Introductory paragraph
Among organisms with chromosome-based mechanisms of sex determination, failure to equalize expression of X-linked genes between the sexes is typically lethal. In C. elegans, XX hermaphrodites halve transcription from each X chromosome to match the output of XO males1. Here, we mapped the binding location of the condensin homolog DPY-27 and the zinc finger protein SDC-3, two components of the C. elegans dosage compensation complex (DCC)2,3. Strong foci of DCC binding were observed on X, around which broader regions of localization were centered. Binding foci, but not adjacent regions of localization, were distinguished by clusters of a stereotypic 10-bp DNA sequence, suggesting a recruitment-and-spreading mechanism for X recognition. In contrast to the Drosophila DCC, the C. elegans DCC was bound preferentially upstream of genes, suggesting modulation of transcriptional initiation and polymerase-coupled spreading. A mechanism for tuning DCC activity at specific loci was indicated by stronger DCC binding upstream of genes with high transcriptional activity. These data provide a basis for understanding how proteins involved in higher-order chromosome dynamics can regulate transcription at individual loci.
Main Text
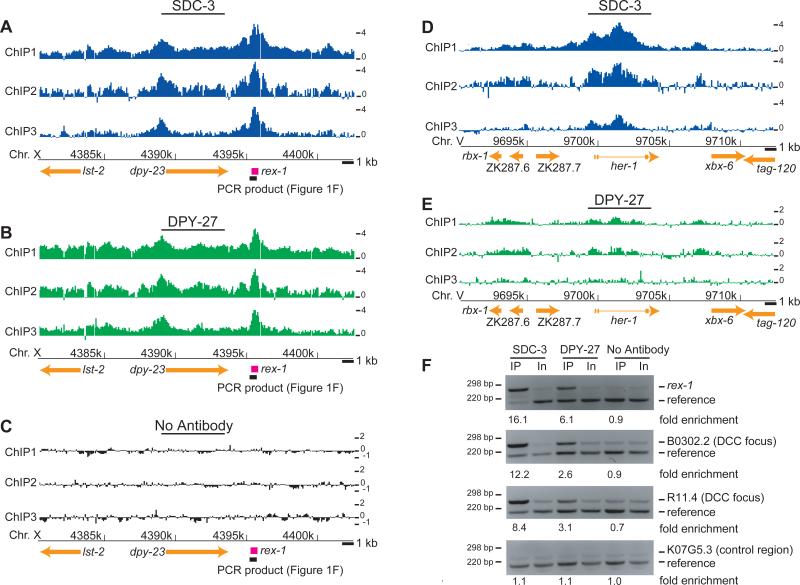
To compensate for differences in X-linked gene dosage between XY males and XX females, mammals inactivate most genes on one of the two female X chromosomes4. In contrast, C. elegans XX hermaphrodites dosage compensate by reducing transcription from each X chromosome by a factor of two to match the expression of XO males1. This mechanism is remarkable in that the subtle two-fold downregulation is imposed upon X-linked genes expressed over a large dynamic range5,6. The dosage compensation complex (DCC) required for C. elegans X-repression is composed of proteins encoded by the genes sdc-1, sdc-2, sdc-3, dpy-21, dpy-26, dpy-27, dpy-28, dpy-30 and mix-17. DPY-26, DPY-27, DPY-28 and MIX-1 are homologous to members of the condensin complex, which is required for chromosome condensation and segregation in organisms ranging from bacteria to humans8. While the molecular parallels to mitotic chromosome condensation broadly suggest a mechanism for reducing X-linked transcription9,10, the features of X that control DCC binding have proven more difficult to investigate. Four loci on X (rex-1-4) sufficient for DCC recruitment were identified recently by using confocal microscopy to detect DCC binding to multiple-copy extrachromosomal transgenic DNA11,12. Here, we take an alternative approach that comprehensively maps the distribution of DCC binding along the natural X chromosomes of wild-type animals, identifies DNA sequence elements enriched at sites of DCC recruitment on chromosomes in vivo, and determines the relationship between DCC binding and genomic organization at high resolution.We performed ChIP (Chromatin ImmunoPrecipitation) of the zinc-finger protein SDC-3 and the condensin subunit homolog DPY-27 and hybridized the enriched DNA fragments to microarrays consisting of 50-bp oligonucleotide probes tiled across the genome with 36-bp spacing (“ChIP-chip”, Methods). To confirm that our reagents and detection methods could identify DCC binding sites, we first examined rex-1, a locus previously shown to recruit the DCC to extrachromosomal DNA in vivo11. Enrichment at rex-1 was consistently strong for SDC-3 (Figure 1A) and DPY-27 (Figure 1B), while the identical procedure performed in the absence of an antibody revealed no enrichment (Figure 1C). As a second validation, we examined the her-1 locus on Chromosome V, the lone autosomal region known to be bound by the DCC13. DCC binding to her-1 is required for the repression of her-1 transcription in XX animals13. SDC-3 exhibited strong binding at the promoter and second intron of her-1 (Figure 1D-E), consistent with published data showing binding to these regions13,14. Finally, the previously identified rex-1 site12 and two new sites discovered from our microarray data were confirmed by locus-specific PCR (Figure 1F). Based on these results, we interpret the ChIP enrichment measured by our high-resolution DNA microarray hybridizations to reflect the binding location and relative local abundance of the C. elegans dosage compensation machinery. To our knowledge, this is the first successful application of ChIP-chip methodology to this important model system.
Figure 1. High-resolution localization of the DCC authenticates binding at putative recruitment elements and reveals new targets.
(A) Raw log2 ratios (ChIP/Input) from each of three SDC-3 ChIP replicates plotted along 25 kb of the X chromosome, with gene annotations below. Arrows indicate direction of transcription. A previously identified recruitment element on X (rex-1)12 downstream of dpy-23 is indicated in pink. (B) Same as (a), but a DPY-27 ChIP (C) Same as (a), but a no antibody control ChIP. (D) SDC-3 localization at the her-1 locus. Introns are indicated by thin lines connecting exons. (E) Same as (d), but for DPY-27. Lower levels of DPY-27 localization are observed at her-1, consistent with the lower requirement indicated by genetic data. (F) Sequence-specific ChIP analysis of rex-112, two newly identified DCC binding foci on X (near B0302.2 and R11.4) and a reference region on chromosome I (K07G5.3).
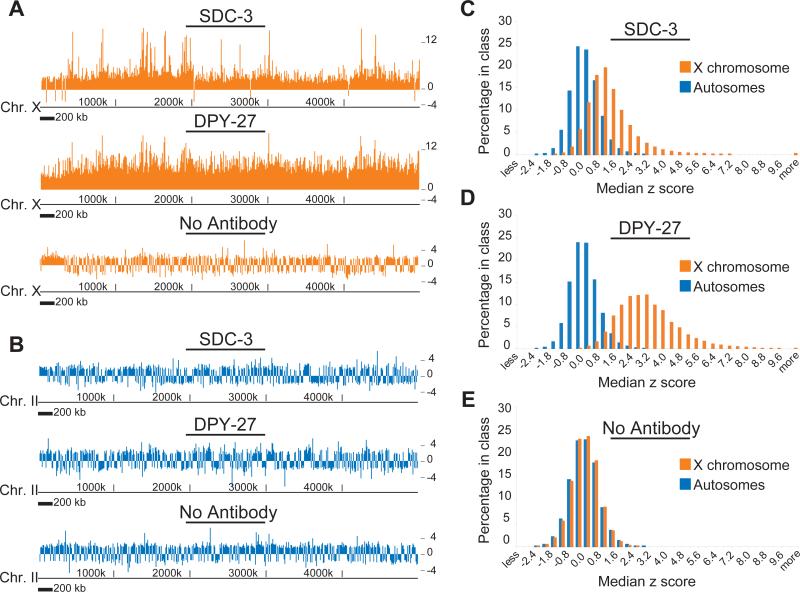
The DCC exhibits a striking preference for binding to the X chromosome relative to autosomes (Figure 2; p < 10−16 for SDC-3 and DPY-27; p = 0.88 for no antibody control), consistent with earlier results obtained by in situ microscopy2,3. For both DPY-27 and SDC-3, we noted a “baseline” level of enrichment on X, punctuated by peaks of greater enrichment (Figure 2A). This led us to speculate that the binding and distribution of the DCC on X involves at least two distinct mechanisms, one that leads to the appearance of distinct peaks, and one that leads to a more uniform distribution. Furthermore, the “baseline” level of binding was higher for DPY-27 than SDC-3 (Figure 2A and B). This, coupled with the observation that SDC-2 can bind to X independently of other DCC components15 and that SDC-2 and SDC-3 bind to X early in the DCC assembly process3, suggests that a protein complex containing SDC-2 and SDC-3 is critical for recruitment of the DCC to specific locations, while a more uniform distribution of the DCC, or possibly a subcomplex containing DPY-27 but not SDC-3, spreads from those points. The baseline binding and putative spreading of the DCC helps explain previous observations in which large regions of X were unable to recruit the DCC as extrachromosomal DNA, but were dosage compensated and bound by the DCC in their natural chromosomal context11.
