Abstract
Endothelium-derived nitric oxide (NO) is an important regulator of vascular function. NO is produced by endothelial NO synthase (eNOS), whose expression is downregulated by tumor necrosis factor (TNF)-α at the post-transcriptional level. To elucidate the molecular basis of TNF-α-mediated eNOS mRNA instability, eNOS 3' untranslated region (3'-UTR) binding proteins were purified by RNA affinity chromatography from cytosolic fractions of TNF-α-stimulated human umbilical vein endothelial cells (HUVECs). The formation of 3'-UTR ribonucleoprotein (RNP) complexes, with molecular weight of 52 and 57 kDa, was increased by TNF-α. MALDI-TOF-MS analysis of the 52 kDa protein identified three peptides that comprise the peptide sequence of translation elongation factor 1-alpha 1(eEF1A1). In HUVECs, TNF-α rapidly increased eEF1A1 expression, which is maximal after 1 hr and persists for up to 48 hr. RNA gel mobility shift and UV cross-linking assays indicated that recombinant GST-eEF1A1 fusion protein specifically binds to an UC-rich sequence in the 3'-UTR of eNOS mRNA. In addition, the domain III of eEF1A1 mediates the binding of eNOS 3'-UTR in eEF1A1. Overexpression of eEF1A1 markedly attenuated the expression of eNOS and luciferase gene fused with eNOS 3'-UTR in both COS-7 cells and bovine aortic endothelial cells (BAECs). Furthermore, adenovirus-mediated overexpression of eEF1A1 increased eNOS mRNA instability, while knockdown of eEF1A1 substantially attenuated TNF-α-induced destabilization of eNOS mRNA and downregulation of eNOS expression in HUVECs. These results indicate that eEF1A1 is a novel eNOS 3'-UTR binding protein that plays a critical role in mediating TNF-α-induced decrease in eNOS mRNA stability.
Keywords: TNF-α, eNOS, mRNA, stability, translation elongation factor 1-alpha
Introduction
eNOS is a key enzyme involved in the regulation of vascular function and abnormality of eNOS activity and/or expression has been shown to cause several vascular diseases 1. Although eNOS was initially considered to be a constitutive enzyme, it was shown later that eNOS expression was regulated by a variety of exogenous stimuli. For instance, in cultured endothelial cells, cytokines, lipopolysaccaride, and oxidized LDLs have been shown to downregulate eNOS expression 2 3. In contrast, shear stress, estrogen, and 3-hydroxy-3-methylglutaryl coenzyme A reductase inhibitors have been demonstrated to upregulate eNOS expression 4–6. For many of these stimuli, modulation of eNOS mRNA stability plays an essential role in the regulation of eNOS expression 7. However, the mechanism(s) responsible for the regulation of eNOS mRNA half-life remains to be determined.
One of the most potent inhibitory stimuli for eNOS expression in vascular endothelial cells is tumor necrosis factor-α (TNF-α). TNF-α mediated inhibition of eNOS expression, via a mechanism of destabilization of eNOS mRNA, has been shown to be associated with vascular dysfunction in several disease states, such as atherosclerosis, diabetes, and heart failure 8, 9. It is becoming increasingly clear that the 3'-UTR of eNOS mRNA plays an important role in the posttranscriptional regulation of mRNA stability 10–12. For example, in BAECs, TNF-α induced eNOS mRNA destabilization is mediated, in part, by specific binding of a cytosolic 60-kDa protein to the 3'-UTR12. Furthermore, a 51-kDa protein was identified to bind to the eNOS 3'-UTR and regulate eNOS mRNA half-life during cell growth 11. Similarly, in HUVECs, the binding of the 53-and 56-kDa proteins to the eNOS 3'-UTR has also been implicated in the modulation of mRNA stability by TNF-α 13. However, the identity of eNOS mRNA 3'-UTR binding proteins and precise mechanism by which these proteins regulate eNOS mRNA stability remain to be determined.
To identify the cytosolic proteins that could be important in regulating eNOS mRNA stability, we used a MS–protein-sequencing approach to identify proteins that copurify with eNOS 3'-UTR from a cytosolic fraction of TNF-α stimulated HUVECs. From these studies, we found that the ribosomal protein translation elongation factor eEF1A1, an actin binding protein, interacts with eNOS 3'-UTR and modulates eNOS mRNA stability in response to TNF-α.
Materials and Methods
For details, please see the Data Supplement available at http://circres.ahajournal.org. Briefly, HUVECs were routinely cultured and treated with TNF-α (20 ng/mL) for 24 hr. eNOS 3'-UTR binding proteins were purified by using RNA affinity chromatography and identified by MALDITOF-MS. The interaction of eEF1A1 with eNOS 3'-UTR was determined by RNA electrophoretic mobility shift assay (R-EMSA) and UV cross-linking assay. Western blotting and Northern Blotting were performed following standard protocols. Adenovirus harboring Flag-tagged eEF1A1 was made using AdMax (Microbix). Gene-specific siRNA was transfected with Gene Silencer® transfecting agent. Luciferase activity was determined using the Dual Luciferase Assay System (Promega).
