Abstract
Mutations in the mouse indicate that quaking gene function is essential for both embryogenesis and for development of the nervous system. Recent isolation of the mouse quaking gene identified a putative RNA-binding protein containing a single KH domain. We have previously isolated the Xenopus homolog of quaking, Xqua, and shown that the sequence is highly conserved through evolution. Here, we report experimental data on the biochemical function of the quaking protein and its role during development. We demonstrate that the quaking protein expressed during early embryogenesis, pXqua357, can bind RNA in vitro, and we have mapped the regions of the protein that are essential for RNA binding. We present evidence that pXqua can form homodimers and that dimerization may be required for RNA binding. Oocyte injection experiments show that pXqua357 is located in both the nucleus and cytoplasm. In the Xenopus embryo, Xqua is first expressed during gastrulation in the organizer region and its derivative, the notochord. In later stage embryos, Xqua is expressed in a number of mesodermal and neural tissues. We demonstrate that disruption of normal Xqua function, by overexpression of a dominant inhibitory form of the protein, blocks notochord differentiation. Xqua function appears to be required for the accumulation of important mRNAs such as Xnot, Xbra, and gsc. These results indicate an essential role for the quaking RNA-binding protein during early vertebrate embryogenesis.
Keywords: quaking, Xqua, KH domain, RNA binding, notochord, Xenopus
The function of the murine quaking gene product is required both during embryogenesis and during later development of the nervous system as indicated by two classes of mutant recessive quaking alleles. Mice that are homozygous for the quaking viable allele (qkv/qkv) have a deficiency of myelin in their nervous system and exhibit a characteristic tremor or “quaking” of the hind quarters (Sidman etal. 1964; Samorajski et al. 1970). The second class of quaking alleles, qke1–4/qke1–4, are embryonic lethal around day 9–10 of gestation (Bode 1984; Justice and Bode 1988; Shedlovsky et al. 1988). The mutant embryos are disorganized and exhibit generalized atrophy, but the precise cause of lethality has not been characterized (Justice and Bode 1988).
The sequence of the recently cloned mouse quaking gene (qkI) (Ebersole et al. 1996) and its human (Hqk) and Xenopus (Xqua) homologs (Zorn et al. 1997) indicate that quaking encodes a KH domain RNA-binding protein. The KH domain is an evolutionarily conserved sequence found in a diversity of proteins, most of which are implicated in some aspect of RNA metabolism (Gibson et al. 1993; Siomi et al. 1993a). Notable examples include the heterogeneous nuclear ribonucleoprotein K (hnRNP K) (Siomi et al. 1993a) and the human Fragile X Syndrome gene, FMR1 (Siomi et al. 1993b). Experimental evidence indicates that the KH domain is directly involved in RNA binding. For example, a particularly severe allele of the Fragile X Syndrome (De Boulle et al. 1993) is caused by a single point mutation in a conserved residue in the second KH domain, resulting in a mutant protein that shows dramatically reduced RNA-binding activity (Siomi et al. 1994). quaking belongs to a new subclass of KH proteins, called GSG domain proteins (Jones and Schedl 1995; Ebersole et al. 1996), based on sequence similarity with GLD-1, a tumor suppressor gene required for germ line development in Caenorhabditis elegans (Jones and Schedl 1995); SAM68, a mammalian phosphoprotein involved in the Src signaling pathways during mitosis (Won et al. 1992; Fumagalli et al. 1994; Taylor and Shalloway 1994; Lock et al. 1996); and GRP33, an hnRNP isolated from brine shrimp (Cruz-Alvarez and Pellicer 1987). GSG proteins are characterized by an ∼200-amino-acid region of sequence similarity centered on a single KH motif. Although the KH domain is highly conserved between GSG family members, it is rather divergent from the KH motifs found in other proteins.