Figure 2. SDC-3 and DPY-27 bind specifically to the X with distinct modes of distribution.
(A) Median z scores of enrichment (calculated from ChIP/Input signals) following ChIP using antibodies to SDC-3, DPY-27 and a no antibody control, plotted along a 5 MB region from the left end of X. (B) Same as (a), plotted along a 5 MB region of chromosome II. (C-E) Histograms of the distribution of z-score ChIP enrichment values of individual probes for autosomes and X chromosomes. (C) SDC-3 ChIP, (D) DPY-27 ChIP and (E) a no antibody control ChIP.
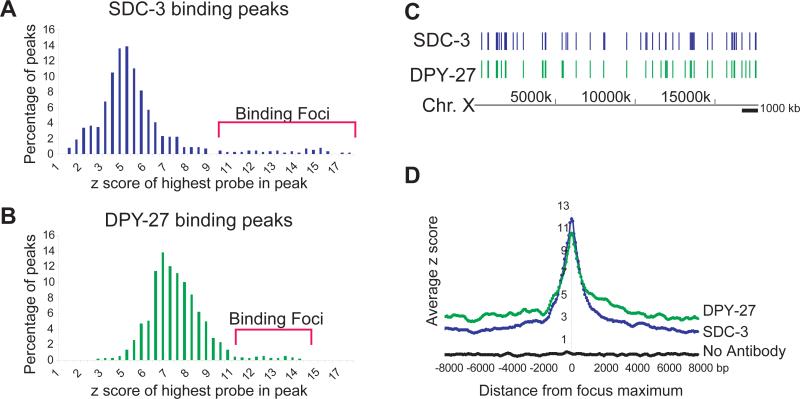
This recruitment-and-spreading hypothesis predicts that at recruitment sites, peaks of DPY-27 and SDC-3 would be largely coincident. We identified 1193 binding peaks for SDC-3 and 1499 for DPY-27 on X, compared to a total of 9 sites of enrichment for each protein on the five autosomes (Methods, Supplementary Table)16. To identify potential recruitment sites, we designated a class of exceptionally high-amplitude peaks as “foci” of DCC binding (Figure 3A-C, Supplementary Figure 1A-B). Fifty-five foci were identified for SDC-3, and 44 for DPY-27 (Supplementary Table). Whereas fewer than two foci would be expected to overlap under the null expectation, 35 of the SDC-3 and DPY-27 foci were coincident (p < 10−100). Three of the four previously discovered rex sites12 were classified as foci, confirming that these sites act as recruitment centers on natural chromosomes.
Figure 3. A distinct class of DCC localization foci along X.
(A) The distribution of maximum amplitudes (z-score) for SDC-3 ChIP peaks. Peaks classified as “Foci” are indicated by brackets (> 2 standard deviations from mean of the distribution). (B) Same as (a) for DPY-27. (C) Distribution of SDC-3 foci and DPY-27 foci along X. (D) Peaks classified as foci were aligned by their maxima, and the rate of signal decline was measured by sliding a window (width of 3 probes, step size of 1 probe) away from the maximum in both directions.
We further hypothesized that the baseline mode of binding (represented in part by the smaller peaks) would emanate from putative recruitment sites, which themselves are distributed randomly along X. Indeed, foci of SDC-3 or DPY-27 binding did not cluster (p = 0.15 and 0.14 respectively; Methods; Supplementary Figure 1C-D), but smaller peaks were strongly clustered (SDC-3 p < 10−25; DPY-27 p < 10−4). Furthermore, small peaks were clustered around foci. For example, 45% of SDC-3 peaks were within 50 kb of a focus (22% expected by chance; p = 2×10−6; Supplementary Figure 1E-F). At foci, SDC-3 binding was high but diminished rapidly to a low baseline, while the signal from DPY-27 maintained a higher baseline along the X (Figure 2A-B, Figure 3D, Supplementary Note). These data further support the hypothesis that complexes at foci are of distinct composition relative to complexes that spread from the foci.
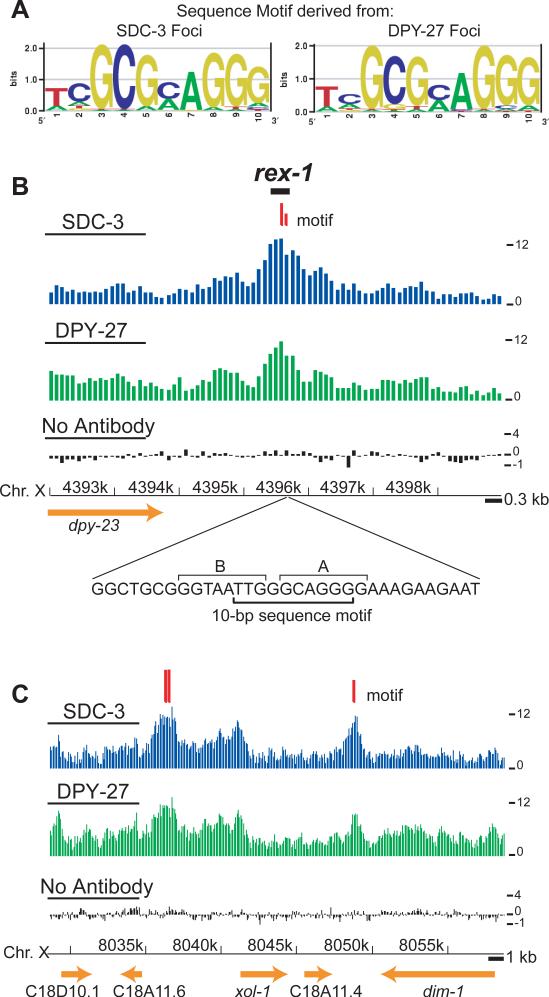
We reasoned that if foci were DCC recruitment centers, they may be specified by DNA sequence motifs. When used independently as input for motif discovery algorithms, DNA sequence defined by SDC-3 and DPY-27 foci yielded nearly identical 10-bp motifs (Figure 4A, Methods). In contrast, no motifs were derived using sequence from the smaller peaks as input, consistent with the hypothesis that foci are DNA-sequence based recruitment centers. The motif we identify using the natural DCC binding data expands one of the two motifs derived from extrachromosomal DNA assays12 by 4 bp and helps explain focused tests of function on the rex sites (Figure 4B, Supplementary Figure 2). For example, at rex-1 a 33-bp fragment sufficient for recruitment includes the motif, and mutations that abolished recruitment activity12 specifically alter the motif identified here (Figure 4B). In addition, the second intron of her-1 contains the motif (Figure 1D). Finally, one of the strongest newly discovered sites of SDC-3 and DPY-27 binding along the X is centered on two closely spaced motifs upstream of the male-specific xol-1 gene17 (Figure 4C). In XX hermaphrodites, xol-1 is repressed both transcriptionally and post-transcriptionally18,19, but the potential contribution of the DCC in directly repressing xol-1 was previously unknown.
Figure 4. A stereotypic 10 bp DNA sequence motif is enriched in foci of DCC binding.
(A) DNA sequence motifs derived from foci of SDC-3 and DPY-27 binding, depicted by sequence logos30. (B) DCC binding and motif occurrence at rex-1. Median z scores are plotted with respect to the chromosome coordinates for SDC-3, DPY-27, and no antibody control ChIPs. Locations of the 10-bp motifs are depicted with red tick marks, with longer marks indicating better matches to the consensus motif (MatrixScan p ≤ 10−5) than shorter marks (MatrixScan p ≤ 10−4). The location and sequence of a 33-bp fragment of rex-1 competent for DCC recruitment is shown below. In this fragment, the motif we identify spans two previously characterized12 motifs (7-bp A and 8-bp B), which were derived from four rex sites. The ChIP data suggest the previously reported “B” motif (TGTAATTG)12 plays a weak role, if any, in DCC recruitment on natural chromosomal substrates (Supplementary Figure 5). (C) DCC binding and motif occurrence near xol-1. Longer marks indicate better matches to the consensus motif (MatrixScan p ≤ 10−6) than shorter marks (MatrixScan p ≤ 10−5).
