Abstract
Toll-like receptor 9 (TLR9) promiscuously binds self and microbial DNA, but only microbial DNA elicits an inflammatory response. How TLR9 discriminates between self and foreign DNA is unclear, but inappropriate localization of TLR9 permits response to self DNA, suggesting that TLR9 localization and trafficking are critical components. The molecular mechanisms controlling the movement of TLR9 may provide new insight into the recognition of DNA in normal and in pathological conditions such as autoimmune systemic lupus erythematosus. We previously showed that TLR9 is retained in the endoplasmic reticulum (ER) and it moves to endolysosomes to recognize CpG DNA. Other studies have suggested that TLR9 bypasses the Golgi complex to access endolysosomes. Here, we demonstrate that TLR9 translocates from ER to endolysosomes via the Golgi complex and that Golgi export is required for optimal TLR9 signaling. Six to thirteen percent of TLR9 constitutively exits the ER, moves through the Golgi complex and resides in LAMP-1 positive vesicles. TLR9 bound to CpG DNA had glycan modifications indicative of Golgi processing confirming that TLR9 travels through the Golgi complex to access CpG DNA in endolysosomes. Together, these data support a model where TLR9 uses traditional secretory pathways and does not bypass the Golgi complex.
Keywords: Brefeldin A, CpG DNA, Endolysosome, Golgi, TLR9, trafficking
INTRODUCTION
Toll-like receptors (TLRs) are innate immune receptors important for host defense against pathogens1. While many TLRs are present at the cell surface, those dedicated to recognition of various nucleic acid structures are retained in intracellular compartments. Nucleic acid structures recognized by TLRs include dsRNA by TLR32, single-stranded RNA by TLR7 and 83 and CpG DNA by TLR94, 5. Notably, these structures are also present in self nucleic acids.
Recognition of DNA by TLR9 occurs in endolysosomes, but TLR9 resides predominantly in the endoplasmic reticulum (ER) in resting cells5–9. Endolysosomal localization is important for TLR9 response to CpG DNA since TLR9 binds some DNA preferentially at low pH, endosomal acidification inhibitors block TLR9 signaling and TLR9 undergoes a conformational change upon CpG DNA binding in endolysosomes10–12. CpG DNAs with different sequence and secondary structure can elicit either production of type I Interferon (IFN-α) or maturation of plasmacytoid dendritic cells (pDCs) depending on the precise location of interaction with TLR9. Those CpG DNAs that induce IFN-α production accumulate in early endosomes while those that induce maturation accumulate in lysosomes13. The mechanism regulating TLR9 translocation from the ER to these different compartments remains unclear.
Intracellular localization of TLRs 3, 7, 8 and 9 may be a key factor to prevent recognition of self nucleic acids. Self nucleic acids are poorly endocytosed and rapidly degraded in the extra-cellular milieu. However, self DNA is recognized by TLR9 when it is presented by chromatin specific autoantibodies or the microbial peptide LL37, or when TLR9 is inappropriately localized. The inappropriate activation of TLR9 by self DNA results in the onset of a severe inflammatory response14–16. Therefore, it is critical to uncover the mechanisms regulating TLR9 localization and trafficking.
Since recent studies have proposed that TLR9 bypasses the Golgi complex during translocation from the ER to early endosomes7, 17, we investigated the importance of Golgi export in TLR9 movement using newly developed assays and traditional biochemical methods. Our results show that a pool of TLR9 constitutively traffics from the ER through the Golgi complex and resides in endolysosomes and that this pool of TLR9 is likely to be involved in signaling. Furthermore, transit of TLR9 through the Golgi complex occurs in response to CpG DNA and is required for optimal signaling. These data shed new light on the pathways of TLR9 movement within the cell. Control of TLR9 movement along these pathways likely regulates discrimination between self and non-self DNA.
RESULTS
TLR9 signaling requires Golgi transport
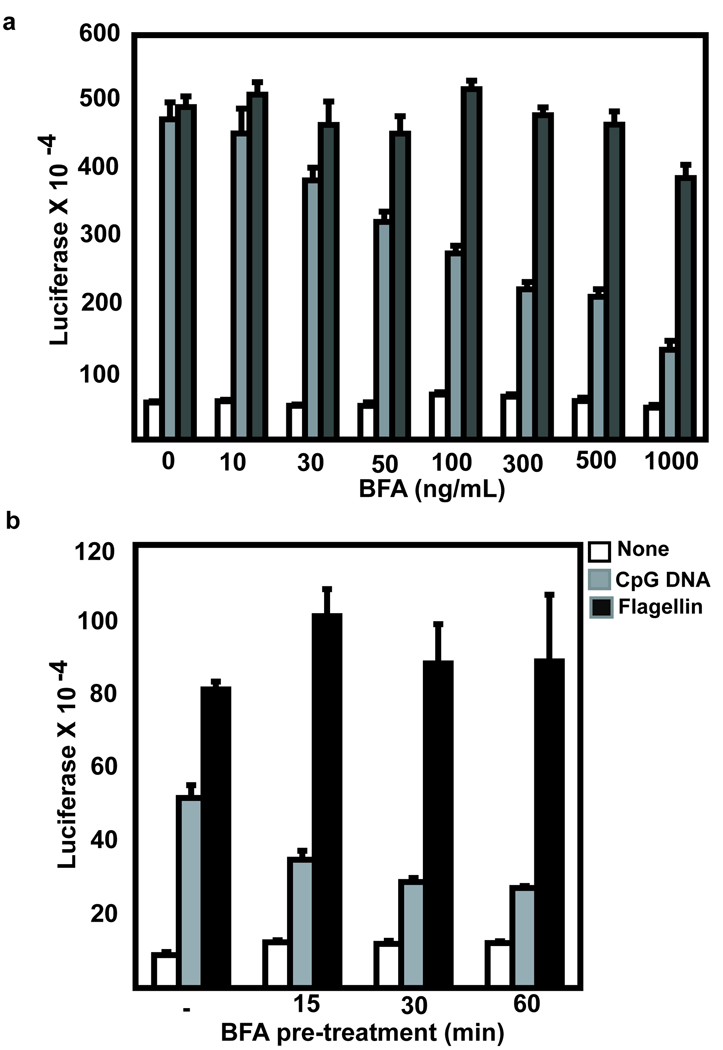
The observation that TLR9 remains sensitive to digestion with endoglycosidase H (EndoH) following CpG DNA stimulation7 (Supplementary Figure 1) has led to a model where TLR9 bypasses the Golgi complex when it translocates from the ER to endolysosomes. However, sensitivity to EndoH digestion is not a definitive marker for ER localization18. EndoH digests both high mannose glycans (found on proteins in the ER) and hybrid glycan modifications (found on proteins in the Golgi complex). To directly test if TLR9 bypasses the Golgi complex, we blocked Golgi export using the fungal metabolite Brefeldin A (BFA). BFA blocks Golgi export by preventing ADP ribosylation factor (ARF) association with the Golgi membrane and the formation of the coatamer complex19–21. Pre-treatment with BFA significantly inhibited TLR9-induced NF-κB activation in a dose dependent manner (Figure 1a). The inhibitory effect of BFA was significant with 15 minutes pre-treatment, an effect that was slightly greater with longer pre-treatment (Figure 1b). Some TLRs, such as TLR2, TLR4 and TLR5 constitutively traffic to and signal from the cell surface6, 9, 22–25. These TLRs should not be inhibited by BFA since they do not depend on Golgi export to initiate signal transduction. TLR5 signaling was not inhibited by pretreatment with BFA, supporting the conclusion that the inhibitory effect on TLR9 signaling was not due to toxic effects (Figure 1a and 1b). Incubation of a human B cell line with fluorescently labeled CpG DNA resulted in binding and endocytosis as measured by an increase in mean fluorescence intensity (MFI) (3.9 with no CpG DNA, binding: 24.3 with labeled CpG DNA at 4°C, and endocytosis: 83.7 with CpG DNA at 37°C) (Supplementary Figure 2). Uptake of labeled CpG DNA was not decreased, but slightly increased (97.9 MFI) upon BFA treatment indicating that the mechanism of inhibition of TLR9 signaling was not due to inhibition of CpG DNA endocytosis (Supplementary Figure 2). Together these data indicate that trafficking through the Golgi complex is required for optimal TLR9 signaling.
Figure 1. Brefeldin A inhibits TLR9 induced NF-κB activity.
(a) HEK293 cells were transfected with TLR9 and pre-treated with the indicated concentration of BFA for 1 hour prior to overnight stimulation with 1 µg/ml CpG DNA or 100 ng/ml flagellin. (Error bars=mean+/−standard deviation of triplicate samples) Data are representative of three experiments. (b) Similar to (a) except that the cells were pre-treated for the indicated time period with 30 ng/mL BFA. (Error bars=mean+/−standard deviation of triplicate samples) Data are representative of three experiments.
