Abstract
Background
Positive selection, sorting, and collection of single cells from within a heterogeneous population are required for many biological studies. We recently demonstrated a miniaturized cell array for this purpose; however, on-chip pre-enrichment and isolation of specific target cells would provide significant value for cell isolation.
Methods
In the current work, mixed cell samples of fewer than 30,000 cells were used for panning by means of on-array antibody-capture to pre-enrich the target population. The cell surface receptors FcεR1, c-Kit, and ErbB2 were used for positive selection of RBL, RBL, and SK-BR-3 cells, respectively, from the mixed population. The capture efficiency, selectivity, and enrichment for the target cells were calculated and compared to fibronectin-coated controls.
Results
As expected, the capture efficiency depended upon the frequency of the target cell in the mixed population over the range of 0.3% to 33%. For a frequency of 5% target cells, the capture efficiency was 39 – 53% for the three conditions, while the selectivity varied between 78 – 98% with 16 – 20 fold enrichment. Furthermore, single-cell cloning studies demonstrated a high cloning efficiency of target cells selectively isolated from the array.
Conclusions
Antibody-based pre-enrichment in combination with micropallet-based cell selection will be a valuable tool for isolation and expansion of rare cells from small heterogeneous populations.
Keywords: Microarray, micropallet, SU-8, antibody panning, IgE, FcεR1, c-Kit, CD117, ErbB2, single cell
Positive selection, sorting, and collection of single or small groups of cells from within a heterogeneous population are common and essential procedures in most areas of biomedical research (1-6). A few examples are selective isolation and cloning of genetically modified cells for genomic and proteomic studies (7), separation of undifferentiated and differentiated cells in stem cell studies (8), and sorting of cells from patient samples for developing new cell lines (9). To achieve these goals, many techniques such as fluorescence-activated cell sorting (FACS), limiting dilution, cloning rings, panning, column chromatography, magnetic sorting (e.g., Dynabeads), and dielectrophoresis have been used successfully for sorting nonadherent cells using both bench-top and microchip platforms (10-16). Unfortunately, sorting of adherent cells is more challenging due to the cells’ natural propensity to attach to surfaces. Although strategies such as disaggregation using enzymes or mechanical forces have been used to create cell suspensions, these techniques provide significant disadvantages for the sorting procedure including loss of cell morphology, removal of cell surface markers, damage to cell membranes, alterations in cellular physiology, and loss of viability (17-22). Cell separation by Histopaque centrifugation can be used to generate enriched populations, but this density-based method is limited when target and nontarget cells possess the same or very similar densities (23). In addition, most of these sorting techniques are tedious and time consuming, and in most cases, the sort must begin with a large number of cells (typically 105-106 for FACS and 107 cells for sorting by magnetic beads) (24,25).
To address these concerns for sorting of adherent cells, new technologies have been developed to select and isolate cells while they remain adherent to their growth substrate (26-28). The reported sorting strategies utilizing laser microdissection (LM) or aspiration of colonies of loosely adherent cells (27,29) have limitations such as applicability primarily for fixed or frozen samples, low-throughput, stressful handling of live cells, or limitation to specific types of adherent cells (27,30,31). To overcome some of the challenges listed above, we recently reported a new strategy using arrays of releasable, microfabricated pallets to isolate and clone cells from a microarray while the cells remain adherent to their growth surface (32-36). The cells were localized to the surfaces of the micropallets by air or poly(ethylene glycol) walls to segregate each micropallet (32,33). The system utilized a 5-ns pulsed laser to selectively release individual micropallets composed of SU-8 or 1002F (32,34). The released micropallets with their attached cells were collected in microwell plates and then clonally expanded after collection (35).
Prior studies also demonstrated the ability to coat micropallet surfaces with substrates appropriate for cell adhesion, such as fibronectin or collagen (32,34,35,37). Other investigators have shown the utility of using antibodies to create surfaces for capturing target cells from a mixed population (38-40). It was reasoned that by using a capture agent directed at a cell-surface protein as a coating on the micropallet, a pre-sort to enrich the numbers of attached target cells could be accomplished on-chip. The goal of the current work was to enhance the utility of the micropallet array in cell cloning by combining surface-directed target cell capture with micropallet-based cell sorting. In this work, antibody-coated microarrays were evaluated for capture and enrichment of target cells from a mixed population prior to the cell isolation step. The micron-scale pallets were fabricated on a glass surface from SU-8, an inexpensive biocompatible polymer. Three different antibody:cell interactions were tested: IgE:rat basophilic leukemia (RBL) cells, anti-c-Kit-IgG:RBL cells and anti-ErbB2-IgG:SK-BR-3 cells were evaluated to establish the feasibility of the concept. Antibodies were immobilized on the micropallet surfaces to capture cells expressing a binding partner for the antibody. Cell capture was then screened on the microarrays, and individual micropallets with their attached target cell were released. The target cells were collected, cultured, and grown into clonal colonies. Quantitative studies of microarray capture efficiency, selectivity and enrichment are reported.
Materials and Methods
Materials
SU-8 photoresist and SU-8 developer were purchased from MicroChem Corp. (Newton, MA). The Sylgard 184 silicone elastomer kit was purchased from Dow Corning (Midland, MI). (Heptadecafluoro-1,1,2,2-tetrahydrodecyl) trichlorosilane was from Gelest Inc. (Morrisville, PA). Alexa Fluor 647 labeled bovine serum albumin (BSA-Alexa 647, excitation 650 nm, emission 668 nm), 2, 4-dinithrophenol conjugated BSA (BSA-DNP), rat IgE against-DNP (anti-DNP IgE), mouse anti-CD117 (anti-c-Kit) specific for the N-terminal Ig domain of the c-Kit receptor, Dulbecco’s Modified Eagle Medium (DMEM), fetal bovine serum (FBS) and penicillin/streptomycin were obtained from Invitrogen (Carlsbad, CA). ATCC-formulated McCoy’s 5a medium was obtained from the American Type Culture Collection, ATCC (Manassas, VA). Cy5 mouse anti-c-Kit (Cy5 anti-c-Kit, excitation 633 nm, emission 670 nm) was purchased from eBioscience (San Diego, CA). Mouse antibody against ErbB2 (anti-ErbB2) and goat-anti-mouse IgG Cy5 (anti-IgG Cy5, excitation 650 nm, emission 667 nm) were purchased from Abcam (Cambridge, MA). L-glutamine and human plasma fibronectin were obtained from Sigma-Aldrich (St. Louis, MO). All other reagents and materials were obtained from Fisher Scientific (Pittsburgh, PA).
Cell Lines
SK-BR-3 (a human breast cancer cell line) and rat basophilic leukemia (RBL) cell lines were purchased from ATCC. 3T3 cells expressing green fluorescence protein (GFP-3T3, excitation 480 nm, emission 535 nm) and HeLa cells stably expressing GFP-tagged histone H1 (GFP-HeLa, excitation 480 nm, emission 535 nm) were the kind gift of Eva Lee at the University of California, Irvine.