It is clear, however, that the identified motif does not specify DCC binding completely. At the most strict motif definition (Matrixscan p ≤ 10−6, Methods), 47% of X-linked motif occurrences were within an SDC-3 binding focus, and 42% of foci on X contained a motif (Supplementary Figure 3A-B). Furthermore, the motif occurs throughout the genome and is only moderately X-enriched (Supplementary Figure 3C-D). Consistent with previous observations12 and the occurrence of two closely spaced motifs upstream of xol-1, clustering of motif occurrences appears to be important for DCC recruitment along X (Supplementary Figure 3E).
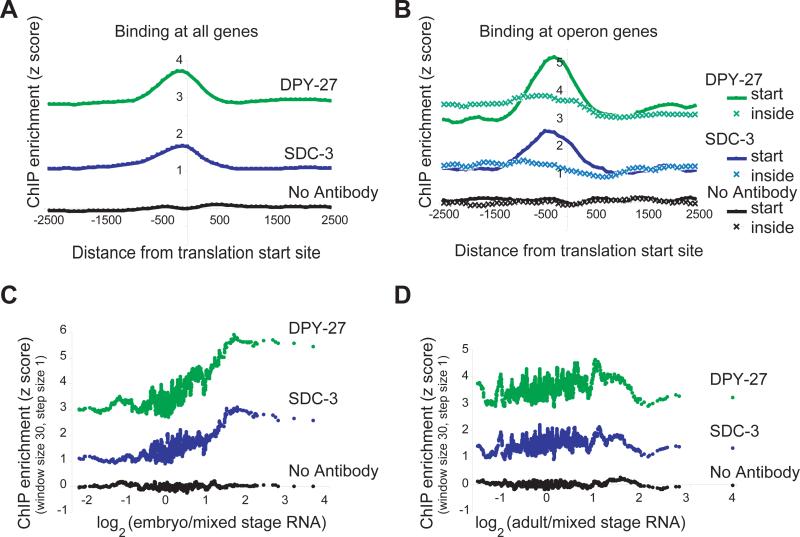
We sought to determine the relationship between DCC binding and gene location (Figure 5A). We found that DCC binding was strongest at a stereotypic distance upstream of the translation start and interpreted the signal to emanate from the transcription start site. Consistent with this interpretation, DCC binding peaks occurred preferentially in intergenic regions, specifically regions annotated to be promoters (Supplementary Figure 4A-B). The C. elegans genome contains numerous operons, in which multiple genes are encoded by a single transcript. If the signal indeed emanates from transcription start sites, signal at operons is predicted to occur upstream of the first translation initiation site, but not upstream of internal translation start sites. Indeed, a DCC binding peak is observed only upstream of genes that are at the start of an operon, supporting our interpretation (Figure 5B). Localization to transcription start sites suggests a mechanism of repression that acts at transcriptional initiation, in contrast to the Drosophila DCC (Supplementary Note). Furthermore, yeast studies have shown that the homologous condensins and the related cohesin complexes are pushed along genes by polymerases20-22, suggesting a spreading mechanism coupled directly to polymerase movement. This could explain the “baseline” X-association of the DCC that occurs in the absence of DNA sequence signals, and the preferential occurrence of foci downstream of convergently transcribed genes (Supplementary Figure 4B).
Figure 5. The DCC preferentially binds upstream of genes and is positively correlated with transcriptional activity.
(A) Data was centered (coordinate 0) according to the predicted translation start sites of each X-linked gene, and the average z score of the probes in a sliding window (width 3 probes, step size 1 probe) was plotted, taking into account the direction of transcription. Translation start sites were used as a proxy for transcription start sites because they are much better-annotated. (B) Translation start sites of X-linked genes located at the start (solid, 56 genes) or inside (hatched, 69 genes) of an operon were centered. As in panel A, DCC binding is plotted as a function of distance from translation start site. (C) A moving average of DCC ChIP enrichment (performed in embryos) plotted as a function of embryonic transcript level of the downstream gene6. The positive correlation between embryonic DCC binding and embryonic transcription persisted when RNA measurements from a second publication were utilized5 (Supplementary Figure 4C). (D) As in panel C, except adult RNA levels are used. Additionally, as expected, DCC binding was not correlated with male- or hermaphrodite-specific expression (Supplementary Figure 4D).
The DCC uniformly down-regulates genes transcribed at diverse frequencies, but how this is achieved is unknown. We asked whether genes associated with abundant transcripts required more DCC binding to achieve down-regulation. Indeed, a positive correlation between DCC binding and the transcript level of downstream genes during embryogenesis was observed (Figure 5C, Supplementary Figure 4C). Furthermore, DCC binding to promoters is likely to be dynamic because embryonic DCC binding was not correlated to adult RNA levels (Figure 5D). This “tuning” of the degree of DCC binding for different levels of gene expression presents a more refined depiction of DCC action than previous models, which suggested a more uniform global condensation or “overwinding” of the entire X10. A possible mechanism for directing the DCC to sites of active transcription is suggested by DPY-30, which is required for the localization of the DCC to X23. DPY-30 is a homolog of S. cerevisiae Saf19, a member of the histone methyltransferase Set1 complex responsible for H3K4 methylation, a mark associated with active transcription24.
Our comprehensive DCC binding data reveal that a non-uniform distribution of a condensin-like protein complex applies a constant, subtle, two-fold level of repression across an entire chromosome. The emerging model involves cooperation of DNA sequence and chromatin signals that target the DCC to the promoters of individual genes, possibly affecting transcriptional initiation. Recruitment, which is tuned to the level of expression, may be followed by a short-range polymerase-coupled spreading mechanism. Detailed tests of the hypotheses suggested here are required to determine the relative contribution of DNA sequence, transcriptional activity, and chromatin modifications to DCC localization, whether DCC composition varies across the X, and the precise mechanism of DCC-mediated repression.
METHODS
Antibodies
Recombinant proteins containing amino acids 1067-1340 of SDC-3 and 1-409 of DPY-27 were cloned and expressed with Novagen's Pet30-EkLIC system. Rabbit polyclonal antibodies were produced at Covance Immunology Services. DNA encoding the SDC-3 and DPY-27 epitopes were cloned into pGEX-5X-2 vector (Amersham Biosciences) and used to affinity purify antibodies.
Chromatin Immunoprecipitation
For extract preparations, N2 adults were grown in standard S-liquid media and embryos were obtained by bleaching25. Embryos were fixed with 2% formaldehyde for 30 minutes at room temperature and washed with 100 mM Tris pH 7.5 once, M9 buffer twice and 10 ml FA buffer (50 mM HEPES/KOH pH 7.5, 1 mM EDTA, 1% Triton X-100, 0.1 % sodium deoxycholate; 150 mM NaCl) with protease inhibitors (Calbiochem) and frozen at −80°C. 500 μl of packed embryos were thawed and resuspended in 2 ml of FA buffer and dounce homogenized on ice with 30 strokes. To shear chromatin (size range 200-800 bp), samples were sonicated with a Branson sonifier at 30% amplitude for 7 cycles of 12 pulses (0.9 sec on, 0.1 sec off), with cooling in dry ice/ethanol bath for 2 seconds between cycles. Cellular debris was removed by microfuge centrifugation at 13,000 g for 15 minutes at 4°C. Immunoprecipitation reactions contained approximately 3 mg of total protein with 3-5 μg of purified antibodies in 500 μl total volume, with 1% sarkosyl. Prior to addition of the antibody, 10% of the material was taken as input. Immunocomplexes were collected with Protein A-sepharose beads (Amersham Biosciences) and washed with 1 ml of the following buffers : FA buffer 2 × 5 min, FA-1M NaCl (FA buffer with 1 M NaCl) 5 min, FA-500 mM NaCl 10 min, TEL buffer (0.25 M LiCl, 1% NP-40, 1% sodium deoxycholate, 1 mM EDTA, 10 mM Tris-HCl, pH 8.0) 10 min., TE pH 8.0 2 × 5 min. Complexes were eluted in 1% SDS in TE with 250 mM NaCl at 65°C 30 minutes. Samples and inputs were treated with Proteinase K for 1 hour and crosslinks were reversed at 65°C overnight. DNA was purified with Qiagen PCR purification columns and amplified by LM-PCR (Supplementary Methods). Locus-specific ChIP PCRs were performed by diluting amplified material 50-fold and querying with locus-specific primers (Supplementary Methods).
