Abstract
Members of the protein kinase C (PKC) family are activated by interferon-γ (IFN-γ) and modulate IFN-γ-induced cellular responses by regulating the activity of transcription factors. We previously reported that PKC-α enhances the ability of IFN regulatory factor-1 to transactivate the class II transactivator (CIITA) promoter IV in IFN-γ-stimulated macrophages. In addition, we showed that IFN-γ induces the nuclear translocation of PKC-α but the mechanisms for this remain to be elucidated. In this study, we sought to identify signalling pathways involved in IFN-γ-induced activation of PKC-α and to characterize their potential roles in modulating IFN-γ-induced responses in macrophages. IFN-γ-mediated nuclear translocation of PKC-α was a Janus activated kinase 2 (JAK2)-independent process, which required phosphatidylinositol 3-kinase (PI3K) and p38 mitogen-activated protein kinase (MAPK). However, PKC-α phosphorylation was independent of PI3K and p38 MAPK, indicating that IFN-γ-induced phosphorylation and nuclear translocation of PKC-α are mediated by distinct mechanisms. In addition, inhibition of PI3K, but not of p38 MAPK, strongly impaired IFN-γ-induced CIITA and MHC II gene expression. Finally, PKC-α associated with signal transducer and activator of transcription 1 (STAT1) and was required for the phosphorylation of STAT1 on serine 727 in IFN-γ-stimulated macrophages. Taken together, our data indicate that PI3K and p38 MAPK modulate IFN-γ-stimulated PKC-α nuclear translocation independently of JAK2 activity and that both PI3K and PKC-α are required for type IV CIITA and MHC II gene expression in IFN-γ-stimulated macrophages.
Keywords: interferon-γ, macrophages, nuclear translocation, phosphatidylinositol 3-kinase, protein kinase C
Introduction
Interferon-γ (IFN-γ) is a potent macrophage activator that plays a key role in host defense. Following binding to its multisubunit receptor, IFN-γ exerts its action by triggering phosphorylation on tyrosine residues of the IFN-γ receptor-associated Janus-activated kinases (JAKs) JAK1 and JAK2. These events are followed by tyrosine and serine phosphorylation, homodimerization and nuclear translocation of the transcription factor signal transducer and activator of transcription 1 (STAT1).1 Binding of STAT1 to specific sequences present in the promoter region of early IFN- γ-responsive genes leads to their rapid expression.2 Among those, the IFN regulatory factor-1 (IRF-1), in synergy with STAT1, activates the transcription of several genes, including the class II transactivator (CIITA), a non-DNA-binding protein essential for IFN-γ-induced expression of major histocompatibility complex (MHC) class II in macrophages.3–5 In addition to the JAK–STAT1 pathway, IFN-γ induces gene expression through STAT1-independent pathways6,7 and modulates various signalling pathways, including phosphatidylinositol 3-kinase (PI3K), MyD88/p38 mitogen-activated protein kinase (MAPK), extracellular-signal regulated kinases 1/2 (ERK1/2) and Jun N-terminal kinase, as well as members of the protein kinase C (PKC) family.8–20
PKC is a family of structurally related serine/threonine-specific protein kinases comprising at least 10 members with distinct requirements for activity.21,22 There is evidence that PKC isoenzymes modulate cellular responses to IFN-γ by regulating the activity of various transcription factors.14–18,20 Hence, full STAT1 transcriptional activity requires serine phosphorylation23, which implicates, among others, the activity of PKC isoenzymes. Depending on the cell type, IFN-γ-induced phosphorylation of STAT1 on serine 727 requires the activity of either PKC-δ or PKC-ε.14,17,24 In addition, IFN-γ stimulates PU.1 phosphorylation through the activation of PKC-α and/or PKC-βI, leading to the association of PU.1 with the promoter of gp91phox and to increased gp91phox expression in human monocytes.16 PKC-α also contributes to the expression of CIITA by enhancing the ability of IRF-1 to transactivate the CIITA promoter IV in IFN-γ-stimulated RAW 264.7 cells.20
Whereas the JAK–STAT1 pathway received a great deal of attention, little is known concerning the mechanisms by which IFN-γ activates PKC isoenzymes. PI3K appears to play an important role in the activation of PKC-ε and PKC-δ in IFN-γ-stimulated mesanglial cells and in promyelocytic leukemia cells, respectively.14,17,25 In epithelial cells, IFN-γ-induced intercellular adhesion molecule-1 (ICAM-1) gene expression is mediated by the activation of phosphatidylinositol phospholipase C-γ2, which is required for the activation of PKC-α, followed by those of c-Src and STAT1.15 In macrophages and monocytes, IFN-γ induces the nuclear translocation of PKC-α and PKC-βI, where they can interact with transcription factors.16,20 In contrast to the majority of nuclear proteins, PKC isoenzymes lack a canonical nuclear localization signal, indicating that other mechanisms are involved in the transport process. In the present study, we sought to identify signalling pathways involved in IFN-γ-induced activation and nuclear translocation of PKC-α in macrophages, and to characterize their roles in IFN-γ-induced gene expression.
Materials and methods
Macrophages
Bone marrow-derived macrophages (BMDM) were obtained by growing bone marrow cells from female BALB/c mice at 37° in 5% CO2 in complete medium [Dulbecco’s Modified Eagle’s minimal essential medium with glutamine (Life Technologies Inc., Burlington, ON, Canada), containing 10% heat-inactivated fetal bovine serum (FBS) (Hyclone, Logan, UT), 10 mm HEPES, pH 7·4, and antibiotics] for 7 days in the presence of 15% (v/v) L929 cell-conditioned medium, a source of colony-stimulating factor-1 (CSF-1).26 BMDM were made quiescent by culturing them in the absence of CSF-1 for 18 hr prior to use. The murine macrophage cell line RAW 264.7 was grown in complete medium.
