve18) and let-7::gfp; 50% hbl-1(RNAi) animals exhibited persistent GFP expression in the hyp7 (Fig. 4O, 4P-U). The precocious and heightened expression, similar to the expression seen in Δhbl-1::gfp indicates that HBL-1 may be involved in inhibiting transcription of let-7 through a 62 bp region containing predicted HBL-1 binding sites.
Figure 4.
Reduced HBL-1 activity causes precocious and sustained let-7::gfp expression similar to Δhbl1::gfp. (A,H,O) N ≥ 30 animals for each stage. (A) When let-7::gfp is crossed with the hbl-1(ve18) loss-of-function mutant or grown on 50% hbl-1(RNAi), there is precocious Δhbl1::gfp expression in the seam cells beginning in L1. (B) GFP image of an L2 let-7::gfp animal showing that there is no GFP expression in the seam cells. (C) Nomarski DIC image showing the stage of the animal in (B). (D) GFP image of an L2 let-7::gfp;hbl-1(ve18) animal showing expression in the seam cells, as indicated by arrows. (E) Nomarski DIC image showing the stage of the animal in (D). (F) GFP image of an L2 let-7::gfp;50% hbl-1(RNAi) animal showing expression in the seam cells, as indicated by arrows. (G) Nomarski DIC image showing the stage of the animal in (F). (H) When let-7::gfp is crossed with the hbl-1(ve18) loss-of-function mutant or grown on 50% hbl-1(RNAi), there is precocious let-7::gfp expression in the VPCs beginning in L2 or eL3, respectively. (I) GFP image of an eL3 let-7::gfp animal showing that there is no GFP expression in the VPCs. (I) Nomarski DIC image showing the stage of the animal in (J). (K) GFP image of an eL3 let-7::gfp;hbl-1(ve18) animal showing expression in the VPCs, as indicated by arrows. (L) Nomarski DIC image showing the stage of the animal in (K). (M) GFP image of an eL3 let-7::gfp;50% hbl-1(RNAi) animal showing expression in the VPCs, as indicated by arrows. (N) Nomarski DIC image showing the stage of the animal in (M). (O) let-7::gfp expression is sustained in the hyp7 in a higher proportion of animals through later larval stages in hbl-1(ve18) loss-of-function mutant or grown on 50% hbl-1(RNAi). It is unknown why there is an increase in GFP detected between let-7::gfp C. elegans fed wild-type bacteria versus bacteria expressing the RNAi empty vector, L4440. Importantly, the same pattern of increased GFP detection was seen when hbl-1 levels were reduced by mutation or RNAi. (P) GFP image of an mL3 let-7::gfp animal showing no GFP expression in the hyp7, indicated by brackets, while there is expression in the seam cells, indicated by asterisks. (Q) Nomarski DIC image showing the stage of the animal in (P). (R,S) GFP image of an mL3 let-7::gfp;hbl-1(ve18) animal showing expression in the hyp7, as indicated by brackets. Seam cells are indicated by asterisks. (S) Nomarski DIC image showing the stage of the animal in (R). (T) GFP image of an mL3 let-7::gfp;50% hbl-1(RNAi) animal showing expression in the hyp7, as indicated by brackets. Seam cells are indicated by asterisks. (U) Nomarski DIC image showing the stage of the animal in (T). Scale bars all represent 20 microns. Anterior is left and dorsal is top on all images.
We speculate that the more penetrant alteration of GFP expression that we observed in the lines containing the deleted cis-region from let-7::gfp compared to the hbl-1 mutants may be due to the fact that both hbl-1(ve18) and 50% hbl-1(RNAi) result in only partial loss-of-function of hbl-1 and not complete abolition. Residual or partially active HBL-1 may be able to maintain some transcriptional inhibition, whereas the removal of the cis-elements from the promoter of the reporter prevented any HBL-1 binding. The null hbl-1 mutant was not used because it is embryonic lethal (National BioResource Program – C. elegans, Japan, unpublished). Nevertheless, the trends observed in the hbl-1(lf) experiments are consistent with the data from Δhbl-1::gfp experiments.