The qkv defect is a classic model for the study of dismyelination, and a wealth of literature describes various aspects of the defective nervous system (Hogan and Greenfield 1984). Despite this intense investigation, the precise role of quaking in neural development remains obscure, and even less is understood about its function during early embryogenesis. The mouse qkI gene produces three transcripts 5 kb, 6 kb, and 7 kb in length, generated by alternative mRNA splicing. The sequences of the proteins resulting from these transcripts are identical except for the extreme carboxy-terminal region (Ebersole et al. 1996). In Xenopus, 5-kb and 5.5-kb mRNAs (analogous to the mouse 5-kb and 6-kb mRNAs) have been observed (Zorn et al. 1997). In both frog and mouse only the 5-kb mRNA is expressed in the early embryo, with the 6-kb form arising later in development. Expression of the 5-kb and 6-kb sequences continues throughout development, and in the adult mouse, the transcripts are abundant in the brain, lung, heart, and testis (Ebersole et al. 1996). Expression of the 7-kb mRNA is apparently restricted to the adult brain. The fact that qkI is expressed in a variety of tissues and that quaking mutations have pleiotropic effects suggests that quaking function is important for the development or maintenance of a number of cell types. Furthermore, the remarkable 94% conservation of the mouse, human, and Xenopus protein sequences (Zorn et al. 1997) suggests that the quaking biochemical pathway is highly conserved through evolution.
A number of KH domain RNA-binding proteins are known to be required during development of invertebrates. These include Bicaudal-C in Drosophila, which is essential for anterior–posterior specification (Mahone et al. 1995); gld-1, which encodes a GSG protein that is essential for germ line development in C. elegans (Jones and Schedl 1995); and mex-3, which regulates early blastomere identity in the C. elegans embryo (Draper et al. 1996). quaking is one of the first examples of a KH domain protein essential for early vertebrate embryogenesis. In an effort to understand the developmental function of the quaking protein, we have characterized the biochemical properties of pXqua357, the Xenopus quaking protein that is expressed during early embryonic development. We have mapped the regions of pXqua357 that are essential for RNA binding and present evidence that pXqua proteins can form homodimers. These results have implications not only for quaking but also for understanding the general biochemical properties of the GSG subfamily of KH domain proteins. Finally, we demonstrate that embryonic overexpression of pXqua357 enhances notochord development, whereas overexpression of a dominant inhibitory pXqua mutant disrupts the endogenous Xqua pathway and blocks notochord development. The normal function of Xqua appears to be essential for the accumulation of important mRNAs such as Xnot, Xbra, and goosecoid (gsc). Our findings provide a possible explanation for the early embryonic lethality of quaking mutant embryos.
Results
pXqua binds RNA in vitro
The presence of a KH motif in the quaking protein (Ebersole et al. 1996; Zorn et al. 1997) suggests that it is an RNA-binding protein. To directly test this possibility, we have examined the in vitro RNA-binding properties of Xenopus quaking proteins pXqua357 and pXqua365, which are identical except for an 8-amino-acid insertion just carboxy-terminal to the GSG domain, generated by alternative splicing (Zorn et al. 1997) (Fig. 1A). In vitro 35S-labeled Xqua proteins were assayed for the ability to bind RNA immobilized on agarose beads (Swanson and Dreyfuss 1988). As shown in Figure 1B, both pXqua357 and pXqua365 bind total Xenopus embryonic RNA but do not bind significantly to agarose alone. We examined whether pXqua had a preference for the type of RNA it would bind by using different RNA homopolymers coupled to agarose. Both pXqua357 and pXqua365 bind preferentially to poly(rG) and poly(rU) RNA, whereas neither binds significantly to poly(rA) or poly(rC) RNA (Fig. 1B), indicating that pXqua has some specificity in its interaction with RNA. Because pXqua357 and pXqua365 exhibit indistinguishable RNA-binding properties in this assay, only pXqua357 was used in subsequent experiments. The strength of the RNA–protein interactions was examined by increasing the salt concentration of the binding assay. As shown in Figure 1C, recombinant pXqua357 binds weakly to total embryonic RNA and poly(rU), neither interaction being stable in 250 mm NaCl. However, pXqua357 shows a much stronger interaction with poly(rG) RNA, retaining some binding up to 500 mm. In additional experiments, pXqua357 exhibited weak binding to single-stranded DNA–agarose, but no detectable binding to double-stranded DNA–agarose (data not shown). The nucleic acid binding properties of pXqua are comparable with those of RNA-binding proteins reported previously (Piñol-Roma et al. 1987; Swanson and Dreyfuss 1988; Siomi et al. 1993b).