Microarrays and Data Extraction
The C. elegans genome tiling arrays were designed and constructed by NimbleGen System Inc. using maskless array synthesis technology, with approximately 650,000 oligonucleotides per array26. Two arrays covered the C. elegans genome at 86-bp resolution as described in the text. We performed three biologically independent experiments for SDC-3, DPY-27 and No Antibody control ChIPs. Amplified samples were labeled and hybridized to high-resolution microarrays by the NimbleGen Service Laboratory as described previously27. Two ChIPs (ChIP 1 and 3) were labeled with Cy5 and the reference materials (Input DNA) were labeled with Cy3, and for one (ChIP 2), the dyes were swapped. Raw intensity data was extracted from scanned images using NimbleScan 2.3 extraction software (NimbleGen Systems Inc.). The peak finding algorithm ChIPOTle16, was used to identify regions of SDC-3 or DPY-27 binding, with a sliding window of 500 bp and 86 bp steps. For details, including data normalization and visualization, see Supplementary Methods.
Motif Discovery
MDscan28 and BioProspector29, were used to identify motifs with the sequence composition of the C. elegans genome used as a background model. Motif widths of up to 15 bp were explored. The 10-bp motif described was most predictive of DCC binding. We used the MatrixScan module from BioProspector to scan the genome for the presence of the 10-bp sequence motif at three cutoffs (p ≤ 10−4, p ≤ 10−5, p ≤ 10−6).
Transcription Analysis
RNA levels in embryos and adults were obtained from the literature6. The transcription level of genes during embryogenesis was determined by averaging the last three time points from a published study5. The average DCC binding z score for each gene was calculated by taking the average value of probes that fall between 500 bp upstream and 200 bp downstream of the translation start site. The list of genes and the translation start coordinates were obtained from UCSC WormBase Genes from the sangerGene table (http://genome.ucsc.edu/). For genes with splice variants, the coordinates for the longest splice variant were used. WormMart (http://www.wormbase.org) was used to determine if a gene was located within or at the start of an operon.
Accession numbers
Raw data are available from GEO (Gene Expression Omnibus), accession number GSE6739. Note: Supplementary information is available on the Nature Genetics website.
Supplementary Material
Supplementary Figure 1. DCC binding peak distribution
(A) The distribution of SDC-3 binding peak widths. Foci are represented by a lighter color. (B) Same as (a) but for DPY-27. (C) The distribution of distances between SDC-3 binding foci. (D) Same as (c) but for DPY-27. (E) For SDC-3 binding peaks (excluding foci) the distance to the nearest focus was calculated, and the distribution of distances is shown. Peaks of the same width were placed at random along the chromosome to estimate the null expectation. (F) Same as (e) but for DPY-27.
Supplementary Figure 2. DCC binding at the previously defined rex loci
DCC binding is shown at three rex sites1 (rex-1 is shown in Figure 4B). Median z scores are plotted along chromosome coordinates for SDC-3, DPY-27 and control IP (no antibody). Locations of the 10-bp motif identified in this study are depicted with red tick marks. Longer tick mark indicate motifs with a closer match to the consensus (p < 10−6 from MatrixScan). (A) rex-2 (B) rex-3, which resides in an exon and was identified as a peak but not a “focus” of binding and (C) rex-4.
(1) McDonel, P., Jans, J., Peterson, B.K. & Meyer, B.J. Clustered DNA motifs mark X chromosomes for repression by a dosage compensation complex. Nature 444, 614-8 (2006).
Supplementary Figure 3. The relationship between DCC binding and the distribution of the putative DCC recruitment motif
(A) The percentage of X-linked motifs that fall under foci or under other DCC binding peaks is shown. The number of motifs was determined by MatrixScan at three different p-value cutoffs, p < 10−4, p < 10−5, and p < 10−6. Lower p-values indicate a closer match to the consensus. The “expected” percentages were calculated by dividing the number of bases covered by each class of peaks by the total number of bases on X. The number of motifs that fall in each peak category is shown below. (B) The percentage of DCC binding peaks that contain a motif is shown. The expected percentage was shown to the right of each bar in lighter color. “Expected” percentages were calculated by assuming all peaks had the average peak width and the motifs were uniformly distributed across X. The number of peaks that contain a motif at each p value are shown below. (C) The relative frequency (motifs per bp / genomic average motifs per bp) of the 10-bp motif derived from SDC-3 and (D) DPY-27. ChIP-chip data is plotted for each chromosome. The number of motifs on each chromosome is shown below. (E) To determine whether motifs were more likely to occur in close proximity on X versus the autosomes, windows of varying length (100 – 10000 bp) were tiled contiguously across each chromosome and the number of motifs in each window was counted. A chi-square statistic was calculated to determine whether the number of windows with 0, 1, or >1 motifs was independent of those windows being from an autosome or X.
Supplementary Figure 4. The DCC preferentially binds to the 5’ region of genes and is positively correlated with transcriptional activity.
(A) Peaks were assigned to a coding sequence (CDS) or intergenic region (INT) according to the genomic position of the probe at the peak maximum. ”Expected” values were determined by calculating the proportion of all X-probes on the array that were annotated to each group. p values reflect the probability that both classes shown deviate from the expected frequencies, and were determined by a chi square test. (B) Among the intergenic peaks, the number of peaks located upstream of two divergently transcribed genes (bidirectional), upstream of one gene and downstream of another (unidirectional) and downstream of two genes (downstream) is shown. Expected values and p-values were calculated as in (a) using all intergenic X-linked probes. (C) A moving average of enrichment of promoters resulting from the indicated ChIP experiment plotted as a function of embryonic transcript level1. Embryonic RNA levels were determined by averaging values from the last three time points reported1. All ChIP experiments were performed from embryo extract. The transcript levels used in this plot were obtained independently from those used in Figure 5C2. The positive correlation between embryonic DCC binding and transcription with these RNA measurements confirms the trend observed in Figure 5C. (D) A moving average of ChIP enrichment plotted as a function of the ratio of RNA level between sexes (adult males/adult hermaphrodites)2.
(1) Baugh, L.R., Hill, A.A., Slonim, D.K., Brown, E.L. & Hunter, C.P. Composition and dynamics of the Caenorhabditis elegans early embryonic transcriptome. Development 130, 889-900 (2003).
(2) Jiang, M. et al. Genome-wide analysis of developmental and sex-regulated gene expression profiles in Caenorhabditis elegans. Proc Natl Acad Sci U S A 98, 218-23 (2001).
Supplementary Figure 5. The predictive value of the 10 bp putative DCC recruitment motif identified in this study versus putative DCC recruitment motifs identified previously.
Receiver Operating Characteristic (ROC) curves were used to estimate the ability of motif occurrence to identify foci of DCC binding (Supplementary Methods). (A) ROC curves for the 10 bp sequence motif found in this study, and those for and “Motif A” and “Motif B”, which were derived from four loci that recruit the DCC as an extrachromosomal array1. With respect to identifying DCC foci on a natural chromosomal substrate, the motif identified here outperforms Motif A, Motif B, or a combination of Motif A and Motif B (see Supplementary Methods for motif scoring). A simple additive model for relations between selfsame or heterologous motifs was assumed here. There are many variations of reasonable but more complex models involving the motifs that may perform better (for example those that heavily weight a very specific spacing between motifs, or those with complex cooperative interactions). (B) We asked whether Motif A and Motif B might identify sequence features that are not captured by the 10 bp motif identified in this study. Shown is the ROC curve for the 10-bp motif alone (blue), or that for the 10-bp motif plus Motif A (red), the 10-bp motif plus Motif B (pink) and the 10-bp motif plus Motif A and Motif B (black). Again, a simple additive model was used. The coincident ROC curves indicate that Motif A and Motif B, in this additive model, do not identify sequence features that are unaccounted for by the 10 bp motif identified in this study. For example, Motif A and Motif B may promote DCC binding, but in this interpretation that effect is already manifest in the 10 bp motif identified here.
(1) McDonel, P., Jans, J., Peterson, B.K. & Meyer, B.J. Clustered DNA motifs mark X chromosomes for repression by a dosage compensation complex. Nature 444, 614-8 (2006).
