Abstract
EH is a recently identified protein–protein interaction domain found in the signal transducers Eps15 and Eps15R and several other proteins of yeast nematode. We show that EH domains from Eps15 and Eps15R bind in vitro to peptides containing an asparagine–proline–phenylalanine (NPF) motif. Direct screening of expression libraries with EH domains yielded a number of putative EH interactors, all of which possessed NPF motifs that were shown to be responsible for the interaction. Among these interactors were the human homolog of NUMB, a developmentally reguated gene of Drosophila, and RAB, the cellular cofactor of the HIV REV protein. We demonstrated coimmunoprecipitation of Eps15 with NUMB and RAB. Finally, in vitro binding of NPF-containing peptides to cellular proteins and EST database screening established the existence of a family of EH-containing proteins in mammals. Based on the characteristics of EH-containing and EH-binding proteins, we propose that EH domains are involved in processes connected with the transport and sorting of molecules within the cell.
Keywords: EH domain, protein interaction, Eps15, internalization, trafficking
Many cellular functions, including proliferation, differentiation, cytoskeleton organization, and apoptosis, are regulated through a complex intracellular network of signal transducers. Specialized protein domains, such as SH2, SH3, PTB, or WW, mediate this vast array of interactions by binding to specific sequences located in target proteins (for review, see Musacchio et al. 1994; Cohen et al. 1995; van der Geer and Pawson 1995 and references therein). Within individual groups, binding domains have diverged to recognize specific consensus sequences in which some positions are obligatory occupied by given amino acids and other can vary. For example the SH2 and PTB domains both recognize phosphotyrosine-containing motifs (Songyang et al. 1993, 1994; Blaikie et al. 1994; Kavanaugh and Williams 1994; Zhou et al. 1995). Specificity appears to be determined by variations in amino acids distal or proximal to the phosphotyrosine residue for SH2 and PTB domains, respectively (for review, see Cohen et al. 1995; van der Geer and Pawson 1995). SH3 domains, on the other hand, interact with proline-rich peptides in target proteins (Cichetti et al. 1992; Gout et al. 1993; Ren et al. 1993; Yu et al. 1994). Screening of biased random libraries allowed the identification of two classes of SH3 ligands, conforming to the general structure RXLPPZP (class 1) and XPPLPXR (class 2), whereas X is any amino acid other than C and Z is either L or R (Feng et al. 1994; Schlessinger 1994).
SH2 and SH3 domains contribute to the propagation of signals by affecting the subcellular localization of the domain-containing proteins or their targets, or by recruting these proteins to specific enzymes (for review, see Cohen et al. 1995). Recent evidence, however, suggests that these domains might not serve solely as passive binding interfaces. In the case of SH3s, in particular, it has been shown that binding to the dynamin GTPase and to phosphatidylinositol–3′ kinase (PI–3K) activates these enzymes in vitro (Gout et al. 1993; Herskovits et al. 1993; Pleiman et al. 1994).
Finally, a phylogenetic hierarchy of interactions might also exist, as witnessed by the presence of some binding domains, such as SH3 and WW in lower eukaryotes like yeast, and the appearance of others, such as SH2 and PTB, concomitantly with the emergence of tyrosine kinase–mediated signaling in multicellular eukaryotes (for review, see Cohen et al. 1995).
We have recently described a novel protein–protein interaction domain, called EH (for Eps15 homology), present in three copies in the amino terminus of the tyrosine kinase substrates Eps15 and Eps15R and shared with other proteins of yeast and nematode (Wong et al. 1995). Except for its protein-binding ability, no other clues to the function(s) of the EH domain are available, as EH-containing proteins appear to serve different roles in cellular homeostasis. One EH-containing protein, End3p, is involved in the internalization of the yeast α-mating factor (Benedetti et al. 1994); another, Pan1p, is required for normal organization of the actin cytoskeleton in yeast (Tang and Cai 1996). Other EH-containing proteins still have unknown functions. However, mutagenesis studies in END3 suggested that the presence of its EH domain is necessary for function (Benedetti et al. 1994). Understanding of the fuctions of EH will ultimately rely on the identification of its binding specificity and of the intracellular targets recruited in a hypothetical EH-based network. Studies described in this paper were undertaken to shed light on these issues.
Results
EH domains from Eps15 and Eps15R bind to NPF-containing peptides
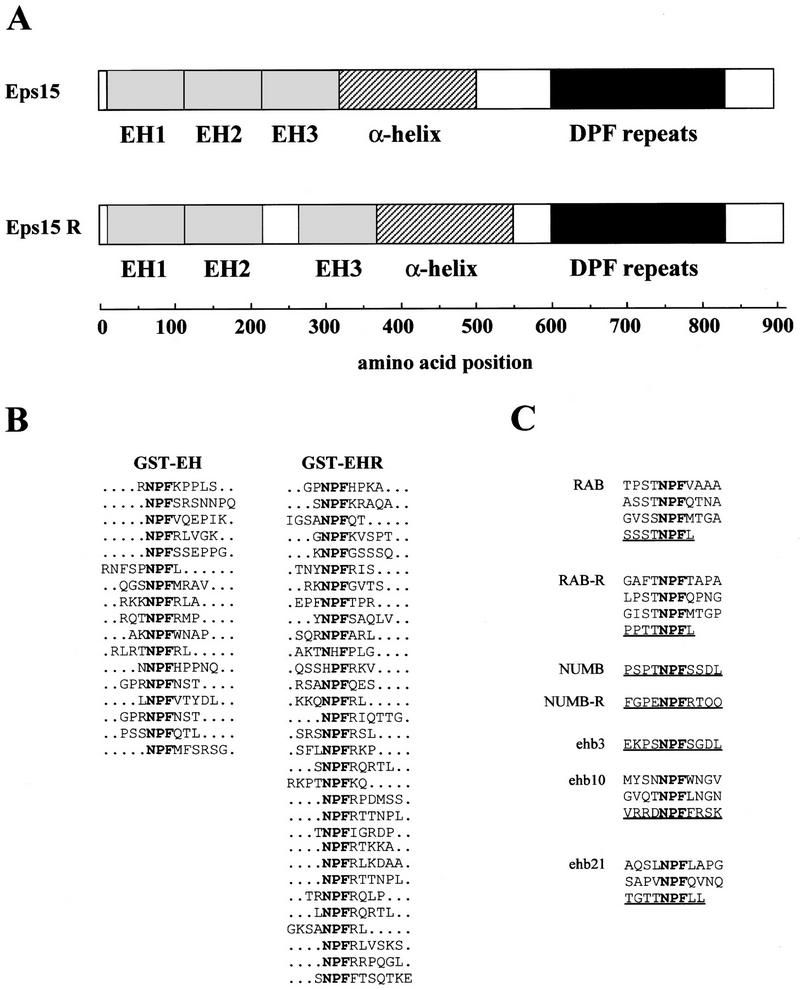
In their amino termini, Eps15 and Eps15R contain three copies of a novel protein–protein interaction domain, which we named EH (Fig. 1A). To understand the binding specificity of EH domains, we engineered glutathione S-transferase (GST) fusion proteins containing the three EH domains of Eps15 (GST–EH) or Eps15R (GST–EHR). These fusion proteins were employed to screen a random phage-displayed peptide library, and the selected phages were sequenced in the region corresponding to the random inserts. In Figure 1B, the conceptually translated peptides are displayed. Of 48 selected peptides, 46 contained the motif NPF (asparagine–proline–phenylalanine). Two additional phages containing either NHF (asparagine–histidine–phenylalanine) or HPF (histidine–proline–phenylalanine) were also selected by GST–EHR (Fig. 1B). Position +1 (with respect to NPF) exhibited a strong preference for basic and basic/hydrophobic residues in peptides selected by GST–EHR and GST–EH, respectively. Of note, negatively charged residues were never present in this position. Positions −1 and −2 displayed a weaker preference for serine or threonine (Fig. 1B).
Figure 1.