Results
Identification of eEF1A1 as an eNOS 3'-UTR binding protein
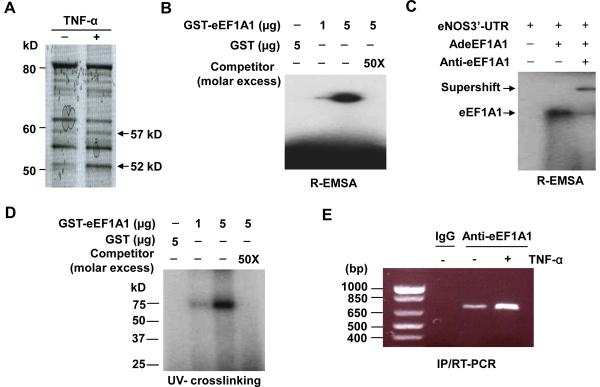
Previous studies indicated that TNF-α decreases the eNOS protein expression through destabilization of eNOS mRNA by cytosolic proteins which binds to the 3'-UTR of eNOS mRNA 10, 12–14. To identify these proteins, eNOS 3'-UTR binding proteins were purified from a cytosolic fraction of TNF-α unstimulated and stimulated HUVECs, using RNA affinity chromatography via biotinylated transcripts coupled to streptavidin-agarose. The binding of two bands, at 57 kDa and 52 kDa, to eNOS 3'-UTR was significantly upregulated in TNF-α stimulated HUVECs (Figure 1A). MALDI-TOF-MS analysis of the 52 kDa band identified three peptides (YYVTIIDAPGHR, EHALLAYTLGVK, VETGVLKPGMVVTFAPVNVTTEVK) that correspond to peptides deduced from translation elongation factor 1-alpha 1 (eEF1A1). The identity of the 57 kDa protein remains to be characterized.
Figure 1. Identification and characterization of eNOS 3'-UTR binding proteins.
(A) eNOS 3'-UTR binding proteins were purified by RNA affinity chromatography from a cytosolic fraction of unstimulated and stimulated HUVECs with 20 ng/mL TNF-α for 24 hr. After extensive washing , proteins bound to the biotinylated eNOS 3'-UTR coupled to streptavidin agarose beads were eluted with SDS-PAGE sample buffer , electrophoretically separated by 12 % SDS-PAGE, and Coomassie-stained. The target protein bands at 57 kD and 52 kD were digested with trypsin followed by MALDI-TOF-MS analysis. (B) R-EMSA was performed by incubating the radiolabeled eNOS 3'-UTR with increasing concentration of GST-eEF1A1 (1μg and 5μg) and a fixed concentration of GST (5μg). Competition with specific competitor (50 molar excess) corresponding to unlabeled eNOS 3'-UTR transcript was also performed in the presence of 5μg GST-eEF1A1. (C) EMSA was performed by incubating the radiolabeled eNOS 3'-UTR with 30 μg cytosolic extracts from HUVECs transduced with AdeEF1A1 for 48 hr in the presence and absence of anti-eEF1A antibody. (D) UV Cross-linking was performed by incubating the radiolabeled eNOS 3'-UTR with increasing concentration of GST-eEF1A1 (1μg and 5μg) and a fixed concentration of GST (5μg). Competition with specific competitor (50 molar excess) corresponding to unlabeled eNOS 3'-UTR transcript was also performed in the presence of 5μg GST-eEF1A1. (E) Immunoprecipitation, followed by RT-PCR assays, showing interaction of eEF1A and eNOS mRNA 3'-UTR in HUVECs treated with or without TNF-α (20 ng/ml) for 1 hr.
To determine whether eEF1A1 interacts with eNOS 3'-UTR, electrophoretic mobility shift assay (EMSA) with in vitro-translated eNOS 3'-UTR was used to further investigate the binding of eEF1A1 to the eNOS 3'-UTR. GST-eEF1A1, but not GST, shifted the radiolabeled eNOS 3'-UTR probe, whereas preincubation with cold eNOS 3'-UTR oligonucleotide completely eliminated this band shift (Figure 1B). EMSA, performed on cytosolic extracts from HUVECs transduced with adenovirus harboring eEF1A1 (AdeEF1A1), also demonstrated a ribonucleoprotein (RNP) complex formation, which was retarded by anti-eEF1A antibody (Figure 1C), providing further evidence of specificity of complex formation between eNOS 3'-UTR with eEF1A1 in HUVECs. In addition, eEF1A1-eNOS 3'-UTR interaction was assayed by UV cross-linking experiments. Complex formation between recombinant GST-eEF1A1 protein and the whole 3'-UTR transcript was observed, whereas the binding activity was not observed with the GST protein alone and was completely competed by excessive cold eNOS 3'-UTR transcript (Figure 1D). Furthermore, immunoprecipitation, followed by reverse transcription-PCR assays, also demonstrated the specific interaction of eEF1A and eNOS mRNA 3'-UTR in both TNF-α unstimulated and stimulated HUVECs. However, TNF-α stimulation markedly increased the levels of eNOS mRNA associated with eEF1A (Figure 1E).
Identification of the eEF1A-binding Site in human eNOS 3'-UTR
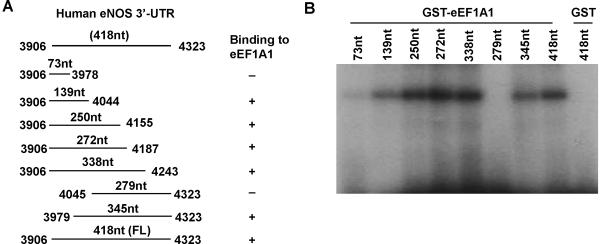
Based on the human eNOS mRNA sequence deposited in the GenBank database (accession no NM_000603), eNOS 3'-UTR is 418 nt long (3906–4323). To localize the eEF1A1-binding site within the eNOS 3'-UTR, a series of riboprobes corresponding to eNOS 3'-UTR deletion mutants were generated (Figure 2A) and used in UV cross-linking experiments. GST-eEF1A displayed a binding activity to riboprobes corresponding to 3906 to 4044 (139nt), 3906 to 4155 (250nt), 3906 to 4187 (272nt), 3906 to 4243 (338nt), 3979 to 4323 (345nt), and full length 3'UTR (418nt) of eNOS mRNA, but not to the fragments of 3906 to 3978 (73nt) and 4045 to 4325 (279nt) of eNOS mRNA 3'-UTR (Figure 2B). Together, these results suggested that the eEF1A1-binding site is located in the region between 3979 to 4044 (66nt), an UC-rich sequence in the 3'-UTR of eNOS mRNA (5'-GCCCCGCUCCUCCCCUCUUGAGGUGGUGCCUUCUCACAUCUGUCCA GAGGCUGCAAGGAUUCAGCAUUA-3').