Figure 1.
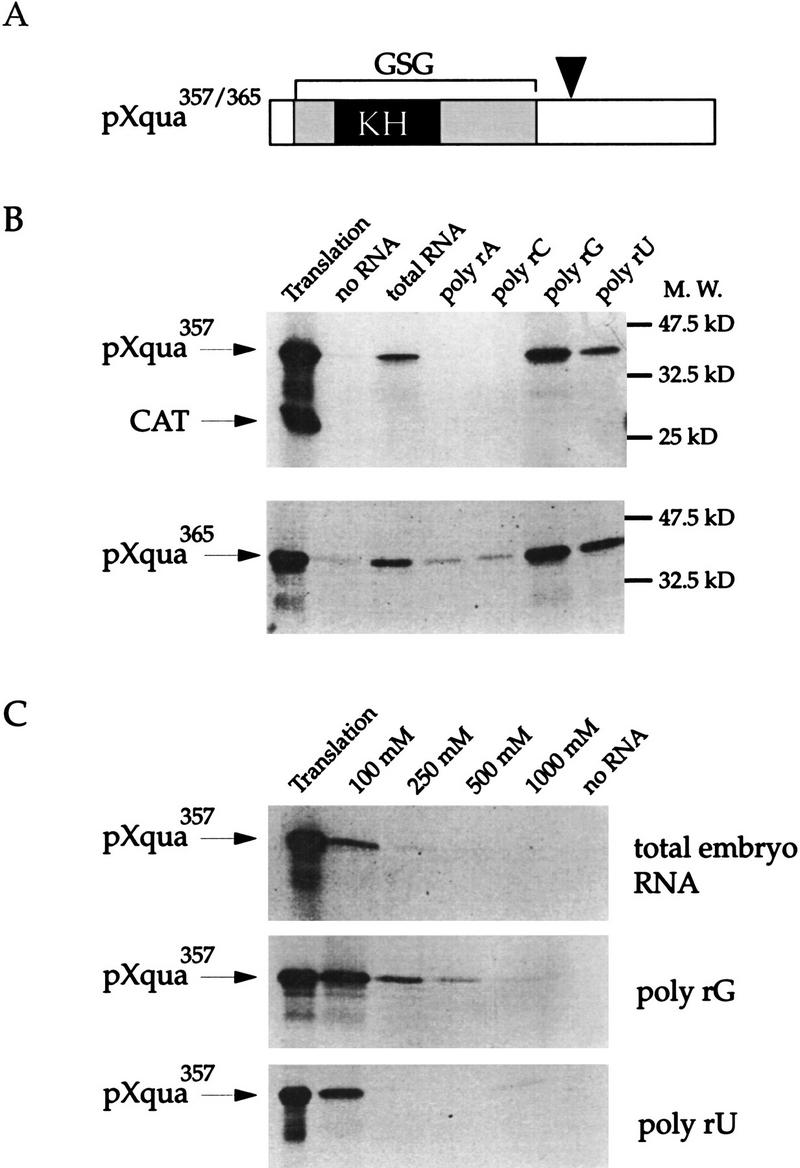
pXqua is an RNA-binding protein. (A) Two alternatively spliced forms of the 5-kb Xqua mRNA are predicted to encode proteins of 357 (pXqua357) and 365 (pXqua365) amino acids, respectively. The position of the alternative exon is indicated by the arrowhead. pXqua belongs to the GSG subfamily of KH domain proteins, characterized by a single KH motif (in black) found in a larger ∼200-amino-acid region of conserved sequence (in grey). To test for RNA-binding activity, 35S-labeled pXqua365, pXqua357, or an irrelevant control protein, CAT, were incubated with total embryonic RNA–agarose, homopolymer RNA–agarose, or agarose with no RNA. Proteins bound to the RNA–agarose were resolved by SDS-PAGE and visualized by fluorography. (B) The RNA-binding properties of pXqua357 and pXqua365 are indistinguishable, binding to total Xenopus embryonic RNA, poly(rG), and poly(rU) RNA but not to poly(rA) or poly(rC). The control CAT protein did not bind to the RNA–agarose. An amount equal to 20% of the protein used in each binding reaction is shown in the track marked translation. (C) The relative RNA-binding strength of pXqua357 to total embryonic RNA, poly(rU) RNA, and poly(rG) RNA was tested by increasing the salt concentrations of the binding reactions from 100 mm to 250 mm, 500 mm, and 1 m NaCl. pXqua357 binds total Xenopus embryonic RNA and poly(rU) at moderate strength but binds most strongly to poly(rG).