Supplementary Note
Sharp peaks of SDC-3 binding suggest variation in the composition of focus-distal dosage compensation complexes
At foci, SDC-3 binding was high but diminished rapidly to a baseline. While the signal from DPY-27 foci also diminished rapidly, it maintained a higher baseline along the X chromosome (Figure 3D, Figure 2A-B and D). This qualitative difference in the binding pattern can be quantitated by measuring the variance of the data, expressed as the ratio of standard deviation to the mean. This quantity is called the coefficient of variation. For probes near foci (within 8 kb on either side), the coefficient of variation for SDC-3 is 61.8%, compared to a value of 36.2% for DPY-27. For the entire X, these values are 106% and 49%, respectively. The fact that the maximal enrichment for SDC-3 is higher than that of DPY-27 for most foci (average maximum log ratio on X for SDC-3 is 5.41 and DPY-27 is 3.87) make a technical explanation (e.g. differences in the specificity or affinity of the antibodies used) for the higher variance unlikely. In other words, if technical factors were at play, the higher focus amplitude observed for SDC-3 might be expected to accompany higher baseline values, but in fact the opposite is observed. In addition, both antibodies produce similar levels of measurement noise based on signal emanating from autosomes (SDC-3 ratio 0.004 ± 0.352; DPY-27 ratio 0.019 ± 0.355). Therefore, the data support a hypothesis in which complexes at foci are of distinct composition relative to those that spread from the foci.
Fine-tuning an entire chromosome: comparison to the Drosophila DCC
While the C. elegans DCC acts to down-regulate each X by a factor of two in XX animals, the Drosophila DCC performs a nearly perfectly reciprocal function by up-regulating transcription from the single X of male flies by a factor of two to match the output from XX females1,2. Our data reveals many interesting parallels and contrasts with the localization and mechanism of the Drosophila DCC.
Drosophila dosage compensation is thought to be achieved by modulating transcriptional elongation, based on the localization of the Drosophila DCC to the 3’ end of coding regions of genes and the observation that X chromosomes do not recruit more polymerase than autosomes3-5. In contrast, the localization of the C. elegans DCC to the 5′ region of genes suggests a mechanism of repression that acts at transcriptional initiation. In both systems, it is clear that DNA sequence alone does not fully determine the distribution of the DCC. However, the identity of the C. elegans DCC immediately suggests models for its recruitment and spreading. It contains novel proteins (SDC-2 and the zinc-finger proteins SDC-1 and SDC-3) that may recognize the sequence motif we identify here; a protein homologous to a member of the yeast Set1 complex (DPY-30) that may be involved in directing the DCC to the 5′ region of actively transcribed genes, and proteins homologous to members of the condensin complexes required for mitotic chromosome condensation (DPY-26, DPY-27, DPY-28, and MIX-1) that may effect dosage compensation by their enrichment at sites of transcriptional initiation. This “tuning” of the level of the DCC for different levels of gene expression again presents a more refined picture of DCC action than previous models that suggested a more uniform global condensation or “overwinding” over the entire X6. Finally, a direct mechanism for spreading is afforded by studies on the homologous cohesin and condensin complexes in yeast, which are “pushed” along genes by RNA polymerase and DNA polymerase respectively7-9. This may explain the general “baseline” X-association we observe. In contrast, the Drosophila DCC binds to approximately 25% of the X, without a baseline level of binding.
REFERENCES
1. Hamada, F.N., Park, P.J., Gordadze, P.R. & Kuroda, M.I. Global regulation of X chromosomal genes by the MSL complex in Drosophila melanogaster. Genes Dev 19, 2289-94 (2005).
2. Nusinow, D.A. & Panning, B. Recognition and modification of seX chromosomes. Curr Opin Genet Dev 15, 206-13 (2005).
3. Buscaino, A., Legube, G. & Akhtar, A. X-chromosome targeting and dosage compensation are mediated by distinct domains in MSL-3. EMBO Rep 7, 531-8 (2006).
4. Alekseyenko, A.A., Larschan, E., Lai, W.R., Park, P.J. & Kuroda, M.I. High-resolution ChIP-chip analysis reveals that the Drosophila MSL complex selectively identifies active genes on the male X chromosome. Genes Dev 20, 848-57 (2006).
5. Gilfillan, G.D. et al. Chromosome-wide gene-specific targeting of the Drosophila dosage compensation complex. Genes Dev 20, 858-70 (2006).
6. Lieb, J.D., Albrecht, M.R., Chuang, P.T. & Meyer, B.J. MIX-1: an essential component of the C. elegans mitotic machinery executes X chromosome dosage compensation. Cell 92, 265-77 (1998).
7. Wang, B.D., Eyre, D., Basrai, M., Lichten, M. & Strunnikov, A. Condensin binding at distinct and specific chromosomal sites in the Saccharomyces cerevisiae genome. Mol Cell Biol 25, 7216-25 (2005).
8. Lengronne, A. et al. Cohesin relocation from sites of chromosomal loading to places of convergent transcription. Nature 430, 573-8 (2004).
9. Glynn, E.F. et al. Genome-wide mapping of the cohesin complex in the yeast Saccharomyces cerevisiae. PLoS Biol 2, E259 (2004).
SUPPLEMENTARY METHODS
Primers for locus-specific PCR (Figure 1F)
rex-1: GCATCAACAAGCCGCAATGC and TTGCTTGTACGCACATATAC
B0302.2: CTCGCTTTGTGACACCTGTA and GCACGCCACGACCTTGCATT
R11.4: CACGTTGAACGATAGGCAGA and AACAAGATCATCCCACTTGTC
K07G5.3: GTTGATTTCGGAAGACCTCATAC and ATCTCATAGCAATAAAGCATTTAC
Reference: TCGTCCTCGACTCTGGAGAT and GTGATCTTACTGATTACCTC
Samples were separated by agarose gel electrophoresis and band intensities were measured by Kodak ID 3.6 software. Fold enrichment was calculated by the following formula: (ChIP locus/ChIP reference)/(Input locus/ Input reference). All values are sums of pixel intensities.
LM-PCR
LM-PCR was performed as in10.
Data Normalization and Visualization
The ratio of intensity from sample/reference channel was transformed to log2 space. Data for each IP was normalized so that the median of the autosomal spots was 0. z scores were obtained by standardizing the data for each IP with the mean and standard deviation of the autosomal spots. In this context, the autosomal probes were assumed to be representative of technical variation with regard to both the ChIP procedure and microarray measurements. We calculated the z scores of each data point in the three replicates and took the median of these for most analyses.
The significance of autosome versus X chromosome distribution was determined by analysis of variance (one way ANOVA of data points classified by being on an autosome or the X chromosome) with R Commander11.
Data was visualized with the UCSC Genome Browser (C. elegans March 2004 http://genome.ucsc.edu). All data is based on WS120 build (March 2004) of the C. elegans genome.
Signal Processing
Since binding for both SDC-3 and DPY-27 was limited almost exclusively to X, to better account for the X-specific variability we analyzed X in isolation for peak detection. We median-centered the z scores of X-chromosome probes. For both SDC-3 and DPY-27, four sets of peaks (one from each of the three biological replicates and a fourth for the median z–score) were called with ChIPOTle12. We removed any peaks from the median z-score set that were not present in at least 2 out of the 3 replicates (250 bp) or that were found in the no-antibody dataset. Sites of enrichment for autosomes were found with ChIPOTle using the median z scores, a 1 kb window, and 86 bp step size.
Analysis of DCC binding peaks on X
For SDC-3 and DPY-27 foci comparisons, foci were considered to be near coincident when their coordinates overlapped. To assess the significance of these relationships, we iteratively sampled a new set of SDC-3 or DPY-27 peaks from X, keeping both the number and widths of the peaks the same. The position of the new peaks on X, were selected such that they were equally likely to occur across the entire chromosome. The overlap of these newly selected peak with the observed peaks was calculated. This process was repeated 200 times. Using the mean and standard deviation of the resulting distribution of overlaps, we calculated the z-score for the proportion of observed peak overlaps and obtained a p-value from the standard normal distribution.
To assess to extent to which peaks were uniformly distributed versus clustered on the X we divided the chromosome into 12 windows and counted the number of peaks in each. We performed a chi-squared test using the counts for uniformly distributed peaks as our expected value.
To determine the extent to which peaks were clustered around the binding foci we calculated the distance from the middle of a peak to the middle of the nearest focus. Then, using a analysis similar to that described above, we sampled a new set of peaks from X with the position of the foci fixed. We repeated this 100 times, each time recording the distance of each peak to the nearest focus. To assess the significance of this relationship, we calculated the probability that the observed distances between foci and peaks were smaller than what would be expected if peaks were uniformly distributed between foci13:
Where τ is the difference between the observed and expected distances and n is the number of peaks.