TLR9 constitutively traffics through the Golgi complex
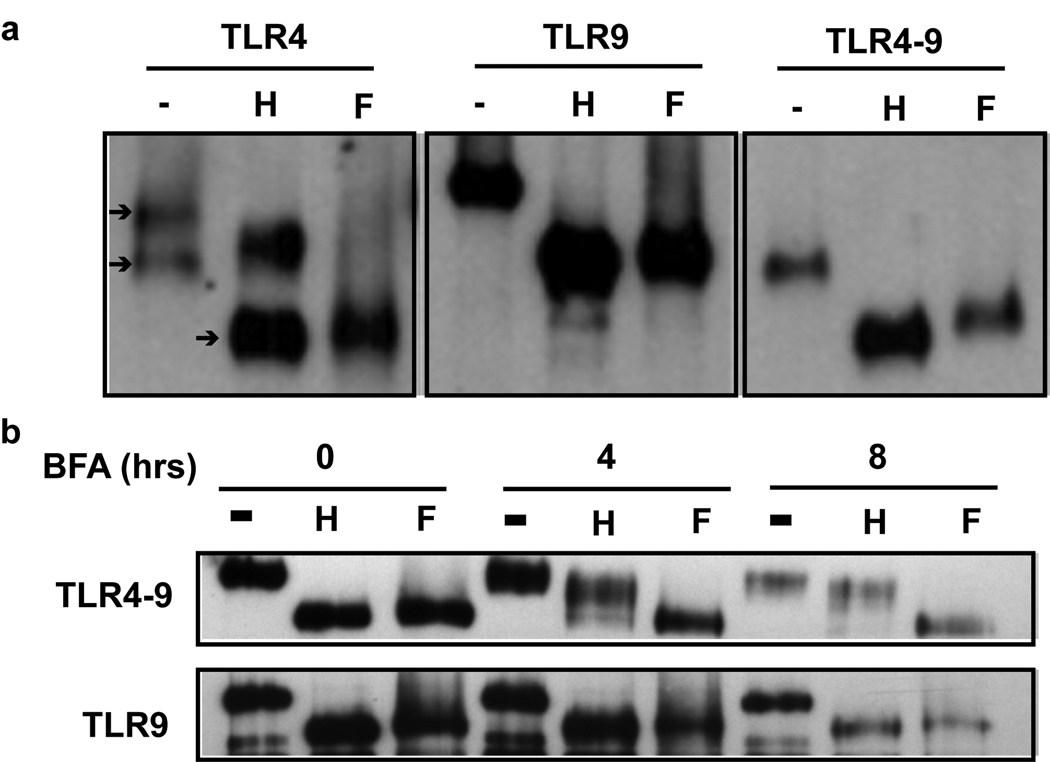
Since we showed that Golgi export is required for TLR9 signaling, we could not explain why TLR9 did not become EndoH resistant following CpG DNA stimulation. We next determined whether TLR9 became resistant to EndoH digestion following BFA treatment. If TLR9 normally remains sensitive to EndoH digestion because it bypasses the Golgi complex, thereby evading the Golgi-resident glycosylation modifying enzymes, then treatment with BFA will change TLR9 glycosylations to complex forms and the protein will become resistant to EndoH digestion. This is because BFA treatment will force co-localization of TLR9 with the Golgi contents. As expected in untreated cells, TLR9 was reduced to the unglycosylated molecular weight by treatment with EndoH digestion or with PNGase F (Figure 2a and 2b)8. TLR4 is synthesized in the ER, transits through the Golgi complex and resides at the cell surface. Therefore, TLR4 normally runs as a doublet, the upper band representing the surface expressed, EndoH resistant form and the lower band representing the immature, EndoH sensitive form (Figure 2a)26. When treated with PNGase F, all of TLR4 was reduced to the unglycosylated molecular weight (Figure 2a). A chimeric fusion protein between the ecto-domain of TLR4 and the transmembrane and cytoplasmic tail of TLR9 (TLR4-9) has been previously shown to localize similarly to TLR98. The TLR4-9 chimera was sensitive to EndoH digestion, but became resistant to EndoH digestion within four hours of treatment with BFA and was completely resistant to digestion by eight hours (Figure 2a and 2b). In contrast, TLR9 remained sensitive to EndoH digestion following treatment with BFA for up to eight hours, the half-life of TLR9 (Figure 2b)9. These data indicate that EndoH sensitivity is not an accurate indicator of TLR9 localization. Despite forced localization with enzymes capable of inducing maturation of the N-linked carbohydrates, the glycosylations on TLR9 remained sensitive to EndoH digestion.
Figure 2. TLR4-9 but not TLR9 is resistant to digestion with EndoH upon treatment with BFA.
(a) GFP immunoprecipitates from HeLa cells transfected with TLR4-GFP, TLR4-9-GFP or TLR9-GFP were either untreated (−) or treated with either EndoH (H), or PNGase F (F) prior to immunoblotting for GFP. Arrows indicate the mature (top), immature (middle) and unglycosylated (lower) glycoforms of TLR4. Data are representative of three experiments. (b) As in (a) except cells were pre-treated with 30 ng/ml Brefeldin A for the indicated time prior to cell lysis. Data are representative of three experiments.
We next developed alternative biochemical assays to detect movement of TLR9 within the cell. Lectins are plant proteins that have high specificity for recognition of carbohydrate structures. Galanthus nivalis (GN) lectin recognizes high mannose glycans found on proteins modified in the ER and hybrid glycans on proteins minimally processed in the Golgi complex. Biotinylated GN lectin bound to TLR9 and to the lower, EndoH sensitive, band of TLR4 in lysates from cells transfected with GFP-tagged TLR9 and TLR4 (Figure 3a). This demonstrated that each contained high mannose or hybrid glycans. Partially degraded TLR9 (lower band in TLR9 lane) also bound to GN lectin (Figure 3a). Importantly, the bands detected upon incubation with the biotinylated lectin were not due to incomplete stripping of the blot since no signal was detected when the stripped blot was developed with enhanced chemiluminescent reagent and exposed to radiographic film. Datura stramonium (DS) lectin specifically recognizes “Galβ1→4GlcNac” structures present on both hybrid and complex glycans, modifications only found on proteins that have moved into the Golgi complex. Biotinylated DS lectin bound to both bands of TLR4 since they represent the hybrid glycosylated and complex/mature glycosylated forms. Biotinylated DS lectin also bound to TLR9 (Figure 3a) suggesting that TLR9’s glycosylations had been processed in the Golgi. This was not an artifact of overexpression of tagged TLR9 since endogenous TLR9 also bound to DS lectin (Figure 3b). DS lectin did not bind to BSA that lacks canonical N- and O-linked glycosylation sites, indicating that binding was specific (Supplementary Figure 3). Also, DS lectin did not bind to the lower molecular weight form of TLR9, which is likely a degraded form that is generated by partial cleavage of the glycosylated ecto-domain (Figure 3a). To determine whether the carbohydrate modifications on TLR9 were hybrid (i.e. able to bind DS lectin, but EndoH sensitive), we treated TLR9 with PNGase F or EndoH prior to blotting with biotinylated lectins. PNGase F digests all N-linked glycan residues and, as expected, treatment of either TLR9 or TLR4 eliminated binding of both GN and DS lectin (Figure 3c). EndoH treatment eliminated GN lectin binding to both TLRs, since the binding specificity of GN lectin correlates with the specificity for glycosidase activity of EndoH as it cleaves high mannose and hybrid glycans and not complex glycans (Figure 3c). However, EndoH digestion prevented DS lectin binding to TLR9, but not to the upper, EndoH resistant, TLR4 band (Figure 3c). Therefore, TLR4 contained complex/mature (DS lectin binding, EndoH resistant) glycosylations while TLR9 contained hybrid (DS lectin binding, EndoH sensitive) glycosylations. The hybrid glycan modifications on TLR9 demonstrate that it had reached the Golgi complex despite remaining sensitive to EndoH digestion.
Figure 3. Biotinylated DS lectin blotting indicates TLR9 glycans are modified in the Golgi complex.
(a) GFP immunoprecipitates from HeLa cells transfected with TLR9-GFP, TLR4-YFP or pEGFP empty vector (V) were probed for GFP. The blot was stripped and probed with biotinylated GN or DS lectin. Arrowheads indicate full length TLR9 and small arrows indicate the upper and lower glycoforms of TLR4. Data are representative of four experiments. (b) Either BJAB cells (left lane), or as a positive control, HeLa cells transfected with TLR9-YFP (Right lane), were lysed and TLR9 immunoprecipitates were immunoblotted for TLR9. The blot was subsequently stripped and probed with biotinylated DS lectin. Data are representative of three experiments. (c) GFP immunoprecipitates from HEK293 cells stably transfected with TLR9YFP or TLR4GFP were left untreated (−) or treated with EndoH (H), or PNGase F (F) prior to immunoblotting with GFP (lower blots). The blots were stripped and probed with biotinylated GN or DS lectin (upper blots). Arrow(head)s are the same as in (a). Data are representative of four experiments.