Figure 2. Analysis of the eEF1A1-binding site in the human eNOS 3'-UTR.
(A) Schematic representation of human eNOS 3'-UTR mutants. (B) UV cross-linking showing the binding of eEF1A1 to the different deletion mutants of human eNOS 3'-UTR.
Identification of the eNOS 3'-UTR binding Site in eEF1A1
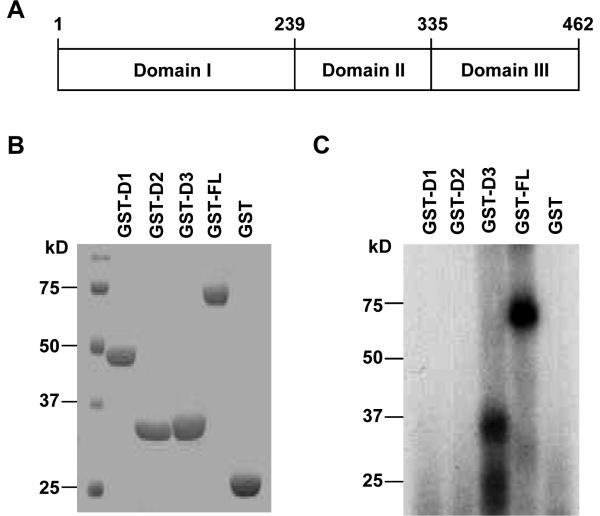
eEF1A is a highly conserved protein in eukaryotic cells, with three functional domains: namely, domain I, domain II and domain III15(Figure 3A). Domain I and domain II have been shown to interact with GDP/GTP and aminoacyl-tRNA, respectively, whereas domain III interacts with actin and is responsible for the non-canonical functions of eEF1A in actin binding and bundling 15, 16. To localize the site(s) essential for eNOS 3'-UTR binding in eEF1A1, we performed UV cross -linking assays by using bacterially expressed GST fusing proteins (Figure 3B). eNOS 3'-UTR strongly interacted with GST fusion proteins bearing the full length of eEF1A1 and eEF1A1 domain III , but not with domain I and domain II of eEF1A1 (Figure 3C). These results suggest that domain III of eEF1A1 is responsible for the eNOS 3'-UTR binding in eEF1A1.
Figure 3. Identification of the eNOS 3'-UTR binding site in eEF1A1.
(A) Schematic representation of human eEF1A1 functional domains. (B) Identification of GST-fusion proteins containing different domains of eEF1A1. GST-fusion proteins were bacterially expressed and purified by glutathione-Sepharose beads. Purified proteins were separated on 12% SDS-PAGE and stained with Coomassie Blue R250. (C) UV cross-linking showing the binding of eEF1A1 domain III (D3) to the human eNOS 3'-UTR. UV cross-linking assays were performed by incubating the radiolabeled eNOS 3'-UTR with equal molar of GST fusion proteins.
Overexpression of eEF1A1 decreases eNOS mRNA stability and inhibits eNOS expression
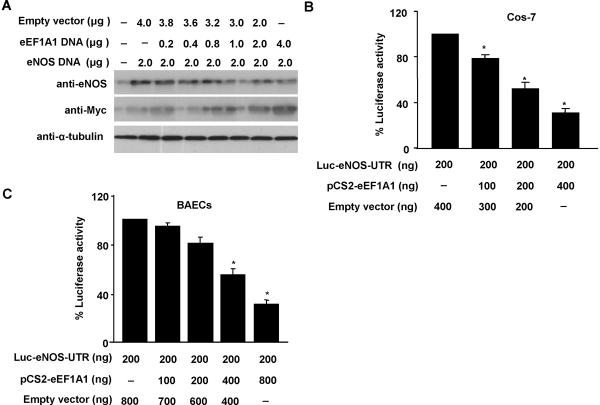
The interaction of eEF1A1 with the eNOS mRNA 3'-UTR prompted us to investigate whether eEF1A1 affects eNOS expression through regulating eNOS mRNA stability. To this end, Myc-tagged eEF1A1 cDNA, together with eNOS cDNA containing its 3'-UTR, were cotransfected into Cos-7 cells, and the expression of eNOS was determined by Western blotting analysis. In a dose dependent manner, cotransfection of eEF1A1 markedly inhibited eNOS expression in cotransfected Cos-7 cells (Figure 4A). In contrast, overexpression eEF1A1 had no effect on eNOS expression, when cotransfected with eNOS cDNA without its 3'-UTR sequence (data not shown). These results suggest that the binding of eEF1A1 to eNOS 3'-UTR may be functionally important for the regulation of eNOS expression.
Figure 4. Effect of eEF1A1 on the expression of eNOS.
(A) Cos-7 cells in 100 mm plate were transfected with eNOS cDNA containing 3'-UTR in combination of either empty vector or myctagged eEF1A1 cDNAs at the concentrations indicated. 48hr after transfection, cell lysates were collected for western blot analysis. (B) Cos-7 cells and (C) BAECs in 6-well plates were cotransfected with Luc-eNOS-UTR reporter plasmid together with either pCS2-Myc-eEF1A1 expression or empty vectors. 48hr after transfection, cell lysates were assayed for luciferase activities. n=5, * p<0.05 vs AdLacZ alone.
To further determine whether the inhibitory effect of eEF1A1 on eNOS expression in intact cells results from its binding to the 3'-UTR, we performed luciferase analyses. We generated reporter vector with the eNOS 3'-UTR cloned behind the firefly luciferase reporter gene. The luciferase reporter construct together with eEF1A1 expression vector or empty vector were transiently transfected into both Cos-7 cells and BAECs. In a concentration dependent manner, overexpression of eEF1A1 decreased luciferase activity in both Cos-7 cells and BAECs (Figure 4B and 4C). Together, these findings strongly suggest that the effect of eEF1A1 on eNOS mRNA is mediated by its binding to the eNOS 3'-UTR sequence.