Reagents
Recombinant mouse IFN-γ (R&D Systems, Minneapolis, MN) was used at a final concentration of 100 U/ml , and lipopolysaccharide (LPS) from Escherichia coli serotype O127:B8 (Sigma, St Louis, MO) was used at a final concentration of 100 ng/ml. The inhibitors and their final concentrations used were: tyrphostin AG-490 (250 μm), Gö-6976 (1 μm), SB203580 (10 μm), PD98059 (30 μm), Ly294002 (100 μm), U-73122 (50 μm), U-73343 (50 μm), PP2 (50 μm) and PP3 (50 μm) (Calbiochem, San Diego, CA). The inhibitors were prepared following the manufacturer’s instructions. BpV(phen) (kindly provided by Dr Martin Olivier, McGill University, Montreal, QC, Canada) was prepared in phosphate-buffered saline (PBS) and used at a final concentration of 10 μm. Those inhibitors are respectively selective for JAK2, conventional PKC, p38 MAPK, ERK1/2, PI3K, phospholipase-C, U-73122 inactive analog, Src-family protein tyrosine kinases, the negative control for PP2 and protein tyrosine phosphatases.
Reverse transcription–polymerase chain reaction
Total RNA was prepared and reverse transcribed as described previously.20,27 For the polymerase chain reaction (PCR), samples were amplified under the following conditions: 30 seconds at 94°, 1 min at 56° and 1 min at 72° (30 cycles). The PCR products were migrated on a 1·3% (w/v) agarose gel and the pictures were acquired using an AlphaImager 3400 (Alpha Innotech Corporation, San Leandro, CA). The primers used for hypoxanthine-guanine phosphoribosyl transferase (HPRT) were AD-55 (forward) 5′-GTTGGATACAGGCCAGACTTTGTTG-3′ and AD-56 (reverse) 5′-GATTCAACTTGCGCTCATCTTAGGC-3′; the primers used for type IV CIITA were AD-267 (forward) 5′-ACAGCCACAGCCGCGACCATA-3′ and AD-268 (reverse) 5′-CTCTGCTCCAATGTGCTCCTA-3′; and the primers used for MHC II were AD-70 (forward) 5′-GGAATTCTGGGAATCTCAGGTTCCCAGTG-3′ and AD-71 (reverse) 5′-GGAATTCTGAACACCATGCTCAGCCTCTG-3′.20
Signalling pathway analyses
BMDM were seeded (3 × 106 cells/well) in six-well plates for 18 hr and incubated with or without selective inhibitors for 1 hr prior to stimulation with 100 U/ml of IFN-γ or 100 ng/ml of LPS for the indicated periods of time.
Western blot analyses
Cells were lysed and Western blot analyses were performed as described previously.28 Rabbit polyclonal antisera against IRF-1 was from Santa Cruz Biotechnology (Santa Cruz, CA), antibody against PKC-α was from BD Transduction Laboratories (Oakville, ON, Canada), antibodies against STAT1 and phospho-STAT1 (Ser727) were from Upstate Laboratories (Lake Placid, NY) and antibodies against p38 MAPK, phospho-p38 (Thr180/Tyr182), Akt, phospho-Akt (Ser473), phospho-PKC-α (Thr638) and phospho-STAT1 (Tyr 701) were from Cell Signaling Technology, Beverly, MA.
Translocation of the PKC-α–green fluorescent protein construct and confocal microscopy
Adherent RAW 264.7 cells were transfected with 0·3 μg of the PKC-α– green fluorescent protein (GFP) construct29 (kindly provided by C. Quittau-Prévostel, INSERM, France) using Fugene-6 (Roche Diagnostics, Laval, QC, Canada). Briefly, cells were seeded (105 cells/well) in 24-well plates containing microscope coverslips for 18 hr and transfected with 100 μl of Fugene-6/DNA mix for 24 hr. Cells were incubated for 1 hr in the presence or the absence of signalling inhibitors and then stimulated for 2 hr with 100 U/ml of IFN-γ. Cells were fixed with 2% formaldehyde, permeabilized for 15 min at 21° with PBS containing 0·1% Triton-X-100 and 1% bovine serum albumin (BSA) and then incubated with PBS containing 1% Alexa Fluor 568 phalloidin (Molecular Probes, Oregon, OR, USA) and 0·2% DRAQ5 (BioStatus Ltd., Shepshed, UK). Coverslips were mounted with Fluoromount-G (Southern Biotechnology Associates Inc., Birmingham, AL). Nuclear translocation of PKC-α was quantified using an oil immersion Nikon Plan Apo 100× objective mounted on a Nikon Eclipse E800 microscope (Nikon, Melville, NY) equipped with a BioRad Radiance 2000 confocal imaging system (BioRad Laboratories, Hercules, CA), and data were analyzed using the LaserSharp 2000 software (BioRad, Hercules, CA). At least 30 transfected cells were counted for each experiment and the results presented are representative of three independent experiments. Percentage of nuclear translocation represents the number of transfected cells with nuclear translocation of human PKC-α–GFP on the total number of transfected cells observed.
Immunofluorescence
Adherent macrophages in 24-well plates containing microscope coverslips (Fisher Scientific, Ottawa, ON, Canada) (105 cells/well for RAW 264.7 cells or 2 × 105 cells/well for BMDM) were stimulated for 2 hr with 100 U/ml of IFN-γ and fixed with 2% formaldehyde in PBS. Cells were permeabilized with 0·1% Triton-X-100 and blocked with 1% BSA and 20% goat serum (Jackson ImmunoResearch Laboratories Inc., West Grove, PA). Cells were then marked with mouse monoclonal anti-PKC-α (1 : 300 dilution) (Cell Signaling Technology) and stained with PBS containing the anti-mouse immunoglobulin coupled to Alexa-488 (1 : 500 dilution) (Molecular Probes, Burlington, ON, Canada), 1% Alexa Fluor 568 phalloïdin (Molecular Probes) and 0·2% DRAQ5 (BioStatus Ltd). Coverslips were mounted and the nuclear localization of PKC-α was assessed using confocal microscopy.