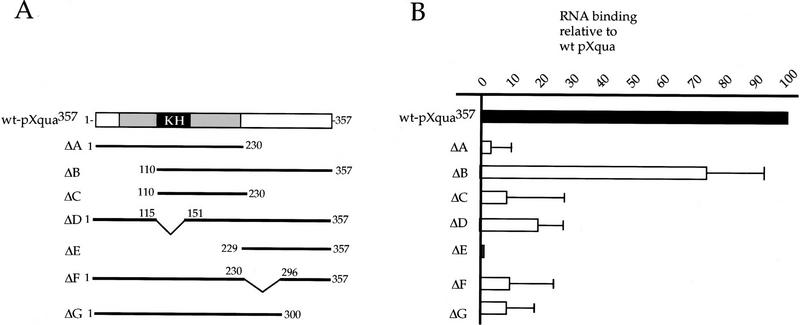
The KH domain and the carboxy-terminal region are required for RNA binding in vitro
quaking and the other members of the GSG subfamily are characterized by a single KH motif. They are therefore distinct from most previously studied KH proteins that contain multiple KH motifs (Gibson et al. 1993; Siomi et al. 1993a,b). Detailed RNA-binding studies of hnRNP K and FMR1, for example, have shown that cooperativity between the multiple KH domains is required for full RNA binding and that deletion of any one KH motif practically abolishes RNA-binding activity (Siomi et al. 1994). In light of this, it is important to determine which regions of pXqua are required for its RNA-binding activity. We therefore generated a series of truncated pXqua proteins (Fig. 2A) and assayed for in vitro RNA-binding activity (Fig. 2B). Truncation of the amino-terminal region up to the KH domain (ΔB) caused only a slight reduction in RNA-binding activity (to 75% of wild-type levels). In contrast, deletion of either the core KH domain (ΔD) or the carboxy-terminal region (ΔA) resulted in severe loss of RNA-binding activity, demonstrating that both of these regions are essential for maximal RNA-binding activity. Consistent with this observation, ΔC, which contains the KH domain but not the carboxy-terminal region, and ΔE, which contains the carboxy-terminal region alone, exhibit no RNA-binding activity in this assay. In an attempt to further define the sequences in the carboxy-terminal region required for RNA binding, two additional truncated proteins, ΔF and ΔG, were tested. Neither of these proteins shows significant RNA-binding activity. We conclude that both the KH domain and sequences in the carboxy-terminal region are essential for the RNA-binding activity of pXqua357. Examination of the carboxy-terminal sequence, however, does not reveal the presence of any previously described RNA-binding motif.
Figure 2.
The KH motif and the carboxy-terminal region of pXqua are required for RNA binding. (A) Schematic representation of pXqua357 and the truncated pXqua357-Δ proteins. (B) 35S-labeled, pXqua-Δ proteins were produced by in vitro translation of synthetic mRNA and assayed for RNA binding to total embryonic RNA–agarose at 100 mm NaCl. Bound proteins were resolved on 12.5% SDS-PAGE, visualized by fluorography, and quantitated using National Institutes of Heath (NIH) image software. The histogram shows relative RNA-binding activities of the different truncated pXqua-Δ proteins averaged from four separate experiments.