Fabrication of SU-8 Microarray on a Glass Substrate
Micropallets composed of SU-8 (50 μm sides, ~20 μm gap, and ~50 μm height) were fabricated on a glass slide as described previously (37). The formation of a hydrophobic perfluoroalkylsilane layer on the silicone oxide surface was carried out in a low-pressure reactor (37). The SU-8 microarrays were stored in the vacuum desiccator until use.
Polydimethylsiloxane (PDMS) O-rings (I.D./O.D.~18/35 mm) were constructed from Sylgard 184 cured in a plastic mold. The mold was made simply by putting a Teflon ring (O.D.~18 mm) inside a polystyrene Petri dish (35 mm diameter). After pouring the PDMS into the mold, the assembly was heated in a 65°C oven for at least 30 min, after which time the assembly was cooled and the solid O-ring extracted from the mold. A culture chamber was constructed by using a thin layer of Sylgard 184 to glue the fabricated O-ring to a microarray. To facilitate attachment, the entire assembly was heated on a 65°C hot plate for ~20 min. The microarray with attached cell chamber contained 44,000 micropallets. The number and size of micropallets were kept constant during experiments. Immediately before use, the microarrays with O-rings were sterilized by rinsing with 95% ethanol, and then dried in a tissue culture hood before use.
Coating of SU-8 Surface with Proteins
In different sets of experiments, 1 mL of BSA-Alexa 647, BSA-DNP, anti-c-Kit, anti-c-Kit-Cy5, and anti-ErbB2 (50 μg/mL) were added to culture chambers containing a microarray and incubated overnight (~16 h). In control experiments used for cell mixtures, 1 mL of fibronectin (20 μg/mL) was added to the culture-chamber/microarray assembly and incubated overnight. Coated arrays were rinsed five times with phosphate buffered saline (PBS) prior to cell culture experiments. The fluorescence intensity of BSA-Alexa 647- and anti-c-Kit-Cy5-coated microarrays was measured using an inverted epifluorescence microscope (TE 300, Nikon, Melville, NY) and a cooled CCD camera (Photometrix Coolsnap FX, Tucson, AZ). Fluorescence data were collected using Metafluor software (Universal Imaging Corporation, Downingtown, PA). For those experiments in which the proteins BSA-DNP and anti-ErbB2 were used to coat the micropallets, the fluorescent secondary antibodies anti-DNP IgE and anti-IgG-Cy5 were used to label the adsorbed protein. In this case, the microarrays were initially coated with the proteins BSA-DNP and anti-ErbB2, respectively, as described above. The arrays were then rinsed with PBS, and the secondary anti-DNP IgE and anti-IgG-Cy5 antibodies were immobilized on the array by incubating with the appropriate antibody (20 μg/mL, 1 mL) for 2 h. Samples were then rinsed five times with PBS before cell culture. All coating steps were performed under sterile conditions in a tissue culture hood.
Cell Culture
All cells were maintained at 37°C in a humidified 5% CO2 atmosphere in their respective culture medium. RBL, GFP-HeLa and GFP-3T3 cells were cultured in DMEM supplemented with 10% FBS, 584 mg/L L-glutamine, 100 units/mL penicillin and 100 μg/mL streptomycin. ATCC-formulated McCoy’s 5a medium with 10% FBS was used to culture SK-BR-3 cell lines. For experiments involving mixtures of SK-BR-3 with other cell types, a 1:1 mixture of the two types of media was used for cell culture after plating of cells on the microarray (see below). For all experiments, a total of 30,000 cells were plated on the array. Plating of this cell number was arrived at empirically to provide a majority of pallets with ≤1 cell per pallet.
Cell Collection
An O-ring identical to that constructed for use with the microarrays was attached to a polystyrene Petri dish (60 × 15 mm) using the protocol described above. This O-ring/Petri dish chamber with polystyrene base was used to collect released pallets and their attached cells (see below). Before use, the chamber was rinsed with ethanol and then five times with PBS. The chamber was then coated with 1 mL of 20 μg/mL fibronectin in PBS overnight immediately prior to use. All manipulations were performed under sterile conditions to ensure cell viability and prevent contamination of the cells under study before and after expansion.
Laser-Assisted Release of Micropallets and Cell Collection
Micropallets containing an attached target cell were selected and released using a laser-based release technology developed previously (32). Briefly, a frequency-doubled Q-switched Nd:YAG laser (Minilite™ II, Continuum, Santa Clara, CA) was used to generate a single laser pulse (5-ns pulse width, TEM00, 532 nm), which was spatially expanded to a 4-mm diameter (Oriel Beam Expander). A polarizer (DP-100-VIS1, Thorlabs, Newton, NJ) was used to vary the beam energy (~2 μJ in the current experiments), and the energy was verified using a laser energy meter (EPM 1000, Molectron, Santa Clara, CA). The beam was directed into the microscope, and was focused at the micropallet-glass interface using a 20X microscope objective (0.5 NA, Nikon).
Prior to laser release, the microarray was rinsed with fresh culture medium five times to remove nonadherent cells, and then 1 mL of cell culture medium was added to the microarray chamber. The collection chamber was rinsed with fresh culture medium, and placed directly above the pallet chamber in a sterile environment with the two O-rings of the collection chamber and the microarray opposed. This assembly was placed on the microscope stage and selected cells/micropallets were released with the pulsed laser. The assembly was then inverted to transfer the media and released micropallets into the collection chamber as described previously (35). The collection chamber and microarray assembly were separated under sterile conditions and the collection chamber was covered and transferred into a tissue culture incubator. The growth of collected cells on released micropallets was then observed over time by microscopy.
Capture Efficiency
Determination of the efficiency of antibody-coated microarrays for capturing RBL and SK-BR-3 cells either alone or as a mixture with GFP-HeLa and GFP-3T3 cells were performed as follows. After preparation of the microarray for cell plating, cells were introduced into the microarray chamber. In each experiment, cells (as a pure or mixed population, see “Results and Discussion”) were plated by placing 1.5 mL of cell suspension in the culture chamber housing the microarray. After being placed on microarrays, cells were allowed to recover and attach for 30 min in the appropriate media under standard tissue culture conditions. Cells cultured on microarrays were microscopically imaged by transillumination and fluorescence. The efficiency and selectivity of microarrays for capturing target cells (i.e., RBL and SK-BR-3) were evaluated by inspecting corresponding optical and fluorescence micrographs. The microarray cell capture efficiency was defined as the number of cells of a particular type captured on the micropallets from the original population. The determination was made from a statistical sampling of random areas on the array after cell attachment and washing. The value was calculated as the number of individual target cells captured on the micropallets (i.e., no accompanying nontarget cell) divided by the total number of target cells added to the array chamber. For each measurement, 875 micropallets were inspected from 25 separate micrographs (both optical and fluorescence modes for each sampling) taken of the array. The field of view of each image included 35 micropallets. The selectivity of cell capture was calculated as 100% multiplied by the number of micropallets containing individual target cells divided by the total number of micropallets with cells. A flow chart of enrichment steps using antibody-coated micropallet arrays is shown in Supplementary Information (Fig. S1).