Co-immunoprecipitations and Western blot analyses
Adherent BMDM were incubated in the absence or in the presence of 100 U/ml of IFN-γ for the indicated periods of time. Cells were then washed with ice-cold PBS and lysed in lysis buffer (50 mm Tris–HCl, pH 7·4; 1 mm EDTA; 1% Nonidet P-40; 150 mm NaCl; 0·25% deoxycholate) containing proteases and phosphatase inhibitors. PKC-α was immunoprecipitated using a rabbit anti-PKC-α (1 : 500 dilution; Transduction Laboratories, San Jose, CA), and immunocomplexes were resolved by sodium dodecyl sulfate–polyacrylamide electrophoresis (SDS–PAGE; 8% gel) and electroblotted onto Hybond-Enhanced Chemiluminescence (ECL) membranes (Amersham Biosciences, Piscataway, NJ). Immunodetection of STAT1 was achieved by enhanced chemiluminescence using a rabbit anti-STAT1 serum (1 : 1000 dilution; Upstate Laboratories). To detect PKC-α, we used a mouse anti-PKC-α immunoglobulin (IgG2b) (1 : 1000 dilution; Transduction Laboratories).
Results
PI3K and p38 MAPK are involved in the IFN-γ-induced nuclear translocation of PKC-α
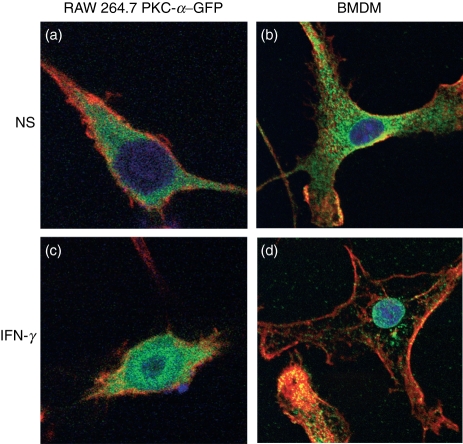
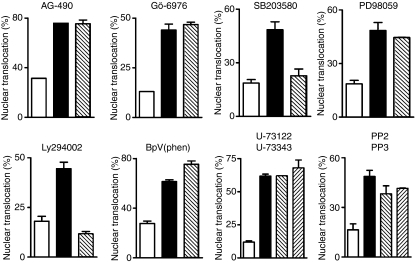
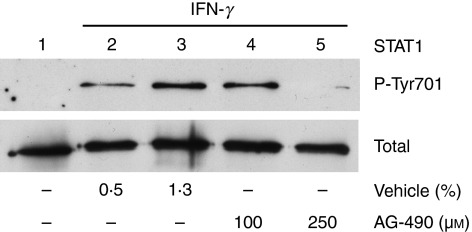
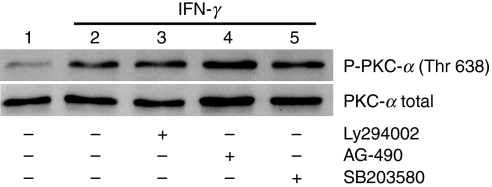
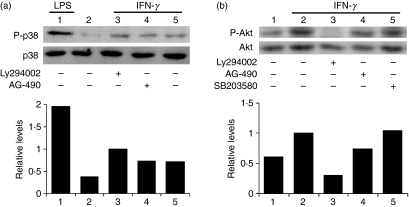
In this study, we used PKC-α–GFP as a reporter to investigate the mechanisms by which IFN-γ stimulates the nuclear translocation of PKC-α.20 As shown in Fig. 1, similarly to endogenous PKC-α in BMDM (Fig. 1b,d), the PKC-α–GFP fusion protein translocated to the nucleus of RAW 264.7 cells following stimulation with IFN-γ (Fig. 1a,c). To identify signalling pathways involved in IFN-γ-induced PKC-α nuclear translocation, we incubated RAW264.7 cells, transiently expressing PKC-α–GFP, in the absence or presence of the indicated pharmacological inhibitors prior to stimulation with IFN-γ. As shown in Fig. 2, inhibition of PI3K (with Ly294002) and p38 MAPK (with SB203580) reduced IFN-γ-induced nuclear translocation of PKC-α–GFP to the levels observed in unstimulated cells. By contrast, inhibition of JAK2 (with AG-490), PKC-α/β1 (with Gö-6976), ERK1/2 (with PD98059), protein tyrosine phosphatases [with BpV(phen)], phospholipase C (with U-73122) and Src-family tyrosine kinases (with PP2) had no significant impact on the nuclear translocation of PKC-α–GFP. These results indicate that IFN-γ-induced nuclear translocation of PKC-α is a process requiring PI3K and p38 MAPK, independently of JAK2 activity. Of note, under the conditions used, the JAK2 inhibitor AG-490 completely abrogated IFN-γ-induced STAT1 tyrosine phosphorylation (Fig. 3). Phosphorylation of PKC on conserved serine and threonine residues of the catalytic domain is necessary for catalytic competence and proper intracellular localization.30,31 We thus determined whether PI3K and p38 MAPK are involved in the phosphorylation of PKC-α by IFN-γ. BMDM were incubated in the absence or in the presence of inhibitors for PI3K (Ly294002), p38 MAPK (SB203580), or JAK2 (AG-490) prior to the addition of 100 U/ml IFN-γ for 30 min, and PKC-α phosphorylation was assessed using Western blot analysis. As shown in Fig. 4, none of the inhibitors tested affected PKC-α phosphorylation in IFN-γ-stimulated BMDM. These results indicate that similarly to its IFN-γ-induced nuclear translocation, PKC-α phosphorylation is independent of JAK2 activity. However, PKC-α phosphorylation was also independent of PI3K and p38 MAPK, suggesting that IFN-γ-induced phosphorylation and nuclear translocation of PKC-α are mediated by distinct mechanisms.
Figure 1.
Interferon-γ (IFN-γ) induces protein kinase C-α (PKC-α) nuclear translocation in RAW264.7 cells and in bone marrow-derived macrophages (BMDM). RAW 264.7 cells transfected with PKC-α–green fluorescent protein (GFP) conjugates (a and c) or BMDM (b and d) were incubated in the absence (a and b) or presence (c and d) of 100 U/ml of IFN-γ for 2 hr and prepared for immunofluorescence analyses as described in the Materials and methods (red, F-actin; green, PKC-α; blue, DNA). NS, non-stimulated.
Figure 2.
