Abstract
The fruit fly, Drosophila melanogaster, is an excellent model system that has a vast set of molecular tools and mutants to dissect the genetic pathways that are responsible for the normal and abnormal cardiac function. While the majority of studies have focused on heart development in the Drosophila embryo, attention has recently focused on the structure and function of the adult fly heart as a model of human heart failure. Here we review strategies to identify novel genes and pathways that cause or modify dilated cardiomyopathy in adult Drosophila.
Introduction
Identifying genes that affect cardiac function is critical in understanding the complex biology that is responsible for dilated cardiomyopathies and human heart failure. Model systems of disease are key components to achieve this goal. In fact, the wealth of information gained from efforts to sequence the genomes of several species now provides the tools to evaluate evolutionarily conserved signaling pathways. For nearly a hundred years, Drosophila has been studied as a model system and has a unique armamentarium of powerful tools to identify mutant genes.[1] These reagents underlie the utility of Drosophila as a model to identify “disease-causing” or “disease-modifying” genes that are responsible for dilated cardiomyopathy and heart failure.
Advantages of the Drosophila as a genetic model
Several model systems based on the mouse, zebrafish, and fruitfly have been developed to investigate the genes that contribute to cardiovascular disease (Table 1). There are numerous mouse models of cardiovascular diseases that have been well-characterized. In general, mouse models of heart disease are based on transgenically-targeted lines and are currently amenable to physiologic studies that include echocardiography and invasive hemodynamic monitoring. More recently, zebrafish has been recognized as a model of cardiovascular disease. For example, the zebrafish is useful for investigating conduction abnormalities that lead to the development of arrhythmias, vascular abnormalities, and the molecular and cellular signaling pathways that lead to heart regeneration after injury [2, 3]. We will focus on Drosophila as a model system of cardiovascular disease.
Table 1.
Comparison of animal models of cardiovascular disease.
| |
Fly |
Fish |
Mouse |
|---|---|---|---|
|
Pros |
Vast array of genetic tools including: - P-element insertion Mutants - Genomic deficiency mutants - siRNA transgenic stocks - chemically and radiation induced mutants Well developed bipartite Gal4/UAS transgene expression system Extremely well annotated genomic databases (flybase.org) and publically available fly stocks (Bloomington Collection) Low cost maintenance Short generation time |
Two chambered heart with distinct atrium and ventricle Well developed genetic tools for generating transgenic mutants and knockdowns (morpholino) Heart has regenerative capacity Transgenic systems |
Four chambered heart that is highly similar to the human heart Well developed transgenic tools Easily amenable to physiologic studies: - Transaortic banding/pressure overload; coronary artery ligation - Ischemic modeling, intracardiac hemodynamic measurements (pressure-volume loops) Amenable to QTL mapping and SNP analysis Publically available stocks (Jackson Labs) and several well-annotated databases (Ensembl, GFN Symatlas) |
|
Cons |
Single chamber heart Ultrastucturally similar to mammalian myocardium but has perforated Z-discs No coronary arteries |
Difficulty imaging cardiac function (ventricle is highly trabeculated ventricle) Cost of maintaining stocks Difficulty mapping newly isolated mutants |
Cost of maintaining stocks Breeding time |
| Best Use of Model | Genetic screens to identify genes and pathways that affect cardiac function |
Genetic screens Model of regeneration after cardiac injury |
Transgenic analysis and physiologic experiments |
Drosophila genetics has a rich history dating back to the early 1900's with the work by Morgan, Sturtevant, and Bridges to map the genetic basis of physical traits [1]. Drosophila has several distinct advantages as a model organism including a short generation time facilitating studies of population genetics, an inexpensive cost associated with maintaining of stocks, and the presence of balancer chromosomes that assist with the chromosomal mapping of traits and the propagation of recessive, often lethal, mutations. Moreover, the high degree of homology to the mammalian genome and the numerous genetic mutants available for study makes the fly a wonderful model system to dissect the genetic basis of disease.