Results and Discussion
Coating Microarrays with Antibodies
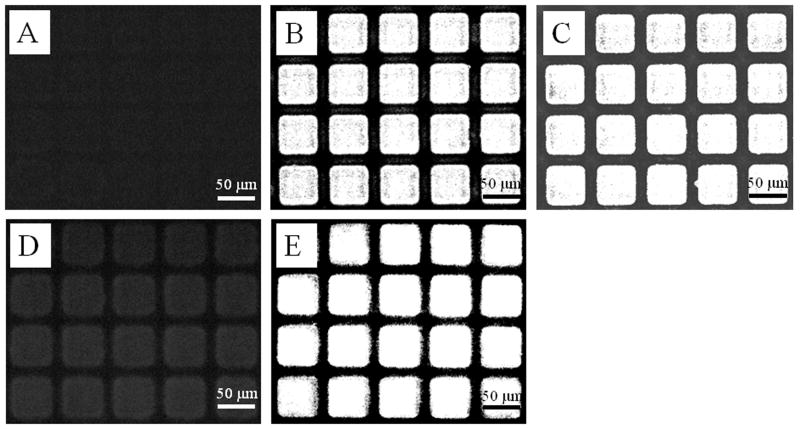
Two different strategies were used to coat antibodies onto the micropallets’ top surfaces. In the first, an antigen (BSA-DNP) was adsorbed to the microarray followed by binding of the corresponding antibody (anti-DNP IgE). This strategy was utilized to select cells competent to bind the Fc portion of IgE through the cell surface Fcε receptor. In the second strategy, the antibody (anti-ErbB2 or anti-c-Kit) was adsorbed to the microarray in order to capture cells expressing the ErbB2 or c-Kit surface receptors. Since BSA-DNP was difficult to detect on the micropallet surfaces, the fluorescent BSA-Alexa 647 was used initially to confirm the binding of BSA. Microarrays were incubated with BSA-Alexa 647, washed and then viewed by fluorescence microscopy. The background corrected average relative fluorescence intensity of micropallets (n = 35) on an uncoated array was 8 ± 1. In contrast, the fluorescence intensity of micropallets coated with BSA-Alexa 647 was 155 ± 3 (n = 35), a 19-fold increase (Fig.1 A&B). This difference was statistically significant as determined by Student’s t-test (α = 0.05 with P < 10-6). The BSA-Alexa 647 was present only on the top surfaces of the micropallets since the side walls were protected by virtual air walls (34,37). These data supported that BSA-DNP would also bind to the micropallets as both the DNP and Alexa 647 moieties are small relative to BSA.
Fig. 1.
Fluorescence micrographs (excitation 650 nm, emission 668 nm) of microarrays after incubation with: (A) untreated control, (B) BSA-Alexa 647, (C) anti-c-Kit-Cy5, (D) anti-IgG-Cy5, (E) anti-ErbB2 and anti-IgG-Cy5. All images were sequentially acquired on the same day. All pallets were 50 μm squares and 50 μm tall with ~20 μm spacing.
In a second series of experiments, coating of arrays with antibodies directed against the cell surface proteins c-Kit and ErbB2 was assessed. Anti-c-Kit-Cy5 was incubated with a microarray overnight and the array was then washed. Micropallets incubated with anti-c-Kit-Cy5 possessed a greater fluorescence intensity (181 ± 9, n = 35) than those on uncoated arrays (P < 10-6, Fig. 1C). For the ErbB2 experiment, the antibody used in this study was nonfluorescent; therefore, a labeled secondary antibody was used to assess the binding of anti-ErbB2. The micropallets were first incubated with anti-ErbB2 IgG followed by incubation with anti-IgG-Cy5. Micropallets (n = 35) treated in this manner demonstrated a significantly greater fluorescence intensity (154 ± 22, P < 10-9, Fig. 1 E) than that of control micropallets incubated only with the anti-IgG-Cy5 (30 ± 6, Fig. 1 D). It should be noted that in experiments involving direct coating of pallets with the two Cy5-labeled antibodies, a higher fluorescence intensity was present for the primary antibody anti-cKit-Cy5-coated array (Fig. 1C) than for the secondary anti-IgG-Cy5-coated array (Fig. 1D). This apparent discrepancy was due to the use of a higher concentration of antibody (50 vs. 20 μg/mL) and longer incubation period (16 vs. 2 h) for coating with anti-c-Kit-Cy5 in comparison to the secondary antibody anti-IgG-Cy5 (see Methods). In conclusion, these fluorescence data demonstrate that the micropallets could be readily coated with the appropriate proteins directed against cell surface ligands.
Capturing RBL Cells with IgE Microarrays
A number of factors must be considered in using a surface immobilized antibody for cell capture. These considerations include the affinity of the antibody for its ligand, the manner in which the antibody is bound to the capture surface (e.g. via its Fc portion or through non-specific adsorption), antibody density on the surface, ligand density, and size of the interacting species (25). In the current experiments, the pallets were coated in a manner that provided near saturation of the pallet surface by adsorption with the capture antibody. In the initial experiment, binding between surface bound antibody and cell ligand was through a cell surface receptor for the Fc portion of the antibody. RBL cells possess FcεR1 receptors on their cell surface which bind the Fc region of IgE in a specific manner (41-43). The binding between IgE and FcεR1 on RBL cells is high affinity with an activation energy of binding of 7.8 kcal/mol (44). To evaluate the feasibility of capturing RBL cells using IgE-coated microarrays and for quantitative comparisons, three sets of experiments were designed using arrays of native SU-8 micropallets, arrays coated with BSA-DNP alone, and arrays coated with BSA-DNP and IgE (n = 875 micropallets analyzed under each condition). When RBL cells were plated on bare SU-8 arrays followed by washing, very few micropallets possessed adherent cells reflecting a capture efficiency of only 1.2 ± 1.0%, Fig. S2. This is consistent with the hydrophobicity of native SU-8, which is reported to be a poor surface for cell attachment and growth (37). When RBL cells were cultured on the BSA-DNP-coated array, a partial improvement in capture efficiency compared to uncoated SU-8 microarrays was seen (22 ± 14% vs. 1.2 ± 1.0%, see Fig. S2). This increase was likely the result of increased surface roughness and hydrophilicity of the BSA-DNP-coated micropallets (45). In contrast, the capture efficiency of RBL cells plated on arrays coated with BSA-DNP and incubated with IgE increased to 96 ± 29%, an increase of 96-fold over that of the uncoated arrays. The large standard deviation is due to the fact that only a subgroup of pallets on an array (875 of 44,000 pallets) was counted. While every effort was made to distribute the cells homogeneously throughout the array, cells were not always perfectly distributed throughout the array. The RBL cells were captured with the highest efficiency on the IgE-coated arrays suggesting that these arrays might prove useful for selecting FcεR1-possessing cells from a mixture of cell types.