EH-mediated binding to peptides and proteins in vitro and in vivo. (A) Schematic of the Eps15 and Eps15R proteins with their EH domains. Amino acids positions are indicated. (B) Predicted amino acid sequence of peptides selected by screening of a random phage-displayed peptide library with GST–EH and GST–EHR, from Eps15 and Eps15R, respectively. NPFs are in boldface type. (C) NPF-containing motifs present in the cDNAs identified by screening of a human fibroblast expression library using GST–EH as a probe. Underlined peptides were used for the in vitro binding experiments described in Figs. 4, 6, and 8.
Cellular proteins binding to EH in vitro invariably contain NPF motifs
We have shown previously that the GST–EH fusion protein is able to specifically bind to a number of cellular proteins, in Far Western assays (Wong et al. 1995). To clone the cDNAs encoding these proteins, we employed the GST–EH fusion protein to screen a prokaryotic expression library from M426 human fibroblasts. We identified several positive plaques that specifically reacted with GST–EH but not with control GST. By cross-hybridization experiments, the positive phages could be assigned to seven groups, corresponding to distinct cDNAs (not shown).
Nucleotide sequence of the longest phage inserts of these cDNAs revealed that they represented partial cDNAs of the human homolog of NUMB, a developmentally regulated gene of Drosophila (Uemura et al. 1989); a NUMB-related gene, which we named NUMB-R; RAB, coding the cellular cofactor of the HIV REV protein (Bogerd et al. 1995; Fritz et al. 1995); and a RAB-related gene, which we named RAB-R (see next section). We also identified three novel genes that we named ehb3, ehb10, and ehb21 (for EH binding, followed by the original plaque identifier; data not shown).
There were no homologies among the seven partial cDNAs (apart from those between NUMB and NUMB-R, and RAB, and RAB-R, respectively), except for the presence of NPF motifs, which frequently are present in multiple copies (Fig. 1C). Alignment of all NPF-containing stretches revealed preference for hydrophobic residues at position +1 (relative to NPF) and for S/T at positions −1 and −2 (Fig. 1C).
Cloning of cDNAs containing entire ORFs for human NUMB, NUMB-R, RAB, and RAB-R and characterization of their predicted products
The cDNAs obtained from the screening of the prokaryotic expression library represented partial cDNAs that did not contain entire open reading frames (ORFs) (data not shown). We therefore screened a human fibroblast cDNA library to obtain the entire ORFs of human RAB, RAV-R, NUMB, and NUMB-R. Several cDNAs were isolated, and the longest ones, representative of each gene, were sequenced. A schematic of the cDNAs containing the entire ORFs of human NUMB and NUMB-R and of human RAB and RAB_R is presented in Figures 2A and 3A, respectively, and details are given in the figure legends. The sequence of human RAB corresponded to that already reported by Bogerd et al. (1995) and Fritz et al. (1995). Conceptual translation of the cDNAs allowed for comparison of human NUMB and NUMB-R (Fig. 2B) and RAB and RAB-R (Fig. 3B).
Figure 2.
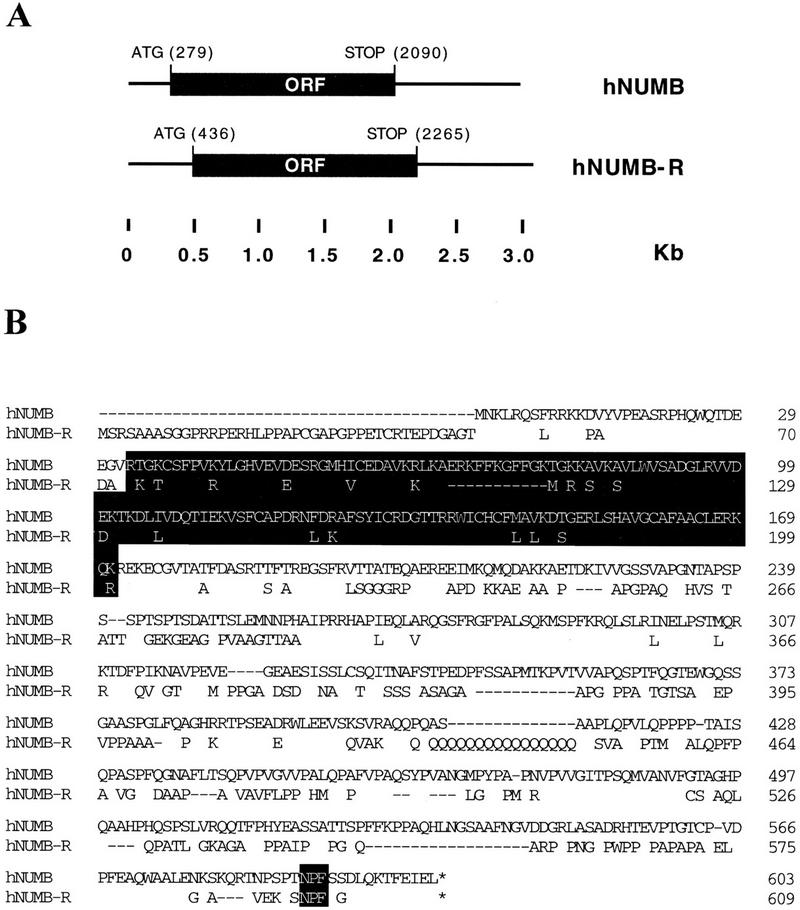
Human NUMB and NUMB-R cDNAs and proteins. (A) The structures of the human NUMB (hNUMB) and NUMB-R (hNUMB-R) cDNAs are depicted. The ORFs are labeled. Positions are indicated in kb. The nucleotide positions of initiator and terminator codons are also indicated. Canonical polyadenylation sites (AATAAA) are at positions 2649, 2823, 2958, and 3025, for hNUMB and hNUMB-R, respectively (not shown). (B) Predicted protein sequences and alignment of human NUMB and NUMB-R. In the hNUMB-R sequence, only nonidentical amino acids are reported, except for the NPF motifs. Dashes indicate gaps introduced to maximize the alignment. Accepted conservations, employed to calculate relatedness, are D, E, N, Q; L, I, V, M; K, R, H; F, Y, W; and A, G, P, S, T. The PTB domains and the NPF motifs are indicated in reverse print.
Figure 3.
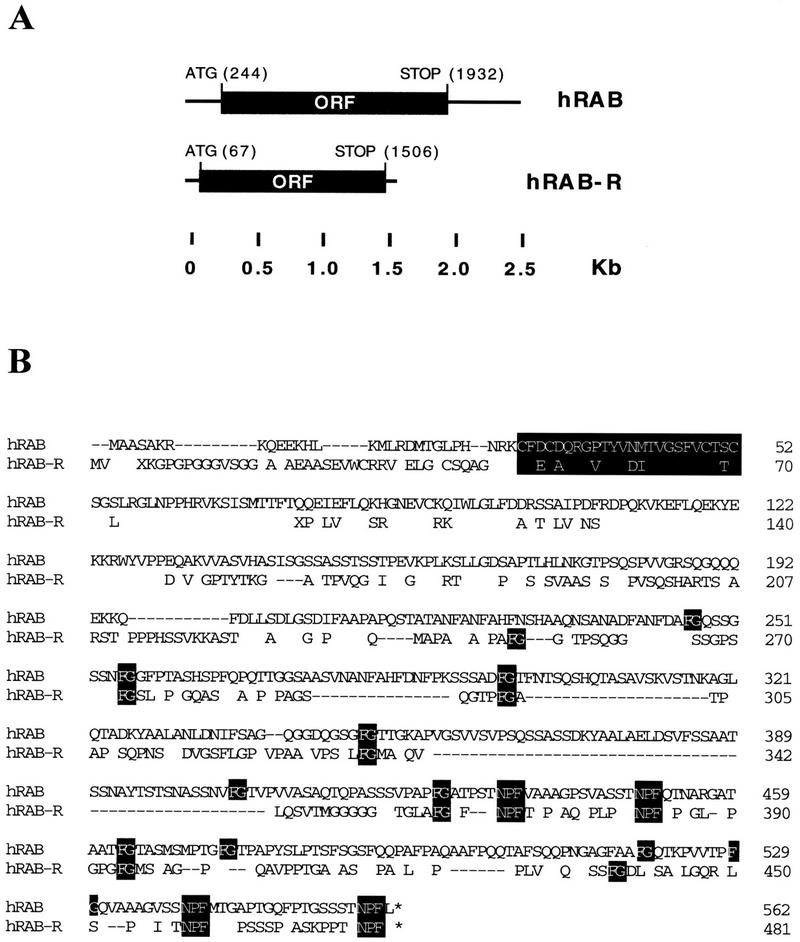
Human RAB and RAB-R cDNAs and proteins. (A) The structures of the human RAB (hRAB) and RAB-R (hRAB-R) cDNAs are depicted. The ORFs are labeled. Positions are indicated in kb. The nucleotide positions of initatior and terminator codons are also indicated. Canonical polyadenylation sites (AATAAA) are at positions 2499, 2542, and 2556 of the RAB sequence; no polyadenylation site was found in the isolated hRAB-R cDNA (not shown). (B) Predicted protein sequences and alignment of human RAB and RAB-R. The sequence of hRAB is identical to that reported by Bogerd et al. (1995) and Fritz et al. (1995). In the hRAB-R sequence, only nonidentical amino acids are reported, except for the FG and NPF motifs. Dashes indicate gaps introduced to maximize the alignment. Accepted conservation, employed to calculate relatedness, are D, E, N, Q; L, I, V, M; K, R, H; F, Y, W; and A, G, P, S, T. The FG, zinc-finger, and NPF motifs are indicated in reverse print.
