Abstract
Using two-dimensional sodium dodecyl sulphate–polyacrylamide gel electrophoresis, we found that copper/zinc superoxide dismutase (Cu/Zn-SOD, SOD-1) was induced in constructed CCR5 stably transfected HEK 293 cells, but not in mock cells, treated with CCL5. CCL5-induced SOD-1 expression was also confirmed in HEK 293-CCR5 cells and CCR5-positive granulocyte–macrophage colony-stimulating factor-induced human macrophages and murine macrophage RAW264.7 cells. CCL5 and CCR5 interaction induced SOD-1 expression mainly via MEK–ERK activation. In addition, we provided evidence that upregulation of SOD-1 by CCL5/CCR5 activation occurred in parallel with the increased release of tumour necrosis factor-α and nitric oxide and production of intracellular reactive oxygen species as well as enhanced nuclear factor-κB transcriptional activity in CCR5-positive RAW264.7 cells. Conversely, the MEK1/2 inhibitor PD98059 significantly inhibited SOD-1 expression with the decrease of these biological responses. More importantly, inhibition of SOD-1 activity by disulfiram also strongly inhibited the CCL5-induced biological effects. These data suggest that SOD-1 mediates CCR5 activation by CCL5 and that pharmacological modulation of SOD-1 may be beneficial to CCR5-associated diseases.
Keywords: CCL5, CCR5, copper/zinc-superoxide dismutase, macrophages
Introduction
The CC chemokine receptor 5 (CCR5) is a G-protein-coupled seven-transmembrane receptor that binds the chemokines monocyte inflammatory protein CCL3 (MIP-1α), CCL4 (MIP-1β) and CCL5 (RANTES).1 It is mainly expressed in memory T cells,2,3 macrophages4 and immature dendritic cells.5–7
CCR5 is the main co-receptor for macrophage (M) and dual (T-cell and M)-tropic immunodeficiency viruses8,9 and is implicated in human immunodeficiency virus infection.10–12 Chemokine interaction with CCR5 plays a crucial role in the trafficking of leucocytes13 and the regulation of effector functions,14,15 and is involved in the development of several inflammatory diseases10 and cancer.16
Activation of CCR5 has been shown to trigger diverse cellular responses including inhibition of cyclic adenosine monophosphate production,13 stimulation of Ca2+ release,17 and activation of phosphatidyl inositol 3-kinase (PI3K)18 and mitogen-activated protein (MAP) kinases,19 as well as other tyrosine kinase cascades.20,21 Although considerable progress regarding CCR5 signal transduction has been made, many downstream events are still poorly understood.
Reactive oxygen species (ROS) are produced by inflammatory macrophages and neutrophils and serve an essential role in host defence probably against viruses, bacteria and tumours.22–24 It is also recognized as an important mediator in the pathogenesis of inflammatory diseases such as asthma,25 rheumatoid arthritis26 and acquired immune deficiency syndrome.27 In addition, ROS and, specifically, hydrogen peroxide have been shown to serve as second messengers at small, non-toxic concentrations.28 They induce the activation and phosphorylation of stress kinases (JNK, ERK, p38) and redox-sensitive transcription factors such as nuclear factor-κB (NF-κB), all of which lead to an increase in the expression of genes of proinflammatory mediators. An increasing body of evidence has demonstrated that cytoplasmic copper/zinc superoxide dismutase (Cu/Zn-SOD; SOD-1), one of the superoxide dismutases that convert ROS into hydrogen peroxide, may be involved in mediating some signal transduction events. The decreased expression of SOD-1 has been observed in the myeloid cells in response to granulocyte colony-stimulating factor29 while increased expression of SOD-1 has been found in the macrophages treated with tumour necrosis factor-α (TNF-α) and lipopolysaccharide (LPS).30 Moreover, transgenic mice overexpressing SOD-1 have a higher angiogenic potential, inflammatory and immune responses.30
In this study, we focused on the effect of CCL5 on CCR5-expressing cells. Proteomic technology revealed that SOD-1 was induced in a CCR5-dependent manner in the constructed HEK 293-CCR5 cells stimulated by CCL5, followed by verification with Western blot analysis in the HEK 293-CCR5 cells, CCR5-positive granulocyte–macrophage colony-stimulating factor (GM-CSF)-induced human macrophages and murine macrophage RAW264.7 cells. We further examined the correlation between SOD-1 level and CCR5/CCL5-mediated responses, such as release of inflammatory mediators like TNF-α and nitric oxide, intracellular ROS formation and NF-κB transcriptional activity, and the possible involvement of signal transduction pathways in CCR5-expressing macrophages to explore SOD-1 as a potential target for pharmacological modulation of CCR5/CCL5-associated diseases.
Materials and methods
Materials
CCL5 was purchased from PeproTech (Rocky Hill, NJ). PD98059 was obtained from New England Biolabs (Beverly, MA) and wortmannin was obtained from Calbiochem (San Diego, CA). Disulfiram (DSF) was purchased from Fluka (Buchs, Switzerland). 2′,7′-dichlorofluorescin diacetate (DCFH-DA) and LPS (0111:B4) were supplied by Sigma (St Louis, MO). Enzyme-linked immunosorbent assay (ELISA) kits for murine TNF-α, recombinant human (rh) GM-CSF, and anti-mCCR5 antibody (Clone: 45531) were purchased from R&D Systems (Minneapolis, MN). Anti-human CCR5 antibody (Clone: C34-3448) was obtained from BD Pharmingen™ (San Diego, CA). Anti-Cu/Zn SOD-specific polyclonal antibody was supplied by abcam (Cambridge, UK), and horseradish peroxidase-coupled secondary antibodies were purchased from Santa Cruz Biotechnology (Santa Cruz, CA).
Cells and culture
HEK 293 cells and murine macrophage RAW264.7 cells were obtained from the American Type Culture Collection (ATCC, Manassas, VA). They were cultured in Dulbecco’s modified Eagle’s minimum essential medium (DMEM) supplemented with 10% (v/v) heat-inactivated fetal calf serum (Invitrogen, Carlsbad, CA), 2 mm l-glutamine, 100 units/ml penicillin and 100 μg/ml streptomycin at 37° in a humidified incubator of 5% CO2.
GM-CSF-induced human macrophages (GHM) were prepared as described.31 In brief, monocytes were isolated by NycoPrep 1.068 (Nycomed, Oslo, Norway) or Ficoll–hypaque density gradient centrifugation from at least three different normal human donors, then cultured at approximately 1 × 107 cells per well in six-well plates with serum-free medium RPMI-1640 at 37° for 4 hr. Then, the culture medium was replaced with RPMI-1640 plus 10% fetal bovine serum and 10 ng/ml rhGM-CSF (R&D Systems) and refreshed with same medium 3 days later. After 7 days of culture, cells were designated as GM-CSF-induced human macrophages and collected for the following experiment.