pri- let-7 and mature let-7 levels are increased with hbl-1 loss-of-function
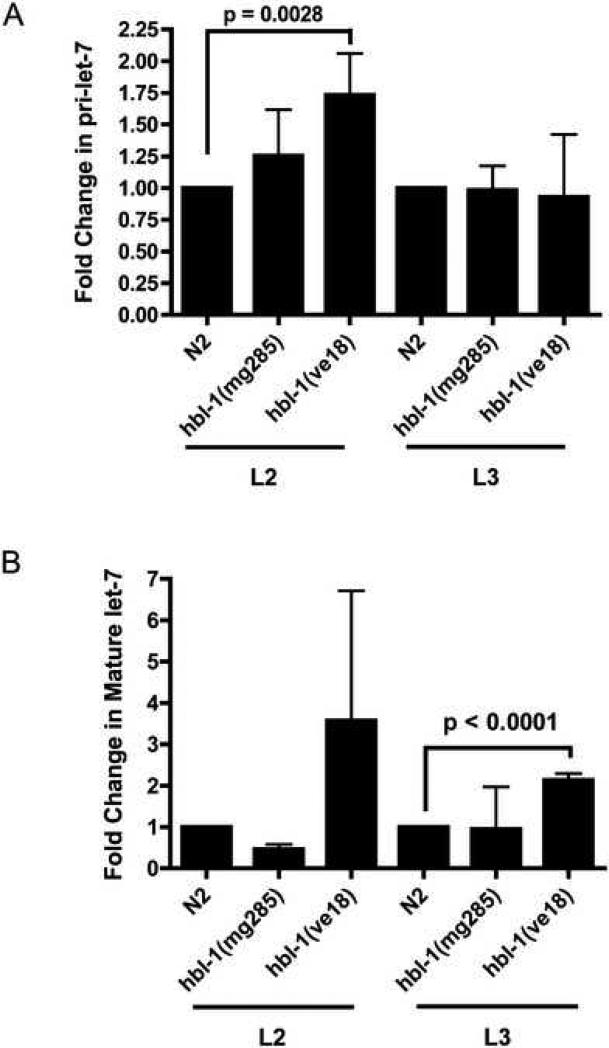
Previous work measuring mature let-7 levels from extracts from the entire body of C. elegans using northern blot analysis showed little effect of the weak hbl-1(mg285) mutation on mature let-7 levels (Lin et al., 2003). However, as our new evidence reveals, there are tissue specific changes in pri-let-7 transcription when HBL-1 activity is decreased. We therefore decided to examine pri-let-7 and mature let-7 levels in hbl-1 loss-of-function mutants. Pri-let-7 levels were measured to more accurately monitor changes in transcription, in contrast to earlier work that only examined mature let-7 levels (Lin et al., 2003). Since C. elegans pri-let-7 is not detectable by northern blot analysis (Bracht et al., 2004), we used qRT-PCR to measure the levels of SL1-spliced pri-let-7 and mature let-7. SL1-spliced pri-let-7 was measured because it is thought to be the form of pri-let-7 that will be processed by Drosha to pre-let-7 (Bracht et al., 2004). The two other pri-let-7 forms are spliced to the SL1 form (Bracht et al., 2004), and so it was assumed that any functional version of these two would be converted to the SL-1 pri-let-7, thus allowing us to measure both forms at once. Using qRT-PCR, we found that pri-let-7 levels are increased at an early larval stage when there is a reduction of HBL-1 activity due to gene mutation (Fig. 5A). Specifically, pri-let-7 levels in hbl-1(ve18) were significantly increased by ∼75% (Fig. 5A) in the L2 stage. While the pri-let-7 levels in hbl-1(mg285) were increased on average by ∼25%, this increase was not statistically significant (Fig. 5A). These modest increases in the amount of pri-let-7 detected are consistent with our observations with the GFP transcriptional reporter showing only a few tissues with changes in pri-let-7 transcription. The increase in pri-let-7 levels combined with precocious expression of the let-7 transcriptional reporter indicate that HBL-1 acts to inhibit let-7 transcription.
Figure 5.