The ORFs of human NUMB and NUMB-R have the capacity of encoding peptides of 603 and 609 amino acids, respectively, with a predicted molecular mass of 66 and 65 kD. The two predicted proteins display an overall relatedness of 74% with 57% identity (Fig. 2B). As already reported for murine and Drosophila (Verdi et al. 1996; Zhong et al. 1996), both human NUMB and NUMB-R contained a phosphotyrosine interaction domain (PID/PTB; van der Geer and Pawson 1995) in their amino terminus (Fig. 2B), and putative SH3-binding sites in their carboxyl terminus. In addition, they both contained a single NPF motif, located a few amino acids before the end of the proteins (Fig. 2B).
The ORFs of human RAB and RAB-R have the capacity of encoding peptides of 562 and 481 amino acids, respectively, with a predicted molecular mass of 58 and 49 kD. The two predicted proteins display an overall relatedness of 71% with 46% identity (Fig. 3B). Conserved features between RAB and RAB-R include a zinc finger region, in the amino terminus of the proteins, and several FG (phenylalanine–glycine) motifs, characteristic of nucleoporin-like proteins (Bogerd et al. 1995, Fritz et al. 1995). In addition, they both contained four NPF motifs, located in the carboxy-terminal half of the molecule (Fig. 3B). It has not been established whether all of the NPF motifs are endowed with EH-binding ability, and if so, whether they bind to different EH-containing proteins.
Binding of various cellular proteins to EH domains in vitro and in vivo
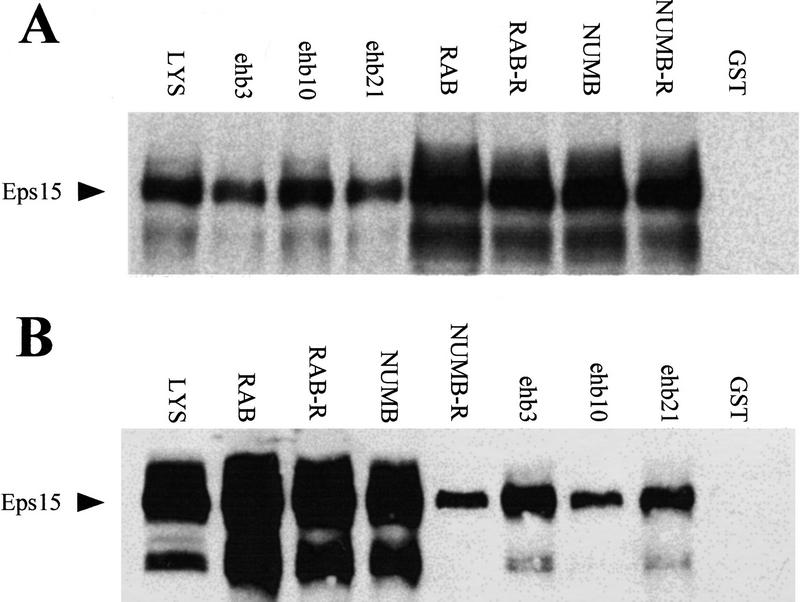
To characterize the binding of the EH interactors to Eps15, we engineered GST fusion proteins encompassing fragments derived from the seven identified EH-binding proteins. In all cases, the original partial cDNAs were used (see Materials and Methods), as they should contain all of the determinants necessary and sufficient for EH binding. As shown in Figure 4A, all of the GST fusion proteins were able to specifically recover native Eps15 from cell lysates in vitro binding experiments.
Figure 4.
In vitro binding of NPF-containing proteins and peptides to Eps15. (A) In vitro binding of Eps15 to Gst fusions of EH-binding proteins. Total cellular lysates from NIH–3T3 cells (1 mg/lane) were incubated with the indicated GST fusion proteins (10 μg) for 1 hr at 4°C. Specifically bound Eps15 was detected by immunoblot with an anti-Eps15 antibody. (B) Binding of Eps15 to GST fusions encompassing NPF-containing peptides. GST fusion proteins were engineered to encompass the NPF-containing peptides underlined in Fig. 1C and are indicated with the names of the proteins from which the peptides were derived. In vitro binding experiments were performed as described in A. Lanes marked LYS in A and B were loaded with 50 μg of total cellular proteins to serve as a reference for the position of Eps15. The position of Eps15 is also indicated.
To address the issue of whether the NPF-containing peptides, present in the EH-binding proteins, were actually responsible for binding to Eps15, we engineered GST fusion proteins encompassing short NPF-containing peptides from the proteins of interest. The peptides engineered as GST fusions are underlined in Figure 1C. Also in this case (Fig. 4B), we evidenced specific binding of the GST peptide fusions to native Eps15 from total cellular lysates. Although the participation of other regions of the EH-interacting proteins to the binding cannot be excluded, the sum of the above results indicates that NPF-containing peptides are sufficient for binding to Eps15.
We then tested whether Eps15 can physically interact with some of the EH-binding proteins in vivo. To this end, we utilized the GST–NUMB and GST–RAB fusion proteins, described in Figure 4A, as immunogens to generate polyconal sera. The anti-RAB and anti-NUMB sera recognized specifically proteins of ∼60 kD and a doublet of ∼70–72 kD, respectively, in several human and mouse cell lines (data not shown). The two sera were then used to coimmunoprecipitate Eps15 from lysates of NIH–3T3 cells (Fig. 5A) or of U2OS cells (not shown). As shown in Figure 5A, Eps15 was recovered in immunoprecipitates obtained with anti-NUMB or anti-RAB sera but not with control sera.
Figure 5.