Construction of CCR5 stable transfectants
The expression vector of human CCR5 constructed in pcDNA3.0 was a kind gift from Michel Samson (Univeristé de Rennes I, France).32 The clone was confirmed by DNA sequencing. HEK 293 cells were transfected with CCR5 constructs and control vector by lipofectamine reagent (Invitrogen). Two days after transfection, selection for stably transfected cells was initiated by the addition of 400 μg/ml G418 (Life Technologies, Inc., Rockville, MD). After 2 weeks in G418 culture medium, the stable transfectants of CCR5 constructs and control vector were obtained and designated as HEK 293-CCR5 cells and mock cells. For all studies, at least two different clones of HEK 293-CCR5 cells were studied and similar results were obtained from both clones.
Flow cytometric analysis of CCR5 expression
HEK 293-CCR5 cells, mock cells, GHM and RAW264.7 cells were incubated with anti-CCR5 antibodies at 4° overnight and washed twice with phosphate-buffered saline (PBS). GM-CSF-induced human macrophages and RAW264.7 were treated with an isotype matched antibody (Pharmingen) as control. Cells were then incubated with the fluorescein isothiocyanate (FITC)-conjugated immunoglobulin at 4° for 30 min and washed with PBS again. Finally, cells were subjected to Flow cytometer (BD company, San Jose, CA) to analyse the cell-surface expression of CCR5.
Two-dimensional analysis
HEK 293-CCR5 cells and mock cells (2 × 106 cells) were treated with CCL5 (50 nm) for 8 hr. Then, cells were lysed with 0·2 ml rehydration/sample buffer [8 m urea, 2% 3-((3-cholamidopropyl)dimethylammonio)-1-propanesulphonic acid (CHAPS), 0·2% pH 3–10 isoelectric pH gradient (IPG) buffer, 50 mm dithiothreitol] with repeated supersonic liberation on ice and centrifuged at 8000 g for 1 hr. The supernatants were collected and the protein concentrations were determined by Bradford protein assay method, using bovine serum albumin as a standard.25 Proteins samples (300 μg total proteins) were mixed with 350 μl rehydration/sample buffer (8 m urea, 2% CHAPS, phenyl methylsulphonyl fluoride 0·2% pH 3–10 IPG buffer, 50 mm dithiothreitol). Isoelectric focusing was carried out on Protean IEF System using pH 3–10 IPG gel strips (Bio-Rad Laboratories Inc., Hercules, CA) at 20°. The strips were then placed on 12% sodium dodecyl sulphate (SDS) polyacrylamide gels and the second direction electrophoresis was run at constant current (15 mA per gel) for 30 min and then run at constant current (50 mA) until bromophenol blue reached the end of the gel, according to the manufacturer’s instructions from Bio-Rad. After being stained with 0·25% silver nitrate, the two-dimensional gels were scanned with GS800 (Bio-Rad) and were analysed using PDQuest software (Version 7.0.0; Bio-Rad) according to the protocols provided by the manufacturer.
MALDI-TOF-MS-MS analysis and database search
Proteins were analysed by matrix-assisted laser desorption/ionization time-of-flight mass spectroscopy (MALDI-TOF-MS/MS) as the reference.33 Briefly, the differential protein spots were excised from the gels, washed, destained with a solution of 15 mm potassium ferricyanide and 50 mm sodium thiosulphate (1 : 1) for 20 min at room temperature, followed by incubation in a digestion buffer containing 20 mm ammonium bicarbonate and 20 ng/ml sequencing-grade modiWed trypsin for 12 hr at 37°. The resulting peptides were then extracted twice with 0·1% trifluoroacetic acid in 50% acetonitrile, dried with N2. The dried peptides were then mixed with 20 mg/ml α-cyano 4-hydroxy cinnamic acid in acetonitrile and loaded onto a MALDI target plate. Spectra were obtained using a mass spectrometer MALDI-TOF-MS/MS (ABI 4700). The search parameters were as follows: NCBI database, human, molecular weight ranged from 700 to 3200, trypsin digest with one missing cleavage, peptide tolerance of ± 0·2, MS/MS ion mass tolerance of ± 0·6, and possible oxidation of methionine. The Mascot score calculated by the software was used as criterion for correct identification.
Western blot analysis of SOD-1 expression
HEK 293-CCR5 cells and mock cells were seeded in six-well plates at a density of 2 × 106 cells/well and incubated with CCL5 (50 nm) for 0, 2, 4, 8, 12 and 16 hr, respectively. Cells were then washed three times with ice-cold PBS and subsequently lysed with 0·2 ml of buffer (Pierce) at room temperature. The supernatants were collected after the centrifugation at 8000 g for 10 min. Samples were quantified with the micro BCA Protein Assay Reagent kit (Pierce, Biotechnology Inc., Rockford, IL). Then, 20 μg of protein extracts were run on 12% SDS–PAGE gels, transferred onto nitrocellulose filter. The filter was incubated with anti-Cu/Zn SOD specific polyclonal antibody (abcam, Cambridge, UK) (1 : 1000), followed by a rabbit anti-sheep-specific antibody (1 : 10000) (Santa Cruz) and lastly developed using chemiluminescent detection methods (ECL Kit, Amersham Pharmacia Biotech, Little Chalfont, UK). In some experiments, GHM and RAW264.7 cells (2 × 106 cells) were treated with CCL5 (50 nm) for the indicated times.
To examine the effects of the MEK1/2 inhibitor or PI3K inhibitor on SOD-1 expression, HEK 293-CCR5 cells and mock cells, and RAW264.7 cells were pretreated with PD98059 (50 μm) or wortmannin (100 nm) for 30 min and then stimulated with CCL5 (50 nm) for 8 hr.
To exclude the possible endotoxin contamination of reagents, CCL5 was heated at 100° for 5 min and the heat-inactivated CCL5 was tested for the induction of SOD-1 in RAW264.7 cells.
Detection of intracellular ROS formation
Intracellular ROS formation was assessed using a redox-sensitive dye 2′, 7′-dichlorofluorescin diacetate (H2DCFDA). RAW264.7 cells (2 × 106 cells/ml) were cultured in 96-well plate and pretreated with CCR5 antibody, PD98059, wortmannin, DSF or left untreated for 30 min followed by stimulation with CCL5 (50 nm) for 12 hr. Fluorescence was measured with a fluorometer (FLUOstar, BMG LabTechnologies, Offenburg, Germany) after the cells were loaded with 10 μm H2DCFDA (Sigma) for 30 min.