Selective Capture of FcεR1-Expressing Cells
The enhanced capture of RBL cells from a pure cell suspension indicated that this approach might be used to enrich a mixed cell population for RBL cells. To test this enrichment method, varying numbers of non-fluorescent RBL cells were mixed with cells from HeLa and 3T3 cell lines both of which stably expressed an enhanced green fluorescent protein (GFP). This mixed population of three cell types was then plated on microarrays coated with BSA-DNP-IgE or a fibronectin control. Fibronectin is commonly used as an extracellular matrix (ECM) protein that enhances the adherence and growth of many cells in tissue culture (46). Since the cell types used in this study possess integrin receptors and bind well to fibronectin, this ECM was used as a control for the selectivity provided by binding with the specific ligand under study.
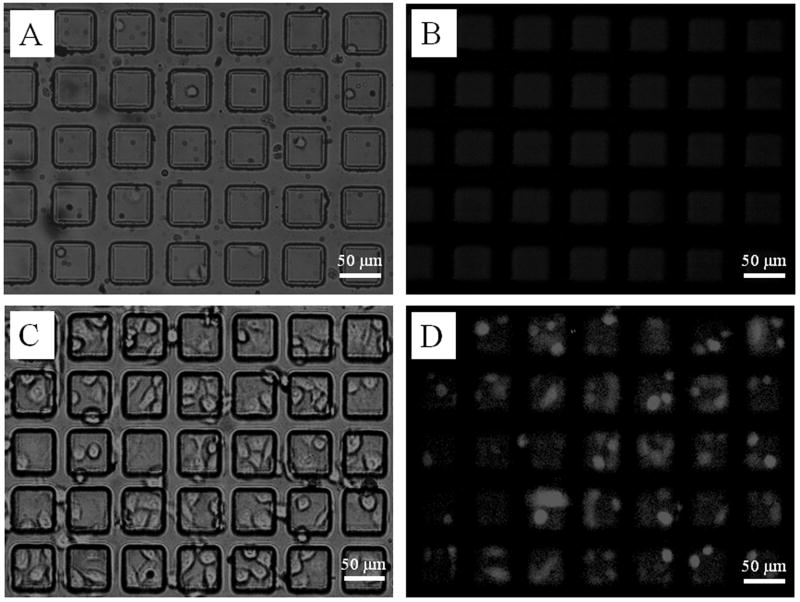
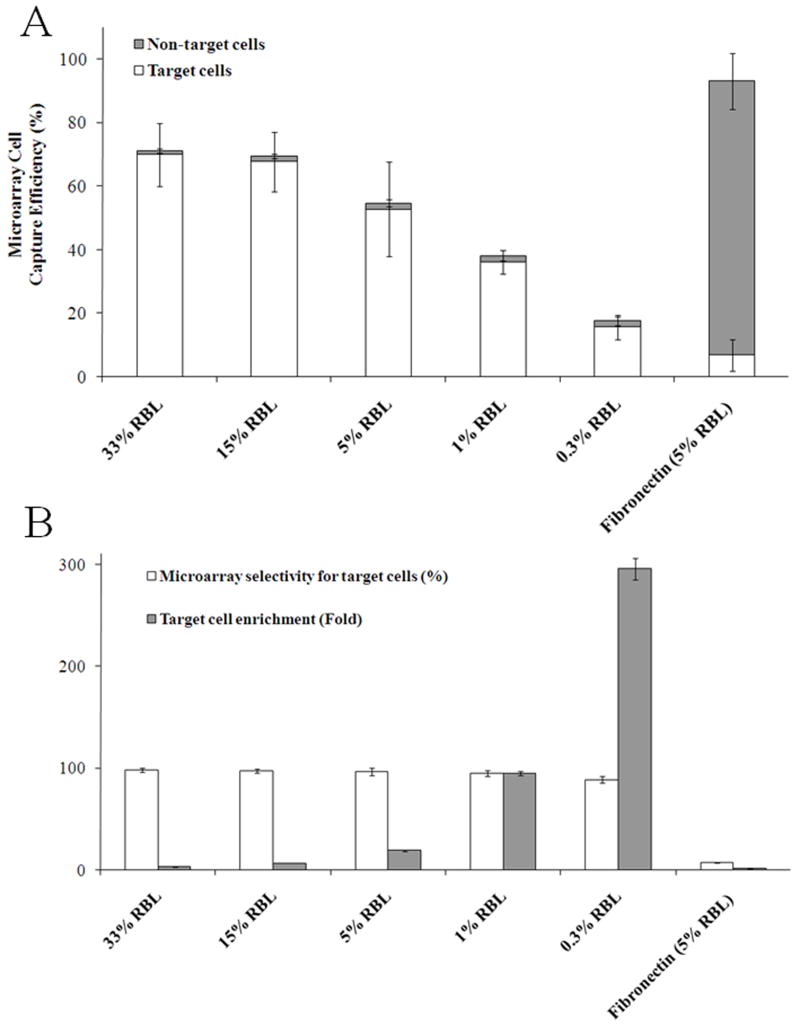
To evaluate the efficiency of antibody-coated arrays for selective capture of FcεR1-expressing cells, initially a cell mixture containing 5% RBL and 95% GFP-HeLa and GFP-3T3 cells (1:1 ratio) were plated on arrays coated with BSA-DNP-IgE or fibronectin (Fig. 2A-D). Inspection of the transmitted light (Fig. 2A, C) and fluorescence images (Fig. 2B, D) was used to determine the microarray cell capture efficiency for target cells (non-fluorescent RBL cells) and non-target cells (fluorescent GFP-HeLa and GFP-3T3) in the captured population (n = 875 pallets). In this experiment, most cells adherent to the array were the target RBL cells with a capture efficiency of 53 ± 15% while the capture efficiency of non-target cells was only 2 ± 1% (Fig. 3A). The capture efficiencies were then determined while varying the percentages of target cells in the cell mixture (Fig. 3A). In these experiments, the cell capture efficiency was diminished compared with that seen in the pure population (96 ± 29%, Fig. S2), and varied between 16 ± 4% with 0.3% RBLs in the plated suspension to 70 ± 10% with a 33% RBL suspension (Fig. 3A). This finding is likely the result of increasing the number of non-target cells that settle on the micropallets upon plating and non-specifically block the coated IgE region available to bind target cells. These loosely adherent cells are then removed from the array by the subsequent washing steps prior to image analysis. In contrast, the fibronectin-coated control array possessed cells on almost all pallets (microarray capture efficiency for all cell types 93 ± 10%), but showed a cell capture efficiency for target cells (7 ± 5%) comparable to that contained in the population of plated cells (i.e., 5%) reflecting the nonselective nature of the fibronectin-mediated cell attachment.
Fig. 2.
Optical (A & C) and fluorescence (B & D) micrographs of mixtures of 5% RBL with 95% GFP-3T3 and GFP-HeLa cells using a BSA-DNP-IgE-coated microarray (A & B), and fibronectin-coated array (C & D) after cell plating.
Fig. 3.
Capture efficiency and selectivity for cell mixtures. (A) Variation of microarray cell capture efficiency for varying percentages of target (RBL) amongst non-target (GFP-HeLa and GFP-3T3) cells. (B) Capture selectivity (open bars) and enrichment (gray bars) for target cells. The data shown are for the IgE and fibronectin-coated microarrays.