Levels of SL1-spliced pri-let-7 and mature let-7 are increased in hbl-1 loss-of-function mutants when measured by qRT-PCR. (A) The amount of SL1-spliced pri-let-7 during the L2 stage was significantly increased by approximately 75% in the hbl-1(ve18) mutant. The hbl-1(mg285) mutant also showed an increase in the amount of SL1-spliced pri-let-7 present in the L2 stage, but this value was not significant. The SL1-spliced pri-let-7 levels in the hbl-1 mutants return to levels similar to those detected in wild-type C. elegans during the L3 stage. The standard deviations are shown. (B) Levels of mature let-7 are significantly increased by approximately 2-fold during the L3 stage in the hbl-1(ve18) mutant. The increase in let-7 in hbl-1(ve18) in the L2 stage is not statistically significant. The standard deviations are shown. Paired t-tests were used to measure statistical significance. Three technical replicates from two independent biological replicates were assayed for each column in Fig. 5.
When mature let-7 expression was measured by qRT-PCR, levels of let-7 were also significantly increased two-fold in hbl-1(ve18) mutants, however this increase was only statistically significant in the L3 stage (Fig. 5B). The increased mature levels in L3 but not in L2 may represent the processing time from pri-let-7 to mature let-7. Consistent with our previous study (Lin et al., 2003), we did not detect increased mature let-7 levels in hbl-1(mg285) mutants.
The putative HBL-1 sites are specifically bound by a protein found in wild-type C. elegans cell extract
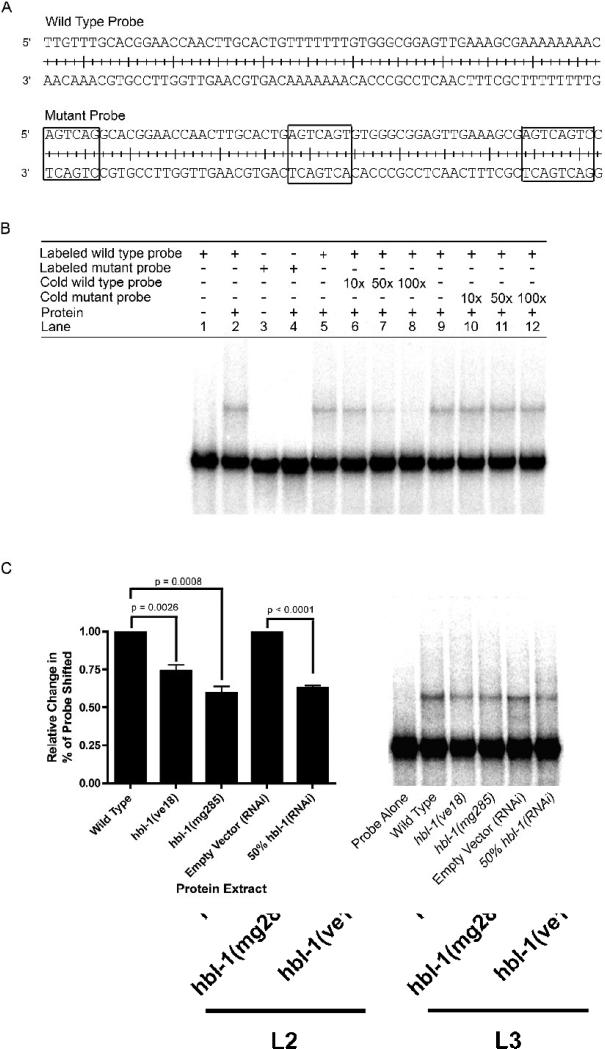
Electrophoretic mobility shift assays (EMSAs) were used to determine if the 62 bp region was specifically bound by a trans-factor in C. elegans whole-cell extract. We found an activity that shifted a radiolabeled 62 nt probe (Fig. 6A and 6B, lanes 1−2). By contrast, a probe with site-specific mutations predicted to disrupt the HBL-1 binding sites did not cause the same shift as the wild-type sequence probe (Fig. 6A and 6B, lanes 3−4). Competition EMSAs were used to show that the binding is specific to the wild-type probe. The wild-type shift can be competed away with increasing amounts of non-radiolabeled, wild-type probe (Fig. 6B, lanes 5−8). However, the non-radiolabeled mutant probe was unable to compete away the shift created with the wild-type probe (Fig. 6B, lanes 9−12), thus showing that the shift seen in the EMSA was specific to the wild-type probe and that the putative HBL-1 binding sites were needed for this shift.
Figure 6.