Assay of NF-κB activity
RAW264.7 cells (2 × 105) were cotransfected with the mixture of pGL3.5XκB-luciferase, and pRL-TK-Renilla-luciferase using lipofectamine reagent. Twenty-four hours after transfection, cells were pretreated with inhibitors for 30 min and then left untreated or were treated with CCL5 (50 nm) for 6 hr. The NF-κB luciferase activities were measured using Dual-Luciferase Reporter Assay System (Promega Corp., Madison, WI) according to the manufacturer’s instructions. Data are normalized for transfection efficiency by dividing firefly luciferase activity with that of Renilla luciferase.
TNF-α assay
The RAW264.7 cells (2 × 106 cells/ml) were pretreated with inhibitors for 30 min and then left untreated or treated with CCL5 (50 nm) for 6 hr. Cell supernatants were collected and assayed for TNF-α activity. The TNF-α was assayed both by ELISA and for biological activity. For ELISA, TNF-α protein was determined using the Quantikine murine TNF-α ELISA kit (R&D Systems) according to the manufacturer’s instructions. The TNF-α biological activity was measured using TNF-α-sensitive L929 cells. L929 (mouse fibrosarcoma) cells were obtained from the ATCC and maintained in DMEM supplemented with 10% fetal calf serum. Briefly, the supernatants were added to cultures of L929 (2 × 104 cells/well) in the presence of actinomycin D (1 μg/ml) for 24 hr, cell viability was determined by MTT uptake as previously described.34
Detection of nitric oxide production
The RAW264.7 cells (2 × 106 cells/ml) were cultured in DMEM without phenol-red (GIBCO Life Technologies). Cells were pretreated with inhibitors for 30 min and then left untreated or treated with CCL5 (50 nm) for 0, 6, 12, 24, 48 hr. Cell supernatants were harvested and the concentrations of nitric oxide in supernatants were measured using the Griess reagent. In brief, Griess reagent (Sigma) was prepared by mixing equal volumes of 1% sulfanilamide and 0·1% naphtylethylenediamide in 2·5% phosphoric acid just before use. One hundred microlitres of Griess reagent was mixed with equal amounts of cell supernatants. After incubation at room temperature for 10 min, the absorbance value was measured using a Bio-Rad (Model 550) microplate reader at 570 nm. Concentration of nitrite was assessed by reference to a sodium nitrites standard curve.
Statistical analysis
All experiments were repeated at least three times. Data are shown as means ± standard deviations. Statistical significance was determined by Student’s t-test with a value of P< 0·05 considered as statistically significant.
Results
CCR5 expression
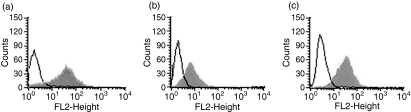
To study the function of CCR5 and its ligands, we first constructed HEK 293-CCR5 cells with HEK 293 cells transfected with pcDNA3-CCR5 constructs and selected with G418 and analysed the expression CCR5 on the cell surface by flow cytometry. Cells were incubated with CCR5 antibody and subsequently FITC-conjugated goat anti-mouse immunoglobulin as secondary antibody. As expected, considerable fluorescence staining was observed in HEK 293-CCR5 cells. Meanwhile, GM-CSF-induced human macrophages prepared from human monocytes by 7 days culture of 10 ng/ml rhGM-CSF, and murine macrophage RAW264.7 cells also showed a log shift in CCR5 fluorescence intensity (Fig. 1). These CCR5-expressing cells were used for the following experiments.
Figure 1.
CCR5 expression on the surface of HEK 293-CCR5 cells, granulocyte–macrophage colony-stimulating factor (GM-CSF)-induced human macrophages and murine macrophage RAW264.7 cells. (a) HEK 293-CCR5 cells (grey area) and mock cells (open area) were incubated with 5 μg/ml monoclonal antibody against CCR5 and fluorescein isothiocyanate-conjugated immunoglobulin. CCR5 expression of 5000 cells was determined by flow cytometry. (b, c) Human macrophages were isolated from different healthy donors and treated with 10 ng/ml recombinant human GM-CSF for 7 days. The GM-CSF-induced human macrophages and murine macrophage RAW264.7 cells were incubated with CCR5 antibody (grey area) or the isotpye matched immunoglobulin (open area). Three separate experiments were performed and a representative one is shown.
Expression of Cu/Zn SOD is upregulated upon the activation of CCR5 with CCL5
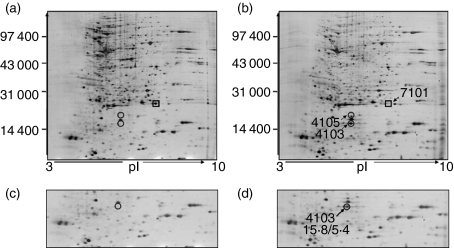
Proteomic technology was performed to explore proteins related with the function of CCR5. Total protein samples from HEK 293-CCR5 cells and mock cells treated with CCL5 were prepared in parallel for three independent experiments. Each protein extract was then separated in triplicate by two-dimensional SDS–polyacrylamide gel electrophoresis (2-D SDS–PAGE). As the typical high resolution silver-stained 2-D map shows in Fig. 2, several unique differentially expressed proteins in HEK 293-CCR5 cells were observed compared with mock cells treated with CCL5. Of particular interest was the identification of one protein (spot 4103), Cu/Zn-SOD, SOD-1, in HEK 293-CCR5 cells, but not in mock cells. Two other proteins were found to be modulated by CCL5, dihydrofolate reductase (spot 7101) and dUTP pyrophosphatase (spot 4105).
Figure 2.
Two-dimensional sodium dodecyl sulphate–polyacrylamide gel electrophoresis of HEK 293-CCR5 cells and mock cells stimulated with CCL5. Total proteins were extracted from HEK 293 mock cells (a, c) and HEK 293-CCR5 cells (b, d) treated with CCL5 (50 nm) for 8 hr. Molecular weight (MW) and isoelectric point (pI) are indicated along the y- and x-axes, respectively. Significant differences in protein expression profile are shown for selected regions. The spots marked with circles were upregulated and those marked with squares were downregulated. The results show that the 15·8/5·4 protein at spot 4103 was identified as superoxide dismutase-1.