Effect of pharmacological inhibitors on the interferon-γ (IFN-γ)-induced nuclear translocation of protein kinase C-α (PKC-α). Adherent RAW 264.7 cells were transfected with PKC-α–green fluorescent protein (GFP) conjugate, incubated with the various pharmacological inhibitors at the concentrations indicated in the Materials and methods for 1 hr, stimulated with 100 U/ml of IFN-γ for 2 hr and then prepared for confocal microscopy analysis. At least 30 PKC-α–GFP transfected cells were counted for each inhibitor tested and the results are representative of three independent experiments. The percentage of nuclear translocation represents the ratio of nuclear PKC-α-positive cells/total transfected cells. White bars, unstimulated control; black bars, IFN-γ-stimulated cells; hatched bars, cells treated with inhibitors prior to IFN-γ stimulation.
Figure 3.
Interferon-γ (IFN-γ)-induced phosphorylation of signal transducer and activator of transcription 1 (STAT1) on Tyr701 requires Janus activated kinase 2 (JAK2) activity. Adherent bone marrow-derived macrophages (BMDM) were incubated for 30 min in the absence or the presence of 100 or 250 μm AG-490 before the addition of 100 U/ml of IFN-γ for 15 min. Vehicle was 100% dimethyl sulphoxide (DMSO). Phosphorylation of STAT1 on Tyr701 (P-Tyr701) was determined by Western blotting, as described in the Materials and methods. Similar results were obtained in two independent experiments.
Figure 4.
Interferon-γ (IFN-γ)-induced phosphorylation of protein kinase C-α (PKC-α) is independent of Janus activated kinase 2 (JAK2), phosphatidylinositol 3-kinase (PI3K) and p38 mitogen-activated protein kinase (MAPK). Adherent bone marrow-derived macrophages (BMDM) were incubated in the absence or the presence of AG-490 (250 μm), Ly294002 (100 μm), or SB203580 (10 μm) for 1 hr prior to the addition of 100 U/ml of IFN-γ for 30 min. The presence of phosphorylated PKC-α (P- PKC-α; top panel) and total PKC-α (PKC-α total; bottom panel) in cell extracts was detected by Western blotting, as described in the Materials and methods. Similar results were obtained in two independent experiments.
Activation of PI3K and p38 MAPK by IFN-γ in BMDM
As IFN-γ-induced PKC-α nuclear translocation involves PI3K and p38 MAPK, we further characterized the mechanisms by which these kinases are activated by IFN-γ in BMDM. As shown in Fig. 5a, p38 MAPK phosphorylation was not significantly affected by the presence of inhibitors of either JAK2 (AG-490) or PI3K (Ly294002), indicating that these kinases are not involved in p38 MAPK activation by IFN-γ. To assess PI3K activation, we determined the phosphorylation status of Akt on serine 473 using Western blotting. Expectedly, IFN-γ-stimulated phosphorylation of Akt on serine 473 was inhibited by the PI3K inhibitor, Ly294002, but it was not affected by the presence either of the JAK2 inhibitor, AG-490, or of the p38 MAPK inhibitor, SB203580 (Fig. 5b). Collectively, our results indicate that in BMDM, activation of PI3K and p38 MAPK by IFN-γ occurs through distinct mechanisms, independently of JAK2 activity.
Figure 5.
Interferon-γ (IFN-γ) induces p38 mitogen-activated protein kinase (MAPK) and Akt phosphorylation in bone marrow-derived macrophages (BMDM). Adherent BMDM were incubated in the absence or the presence of AG-490 (250 μm), Ly294002 (100 μm), or SB203580 (10 μm) for 1 hr before the addition of 100 U/ml of IFN-γ for 5 min (a) or 30 min (b). Controls consisted of BMDM incubated for 15 min with 100 ng/ml of lipopolysaccharide (LPS). Cell extracts were analyzed by Western blotting for the presence of (a) phosphorylated (P-p38; top panel) and total p38 MAPK (bottom panel) and (b) phosphorylated (P-Akt; top panel) and total Akt (bottom panel). Similar results were obtained in two independent experiments.
IFN-γ-induced type IV CIITA and MHC II gene expression require PI3K but not p38 MAPK
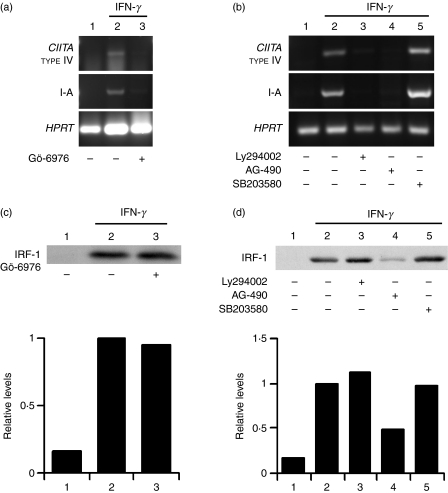
We previously reported that PKC-α activity regulates IFN-γ-induced CIITA and MHC II gene expression by modulating the ability of IRF-1 to transactivate the CIITA promoter IV.20 Consistent with these observations, IFN-γ-stimulated type IV CIITA and MHC II (I-A) gene expression were strongly inhibited by the PKC-α inhibitor, Gö-6976 (Fig. 6a). We next determined whether PI3K and p38 MAPK contribute to the expression of type IV CIITA and MHC II in IFN-γ-stimulated BMDM. To this end, we compared the effect of the PI3K inhibitor, Ly294002, and the p38 MAPK inhibitor, SB203580, on IFN-γ-induced type IV CIITA and MHC II gene expression. We included the JAK2 inhibitor, AG-490, as a control. As expected, inhibition of JAK2 completely abrogated the expression of both type IV CIITA and MHC II. Similar results were obtained with the PI3K inhibitor, Ly294002, indicating that PI3K plays an important role in the regulation of IFN-γ-induced type IV CIITA and MHC II gene expression (Fig. 6b). By contrast, as previously reported,19 the p38 MAPK inhibitor, SB203580, had no significant effect on type IV CIITA and MHC II gene expression.
Figure 6.