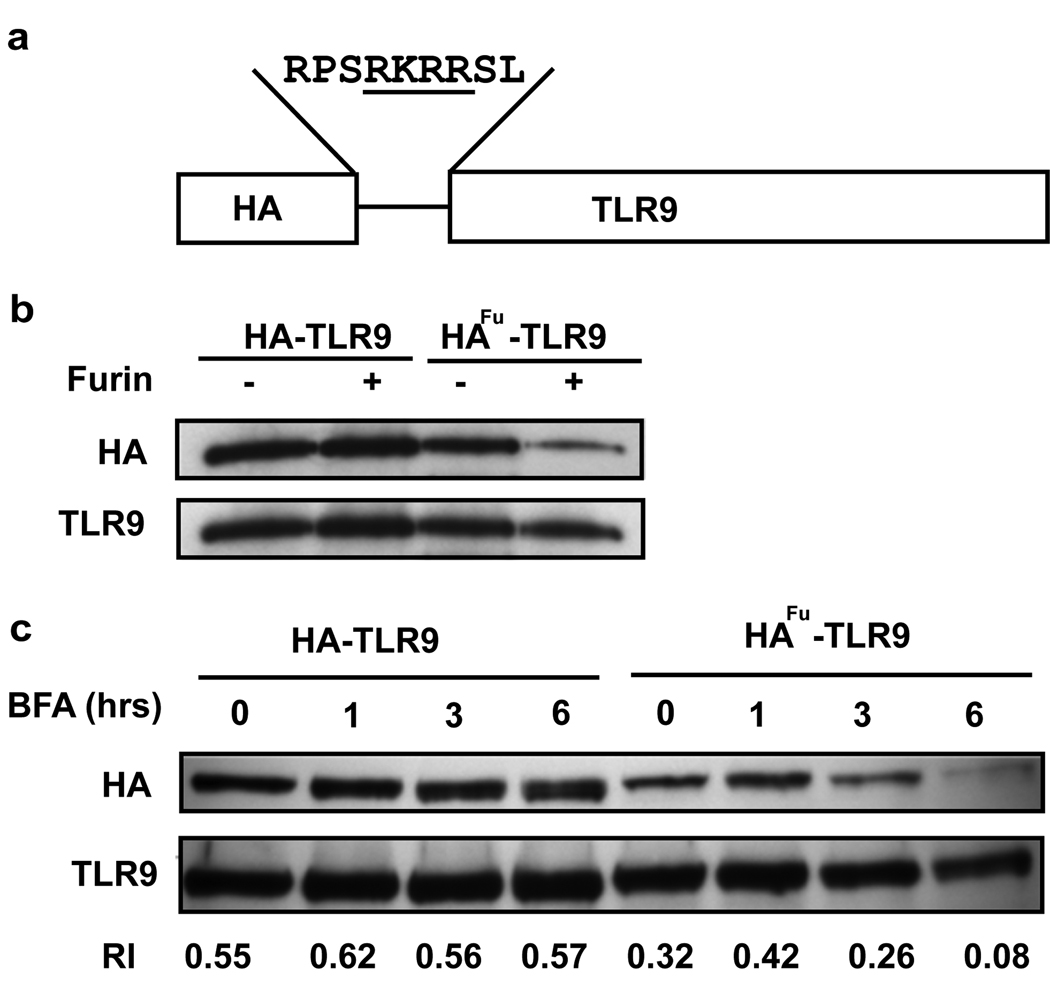
To confirm the importance of Golgi export in TLR9 movement, we developed a furin protease tag cleavage assay. Furin is a protease present in the trans-Golgi network and at the cell surface and cleaves at RXRR sequences (R=arginine, X=amino acid)27, 28. A furin cleavage site was engineered into HA-tagged TLR9 (HAfu-TLR9) such that the HA tag is cleaved from TLR9 when it is exposed to furin enzyme, thereby identifying the pool of TLR9 that has transited through the Golgi complex (Figure 4a). HA-tagged TLR9 and HAfu-TLR9 retained their activity when tested by NF-κB luciferase reporter assay (Brooks J, 2007 unpubl. data). In vitro incubation with recombinant furin resulted in loss of the HA tag from the HAfu-TLR9 protein, but not from the HA-TLR9 protein, confirming that the engineered cleavage site was functional (Figure 4b). In transfected cells, treatment with BFA induced a time-dependent loss of the HA tag from HAfu-TLR9 but not HA-TLR9 when compared to total TLR9 levels, demonstrating that furin cleavage occurred in cells (Figure 4c). When the bands were analyzed by densitometry, the relative intensity (RI) of the HA tag to TLR9 from HA-TLR9 cells was constant over six hours treatment with BFA (0.55 to 0.62). In contrast, the RI of the HA tag to TLR9 in HAfu-TLR9 cells decreased dramatically over the six hour treatment with BFA (0.32 RI down to 0.08 RI). Therefore the HA tag from HAfu-TLR9 was cleaved inside cells by endogenous furin. Consistent with our ability to detect DS lectin binding to TLR9, the basal level of the HA tag on HAfu-TLR9 was lower than on HA-TLR9 when compared to total TLR9 levels (0.55 RI for HA-TLR9 versus 0.32 RI for HAfu-TLR9) (Figure 4c). This suggested that a pool of HAfu-TLR9 constitutively translocates through the Golgi complex and is processed by the endogenous furin enzyme. Taken together, our studies using BFA treatment, lectin blotting and the furin cleavage assay support a model where TLR9 constitutively traffics to the Golgi complex and where export from the Golgi complex is required for optimal signaling.
Figure 4. HA-TLR9 furin cleavage assay.
(a) Schematic representation of the engineered furin cleavage site in HA-TLR9. (b) HA immunoprecipitates from HeLa cells transfected with HA-TLR9 or HAfu-TLR9 were treated with recombinant furin in-vitro for 2 hrs prior to immunoblotting for HA and TLR9. Data are representative of two experiments. (c) HeLa cells transfected with HA-TLR9 and HAfu-TLR9 were pretreated with BFA for the indicated times prior to immunoprecipitation with TLR9 and immunoblotting for HA and TLR9. Relative intensities (RI) were calculated as described in the methods. Data are representative of three experiments.
A pool of TLR9 constitutively localizes in endolysosomes
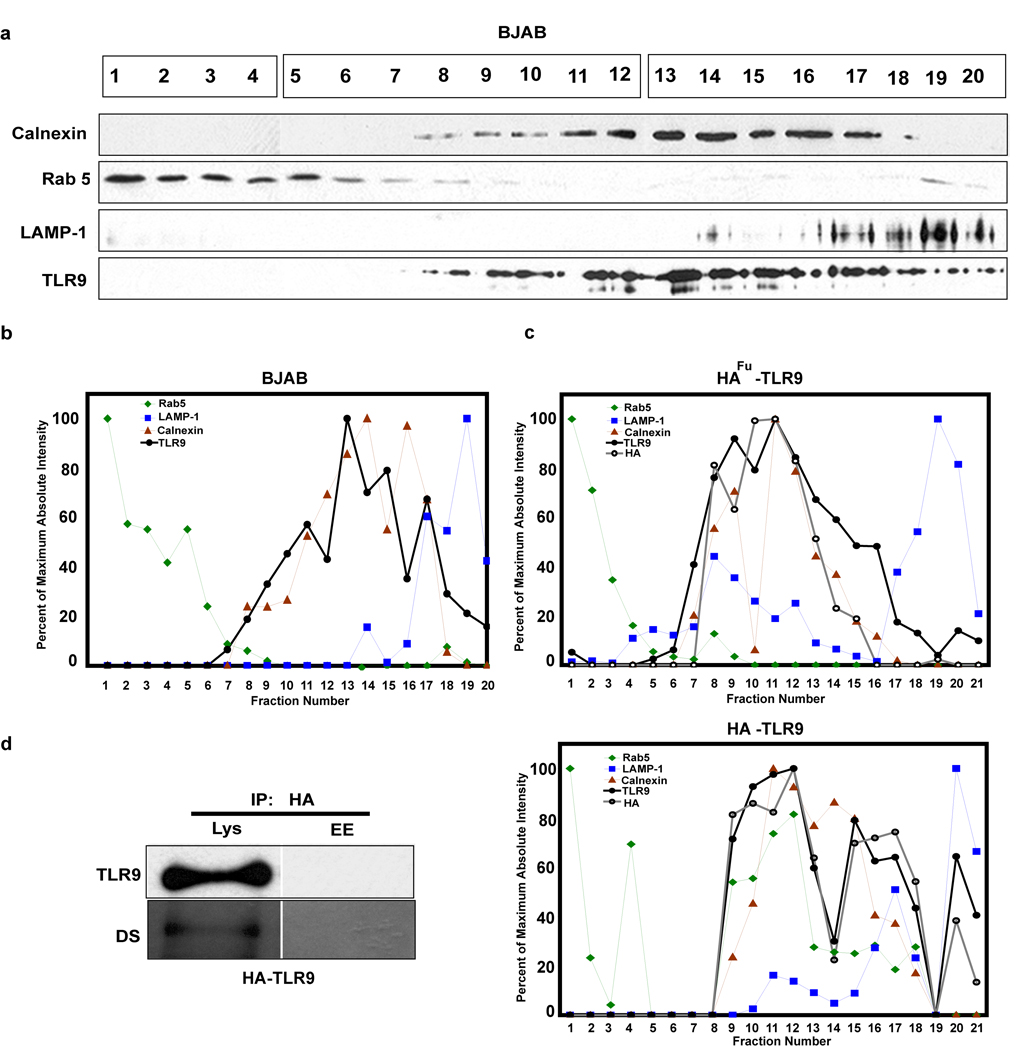
We next asked where the TLR9 that constitutively exits the ER is localized. Centrifugation of cell homogenates on Percoll-sucrose, self-forming, gradients allowed separation of Rab 5 positive early endosomes, GM130 positive Golgi complex, calnexin positive ER and LAMP-1 positive lysosomes (Figure 5a, b, c and Supplementary Figure 4). In unstimulated cells, both endogenous (Figure 5a and 5b) and stably expressed (Figure 5c) TLR9 were detected in the ER fractions, as expected. However, 5.96 percent of the total endogenous TLR9 was detected in the LAMP-1 positive fractions. Similarly, in cells stably expressing HA-TLR9, 13.82 percent of total TLR9 was detected in the LAMP-1 positive fractions.