The past decades have witnessed tremendous growth in the resources available to the Drosophila community. First, large collections of Drosophila stocks are available to investigate specific phenotypes and identify genetic mutations. These stocks include, but not limited to: ∼19,000 P-elements (“foreign pieces of DNA”) that are inserted throughout the fly genome that potentially disrupt gene function[4]; collections of molecularly-defined genomic deficiencies[5, 6]; chemically and radiation-induced mutants; and a collection of ∼22,000 transgenic fly mutants that harbor specific small interfering RNAs (si-RNA) have been generated to examine the effects of tissue specific gene knockdown [7]. Second, the Drosophila genome has been successfully sequenced and is extremely well-annotated (www.ensembl.org or www.flybase.org). Drosophila melanogaster has five chromosomes (X, 2L, 2R, 3L, 3R, and 4) that contains ∼125 million basepairs of DNA encoding ∼14,000 predicted and confirmed genes[8-10]. Third, single nucleotide polymorphism (SNP) maps have been generated to facilitate the mapping of strains and identification of modifier genes [10]. Fourth, the Drosophila genetics community has an extremely well-annotated bioinformatic database (www.flybase.org) and repository of publically-available mutants (Bloomington Drosophila Stock Center at Indiana University (www.flystocks.bio.indiana.edu). Fifth, tissue and temporal specific transgene expression is readily achieved through a bipartite Gal4-driver and yeast upstream activating (UAS)-transgene system. Additionally, knockout and gene replacement strategies based on homologous recombination have been established. Finally, strategies have been developed and successfully used to conduct large genome-wide forward genetic screens including suppressor and enhancer screens in the context of a sensitized genetic background [11]. These resources make Drosophila ideally suited as a genetic model to investigate human disease.
The adult Drosophila cardiac system
The embryonic dorsal vessel is the precursor of the adult fly heart and develops through an orchestrated series of morphogenic signals within the mesoderm. Drosophila embryonic heart development requires an interplay among critical transcription factors that have been well described (see reviews by Cripps, Olsen, Bodmer, and Frasch [12-15]). Many of the signaling molecules and transcription factors are conserved among species, including humans. For example, the mutant, tinman, lacks a properly formed dorsal vessel in Drosophila embryos and mutations in the human orthologue, NKX2.5, are associated with human congenital heart disease.[16]
The fly heart undergoes a significant morphogenetic change during development from the late larval state through the pupal state and subsequent eclosion of the adult fly from the pupal case. Molina and Cripps elegantly described the cellular changes that occur as the larval heart transitions to produce a set of functionally distinct muscle cells that become the adult circulatory system. Subsequent studies by Perrin's laboratory have described edycosone-mediated alterations that occur in specific gene transcription expression during the transition form the larval to adult heart [17-19]. The adult fly has an open circulatory system with the cardiac chamber located directly beneath the cuticle along the dorsal aspect between the thorax and the abdomen [20]. Electron microscopy of the adult Drosophila heart demonstrated several similarities compared to mammalian myocardium with the exception that the Z-disc is perforated, consistent with the supracontractile nature of the insect heart [21].
The innervations of the adult fly heart is complex and was elegantly studied by Dulcis and Levine [22]. In the larva, the dorsal vessel lacks innervations and is appears to have a completely myogenic cardiac impulse. During metamorphosis the myocardium develops neuronal inputs via a process that establishes bilateral neuronal connections in a segmental distribution along the cardiac chamber and alary muscles. The longitudinal muscles of the conical chamber have unique synaptic structures that have been suggested to provide a mechanism that is responsible for the retrograde heart beating that is observed in the adult fly. Recently, Wasserthal has performed an series of studies that examined the ultrastructure of the adult fly heart and demonstrated that the heart has a set of first set of valves, called ostia, that connect an extracardiac venous space to the heart in the A1 segment. The fly heart also has ostia located along the abdominal aspects of the circulatory system in the A2-A5 segments. Furthermore, the use of an infrared-light beam and a linear optosensor chip system to study the contractile properties of the adult Drosophila heart had supported the concept that the fly heart has both anterograde and retrograde beating. During retrograde beating, hemolymph flows through the first set of ostia from the venous space into the cardiac chamber and during anterograde beating, the cardiac chamber receives hemolymph from all the ostia and then is pumped forward through the aorta [23].
Therefore, the evolutionarily conserved signaling pathways and physical properties of the Drosophila heart are well-suited to identify genes that may be involved in vertebrate cardiac function.