Overexpression of eEF1A1 mimics destabilization of eNOS mRNA by TNF-α in HUVECs
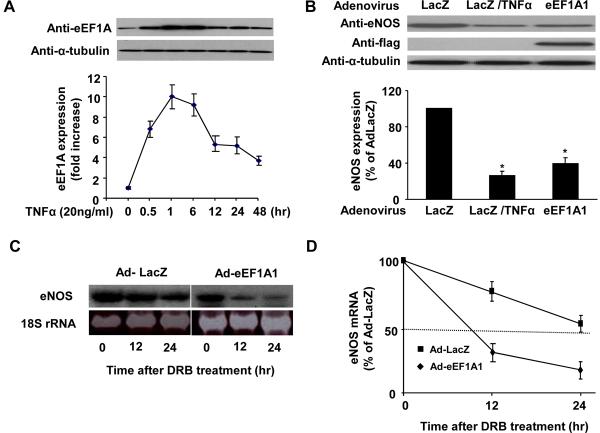
Because the binding of eEF1A1 to the eNOS 3'-UTR was significantly increased by TNF-α treatment in HUVECs (Figure 1A), we sought to determine the role of eEF1A1 in TNF-α induced destabilization of eNOS mRNA. In a time dependent manner, treatment of HUVECs with TNF-α (20 ng/ml) rapidly increased eEF1A expression, which is maximal after 1 hr and persists for up to 48 hr (Figure 5A). To determine whether overexpression of eEF1A1 can mimic the inhibitory effect of TNF-α on eNOS expression, preconfluent HUVECs were infected with AdeEF1A1 for 48 hours. The eNOS expression was then determined by Western blotting analysis. Indeed, overexpression of eEF1A1 markedly reduced eNOS protein expression by approximately 60% in HUVECs, compared to that observed with TNF-α treatment (i.e.75% reduction) (Figure 5B).
Figure 5. Overexpression of eEF1A1 mimics the downregulation of eNOS expression by TNF-α.
(A) TNF-α increases the expression of eEF1A in HUVECs. Cells were treated with TNF-α (20 ng/ml) for various time points and cell lysates were then collected for western blot analysis using anti-eEF1A and anti-α-tubulin antibodies. (B) Overexpression of Flag-tagged eEF1A1 inhibits the expression of eNOS in HUVECs. HUVECs were transduced with Ad-LacZ and AdeEF1A1 at the MOI of 100. 48 hr after transduction, cells were then treated with TNF-α (20 ng/ml) for 24 hr. The expression of eNOS was detected by western blot analysis and quantitated by densitometry. *p<0.05 vs Ad-LacZ. (C) Overexpression of eEF1A1 decreases eNOS mRNA stability. Preconfluent HUVECs were transduced with Ad-Lacz and Ad-eEF1A1 at the MOI of 100 for 48 hr. 20 μg/mL DRB was then added and eNOS mRNA half-life was determined by Northern blot analysis. A representative Northern blot is shown. (D) Combined results from 5 different experiments of eNOS mRNA half-life determination by Northern blot analysis.
The posttranscriptional regulation of eNOS mRNA by eEF1A1 was determined in the presence of the transcriptional inhibitor 5,6 -dichloro-1-β-D-ribofuranosylbenzimidazole (DRB) (20μg/ml). HUVECs were transduced with either AdeEF1A1 or AdLacZ for 48 h. DRB were then added to stop transcription, and total RNAs were prepared 0–24 hours thereafter. Relative eNOS mRNA amounts were performed by Northern blotting analysis. Overexpression of eEF1A1 in HUVECs decreased the half-life of eNOS mRNA from 29 ± 3 to 10 ± 2 hours (P< 0.05, n=5) (Figure 5C and 5D). Together, these results suggest that eEF1A1 is involved in the posttranscriptional regulation of eNOS mRNA stability.
eEF1A1 Knockdown prevents destabilization of eNOS mRNA by TNF-α
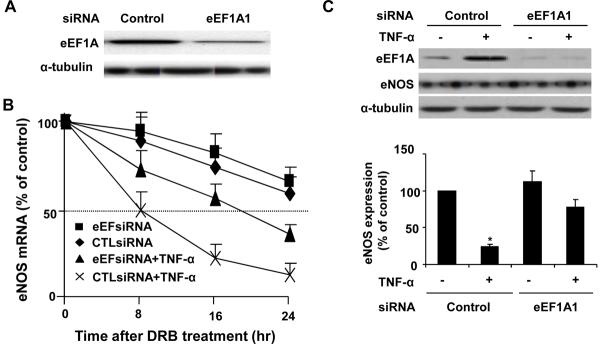
To further substantiate the functional significance of eEF1A1 in the regulation of eNOS mRNA stability, we performed loss of function studies, using the RNA interference technique. Transfection of eEF1A1 siRNA substantially inhibited eEF1A1 expression by approximately 78% (Figure 6A). Analysis of the half-life of eNOS mRNA in these cells showed that TNF-α induced destabilization of eNOS mRNA was clearly attenuated by eEF1A1 knockdown, as demonstrated in Figure 6B. Compare with control siRNA (CTLsiRNA), eEF1A specific siRNA (eEFsiRNA) modestly prolonged the half-life of eNOS mRNA under basal conditions. In HUVECs transfected with control siRNA, TNF-α (20 ng/mL) shortened the half-life of eNOS mRNA from 32±3 to 8±2 hours (P< 0.05, n=5). In contrast, knockdown of eEF1A1 by eEFsiRNA substantially blocked the decrease in eNOS mRNA half-life by TNF-α treatment. (8±2 to 19±2 hours, P< 0.05, n=5). To determine whether the modulation of eNOS mRNA half-life by eEFsiRNA corresponds to eNOS mRNA expression, we transfected HUVECs with control or eEF1A1 siRNA before stimulation with TNF-α and measured the protein levels of eEF1A1 and eNOS. eEF1A1 siRNA substantially prevented TNF-α-induced upregulation of eEF1A1 and subsequent downregulation of eNOS protein expression in HUVECs (Figure 6C). These results indicate that the regulation of eNOS mRNA stability and expression by TNF-α is specifically mediated, in part, by eEF1A1.