In vivo binding of NPF-containing proteins to Eps15. (A) Coimmunoprecipitation of Eps15 with endogenously expressed NUMB and RAB. Total cellular proteins from NIH–3T3 cells were obtained under mild lysis conditions to preserve protein–protein interactions, and 5 mg of protein was immunoprecipitated with an anti-RAB (Ip RAB), an anti-NUMB (Ip NUMB), or a control serum (C). After SDS–PAGE, coimmunoprecipitating Eps15 was detected by immunoblot with an anti-Eps15 antibody. (B) Eps15 coimmunoprecipitation with HA-tagged RAB and NUMB proteins. (Top) C33A cells were transfected with expression vectors engineered to express HA-tagged RAB or NUMB proteins (HA–RAB or HA–NUMB lanes, respectively) or mock transfected (− lanes). Total cellular lysates (100 μg) were immunoblotted with an anti-HA antibody. (Bottom) Five milligrams of total cellular proteins from HA–RAB or HA–NUMB transfectants (HA–RAB Tfx and HA–NUMB Tfx, respectively) obtained as in A were immunoprecipitated with the anti-HA antibody (HA lanes) or with a control serum (C lanes) and detected by immunoblot with an anti-eps15 antibody. (C) Eps15 coimmunoprecipitation with HA-tagged NUMB and NUMB mutant. C33A cells were transfected with expression vectors encoding HA-tagged NUMB or an HA-tagged NUMB mutant in which the NPF motif was mutagenized to NAA (lanes HA–NUMB and HA–NUMB/NAA, respectively), or mock transfected (− lane). (Top) Total cellular lysates (100 μg) were immunoblotted with an anti-HA antibody; (middle) 5 mg of total cellular proteins obtained as in A was immunoprecipitated with an anti-HA antibody and detected by immunoblot with an anti-Eps15 serum; (bottom) total cellular lysates (100 μg) were immunoblotted with an anti-Eps15 antibody. Lanes marked LYS in A–C were loaded with 50 μg of total cellular proteins to serve as a reference for the position of Eps15. The positions of Eps15, NUMB, and RAB are also indicated.
The amount of Eps15 coimmunoprecipitated by NUMB was higher than that coimmunoprecipitated by RAB. Although this might be attributable to a lower affinity of the RAB/Eps15 interaction, we favor the possibility that it simply reflects less efficient immunoprecipitation of RAB by the anti-RAB antibody. In another set of experiments, we engineered eukaryotic expression vectors encoding for either NUMB or RAB fused to a hemagglutinin (HA) epitope, at their amino termini. C33A cells were then transiently transfected with vectors coding HA–NUMB or HA–RAB. As shown in Figure 5B (top) HA–NUMB and HA–RAB were expressed at similar levels. Under these conditions, Eps15 was readily and comparably recoverable by immunoprecipitation with an anti-HA antibody but not by control antibodies, in both HA–NUMB or HA–RAB transfectants (Fig. 5B, bottom). The sum of the above results strongly argues that Eps15 can interact with NUMB and RAB in vivo.
To prove that the in vivo interaction of Eps15 with a representative EH-binding protein protein was attributable to a NPF-mediated interaction, we engineered a HA-tagged mutant of NUMB (HA–NUMB/NAA) in which its unique NPF motif was mutageneized to NAA (asparagine–alanine–alanine). Upon transfection in C33A cells, both the HA–NUMB/NAA and the wild-type HA–NUMB mutants were expressed at compatable levels (Fig. 5C, top). In HA–NUMB/NAA transfectants, however, little if any Eps15 was visible in anti-HA immunoprecipitates, whereas it was readily detectable in similar immunoprecipitates obtained from wild-type HA–NUMB transfectants (Fig. 5C, middle). These differences were not attributable to the levels of expression of endogenous Eps15, which appeared comparable in all transfectants (Fig. 5C, bottom). Thus, the in vivo binding of Eps15 to NUMB is most likely mediated by an EH–NPF interaction.
Characterization of the NPF–EH interaction
To assess the relevance of the NPF motif and of the surrounding positions for binding to EH, we selected the peptide SSSTNPFL, which is present in RAB and strongly binds to Eps15 and mutated individual amino acid positions in a GST fusion protein background. The various GST fusion proteins depicted in Figure 6 (left panels) were then tested for their ability to recover native Eps15 from cellular lysates in in vitro binding assays.
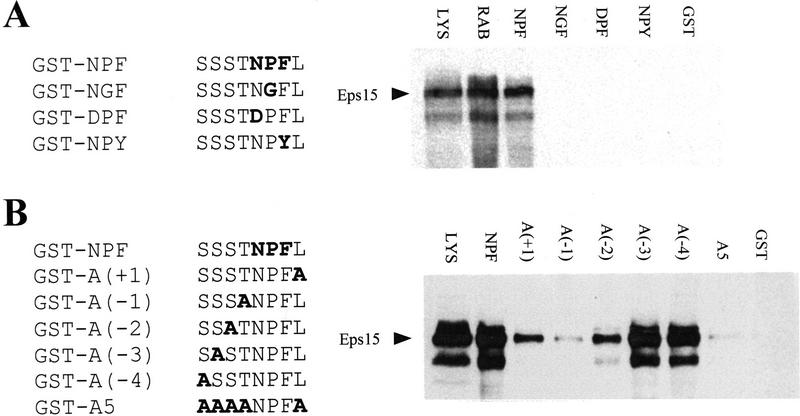
Figure 6.
Requirement for the NPF motif and surrounding positions for binding to Eps15. (A) Binding to Eps15 of mutant peptides containing mutations in the NPF sequence. Peptides engineered in GST fusion proteins are indicated on the left. GST–NPF corresponds to the sequence of a NPF-containing peptide derived from the sequence of RAB (underlined in Fig. 1C). Mutant peptides (GST–NGF, GST–DPF, GST–NPY) are also indicated. The in vitro bindings to Eps15, obtained as described in Fig. 4, are shown on the right. The lane marked RAB represents an in vitro binding obtained with the GST–RAB protein (Fig. 4A) to serve as a positive control. (B) Alanine scanning. Positions surrounding the NPF motif in the RAB peptide were alanine scanned as indicated on the left. The in vitro binding to Eps15, obtained as described in Fig. 4, are shown on the right. Lanes marked LYS were loaded with 50 μg of total cellular proteins to serve as a reference for the position of Eps15 (also indicated). Amino acids corresponding to mutagenized codons are shown in boldface type.
Mutations in the NPF motif to NGF, DPF, and NPY abolish binding completely (Fig. 6A, right). We then performed alanine scanning of the positions surrounding the NPF motif. This unconvered a hierarchy of contribution to binding, as A(−1), A(−2), and A(+1) mutants, containing mutations to alanine at positions −1, −2, and +2 respective to the NPF motif, displayed binding reduced by 95%, 60%, and 55%, respectively (Fig. 6B, right). Mutations of residues −3 and −4 to alanine did not appreciably affect binding (Fig. 6B, right). Finally a GST peptide bearing five simultaneous mutations to alanine (A5 mutant), leaving the NPF motif intact, displayed little if any detectable binding (Fig. 6B, right). These results indicate that an intact NPF is necessary but not sufficient for binding and that optimal binding is conditioned by the presence of certain amino acids in the surrounding positions. Whether this is attributable to impact on conformation of directly on binding remains to be established.
A single EH is necessary and sufficient for interaction with an EH-binding protein
Many EH-containing proteins display repeated copies of the binding module (Wong et al. 1995); this observation raises the question as to whether a single EH is endowed with binding ability or whether multiple copies are required. Our screenings of phage-displayed and bacterial expression libraries were performed with GST fusion proteins containing the three EH domains of Eps15 and Eps15R, to maximize the chance of detecting protein–protein interactions. To design a strategy to engineer GST fusions containing single EH domains, we had to take into account the fact that the EH domains of Eps15 are contiguous. Thus, definition of the exact boundaries and of the structural consequences of extrapolating sequences from their natural context was difficult to predict.