Imaging strategies to evaluate the adult Drosophila heart
The ability to accurately image the adult Drosophila heart is critical for the use of the vast resources that are available in Drosophila genetics to design screens and identify genes that are important in cardiac chamber structure and function. The phenotyping strategies are represented in Table 2 and can be generally divided into an examination of structure or function.
Table 2.
Comparison of Drosophila cardiac phenotyping methods
| |
Whole mount staining/histology |
Edge-effect microscopy |
Optical Coherence Tomography |
|---|---|---|---|
|
Pros |
-Detailed structural information -Gene expression |
-Detailed information about heart rate and periodicity |
-Performed on awake, adult specimens -Non-invasive, non-destructive -Serial measurements from same fly -provides images that are similar to echocardiography in humans -Can visualize the heart during the entire cardiac cycle -Can readily visualize the endocardium and calculated EDD, ESD without significant post-image processing -Can image individual flies quickly (∼3 minutes per fly) |
| Cons |
-Dissected/fixed specimens -Does not provide information about cardiac function |
-Requires significant post-imaging analysis -Requires assumptions about the identity of the endocardial edge thus making evaluation of EDD/ESD difficult -Usually performed with dissected specimens |
-Requires custom built microscopy system -Resolution to ∼10-20 microns |
Structural imaging consists of whole mount and immunohistochemical staining. These studies form the basis for the examination of the embryonic dorsal vessel and are used to examine the processes involved in mesodermal signaling during dorsal vessel formation in the developing embryo. Whole mount and immunohistochemical staining has been applied to large scale genetic screens to identify genes that disrupt dorsal vessel development. For example, Kim et. al. have conducted a genomic screen for cardiac genes using RNA interference with a phenotyping strategy based on lacZ staining of the dorsal vessel cardial prescursor cells in embryo whole mounts [24]. In general, these approaches provide significant insight into organ anatomy and the temporal relationships of gene expression.
Several functionally-based imaging strategies have been developed to examine the Drosophila heart. Cardiac chamber edge-detection methods using Nomarski differential interference contrast (DIC) microscopy were initially used by Paternostro et.al. to examine heart rate in the adult fly [25]. DIC microscopy does not require staining of the sample and uses a polarized light source to detect differences in the refractive index of the specimen thereby allowing visualization of the external edges of the fly heart. This method provides information about heart rate under baseline conditions and in response to external electric stimulation. The authors also employed confocal microscopy to examine cardiac function and estimate cardiac chamber dimensions in adult transgenic flies expressing GFP under control of an actin promoter [25].
To investigate the contractile function and electrophysiologic properties of the Drosophila circulatory system, the Bodmer laboratory has developed several approaches [26-32]. These approaches include the development of an edge-detection imaging strategy in pupae and adult flies. Furthermore, Bodmer and collegues have conducted elegant studies to examine age-related affects on heart rate and response to external stress [30-32]. These methods examine the ability of the fly heart to respond to stimulation from an external electric field and define heart failure as the percentage of flies that either fibrillate or arrest immediately after stimulation. Using this approach, the Bodmer laboratory has identified alterations in insulin-like signaling peptides and mutations in the insulin-IFG receptors as a regulatory mechanism that control age-related cahngesin Drsophila cardiac function [32].
More recently, the Bodmer laboratory has developed a whole-field optical analysis strategy to analyze adult Drosophila cardiac function [28]. The fly circulatory system is examined in specimens that have all internal organs removed except for the heart and abdominal fat and bathed in artificial hemolymph. Images of cardiac contractions are obtained via a high-speed digital camera and processed to provide an assessment of changes in chamber size during systole and diastole as well as heart rate and rhythm. This strategy has been successfully used to identify mutations in the KCNQ potassium channel that cause cardiac arrhythmias and suggests that age-related changes in cardiac chamber relaxation contribute to the cardiac dysfunction observed in older flies [28].
Optical coherence tomography (OCT) has been successfully employed to examine the cardiac function in awake, adult Drosophila [21]. OCT is a non-destructive, non-invasive imaging strategy that is similar in principle to echocardiography in humans. OCT is based on the reflectivity of near-infrared light and has a special resolution of ∼20 microns [21, 33]. Since OCT imaging is non-invasive, serial OCT measurements can be performed on the same fly to examine changes in cardiac function over time. OCT directly visualizes the cardiac chamber throughout the entire cardiac cycle and therefore provides direct measurements of end-diastolic and end-systolic dimension as well as heart rate. (Figure 1) Due to the non-invasive properties of OCT, consecutive imaging can be serially on non-anesthetized, non-dissected individual flies. Furthermore, OCT imaging can provide a strategy to readily distinguish between normal and abnormal cardiac function in a rapid manner, a requirement for high-throughput genetic screening for genes that cause or modify heart function [21, 34].