For BSA-DNP-IgE-coated microarrays, the enrichment of target cells in the plated population increased with decreasing percentages of RBL cells present in the suspension (Fig. 3B). When the suspension contained 33% RBL cells, only 3.0 ± 0.1 fold enrichment was seen, but enrichment was increased to 295 ± 11 fold when RBL cells were present at only 0.3% in the original population. The selectivity of the microarray approach for pre-sorting RBL cells was assessed after washing the array by determining the percentage of target cells present in the mixed cell populations plated on the BSA-DNP-IgE microarrays. At 15% and 33% RBLs in the cell suspension, the selectivity was 97 ± 2% and 98 ± 2%, respectively. For the 5% RBL cell suspension, the selectivity diminished slightly to 96 ± 4%, and was 89 ± 3% using a 0.3% RBL cell suspension. The enrichment and selectivity results highlight that this approach is most amenable for pre-sorting when target cells make up a very low percentage of the sample population.
Selective Sorting of Target Cells using Anti-c-Kit and Anti-ErbB2 Coated Arrays
To broaden the applicability of micropallet arrays for presorting mixed cell populations, antibodies with FAB regions directed against specific cell-surface epitopes were tested. Arrays with immobilized antibodies to c-Kit and ErbB2 were used to enrich mixed populations containing RBL and SK-BR-3 cells, respectively. In these experiments, RBL cell lines are known to express the c-Kit protein on their cell surface, while the SK-BR-3 cell line is a popular model system for the study of the ErbB2 receptor in breast cancer (47,48). These cell surface proteins were chosen by virtue of their broad interest in clinical oncology and biological research, and their common use as cell surface markers for cell analysis and sorting (49-52). The design of the following experiments was similar to that described above with non-fluorescent target cells being mixed with fluorescent HeLa and 3T3 cells, which were then plated on microarrays pre-coated with a specific antibody or a fibronectin control. For these experiments, mixed cell populations containing 5% target cells (RBL or SK-BR-3) were used.
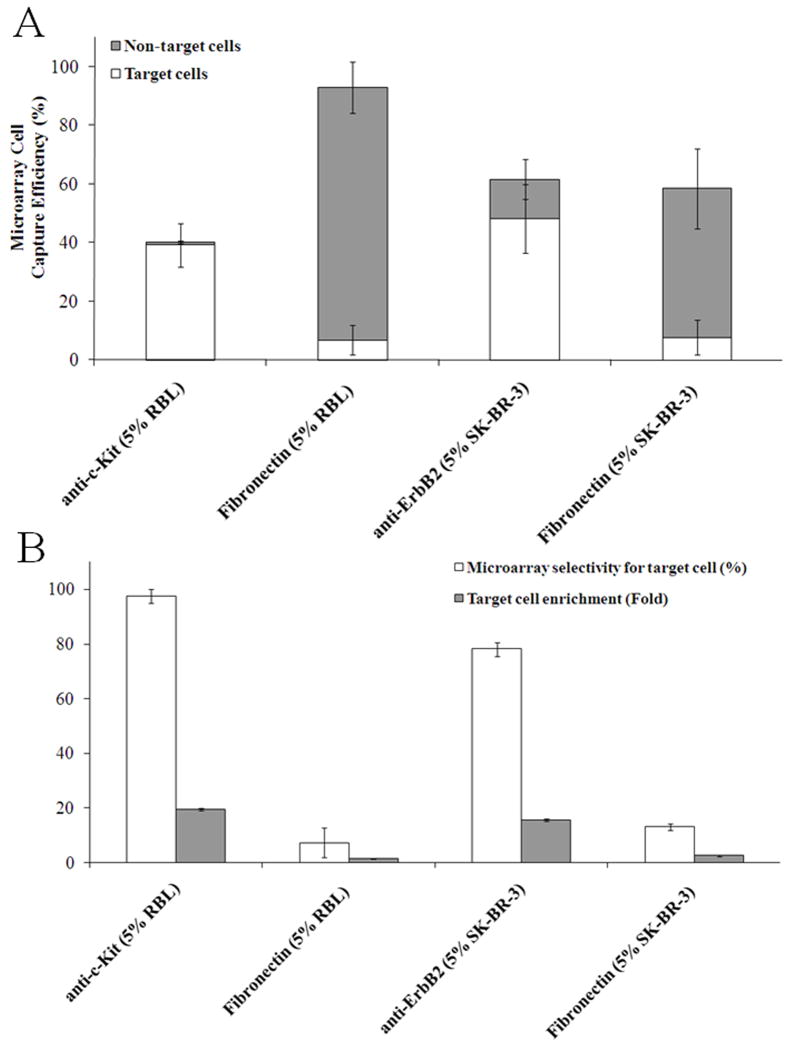
Transmitted light and fluorescent photomicrographs of arrays with immobilized anti-c-Kit or anti-ErbB2 and plated with a mixed suspension of RBL or SK-BR-3 cells, respectively, revealed similar results to that for the BSA-DNP-IgE-coated arrays (Fig. S3). Both anti-c-Kit and anti-ErbB2-coated arrays showed high capture efficiency in sorting target cells from a mixed population when the corresponding antibody was adsorbed to the micropallets (Fig. 4A). Most cells captured by the arrays were target cells (capture efficiencies of 39 ± 7% for the anti-c-Kit array and 48 ± 12% for anti-ErbB2) with significantly lower capture efficiencies for non-target cells (1.0 ± 0.5% for the anti-c-Kit array and 14 ± 7% for the anti-ErbB2 array). In contrast, the control fibronectin-coated array captured both target and non-target cells in the same proportions as present in the mixed population. The selectivity for captured target cells was also excellent (97 ± 3% for the anti-c-Kit-coated array and 78 ± 3% for the anti-ErbB2-coated array) relative to the fibronectin-coated pallets (Fig. 4B). The corresponding enrichment values were 19.5 ± 0.5 fold for the anti-c-Kit-coated array and 15.6 ± 0.5 fold for the anti-ErbB2-coated array when compared to fibronectin controls.
Fig. 4.
Capture of cells on arrays coated with antibodies to c-Kit and ErbB2. (A) Capture efficiency for mixtures of cells possessing 5% target (RBL or SK-BR-3) and 95% non-target (GFP-HeLa and GFP-3T3) cells. Anti-c-Kit- or anti-ErbB2-coated arrays were used for the RBL and SK-BR-3 target cells, respectively. (B) Selectivity and fold enrichment for target cells under the conditions in panel (A).
These data support that cell capture on the array as a first stage in cell isolation compares favorably to alternative cell isolation methods such as FACS and magnetic beads. Direct comparisons among these methods based on the literature are difficult as reported efficiencies will depend on various factors such as the application, antibody affinity/specificity, and cell type. Examples of the sorting efficiencies for isolation of CD34+ and CD133+ cells using Dynabeads and FACS have been reported as ~5% and 60%, with reported purities of ~30% and 88%, respectively (25). However, the pallet array technique should be considered a complementary rather than a replacement approach to these high-throughput/bulk procedures for cell isolation. As will be seen in the following section, the strength of the pallet array is in the isolation of selected single cells with high viability to generate clonal populations.