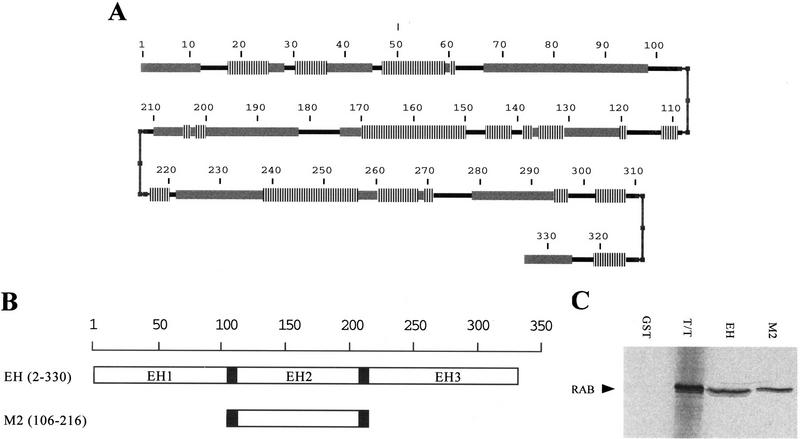
We thought, therefore, to take advantage of secondary structure predictions of the amino terminus of Eps15. As shown in Figure 7A, a Chous–Fasman–Rose algorithm predicted that the three EH domains of Eps15 are flanked by regions with high propensity for turns, possibly underlying the requirement for domain exposure, to achieve binding. Thus, we engineered a GST fusion protein containing residues 106–216 of Eps15, which encompass the second EH flanked by the predicted amino and carboxy-terminal turn (M2 protein, Fig. 7B). This protein displayed strong binding to RAB, obtained by in vitro transcription/translation (Fig. 7C). In addition, binding of GST–M2 to RAB was quantitative and of comparable intensity to that obtained with GST–EH (Fig. 7C), thus establishing that an individual EH domain is endowed with binding ability.
Figure 7.
Mapping of the minimal region of eps15 required for binding to NPF-containing proteins. (A) Secondary structure prediction of the amino-terminal region of Eps15 (containing the three EH domains) by a Chou–Fasman–Rose algorithm. (Vertical lines) α-Helices; (thick shaded bars) β-sheets; (thinner solid bars) coils; (small solid boxes) turn. Amino acid positions are also indicated. (B) Schematic representation of the Eps15 amino-terminal domain and of the GST fusion proteins engineered, with predicted turns indicated by solid boxes. The indicated fragments of Eps15 were engineered into GST fusion proteins and used for in vitro binding experiments. The EH construct contains all three EH domains. The M2 construct contains the region encompassing the second EH domain flanked by the natural regions pedicting the turns shown in A. Amino acid positions are also indicated in parentheses. (C) In vitro bindings. The GST fusions shown in B were used to bind the 35S-labeled RAB protein, obtained by in vitro transcription/translation of the RAB cDNA. Detection was by autoradiography. The lane marked T/T was loaded with the primary product of the in vitro transcription/translation to serve as a reference. The positions of RAB are also indicated.
A family of EH-containing proteins in mammals
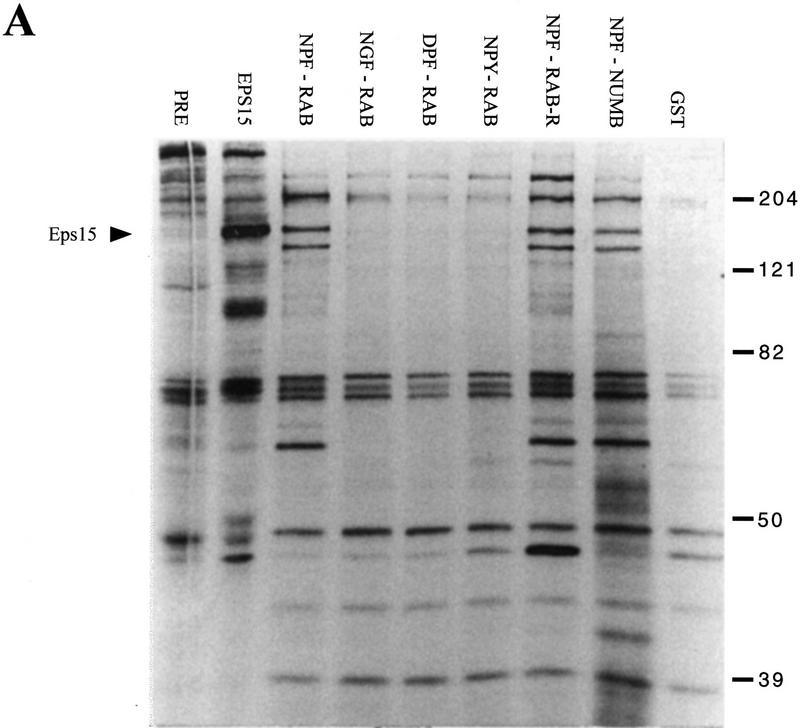
Eps15 and Eps15R are the only EH-containing proteins known in mammals. The identification of peptide sequences that bind to EH allowed for the identification of putative EH-containing proteins in mammalian cells. GST fusion proteins, encompassing NPF-containing peptides from NUMB, RAB, and RAB-R were challenged with 35S-labeled lysates from NIH–3T3 cells, and several cellular proteins were specifically recovered (Fig. 8A). In the case of the RAB peptide, mutations of NPF to DPF, NGF, or NPY totally abolished recognition, arguing strongly that the identified proteins represent EH-containing species (Fig. 8A). In addition, individual NPF-containing peptides were able to bind to both common and different proteins, thus indicating a certain degree of specificity for EH-mediated interactions.
Figure 8.
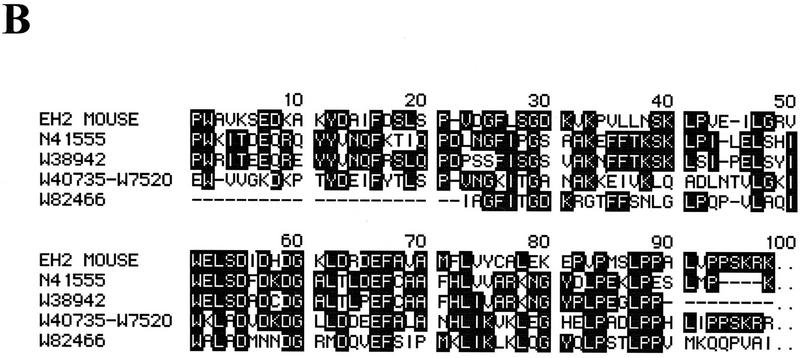
Identification of a family of EH-containing proteins in mammals. (A) 35S-Labeled lysates from NIH–3T3 (50 × 106 TCA-precipitable counts) were incubated with the indicated GST fusion proteins displaying NPF-containing peptides (NPF–RAB, NPF–RAB-R, and NPF–NUMB, underlined in Fig. 1C), mutant peptides (NGR–RAB, DPF–RAB, and NPY–RAB, shown in Fig. 6A), or with GST. The first two lanes represent the same lysate immunoprecipitated with anti-eps15 antibody (EPS15) or with a control serum (PRE). Specifically bound proteins were fractionated by SDS–PAGE and visualized by autoradiography. The position of Eps15 is indicated. Molecular mass markers are indicated in kD. (B) Identification of four novel EH-containing mammalian sequences. The EST db was screened with the TBLASTN algorithm, using the three EH domains of Eps15 as a query. Only sequences displaying a P > 10−5 were considered for further analysis, and they are indicated with their database accession numbers. The alignment of these sequences to the second EH domain of Eps15 is shown, as obtained by a CLUSTAL4 algorithm modified by visual inspection. The W40735–W7520 sequence derives from a merging of two shorter EST sequences of the same gene.
The existence of a family of EH-containing protein in mammals was confirmed by screening the Expressed Sequence Tags Database (dbEST) with the EH domains of Eps15 and Eps15R. Four novel mammalian sequences, two from Homo sapiens and two from Mus musculus, were identified that contained EH domains (Fig. 8B).
Discussion
Data presented in this study establish the existence of an expanding family of EH-containing proteins and of a network of EH-mediated interactions. Furthermore, they define the molecular basis of EH-mediated interactions of two members of this family, Eps15 and Eps15R, and their binding to NPF-containing peptides. We do not know whether other EH domains will display similar binding preferences. By analogy to other binding modules, it is conceivable that critical binding residues will be invariant or conserved. However, a certain degree of variation is expected, as EH domains are only ∼40% homologous. SH3 domains, by analogy, are 30% homologous, and their binding preferences vary (Sparks et al. 1996).
With regard to Eps15, several proteins are known to date that interact with it, including Crk, through its SH3 domain (Schumacher et al. 1995), the coated-pits adapter AP-2 (Benmerah et al. 1995, 1996; Iannolo et al. 1997), and the EH-binding proteins described in this paper. Thus, Eps15 is a multibinding protein that might function in the assembling of macromolecular complexes. We do not know whether all of the Eps15 targets interact simultaneously with it or whether a hierarchy of interaction exists in vivo.