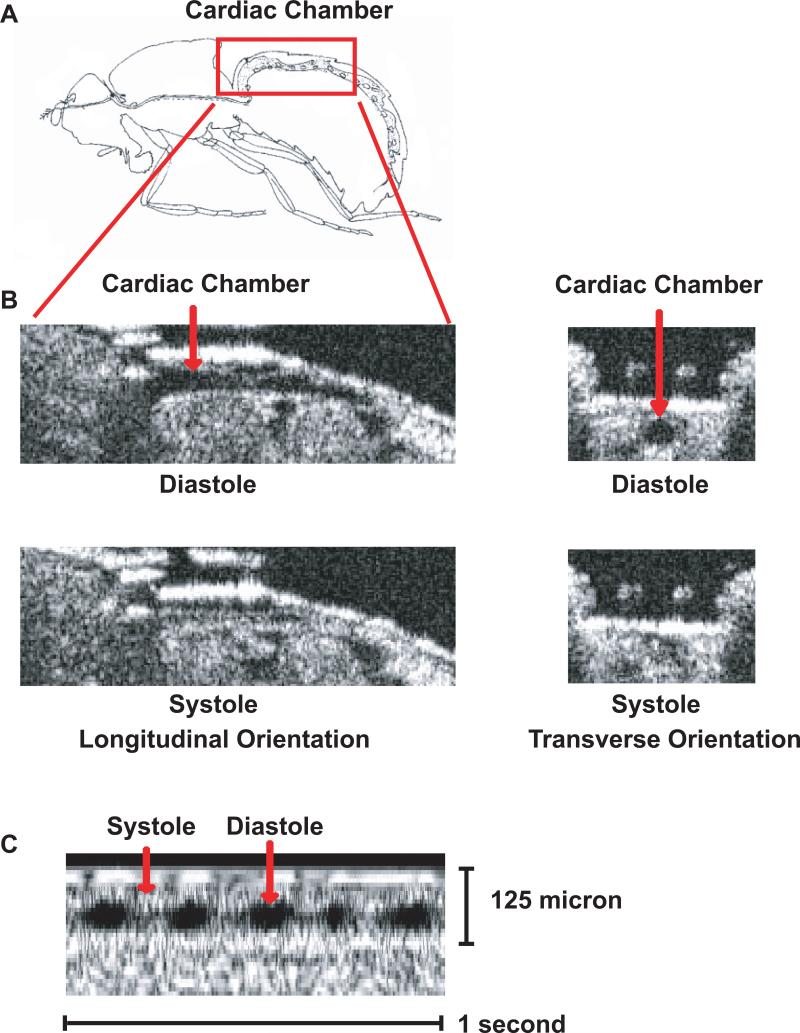
Figure 1.
OCT images of cardiac chamber in adult w1118 Drosophila. (A) Schematic representation of the cardiac chamber (red box) of an adult Drosophila based on Miller [45]. (B) Longitudinal and transverse B-mode OCT images of the cardiac chamber in adult Drosophila during diastole and systole. (C) Representative m-mode OCT image demonstrating the cardiac cycle with diastole and systole denoted by the red arrows.
OCT has limitations that are based on the properties of the imaging system. First, OCT special resolution is in the range of 10 to 20 microns and therefore cannot provide reliable information concerning cardiac wall thickness or surrounding structures. Of note, the conical portion of the cardiac chamber in the adult heart is ∼80 microns at end-diastole and nearly cavity obliterates at end-systole in wild-type adult Drosophila. Second, while most of the adult circulatory system can be visualized in B-mode the most reliable images are obtained from the conical chamber located in the A1-A2 segments. Finally, similar to echocardiography in humans and small animals OCT provides an assessment of cardiac function in the intact, non-anesthetized fly and therefore cardiac function is a combination of both myogenic and nervous system function.