pXqua can form homodimers
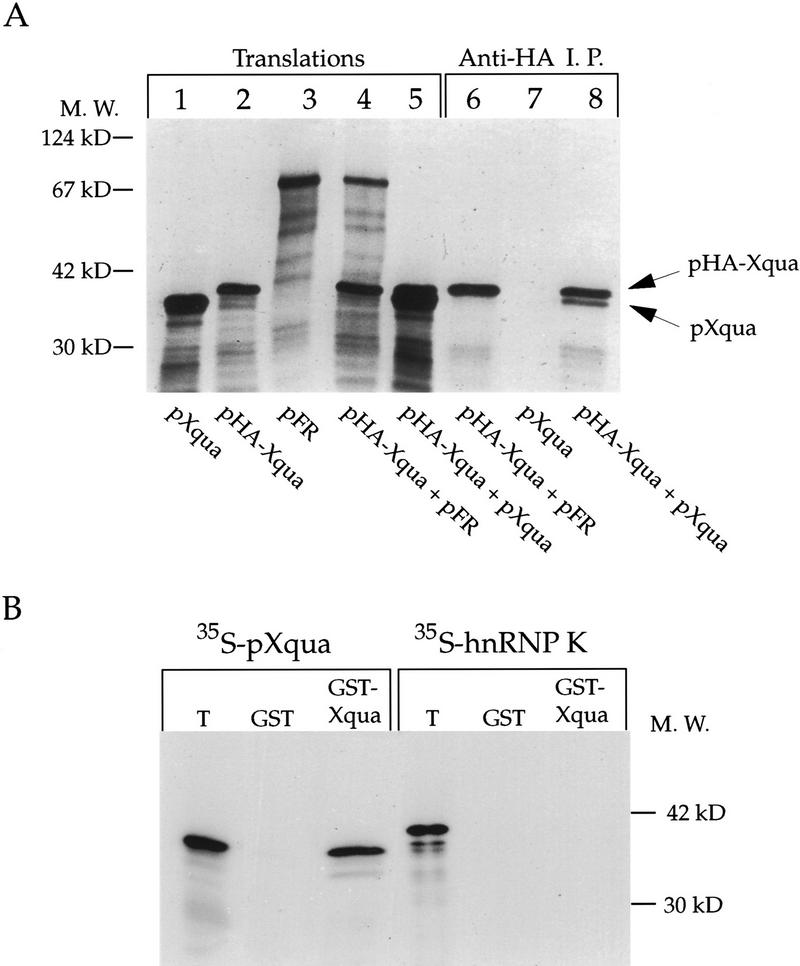
How is it possible for pXqua, with only one KH domain, to bind RNA, whereas other KH proteins such as hnRNP K and FMR1 need multiple KH motifs? One possibility is that two or more pXqua monomers might physically interact, thus allowing multiple KH domains to cooperate in RNA binding. We therefore tested the ability of pXqua357 monomers to physically interact, using two different experimental approaches: first, coimmunoprecipitation (Fig. 3A) and, second, binding reactions using bacterial glutathione S-transferase (GST) fusion proteins (Fig. 3B). For immunoprecipitation experiments, synthetic mRNAs encoding a hemagglutinin (HA) epitope-tagged version of pXqua, pHA–Xqua357 and pXqua357 (with no HA tag) were cotranslated in a reticulocyte lysate (Fig. 3A, lane 5). Immunoprecipitation using an anti-HA antibody resulted in the precipitation of pHA–Xqua357 and the coprecipitation of pXqua357 (Fig. 3A, lane 8), indicating that pHA–Xqua357 and pXqua357 monomers are physically associated. Control immunoprecipitation of translations containing pXqua357 alone did not result in a detectable product (lane 7). Furthermore, when Anti-HA immunoprecipitations were performed on the cotranslation of pHA–Xqua357 and an irrelevant protein (pFR), only pHA–Xqua357 was detected (lanes 4 and 6). These immunoprecipitation results were confirmed using GST–Xqua protein binding experiments. Purified GST or GST–Xqua was immobilized on glutathione–Sepharose and then incubated with 35S-labeled pXqua357 or the control KH domain protein hnRNP K. After washing, the bound proteins were analyzed by SDS-PAGE. As shown in Figure 3B, labeled pXqua357 binds to the GST–Xqua fusion protein but not to GST alone. As expected, the hnRNP K control protein does not bind to either GST or GST–Xqua. Overall, these results demonstrate that pXqua357 monomers are capable of forming physical complexes. The simplest interpretation is that two pXqua monomers interact to form a homodimer, but these experiments do not exclude the possibility of oligomeric complexes containing more than two pXqua monomers.