A wealth of evidence now supports the notion that EH domains are involved in receptor-mediated internalization. The first indication derived from the identification of EH domains in End3p, a yeast protein essential for internalization of the α-mating factor and of its receptor Ste2p (Burkholder and Hartwell 1985; Nakayama et al. 1985; Benedetti et al. 1994; Wong et al. 1995). Another EH-containing yeast protein, Pan1p, also appears to regulate endocytosis (Wendland et al. 1996). Implication of eps15 in this pathway derived from the discovery of its constitutive association with the clathrin adaptor protein complex AP-2 (Benmerah et al. 1995). The related Eps15R protein is also associated in vivo to AP-2 (Iannolo et al. 1997; A. Salcini, P.G. Pelicci, and P. diFiore, unpubl.). Further characterization revealed that the carboxyl termini of Eps15 and Eps15R and the α-subunit of AP-2 are involved in the interaction (Benmerah et al. 1996; Iannolo et al. 1997). In addition, immunomorphological analysis demostrated colocalization of Eps15 and Eps15R with markers of the plasma membrane clathrin-coated pits and vesicles (Tebar et al. 1996; A. Salcini, P.G. Pelicci, and P. diFiore, unpubl.).
Our present results might provide a molecular basis for the understanding of the protein–protein interactions involved in receptor-mediated internalization, especially if viewed in the light of recent findings by Tan et al. (1996). This group demonstrated that the sequence NPFXD constitutes an internalization signal in Saccharomyces cerevisiae. In particular, they showed that NPF-based sequences in a Ste2p/Kex2p chimera and in Ste3p, the receptor for the a-mating factor (Nakayama et al. 1985), are essential for endocytosis (Tan et al. 1996). Thus, the possibility that receptor-mediated endocytosis is regulated by EH–NPF interactions appears to be more than just a hypothesis.
The identification of an EH-mediated network of interactions sheds additional light on the function of the EH domain, in that EH-containing and EH-binding proteins are involved in more functions than internalization. Pan1p, for instance, is required in yeast to maintain proper organization of the actin cytoskeleton (Tang and Cai 1996). NUMB, identified in this study as an EH-binding protein, is involved in cell fate determination through asymmetrical distribution as mitosis (Uemura et al. 1989; Rhyu et al. 1994; Spana et al. 1995), a function that requires precise sorting within the dividing cell. Thus, the EH-based network appears to be involved in processes connected with sorting and organization of molecules within the cell.
This contention is further strengthened by the identification of additional candidate EH-binding proteins. We searched databases for proteins containing multiple NPFs, as a characteristic of many of the EH-binding proteins identified in this study is the presence of multiple NPF-containing binding motifs, a feature that mirrors the frequent presence of EH domains in multiple copies. This search yielded proteins such as synaptojanin (three NPFs), which participates in synaptic vesicle recycling (McPherson et al.1996); SCAMP 37 (three NPFs), which is part of a family of molecules that are components of membranes functioning in cell surface recycling (Brand and Castle 1993); and the Drosophila Dorsal protein (three NPFs), which establishes a nuclear vemtral-to-dorsal gradient that is altered or absent in dorsalized or ventralized embryos, thus probably determining cell fate along the dorsal–ventral axis (Steward 1987). Other proteins containing multiple NPFs include the Drosophila Suppressor of sable protein (Voelker et al. 1991), the mammalian mitogen-responsive proteins p97, p93, and p67 (Xu et al. 1995), and the Stn-b protein of the Drosophila Stoned locus (GenBank accession no. U54982). Although the EH-binding properties of these proteins will have to be validated experimentally, the hypothesis that the EH network is implicated in processes connected with transport, protein sorting, and organization of subcellular structures clearly appears worthy of further investigation.
Binding of Eps15 to NPF-containing targets did not appear to require tyrosine phosphorylation of the former, in that it was readily observed in in vitro bindings performed on serum-starved cell lysates, in which Eps15 is not phosphorylated. In addition, stoichiometry of binding was not improved under conditions in which a sizable fraction of Eps15 was tyrosine phosphorylated, as achieved in lysates from epidermal growth factor (EGF)-stimulated cells (data not shown). None of the Eps15-binding abilities known so far, including binding to Crk and AP-2, appears to be phosphotyrosine-dependent (Benmerah et al. 1995, 1996; Schumacher et al. 1995; Iannolo et al. 1997). This raises the question of how the functional coupling of Eps15 to receptor tyrosine kinase (RTK)-activated pathways (Fazioli et al 1993; Wong et al. 1994) influences its downstream function(s), with particular regard to its EH-mediated interactions. One attractive possibility is that a constitutive Eps15/target complex, such as Eps15/NUMB or Eps15/RAB, can be directed to a proper subcellular location by tyrosine phosphorylation. In this framework, tyrosine phosphorylation would act as a molecular switch through which RTKs can condition the targeting of components of the EH network to their active site within the cell.
Materials and methods
Screening of phage-display libraries
The peptide library utilized and the panning conditions were essentially as described by Felici et al. (1991), the main difference being that the target EH domains (3 μg) were immobilized by binding to 20 μl of GST–Sepharose matrix (Pharmacia). The peptide library was screened with either the GST–EH or the GST–EHR fusion protein, designed to encompass amino acid positions 2–330 and 15–368 of mouse Eps15 and Eps15R, respectively. Two panning cycles were carried out for each domain before the sequencing of the clones, which were enriched during the selection procedure.
Isolation of cDNAs encoding EH-binding proteins
A GST fusion protein containing the three EH domains of Eps15 (GST–EH; Wong et al. 1995) was used to screen a pCEV–LAC-based prokaryotic expression library (a kind gift of T. Miki, National Institutes of Health, Bethesda, MD) from M426 human fibroblasts. Recombinant plaques (5 × 106) were screened, after induction with IPTG, using a modification for the Far Western assay developed to identify proteins interacting with GST fusions (Matoskova et al. 1996a,b). Briefly, filters were blocked in 2% bovine serum albumin (BSA) in TTBS (20 mm Tris-HCl at pH 7.5, 150 mm NaCl, 0.05% Tween 20) for at least 2 hr at room temperature and then in reduced glutathione (Sigma, 3 μm) in TTBS with 0.5% (wt/vol) BSA for 1 hr at room temperature. This latter step substantially reduced background (not shown). Blots were then incubated with GST–EH (10 nm) in TTBS in the presence of reduced glutathione (3 μm) and BSA (0.5% wt/vol) for 1 hr at room temperature. After extensive washing in TTBS, blots were detected with an affinity-purified anti-GST antibody, as described (Matoskova et al. 1996a,b).
Positive phages that specifically reacted with GST–EH but not with control GST were subjected to cross-hybridization experiments and assigned to seven groups, corresponding to distinct cDNAs (not shown). For each group, phages containing the longest inserts were sequenced by the dideoxy-termination method on both strands of the cDNAs, using a commercial kit (Sequenase).
To obtain cDNA clones containing the entire ORFs of human NUMB, NUMB-R, RAB, and RAB-R, a pCEV-29-based eukaryotic expression library from M426 human fibroblasts (a kind gift of T. Miki) were screened using standard techniques. Screening of the GenBank, EMBL, and EST databases was performed by the BLAST program (Altschul et al. 1990). Alignments of peptide sequences were performed by a CLUSTAL4 algorithm on MacDNASIS software.
Production of recombinant proteins
GST fusion proteins containing large fragments of the proteins of interest were obtained by recombinant PCR of the appropriate fragment from the murine Eps15 and Eps15R cDNAs or from the cDNAs of the EH-binding proteins, followed by cloning in the pGEX expression vector, in-frame with the GST moiety.
GST fusions containing short peptides from the proteins of interest were obtained by annealing in vitro complementary oligonucleotides with the appropriate sequence followed by cloning in-frame with the GST moiety in a gGEX-KT vector. Sequences of the oligonucleotides and primers used are available on request. Purification of the GST fusion protein onto agarose-glutathione and in vitro binding experiments were performed as described (Wong et al. 1995; Matoskova et al. 1996a,b).