CCR5/CCL5 induced Cu/Zn-SOD expression via the MEK-ERK pathway
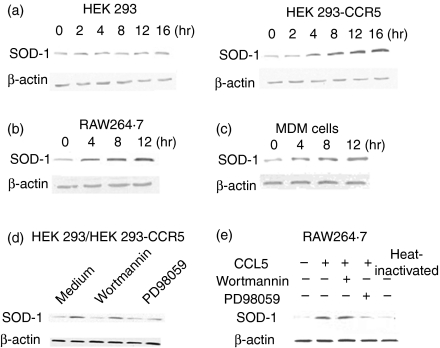
To confirm the regulation of SOD-1 by CCR5, Western blot analysis was conducted to investigate SOD-1 expression in HEK 293-CCR5 cells, GHM and RAW264.7 cells stimulated with CCL5 for various periods of time. As shown in Fig. 3(a–c), the activation of CCR5 by CCL5 markedly stimulated SOD-1 expression in HEK 293-CCR5 cells with maximal effects between 8 and 12 hr. Similarly, SOD-1 expression was also noticeably induced in GHM and RAW264.7 cells. However, in the mock cells there was no change of SOD-1 expression. The results suggest that CCL5 induces SOD-1 expression in a CCR5-dependent manner.
Figure 3.
CCR5/CCL5 regulated the expression of superoxide dismutase-1 (SOD-1) mainly via MEK-ERK signal transduction. (a–c) HEK 293-CCR5 cells and HEK 293 mock cells, RAW264.7 cells and granulocyte–macrophage colony-stimulating factor (GM-CSF)-induced human macrophages were stimulated with CCL5 (50 nm) for the indicated times. (d) HEK 293 mock cells (left) and HEK 293-CCR5 cells (right) were pretreated with 50 μm PD98059 or 100 nm wortmannin or medium with an equal volume of drug solvent for 30 min followed by CCL5 (50 nm) for 8 hr. (e) RAW264.7 cells were pretreated with PD98059 or wortmannin or medium with an equal volume of drug solvent for 30 min followed by CCL5 (50 nm) for 8 hr. Meanwhile heat-inactivated CCL5 (50 nm) was used to exclude the endotoxin contaminant of reagent. Then, the cell lysates were prepared and SOD-1 level was determined by Western blot analysis.
Since binding of CCR5 with its ligands stimulates numerous kinases including PI3-kinase and MAP kinases, MEK1/2 inhibitor (PD98059) and PI3K inhibitor (wortmannin) were used to investigate the possible mechanisms involved in the regulation of CCL5/CCR5 on SOD-1expression. As shown in Fig. 3(d), PD98059 noticeably inhibited the expression of SOD-1 protein in HEK 293-CCR5 cells stimulated with CCR5/CCL5, while wortmannin had no effect. Similar results were also observed in RAW264.7 cells (Fig. 3e) and GHM treated with CCL5 (data not shown). The results indicate that CCR5/CCL5 induces SOD-1 expression mainly via MEK-ERK signal transduction. On the other hand, the heat-inactivated CCL5 appeared to have no effect on SOD-1 expression.
CCL5 induces TNF-α and NO production from macrophages
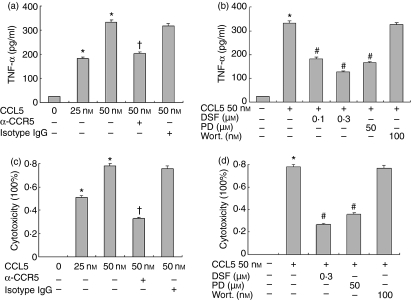
Macrophage activation results in the release of various proinflammatory mediators. To examine the correlation between SOD-1 level and the production of proinflammatory mediators, we analysed CCL5-induced TNF-α production from RAW264.7 cells. CCL5 significantly induced the release of TNF-α in RAW264.7 cells, as measured by biological activity (Fig. 4a); Similar results were demonstrated for TNF-α protein by ELISA (Fig. 4c). This effect was substantially attenuated by a blocking antibody against CCR5, indicating that CCR5 receptors mediate CCL5 signalling in macrophages. Consistent with the observation that CCL5-induced SOD-1 expression via MEK-ERK activation, the release of TNF-α from CCL5-stimulated RAW264.7 cells was also markedly blunted by incubation with PD98059 (25–50 μm) but not by wortmanin (100 nm). Importantly, the SOD-1 inhibitor DSF (0·1–0·3 μm) also dose-dependently inhibited TNF-α release from activated RAW264.7 cells by 29·3 and 50·3% (Fig. 4b). These data indicate that SOD-1 is involved in the CCL5-induced TNF-α release from macrophages. To exclude the possibility that the inhibition was the result of direct toxicity of DSF to the cells, we evaluated cell toxicity of DSF using trypan blue staining in parallel experiments. Our results showed over 99% macrophage viability following the treatment of DSF at the concentrations and time used in the experiments.
Figure 4.
CCL5 induces tumour necrosis factor-α (TNF-α) secretion via MEK-ERK activation and disulfiram (DSF) inhibits release of TNF-α from RAW264.7 cells. (a, c) RAW264.7 cells were pretreated with 5 μg/ml neutralizing anti-CCR5 antibody or isotype-matched immunoglobulin G (IgG). (b, d) Cells were pretreated with PD98059 (50 μm), wortmannin (100 nm) and DSF (0·1–0·3 μm) or medium with an equal volume of drug solvent. After 30 min, CCL5 (50 nm) was added for another 6 hr. TNF-α in the supernatants was determined by enzyme-linked immunosorbent assay (a, b) and by biological activity (c, d), as described in the Materials and methods. Results are expressed as the mean ± SD of three independent experiments. *P< 0·05 compared with medium, †P< 0·05 compared with CCL5 and isotype-matched IgG, #P< 0·05 compared with CCL5 (50 nm) stimulation.
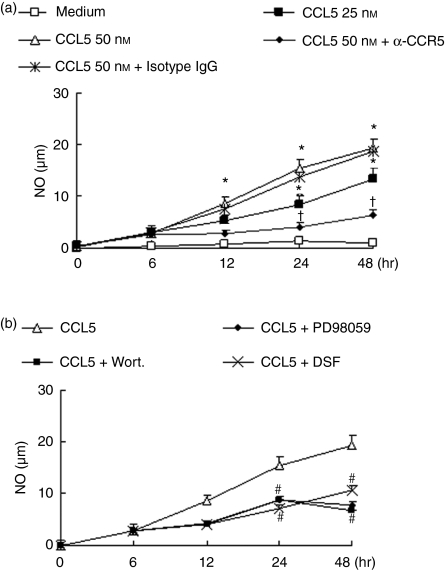
We also analysed nitric oxide production by RAW264.7 cells stimulated with CCL5 and its possible mechanisms. CCL5 caused a marked increase in NO production in a concentration-dependent and time-dependent manner (Fig. 5a). CCR5 blockade also inhibited CCL5-induced NO production, further demonstrating a role for this receptor in mediating CCL5 effects in macrophages. In addition, incubation of cells with PD98059 or wortmannin prevented the release of NO from CCL5-stimulated RAW264.7 cells (Fig. 5b). These results indicates that CCL5-induced NO production involves both MEK-ERK and PI3K pathways. The inhibiting role of DSF (0·3 μm) was observed on NO secretion. It also indicated that SOD-1 is involved in the CCL5-induced NO release from macrophages.