Figure 5. TLR9 trafficking to endolysosomes requires Golgi transport.
(a) Fractions from BJAB cells separated on Percoll-sucrose gradients were immunoblotted for early endosomes (Rab 5), ER (Calnexin), lysosomes (LAMP-1) and TLR9. Fractions were run on three gels, which are indicated with boxes over the fraction numbers, but were immunoblotted and developed simultaneously. (b) Each immunoblot in (a) was quantified to determine absolute intensity. Percent of maximum values were calculated from the absolute intensities and graphed on the same graph to normalize the data. Data are representative of three experiments. (c) Similar to (b) except that the cells were HEK293 cells stably expressing HAfu-TLR9 (upper) or HA-TLR9 (lower) and were additionally immunoblotted for HA. Data are representative of three experiments. (d) HA immunoprecipitates from combined fractions that were LAMP-1 (Lys, fractions 17–20) or Rab 5 (EE, fractions 1–4) positive collected from HA-TLR9 cells were immunoblotted for TLR9, stripped and probed with biotinylated DS lectin. Data are representative of two experiments.
To determine if the constitutive movement of TLR9 to the endolysosomal compartment requires transit through the Golgi complex, cells stably expressing HAfu-TLR9 or HA-TLR9 lacking the furin protease cleavage site were fractionated and tested for loss of the HA tag (indicating cleavage by furin in the Golgi complex). TLR9 was present both in the ER and lysosomal fractions from both HAfu-TLR9 and HA-TLR9 expressing cells (Figure 5c). More than 6.7 percent of the HA tag was detected in lysosomal fractions of HA-TLR9 expressing cells, but less than 0.5 percent of the HA tag was detected in those same fractions from cells expressing HAfu-TLR9 containing the furin cleavage site (Figure 5c). Therefore, TLR9 constitutively localizes in the LAMP-1 positive fractions and arrives there via the Golgi complex. DS lectin bound to HA-tagged and endogenous TLR9 from LAMP-1 positive fractions indicating that this pool of TLR9 had transited through the Golgi complex (Figure 5d and Supplementary Figure 5). TLR9 was not detected, even by immunoprecipitation, in early endosomal fractions of unstimulated cells (Figure 5d). The level of DS lectin binding to endogenous TLR9 in lysosomes was similar to DS lectin binding to total TLR9 suggesting that the lysosomal pool of TLR9 accounted for most, if not all of the TLR9 in the cell that had transited through the Golgi complex (Supplementary Figure 5). The pool of LAMP-1 positive fractions was not contaminated with ER or early endosomes, since it was devoid of calnexin and Rab 5 (Chockalingam A, 2007 unpubl. data). We conclude that a small pool of TLR9 constitutively translocates from the ER, through the Golgi complex and resides in a LAMP-1 positive compartment.
MyD88 constitutively binds to endolysosomal TLR9 that has transited through the Golgi complex
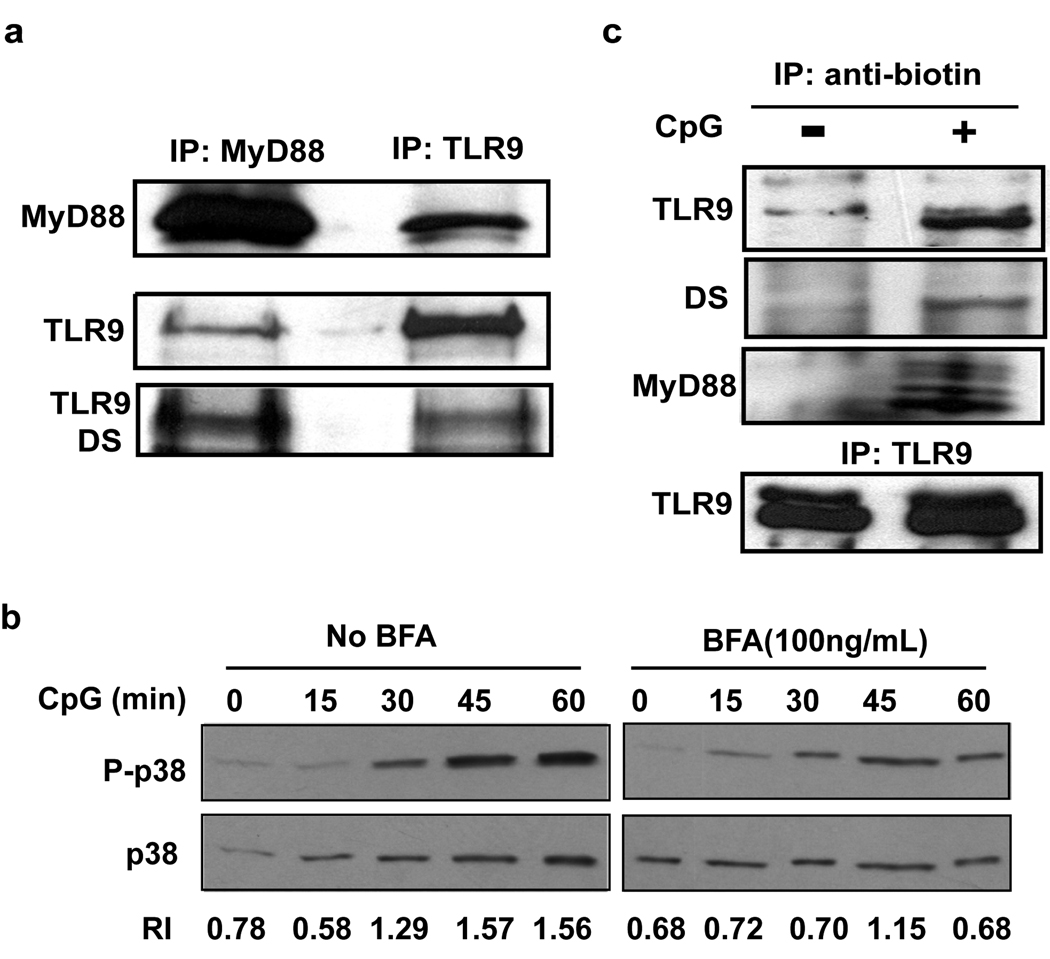
MyD88 is an essential adapter protein that initiates TLR9 signaling in response to CpG DNA4, 29. Previous studies have demonstrated ligand-dependent recruitment of MyD88 to exogenously expressed and tagged TLR9 30, 31. Next we wanted to detect the glycosylation pattern of TLR9 associated with MyD88 in response to CpG DNA in order to determine if the TLR9 that initiates signaling also traffics through the Golgi complex. Unexpectedly, endogenous TLR9 co-immunoprecipitated with endogenous MyD88 in unstimulated cells (Figure 6a). No further increase in MyD88 association was detected upon exposure to CpG DNA (Supplementary Figure 6). The pool of TLR9 that associated with MyD88 also bound to DS lectin indicating that it had transited through the Golgi complex (Figure 6a). MyD88 also co-immunoprecipitated only TLR9 lacking the HA tag from cells expressing HAfu-TLR9 (Chockalingam A, 2008 unpubl. data). We conclude that a pool of TLR9 is constitutively associated with MyD88 and that this pool has exited the ER since it has biochemical signatures of Golgi processing. Given our previous findings, we infer that this MyD88 associated pool of TLR9 is located in the lysosomal compartment although we could not immunoprecipitate sufficient amounts of TLR9 or MyD88 from lysosomal fractions to determine co-association.