Interferon-γ (IFN-γ)-induced class II transactivator (CIITA) and major histocompatibility complex (MHC) class II (I-A) gene expression require phosphatidylinositol 3-kinase (PI3K), but not p38 mitogen-activated protein kinase (MAPK) (a and b). Bone marrow-derived macrophages (BMDM) were incubated for 1 hr in the absence or the presence of Gö-6976 (1 μm) (a), Ly294002 (100 μm), AG-490 (250 μm) or SB203580 (10 μm) (b), and stimulated for 6 hr with 100 U/ml of IFN-γ. Total RNA was extracted and subjected to reverse transcription–polymerase chain reaction (RT-PCR) using specific primers for type IV CIITA, MHC II (I-A) and Hypoxanthine-guanine phosphoribosyl transferase (HPRT) (c and d). IFN-γ-induced IFN regulatory factor-1 (IRF-1) protein expression is independent of protein kinase C (PKC), PI3K and p38 MAPK. Adherent BMDM were incubated for 1 h with or without Gö-6976 (1 μm) (c), Ly294002 (100 μm), AG-490 (250 μm) or SB203580 (10 μm) (d), and stimulated for 4 hr with IFN-γ. IRF-1 expression was assessed by Western blotting, as described in the Materials and methods. Similar results were obtained in two independent experiments.
Together with STAT1, IRF-1 plays an essential role in the regulation of IFN-γ-induced type IV CIITA expression.32,33 It was thus of interest to determine whether inhibition of type IV CIITA expression by the PKC-α and the PI3K inhibitors was the consequence of defective IRF-1 expression. Consistent with its dependence on the JAK-STAT pathway,13,34 IRF-1 expression was inhibited by the JAK2 inhibitor, AG-490 (Fig. 6d). However, our results indicate that PKC-α, PI3K and p38 MAPK are not involved in the regulation of IFN-γ-induced IRF-1 expression (Fig. 6c,d).
IFN-γ induces the phosphorylation of STAT1 on serine 727 and its association with PKC-α
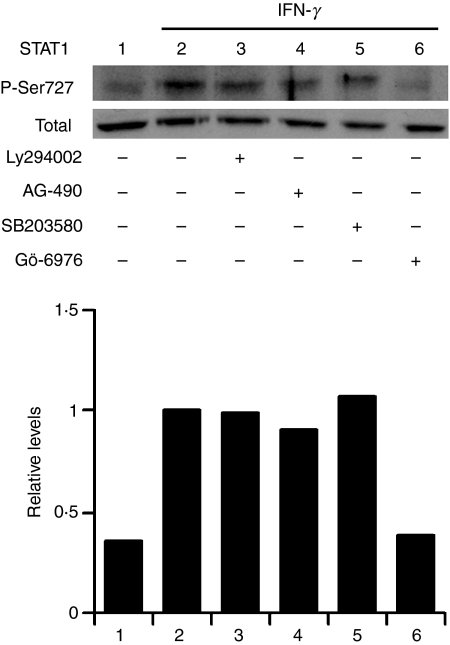
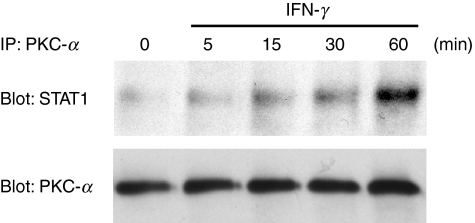
In addition to tyrosine phosphorylation, the complete transcriptional activity of STAT1 requires the phosphorylation of serine 727, and members of the PKC family have been implicated in this response.10,14,25 To verify the possible requirement for PKC-α, we assessed, using Western blotting, the IFN-γ-induced phosphorylation of STAT1 on serine 727 in the presence or the absence of the PKC-α inhibitor, Gö-6976. We also included the PI3K inhibitor, Ly294002, the p38 MAPK inhibitor, SB203580, and the JAK2 inhibitor, AG-490. As shown in Fig. 7, IFN-γ-induced phosphorylation of STAT1 on serine 727 in BMDM was impaired by the PKC-α inhibitor Gö-6976 but remained unaffected by the inhibitors of PI3K, p38 MAPK and JAK2. The finding that Gö-6976 inhibited the IFN-γ-stimulated phosphorylation of STAT1 on serine 727 led us to determine whether PKC-α associates with STAT1 in response to IFN-γ. BMDM were stimulated with IFN-γ and, at various time-points, cells lysates were immunoprecipitated with an antiserum against PKC-α. Immunoprecipitated proteins were separated by SDS–PAGE and the presence of STAT1 was assessed by Western blotting. As shown in Fig. 8, PKC-α and STAT1 associated in an IFN-γ-dependent manner in BMDM.
Figure 7.
Interferon-γ (IFN-γ)-induced phosphorylation of signal transducer and activator of transcription 1 (STAT1) on serine 727 requires the activity of protein kinase C-α (PKC-α) but remains unaffected by the inhibition of phosphatidylinositol 3-kinase (PI3K) and p38 mitogen-activated protein kinase (MAPK). Adherent bone marrow-derived macrophages (BMDM) were incubated for 30 min in the absence or the presence of Ly294002 (100 μm), AG-490 (250 μm), SB203580 (10 μm), or Gö-6976 (1 μm) before the addition of 100 U/ml of IFN-γ for 30 min. Phosphorylation of STAT1 on serine 727 was determined by Western blotting, as described in the Materials and methods. Similar results were obtained in two independent experiments.
Figure 8.
Interferon-γ (IFN-γ) induces the association of protein kinase C-α (PKC-α) with signal transducer and activator of transcription 1 (STAT1). Adherent bone marrow-derived macrophages (BMDM) were incubated in the absence or presence of 100 U/ml of IFN-γ for the indicated periods of time. PKC-α was immunoprecipitated and the presence of STAT1 in immunoprecipitates was determined by Western blot analysis, as described in the Materials and methods. The blot was stripped and reprobed with an anti-PKC-α immunoglobulin (IgG2b) to ensure that equal amounts of PKC-α were immunoprecipitated in each sample. Similar results were obtained in two independent experiments.
Discussion
Macrophage responses to IFN-γ are mediated, to a large extent, by the JAK-STAT pathway.6,7 Nevertheless, the fact that IFN-γ regulates the expression of a large number of genes in cells lacking STAT1, and activates PI3K and ERK1/2 in cells deficient in either JAK1 or JAK2,7,10 is consistent with the notion that additional signalling pathways contribute to the modulation of cellular responses by IFN-γ.10,35 The participation of PKC isoenzymes in the regulation of IFN-γ-induced responses has been reported in various systems.14–18,20 In macrophages, we previously showed that PKC-α translocates to the nucleus in response to IFN-γ, and that PKC-α activity regulates IFN-γ-induced MHC II expression by modulating the ability of IRF-1 to transactivate the CIITA promoter IV.20 In this study, we found that both PI3K and p38 MAPK are involved in the activation of PKC-α in macrophages stimulated with IFN-γ, and we further characterized the roles of these kinases in the regulation of IFN-γ-induced responses in macrophages.