Figure 3.
pXqua can form homodimers in vitro. (A) Synthetic mRNA encoding pXqua357 (lane 1); pHA–Xqua357, a HA epitope-tagged version of pXqua357 (lane 2); or an irrelevant control protein, pFR (lane 3), was translated in reticulocyte lysates, resolved on SDS-PAGE, and visualized by fluorography. pXqua357 and pHA–Xqua357 are distinguished from each other on the basis of the difference in their molecular mass (cf. lanes 1 and 2). To test for a physical interaction between pXqua monomers, pXqua357 and pHA–Xqua357 were cotranslated (lane 5), and pFR and pHA–Xqua357 were also cotranslated as a control (lane 4). An equivalent portion of each reaction was immunoprecipitated with the anti-HA monoclonal antibody 12CA5, and precipitated proteins were resolved on 10% SDS-PAGE and visualized by fluorography. The untagged pXqua was coprecipitated with pHA–Xqua (arrows) from the pXqua/pHA–Xqua cotranslation (lane 8), indicating that pXqua was bound to the HA–Xqua protein. pXqua357 alone was not precipitated (lane 7), nor was the control protein pFR coprecipitated from the pFR/pHA–Xqua cotranslation (lane 6). (B) pXqua dimerization demonstrated using GST-fusion protein binding assays. 35S-labeled pXqua357 or a control KH protein, hnRNP K, was incubated with either 2 μg of GST alone or a GST–Xqua fusion protein, coupled to glutathione–Sepharose. The bound proteins were resolved on 10% SDS-PAGE and visualized by fluorography. The lane marked T represents 20% of the input translation.
pXqua357 is both cytoplasmic and nuclear
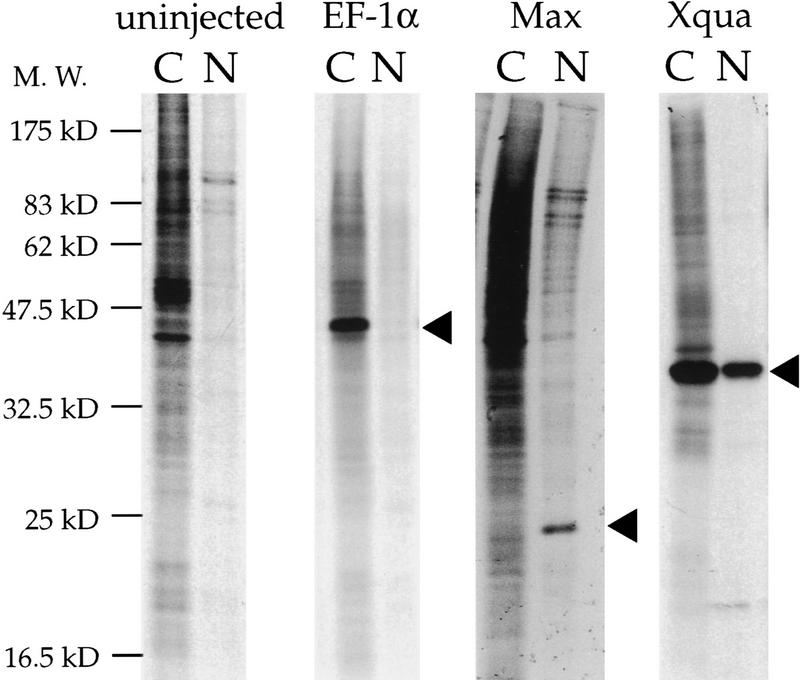
A role for KH domain proteins has been suggested at many levels of RNA metabolism from pre-mRNA processing to translational regulation (Gibson et al. 1993). To obtain some information concerning the possible function of pXqua357, we examined its subcellular location using the well-characterized Xenopus oocyte expression system. Numerous experiments have demonstrated that oocytes translate microinjected mRNAs and faithfully compartmentalize the proteins to the correct subcellular location (Colman 1984). Synthetic capped mRNA encoding pXqua357, or control sequences encoding cytoplasmic EF-1α (Krieg et al. 1989) or nuclear XMax (Tonissen and Krieg 1994), were microinjected into Xenopus oocytes. The results of these experiments (Fig. 4) show that pXqua357 is located in both the cytosolic and nuclear compartments. In each of 10 separate injection experiments, ∼60% of the pXqua357 is cytoplasmic, whereas 40% resides in the nucleus. Examination of controls indicates that 91% of the total EF-1α protein is in the cytoplasmic fraction, whereas 80% of the XMax protein is translocated to the nucleus. Because the oocyte nucleus represents ∼4% of the volume of the whole cell, pXqua is concentrated ∼10-fold in the nucleus, relative to a random distribution. The same nuclear/cytoplasmic ratio is observed in each experiment, independent of the overall level of pXqua expression in the oocyte (data not shown), suggesting that its subcellular distribution is regulated. These experiments suggest that pXqua357 may have a biological function in both the nuclear and cytoplasmic compartments.