A probe using the 62 bp region containing putative HBL-1 binding sites is specifically shifted in EMSAs by wild-type C. elegans protein extracts, and this shift is significantly decreased when extracts from hbl-1 loss-of-function mutants and 50% hbl-1(RNAi) are used. (A) Wild-type and mutant probe sequences used in EMSAs. Boxed sequences in the mutant probe indicate the mutations and location of potential HBL-1 sites. (B) Lanes 1−4, the wild-type probe, but not a probe with mutated HBL-1 binding sites, is shifted by C. elegans protein extract. Lanes 5−8, increasing amount of non-radiolabeled, wild-type probe (10, 50 and 100× greater than labeled probe) can compete away the labeled probe's shift. Lanes 9−12, increasing amounts of non-radiolabeled mutant probe cannot compete away wild-type probe's shift. (C) EMSAs using protein extracts from hbl-1(ve18), hbl-1(mg285) and 50% hbl-1(RNAi), have a significant decrease in the amount of probe shifted. n = 5, a representative gel is shown. Paired t-tests were used to measure statistical significance.
When protein extracts from hbl-1 loss-of-function mutants, ve18 and mg285, or animals grown on 50% hbl-1(RNAi) were used in EMSAs, the loss of hbl-1 caused a significant decrease (26% for hbl-1(ve18),p=0.0026; 41% for hbl-1(mg285), p=0.0008; 37% for 50% hbl-1(RNAi), p<0.0001) in the amount of shift observed (Fig. 6C). Since these extracts were not from null alleles, it is not surprising that there was not a complete loss of the shift. In fact, the hbl-1(ve18) and hbl-1(mg285) mutants are still predicted to have at least some of their zinc-finger domains intact (Abrahante et al., 2003; Lin et al., 2003) The 50% RNAi was also not expected to have a complete loss of shift since RNAi does not cause 100% loss of a protein and the RNAi used in these experiments was diluted to decrease the effect of the RNAi. Interestingly, using a diluted hbl-1(RNAi) led to a similar decrease in levels of probe shifted as the partial loss-of-function hbl-1 mutants suggesting that using 50% hbl-1(RNAi) phenocopies the partial loss-of-function mutants.
Discussion
We identified a region in the C. elegans let-7 promoter that is conserved in other nematode species and contains potential HBL-1 binding sites. This region was shown to be important for inhibition of expression of a reporter construct driven by the let-7 promoter in the early larval stages. Removal of this region from the let-7 rescue construct leads to precocious alae formation, thus indicating that the deleted region has biological importance. Reduction of hbl-1 activity leads to similar precocious reporter expression and precocious alae formation. Reduction of hbl-1 also leads to an increase in the levels of pri-let-7 and mature let-7 when measured by qRT-PCR. Furthermore, this element is specifically bound by a protein in C. elegans cell extract in an EMSA. The amount of probe shifted in the EMSA is significantly decreased when hbl-1 loss-of-function extracts are used. Taken together, the data presented in this study strongly suggest that HBL-1 is the transcription factor that inhibits let-7 expression in early larval stages and that it does so by binding a 62 bp transcriptional regulatory region in its promoter. An alternative explanation of the data is that HBL-1 affects some other transcription factor with similar binding sites to itself.
The results presented here indicate that let-7's target in the ventral cord neurons and anterior nerve ring, HBL-1 (Abrahante et al., 2003; Lin et al., 2003) acts to transcriptionally inhibit the expression of let-7 in the hypodermis of C. elegans. This inhibition helps to control the expression of let-7 in tissues where let-7 has a temporally regulated expression pattern. This control ensures that let-7 is expressed at the correct transition times for normal progression of cell fates into the later larval stages and adulthood. The evidence suggests that HBL-1 directly binds to the let-7 promoter, and we propose that let-7 is thus the first transcriptional target identified for HBL-1 in C. elegans. Furthermore, the data presented here combined with that in previous studies present an interesting model for temporal regulation in the hypodermis where let-7 family members mir-48, -84, and -241 down-regulate hbl-1 in the L3 stage, which then removes HBL- 1's transcriptional inhibition of another family member, let-7 (Abbott et al., 2005). As let-7 is expressed, it then promotes the transition from L4 to adulthood in the hypodermis (Reinhart et al., 2000).