Protein studies
Expression vectors for HA-tagged RAB and NUMB proteins were engineered in the pMT2 eukaryotic expression vector by inserting (by insertional overlapping PCR) the sequence encoding the HA epitope (YDVPDYASLP) between codons 1 and 2 of the ORF of the RAB and NUMB cDNAs to obtain pMT2–HA–RAB and pMT2–HA–NUMB, respectively. The HA–NUMB/NAA mutant was generated by changing the sequence coding for the unique NPF motif to a sequence coding for NAA by PCR-based oligonucleotide-directed mutagenesis (Ho et al. 1989). Transient transfection of C33A cells by calcium phosphate was performed as described previously for NIH–3T3 cells (Fazioli et al. 1993). Immunoprecipitation and immunoblotting experiments were performed as described previously (Fazioli et al. 1993; Matoskova et al. 1996a). Typically, we employed 50–100 μg of total cellular proteins for direct immunoblot analysis and 3–5 mg of total cellular proteins for immunoprecipitation/immunoblotting experiments. For coimmunopreciptiation experiments total cellular proteins were obtained in mild lysis conditions, in the absence of ionic detergents, to preserve protein–protein interactions, as described (Fazioli et al. 1993).
[35S]methionine-labeled RAB protein, employed in the experiments in Figure 7, was synthesized by in vitro transcription–translation using a commercial kit (Promega) and the full-length RAB cDNA. Metabolic labeling with [35S]methionine of NIH–3T3 cells was performed as described previously (Fazioli et al. 1993).
Acknowledgments
This work was supported in part by grants from the Associazione Italiana Ricerca sul Cancro to P.P.D.F., P.G.P, and G.C. and from the Consiglio Nazionale delle Ricerche to P.P.D.F. and P.G.P. The support of the European Community (BIOMED-2 Programme) to P.P.D.F. is also acknowledged. A.E.S. and E.S. are recipients of fellowships from the Fondazione Italiana Ricerca sul Cancro. M.D. is the recipient of a fellowship from the Fondazione Vollaro.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL pdifiore@ieo.cilea.it; FAX 39-2-57489851.
References
- Alschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Benedetti H, Raths S, Crausaz F, Riezman H. The END3 gene encodes a protein that is required for the internalization step of endocytosis and for actin cytoskeleton organization in yeast. Mol Biol Cell. 1994;5:1023–1037. doi: 10.1091/mbc.5.9.1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benmerah A, Gagnon J, Begue B, Megarbane B, Dautry-Varsat A, Cerf-Bensussan N. The tyrosine kinase substrate eps15 is constitutively associated with the plasma membrane adaptor AP-2. J Cell Biol. 1995;131:1831–1838. doi: 10.1083/jcb.131.6.1831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benmerah A, Begue B, Dautry-Varsat A, Cerf-Bensussan N. The ear of α-Adaptin interacts with the COOH-terminal domain of the Eps15 protein. J Biol Chem. 1996;271:12111–12116. doi: 10.1074/jbc.271.20.12111. [DOI] [PubMed] [Google Scholar]
- Blaikie P, Immanuel D, Wu J, Li N, Yainik V, Margolis B. A region in shc distinct from the SH2 domain can bind tyrosine-phosphorylated growth factor receptors. J Biol Chem. 1994;269:32031–32034. [PubMed] [Google Scholar]
- Bogerd HP, Fridell RA, Madore S, Cullen BR. Identification of a novel cellular cofactor for the Rev/Rex class of retroviral regulatory proteins. Cell. 1995;82:485–494. doi: 10.1016/0092-8674(95)90437-9. [DOI] [PubMed] [Google Scholar]
- Brand SH, Castle JD. SCAMP 37, a new marker within the general cell surface recycling system. EMBO J. 1993;12:3753–3761. doi: 10.1002/j.1460-2075.1993.tb06053.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burkholder AC, Hartwell LH. The yeast alpha-factor receptor: Structural properties deduced from the sequence of the STE2 gene. Nucleic Acids Res. 1985;13:8463–8475. doi: 10.1093/nar/13.23.8463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cicchetti P, Mayer BJ, Thiel G, Baltimore D. Identification of a protein that binds to the SH3 region of Abl and is similar to Bcr and GAP-rho. Science. 1992;257:803–806. doi: 10.1126/science.1379745. [DOI] [PubMed] [Google Scholar]
- Cohen GB, Ren R, Baltimore D. Modular binding domains in signal transduction proteins. Cell. 1995;80:237–248. doi: 10.1016/0092-8674(95)90406-9. [DOI] [PubMed] [Google Scholar]
- Fazioli F, Minichiello L, Matoskova B, Wong WT, Di Fiore PP. Eps15, a novel tyrosine kinase substrate, exhibits transforming activity. Mol Cell Biol. 1993;13:5814–5828. doi: 10.1128/mcb.13.9.5814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felici F, Castagnoli L, Musacchio A, Jappelli R, Cesareni G. Selection of antibody ligands from a large library of oligopeptides expressed on a multivalent exposition vector. J Mol Biol. 1991;222:301–310. doi: 10.1016/0022-2836(91)90213-p. [DOI] [PubMed] [Google Scholar]
- Feng S, Chen JK, Yu H, Simon JA, Schreiber SL. Two binding orientations for peptides to the Src SH3 domain: Development of a general model for SH3-ligand interactions. Science. 1994;266:1241–1247. doi: 10.1126/science.7526465. [DOI] [PubMed] [Google Scholar]
- Fritz CC, Zapp ML, Green MR. A human nucleoporin-like protein that specifically interacts with HIV Rev. Nature. 1995;376:530–533. doi: 10.1038/376530a0. [DOI] [PubMed] [Google Scholar]
- Gout I, Dhand R, Hiles ID, Fry MJ, Panayotou G, Das P, Truong O, Totty NF, Hsuan J, Booker GW. The GTPase dynamin binds to and is activated by a subset of SH3 domains. Cell. 1993;75:25–36. [PubMed] [Google Scholar]
- Herskovits JS, Shpetner HS, Burgess CC, Vallee RB. Microtubules and Src homology 3 domains stimulate the dynamin GTPase via its C-terminal domain. Proc Natl Acad Sci. 1993;90:11468–11472. doi: 10.1073/pnas.90.24.11468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho SN, Hunt HD, Horton RM, Pullen JK, Pease LR. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene. 1989;77:51–59. doi: 10.1016/0378-1119(89)90358-2. [DOI] [PubMed] [Google Scholar]
- Iannolo G, Salcini AE, Gaidarov I, Goodman Jr OB, Pelicci PG, Di Fiore PP, Keen JH. Mapping of the molecular determinants involved in the interaction between eps15 and AP-2. Cancer Res. 1997;57:240–245. [PubMed] [Google Scholar]
- Kavanaugh WM, Williams LT. An alternative to SH2 domains for binding tyrosine phosphorylated proteins. Science. 1994;266:1862–1865. doi: 10.1126/science.7527937. [DOI] [PubMed] [Google Scholar]
- Matoskova B, Wong WT, Seki N, Nagase T, Nomura N, Robbins KC, Di Fiore PP. RN-tre identifies a family of tre-related proteins displaying a novel potential protein binding domain. Oncogene. 1996a;12:2563–2571. [PubMed] [Google Scholar]
- Matoskova B, Wong WT, Nomura N, Robbins KC, Di Fiore PP. RN-tre specifically binds to the SH3 domain of eps8 with high affinity and confers growth advantage to NIH3T3 upon carboxy-terminal truncation. Oncogene. 1996b;12:2679–2688. [PubMed] [Google Scholar]