Annotation of DCC binding peaks on X
We annotated each peak to a feature by determining where the maximum value probe within each peak falls in the genome. Translation start and stop coordinates were derived from the WS120 (March 2004) pre-built MySql database obtained from http://www.wormbase.org. Probe coordinates were assigned as overlapping annotated CDS sequences or the intergenic space between them. We performed chi-squared tests by calculating expected values based on the proportion of probes on the array that were annotated to the feature of interest.
Receiver Operating Characteristic (ROC) curves and motif analysis
To generate Motif A and Motif B position weight matrices, we used the sequences provided by Supplementary Table 1 of McDonel et al 14 as input for MDscan15. This procedure generated a matrix that accurately reflected the reported motifs. We divided the genome in 12,000 bp segments for input to MatrixScan, which assessed the presence and strength of the given sequence matrix and produced a motif score for each genomic segment. We designated segments that overlapped with coordinates of SDC-3 foci to be “true positives targets”, used these to create the ROC curves in Supplementary Figure 5. To test for additive effects of motifs, we simply summed the motif scores of interest for each segment.
ACKNOWLEDGMENTS
We thank Andrew B. Nobel (Professor of Statistics, University of North Carolina) for providing statistical guidance, especially regarding the clustering of DCC binding peaks. This work was supported by the Carolina Center for Genome Sciences and by a grant awarded to J.D.L. from The V Foundation for Cancer Research.
Footnotes
AUTHOR CONTRIBUTIONS
This study was designed by SE and JDL. SE conducted the experiments. SE, PGG, CMW, and JDL conducted the data analysis. XZ and RDG designed, manufactured, and hybridized the DNA microarrays. SE and JDL wrote the paper.
COMPETING INTERESTS STATEMENT
The authors declare that they have no competing financial interests.
REFERENCES
- 1.Meyer BJ, Casson LP. Caenorhabditis elegans compensates for the difference in X chromosome dosage between the sexes by regulating transcript levels. Cell. 1986;47:871–81. doi: 10.1016/0092-8674(86)90802-0. [DOI] [PubMed] [Google Scholar]
- 2.Chuang PT, Albertson DG, Meyer BJ. DPY-27:a chromosome condensation protein homolog that regulates C. elegans dosage compensation through association with the X chromosome. Cell. 1994;79:459–74. doi: 10.1016/0092-8674(94)90255-0. [DOI] [PubMed] [Google Scholar]
- 3.Davis TL, Meyer BJ. SDC-3 coordinates the assembly of a dosage compensation complex on the nematode X chromosome. Development. 1997;124:1019–31. doi: 10.1242/dev.124.5.1019. [DOI] [PubMed] [Google Scholar]
- 4.Plath K, Mlynarczyk-Evans S, Nusinow DA, Panning B. Xist RNA and the mechanism of X chromosome inactivation. Annu Rev Genet. 2002;36:233–78. doi: 10.1146/annurev.genet.36.042902.092433. [DOI] [PubMed] [Google Scholar]
- 5.Baugh LR, Hill AA, Slonim DK, Brown EL, Hunter CP. Composition and dynamics of the Caenorhabditis elegans early embryonic transcriptome. Development. 2003;130:889–900. doi: 10.1242/dev.00302. [DOI] [PubMed] [Google Scholar]
- 6.Jiang M, et al. Genome-wide analysis of developmental and sex-regulated gene expression profiles in Caenorhabditis elegans. Proc Natl Acad Sci U S A. 2001;98:218–23. doi: 10.1073/pnas.011520898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Meyer BJ, Community, T.C.e.R. X-Chromosome dosage compensation. WormBook. 2005 doi: 10.1895/wormbook.1.8.1. WormBook doi/10.1895/wormbook.1.8.1, http://www.wormbook.org. [DOI] [PMC free article] [PubMed]
- 8.Hirano T. Condensins: organizing and segregating the genome. Curr Biol. 2005;15:R265–75. doi: 10.1016/j.cub.2005.03.037. [DOI] [PubMed] [Google Scholar]
- 9.Hagstrom KA, Holmes VF, Cozzarelli NR, Meyer BJ. C. elegans condensin promotes mitotic chromosome architecture, centromere organization, and sister chromatid segregation during mitosis and meiosis. Genes Dev. 2002;16:729–42. doi: 10.1101/gad.968302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lieb JD, Albrecht MR, Chuang PT, Meyer BJ. MIX-1: an essential component of the C. elegans mitotic machinery executes X chromosome dosage compensation. Cell. 1998;92:265–77. doi: 10.1016/s0092-8674(00)80920-4. [DOI] [PubMed] [Google Scholar]
- 11.Csankovszki G, McDonel P, Meyer BJ. Recruitment and spreading of the C. elegans dosage compensation complex along X chromosomes. Science. 2004;303:1182–5. doi: 10.1126/science.1092938. [DOI] [PubMed] [Google Scholar]
- 12.McDonel P, Jans J, Peterson BK, Meyer BJ. Clustered DNA motifs mark X chromosomes for repression by a dosage compensation complex. Nature. 2006;444:614–8. doi: 10.1038/nature05338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chu DS, et al. A molecular link between gene-specific and chromosome-wide transcriptional repression. Genes Dev. 2002;16:796–805. doi: 10.1101/gad.972702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li W, Streit A, Robertson B, Wood WB. Evidence for multiple promoter elements orchestrating male-specific regulation of the her-1 gene in Caenorhabditis elegans. Genetics. 1999;152:237–48. doi: 10.1093/genetics/152.1.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dawes HE, et al. Dosage compensation proteins targeted to X chromosomes by a determinant of hermaphrodite fate. Science. 1999;284:1800–4. doi: 10.1126/science.284.5421.1800. [DOI] [PubMed] [Google Scholar]
- 16.Buck MJ, Nobel AB, Lieb JD. ChIPOTle: a user-friendly tool for the analysis of ChIP-chip data. Genome Biol. 2005;6:R97. doi: 10.1186/gb-2005-6-11-r97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Miller LM, Plenefisch JD, Casson LP, Meyer BJ. xol-1: a gene that controls the male modes of both sex determination and X chromosome dosage compensation in C. elegans. Cell. 1988;55:167–83. doi: 10.1016/0092-8674(88)90019-0. [DOI] [PubMed] [Google Scholar]
- 18.Hodgkin J, Zellan JD, Albertson DG. Identification of a candidate primary sex determination locus, fox-1, on the X chromosome of Caenorhabditis elegans. Development. 1994;120:3681–9. doi: 10.1242/dev.120.12.3681. [DOI] [PubMed] [Google Scholar]
- 19.Nicoll M, Akerib CC, Meyer BJ. X-chromosome-counting mechanisms that determine nematode sex. Nature. 1997;388:200–4. doi: 10.1038/40669. [DOI] [PubMed] [Google Scholar]
- 20.Wang BD, Eyre D, Basrai M, Lichten M, Strunnikov A. Condensin binding at distinct and specific chromosomal sites in the Saccharomyces cerevisiae genome. Mol Cell Biol. 2005;25:7216–25. doi: 10.1128/MCB.25.16.7216-7225.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lengronne A, et al. Cohesin relocation from sites of chromosomal loading to places of convergent transcription. Nature. 2004;430:573–8. doi: 10.1038/nature02742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Glynn EF, et al. Genome-wide mapping of the cohesin complex in the yeast Saccharomyces cerevisiae. PLoS Biol. 2004;2:E259. doi: 10.1371/journal.pbio.0020259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hsu DR, Chuang PT, Meyer BJ. DPY-30, a nuclear protein essential early in embryogenesis for Caenorhabditis elegans dosage compensation. Development. 1995;121:3323–34. doi: 10.1242/dev.121.10.3323. [DOI] [PubMed] [Google Scholar]
- 24.Nagy PL, Griesenbeck J, Kornberg RD, Cleary ML. A trithorax-group complex purified from Saccharomyces cerevisiae is required for methylation of histone H3. Proc Natl Acad Sci U S A. 2002;99:90–4. doi: 10.1073/pnas.221596698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lewis JA, Fleming JT. Basic culture methods. Methods Cell Biol. 1995;48:3–29. [PubMed] [Google Scholar]
- 26.Singh-Gasson S, et al. Maskless fabrication of light-directed oligonucleotide microarrays using a digital micromirror array. Nat Biotechnol. 1999;17:974–8. doi: 10.1038/13664. [DOI] [PubMed] [Google Scholar]
- 27.Selzer RR, et al. Analysis of chromosome breakpoints in neuroblastoma at subkilobase resolution using fine-tiling oligonucleotide array CGH. Genes Chromosomes Cancer. 2005;44:305–19. doi: 10.1002/gcc.20243. [DOI] [PubMed] [Google Scholar]
- 28.Liu XS, Brutlag DL, Liu JS. An algorithm for finding protein-DNA binding sites with applications to chromatin-immunoprecipitation microarray experiments. Nat Biotechnol. 2002;20:835–9. doi: 10.1038/nbt717. [DOI] [PubMed] [Google Scholar]
- 29.Liu X, Brutlag DL, Liu JS. BioProspector: discovering conserved DNA motifs in upstream regulatory regions of co-expressed genes. Pac Symp Biocomput. 2001:127–38. [PubMed] [Google Scholar]
- 30.Workman CT, et al. enoLOGOS: a versatile web tool for energy normalized sequence logos. Nucleic Acids Res. 2005;33:W389–92. doi: 10.1093/nar/gki439. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1. DCC binding peak distribution
(A) The distribution of SDC-3 binding peak widths. Foci are represented by a lighter color. (B) Same as (a) but for DPY-27. (C) The distribution of distances between SDC-3 binding foci. (D) Same as (c) but for DPY-27. (E) For SDC-3 binding peaks (excluding foci) the distance to the nearest focus was calculated, and the distribution of distances is shown. Peaks of the same width were placed at random along the chromosome to estimate the null expectation. (F) Same as (e) but for DPY-27.