hbl-1 is transcribed in the seam cells, but one complication in this model is the report, using a hbl-1p::gfp::hbl-1 reporter construct, that hbl-1 expression decreases after the middle of the L1 stage (Abbott et al., 2005; Abrahante et al., 2003; Fay et al., 1999; Lin et al., 2003), at least 1 stage before let-7 is expressed in seam cells. One possibility is that some additional protein, for example one binding to the TRE, is also required for full let-7 expression. However, there is evidence that HBL-1 functions in seam cells after the L1 stage to regulate division of the seam cells at the L2 to L3 transition (Abbott et al., 2005; Abrahante et al., 2003; Fay et al., 1999). This evidence suggests that there is HBL-1 protein present in seam cells, which is not accurately revealed by the transcriptional reporter. Alternatively, since HBL-1 is expressed into the L3 stage in the hyp7 (Abbott et al., 2005; Abrahante et al., 2003; Lin et al., 2003), it has also been proposed that HBL-1 can act non-cell autonomously from the hyp7 to affect the seam cells and vulval precursor cells (Abbott et al., 2005; Abrahante et al., 2003; Fay et al., 1999).
hbl-1 inhibiting let-7 transcription could explain the mixed heterochronic phenotypes of hbl-1 and why loss of hbl-1 will partially suppress let-7 hypodermal phenotypes even though let-7 does not appear to regulate hbl-1 in the hypodermis (Abrahante et al., 2003; Lin et al., 2003). Our research indicates that let-7 and hbl-1 could regulate each other through a negative feedback loop mechanism, which has been proposed as a mechanism for other miRNAs and their targets (Johnston and Hobert, 2005; Martinez et al., 2008), to control later larval development. Higher levels of let-7 earlier in development seen in the hbl-1 mutants could explain the precocious alae phenotype seen here and perhaps in the transgenic lines expressing the let-7 rescue fragment with the putative HBL-1 binding sites deleted (Fig. 3).
Our data also provides a genetic connection between the earlier and later stages of the heterochronic gene pathway. hbl-1 is down-regulated in early larval stages in hyp-7 by let-7 family members in what is referred to as the “early timer” (Abbott et al., 2005) and is again down-regulated in neurons at late larval stages by let-7 itself (Abrahante et al., 2003; Lin et al., 2003) as part of the “late timer”. Until now what connected these timers during the L3 stage was unknown. This work demonstrates that hbl-1 regulation of let-7 expression could be the link between the two timers.
The high conservation of the heterochronic gene pathway between species suggests that HBL-1's transcriptional regulation of let-7 may be a mechanism utilized in other organisms. In fact, there are several potential hb binding sites upstream of Drosophila dme-let-7 (data not shown). Based on the data presented in this study, it would be interesting to see if HBL-1 and these sites are also important for dme-let-7 expression.
Acknowledgements
The authors would like to thank R. Niwa, G. Stefani, S. Chan, and M. Turner for many helpful discussions, reagents, and critical reading of the manuscript. FJS was supported by NIH (R01-GM064701).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abbott AL, Alvarez-Saavedra E, Miska EA, Lau NC, Bartel DP, Horvitz HR, Ambros V. The let-7 MicroRNA family members mir-48, mir-84, and mir-241 function together to regulate developmental timing in Caenorhabditis elegans. Dev Cell. 2005;9:403–14. doi: 10.1016/j.devcel.2005.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abrahante JE, Daul AL, Li M, Volk ML, Tennessen JM, Miller EA, Rougvie AE. The Caenorhabditis elegans hunchback-like gene lin-57/hbl-1 controls developmental time and is regulated by microRNAs. Dev Cell. 2003;4:625–37. doi: 10.1016/s1534-5807(03)00127-8. [DOI] [PubMed] [Google Scholar]