- McPherson PS, Garcia EP, Slepnev VI, David C, Zhang X, Grabs D, Sossin WS, Bauerfeind R, Nemoto Y, De Camilli P. A presynaptic inositol-phosphatase. Nature. 1996;379:353–357. doi: 10.1038/379353a0. [DOI] [PubMed] [Google Scholar]
- Musacchio A, Wilmanns M, Saraste M. Structure and function of the SH3 domain. Prog Biophys Mol Biol. 1994;61:283–297. doi: 10.1016/0079-6107(94)90003-5. [DOI] [PubMed] [Google Scholar]
- Nakayama N, Miyajima A, Arai K. Nucleotide sequences of STE2 and STE3, cell type-specific sterile genes from Saccharomyces cerevisiae. EMBO J. 1985;4:2643–2648. doi: 10.1002/j.1460-2075.1985.tb03982.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pleiman CM, Hertz WM, Cambier JC. Activation of phosphatidylinositol-3′ kinase by Src-family kinase SH3 binding to the p85 subunit. Science. 1994;263:1609–1612. doi: 10.1126/science.8128248. [DOI] [PubMed] [Google Scholar]
- Ren R, Mayer BJ, Cicchetti P, Baltimore D. Identification of a ten-amino acid proline-rich SH3 binding site. Science. 1993;259:1157–1161. doi: 10.1126/science.8438166. [DOI] [PubMed] [Google Scholar]
- Rhyu MS, Jan LY, Jan YN. Asymmetric distribution of numb protein during division of the sensory organ precursor cell confers distinct fates to daughter cells. Cell. 1994;76:477–491. doi: 10.1016/0092-8674(94)90112-0. [DOI] [PubMed] [Google Scholar]
- Schlessinger J. SH2/SH3 signaling proteins. Curr Opin Genet Dev. 1994;4:25–30. doi: 10.1016/0959-437x(94)90087-6. [DOI] [PubMed] [Google Scholar]
- Schumacher C, Knudsen BS, Ohuchi T, Di Fiore PP, Glassman RH, Hanafusa H. The SH3 domain of Crk binds specifically to a conserved proline-rich motif in Eps15 and Eps15R. J Biol Chem. 1995;270:15341–15347. doi: 10.1074/jbc.270.25.15341. [DOI] [PubMed] [Google Scholar]
- Songyang Z, Shoelson SE, Chaudhuri M, Gish G, Pawson T, Haser WG, King F, Roberts T, Ratnofsky S, Lechleider RJ. SH2 domains recognize specific phosphopeptide sequences. Cell. 1993;72:767–778. doi: 10.1016/0092-8674(93)90404-e. [DOI] [PubMed] [Google Scholar]
- Songyang Z, Shoelson SE, McGlade J, Oliver P, Pawson T, Bustelo XR, Barbacid M, Sabe H, Hanafusa H, Yi T. Specific motifs recognized by the SH2 domains of Csk, 3BP2, fps/fes, GRB-2, HCP, SHC, Syk, and Vav. Mol Cell Biol. 1994;14:2777–2785. doi: 10.1128/mcb.14.4.2777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spana EP, Kopczynski C, Goodman CS, Doe CQ. Asymmetric localization of numb autonomously determines sibling neuron identity in the Drosophila CNS. Development. 1995;121:3489–3494. doi: 10.1242/dev.121.11.3489. [DOI] [PubMed] [Google Scholar]
- Sparks A, Rider J, Hoffman N, Fowlkes D, Quilliam L, Kay B. Distinct ligand preferences of Src homology 3 domains from Src, Yes, Abl, Cortactin, p53bp2, PLCgamma, Crk, and Grb2. Proc Natl Acad Sci. 1996;93:1540–1544. doi: 10.1073/pnas.93.4.1540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steward R. Dorsal, an embryonic polarity gene in Drosophila, is homologous to the vertebrate proto-oncogene, c-rel. Science. 1987;238:692–694. doi: 10.1126/science.3118464. [DOI] [PubMed] [Google Scholar]
- Tan PK, Howard JP, Payne GS. The sequence NPFXD defines a new class of endocytosis signal in Saccharomyces cerevisae. J Cell Biol. 1996;135:1789–1800. doi: 10.1083/jcb.135.6.1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang H-Y, Cai M. The EH-containing protein Pan1 is required for normal organization of the actin cytoskeleton in Saccaromyces cerevisiae. Mol Cell Biol. 1996;16:4897–4914. doi: 10.1128/mcb.16.9.4897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tebar F, Sorkina T, Sorkin A, Ericsson M, Kirchhausen T. Eps15 is a component of clathrin-coated pits and vesicles and is located at the rim of coated pits. J Biol Chem. 1996;271:28727–28730. doi: 10.1074/jbc.271.46.28727. [DOI] [PubMed] [Google Scholar]
- Uemura T, Shepard S, Ackerman L, Jan LY, Jan YN. Numb, a gene required in determination of cell fate during sensory organ formation in Drosophila embryos. Cell. 1989;58:349–360. doi: 10.1016/0092-8674(89)90849-0. [DOI] [PubMed] [Google Scholar]
- van der Geer P, Pawson T. The PTB domain: A new protein module implicated in signal transduction. Trends Biochem Sci. 1995;20:277–280. doi: 10.1016/s0968-0004(00)89043-x. [DOI] [PubMed] [Google Scholar]
- Verdi JM, Schmandt R, Bashirullah A, Jacob S, Salvino R, Craig CG, Program AE, Lipshitz HD, McGlade CJ. Mammalian NUMB is an evolutionarily conserved signaling adapter protein that specifies cell fate. Curr Biol. 1996;6:1134–1145. doi: 10.1016/s0960-9822(02)70680-5. [DOI] [PubMed] [Google Scholar]
- Voelker RA, Gibson W, Graves JP, Sterling JF, Eisenberg MT. The Drosophila suppressor of sable gene encodes a polypeptide with regions similar to those of RNA-binding proteins. Mol Cell Biol. 1991;11:894–905. doi: 10.1128/mcb.11.2.894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wendland B, McCaffery JM, Xiao Q, Emr SD. A novel fluorescence-activated cell sorter-based screen for yeast endocytosis mutants identifies a yeast homologue of mammalian eps15. J Cell Biol. 1996;135:1485–1500. doi: 10.1083/jcb.135.6.1485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong WT, Kraus MH, Carlomagno F, Zelano A, Druck T, Croce CM, Huebner K, Di Fiore PP. The human eps15 gene, encoding a tyrosine kinase substrate, is conserved in evolution and maps to 1p31-p32. Oncogene. 1994;9:1591–1597. [PubMed] [Google Scholar]
- Wong WT, Schumacher C, Salcini AE, Romano A, Castagnino P, Pelicci PG, Di Fiore PP. A protein-binding domain, EH, identified in the receptor tyrosine kinase substrate Eps15 and conserved in evolution. Proc Natl Acad Sci. 1995;92:9530–9534. doi: 10.1073/pnas.92.21.9530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H, Chen JK, Feng S, Dalgarno DC, Brauer AW, Schreiber SL. Structural basis for the binding of proline-rich peptides to SH3 domains. Cell. 1994;76:933–945. doi: 10.1016/0092-8674(94)90367-0. [DOI] [PubMed] [Google Scholar]
- Xu XX, Yang W, Jackowski S, Rock CO. Cloning of a novel phosphoprotein regulated by colony-stimulating factor 1 shares a domain with the Drosophila disabled gene product. J Biol Chem. 1995;270:14184–14191. doi: 10.1074/jbc.270.23.14184. [DOI] [PubMed] [Google Scholar]
- Zhong W, Feder JN, Jang MM, Jan LY, Jan YN. Asymmetric localization of a mammalian numb homolog during mouse cortical neurogenesis. Neuron. 1996;17:43–53. doi: 10.1016/s0896-6273(00)80279-2. [DOI] [PubMed] [Google Scholar]
- Zhou M-M, Ravichandran KS, Olejniczak ET, Petros AM, Meadows RP, Sattler M, Harlan JE, Wade WS, Burakoff SJ, Fesik SW. Structure and ligand recognition of the phosphotyrosine binding domain of shc. Nature. 1995;378:584–592. doi: 10.1038/378584a0. [DOI] [PubMed] [Google Scholar]