Figure 6. eEF1A1 knockdown prevents TNF-α-induced destabilization of eNOS mRNA stability.
(A) Western blotting showing the expression of eEF1A1 in HUVECs transfected with eEF1A1 specific small interference RNA (siRNA) and control siRNA. (B) Knockdown of eEF1A1 attenuates TNF-α induced destabilization of eNOS mRNA. HUVECs were transfected with eEF1A1 specific siRNA (eEFsiRNA) and control siRNA (CTLsiRNA). 72 hr after transfection, HUVECs were treated with TNF-α (20ng/ml) and eNOS mRNA half-life was determined by Northern blot analysis. The data are representative of 5 independent experiments. (C) Representative Western blots showing the effects of control and eEF1A1 siRNA on eEF1A and eNOS expression in the presence or absence of TNF-α (20 ng/ml). The data represents 3 independent experiments. *P<0.05 vs control siRNA alone or eEF1A siRNA + TNF-α.
Discussion
The regulation of eNOS occurs at multiple levels, including gene transcription, posttranscriptional and posttranslational mechanisms, and protein-protein interactions. In response to a variety of stimuli, including cytokines, lipopolysaccaride, and oxidized LDLs, the modulation of eNOS mRNA stability plays an important role in the regulation of eNOS expression 7. Previous studies suggest that the binding of cytosolic proteins to the cis-acting sequences within eNOS mRNA 3'-UTR was involved in the regulation of eNOS mRNA stability 10–13. However, the identity and function of these eNOS mRNA 3'-UTR binding proteins remain to be determined. In the present study, using RNA affinity chromatography to identify trans-acting factors that might regulate eNOS mRNA, we, for the first time, identified eEF1A1 as a cytosolic protein that binds to the eNOS 3'-UTR in HUVECs in response to TNF-α stimulation. The interaction of eEF1A1 with eNOS 3'-UTR was confirmed by immunocolocalization, EMSA, and UV cross-linking assays. Furthermore, adenovirus mediated overexpression of eEF1A1 and siRNA specific knockdown led to alterations in eNOS mRNA half-life that were consistent with eEF1A1 being part of destabilization mechanism for eNOS mRNA. These results suggest that eEF1A1 may be an important regulator of vascular function through its effects on eNOS expression.
eEF1A is an abundant and highly conserved protein in eukaryotic cells and is one of the most extensively characterized proteins of the translational machinery17 18. Two eEF1A isoforms, including eEF1A1 and eEF1A2, exist in humans, and these two proteins share 92% similarity 19. Because of its role in protein translation, eEF1A possesses a strong RNA binding activity. The canonical role of eEF1A during protein synthesis is to bind and transport aa-tRNA to the A site of the ribosome during translation in a GTP-dependent mechanism 20. During its normal cell functions, eEF1A interacts specifically with the other elongation complex proteins, tRNAs, and rRNA. Interestingly, eEF1A has been recently shown to bind to the 3-'UTR regions of several viruses, including tobacco mosaic virus, turnip yellow mosaic virus, dengue 4 virus, and west nile virus, and these RNA protein interactions may play important roles in virus growth or viral RNA synthesis21–24. At present, little is known about the function of eEF1A1 in vascular endothelial cells, besides its role in the translation. Our findings indicate that eEF1A1 is a novel eNOS 3'-UTR binding protein that mediates TNF-α induced destabilization of eNOS mRNA in HUVECs. Whether eEF1A2 interacts with eNOS 3'-UTR requires further investigation.
The mechanism by which eEF1A1 destabilizes eNOS mRNA remains elusive. Recent studies suggest that the anchoring of mRNAs to the actin cytoskeleton and their colocalization with ribosomes and RNA-binding protein complexes are necessary for their stability and translational expression25, 26. Importantly, the role of the endothelial actin cytoskeleton in the post-transcriptional stabilization of eNOS mRNA has been described 27 28. Hypoxia, oxidized LDL, and cytokines such as TNF-α have been shown to decrease eNOS expression by reducing eNOS mRNA stability 2, 29–31 13. In contrast, inhibition of Rho and actin cytoskeletal inhibitors have been shown to prolong eNOS mRNA half-life and prevent the downregulation of eNOS by oxidized LDL and TNF-α under hypoxic conditions 27, 30, 32–34. Moreover, the interaction of actin with eNOS 3'-UTR has been shown to play an essential role in the posttranscriptional regulation of eNOS during cell growth 28. These studies highlight the importance of actin and/or actin-binding proteins in the regulation of eNOS mRNA stability. Interestingly, beyond its conventional function in translation, eEF1A has been shown to play a key role in regulating cytoskeleton organization 16, 35. The association of eEF1A with the cytoskeleton has been well established across species from yeast to mammals16, 35–37. It has been suggested that >60% of eEF1A in cells is predicted to be associated with the actin cytoskeleton38. Because eEF1A has been shown to cross-link actin filaments with a unique bonding rule that excludes other proteins that cross-link F-actin 39, the resulting actin structure could be the scaffold for the transport and/or anchorage of mRNA 40. The binding of eEF1A to actin is now regarded as a function that may link two distinct cellular processes, cytoskeletal organization and gene expression 15. Thus, an intriguing possibility is that the association of eEF1A1 with eNOS mRNA 3'-UTR may affect eNOS mRNA stability by regulating the cytoskeletal-mediated eNOS mRNA transport, localization, and translation. In this regard, it would be interesting to investigate whether the dominant negative mutants of eEF1A1, such as domain III of eEF1A1, which has been shown to inhibit the interaction of eEF1A1 with F-actin 15, could attenuate TNF-α induced destabilization of eNOS mRNA.