- Ambros V. A hierarchy of regulatory genes controls a larva-to-adult developmental switch in C. elegans. Cell. 1989;57:49–57. doi: 10.1016/0092-8674(89)90171-2. [DOI] [PubMed] [Google Scholar]
- Baskerville S, Bartel DP. Microarray profiling of microRNAs reveals frequent coexpression with neighboring miRNAs and host genes. Rna. 2005;11:241–7. doi: 10.1261/rna.7240905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohnsack MT, Czaplinski K, Gorlich D. Exportin 5 is a RanGTP-dependent dsRNA-binding protein that mediates nuclear export of pre-miRNAs. Rna. 2004;10:185–91. doi: 10.1261/rna.5167604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bracht J, Hunter S, Eachus R, Weeks P, Pasquinelli AE. Trans-splicing and polyadenylation of let-7 microRNA primary transcripts. Rna. 2004;10:1586–94. doi: 10.1261/rna.7122604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang TC, Yu D, Lee YS, Wentzel EA, Arking DE, West KM, Dang CV, Thomas-Tikhonenko A, Mendell JT. Widespread microRNA repression by Myc contributes to tumorigenesis. Nat Genet. 2008;40:43–50. doi: 10.1038/ng.2007.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang TC, Zeitels LR, Hwang HW, Chivukula RR, Wentzel EA, Dews M, Jung J, Gao P, Dang CV, Beer MA, Thomas-Tikhonenko A, Mendell JT. Lin-28B transactivation is necessary for Myc-mediated let-7 repression and proliferation. Proc Natl Acad Sci U S A. 2009;106:3384–9. doi: 10.1073/pnas.0808300106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denli AM, Tops BB, Plasterk RH, Ketting RF, Hannon GJ. Processing of primary microRNAs by the Microprocessor complex. Nature. 2004;432:231–5. doi: 10.1038/nature03049. [DOI] [PubMed] [Google Scholar]
- Esquela-Kerscher A, Slack FJ. Oncomirs - microRNAs with a role in cancer. Nat Rev Cancer. 2006;6:259–69. doi: 10.1038/nrc1840. [DOI] [PubMed] [Google Scholar]
- Fay DS, Stanley HM, Han M, Wood WB. A Caenorhabditis elegans homologue of hunchback is required for late stages of development but not early embryonic patterning. Dev Biol. 1999;205:240–53. doi: 10.1006/dbio.1998.9096. [DOI] [PubMed] [Google Scholar]
- Gregory RI, Yan KP, Amuthan G, Chendrimada T, Doratotaj B, Cooch N, Shiekhattar R. The Microprocessor complex mediates the genesis of microRNAs. Nature. 2004;432:235–40. doi: 10.1038/nature03120. [DOI] [PubMed] [Google Scholar]
- Han M. Wormbase WS198. 2008 [Google Scholar]
- Hayes GD, Ruvkun G. Misexpression of the Caenorhabditis elegans miRNA let-7 is sufficient to drive developmental programs. Cold Spring Harb Symp Quant Biol. 2006;71:21–7. doi: 10.1101/sqb.2006.71.018. [DOI] [PubMed] [Google Scholar]
- Heo I, Joo C, Cho J, Ha M, Han J, Kim VN. Lin28 mediates the terminal uridylation of let-7 precursor MicroRNA. Mol Cell. 2008;32:276–84. doi: 10.1016/j.molcel.2008.09.014. [DOI] [PubMed] [Google Scholar]
- Hoch M, Seifert E, Jackle H. Gene expression mediated by cis-acting sequences of the Kruppel gene in response to the Drosophila morphogens bicoid and hunchback. EMBO J. 1991;10:2267–78. doi: 10.1002/j.1460-2075.1991.tb07763.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isshiki T, Pearson B, Holbrook S, Doe CQ. Drosophila neuroblasts sequentially express transcription factors which specify the temporal identity of their neuronal progeny. Cell. 2001;106:511–21. doi: 10.1016/s0092-8674(01)00465-2. [DOI] [PubMed] [Google Scholar]
- Johnson SM, Lin SY, Slack FJ. The time of appearance of the C. elegans let-7 microRNA is transcriptionally controlled utilizing a temporal regulatory element in its promoter. Dev Biol. 2003;259:364–79. doi: 10.1016/s0012-1606(03)00202-1. [DOI] [PubMed] [Google Scholar]