Figure 6. The TLR9 that initiates signaling has trafficked through the Golgi complex.
(a) MyD88 or TLR9 immunoprecipitates from BJAB cells were immunoblotted for MyD88 and TLR9, stripped and the TLR9 portion of the blot was probed with biotinylated DS lectin. Data are representative of three experiments. (b) BJAB cells were pretreated with media or 100 ng/ml BFA for 2 hours prior to stimulation with 5 µg/ml CpG DNA 2006 for the indicated times. Whole cell lysates were immunoblotted for phospho-p38 (P-p38) and total p38 (p38). Relative intensities were calculated as described in the methods. Data are representative of two experiments. (c) BJAB cells were treated with media or 3’ biotinylated CpG 2006 for 15 minutes, lysed and immunoprecipitated for biotin or TLR9 prior to immunoblotting for MyD88 and TLR9. The TLR9 blot was stripped and subsequently probed with biotinylated DS lectin. Note that there is a non-specific band of slightly higher molecular weight than TLR9 that is detected in the TLR9 blot of the anti-biotin IP from cells incubated with or without biotinylated CpG DNA. Data are representative of three experiments.
Next we determined if the pool of TLR9 present outside the Golgi could initiate signaling. CpG DNA treatment of cells induced p38 phosphorylation as early as 30 minutes, which increased up to 60 minutes. When the ratio of phosphorylated p38 to total p38 was calculated (relative intensity ratio), the ratio increased from 0.78 in unstimulated cells to 1.29 at 30 minutes. The relative ratio increased further to 1.57 and remained at that level until 60 minutes (Figure 6b). In the presence of BFA, CpG DNA induced phosphorylation of p38 was delayed and reduced, but still occurred. The relative ratio of phosphorylated p38 to total p38 remained at 0.68–0.70 for 30 minutes, increased to 1.15 at 45 minutes and reduced back to 0.68 at 60 minutes. This indicated that Golgi export was not absolutely required to initiate signaling, but was required for optimal and sustained signaling (Figure 6b). These data are consistent with a model where TLR9 is constitutively present in endolysosomes, where it initiates early signaling in response to CpG DNA. This initial signaling event is followed by additional TLR9 translocation through the Golgi complex to endolysosomes and results in optimal TLR9 signaling as previously described.
The TLR9 that initiates signaling upon CpG DNA binding has translocated through the Golgi complex
We next determined whether the TLR9 that binds to CpG DNA and initiates signal transduction had trafficked through the Golgi complex. Since we were unable to detect an increase in association between MyD88 and TLR9 upon CpG DNA stimulation (Supplementary Figure 6) we could not use the recruitment of MyD88 to TLR9 to monitor the TLR9 involved in signaling. Instead, we used biotinylated CpG DNA to pull down active TLR9 as previously described7. The TLR9 that precipitated with CpG DNA was associated with MyD88 (Figure 6c) and was capable of binding to DS lectin, suggesting its glycans were modified in the Golgi complex (Figure 6c). These data indicate that the TLR9 actually involved in initiating signal transduction upon CpG DNA binding has translocated through the Golgi complex. It is possible that this pool of “signaling TLR9” was already present in endolysosomes, or it may represent newly trafficked TLR9 that arrived via the Golgi complex to endolysosomes after exposure to CpG DNA.
DISCUSSION
Our current studies provide compelling evidence that TLR9 transits through the Golgi complex and that Golgi export is critical for optimal response to CpG DNA. Synthesis of transmembrane and secreted proteins begins on rough ER where the nascent protein is concurrently translated and transferred through the ER membrane. For proteins with N-linked glycosylations, a preformed oligosaccharide precursor containing several mannose residues is then transferred to an asparagine (in the sequence Asn-X-Ser/Thr) on the nascent protein (high mannose Glycoform). When the protein is ready for export, it transits through the Golgi complex where modification of the oligosaccharide is completed. These modifications include trimming of mannose residues (hybrid Glycoform) and the incorporation of N-acetylglucosamine, galactose and N-acetylneuraminic acid (complex Glycoform). Here we show that TLR9 is glycosylated in the ER, since it binds GN, a lectin specific for high mannose and hybrid glycoforms (Figure 3). TLR9 also binds to DS, a lectin with high affinity for hybrid and complex glycoforms, demonstrating it contains Golgi-dependent glycan modifications. We further show, using a newly developed furin cleavage assay, that TLR9 is exposed to the Golgi-resident protease furin during intracellular movement (Figure 4). These data are in contrast to the current model, which proposes that TLR9 translocates directly from the ER to endosomes by a mechanism similar to ER-mediated phagocytosis. This model was based on the observation that glycans on TLR9 remained sensitive to digestion with EndoH despite TLR9’s movement to endosomes. We now clearly show that the glycans on TLR9 remain sensitive to EndoH digestion upon treatment with BFA despite a change in EndoH sensitivity of the fusion chimera TLR4-9. Furthermore, optimal TLR9 signaling is blocked by pre-treatment of cells with BFA demonstrating that TLR9 not only passes through the Golgi complex, but that Golgi export is critical for TLR9 signaling (Figure 1). These results are important for defining the intracellular trafficking pathway of TLR9, a pathway that may be amenable to interruptive drug treatment to prevent TLR9 signaling in diseases such as systemic lupus erythematosus (SLE).
Another important finding of our studies is that TLR9 is not fully retained in the ER, but constitutively moves through the cell and reaches the lysosomal compartment (Figure 5). These data are consistent with previous studies, which have suggested that under certain conditions that some TLR9 can be detected in an endosomal compartment6, 32. There are two possible explanations as to why TLR9 is detected in the lysosomal compartment of untreated cells. It could be a mechanism for continuous turnover of TLR9, or it could provide a pool of TLR9 immediately ready to respond to endocytosed CpG DNA. High levels of TLRs in general lead to constitutive activation and constant turnover in the lysosome may help to regulate these levels. However, we provide data demonstrating that the pool of TLR9 that has exited the Golgi is constitutively associated with MyD88 and thus likely contributes to signaling (Figure 6). Therefore, lysosomal TLR9 is poised to rapidly respond to an influx of CpG DNA. Early signaling events such as the phosphorylation of p38, although delayed, still occur in BFA treated cells. Thus, we concluded that the extra-Golgi TLR9 can respond to CpG DNA. Since our organelle separation studies demonstrate that the TLR9 that exits the Golgi complex moves to lysosomes, it is likely that signaling can be initiated from this compartment. What role this initial signaling plays in the optimal TLR9 signaling, which requires additional TLR9 translocation from the ER to endolysosomes, is not clear. TLR9 translocation to endolysosomes in response to CpG DNA still occurs in MyD88 deficient dendritic cells, so it is unlikely that signaling initiated by the small pool of TLR9 in endolysosomes is responsible for inducing the additional movement of TLR97. Further studies are necessary to decipher the contribution of lysosomal TLR9 in signaling.
Recently, a partially cleaved form of TLR9 that contains approximately half of the ecto-domain and all of the transmembrane domain and cytoplasmic tail has been described31. This form is enriched in phagosomes and may be the functional receptor. We have detected a lower molecular weight form of TLR9 in some experiments using either transient or stable cell lines expressing tagged or untagged TLR9. Surprisingly, we detect this lower molecular weight form with an antibody that recognizes amino acids 273–288, an epitope that would be lost based on the predicted cleavage site31. However, we have failed to consistently detect this form in the lysosomal fractions from either HEK293 cells expressing HA-TLR9 (detected with antibody to TLR9) or BJAB cells expressing endogenous TLR9 (Chockalingam A, 2008 unpubl. data). It is possible that cell lineage, growth conditions or endocytosis/phagocytosis influence the amount of cleaved TLR9 detected in unstimulated cells.