Using RNA affinity chromatography with a riboprobe encoding the entire human eNOS 3'-UTR as a matrix, we showed that the formation of two RNPs, with molecular masses of 52 and 57 kDa, were upregulated by TNF-α. In this regard, our finding are consistent with a previous notion showing that the formation of the 53- and 56-kDa RNPs were upregulated in UV cross-linking assays of the human eNOS 3'-UTR with protein extracts from HUVECs treated with TNF-α13. However, the identity of the 53 and 56-kDa factors was unknown. In this study, we have identified the 52 kDa protein as eEF1A1 by mass spectrometric analysis. Since the expression of eEF1A was rapidly induced by TNF-α, the changes in eEF1A binding to eNOS 3'-UTR may be attributed to the relative abundance of eEF1A in endothelial cells. However, at this point, we can not exclude the possibility that the potential changes in the binding affinity, due to posttranslational modification, may also contribute to the increased binding of eEF1A to eNOS mRNA by TNF-α stimulation. Indeed, eEF1A has been shown to undergo phosphorylation and methylation 18 41. Interestingly, Rho kinase has been shown to phosphorylate eEF1A1 and regulate its binding to F-actin 42. Since TNF-α has been shown to activate Rho kinase in vascular endothelial cells 43 44 45, it remains to be determined whether Rho kinase mediated phosphorylation of eEF1A1 affects its interaction with eNOS 3'-UTR, thereby modulating the eNOS mRNA stability by TNF-α.
In summary, our study provides the first evidence to our knowledge that eEF1A1 is a novel cytosolic protein that binds to the 3'-UTR sequence of eNOS mRNA. This interaction is specific and leads to a reduction in eNOS mRNA stability and eNOS mRNA expression. Further studies, however, are required to determine the pathophysiological role of eEF1A1 in vascular disease. Nevertheless, since eEF1A1 also functions as an actin binding protein, these results provide a paradigm that links the control of the endothelial actin cytoskeleton to the regulation of gene expression, and suggest a therapeutic target that could improve endothelial function.
Supplementary Material
Acknowledgements
We thank Lauren Danridge for advice during the preparation of the manuscript.
Sources of Funding Section This work was supported by AHA Scientist Development Grant 0630047N (to J.S.), and NIH grants HL52233, HL70274 and DK62729 (to J.K.L.).
Footnotes
Disclosures None
References
- 1.Alderton WK, Cooper CE, Knowles RG. Nitric oxide synthases: structure, function and inhibition. Biochem J. 2001;357:593–615. doi: 10.1042/0264-6021:3570593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yoshizumi M, Perrella MA, Burnett JC, Jr., Lee ME. Tumor necrosis factor downregulates an endothelial nitric oxide synthase mRNA by shortening its half-life. Circ Res. 1993;73:205–209. doi: 10.1161/01.res.73.1.205. [DOI] [PubMed] [Google Scholar]
- 3.Liao JK, Shin WS, Lee WY, Clark SL. Oxidized low-density lipoprotein decreases the expression of endothelial nitric oxide synthase. J Biol Chem. 1995;270:319–324. doi: 10.1074/jbc.270.1.319. [DOI] [PubMed] [Google Scholar]
- 4.Endres M, Laufs U, Huang Z, Nakamura T, Huang P, Moskowitz MA, Liao JK. Stroke protection by 3-hydroxy-3-methylglutaryl (HMG)-CoA reductase inhibitors mediated by endothelial nitric oxide synthase. Proc Natl Acad Sci U S A. 1998;95:8880–8885. doi: 10.1073/pnas.95.15.8880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sumi D, Hayashi T, Jayachandran M, Iguchi A. Estrogen prevents destabilization of endothelial nitric oxide synthase mRNA induced by tumor necrosis factor alpha through estrogen receptor mediated system. Life Sci. 2001;69:1651–1660. doi: 10.1016/s0024-3205(01)01251-6. [DOI] [PubMed] [Google Scholar]
- 6.Drummond GR, Cai H, Davis ME, Ramasamy S, Harrison DG. Transcriptional and posttranscriptional regulation of endothelial nitric oxide synthase expression by hydrogen peroxide. Circ Res. 2000;86:347–354. doi: 10.1161/01.res.86.3.347. [DOI] [PubMed] [Google Scholar]
- 7.Searles CD. Transcriptional and posttranscriptional regulation of endothelial nitric oxide synthase expression. Am J Physiol Cell Physiol. 2006;291:C803–816. doi: 10.1152/ajpcell.00457.2005. [DOI] [PubMed] [Google Scholar]
- 8.Picchi A, Gao X, Belmadani S, Potter BJ, Focardi M, Chilian WM, Zhang C. Tumor necrosis factor-alpha induces endothelial dysfunction in the prediabetic metabolic syndrome. Circ Res. 2006;99:69–77. doi: 10.1161/01.RES.0000229685.37402.80. [DOI] [PubMed] [Google Scholar]
- 9.Agnoletti L, Curello S, Bachetti T, Malacarne F, Gaia G, Comini L, Volterrani M, Bonetti P, Parrinello G, Cadei M, Grigolato PG, Ferrari R. Serum from patients with severe heart failure downregulates eNOS and is proapoptotic: role of tumor necrosis factor-alpha. Circulation. 1999;100:1983–1991. doi: 10.1161/01.cir.100.19.1983. [DOI] [PubMed] [Google Scholar]