Activation of PI3K by both type I and type II IFNs has been reported in several cell types.8,10,14,17,36,37 One of the main consequences of PI3K activation is the phosphorylation of STAT1 on serine 727, which has been shown to be mediated by either PKC-δ in human acute promyelocytic leukemia cells 14 or PKC-ε in rat mesanglial cells.17 In BMDM, we found that IFN-γ-induced STAT1 serine 727 phosphorylation required PKC-α but was independent of PI3K. This is consistent with the finding that PI3K had no effect on the phosphorylation of PKC-α. The finding that PI3K was required for IFN-γ-stimulated nuclear translocation of PKC-α suggests the existence of at least two distinct steps in the regulation of PKC-α activation in IFN-γ-stimulated BMDM. Hence, the initial step of PKC-α maturation (phosphorylation)30 is regulated by a yet-unidentified pathway(s), whereas the targeting step (nuclear localization) requires PI3K.
Although both type I and type II IFNs activate p38 MAPK, the role of this kinase in the modulation of IFN responses remains poorly understood. Whereas p38 MAPK is required for STAT1-driven gene expression in response to IFN-α and IFN-β, it plays an important role in the regulation of the expression of chemokines and cytokines in IFN-γ-stimulated macrophages.11,19 Consistent with previous reports,19,38,39 we found that, in BMDM, p38 MAPK is not required for IFN-γ-induced phosphorylation of STAT1 on serine 727 or for the expression of IRF-1, CIITA and MHC II. These events are thus clearly distinct from IFN-γ-induced chemokine and cytokine expression. As p38 MAPK was involved in the nuclear translocation of PKC-α, it is tempting to speculate that PKC-α may participate in the regulation of chemokine and cytokine expression. As p38 MAPK was not involved in the initial step of PKC-α activation (phosphorylation) by IFN-γ, the mechanism(s) by which p38 MAPK modulates PKC-α nuclear translocation remains to be identified.
We previously reported that PKC-α activity regulates IFN-γ-induced MHC II expression by modulating the ability of IRF-1 to transactivate the CIITA promoter IV.20 Consistent with these previous findings, IFN-γ-stimulated type IV CIITA and MHC II expression was strongly inhibited by Gö-6976. Our data also indicated that whereas PI3K and p38 MAPK regulated IFN-γ-induced nuclear translocation of PKC-α, they had no significant impact on IFN-γ-stimulated PKC-α phosphorylation and on STAT1-mediated IRF-1 expression. In the light of these observations, it was of interest to determine the contribution of these pathways to the expression of type IV CIITA and MHC II in IFN-γ-stimulated BMDM. As recently shown,19 inhibition of p38 MAPK had no significant effect on the gene expression of either CIITA or MHC II. By contrast, inhibition of PI3K strongly impaired the expression of type IV CIITA and MHC II genes, indicating an important role for PI3K in the modulation of IFN-γ-induced responses in BMDM. Taken together, these data suggest that although PKC-α activity is essential, its nuclear translocation is not required for IFN-γ-stimulated type IV CIITA and MHC II gene expression, and point to the existence of yet-unidentified regulatory mechanisms involving PI3K. Clearly, additional investigation will be required to understand in greater detail the regulation of CIITA expression.
In contrast to the expression of IRF-1, CIITA and MHC II, which depends on the JAK-STAT pathway, activation of PKC-α, PI3K and p38 MAPK occurred independently of JAK2 in IFN-γ-stimulated BMDM. These findings raise the issue of how IFN-γ activates signalling pathways independently of JAK. A recent study provided evidence that IFN-γ induces a rapid physical association between IFN-γ receptor 1 and the adaptor protein MyD88, as well as the formation of a signalosome containing p38 MAPK, mixed-lineage kinase (MLK) and c-Jun N-terminal kinase-interacting protein (JIP)-1.11 A similar mechanism may account for the activation of PI3K, which was found to associate with MyD88 in LPS-stimulated macrophages.40 Whether this is also the case for the activation of PKC-α by IFN-γ remains to be demonstrated. Clearly, additional investigation will be required to address the potential role of MyD88 in mediating the activation of PKC-α and PI3K in macrophages stimulated by IFN-γ.
The finding that IFN-γ-stimulated phosphorylation of STAT1 on serine 727 is inhibited by Gö6976 suggests that PKC-α acts as a kinase for this phosphorylation event and is consistent with the observation that IFN-γ induces the association of PKC-α with STAT1. A similar role for PKC-δ has been reported in acute promyelocytic leukemia cells, where PKC-δ was shown to associate with and to phosphorylate STAT1.14 In contrast to the majority of nuclear proteins, PKC isoenzymes lack a canonical nuclear localization signal,41 indicating that other mechanisms are involved in the transport process. As phosphorylated STAT1 dimers translocate to the nucleus, it is possible that PKC-α uses STAT1 as a shuttle to access the nucleus.
Acknowledgments
We are grateful to Marcel Desrosiers and Dr Robert Lodge for their help with confocal microscopy and immunofluorescence. This work was supported by Canadian Institutes of Health Research grant MOP-12933. Albert Descoteaux is the holder of a Canada Research Chair and was a Chercheur-boursier from the FRSQ. Tamsir O. Diallo was partly supported by a postdoctoral fellowship from the Fondation Pasteur/Fondation Armand-Frappier. Christine Matte was supported by a studentship from the Canadian Institutes of Health Research.
Disclosures
The authors declare no conflict of interest or financial interests.