Figure 4.
pXqua357 is both nuclear and cytosolic. The subcellular distribution of pXqua357 was determined using the Xenopus oocyte injection assay. Synthetic mRNAs encoding pXqua357 or control sequences encoding the cytoplasmic protein EF-1α or the nuclear protein XMax were injected into the cytoplasm of Xenopus oocytes. Synthetic RNAs were translated in the oocytes in the presence of [35S]methionine. Oocytes were manually dissected into cytosolic (C) and nuclear (N) fractions. Protein extracts equivalent to one-fourth of each fraction from a single oocyte were resolved on SDS-PAGE and visualized by fluorography. Results of a representative experiment show that control proteins EF-1α and XMax localize to the cytoplasmic and nuclear fractions, respectively. pXqua357 is present in both cytoplasmic and nuclear fractions. Arrowheads indicate the position of the relevant proteins.
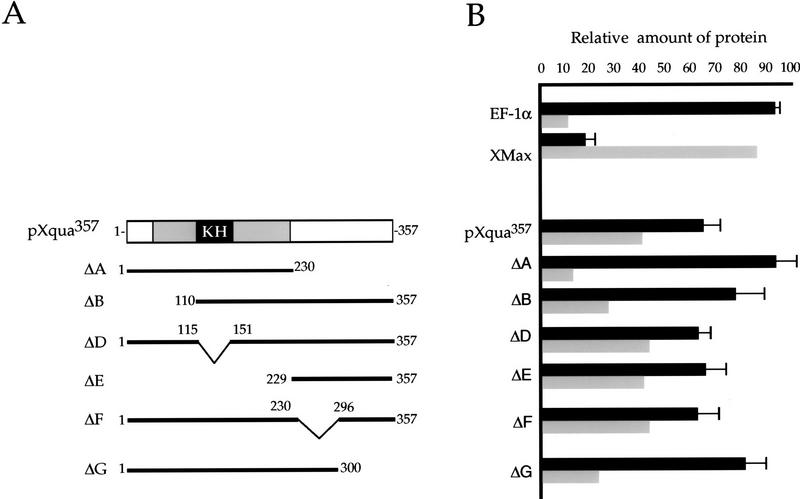
The carboxy-terminal region of pXqua is essential for translocation to the nucleus
Nuclear import/export is a dynamic process that is regulated tightly (Golbfarb and Michaud 1991; Powers and Forbes 1994), and therefore, it is of interest to determine which regions of pXqua357 are responsible for its subcellular location. To investigate this, we examined the nuclear/cytoplasmic distribution of several pXqua deletion constructions (Fig. 5A). As shown in Figure 5B, deletion of the KH domain (ΔD) does not affect the nuclear/cytoplasmic ratio, and deletion of amino-terminal residues 1–110 (ΔB) only moderately impairs the translocation to the nucleus. The most dramatic effect on the nuclear/cytoplasmic distribution is observed when the carboxyl terminus of the protein is deleted. In this case, virtually all of ΔA is retained in the cytoplasm, approximately equivalent to the cytoplasmic control protein EF-1α. Consistent with this observation, ΔE, which only contains carboxyl residues 229–357, behaves identically to the wild-type protein, indicating that the elements controlling nuclear import are found within this region. To further define the sequences responsible for nuclear translocation, ΔF and ΔG were also expressed in oocytes. The truncated protein ΔF showed a distribution equivalent to the wild-type pXqua, whereas deletion of the last 60 amino acids (ΔG) caused a significant reduction of protein in the nucleus. We note, however, that ΔG was not as severely impaired as pXqua ΔA in its ability to translocate to the nucleus, suggesting that perhaps not all of the sequences required for the correct nuclear/cytoplasmic distribution of pXqua357 are contained in the carboxy-terminal 60 amino acids. In no case do we find a significant increase in nuclear pXqua, a result that would be consistent with the deletion of a nuclear export or a cytoplasmic retention signal.