A remaining question is what regulates TLR9 translocation from the ER through the Golgi complex to endolysosomes. Several proteins have been described to control response of TLR9 to CpG DNA including, Hsp90, cathepsins HMGB1, and UNC93B117, 30, 33–36. A macrophage specific Hsp90 deficiency in mouse resulted in loss of response to all TLRs, suggesting that this chaperone is a master regulator of TLR signaling35. Other studies have suggested that the function of Hsp90 is to control intracellular TLR trafficking37. Cathepsins have also been implicated in regulating TLR9 response to CpG DNA. Given the cleavage product of TLR9 described above31, it is interesting to note that dendritic cells treated with cathepsin K inhibitors or deficient in cathepsin K failed to respond to stimulation with CpG DNA36. However, the role of cathepsin K in regulating TLR9 signaling is unclear since several recent papers provide conflicting data on the role of different cathepsins in regulating TLR9 signaling31, 36, 38. HMGB1 also regulates TLR9 signaling and can directly bind to both TLR9 and immune complexes containing DNA that are found in SLE serum30. TLR9 directly associates with the HMGB1 receptor, RAGE and this association regulates TLR9 response. An Fc-RAGE fusion blocks TLR9 response to DNA; however, we do not know if this regulation is at the level of receptor translocation. UNC93B1 is an ER resident protein that interacts with nucleic acid recognizing TLRs via their transmembrane domains and thus regulates their intracellular trafficking17, 33. UNC93B1 deficient mice fail to respond to CpG DNA, as well as TLR7 and TLR3 ligands17, 33, 34. UNC93B1 is likely required for TLR9 to exit the ER, but it is unclear if it plays any additional role in regulating TLR9 movement. Here we have established that TLR9 traffics through Golgi complex and that Golgi export is required for optimal TLR9 signaling. It will be interesting to determine how these TLR-interacting proteins regulate TLR9 movement from the ER to endolysosomes via the Golgi complex.
In summary, we have demonstrated that TLR9 does not bypass the Golgi complex during translocation to endolysosomes, and that Golgi export is required for optimal signaling. A small pool of TLR9 is constitutively localized in endolysosomes and this pool is bound to MyD88 and likely initiates signaling. Elucidating the mechanisms regulating TLR9 movement within cells is critical to understand how TLR9 normally remains quiescent when cells are exposed to self DNA. A better understanding of the molecular mechanisms controlling these events will allow in identification and testing of inhibitors that target the regulatory pathways of TLR9 trafficking and prevent recognition of self DNA in autoimmune diseases such as SLE. It is intriguing to think that some of these pathways may be shared with other TLRs, such as TLR7, that also contribute to SLE. Therefore, manipulating the translocation of nucleic acid recognizing TLRs could be a novel approach to treat debilitating autoimmune inflammation.
METHODS
Reagents and Plasmids
BFA, TLR9 antibody and avidin-HRP were from eBioscience. GFP antibody for immunoprecipitation was from Invitrogen/Molecular Probes and for immunoblotting was from BD Clontech. Antibodies to phosphorylated and total p38 were from Cell Signaling Technologies. High affinity antibody to hemaglutinin (anti-HA) was from Roche. Recombinant furin, EndoH and PNGase F were from New England Biolabs. Antibodies to LAMP-1, Rab 5 and Calnexin were from BD-transduction labs. MyD88 antibodies for immunoprecipitation and immunoblotting were from Santa Cruz Biotechnology. CpG DNAs were from Coley Pharmaceutical (10104 5’-TCGTCGTTTCGTCGTTTTGTCGTT-3’) or synthesized by IDT (2006 5’-TCGTCGTTTTGTCGTTTTGTCGTT-3’). HAfu-TLR9 was generated by site directed mutagenesis (Stratagene) of HA-TLR95 to change PARS (single amino acid code) to RXRR in the intervening sequence between HA tag and TLR9. Sequences of mutagenesis primers were 5’-GATTATGCTGGGGCCCAGCGGGCCAGACGTCTGGCCACCTTGCCTG-3’ and 5’-CAGGCAAGGTGGCCAGACGTCTGGCCCGCTGGGCCCCAGCATAATC -3’.TLR9GFP, TLR4GFP and TLR4-9GFP have been previously described7–9. 5xNF-κB-luciferase and pSV-β-galactosidase were from Stratagene and Promega respectively. All plasmids were prepared using Qiagen Endo free kits.
Cell culture and transfection
HeLa and Human Embryonic Kidney 293 (HEK293) cells were cultured in DMEM with 2 mM L-glutamine, 50 U/ml penicillin, 50 µg/ml streptomycin, 10 mM Hepes, 1 mM sodium pyruvate and 10% low endotoxin FBS (complete DMEM). BJAB cells were cultured in complete RPMI 1640. Cells routinely tested negative for mycoplasma. Stable cells were generated in HEK293 cells by transfection with 24 µg DNA in a 10 cm tissue culture dish with 48 µl TransIT (Mirus). After 24 hours, G418 (1 mg/ml) was added to the medium and the cells were selected under G418 with media change every 2 days. Cells were then subjected to single cell selection. All TLR9 stable cell lines responded to CpG DNA as detected by 5x NF-κB luciferase reporter assay in a transient transfection system.
HAfu-TLR9 cleavage assay
HeLa cells were transiently transfected or HEK293 cells were stably transfected with HAfu-TLR9 or HA-TLR9. For in vitro assays, cells were lysed (Lysis buffer: 137 mM NaCl, 20 mM Tris pH7.4, 1 mM EDTA, 0.5% Triton X-100, protease inhibitor cocktail (Sigma) and 100 mM phenylmethylsulphonyl fluoride), TLR9 was immunoprecipitated with anti-HA antibody and protein A/G sepharose, washed 3x with lysis buffer and resuspended in cleavage buffer (100 mM Hepes pH 7.5, 1 mM CaCl2, 0.5% Triton X-100) with or without furin for 2 hours at 30°C. Reducing, denaturing sample buffer was added and samples were boiled for 5 minutes prior to separation by SDS-PAGE. After transferring to nitrocellulose, blots were probed with anti-TLR9 and anti-HA antibodies. For in vivo cleavage, TLR9 was precipitated with anti-TLR9, separated by SDS-PAGE, and the western blot was sequentially probed with anti-HA and anti-TLR9 antibodies. HA-TLR9 without a furin cleavage site was used as a control for each condition.
Lectin blotting
Lectins directly conjugated to biotin can be used to detect specific glycosylation pattern on proteins similarly to immunoblotting using avidin HRP as a secondary detection reagent. Lysates from transiently transfected HeLa cells or stably transfected HEK293 cells expressing TLR9-YFP and TLR4-YFP were immunoprecipitated with anti-GFP, separated by SDS-PAGE and transferred to nitrocellulose. Membranes were blocked in 1% BSA (fraction V) in PBS plus 0.05% Tween, (PBST) and incubated for 40 minutes with 2 µg/ml biotinylated lectin. After 3 washes of 15 minutes each in PBST, membranes were incubated with 1: 10,000 dilution of avidin-HRP for 30 minutes. Membranes were washed 3 times for 60 minutes prior to developing with ECL substrate (Pierce).
HEK293 TLR9 luciferase reporter assay
Cells were transiently transfected in 96 well plates using TransIT (Mirus, Madison WI) with TLR encoding plasmids, 5X-NF-κB-luciferase and empty vector to equal 200 ng/well of total DNA. After overnight culture, the cells were treated with CpG DNA for 18 hours. Cell lysates (5X Reporter Lysis buffer, Promega) were assayed for luciferase activity using the luciferase substrate (20 mM tricine, 2.67 mM MgSO4·7 H2O, 33.3 mM DTT, 100 µM EDTA, 530 µM ATP, 270 µM Acetyl CoA, 132 µg/ml luciferin, 5 mM NaOH, 265 µM magnesium carbonate hydroxide) on a Veritas luminometer with injector.
EndoH sensitivity assay
Transfected HeLa cells were lysed in lysis buffer and immunoprecipitated for the respective TLRs. The immunoprecipitates were divided equally into three and were either left untreated or treated with EndoH or PNGase F according to the manufacturer instructions (New England Biolabs). Samples were digested for two hours at 37°C and the reactions were stopped by adding SDS-PAGE sample buffer. All treated samples were separated by SDS-PAGE, transferred to nitrocellulose and probed with indicated antibodies.
Organelle fractionation
BJAB, a human B cell line expressing endogenous TLR9, or HEK293 cells stably expressing HAfu-TLR9 or HA-TLR9 were incubated in homogenization buffer (HB: 0.25M Sucrose, 1 mM EDTA, 10 mM Tris, pH 7.4) for 15 minutes on ice and homogenized with a loose fitting 2 ml glass dounce homogenizer for 20 strokes, and passed 10 times through a 25G needle attached to a 3cc syringe. The homogenate was spun at 900 × g for 10 minutes at 4°C and the post-nuclear supernatant (PNS) was transferred to a fresh tube. The pellet was washed with additional HB, spun at 4°C and the combined PNS was layered on Percoll in HB (density 1.05 g/ml) and spun for 60 minutes at 34,000 × g max with a 50Ti rotor in a Beckman L-8M ultracentrifuge. Fractions (500 µl each) were collected from the top using a gradient unloader (Haakebuchler AutoDensi-Flow IIC). Each fraction was tested for refractive index (Reichert Abbe Mark II Refractometer) and protein concentration (DC-BioRad assay) to confirm separation. SDS-PAGE sample buffer was added to each fraction and multiple 10% SDS-PAGE gels were run. After transfer, nitrocellulose membranes were probed with antibodies to Rab 5, Calnexin, GM130, LAMP-1, HA and TLR9.