Screening the Drosophila genome to identify genes that cause dilated cardiomyopathy
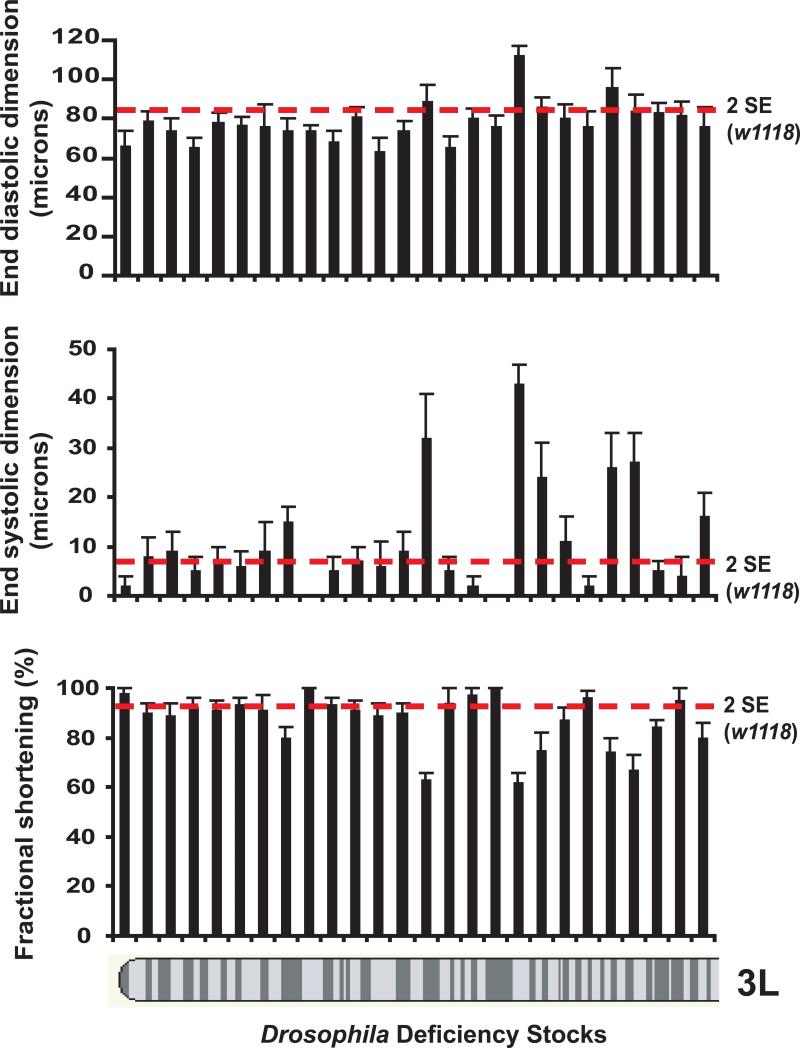
Drosophila has been successfully used to conduct large scale forward genetic screens to identify genes involved in signaling pathways. The development of sophisticated imaging strategies of the adult fly heart now allows the ability to harness the resources available within the field of the Drosophila genetics to identify genes mutations that cause or influence dilated cardiomyopathy and heart failure. OCT imaging provides a robust method to rapidly detect mutant Drosophila that have dilated cardiomyopathy, defined as an enlarged end-diastolic and end-systole dimension or pure systole impairment defined as an enlarged end-systolic dimension with preserved diastolic dimensions. The cardiac function in a stock of ten to fifteen adult flies can be evaluated in ∼45 to 60 minutes translating to ∼3 minutes per fly. Therefore, large collections of fly mutants can be screened for abnormalities in cardiac function. For example, the evaluation of cardiac function in adult Drosophila mutants with molecularly-defined genomic deficiencies demonstrates that we can readily identify mutants with dilated cardiomyopathy (Figure 2). This genetic screen has the potential to identify novel genes that are responsible for human heart failure.
Figure 2.
Screening cardiac function in adult Drosophila by OCT identifies mutants with dilated cardiomyopathy. End-diastolic dimension (EDD), end-systolic fraction (ESD), and fractional shortening defined as (EDD-ESD)/EDD are shown for a series of mutants with deficiencies along the 3L chromosome. The red lines represent measurements that are two standard error units outside the mean for w1118 flies cardiac measurements that were used as controls. A series of mutants with dilated cardiomyopathy or impaired systolic function with preserved diastolic dimensions can be readily identified.