Selective Release and Clonal Expansion of Target Cells from Immobilized Antibody-coated Arrays
To demonstrate that enrichment by cell capture using a surface immobilized antibody:ligand interaction is compatible with the isolation of viable cells utilizing the micropallet arrays, single-cell cloning experiments were undertaken. Micropallets containing single target cells were released and collected from cell-capture arrays with the following capture agents: 1) BSA-DNP-IgE, 2) anti-c-Kit, 3) anti-ErbB2, and 4) fluorescent non-target cells from a fibronectin array. The viability of the collected cells was assessed by placing the collected pallet:cell in culture and following cell growth over time as previously described (34,35). In each series, 10 micropallets were released with their attached cell and collected. Within 10 days following release, colonies of cells possessing the expected characteristic (absence or presence of fluorescence) were present in the collection culture wells. Despite the presence of the capture antibody, as the progeny of the original cell divided, they migrated off of the pallet onto the tissue culture surface (Fig. 5). Thus, detachment from the antibody-coated pallet of the captured cell by trypsinization or other method was not necessary. Cell viability was defined as the clonal expansion of a single cell into a colony of cells within 10 days of culture. The results for each experiment were: 90% viability of RBLs captured on the BSA-DNP-IgE-coated and on the antic-Kit-coated arrays; 80% viability of SK-BR-3 cells captured on the anti-ErbB2-coated array; and 90% viability of cells captured on the fibronectin-coated array. These data demonstrate the feasibility of collecting and cloning captured target cells after enrichment of a mixed cell population using arrays of micropallets with immobilized antibodies.
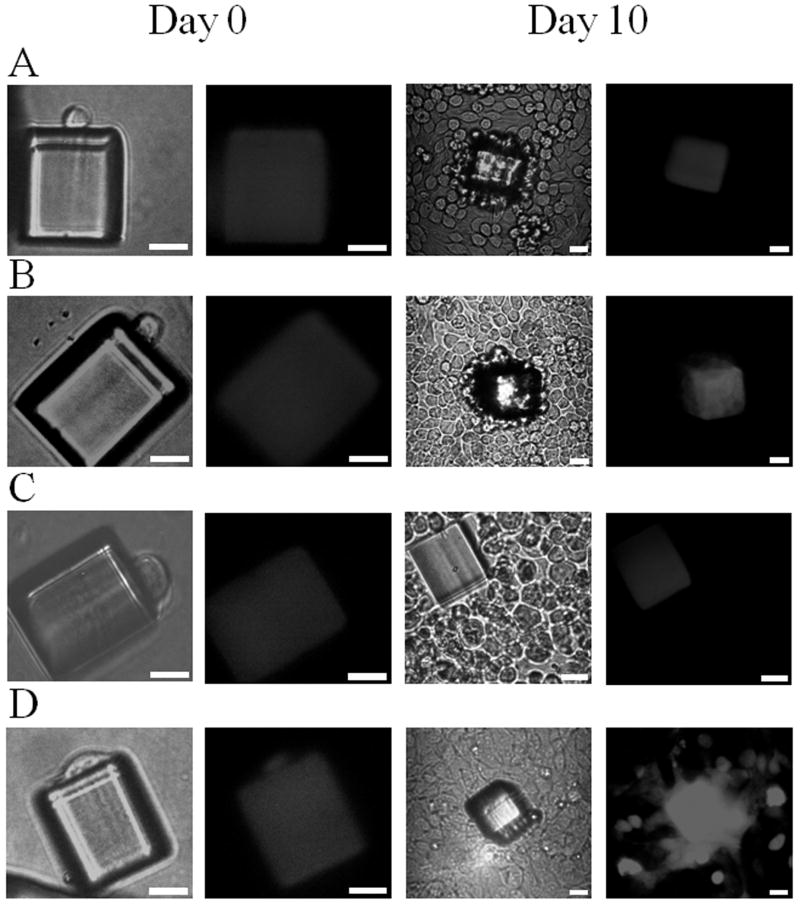
Fig. 5.
Optical (left) and fluorescence (right) micrographs of individual released pallets containing a single captured cell on the day of release and after 10 days in culture. (A) Pallet with RBL target cell (nonfluorescent) released from a BSA-DNP-IgE-coated microarray. (B) Pallet with RBL target cell released from an anti-c-Kit-coated microarray. (C) Pallet with an SK-BR-3 cell (nonfluorescent) released from an anti-ErbB2-coated microarray. (D) Pallet possessing a fluorescent cell (HeLa or 3T3) released from a fibronectin-coated microarray. Scale bars are 25 μm in all micrographs.
Conclusion
In the current work we have demonstrated the feasibility of combining an enrichment step with the micropallet-based cell sorting technique. The applicability of this approach was demonstrated by implementing three different antibody:ligand interactions to target specific cell types in a mixed cell population. The cell types for this developmental work were chosen by virtue of their well established expression of non-overlapping cell surface markers. Extension of this work to a heterogeneous population, such as that obtained from a surgical biopsy specimen, will be an important next step. While in the initial step for cell capture, the detrimental effects of placing cells in suspension persist, the technique allows cells to recover prior to isolation. The cells are cultured for hours to days on the array after capture during which time cell surface proteins are re-expressed and the cells recover from other stresses imposed by removal from their culture surface. At this later time, cells can be again screened for cell surface ligands or other characteristics that would otherwise not be assessable immediately after enzymatic or mechanical treatment. Furthermore, the combination of two screening steps will increase the number of characteristics available for cell sorting. With extensive numbers of high affinity antibodies targeting large numbers of cell surface ligands now available commercially, the approach is readily applicable to many cellular targets of interest for biological and biomedical research. In the experiments, 295-fold enrichment was achieved when target cells comprised 3 per 1,000 cells. This enrichment is likely to be even greater when the target cell population represents an even smaller percentage of the bulk population. The size and geometry of the array can be modified to enable use with samples of fewer than 5,000 to more than 100,000 cells (32-35,37,53). As a result, this technique can be used for sorting and enrichment of target cells in complicated biological samples composed of only relatively small numbers of cells. The micropallet arrays modified with immobilized antibodies are easily generated simply by incubating the array in a solution of the targeting antibody and can be used immediately, or stored in a refrigerator under sterile conditions for at least one week (the longest time yet tested). The enrichment step can be performed in a matter of minutes (e.g., 30 min in the current work) as cells are captured and adhere quickly with the use of high affinity antibodies. Finally, targeted cells are isolated and placed in culture in a single step leading to high efficiency cloning. These attributes are valuable in terms of reducing the labor, time, and costs required to perform the isolation and expansion. The flexibility coupled with the ability to enrich, sort and clone cells of interest efficiently and with high success provides a powerful platform technology for the biomedical investigator.
Supplementary Material
Acknowledgments
This work was supported by the National Institutes of Health (EB07612). We thank Grace Young for assisting in the initial steps of this project. We also acknowledge Dr. Eva Lee at the University of California, Irvine for kindly providing the GFP-expressing cell lines. NLA and CES disclose a financial interest in Intellego, LLC.