Co-immunoprecipitation
BJAB or HEK293 cells stably expressing HAfu-TLR9 were left untreated or treated with CpG 2006 (5 µg/ml), lysed and immunoprecipitated for MyD88 or TLR9 for four hours prior to adding protein A/G sepharose beads and incubating for additional 12 hours at 4°C. Some cells were treated with 25 µg/ml biotinylated CpG 2006 prior to cell lysis and immunoprecipitated for biotin. After three washes in lysis buffer, 1x SDS sample buffer was added and proteins were separated in SDS-PAGE and immunoblotted for MyD88 and TLR9. The TLR9 blot was stripped and probed with biotinylated DS lectin.
Densitometry analysis
Each protein band was quantified by outlining using Photoshop. Briefly, after inverting the raw image, each band was outlined using the lasso tool and the mean intensity and pixels were determined. The absolute intensity (AI) was calculated by multiplying the mean by the pixels. Where there was no detectable protein band to outline, an absolute intensity of zero was assigned. For Figure 4c, the AI for HA was divided by the AI for TLR9 to obtain the relative intensity (RI). Similarly, for Figure 6b, the RI was calculated by dividing the AI of phosphorylated p38 by the AI of total p38. In Figure 5, the absolute intensity of each band was divided by the highest absolute intensity to determine the percent maximum absolute intensity.
Supplementary Material
TLR9 glycosylation is not modified following CpG DNA stimulation. Ramos (a human B cell line) cells expressing endogenous TLR9 were treated with 5 µg/ml CpG DNA or control DNA for 2 hours. TLR9 was immunoprecipitated and left untreated (−) or treated with EndoH (H) or PNGase F (F), separated by SDS-PAGE, transferred to nitrocellulose and immunoblotted for HA. Data are representative of four experiments.
CpG DNA uptake is not inhibited by BFA treatment. HEK293 cells were pretreated with media or 100ng/mL BFA for 2 hours prior to incubation with 5µg/mL CpG 2006 3’Cy3 for 60 minutes at 37°C except the top right histogram where cells were incubated at 4°C. Cells were analyzed by flow cytometry. MFIs are indicated in each box. Data are representative of two experiments.
DS lectin does not bind to BSA. BSA (Fraction V), or TLR9 immunoprecipitates from TLR9-YFP transfected HEK293 cells, were blotted for TLR9 and DS lectin as described in Figure 3. The presence of BSA on the blot was confirmed by Ponceau S staining (not shown). Data are representative of four experiments.
Golgi fractions separated from ER and lysosomes on Percoll-sucrose gradients. Fractions from HEK293 cells stably expressing HA-TLR9 separated on Percoll-sucrose gradients were immunoblotted for ER (Calnexin), Golgi (GM130), early endosomes (Rab 5) and lysosomes (LAMP-1). This experiment was performed one time to determine the location of the Golgi complex.
Endogenous TLR9 in endolysosomes binds to DS lectin. TLR9 immunoprecipitates of total cells (WCL) or combined LAMP-1 positive fractions (Lys) from BJABs cells were immunoblotted for TLR9 (TLR9), stripped and probed with biotinylated DS lectin (DS). Data are representative of three experiments.
Endogenous MyD88 association with endogenous TLR9 does not increase upon stimulation with CpG DNA. BJAB cells were untreated or treated with 5µg/mL CpG 10104 for 1 hour prior to immunoprecipitation and immunoblotting for MyD88 or TLR9. Data are representative of three experiments
ACKNOWLEDGMENTS
This work was supported by the National Institutes of Health, NCI transition career development award to C.L. (5K22CA113705), and startup funds from the Department of Microbiology and Immunology, and Cornell University College of Veterinary Medicine. We thank R. Corley for his discussion, sharing unpublished data and helpful suggestions.
Footnotes
COMPETING INTERESTS STATEMENT
The authors declare no competing financial interests.
Supplementary information is available at Immunology and Cell Biology’s website.
REFERENCES CITED
- 1.Takeda K, Takeuchi O, Akira S. Recognition of lipopeptides by Toll-like receptors. J. Endotoxin Res. 2002;8:459–463. doi: 10.1179/096805102125001073. [DOI] [PubMed] [Google Scholar]
- 2.Alexopoulou L, Holt AC, Medzhitov R, Flavell RA. Recognition of double-stranded RNA and activation of NF-κB by Toll-like receptor 3. Nature. 2001;413:732–738. doi: 10.1038/35099560. [DOI] [PubMed] [Google Scholar]
- 3.Heil F, Hemmi H, Hochrein H, Ampenberger F, Kirschning C, Akira S, et al. Species-specific recognition of single-stranded RNA via Toll-like receptor 7 and 8. Science. 2004;303:1526–1529. doi: 10.1126/science.1093620. [DOI] [PubMed] [Google Scholar]
- 4.Hemmi H, Takeuchi O, Kawai T, Kaisho T, Sato S, Sanjo H, et al. A Toll-like receptor recognizes bacterial DNA. Nature. 2000;408:740–745. doi: 10.1038/35047123. [DOI] [PubMed] [Google Scholar]
- 5.Takeshita F, Leifer CA, Gursel I, Ishii KJ, Takeshita S, Gursel M, et al. Cutting edge: Role of Toll-like receptor 9 in CpG DNA-induced activation of human cells. J. Immunol. 2001;167:3555–3558. doi: 10.4049/jimmunol.167.7.3555. [DOI] [PubMed] [Google Scholar]
- 6.Ahmad-Nejad P, Hacker H, Rutz M, Bauer S, Vabulas RM, Wagner H. Bacterial CpG-DNA and lipopolysaccharides activate Toll-like receptors at distinct cellular compartments. Eur. J. Immunol. 2002;32:1958–1968. doi: 10.1002/1521-4141(200207)32:7<1958::AID-IMMU1958>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 7.Latz E, Schoenemeyer A, Visintin A, Fitzgerald KA, Monks BG, Knetter CF, et al. TLR9 signals after translocating from the ER to CpG DNA in the lysosome. Nat. Immunol. 2004;5:190–198. doi: 10.1038/ni1028. [DOI] [PubMed] [Google Scholar]
- 8.Leifer CA, Brooks JC, Hoelzer K, Lopez JL, Kennedy MN, Mazzoni A, et al. Cytoplasmic targeting motifs control localization of Toll-like receptor 9. J. Biol. Chem. 2006;281:35585–35592. doi: 10.1074/jbc.M607511200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Leifer CA, Kennedy MN, Mazzoni A, Lee C, Kruhlak MJ, Segal DM. TLR9 is localized in the endoplasmic reticulum prior to stimulation. J. Immunol. 2004;173:1179–1183. doi: 10.4049/jimmunol.173.2.1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hacker H, Mischak H, Miethke T, Liptay S, Schmid R, Sparwasser T, et al. CpG-DNA-specific activation of antigen-presenting cells requires stress kinase activity and is preceded by non-specific endocytosis and endosomal maturation. EMBO J. 1998;17:6230–6240. doi: 10.1093/emboj/17.21.6230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rutz M, Metzger J, Gellert T, Luppa P, Lipford GB, Wagner H, et al. Toll-like receptor 9 binds single-stranded CpG-DNA in a sequence- and Ph-dependent manner. Eur. J. Immunol. 2004;34:2541–2550. doi: 10.1002/eji.200425218. [DOI] [PubMed] [Google Scholar]
- 12.Latz E, Verma A, Visintin A, Gong M, Sirois CM, Klein DC, et al. Ligand-induced conformational changes allosterically activate Toll-like receptor 9. Nat. Immunol. 2007;8:772–779. doi: 10.1038/ni1479. [DOI] [PubMed] [Google Scholar]
- 13.Guiducci C, Ott G, Chan JH, Damon E, Calacsan C, Matray T, et al. Properties regulating the nature of the plasmacytoid dendritic cell response to Toll-like receptor 9 activation. J. Exp. Med. 2006;203:1999–2008. doi: 10.1084/jem.20060401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Barton GM, Kagan JC, Medzhitov R. Intracellular localization of Toll-like receptor 9 prevents recognition of self DNA but facilitates access to viral DNA. Nat. Immunol. 2006;7:49–56. doi: 10.1038/ni1280. [DOI] [PubMed] [Google Scholar]
- 15.Lande R, Gregorio J, Facchinetti V, Chatterjee B, Wang YH, Homey B, et al. Plasmacytoid dendritic cells sense self-DNA coupled with antimicrobial peptide. Nature. 2007;449:564–569. doi: 10.1038/nature06116. [DOI] [PubMed] [Google Scholar]
- 16.Leadbetter EA, Rifkin IR, Hohlbaum AM, Beaudette BC, Shlomchik MJ, Marshak-Rothstein A. Chromatin-IgG complexes activate B cells by dual engagement of IgM and Toll-like receptors. Nature. 2002;416:603–607. doi: 10.1038/416603a. [DOI] [PubMed] [Google Scholar]