References
- 1.Darnell JE., Jr STATs and gene regulation. Science. 1997;277:1630–5. doi: 10.1126/science.277.5332.1630. [DOI] [PubMed] [Google Scholar]
- 2.Stark GR, Kerr IM, Williams BR, Silverman RH, Schreiber RD. How cells respond to interferons. Annu Rev Biochem. 1998;67:227–64. doi: 10.1146/annurev.biochem.67.1.227. [DOI] [PubMed] [Google Scholar]
- 3.Steimle V, Siegrist CA, Mottet A, Lisowska-Grospierre B, Mach B. Regulation of MHC class II expression by interferon-gamma mediated by the transactivator gene CIITA. Science. 1994;265:106–9. doi: 10.1126/science.8016643. [DOI] [PubMed] [Google Scholar]
- 4.Cullell-Young M, Barrachina M, Lopez-Lopez C, Gonalons E, Lloberas J, Soler C, Celada A. From transcription to cell surface expression, the induction of MHC class II I-A alpha by interferon-gamma in macrophages is regulated at different levels. Immunogenetics. 2001;53:136–44. doi: 10.1007/s002510100312. [DOI] [PubMed] [Google Scholar]
- 5.Kroger A, Koster M, Schroeder K, Hauser H, Mueller PP. Activities of IRF-1. J Interferon Cytokine Res. 2002;22:5–14. doi: 10.1089/107999002753452610. [DOI] [PubMed] [Google Scholar]
- 6.Ramana CV, Gil MP, Han Y, Ransohoff RM, Schreiber RD, Stark GR. Stat1-independent regulation of gene expression in response to IFN-gamma. Proc Natl Acad Sci USA. 2001;98:6674–9. doi: 10.1073/pnas.111164198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gil MP, Bohn E, O’Guin AK, Ramana CV, Levine B, Stark GR, Virgin HW, Schreiber RD. Biologic consequences of Stat1-independent IFN signaling. Proc Natl Acad Sci USA. 2001;98:6680–5. doi: 10.1073/pnas.111163898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Navarro A, Anand-Apte B, Tanabe Y, Feldman G, Larner AC. A PI-3 kinase-dependent, Stat1-independent signaling pathway regulates interferon-stimulated monocyte adhesion. J Leukoc Biol. 2003;73:540–5. doi: 10.1189/jlb.1002508. [DOI] [PubMed] [Google Scholar]
- 9.Platanias LC. Mechanisms of type-I- and type-II-interferon-mediated signalling. Nat Rev Immunol. 2005;5:375–86. doi: 10.1038/nri1604. [DOI] [PubMed] [Google Scholar]
- 10.Nguyen H, Ramana CV, Bayes J, Stark GR. Roles of phosphatidylinositol 3-kinase in interferon-gamma-dependent phosphorylation of STAT1 on serine 727 and activation of gene expression. J Biol Chem. 2001;276:33361–8. doi: 10.1074/jbc.M105070200. [DOI] [PubMed] [Google Scholar]
- 11.Sun D, Ding A. MyD88-mediated stabilization of interferon-gamma-induced cytokine and chemokine mRNA. Nat Immunol. 2006;7:375–81. doi: 10.1038/ni1308. [DOI] [PubMed] [Google Scholar]
- 12.Yao Y, Xu Q, Kwon MJ, Matta R, Liu Y, Hong SC, Chang CH. ERK and p38 MAPK signaling pathways negatively regulate CIITA gene expression in dendritic cells and macrophages. J Immunol. 2006;177:70–6. doi: 10.4049/jimmunol.177.1.70. [DOI] [PubMed] [Google Scholar]
- 13.Ramana CV, Gil MP, Schreiber RD, Stark GR. Stat1-dependent and -independent pathways in IFN-gamma-dependent signaling. Trends Immunol. 2002;23:96–101. doi: 10.1016/s1471-4906(01)02118-4. [DOI] [PubMed] [Google Scholar]
- 14.Deb DK, Sassano A, Lekmine F, et al. Activation of protein kinase C delta by IFN-gamma. J Immunol. 2003;171:267–73. doi: 10.4049/jimmunol.171.1.267. [DOI] [PubMed] [Google Scholar]
- 15.Chang YJ, Holtzman MJ, Chen CC. Interferon-gamma-induced epithelial ICAM-1 expression and monocyte adhesion. Involvement of protein kinase C-dependent c-Src tyrosine kinase activation pathway. J Biol Chem. 2002;277:7118–26. doi: 10.1074/jbc.M109924200. [DOI] [PubMed] [Google Scholar]
- 16.Mazzi P, Donini M, Margotto D, Wientjes F, Dusi S. IFN-gamma induces gp91phox expression in human monocytes via protein kinase C-dependent phosphorylation of PU.1. J Immunol. 2004;172:4941–7. doi: 10.4049/jimmunol.172.8.4941. [DOI] [PubMed] [Google Scholar]
- 17.Choudhury GG. A linear signal transduction pathway involving phosphatidylinositol 3-kinase, protein kinase cepsilon, and MAPK in mesangial cells regulates interferon-gamma-induced STAT1alpha transcriptional activation. J Biol Chem. 2004;279:27399–409. doi: 10.1074/jbc.M403530200. [DOI] [PubMed] [Google Scholar]
- 18.Ivaska J, Bosca L, Parker PJ. PKCepsilon is a permissive link in integrin-dependent IFN-gamma signalling that facilitates JAK phosphorylation of STAT1. Nat Cell Biol. 2003;5:363–9. doi: 10.1038/ncb957. [DOI] [PubMed] [Google Scholar]
- 19.Valledor AF, Sanchez-Tillo E, Arpa L, Park JM, Caelles C, Lloberas J, Celada A. Selective roles of MAPKs during the macrophage response to IFN-gamma. J Immunol. 2008;180:4523–9. doi: 10.4049/jimmunol.180.7.4523. [DOI] [PubMed] [Google Scholar]
- 20.Giroux M, Schmidt M, Descoteaux A. IFN-gamma-induced MHC class II expression: transactivation of class II transactivator promoter IV by IFN regulatory factor-1 is regulated by protein kinase C-alpha. J Immunol. 2003;171:4187–94. doi: 10.4049/jimmunol.171.8.4187. [DOI] [PubMed] [Google Scholar]