- Johnston RJ, Jr., Hobert O. A novel C. elegans zinc finger transcription factor, lsy-2, required for the cell type-specific expression of the lsy-6 microRNA. Development. 2005;132:5451–60. doi: 10.1242/dev.02163. [DOI] [PubMed] [Google Scholar]
- Kamath RS, Fraser AG, Dong Y, Poulin G, Durbin R, Gotta M, Kanapin A, Le Bot N, Moreno S, Sohrmann M, Welchman DP, Zipperlen P, Ahringer J. Systematic functional analysis of the Caenorhabditis elegans genome using RNAi. Nature. 2003;421:231–7. doi: 10.1038/nature01278. [DOI] [PubMed] [Google Scholar]
- Kloosterman WP, Plasterk RH. The diverse functions of microRNAs in animal development and disease. Dev Cell. 2006;11:441–50. doi: 10.1016/j.devcel.2006.09.009. [DOI] [PubMed] [Google Scholar]
- Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–54. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- Lee Y, Kim M, Han J, Yeom KH, Lee S, Baek SH, Kim VN. MicroRNA genes are transcribed by RNA polymerase II. EMBO J. 2004;23:4051–60. doi: 10.1038/sj.emboj.7600385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehmann R, Nusslein-Volhard C. hunchback, a gene required for segmentation of an anterior and posterior region of the Drosophila embryo. Dev Biol. 1987;119:402–17. doi: 10.1016/0012-1606(87)90045-5. [DOI] [PubMed] [Google Scholar]
- Lin SY, Johnson SM, Abraham M, Vella MC, Pasquinelli A, Gamberi C, Gottlieb E, Slack FJ. The C elegans hunchback homolog, hbl-1, controls temporal patterning and is a probable microRNA target. Dev Cell. 2003;4:639–50. doi: 10.1016/s1534-5807(03)00124-2. [DOI] [PubMed] [Google Scholar]
- Martinez NJ, Ow MC, Barrasa MI, Hammell M, Sequerra R, Doucette-Stamm L, Roth FP, Ambros VR, Walhout AJ. A C. elegans genome-scale microRNA network contains composite feedback motifs with high flux capacity. Genes Dev. 2008;22:2535–49. doi: 10.1101/gad.1678608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moss EG. Heterochronic genes and the nature of developmental time. Curr Biol. 2007;17:R425–34. doi: 10.1016/j.cub.2007.03.043. [DOI] [PubMed] [Google Scholar]
- Newman MA, Thomson JM, Hammond SM. Lin-28 interaction with the Let-7 precursor loop mediates regulated microRNA processing. Rna. 2008 doi: 10.1261/rna.1155108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasquinelli AE, Reinhart BJ, Slack F, Martindale MQ, Kuroda MI, Maller B, Hayward DC, Ball EE, Degnan B, Muller P, Spring J, Srinivasan A, Fishman M, Finnerty J, Corbo J, Levine M, Leahy P, Davidson E, Ruvkun G. Conservation of the sequence and temporal expression of let-7 heterochronic regulatory RNA. Nature. 2000;408:86–9. doi: 10.1038/35040556. [DOI] [PubMed] [Google Scholar]
- Pearson BJ, Doe CQ. Regulation of neuroblast competence in Drosophila. Nature. 2003;425:624–8. doi: 10.1038/nature01910. [DOI] [PubMed] [Google Scholar]
- Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29:e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piskounova E, Viswanathan SR, Janas M, Lapierre RJ, Daley GQ, Sliz P, Gregory RI. Determinants of microRNA processing inhibition by the developmentally regulated RNA-binding protein Lin28. J Biol Chem. 2008 doi: 10.1074/jbc.C800108200. [DOI] [PubMed] [Google Scholar]
- Qian S, Capovilla M, Pirrotta V. The bx region enhancer, a distant cis-control element of the Drosophila Ubx gene and its regulation by hunchback and other segmentation genes. EMBO J. 1991;10:1415–25. doi: 10.1002/j.1460-2075.1991.tb07662.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhart BJ, Slack FJ, Basson M, Pasquinelli AE, Bettinger JC, Rougvie AE, Horvitz HR, Ruvkun G. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature. 2000;403:901–6. doi: 10.1038/35002607. [DOI] [PubMed] [Google Scholar]
- Rodriguez A, Griffiths-Jones S, Ashurst JL, Bradley A. Identification of mammalian microRNA host genes and transcription units. Genome Res. 2004;14:1902–10. doi: 10.1101/gr.2722704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roush S, Slack FJ. The let-7 family of microRNAs. Trends Cell Biol. 2008;18:505–16. doi: 10.1016/j.tcb.2008.07.007. [DOI] [PubMed] [Google Scholar]