Model Comparison
Drosophila genetics has unique resources that are not currently available to other model of human disease; however, several disadvantages exist when using Drosophila to model cardiovascular disease. First, the fly has a single cardiac chamber that functions as a heart in the context of an open circulatory system. Second, the fly myocardium receives oxygenation through diffusion and does not have coronary arteries. Furthermore, ultrastructural examination demonstrated that the myocytes have perforated Z-discs, allowing for supracontractile properties that lead to near cavity obliteration during cardiac systole. Nonetheless, Drosophila provides the ability to perform large-scale forward genetic screens to examine fundamental questions regarding cardiac development during embryogenesis and cardiac dysfunction in the adult [21, 35].
The zebrafish has a two chambered heart with a closed circulatory system and more closely resembles a mammalian heart compared to Drosophila [36]. Similar to Drosophila, the zebrafish model is also useful for studies based on forward genetic screens but currently lacks many of the genetic resources that are necessary to facilitate rapid mapping of mutations. Mouse models of cardiovascular disease most closely resemble human disease since the mouse has a four chambered and is evolutionally more closely related to humans as compared to the fly or zebrafish. Mouse models have been used to conduct forward genetic screens; however, these studies are complicated by the expense associated with maintaining mouse colonies and mapping mutations.
Model Translation to Humans
Although the fly genome is smaller that vertebrate genomes, including humans, Drosophila and humans share many common genes and signaling pathways. In fact, investigations by Bier et. al. have demonstrated that ∼80% of human diseases in which the disease gene has been identified have an analogous gene in Drosophila [37-39]. Drosophila has been successfully used to model human diseases including neurodegenerative diseases (i.e., Parkinson's Disease, Fragile-X syndrome, and Alzheimer's disease), cancers, infectious diseases, diabetes, and cardiovascular disease [21, 40-44]. Furthermore, transgenic Drosophila that harbor human gene mutations associated with specific human disease can recapitulate disease phenotypes [21]. These striking findings and the evolutionary conservation of genomic information highlights the utility of the fruit fly as a model of human diseases.
Conclusions
The wealth of molecular reagents and advances in imaging technologies has made Drosophila a valuable emerging model to study genes that cause or modify cardiac function. Furthermore, the identification of novel genes and pathways that result in the development of dilated cardiomyopathy in adult Drosophila holds the potential to develop new therapies to treat heart disease in humans.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Beller M, Oliver B. One hundred years of high-throughput Drosophila research. Chromosome Res. 2006;14(4):349–62. doi: 10.1007/s10577-006-1065-2. [DOI] [PubMed] [Google Scholar]
- 2.Chi NC, et al. Genetic and physiologic dissection of the vertebrate cardiac conduction system. PLoS Biol. 2008;6(5):e109. doi: 10.1371/journal.pbio.0060109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Poss KD, Wilson LG, Keating MT. Heart regeneration in zebrafish. Science. 2002;298(5601):2188–90. doi: 10.1126/science.1077857. [DOI] [PubMed] [Google Scholar]
- 4.Bellen HJ, et al. The BDGP gene disruption project: single transposon insertions associated with 40% of Drosophila genes. Genetics. 2004;167(2):761–81. doi: 10.1534/genetics.104.026427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Parks AL, et al. Systematic generation of high-resolution deletion coverage of the Drosophila melanogaster genome. Nat Genet. 2004;36(3):288–92. doi: 10.1038/ng1312. [DOI] [PubMed] [Google Scholar]