- 21.Mellor H, Parker PJ. The extended protein kinase C superfamily. Biochem J. 1998;332:281–92. doi: 10.1042/bj3320281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ohno S, Nishizuka Y. Protein kinase C isotypes and their specific functions: prologue. J Biochem (Tokyo) 2002;132:509–11. doi: 10.1093/oxfordjournals.jbchem.a003249. [DOI] [PubMed] [Google Scholar]
- 23.Wen Z, Zhong Z, Darnell JE., Jr Maximal activation of transcription by Stat1 and Stat3 requires both tyrosine and serine phosphorylation. Cell. 1995;82:241–50. doi: 10.1016/0092-8674(95)90311-9. [DOI] [PubMed] [Google Scholar]
- 24.Kwon MJ, Yao Y, Walter MJ, Holtzman MJ, Chang CH. Role of PKCdelta in IFN-gamma-inducible CIITA gene expression. Mol Immunol. 2007;44:2841–9. doi: 10.1016/j.molimm.2007.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Venkatesan BA, Mahimainathan L, Ghosh-Choudhury N, Gorin Y, Bhandari B, Valente AJ, Abboud HE, Choudhury GG. PI 3 kinase-dependent Akt kinase and PKCepsilon independently regulate interferon-gamma-induced STAT1alpha serine phosphorylation to induce monocyte chemotactic protein-1 expression. Cell Signal. 2006;18:508–18. doi: 10.1016/j.cellsig.2005.05.022. [DOI] [PubMed] [Google Scholar]
- 26.Descoteaux A, Matlashewski G. Regulation of tumor necrosis factor gene expression and protein synthesis in murine macrophages treated with recombinant tumor necrosis factor. J Immunol. 1990;145:846–53. [PubMed] [Google Scholar]
- 27.Flandin JF, Chano F, Descoteaux A. RNA interference reveals a role for TLR2 and TLR3 in the recognition of Leishmania donovani promastigotes by interferon-gamma-primed macrophages. Eur J Immunol. 2006;36:411–20. doi: 10.1002/eji.200535079. [DOI] [PubMed] [Google Scholar]
- 28.Prive C, Descoteaux A. Leishmania donovani promastigotes evade the activation of mitogen-activated protein kinases p38, c-Jun N-terminal kinase, and extracellular signal-regulated kinase-1/2 during infection of naive macrophages. Eur J Immunol. 2000;30:2235–44. doi: 10.1002/1521-4141(2000)30:8<2235::AID-IMMU2235>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- 29.Quittau-Prevostel C, Delaunay N, Collazos A, Vallentin A, Joubert D. Targeting of PKCalpha and epsilon in the pituitary: a highly regulated mechanism involving a GD(E)E motif of the V3 region. J Cell Sci. 2004;117:63–72. doi: 10.1242/jcs.00832. [DOI] [PubMed] [Google Scholar]
- 30.Shirai Y, Saito N. Activation mechanisms of protein kinase C: maturation, catalytic activation, and targeting. J Biochem (Tokyo) 2002;132:663–8. doi: 10.1093/oxfordjournals.jbchem.a003271. [DOI] [PubMed] [Google Scholar]
- 31.Newton AC. Protein kinase C: structural and spatial regulation by phosphorylation, cofactors, and macromolecular interactions. Chem Rev. 2001;101:2353–64. doi: 10.1021/cr0002801. [DOI] [PubMed] [Google Scholar]
- 32.Meraz MA, White JM, Sheehan KC, et al. Targeted disruption of the Stat1 gene in mice reveals unexpected physiologic specificity in the JAK-STAT signaling pathway. Cell. 1996;84:431–42. doi: 10.1016/s0092-8674(00)81288-x. [DOI] [PubMed] [Google Scholar]
- 33.Muhlethaler-Mottet A, Di Berardino W, Otten LA, Mach B. Activation of the MHC class II transactivator CIITA by interferon-gamma requires cooperative interaction between Stat1 and USF-1. Immunity. 1998;8:157–66. doi: 10.1016/s1074-7613(00)80468-9. [DOI] [PubMed] [Google Scholar]
- 34.Harada H, Fujita T, Miyamoto M, Kimura Y, Maruyama M, Furia A, Miyata T, Taniguchi T. Structurally similar but functionally distinct factors, IRF-1 and IRF-2, bind to the same regulatory elements of IFN and IFN-inducible genes. Cell. 1989;58:729–39. doi: 10.1016/0092-8674(89)90107-4. [DOI] [PubMed] [Google Scholar]
- 35.Briscoe J, Rogers NC, Witthuhn BA, et al. Kinase-negative mutants of JAK1 can sustain interferon-gamma-inducible gene expression but not an antiviral state. EMBO J. 1996;15:799–809. [PMC free article] [PubMed] [Google Scholar]
- 36.Platanias LC, Fish EN. Signaling pathways activated by interferons. Exp Hematol. 1999;27:1583–92. doi: 10.1016/s0301-472x(99)00109-5. [DOI] [PubMed] [Google Scholar]
- 37.Kaur S, Uddin S, Platanias LC. The PI3’ kinase pathway in interferon signaling. J Interferon Cytokine Res. 2005;25:780–7. doi: 10.1089/jir.2005.25.780. [DOI] [PubMed] [Google Scholar]
- 38.Li Y, Sassano A, Majchrzak B, et al. Role of p38alpha Map kinase in Type I interferon signaling. J Biol Chem. 2004;279:970–9. doi: 10.1074/jbc.M309927200. [DOI] [PubMed] [Google Scholar]
- 39.Ramsauer K, Sadzak I, Porras A, Pilz A, Nebreda AR, Decker T, Kovarik P. p38 MAPK enhances STAT1-dependent transcription independently of Ser-727 phosphorylation. Proc Natl Acad Sci USA. 2002;99:12859–64. doi: 10.1073/pnas.192264999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ojaniemi M, Glumoff V, Harju K, Liljeroos M, Vuori K, Hallman M. Phosphatidylinositol 3-kinase is involved in Toll-like receptor 4-mediated cytokine expression in mouse macrophages. Eur J Immunol. 2003;33:597–605. doi: 10.1002/eji.200323376. [DOI] [PubMed] [Google Scholar]
- 41.Nakashima S. Protein kinase C alpha (PKC alpha): regulation and biological function. J Biochem (Tokyo) 2002;132:669–75. doi: 10.1093/oxfordjournals.jbchem.a003272. [DOI] [PubMed] [Google Scholar]