Supplementary Figure 2. DCC binding at the previously defined rex loci
DCC binding is shown at three rex sites1 (rex-1 is shown in Figure 4B). Median z scores are plotted along chromosome coordinates for SDC-3, DPY-27 and control IP (no antibody). Locations of the 10-bp motif identified in this study are depicted with red tick marks. Longer tick mark indicate motifs with a closer match to the consensus (p < 10−6 from MatrixScan). (A) rex-2 (B) rex-3, which resides in an exon and was identified as a peak but not a “focus” of binding and (C) rex-4.
(1) McDonel, P., Jans, J., Peterson, B.K. & Meyer, B.J. Clustered DNA motifs mark X chromosomes for repression by a dosage compensation complex. Nature 444, 614-8 (2006).
Supplementary Figure 3. The relationship between DCC binding and the distribution of the putative DCC recruitment motif
(A) The percentage of X-linked motifs that fall under foci or under other DCC binding peaks is shown. The number of motifs was determined by MatrixScan at three different p-value cutoffs, p < 10−4, p < 10−5, and p < 10−6. Lower p-values indicate a closer match to the consensus. The “expected” percentages were calculated by dividing the number of bases covered by each class of peaks by the total number of bases on X. The number of motifs that fall in each peak category is shown below. (B) The percentage of DCC binding peaks that contain a motif is shown. The expected percentage was shown to the right of each bar in lighter color. “Expected” percentages were calculated by assuming all peaks had the average peak width and the motifs were uniformly distributed across X. The number of peaks that contain a motif at each p value are shown below. (C) The relative frequency (motifs per bp / genomic average motifs per bp) of the 10-bp motif derived from SDC-3 and (D) DPY-27. ChIP-chip data is plotted for each chromosome. The number of motifs on each chromosome is shown below. (E) To determine whether motifs were more likely to occur in close proximity on X versus the autosomes, windows of varying length (100 – 10000 bp) were tiled contiguously across each chromosome and the number of motifs in each window was counted. A chi-square statistic was calculated to determine whether the number of windows with 0, 1, or >1 motifs was independent of those windows being from an autosome or X.
Supplementary Figure 4. The DCC preferentially binds to the 5’ region of genes and is positively correlated with transcriptional activity.
(A) Peaks were assigned to a coding sequence (CDS) or intergenic region (INT) according to the genomic position of the probe at the peak maximum. ”Expected” values were determined by calculating the proportion of all X-probes on the array that were annotated to each group. p values reflect the probability that both classes shown deviate from the expected frequencies, and were determined by a chi square test. (B) Among the intergenic peaks, the number of peaks located upstream of two divergently transcribed genes (bidirectional), upstream of one gene and downstream of another (unidirectional) and downstream of two genes (downstream) is shown. Expected values and p-values were calculated as in (a) using all intergenic X-linked probes. (C) A moving average of enrichment of promoters resulting from the indicated ChIP experiment plotted as a function of embryonic transcript level1. Embryonic RNA levels were determined by averaging values from the last three time points reported1. All ChIP experiments were performed from embryo extract. The transcript levels used in this plot were obtained independently from those used in Figure 5C2. The positive correlation between embryonic DCC binding and transcription with these RNA measurements confirms the trend observed in Figure 5C. (D) A moving average of ChIP enrichment plotted as a function of the ratio of RNA level between sexes (adult males/adult hermaphrodites)2.
(1) Baugh, L.R., Hill, A.A., Slonim, D.K., Brown, E.L. & Hunter, C.P. Composition and dynamics of the Caenorhabditis elegans early embryonic transcriptome. Development 130, 889-900 (2003).
(2) Jiang, M. et al. Genome-wide analysis of developmental and sex-regulated gene expression profiles in Caenorhabditis elegans. Proc Natl Acad Sci U S A 98, 218-23 (2001).
Supplementary Figure 5. The predictive value of the 10 bp putative DCC recruitment motif identified in this study versus putative DCC recruitment motifs identified previously.
Receiver Operating Characteristic (ROC) curves were used to estimate the ability of motif occurrence to identify foci of DCC binding (Supplementary Methods). (A) ROC curves for the 10 bp sequence motif found in this study, and those for and “Motif A” and “Motif B”, which were derived from four loci that recruit the DCC as an extrachromosomal array1. With respect to identifying DCC foci on a natural chromosomal substrate, the motif identified here outperforms Motif A, Motif B, or a combination of Motif A and Motif B (see Supplementary Methods for motif scoring). A simple additive model for relations between selfsame or heterologous motifs was assumed here. There are many variations of reasonable but more complex models involving the motifs that may perform better (for example those that heavily weight a very specific spacing between motifs, or those with complex cooperative interactions). (B) We asked whether Motif A and Motif B might identify sequence features that are not captured by the 10 bp motif identified in this study. Shown is the ROC curve for the 10-bp motif alone (blue), or that for the 10-bp motif plus Motif A (red), the 10-bp motif plus Motif B (pink) and the 10-bp motif plus Motif A and Motif B (black). Again, a simple additive model was used. The coincident ROC curves indicate that Motif A and Motif B, in this additive model, do not identify sequence features that are unaccounted for by the 10 bp motif identified in this study. For example, Motif A and Motif B may promote DCC binding, but in this interpretation that effect is already manifest in the 10 bp motif identified here.
(1) McDonel, P., Jans, J., Peterson, B.K. & Meyer, B.J. Clustered DNA motifs mark X chromosomes for repression by a dosage compensation complex. Nature 444, 614-8 (2006).
Supplementary Note
Sharp peaks of SDC-3 binding suggest variation in the composition of focus-distal dosage compensation complexes
At foci, SDC-3 binding was high but diminished rapidly to a baseline. While the signal from DPY-27 foci also diminished rapidly, it maintained a higher baseline along the X chromosome (Figure 3D, Figure 2A-B and D). This qualitative difference in the binding pattern can be quantitated by measuring the variance of the data, expressed as the ratio of standard deviation to the mean. This quantity is called the coefficient of variation. For probes near foci (within 8 kb on either side), the coefficient of variation for SDC-3 is 61.8%, compared to a value of 36.2% for DPY-27. For the entire X, these values are 106% and 49%, respectively. The fact that the maximal enrichment for SDC-3 is higher than that of DPY-27 for most foci (average maximum log ratio on X for SDC-3 is 5.41 and DPY-27 is 3.87) make a technical explanation (e.g. differences in the specificity or affinity of the antibodies used) for the higher variance unlikely. In other words, if technical factors were at play, the higher focus amplitude observed for SDC-3 might be expected to accompany higher baseline values, but in fact the opposite is observed. In addition, both antibodies produce similar levels of measurement noise based on signal emanating from autosomes (SDC-3 ratio 0.004 ± 0.352; DPY-27 ratio 0.019 ± 0.355). Therefore, the data support a hypothesis in which complexes at foci are of distinct composition relative to those that spread from the foci.