- 10.Sanchez de Miguel L, Alonso J, Gonzalez-Fernandez F, de la Osada J, Monton M, Rodriguez-Feo JA, Guerra JI, Arriero MM, Rico L, Casado S, Lopez-Farre A. Evidence that an endothelial cytosolic protein binds to the 3'-untranslated region of endothelial nitric oxide synthase mRNA. J Vasc Res. 1999;36:201–208. doi: 10.1159/000025643. [DOI] [PubMed] [Google Scholar]
- 11.Searles CD, Miwa Y, Harrison DG, Ramasamy S. Posttranscriptional regulation of endothelial nitric oxide synthase during cell growth. Circ Res. 1999;85:588–595. doi: 10.1161/01.res.85.7.588. [DOI] [PubMed] [Google Scholar]
- 12.Alonso J, Sanchez de Miguel L, Monton M, Casado S, Lopez-Farre A. Endothelial cytosolic proteins bind to the 3' untranslated region of endothelial nitric oxide synthase mRNA: regulation by tumor necrosis factor alpha. Mol Cell Biol. 1997;17:5719–5726. doi: 10.1128/mcb.17.10.5719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lai PF, Mohamed F, Monge JC, Stewart DJ. Downregulation of eNOS mRNA expression by TNFalpha: identification and functional characterization of RNA-protein interactions in the 3'UTR. Cardiovasc Res. 2003;59:160–168. doi: 10.1016/s0008-6363(03)00296-7. [DOI] [PubMed] [Google Scholar]
- 14.Jimenez A, Arriero MM, Lopez-Blaya A, Gonzalez-Fernandez F, Garcia R, Fortes J, Millas I, Velasco S, Sanchez De Miguel L, Rico L, Farre J, Casado S, Lopez-Farre A. Regulation of endothelial nitric oxide synthase expression in the vascular wall and in mononuclear cells from hypercholesterolemic rabbits. Circulation. 2001;104:1822–1830. doi: 10.1161/hc3901.095769. [DOI] [PubMed] [Google Scholar]
- 15.Liu G, Grant WM, Persky D, Latham VM, Jr., Singer RH, Condeelis J. Interactions of elongation factor 1alpha with F-actin and beta-actin mRNA: implications for anchoring mRNA in cell protrusions. Mol Biol Cell. 2002;13:579–592. doi: 10.1091/mbc.01-03-0140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gross SR, Kinzy TG. Translation elongation factor 1A is essential for regulation of the actin cytoskeleton and cell morphology. Nat Struct Mol Biol. 2005;12:772–778. doi: 10.1038/nsmb979. [DOI] [PubMed] [Google Scholar]
- 17.Negrutskii BS, El'skaya AV. Eukaryotic translation elongation factor 1 alpha: structure, expression, functions, and possible role in aminoacyl-tRNA channeling. Prog Nucleic Acid Res Mol Biol. 1998;60:47–78. doi: 10.1016/s0079-6603(08)60889-2. [DOI] [PubMed] [Google Scholar]
- 18.Lamberti A, Caraglia M, Longo O, Marra M, Abbruzzese A, Arcari P. The translation elongation factor 1A in tumorigenesis, signal transduction and apoptosis: review article. Amino Acids. 2004;26:443–448. doi: 10.1007/s00726-004-0088-2. [DOI] [PubMed] [Google Scholar]
- 19.Thornton S, Anand N, Purcell D, Lee J. Not just for housekeeping: protein initiation and elongation factors in cell growth and tumorigenesis. Journal of molecular medicine (Berlin, Germany) 2003;81:536–548. doi: 10.1007/s00109-003-0461-8. [DOI] [PubMed] [Google Scholar]
- 20.Carvalho MD, Carvalho JF, Merrick WC. Biological characterization of various forms of elongation factor 1 from rabbit reticulocytes. Arch Biochem Biophys. 1984;234:603–611. doi: 10.1016/0003-9861(84)90310-2. [DOI] [PubMed] [Google Scholar]
- 21.Davis WG, Blackwell JL, Shi PY, Brinton MA. Interaction between the cellular protein eEF1A and the 3'-terminal stem-loop of West Nile virus genomic RNA facilitates viral minus-strand RNA synthesis. J Virol. 2007;81:10172–10187. doi: 10.1128/JVI.00531-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yamaji Y, Kobayashi T, Hamada K, Sakurai K, Yoshii A, Suzuki M, Namba S, Hibi T. In vivo interaction between Tobacco mosaic virus RNA-dependent RNA polymerase and host translation elongation factor 1A. Virology. 2006;347:100–108. doi: 10.1016/j.virol.2005.11.031. [DOI] [PubMed] [Google Scholar]
- 23.Matsuda D, Yoshinari S, Dreher TW. eEF1A binding to aminoacylated viral RNA represses minus strand synthesis by TYMV RNA-dependent RNA polymerase. Virology. 2004;321:47–56. doi: 10.1016/j.virol.2003.10.028. [DOI] [PubMed] [Google Scholar]
- 24.Zeenko VV, Ryabova LA, Spirin AS, Rothnie HM, Hess D, Browning KS, Hohn T. Eukaryotic elongation factor 1A interacts with the upstream pseudoknot domain in the 3' untranslated region of tobacco mosaic virus RNA. J Virol. 2002;76:5678–5691. doi: 10.1128/JVI.76.11.5678-5691.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nasmyth K, Jansen RP. The cytoskeleton in mRNA localization and cell differentiation. Curr Opin Cell Biol. 1997;9:396–400. doi: 10.1016/s0955-0674(97)80013-0. [DOI] [PubMed] [Google Scholar]
- 26.Czaplinski K, Singer RH. Pathways for mRNA localization in the cytoplasm. Trends Biochem Sci. 2006;31:687–693. doi: 10.1016/j.tibs.2006.10.007. [DOI] [PubMed] [Google Scholar]