- Rybak A, Fuchs H, Smirnova L, Brandt C, Pohl EE, Nitsch R, Wulczyn FG. A feedback loop comprising lin-28 and let-7 controls pre-let-7 maturation during neural stem-cell commitment. Nat Cell Biol. 2008 doi: 10.1038/ncb1759. [DOI] [PubMed] [Google Scholar]
- Sampson VB, Rong NH, Han J, Yang Q, Aris V, Soteropoulos P, Petrelli NJ, Dunn SP, Krueger LJ. MicroRNA let-7a down-regulates MYC and reverts MYC-induced growth in Burkitt lymphoma cells. Cancer Res. 2007;67:9762–70. doi: 10.1158/0008-5472.CAN-07-2462. [DOI] [PubMed] [Google Scholar]
- Schug J. a. O., G. C. TESS: Transcription Element Search Software on the WWW, Technical Report CBIL-TR-1997−1001-v0.0. University of Pennsylvania; 1997. [Google Scholar]
- Schulz C, Tautz D. Autonomous concentration-dependent activation and repression of Kruppel by hunchback in the Drosophila embryo. Development. 1994;120:3043–9. doi: 10.1242/dev.120.10.3043. [DOI] [PubMed] [Google Scholar]
- Shi XB, Tepper CG, White RW. Cancerous miRNAs and their regulation. Cell Cycle. 2008:7. doi: 10.4161/cc.7.11.5977. [DOI] [PubMed] [Google Scholar]
- Small S, Kraut R, Hoey T, Warrior R, Levine M. Transcriptional regulation of a pair-rule stripe in Drosophila. Genes Dev. 1991;5:827–39. doi: 10.1101/gad.5.5.827. [DOI] [PubMed] [Google Scholar]
- Stanojevic D, Hoey T, Levine M. Sequence-specific DNA-binding activities of the gap proteins encoded by hunchback and Kruppel in Drosophila. Nature. 1989;341:331–5. doi: 10.1038/341331a0. [DOI] [PubMed] [Google Scholar]
- Stanojevic D, Small S, Levine M. Regulation of a segmentation stripe by overlapping activators and repressors in the Drosophila embryo. Science. 1991;254:1385–7. doi: 10.1126/science.1683715. [DOI] [PubMed] [Google Scholar]
- Stefani G, Slack FJ. Small non-coding RNAs in animal development. Nat Rev Mol Cell Biol. 2008;9:219–30. doi: 10.1038/nrm2347. [DOI] [PubMed] [Google Scholar]
- Struhl G, Johnston P, Lawrence PA. Control of Drosophila body pattern by the hunchback morphogen gradient. Cell. 1992;69:237–249. doi: 10.1016/0092-8674(92)90405-2. [DOI] [PubMed] [Google Scholar]
- Timmons L, Court DL, Fire A. Ingestion of bacterially expressed dsRNAs can produce specific and potent genetic interference in Caenorhabditis elegans. Gene. 2001;263:103–12. doi: 10.1016/s0378-1119(00)00579-5. [DOI] [PubMed] [Google Scholar]
- Treisman J, Desplan C. The products of the Drosophila gap genes hunchback and Kruppel bind to the hunchback promoters. Nature. 1989;341:335–7. doi: 10.1038/341335a0. [DOI] [PubMed] [Google Scholar]
- Vasudevan S, Tong Y, Steitz JA. Switching from repression to activation: microRNAs can up-regulate translation. Science. 2007;318:1931–4. doi: 10.1126/science.1149460. [DOI] [PubMed] [Google Scholar]
- Vasudevan S, Tong Y, Steitz JA. Cell-cycle control of microRNA-mediated translation regulation. Cell Cycle. 2008;7:1545–9. doi: 10.4161/cc.7.11.6018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viswanathan SR, Daley GQ, Gregory RI. Selective blockade of microRNA processing by Lin28. Science. 2008;320:97–100. doi: 10.1126/science.1154040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi R, Qin Y, Macara IG, Cullen BR. Exportin-5 mediates the nuclear export of pre-microRNAs and short hairpin RNAs. Genes Dev. 2003;17:3011–6. doi: 10.1101/gad.1158803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo P, Stanojevic D, Colgan J, Han K, Levine M, Manley JL. Activation and repression of transcription by the gap proteins hunchback and Kruppel in cultured Drosophila cells. Genes Dev. 1991;5:254–64. doi: 10.1101/gad.5.2.254. [DOI] [PubMed] [Google Scholar]