Literature Cited
- 1.Taylor TB, Nambiar PR, Raja R, Cheung E, Rosenberg DW, Anderegg B. Microgenomics: identification of new expression profiles via small and single-cell sample analyses. Cytometry, Part A. 2004;59A:254–261. doi: 10.1002/cyto.a.20051. [DOI] [PubMed] [Google Scholar]
- 2.Kaern M, Elston TC, Blake WJ, Collins JJ. Stochasticity in gene expression: From theories to phenotypes. Nature Reviews Genetics. 2005;6:451–464. doi: 10.1038/nrg1615. [DOI] [PubMed] [Google Scholar]
- 3.Davis JE, Eberwine JH, Hinkle DA, Marciano PG, Meaney DF, McIntosh TK. Methodological considerations regarding single-cell gene expression profiling for brain injury. Neurochemical Research. 2004;29:1113–1121. doi: 10.1023/b:nere.0000023598.04937.83. [DOI] [PubMed] [Google Scholar]
- 4.Hack CJ. Integrated transcriptome and proteome data: the challenges ahead. Briefings in Functional Genomics & Proteomics. 2004;3:212–219. doi: 10.1093/bfgp/3.3.212. [DOI] [PubMed] [Google Scholar]
- 5.Fend F, Raffeld M. Laser capture microdissection in pathology. Journal of clinical pathology. 2000;53:666–72. doi: 10.1136/jcp.53.9.666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lefievre L, Barratt CLR, Harper CV, Conner SJ, Flesch FM, Deeks E, Moseley FLC, Pixton KL, Brewis IA, Publicover SJ. Physiological and proteomic approaches to studying prefertilization events in the human. Reproductive BioMedicine Online. 2003;7:419–427. doi: 10.1016/s1472-6483(10)61885-8. [DOI] [PubMed] [Google Scholar]
- 7.Sambrook JF, Russell DW, editors. Molecular cloning: a laboratory manual. third edition. 2000. p. 2300. [Google Scholar]
- 8.Welm B, Behbod F, Goodell MA, Rosen JM. Isolation and characterization of functional mammary gland stem cells. Cell Proliferation. 2003;36:17–32. doi: 10.1046/j.1365-2184.36.s.1.3.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hay RJ. Human cells and cell cultures: availability, authentication and future prospects. Human Cell. 1996;9:143–52. [PubMed] [Google Scholar]
- 10.Sims CE, Bachman M, Li GP, Allbritton NL. Choosing one from the many: selection and sorting strategies for single adherent cells. Analytical and Bioanalytical Chemistry. 2007;387:5–8. doi: 10.1007/s00216-006-0612-1. [DOI] [PubMed] [Google Scholar]
- 11.Patel D. Separating Cells. Oxford: Springer; 2001. p. 156. [Google Scholar]
- 12.Fu AY, Chou H-P, Spence C, Arnold FH, Quake SR. An integrated microfabricated cell sorter. Analytical Chemistry. 2002;74:2451–2457. doi: 10.1021/ac0255330. [DOI] [PubMed] [Google Scholar]
- 13.Wolff A, Perch-Nielsen IR, Larsen UD, Friis P, Goranovic G, Poulsen CR, Kutter JP, Telleman P. Integrating advanced functionality in a microfabricated high-throughput fluorescent-activated cell sorter. Lab on a Chip. 2003;3:22–27. doi: 10.1039/b209333b. [DOI] [PubMed] [Google Scholar]
- 14.Young SM, Curry MS, Ransom JT, Ballesteros JA, Prossnitz ER, Sklar LA, Edwards BS. High-throughput microfluidic mixing and multiparametric cell sorting for bioactive compound screening. Journal of Biomolecular Screening. 2004;9:103–111. doi: 10.1177/1087057103262335. [DOI] [PubMed] [Google Scholar]
- 15.Sohn LL, Saleh OA, Facer GR, Beavis AJ, Allan RS, Notterman DA. Capacitance cytometry: measuring biological cells one by one. Proceedings of the National Academy of Sciences of the United States of America. 2000;97:10687–10690. doi: 10.1073/pnas.200361297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang X-B, Yang J, Huang Y, Vykoukal J, Becker FF, Gascoyne PRC. Cell separation by dielectrophoretic field-flow-fractionation. Analytical Chemistry. 2000;72:832–839. doi: 10.1021/ac990922o. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Burridge K, Chrzanowska-Wodnicka M. Focal adhesions, contractility, and signaling. Annual Review of Cell and Developmental Biology. 1996;12:463–518. doi: 10.1146/annurev.cellbio.12.1.463. [DOI] [PubMed] [Google Scholar]
- 18.Chiquet M, Matthisson M, Koch M, Tannheimer M, Chiquet-Ehrismann R. Regulation of extracellular matrix synthesis by mechanical stress. Biochemistry and Cell Biology. 1996;74:737–744. doi: 10.1139/o96-080. [DOI] [PubMed] [Google Scholar]
- 19.Ingber DE. Tensegrity: the architectural basis of cellular mechanotransduction. Annual Review of Physiology. 1997;59:575–599. doi: 10.1146/annurev.physiol.59.1.575. [DOI] [PubMed] [Google Scholar]
- 20.Seidl J, Knuechel R, Kunz-Schughart LA. Evaluation of membrane physiology following fluorescence activated or magnetic cell separation. Cytometry. 1999;36:102–11. doi: 10.1002/(sici)1097-0320(19990601)36:2<102::aid-cyto3>3.3.co;2-4. [DOI] [PubMed] [Google Scholar]
- 21.Piercy KT, Donnell RL, Kirkpatrick SS, Mundy BL, Stevens SL, Freeman MB, Goldman MH. Effect of harvesting and sorting on b-1 integrin in canine microvascular cells. Journal of Surgical Research. 2001;100:211–216. doi: 10.1006/jsre.2001.6247. [DOI] [PubMed] [Google Scholar]
- 22.Mackie EJ, Pagel CN, Smith R, De Niese MR, Song SJ, Pike RN. Protease-activated receptors: a means of converting extracellular proteolysis into intracellular signals. IUBMB Life. 2002;53:277–281. doi: 10.1080/15216540213469. [DOI] [PubMed] [Google Scholar]
- 23.Wada S, Kitagawa M. Method of separation and concentration of fetal nucleated red blood cells in maternal blood and its application to fetal diagnosis. Congenital Anomalies. 2004;44:72–8. doi: 10.1111/j.1741-4520.2004.00011.x. [DOI] [PubMed] [Google Scholar]
- 24.Shapiro HM. Practical flow cytometry. Hoboken: John Wiley & Sons, Inc.; 2003. p. 681. [Google Scholar]
- 25.Scheper T. Advances in biochemical engineering/biotechnology: cell separation. Vol. 106. Springer, Inc.; 2007. p. 202. [Google Scholar]
- 26.http://www.zeiss.com
- 27.Burgess DS. Laser microdissection: making inroads in research. Biophotonics International. 2004;11:46–49. [Google Scholar]
- 28.Schutze K, Posl H, Lahr G. Laser micromanipulation systems as universal tools in cellular and molecular biology and in medicine. Cellular and Molecular Biology. 1998;44:735–746. [PubMed] [Google Scholar]