- 27.Laufs U, Endres M, Stagliano N, Amin-Hanjani S, Chui DS, Yang SX, Simoncini T, Yamada M, Rabkin E, Allen PG, Huang PL, Bohm M, Schoen FJ, Moskowitz MA, Liao JK. Neuroprotection mediated by changes in the endothelial actin cytoskeleton. J Clin Invest. 2000;106:15–24. doi: 10.1172/JCI9639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Searles CD, Ide L, Davis ME, Cai H, Weber M. Actin cytoskeleton organization and posttranscriptional regulation of endothelial nitric oxide synthase during cell growth. Circ Res. 2004;95:488–495. doi: 10.1161/01.RES.0000138953.21377.80. [DOI] [PubMed] [Google Scholar]
- 29.Zhang J, Patel JM, Li YD, Block ER. Proinflammatory cytokines downregulate gene expression and activity of constitutive nitric oxide synthase in porcine pulmonary artery endothelial cells. Res Commun Mol Pathol Pharmacol. 1997;96:71–87. [PubMed] [Google Scholar]
- 30.Takemoto M, Sun J, Hiroki J, Shimokawa H, Liao JK. Rho-kinase mediates hypoxia-induced downregulation of endothelial nitric oxide synthase. Circulation. 2002;106:57–62. doi: 10.1161/01.cir.0000020682.73694.ab. [DOI] [PubMed] [Google Scholar]
- 31.Cardaropoli S, Silvagno F, Morra E, Pescarmona GP, Todros T. Infectious and inflammatory stimuli decrease endothelial nitric oxide synthase activity in vitro. J Hypertens. 2003;21:2103–2110. doi: 10.1097/00004872-200311000-00020. [DOI] [PubMed] [Google Scholar]
- 32.Laufs U, Liao JK. Post-transcriptional regulation of endothelial nitric oxide synthase mRNA stability by Rho GTPase. J Biol Chem. 1998;273:24266–24271. doi: 10.1074/jbc.273.37.24266. [DOI] [PubMed] [Google Scholar]
- 33.Takeuchi S, Kawashima S, Rikitake Y, Ueyama T, Inoue N, Hirata K, Yokoyama M. Cerivastatin suppresses lipopolysaccharide-induced ICAM-1 expression through inhibition of Rho GTPase in BAEC. Biochem Biophys Res Commun. 2000;269:97–102. doi: 10.1006/bbrc.2000.2238. [DOI] [PubMed] [Google Scholar]
- 34.Laufs U. Beyond lipid-lowering: effects of statins on endothelial nitric oxide. Eur J Clin Pharmacol. 2003;58:719–731. doi: 10.1007/s00228-002-0556-0. [DOI] [PubMed] [Google Scholar]
- 35.Munshi R, Kandl KA, Carr-Schmid A, Whitacre JL, Adams AE, Kinzy TG. Overexpression of translation elongation factor 1A affects the organization and function of the actin cytoskeleton in yeast. Genetics. 2001;157:1425–1436. doi: 10.1093/genetics/157.4.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Edmonds BT, Wyckoff J, Yeung YG, Wang Y, Stanley ER, Jones J, Segall J, Condeelis J. Elongation factor-1 alpha is an overexpressed actin binding protein in metastatic rat mammary adenocarcinoma. J Cell Sci. 1996;109:2705–2714. doi: 10.1242/jcs.109.11.2705. [DOI] [PubMed] [Google Scholar]
- 37.Murray JW, Edmonds BT, Liu G, Condeelis J. Bundling of actin filaments by elongation factor 1 alpha inhibits polymerization at filament ends. J Cell Biol. 1996;135:1309–1321. doi: 10.1083/jcb.135.5.1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Edmonds BT, Murray J, Condeelis J. pH regulation of the F-actin binding properties of Dictyostelium elongation factor 1 alpha. J Biol Chem. 1995;270:15222–15230. doi: 10.1074/jbc.270.25.15222. [DOI] [PubMed] [Google Scholar]
- 39.Owen CH, DeRosier DJ, Condeelis J. Actin crosslinking protein EF-1a of Dictyostelium discoideum has a unique bonding rule that allows square-packed bundles. J Struct Biol. 1992;109:248–254. doi: 10.1016/1047-8477(92)90037-b. [DOI] [PubMed] [Google Scholar]
- 40.Liu G, Edmonds BT, Condeelis J. pH, EF-1alpha and the cytoskeleton. Trends Cell Biol. 1996;6:168–171. doi: 10.1016/0962-8924(96)20013-3. [DOI] [PubMed] [Google Scholar]
- 41.Ejiri S. Moonlighting functions of polypeptide elongation factor 1: from actin bundling to zinc finger protein R1-associated nuclear localization. Bioscience, biotechnology, and biochemistry. 2002;66:1–21. doi: 10.1271/bbb.66.1. [DOI] [PubMed] [Google Scholar]
- 42.Izawa T, Fukata Y, Kimura T, Iwamatsu A, Dohi K, Kaibuchi K. Elongation factor-1 alpha is a novel substrate of rho-associated kinase. Biochem Biophys Res Commun. 2000;278:72–78. doi: 10.1006/bbrc.2000.3772. [DOI] [PubMed] [Google Scholar]
- 43.Mong PY, Petrulio C, Kaufman HL, Wang Q. Activation of Rho kinase by TNF-alpha is required for JNK activation in human pulmonary microvascular endothelial cells. J Immunol. 2008;180:550–558. doi: 10.4049/jimmunol.180.1.550. [DOI] [PubMed] [Google Scholar]
- 44.McKenzie JA, Ridley AJ. Roles of Rho/ROCK and MLCK in TNF-alpha-induced changes in endothelial morphology and permeability. Journal of cellular physiology. 2007;213:221–228. doi: 10.1002/jcp.21114. [DOI] [PubMed] [Google Scholar]
- 45.Kishore R, Qin G, Luedemann C, Bord E, Hanley A, Silver M, Gavin M, Yoon YS, Goukassian D, Losordo DW. The cytoskeletal protein ezrin regulates EC proliferation and angiogenesis via TNF-alpha-induced transcriptional repression of cyclin A. J Clin Invest. 2005;115:1785–1796. doi: 10.1172/JCI22849. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.