Figure 5.
CCL5 induces release of nitric oxide via MEK-ERK and phosphatidyl inositol 3-kinase (PI3K) activation and disulfiram (DSF) inhibits the release of NO from RAW264.7 cells. (a) RAW264.7 cells were treated with 5 μg/ml neutralizing anti-CCR5 antibody or isotype matched immunoglobulin G (IgG). (b) Cells were treated with PD98059 (50 μm), wortmannin (100 nm) and DSF (0·1–0·3 μm) or medium with equal volume of drug solvent. 30 min later, CCL5 (50 nm) was added for the indicated times. Supernatants were collected and NO production was measured by the Griess method. Results are expressed as the mean ± SD of three independent experiments. *P< 0·05 compared with medium, †P< 0·05 compared with CCL5 and isotype-matched IgG, #P< 0·05 compared with CCL5 (50 nm) stimulation.
CCL5 regulated intracellular ROS level
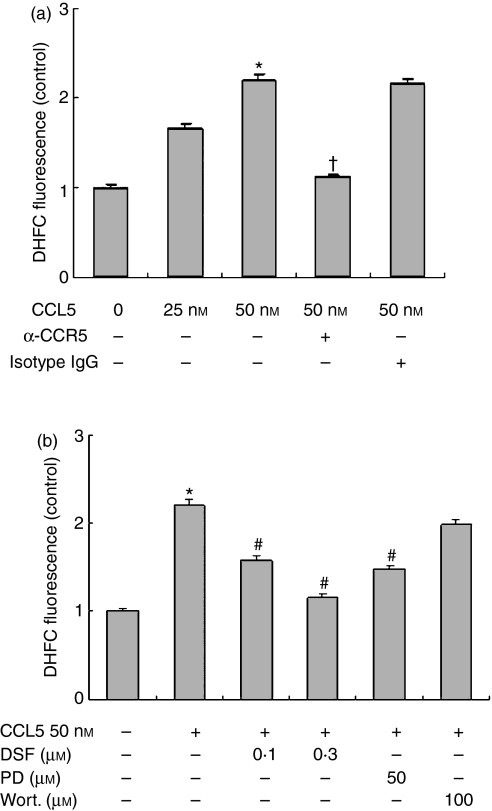
Based on our observation of an association of SOD-1 expression and activity with TNF-α release in CCL5-treated macrophages and the role of SOD-1 in converting superoxide anion into hydrogen peroxide, we examined whether CCL5 affected intracellular ROS formation. As shown in Fig. 6(a), CCL5 induced approximately 1·6-fold and 2·2-fold increases in intracellular ROS level in RAW264.7 at the concentrations of 25 and 50 nm, as measured by DCFDA fluorescence. Similar to our observation on SOD-1 expression and TNF-α release, this effect was attenuated by a CCR5 blocking antibody, by PD98059 but not by wortmannin. These results indicated that CCL5-induced intracellular ROS production is mediated through CCR5 receptors and MEK-ERK activation. In addition, pretreatment of cells with the SOD-1 inhibitor DSF also markedly inhibited intracellular ROS formation from CCL5-stimulated RAW264.7 cells (Fig. 6b), suggesting that the induction of SOD-1 by CCL5 may scavenge of superoxide radicals and cause increased cellular production of hydrogen peroxide.
Figure 6.
Effects of anti-CCR5 antibody, PD98059, wortmannin and disulfiram (DSF) on CCL5-induced intracellular reactive oxygen species production in RAW264.7. (a) RAW264.7 cells were treated with 5 μg/ml neutralizing anti-CCR5 antibody or isotype-matched immunoglobulin G (IgG). (b) Cells were treated with PD98059 (50 μm), wortmannin (100 nm) and DSF (0·1–0·3 μm) or medium with equal volume of drug solvent; 30 min later, CCL5 (50 nm) was added for 12 hr. Cells were then loaded with the redox-sensitive dye H2DCFDA (10 μm) for 30 min and intracellular reactive oxygen species production was measured by a fluorometer. *P< 0·05 compared with medium, †P< 0·05 compared with CCL5 and isotype matched IgG, #P< 0·05 compared with CCL5 (50 nm) stimulation.
CCL5 induced NF-κB activation
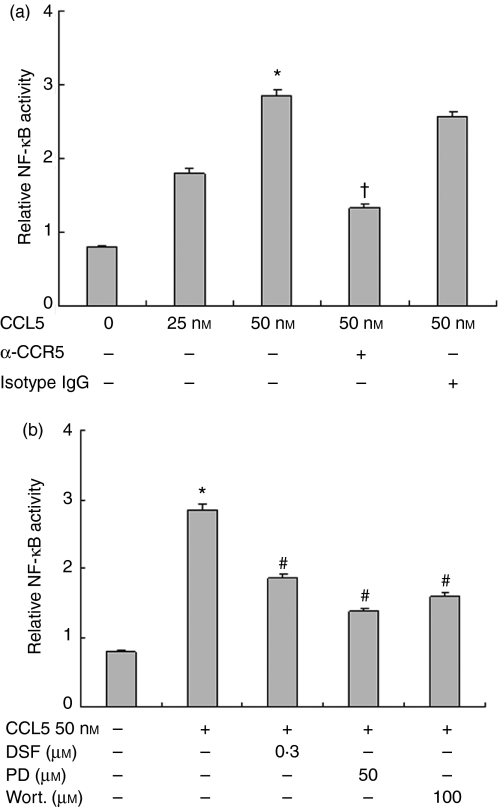
NF-κB is a redox-sensitive transcriptional factor that regulates a number of proinflammatory genes including TNF-α as well as SOD in macrophages. We assessed CCL5-induced NF-κB activity in RAW264.7 cells. As shown in Fig. 7, CCL5 significantly increased NF-κB transcriptional activity in macrophages compared with the control group, as measured by luciferase reporter gene assay. DSF (0·3 μm) markedly inhibited this effect, suggesting that SOD-1 is involved in the CCL5/CCR5-induced NF-κB transcription activity. In contrast, incubation of cells with CCR5 antibody significantly reduced CCL5-induced NF-κB activity. The inhibition of PD98059 (PD, 50 μm) and wortmannin (100 nm) on NF-κB activity showed that both of MEK-ERK and PI3K signal transduction were required for NF-κB activation in macrophages.