- 6.Thibault ST, et al. A complementary transposon tool kit for Drosophila melanogaster using P and piggyBac. Nat Genet. 2004;36(3):283–7. doi: 10.1038/ng1314. [DOI] [PubMed] [Google Scholar]
- 7.Dietzl G, et al. A genome-wide transgenic RNAi library for conditional gene inactivation in Drosophila. Nature. 2007;448(7150):151–6. doi: 10.1038/nature05954. [DOI] [PubMed] [Google Scholar]
- 8.Celniker SE, Rubin GM. The Drosophila melanogaster genome. Annu Rev Genomics Hum Genet. 2003;4:89–117. doi: 10.1146/annurev.genom.4.070802.110323. [DOI] [PubMed] [Google Scholar]
- 9.Rubin GM, et al. Comparative genomics of the eukaryotes. Science. 2000;287(5461):2204–15. doi: 10.1126/science.287.5461.2204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen D, et al. High-resolution, high-throughput SNP mapping in Drosophila melanogaster. Nat Methods. 2008;5(4):323–9. doi: 10.1038/nmeth.1191. [DOI] [PubMed] [Google Scholar]
- 11.St Johnston D. The art and design of genetic screens: Drosophila melanogaster. Nat Rev Genet. 2002;3(3):176–88. doi: 10.1038/nrg751. [DOI] [PubMed] [Google Scholar]
- 12.Bodmer R, Venkatesh TV. Heart development in Drosophila and vertebrates: conservation of molecular mechanisms. Dev Genet. 1998;22(3):181–6. doi: 10.1002/(SICI)1520-6408(1998)22:3<181::AID-DVG1>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 13.Cripps RM, Olson EN. Control of cardiac development by an evolutionarily conserved transcriptional network. Dev Biol. 2002;246(1):14–28. doi: 10.1006/dbio.2002.0666. [DOI] [PubMed] [Google Scholar]
- 14.Zaffran S, Frasch M. Early signals in cardiac development. Circ Res. 2002;91(6):457–69. doi: 10.1161/01.res.0000034152.74523.a8. [DOI] [PubMed] [Google Scholar]
- 15.Zaffran S, et al. Cardiogenesis in the Drosophila model: control mechanisms during early induction and diversification of cardiac progenitors. Cold Spring Harb Symp Quant Biol. 2002;67:1–12. doi: 10.1101/sqb.2002.67.1. [DOI] [PubMed] [Google Scholar]
- 16.Schott JJ, et al. Congenital heart disease caused by mutations in the transcription factor NKX2-5. Science. 1998;281(5373):108–11. doi: 10.1126/science.281.5373.108. [DOI] [PubMed] [Google Scholar]
- 17.Monier B, et al. Steroid-dependent modification of Hox function drives myocyte reprogramming in the Drosophila heart. Development. 2005;132(23):5283–93. doi: 10.1242/dev.02091. [DOI] [PubMed] [Google Scholar]
- 18.Perrin L, et al. Drosophila cardiac tube organogenesis requires multiple phases of Hox activity. Dev Biol. 2004;272(2):419–31. doi: 10.1016/j.ydbio.2004.04.036. [DOI] [PubMed] [Google Scholar]
- 19.Zeitouni B, et al. Signalling pathways involved in adult heart formation revealed by gene expression profiling in Drosophila. PLoS Genet. 2007;3(10):1907–21. doi: 10.1371/journal.pgen.0030174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Curtis NJ, Ringo JM, Dowse HB. Morphology of the pupal heart, adult heart, and associated tissues in the fruit fly, Drosophila melanogaster. J Morphol. 1999;240(3):225–35. doi: 10.1002/(SICI)1097-4687(199906)240:3<225::AID-JMOR2>3.0.CO;2-V. [DOI] [PubMed] [Google Scholar]
- 21.Wolf MJ, et al. Drosophila as a model for the identification of genes causing adult human heart disease. Proc Natl Acad Sci U S A. 2006;103(5):1394–9. doi: 10.1073/pnas.0507359103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dulcis D, Levine RB. Innervation of the heart of the adult fruit fly, Drosophila melanogaster. J Comp Neurol. 2003;465(4):560–78. doi: 10.1002/cne.10869. [DOI] [PubMed] [Google Scholar]
- 23.Wasserthal LT. Drosophila flies combine periodic heartbeat reversal with a circulation in the anterior body mediated by a newly discovered anterior pair of ostial valves and ‘venous’ channels. J Exp Biol. 2007;210(Pt 21):3707–19. doi: 10.1242/jeb.007864. [DOI] [PubMed] [Google Scholar]