- 29.Langer S, Geigl JB, Ehnle S, Gangnus R, Speicher MR. Live cell catapulting and recultivation does not change the karyotype of HCT116 tumor cells. Cancer Genetics and Cytogenetics. 2005;161:174–177. doi: 10.1016/j.cancergencyto.2005.01.013. [DOI] [PubMed] [Google Scholar]
- 30.Burgemeister R. New aspects of laser microdissection in research and routine. Journal of Histochemistry and Cytochemistry. 2005;53:409–412. doi: 10.1369/jhc.4B6421.2005. [DOI] [PubMed] [Google Scholar]
- 31.Todd R, Lingen MW, Kuo WP. Gene expression profiling using laser capture microdissection. Expert Review of Molecular Diagnostics. 2002;2:497–507. doi: 10.1586/14737159.2.5.497. [DOI] [PubMed] [Google Scholar]
- 32.Salazar GTa, Wang Y, Young G, Bachman M, Sims CE, Li GP, Allbritton NL. Micropallet arrays for the separation of single, adherent cells. Analytical Chemistry. 2007;79:682–687. doi: 10.1021/ac0615706. [DOI] [PubMed] [Google Scholar]
- 33.Wang Y, Salazar GTa, Pai J-H, Shadpour H, Sims CE, Allbritton NL. Micropallet arrays with poly(ethylene glycol) walls. Lab on a Chip. 2008;8:734–740. doi: 10.1039/b800286j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pai J-H, Wang Y, Salazar GTA, Sims CE, Bachman M, Li GP, Allbritton NL. Photoresist with low fluorescence for bioanalytical applications. Analytical Chemistry. 2007;79:8774–8780. doi: 10.1021/ac071528q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang Y, Young G, Bachman M, Sims CE, Li GP, Allbritton NL. Collection and expansion of single cells and colonies released from a micropallet array. Analytical Chemistry. 2007;79:2359–2366. doi: 10.1021/ac062180m. [DOI] [PubMed] [Google Scholar]
- 36.Shadpour H, Sims CE, Thresher RJ, Allbritton NL. Sorting and expansion of murine embryonic stem cell colonies using micropallet arrays. Cytometry Part A. 2009;75A:121–129. doi: 10.1002/cyto.a.20672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang Y, Sims CE, Marc P, Bachman M, Li GP, Allbritton NL. Micropatterning of living cells on a heterogeneously wetted surface. Langmuir. 2006;22:8257–8262. doi: 10.1021/la061602k. [DOI] [PubMed] [Google Scholar]
- 38.Neurauter AA, Bonyhadi M, Lien E, Noekleby L, Ruud E, Camacho S, Aarvak T. Cell isolation and expansion using Dynabeads. Advances in Biochemical Engineering/Biotechnology. 2007;106:41–73. doi: 10.1007/10_2007_072. [DOI] [PubMed] [Google Scholar]
- 39.Belov L, Huang P, Barber N, Mulligan SP, Christopherson RI. Identification of repertoires of surface antigens on leukemias using an antibody microarray. Proteomics. 2003;3:2147–2154. doi: 10.1002/pmic.200300599. [DOI] [PubMed] [Google Scholar]
- 40.Tai C-K, Logg CR, Park JM, Anderson WF, Press MF, Kasahara N. Antibody-mediated targeting of replication-competent retroviral vectors. Human Gene Therapy. 2003;14:789–802. doi: 10.1089/104303403765255174. [DOI] [PubMed] [Google Scholar]
- 41.Orida N, Feldman JD, Katz DH, Liu FT. IgE-mediated chemotaxis of rat basophilic leukemia cells towards specific antigen. Journal of Experimental Medicine. 1983;157:2166–71. doi: 10.1084/jem.157.6.2166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Carson DA, Metzger H. Interaction of IgE with rat basophilic leukemia cells. IV. Antibody-induced redistribution of IgE receptors. Journal of Immunology. 1974;113:1271–7. [PubMed] [Google Scholar]
- 43.Kulczycki A, Jr, Isersky C, Metzger H. Interaction of IgE with rat basophilic leukemia cells. I. Evidence for specific binding of IgE. Journal of Experimental Medicine. 1974;139:600–16. doi: 10.1084/jem.139.3.600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kulczycki A, Jr, Metzger H. The interaction of IgE with rat basophilic leukemia cells. II. Quantitative aspects of the binding reaction. The Journal of Experimental Medicine. 1974;140:1676–95. doi: 10.1084/jem.140.6.1676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shadpour H, Musyimi H, Chen J, Soper SA. Physiochemical properties of various polymer substrates and their effects on microchip electrophoresis performance. Journal of Chromatography, A. 2006;1111:238–251. doi: 10.1016/j.chroma.2005.08.083. [DOI] [PubMed] [Google Scholar]
- 46.Lark MW, Laterra J, Culp LA. Close and focal contact adhesions of fibroblasts to a fibronectin-containing matrix. Federation Proceedings. 1985;44:394–403. [PubMed] [Google Scholar]
- 47.Ott VL, Moffitt LA, Cambier JC. Study of SHIP-binding cell surface proteins suggests c-kit as a SHIP-interacting receptor in mast cells. Signal Transduction. 2005;5:28–39. [Google Scholar]
- 48.Li H, Sanchez-Torres J, del carpio A, Salas V, Villalobo A. The ErbB2/Neu/HER2 receptor is a new calmodulin-binding protein. Biochemical Journal. 2004;381:257–266. doi: 10.1042/BJ20040515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Muthuswamy SK, Li D, Lelievre S, Bissell MJ, Brugge JS. ErbB2, but not ErbB1, reinitiates proliferation and induces luminal repopulation in breast epithelial acini. Nature Cell Biology. 2001;3:785–792. doi: 10.1038/ncb0901-785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Legitimo A, Consolini R, Cocito MG, Buffoni R, Basso G, Macchia P. The c-kit receptor and its ligand stem cell factor in childhood malignant lymphoid precursors. Journal of Interferon & Cytokine Research. 1999;19:981–7. doi: 10.1089/107999099313172. [DOI] [PubMed] [Google Scholar]
- 51.Rao RR, Johnson AV, Stice SL. Cell surface markers in human embryonic stem cells. Methods in Molecular Biology. 2007;407:51–61. doi: 10.1007/978-1-59745-536-7_5. [DOI] [PubMed] [Google Scholar]
- 52.Draper JS, Andrews PW. Surface antigen markers. Handbook of Stem Cells. 2004;1:565–571. [Google Scholar]
- 53.Wang Y, Young G, Aoto Phillip C, Pai J-H, Bachman M, Li GP, Sims Christopher E, Allbritton Nancy L. Broadening cell selection criteria with micropallet arrays of adherent cells. Cytometry Part A. 2007;71:866–74. doi: 10.1002/cyto.a.20424. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.