Figure 7.
Effects of anti-CCR5 antibody, PD98059, wortmannin and disulfiram (DSF), on CCL5-induced nuclear factor-κB (NF-κB) transcriptional activity in RAW264.7 cells. RAW264.7 cells (2 × 105) were cotransfected with the mixture of pGL3.5XκB-luciferase, and pRL-TK-Renilla-luciferase for 24 hr. (a) RAW264.7 cells were treated with 5 μg/ml neutralizing anti-CCR5 antibody or isotype-matched immunoglobulin G (IgG). (b) Cells were treated with PD98059 (50 μm), wortmannin (100 nm) and DSF (0·1–0·3 μm) or medium with equal volume of drug solvent; 30 min later, CCL5 (50 nm) was added for 6 hr. The NF-κB luciferase activity was measured using Dual-Luciferase Reporter Assay System and normalized with Renilla luciferase activity. *P< 0·05 compared with medium, †P< 0·05 compared with CCL5 and isotype matched IgG, #P< 0·05 compared with CCL5 (50 nm) stimulation.
Discussion
In this study, we report on the discovery of a novel downstream event linking CCL5-induced activation of the chemokine receptor CCR5 resulting in several biological responses to an induction in SOD-1 expression in CCR5-positive macrophages. Moreover, induction of SOD-1 is CCR5-dependent and mainly via MEK-ERK activation. In addition, DSF, an inhibitor of SOD-1, strongly inhibited some CCL5-induced biological responses. These data suggest that SOD-1 may mediate CCR5 activation by CCL5 and pharmacological modulation of SOD-1 may alter CCR5-associated diseases.
SOD-1, found mainly in the cell cytoplasm, is one of the SODs that converts superoxide anion into hydrogen peroxide.35 Interestingly, we found that SOD-1 was markedly increased by CCL5 in CCR5 stably transfected HEK 293 cells and CCR5-positive human monocyte-derived macrophages and RAW264.7 cells. Moreover, treatment of HEK 293 mock cells with CCL5 had no effect on SOD-1 expression, indicating that elevation of SOD-1 by CCL5 is CCR5-dependent. CCL5–CCR5 interaction has been shown to be involved not only in chemotaxis but also in leucocyte activation.36,37 In accord with previous studies, we found that binding of CCR5 with CCL5 resulted in significant macrophage activation, as shown by increased release of TNF-α and nitric oxide and production of intracellular ROS as well as enhanced NF-κB activity in CCR5-positive RAW264.7 cells. Activation of macrophages by LPS and TNF-α has been reported to increase SOD-1 levels in cultured peritoneal macrophages.30 Taken together, these findings suggest that elevation of SOD-1 may be one of the common pathways of macrophage activation.
Ordinarily, enhanced expression of SOD-1 is considered one of the first lines of antioxidant defence to lower the excess ROS generated when macrophages are activated, thereby providing a mechanism for protection of the cell from injury and apoptosis. Therefore, the overexpression of SOD-1 found in this study may arise from the increase in oxidation that was induced by CCL5/CCR5 activation to maintain lower levels of superoxide anions in the cell. Alternatively, SOD-1 may also be involved in mediating some signal transduction events of CCL5–CCR5 interaction because ROS and specifically hydrogen peroxide at relatively low concentrations serve as second messengers. We provide evidence that several CCL5-induced biological responses in CCR5-positive murine macrophage RAW264.7 cells may arise from changes in SOD-1 activity: (i) the stimulatory effects of CCL5/CCR5 activation on NF-κB activation and TNF-α production described above were in parallel with upregulation of SOD-1; (ii) MEK1/2 inhibitor PD98059 significantly inhibited SOD-1 expression with the decrease of these biological responses; and (iii) importantly, inhibition of SOD-1 activity by DSF also produced similar effects. It has been well documented that ROS, especially hydrogen peroxide, serve as messengers mediating directly or indirectly the activation of transcription factors such as NF-κB, and as a result the induction of various proinflammatory genes.38,39 As we have observed in this study, CCL5/CCR5 activation induced the intracellular ROS formation and elevated SOD-1 expression, which may scavenge superoxide radicals and cause increased cellular production of hydrogen peroxide, and consequently promote NF-κB activation and TNF-α release. Moreover, the inhibitory effect of DSF on NF-κB activation and TNF-α release suggests the involvement of increased production of ROS and hydrogen peroxide in macrophages.
Activation of CCR5 by its ligands has been shown to stimulate numerous kinases including PI3K18 and MAP kinases.19 Indeed, preincubation of the CCR5 stably transfected HEK 293 cells with the MEK1/2 inhibitor PD98059 completely prevented the induction of SOD-1 by CCL5. PD98059 also significantly inhibited CCL5-induced SOD-1 expression in the CCR5-positive GM-CSF-induced human macrophages and murine macrophage RAW264.7 cells. However, the PI3K inhibitor wortmannin failed to inhibit this effect. These data suggest that MEK-ERK is involved in the CCR5-dependent SOD-1 expression. Similarly, CCL5 induced intracellular ROS production and TNF-α release mainly via MEK-ERK but not PI3K activation. Interestingly, in the same condition, CCL5-stimulated NF-κB activation and nitric oxide release in RAW264.7 macrophages involved both MEK-ERK and PI3K activation.
There exist four types of SOD based on the metal species at the active site of the enzyme: Cu/Zn-SOD, Mn-SOD, Fe-SOD and Ni-SOD. Based on the proteomic results this study has examined only Cu/Zn-SOD. It is well-known that the chemokine receptor CCR5 is shared by at least three chemokine ligands, CCL3, CCL4 and CCL5 but we only used CCL5 as a stimulator in this study. Despite these limitations, this study can clearly indicate that SOD-1 plays an important role in CCL5/CCR5 activation. In addition, it seems unlikely that all CCL5 effects can be explained by increased SOD-1 activity, because treatment with DSF failed to completely abolish CCL5-induced responses. Moreover, our result showed the approximately 50% inhibition of TNF-α release by the neutralizing CCR5 antibody or PD98059 compound. Although these systems are probably involved, there appears to be related binding of CCL5 to its receptors or yet unknown complex signal transduction pathways. CCL5 has been shown to interact with the G-coupled protein receptors CC chemokine receptors (CCR) 1, 3 and 5.19 The diverse roles of CCL5 might be a result of the large family of receptors through which it can signal. We showed in this study that both the MEK-ERK and PI-3K pathways were involved in some CCL5-induced biological responses and blocking of CCR5 blunted 50% of CCL5-induced signalling events. The possibility that other chemokine receptors expressed in macrophages mediate the remaining effects of CCL5 cannot be excluded and whether SOD-1 transduces downstream events of other chemokine receptors remains to be determined.