Fine-tuning an entire chromosome: comparison to the Drosophila DCC
While the C. elegans DCC acts to down-regulate each X by a factor of two in XX animals, the Drosophila DCC performs a nearly perfectly reciprocal function by up-regulating transcription from the single X of male flies by a factor of two to match the output from XX females1,2. Our data reveals many interesting parallels and contrasts with the localization and mechanism of the Drosophila DCC.
Drosophila dosage compensation is thought to be achieved by modulating transcriptional elongation, based on the localization of the Drosophila DCC to the 3’ end of coding regions of genes and the observation that X chromosomes do not recruit more polymerase than autosomes3-5. In contrast, the localization of the C. elegans DCC to the 5′ region of genes suggests a mechanism of repression that acts at transcriptional initiation. In both systems, it is clear that DNA sequence alone does not fully determine the distribution of the DCC. However, the identity of the C. elegans DCC immediately suggests models for its recruitment and spreading. It contains novel proteins (SDC-2 and the zinc-finger proteins SDC-1 and SDC-3) that may recognize the sequence motif we identify here; a protein homologous to a member of the yeast Set1 complex (DPY-30) that may be involved in directing the DCC to the 5′ region of actively transcribed genes, and proteins homologous to members of the condensin complexes required for mitotic chromosome condensation (DPY-26, DPY-27, DPY-28, and MIX-1) that may effect dosage compensation by their enrichment at sites of transcriptional initiation. This “tuning” of the level of the DCC for different levels of gene expression again presents a more refined picture of DCC action than previous models that suggested a more uniform global condensation or “overwinding” over the entire X6. Finally, a direct mechanism for spreading is afforded by studies on the homologous cohesin and condensin complexes in yeast, which are “pushed” along genes by RNA polymerase and DNA polymerase respectively7-9. This may explain the general “baseline” X-association we observe. In contrast, the Drosophila DCC binds to approximately 25% of the X, without a baseline level of binding.
REFERENCES
1. Hamada, F.N., Park, P.J., Gordadze, P.R. & Kuroda, M.I. Global regulation of X chromosomal genes by the MSL complex in Drosophila melanogaster. Genes Dev 19, 2289-94 (2005).
2. Nusinow, D.A. & Panning, B. Recognition and modification of seX chromosomes. Curr Opin Genet Dev 15, 206-13 (2005).
3. Buscaino, A., Legube, G. & Akhtar, A. X-chromosome targeting and dosage compensation are mediated by distinct domains in MSL-3. EMBO Rep 7, 531-8 (2006).
4. Alekseyenko, A.A., Larschan, E., Lai, W.R., Park, P.J. & Kuroda, M.I. High-resolution ChIP-chip analysis reveals that the Drosophila MSL complex selectively identifies active genes on the male X chromosome. Genes Dev 20, 848-57 (2006).
5. Gilfillan, G.D. et al. Chromosome-wide gene-specific targeting of the Drosophila dosage compensation complex. Genes Dev 20, 858-70 (2006).
6. Lieb, J.D., Albrecht, M.R., Chuang, P.T. & Meyer, B.J. MIX-1: an essential component of the C. elegans mitotic machinery executes X chromosome dosage compensation. Cell 92, 265-77 (1998).
7. Wang, B.D., Eyre, D., Basrai, M., Lichten, M. & Strunnikov, A. Condensin binding at distinct and specific chromosomal sites in the Saccharomyces cerevisiae genome. Mol Cell Biol 25, 7216-25 (2005).
8. Lengronne, A. et al. Cohesin relocation from sites of chromosomal loading to places of convergent transcription. Nature 430, 573-8 (2004).
9. Glynn, E.F. et al. Genome-wide mapping of the cohesin complex in the yeast Saccharomyces cerevisiae. PLoS Biol 2, E259 (2004).
SUPPLEMENTARY METHODS
Primers for locus-specific PCR (Figure 1F)
rex-1: GCATCAACAAGCCGCAATGC and TTGCTTGTACGCACATATAC
B0302.2: CTCGCTTTGTGACACCTGTA and GCACGCCACGACCTTGCATT
R11.4: CACGTTGAACGATAGGCAGA and AACAAGATCATCCCACTTGTC
K07G5.3: GTTGATTTCGGAAGACCTCATAC and ATCTCATAGCAATAAAGCATTTAC
Reference: TCGTCCTCGACTCTGGAGAT and GTGATCTTACTGATTACCTC
Samples were separated by agarose gel electrophoresis and band intensities were measured by Kodak ID 3.6 software. Fold enrichment was calculated by the following formula: (ChIP locus/ChIP reference)/(Input locus/ Input reference). All values are sums of pixel intensities.
LM-PCR
LM-PCR was performed as in10.
Data Normalization and Visualization
The ratio of intensity from sample/reference channel was transformed to log2 space. Data for each IP was normalized so that the median of the autosomal spots was 0. z scores were obtained by standardizing the data for each IP with the mean and standard deviation of the autosomal spots. In this context, the autosomal probes were assumed to be representative of technical variation with regard to both the ChIP procedure and microarray measurements. We calculated the z scores of each data point in the three replicates and took the median of these for most analyses.
The significance of autosome versus X chromosome distribution was determined by analysis of variance (one way ANOVA of data points classified by being on an autosome or the X chromosome) with R Commander11.
Data was visualized with the UCSC Genome Browser (C. elegans March 2004 http://genome.ucsc.edu). All data is based on WS120 build (March 2004) of the C. elegans genome.
Signal Processing
Since binding for both SDC-3 and DPY-27 was limited almost exclusively to X, to better account for the X-specific variability we analyzed X in isolation for peak detection. We median-centered the z scores of X-chromosome probes. For both SDC-3 and DPY-27, four sets of peaks (one from each of the three biological replicates and a fourth for the median z–score) were called with ChIPOTle12. We removed any peaks from the median z-score set that were not present in at least 2 out of the 3 replicates (250 bp) or that were found in the no-antibody dataset. Sites of enrichment for autosomes were found with ChIPOTle using the median z scores, a 1 kb window, and 86 bp step size.
Analysis of DCC binding peaks on X
For SDC-3 and DPY-27 foci comparisons, foci were considered to be near coincident when their coordinates overlapped. To assess the significance of these relationships, we iteratively sampled a new set of SDC-3 or DPY-27 peaks from X, keeping both the number and widths of the peaks the same. The position of the new peaks on X, were selected such that they were equally likely to occur across the entire chromosome. The overlap of these newly selected peak with the observed peaks was calculated. This process was repeated 200 times. Using the mean and standard deviation of the resulting distribution of overlaps, we calculated the z-score for the proportion of observed peak overlaps and obtained a p-value from the standard normal distribution.
To assess to extent to which peaks were uniformly distributed versus clustered on the X we divided the chromosome into 12 windows and counted the number of peaks in each. We performed a chi-squared test using the counts for uniformly distributed peaks as our expected value.
To determine the extent to which peaks were clustered around the binding foci we calculated the distance from the middle of a peak to the middle of the nearest focus. Then, using a analysis similar to that described above, we sampled a new set of peaks from X with the position of the foci fixed. We repeated this 100 times, each time recording the distance of each peak to the nearest focus. To assess the significance of this relationship, we calculated the probability that the observed distances between foci and peaks were smaller than what would be expected if peaks were uniformly distributed between foci13:
Where τ is the difference between the observed and expected distances and n is the number of peaks.
Annotation of DCC binding peaks on X
We annotated each peak to a feature by determining where the maximum value probe within each peak falls in the genome. Translation start and stop coordinates were derived from the WS120 (March 2004) pre-built MySql database obtained from http://www.wormbase.org. Probe coordinates were assigned as overlapping annotated CDS sequences or the intergenic space between them. We performed chi-squared tests by calculating expected values based on the proportion of probes on the array that were annotated to the feature of interest.
Receiver Operating Characteristic (ROC) curves and motif analysis
To generate Motif A and Motif B position weight matrices, we used the sequences provided by Supplementary Table 1 of McDonel et al 14 as input for MDscan15. This procedure generated a matrix that accurately reflected the reported motifs. We divided the genome in 12,000 bp segments for input to MatrixScan, which assessed the presence and strength of the given sequence matrix and produced a motif score for each genomic segment. We designated segments that overlapped with coordinates of SDC-3 foci to be “true positives targets”, used these to create the ROC curves in Supplementary Figure 5. To test for additive effects of motifs, we simply summed the motif scores of interest for each segment.