- 24.Kim YO, et al. A functional genomic screen for cardiogenic genes using RNA interference in developing Drosophila embryos. Proc Natl Acad Sci U S A. 2004;101(1):159–64. doi: 10.1073/pnas.0307205101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Paternostro G, et al. Age-associated cardiac dysfunction in Drosophila melanogaster. Circ Res. 2001;88(10):1053–8. doi: 10.1161/hh1001.090857. [DOI] [PubMed] [Google Scholar]
- 26.Ocorr K, Akasaka T, Bodmer R. Age-related cardiac disease model of Drosophila. Mech Ageing Dev. 2007;128(1):112–6. doi: 10.1016/j.mad.2006.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ocorr K, et al. Genetic control of heart function and aging in Drosophila. Trends Cardiovasc Med. 2007;17(5):177–82. doi: 10.1016/j.tcm.2007.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ocorr K, et al. KCNQ potassium channel mutations cause cardiac arrhythmias in Drosophila that mimic the effects of aging. Proc Natl Acad Sci U S A. 2007;104(10):3943–8. doi: 10.1073/pnas.0609278104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ocorr KA, et al. Genetic variation for cardiac dysfunction in Drosophila. PLoS ONE. 2007;2(7):e601. doi: 10.1371/journal.pone.0000601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wessells RJ, Bodmer R. Screening assays for heart function mutants in Drosophila. Biotechniques. 2004;37(1):58–60. 62, 64. doi: 10.2144/04371ST01. passim. [DOI] [PubMed] [Google Scholar]
- 31.Wessells RJ, Bodmer R. Age-related cardiac deterioration: insights from Drosophila. Front Biosci. 2007;12:39–48. doi: 10.2741/2047. [DOI] [PubMed] [Google Scholar]
- 32.Wessells RJ, et al. Insulin regulation of heart function in aging fruit flies. Nat Genet. 2004;36(12):1275–81. doi: 10.1038/ng1476. [DOI] [PubMed] [Google Scholar]
- 33.Huang D, et al. Optical coherence tomography. Science. 1991;254(5035):1178–81. doi: 10.1126/science.1957169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Allikian MJ, et al. Reduced life span with heart and muscle dysfunction in Drosophila sarcoglycan mutants. Hum Mol Genet. 2007;16(23):2933–43. doi: 10.1093/hmg/ddm254. [DOI] [PubMed] [Google Scholar]
- 35.Serluca FC, Fishman MC. Big, bad hearts: from flies to man. Proc Natl Acad Sci U S A. 2006;103(11):3947–8. doi: 10.1073/pnas.0600900103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.MacRae CA, Fishman MC. Zebrafish: the complete cardiovascular compendium. Cold Spring Harb Symp Quant Biol. 2002;67:301–7. doi: 10.1101/sqb.2002.67.301. [DOI] [PubMed] [Google Scholar]
- 37.Chien S, et al. Homophila: human disease gene cognates in Drosophila. Nucleic Acids Res. 2002;30(1):149–51. doi: 10.1093/nar/30.1.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fortini ME, et al. A survey of human disease gene counterparts in the Drosophila genome. J Cell Biol. 2000;150(2):F23–30. doi: 10.1083/jcb.150.2.f23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Reiter LT, et al. A systematic analysis of human disease-associated gene sequences in Drosophila melanogaster. Genome Res. 2001;11(6):1114–25. doi: 10.1101/gr.169101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Baker KD, Thummel CS. Diabetic larvae and obese flies-emerging studies of metabolism in Drosophila. Cell Metab. 2007;6(4):257–66. doi: 10.1016/j.cmet.2007.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bier E, Bodmer R. Drosophila, an emerging model for cardiac disease. Gene. 2004;342(1):1–11. doi: 10.1016/j.gene.2004.07.018. [DOI] [PubMed] [Google Scholar]
- 42.Bonini NM, Fortini ME. Human neurodegenerative disease modeling using Drosophila. Annu Rev Neurosci. 2003;26:627–56. doi: 10.1146/annurev.neuro.26.041002.131425. [DOI] [PubMed] [Google Scholar]
- 43.Marsh JL, Thompson LM. Drosophila in the study of neurodegenerative disease. Neuron. 2006;52(1):169–78. doi: 10.1016/j.neuron.2006.09.025. [DOI] [PubMed] [Google Scholar]
- 44.Vidal M, Cagan RL. Drosophila models for cancer research. Curr Opin Genet Dev. 2006;16(1):10–6. doi: 10.1016/j.gde.2005.12.004. [DOI] [PubMed] [Google Scholar]
- 45.Demerec M. Biology of Drosophila. Wiley; New York: 1950. p. x.p. 632. [Google Scholar]