- 17.Kim YM, Brinkmann MM, Paquet ME, Ploegh HL. UNC93B1 delivers nucleotide-sensing Toll-like receptors to endolysosomes. Nature. 2008;452:234–238. doi: 10.1038/nature06726. [DOI] [PubMed] [Google Scholar]
- 18.Uemura K, Yokota Y, Kozutsumi Y, Kawasaki T. A unique CD45 glycoform recognized by the serum mannan-binding protein in immature thymocytes. J. Biol. Chem. 1996;271:4581–4584. doi: 10.1074/jbc.271.9.4581. [DOI] [PubMed] [Google Scholar]
- 19.Peyroche A, Antonny B, Robineau S, Acker J, Cherfils J, Jackson CL. Brefeldin A acts to stabilize an abortive ARF-GDP-Sec7 domain protein complex: Involvement of specific residues of the Sec7 domain. Mol. Cell. 1999;3:275–285. doi: 10.1016/s1097-2765(00)80455-4. [DOI] [PubMed] [Google Scholar]
- 20.Presley JF, Ward TH, Pfeifer AC, Siggia ED, Phair RD, Lippincott-Schwartz J. Dissection of COP1 and ARF1 dynamics in vivo and role in Golgi membrane transport. Nature. 2002;417:187–193. doi: 10.1038/417187a. [DOI] [PubMed] [Google Scholar]
- 21.Sciaky N, Presley J, Smith C, Zaal KJ, Cole N, Moreira JE, et al. Golgi tubule traffic and the effects of Brefeldin A visualized in living cells. J. Cell. Biol. 1997;139:1137–1155. doi: 10.1083/jcb.139.5.1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kagan JC, Medzhitov R. Phosphoinositide-mediated adaptor recruitment controls Toll-like receptor signaling. Cell. 2006;125:943–955. doi: 10.1016/j.cell.2006.03.047. [DOI] [PubMed] [Google Scholar]
- 23.Gewirtz AT, Navas TA, Lyons S, Godowski PJ, Madara JL. Cutting edge: Bacterial flagellin activates basolaterally expressed TLR5 to induce epithelial proinflammatory gene expression. J. Immunol. 2001;167:1882–1885. doi: 10.4049/jimmunol.167.4.1882. [DOI] [PubMed] [Google Scholar]
- 24.Nishiya T, DeFranco AL. Ligand-regulated chimeric receptor approach reveals distinctive subcellular localization and signaling properties of the Toll-like receptors. J. Biol. Chem. 2004;279:19008–19017. doi: 10.1074/jbc.M311618200. [DOI] [PubMed] [Google Scholar]
- 25.Underhill DM, Ozinsky A, Smith KD, Aderem A. Toll-like receptor-2 mediates mycobacteria-induced proinflammatory signaling in macrophages. Proc. Natl. Acad. Sci. USA. 1999;96:14459–14463. doi: 10.1073/pnas.96.25.14459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Visintin A, Mazzoni A, Spitzer JA, Segal DM. Secreted MD-2 is a large polymeric protein that efficiently confers lipopolysaccharide sensitivity to Toll-like receptor 4. Proc. Natl. Acad. Sci. USA. 2001;98:12156–12161. doi: 10.1073/pnas.211445098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Molloy SS, Thomas L, VanSlyke JK, Stenberg PE, Thomas G. Intracellular trafficking and activation of the furin proprotein convertase: Localization to the TGN and recycling from the cell surface. EMBO J. 1994;13:18–33. doi: 10.1002/j.1460-2075.1994.tb06231.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nakayama K. Furin: A mammalian subtilisin/kex2p-like endoprotease involved in processing of a wide variety of precursor proteins. Biochem. J. 1997;327:625–635. doi: 10.1042/bj3270625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hacker H, Vabulas RM, Takeuchi O, Hoshino K, Akira S, Wagner H. Immune cell activation by bacterial CpG-DNA through myeloid differentiation marker 88 and tumor necrosis factor receptor-associated factor (TRAF)6. J. Exp. Med. 2000;192:595–600. doi: 10.1084/jem.192.4.595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tian J, Avalos AM, Mao SY, Chen B, Senthil K, Wu H, et al. Toll-like receptor 9-dependent activation by DNA-containing immune complexes is mediated by HMGB1 and Rage. Nat. Immunol. 2007;8:487–496. doi: 10.1038/ni1457. [DOI] [PubMed] [Google Scholar]
- 31.Ewald SE, Lee BL, Lau L, Wickliffe KE, Shi GP, Chapman HA, et al. The ectodomain of Toll-like receptor 9 is cleaved to generate a functional receptor. Nature. 2008 doi: 10.1038/nature07405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chaturvedi A, Dorward D, Pierce SK. The B cell receptor governs the subcellular location of Toll-like receptor 9 leading to hyperresponses to DNA-containing antigens. Immunity. 2008;28:799–809. doi: 10.1016/j.immuni.2008.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Brinkmann MM, Spooner E, Hoebe K, Beutler B, Ploegh HL, Kim YM. The interaction between the er membrane protein UNC93B and TLRs3, 7, and 9 is crucial forTLR signaling. J Cell Biol. 2007;177:265–275. doi: 10.1083/jcb.200612056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tabeta K, Hoebe K, Janssen EM, Du X, Georgel P, Crozat K, et al. The UNC93B1 mutation 3d disrupts exogenous antigen presentation and signaling via Toll-like receptors 3, 7 and 9. Nat Immunol. 2006;7:156–164. doi: 10.1038/ni1297. [DOI] [PubMed] [Google Scholar]
- 35.Yang Y, Liu B, Dai J, Srivastava PK, Zammit DJ, Lefrancois L, et al. Heat shock protein gp96 is a master chaperone for Toll-like receptors and is important in the innate function of macrophages. Immunity. 2007;26:215–226. doi: 10.1016/j.immuni.2006.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Asagiri M, Hirai T, Kunigami T, Kamano S, Gober HJ, Okamoto K, et al. Cathepsin K-dependent Toll-like receptor 9 signaling revealed in experimental arthritis. Science. 2008;319:624–627. doi: 10.1126/science.1150110. [DOI] [PubMed] [Google Scholar]
- 37.Reed RC, Berwin B, Baker JP, Nicchitta CV. Grp94/gp96 elicits ERK activation in murine macrophages. A role for endotoxin contamination in NF-κB activation and nitric oxide production. J Biol Chem. 2003;278:31853–31860. doi: 10.1074/jbc.M305480200. [DOI] [PubMed] [Google Scholar]
- 38.Matsumoto F, Saitoh S, Fukui R, Kobayashi T, Tanimura N, Konno K, et al. Cathepsins are required for Toll-like receptor 9 responses. Biochem Biophys Res Commun. 2008;367:693–699. doi: 10.1016/j.bbrc.2007.12.130. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
TLR9 glycosylation is not modified following CpG DNA stimulation. Ramos (a human B cell line) cells expressing endogenous TLR9 were treated with 5 µg/ml CpG DNA or control DNA for 2 hours. TLR9 was immunoprecipitated and left untreated (−) or treated with EndoH (H) or PNGase F (F), separated by SDS-PAGE, transferred to nitrocellulose and immunoblotted for HA. Data are representative of four experiments.
CpG DNA uptake is not inhibited by BFA treatment. HEK293 cells were pretreated with media or 100ng/mL BFA for 2 hours prior to incubation with 5µg/mL CpG 2006 3’Cy3 for 60 minutes at 37°C except the top right histogram where cells were incubated at 4°C. Cells were analyzed by flow cytometry. MFIs are indicated in each box. Data are representative of two experiments.
DS lectin does not bind to BSA. BSA (Fraction V), or TLR9 immunoprecipitates from TLR9-YFP transfected HEK293 cells, were blotted for TLR9 and DS lectin as described in Figure 3. The presence of BSA on the blot was confirmed by Ponceau S staining (not shown). Data are representative of four experiments.
Golgi fractions separated from ER and lysosomes on Percoll-sucrose gradients. Fractions from HEK293 cells stably expressing HA-TLR9 separated on Percoll-sucrose gradients were immunoblotted for ER (Calnexin), Golgi (GM130), early endosomes (Rab 5) and lysosomes (LAMP-1). This experiment was performed one time to determine the location of the Golgi complex.
Endogenous TLR9 in endolysosomes binds to DS lectin. TLR9 immunoprecipitates of total cells (WCL) or combined LAMP-1 positive fractions (Lys) from BJABs cells were immunoblotted for TLR9 (TLR9), stripped and probed with biotinylated DS lectin (DS). Data are representative of three experiments.
Endogenous MyD88 association with endogenous TLR9 does not increase upon stimulation with CpG DNA. BJAB cells were untreated or treated with 5µg/mL CpG 10104 for 1 hour prior to immunoprecipitation and immunoblotting for MyD88 or TLR9. Data are representative of three experiments