In conclusion, this study identifies SOD-1 as mediating CCR5 activation by CCL5 in CCR5-positive macrophages. Chemokine CCL5 and chemokine receptor CCR5 are known to play a vital role in regulating leucocyte trafficking, engendering the adaptive immune response and contributing to the pathogenesis of a variety of diseases so SOD-1 may be a potential target for pharmacological modulation of CCR5-associated diseases.
Acknowledgments
We thank Dr Samson (Université de Rennes I, Campus de Beaulieu, Bretagne, France) for kindly providing the recombinant plasmid pcDNA3·0 construct with huCCR5 gene for transfecting experiment in this study. This work was supported by the National Natural Science Foundation of China, No30472032.
References
- 1.Samson M, Labbe O, Mollereau C, Vassart G, Parmentier M. Molecular cloning and functional expression of a new human CC-chemokine receptor gene. Biochemistry. 1996;35:3362–7. doi: 10.1021/bi952950g. [DOI] [PubMed] [Google Scholar]
- 2.Bleul CC, Wu L, Hoxie JA, Springer TA, Mackay CR. The HIV coreceptors CXCR4 and CCR5 are differentially expressed and regulated on human T lymphocytes. Proc Natl Acad Sci USA. 1997;94:1925–30. doi: 10.1073/pnas.94.5.1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wu L, Paxton WA, Kassam N, et al. CCR5 levels and expression pattern correlate with infectability by macrophage-tropic HIV-1, in vitro. J Exp Med. 1997;185:1681–91. doi: 10.1084/jem.185.9.1681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zaitseva M, Blauvelt A, Lee S, Lapham CK, Klaus-Kovtun V, Mostowski H, Manischewitz J, Golding H. Expression and function of CCR5 and CXCR4 on human Langerhans cells and macrophages: implications for HIV primary infection. Nat Med. 1997;3:1369–75. doi: 10.1038/nm1297-1369. [DOI] [PubMed] [Google Scholar]
- 5.de Wynter EA, Durig J, Cross MA, Heyworth CM, Testa NG. Differential response of CD34+ cells isolated from cord blood and bone marrow to MIP-1 alpha and the expression of MIP-1 alpha receptors on these immature cells. Stem Cells. 1998;16:349–56. doi: 10.1002/stem.160349. [DOI] [PubMed] [Google Scholar]
- 6.Lee B, Sharron M, Montaner LJ, Weissman D, Doms RW. Quantification of CD4, CCR5, and CXCR4 levels on lymphocyte subsets, dendritic cells, and differentially conditioned monocyte-derived macrophages. Proc Natl Acad Sci USA. 1999;96:5215–20. doi: 10.1073/pnas.96.9.5215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Granelli-Piperno A, Moser B, Pope M, et al. Efficient interaction of HIV-1 with purified dendritic cells via multiple chemokine coreceptors. J Exp Med. 1996;184:2433–8. doi: 10.1084/jem.184.6.2433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu R, Paxton WA, Choe S, et al. Homozygous defect in HIV-1 coreceptor accounts for resistance of some multiply-exposed individuals to HIV-1 infection. Cell. 1996;86:367–77. doi: 10.1016/s0092-8674(00)80110-5. [DOI] [PubMed] [Google Scholar]
- 9.Samson M, Libert F, Doranz BJ, et al. Resistance to HIV-1 infection in caucasian individuals bearing mutant alleles of the CCR-5 chemokine receptor gene. Nature. 1996;382:722–5. doi: 10.1038/382722a0. [DOI] [PubMed] [Google Scholar]
- 10.Charo IF, Ransohoff RM. The many roles of chemokines and chemokine receptors in inflammation. N Eng J Med. 2006;354:610–21. doi: 10.1056/NEJMra052723. [DOI] [PubMed] [Google Scholar]
- 11.Murdoch C, Finn A. Chemokine receptors and their role in inflammation and infectious diseases. Blood. 2000;95:3032–43. [PubMed] [Google Scholar]
- 12.Lederman MM, Penn-Nicholson A, Cho M, Mosier D. Biology of CCR5 and its role in HIV infection and treatment. JAMA. 2006;296:815–26. doi: 10.1001/jama.296.7.815. [DOI] [PubMed] [Google Scholar]
- 13.Blanpain C, Wittamer V, Vanderwinden JM, et al. Palmitoylation of CCR5 is critical for receptor trafficking and efficient activation of intracellular signaling pathways. J Biol Chem. 2001;276:23795–804. doi: 10.1074/jbc.M100583200. [DOI] [PubMed] [Google Scholar]
- 14.Skwor TA, Cho H, Cassidy C, Yoshimura T, McMurray DN. Recombinant guinea pig CCL5 (RANTES) differentially modulates cytokine production in alveolar and peritoneal macrophages. J Leukoc Biol. 2004;76:1229–39. doi: 10.1189/jlb.0704414. [DOI] [PubMed] [Google Scholar]
- 15.Pervushina O, Scheuerer B, Reiling N, et al. Platelet factor 4/CXCL4 induces phagocytosis and the generation of reactive oxygen metabolites in mononuclear phagocytes independently of Gi protein activation or intracellular calcium transients. J Immunol. 2004;173:2060–7. doi: 10.4049/jimmunol.173.3.2060. [DOI] [PubMed] [Google Scholar]
- 16.Kakinuma T, Hwang ST. Chemokines, chemokine receptors, and cancer metastasis. J Leukoc Biol. 2006;79:639–51. doi: 10.1189/jlb.1105633. [DOI] [PubMed] [Google Scholar]
- 17.Lee C, Liu QH, Tomkowicz B, Yi Y, Freedman BD, Collman RG. Macrophage activation through CCR5- and CXCR4-mediated gp120-elicited signaling pathways. J Leukoc Biol. 2003;74:676–82. doi: 10.1189/jlb.0503206. [DOI] [PubMed] [Google Scholar]
- 18.Gomez-Mouton C, Lacalle RA, Mira E, Jimenez-Baranda S, Barber DF, Carrera AC, Martinez AC, Manes S. Dynamic redistribution of raft domains as an organizing platform for signaling during cell chemotaxis. J Cell Biol. 2004;164:759–68. doi: 10.1083/jcb.200309101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kraft K, Olbrich H, Majoul I, Mack M, Proudfoot A, Oppermann M. Characterization of sequence determinants within the carboxyl-terminal domain of chemokine receptor CCR5 that regulate signaling and receptor internalization. J Biol Chem. 2001;276:34408–18. doi: 10.1074/jbc.M102782200. [DOI] [PubMed] [Google Scholar]
- 20.Bacon KB, Szabo MC, Yssel H, Bolen JB, Schall TJ. RANTES induces tyrosine kinase activity of stably complexed p125FAK and ZAP-70 in human T cells. J Exp Med. 1996;184:873–82. doi: 10.1084/jem.184.3.873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ganju RK, Dutt P, Wu L, Newman W, Avraham H, Avraham S, Groopman JE. Beta-chemokine receptor CCR5 signals via the novel tyrosine kinase RAFTK. Blood. 1998;91:791–7. [PubMed] [Google Scholar]
- 22.Nathan C. Nitric oxide as a secretory product of mammalian cells. FASEB J. 1992;6:3051–64. [PubMed] [Google Scholar]
- 23.Swindle EJ, Hunt JA, Coleman JW. A comparison of reactive oxygen species generation by rat peritoneal macrophages and mast cells using the highly sensitive real-time chemiluminescent probe pholasin: inhibition of antigen-induced mast cell degranulation by macrophage-derived hydrogen peroxide. J Immunol. 2002;169:5866–73. doi: 10.4049/jimmunol.169.10.5866. [DOI] [PubMed] [Google Scholar]
- 24.Lander HM. An essential role for free radicals and derived species in signal transduction. FASEB J. 1997;11:118–24. [PubMed] [Google Scholar]
- 25.Wood LG, Gibson PG, Garg ML. Biomarkers of lipid peroxidation, airway inflammation and asthma. Eur Respir J. 2003;21:177–86. doi: 10.1183/09031936.03.00017003a. [DOI] [PubMed] [Google Scholar]
- 26.Hultqvist M, Backlund J, Bauer K, Gelderman KA, Holmdahl R. Lack of reactive oxygen species breaks T cell tolerance to collagen type II and allows development of arthritis in mice. J Immunol. 2007;179:1431–7. doi: 10.4049/jimmunol.179.3.1431. [DOI] [PubMed] [Google Scholar]
- 27.Liu K, Chi DS, Li C, Hall HK, Milhorn DM, Krishnaswamy G. HIV-1 Tat protein-induced VCAM-1 expression in human pulmonary artery endothelial cells and its signaling. Am J Physiol Lung Cell Mol Physiol. 2005;289:L252–60. doi: 10.1152/ajplung.00200.2004. [DOI] [PubMed] [Google Scholar]
- 28.Forman HJ, Fukuto JM, Torres M. Redox signaling: thiol chemistry defines which reactive oxygen and nitrogen species can act as second messengers. Am J Physiol Cell Physiol. 2004;287:C246–56. doi: 10.1152/ajpcell.00516.2003. [DOI] [PubMed] [Google Scholar]
- 29.Niwa M, Yousif AE, Kohno K, Kanamori Y, Matsuno M, Abe A, Uematsu T. The loss of recombinant human granulocyte colony-stimulating factor and recombinant human TNF-alpha priming effects on the superoxide-generating response in exudated neutrophils is associated with a decrease in their receptor affinities. J Immunol. 1996;157:4147–53. [PubMed] [Google Scholar]
- 30.Marikovsky M, Nevo N, Vadai E, Harris-Cerruti C. Cu/Zn superoxide dismutase plays a role in angiogenesis. Int J Cancer. 2002;97:34–41. doi: 10.1002/ijc.1565. [DOI] [PubMed] [Google Scholar]
- 31.Kiener PA, Davis PM, Starling GC, Mehlin C, Klebanoff SJ, Ledbetter JA, Liles WC. Differential induction of apoptosis by Fas-Fas ligand interactions in human monocytes and macrophages. J Exp Med. 1997;185:1511–6. doi: 10.1084/jem.185.8.1511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bergmans AM, Coenen JL, Bakhuizen R, Mooi BW, Ramdat Misier AR, Wilbrink B, L MS, Wolfhagen MJ. Endocarditis in a Dutch patient caused by Bartonella quintana. Clin Microbiol Infect. 1997;3:692–5. doi: 10.1111/j.1469-0691.1997.tb00480.x. [DOI] [PubMed] [Google Scholar]
- 33.Zhou H, Liu Y, Chui J, et al. Investigation on glycosylation patterns of proteins from human liver cancer cell lines based on the multiplexed proteomics technology. Arch Biochem Biophys. 2007;459:70–8. doi: 10.1016/j.abb.2006.10.027. [DOI] [PubMed] [Google Scholar]
- 34.Mosmann T. Rapid colorimetric assay for cellular growth and survival: application to proliferation and cytotoxicity assays. J Immunol Methods. 1983;65:55–63. doi: 10.1016/0022-1759(83)90303-4. [DOI] [PubMed] [Google Scholar]
- 35.Fridovich I. The biology of oxygen radicals. Science. 1978;201:875–80. doi: 10.1126/science.210504. [DOI] [PubMed] [Google Scholar]
- 36.Olson TS, Ley K. Chemokines and chemokine receptors in leukocyte trafficking. Am J Physiol Regul Integr Comp Physiol. 2002;283:R7–28. doi: 10.1152/ajpregu.00738.2001. [DOI] [PubMed] [Google Scholar]
- 37.Ajuebor MN, Carey JA, Swain MG. CCR5 in T cell-mediated liver diseases: what’s going on? J Immunol. 2006;177:2039–45. doi: 10.4049/jimmunol.177.4.2039. [DOI] [PubMed] [Google Scholar]
- 38.Schreck R, Baeuerle PA. A role for oxygen radicals as second messengers. Trends Cell Biol. 1991;1:39–42. doi: 10.1016/0962-8924(91)90072-h. [DOI] [PubMed] [Google Scholar]
- 39.Schreck R, Rieber P, Baeuerle PA. Reactive oxygen intermediates as apparently widely used messengers in the activation of the NF-kappa B transcription factor and HIV-1. EMBO J. 1991;10:2247–58. doi: 10.1002/j.1460-2075.1991.tb07761.x. [DOI] [PMC free article] [PubMed] [Google Scholar]