Figure 5.
The carboxy-terminal region of pXqua357 is required for translocation to the nucleus. The regions of pXqua357 that regulate its subcellular distribution were mapped by expression in Xenopus oocytes. (A) Schematic representation of pXqua357 and truncated pXqua357-Δ proteins. (B) Synthetic mRNAs encoding pXqua357, the truncated pXqua proteins ΔA–ΔG, or control proteins EF1-α and XMax were injected into the cytoplasm of Xenopus oocytes and translated in the presence of [35S]methionine. After manual enucleation, protein extracts from the cytosolic and nuclear fractions were resolved on SDS-PAGE and visualized by fluorography, and the relative proportion of the test protein in either fraction was determined using NIH image software. The histogram shows the relative amount of each test protein in the cytoplasmic (black) and nuclear (shaded) fraction, presented as a percentage of the total foreign protein expressed in the oocyte. The results were averaged from 5–15 separate injection experiments.
Initial Xqua expression is localized to the dorsal blastopore lip
In mouse, expression of qkI mRNA is first detected at day 7.5 of gestation, and qke homozygous mutant embryos die by day 9 of gestation, indicating that early qkI expression is essential (Justice and Bode 1988; Ebersole et al. 1996). The mutant embryos are smaller than normal and disorganized, but the exact nature of the defect is not clear. In Xenopus, embryonic expression of Xqua commences during early gastrulation (Zorn et al. 1997), suggesting that Xqua may play a role very early during development.
Using whole-mount in situ hybridization, we have examined the spatial pattern of Xqua expression in the frog embryo. As shown in Figure 6A, the earliest Xqua expression is detected in the chordamesoderm of the dorsal blastopore lip of the midgastrula embryo (stage 11). This tissue, which is analogous to the node in the mouse embryo, is of particular interest because it differentiates into the notochord, has embryonic organizing activity, has neural inducing capability, and plays an integral role in the morphogenesis of the embryo (for review, see Hamburger 1988). To our knowledge, this is the first description of a RNA-binding protein that is localized to the blastopore lip. In late gastrula embryos (stage 13), high levels of Xqua expression are evident in the notochord, and the original expression domain expands to include the tissue surrounding the blastopore (Fig. 6B). In neurula embryos, Xqua expression is maintained in the notochord and the circumblastoporal region and now also extends to the paraxial mesoderm and the neuroectoderm (Fig. 6C–E). By the tailbud stage, Xqua is expressed in various mesodermal and neural tissues (Fig. 6F,G). High levels of Xqua mRNA are found in the brain and the neural tube. Expression in the brain is restricted to the proliferative ventricular layer and the marginal zone where active differentiation takes place (Fig. 6H). High levels of Xqua are also detected in the branchial arches and in the developing heart. The tail blastema, which is known to maintain some embryonic organizer activity (Gont et al. 1993), also expresses significant levels of Xqua mRNA. During this period of development, expression in the notochord and the somites declines, and by the hatched tadpole stage, Xqua expression is almost exclusively restricted to the head and heart (Fig. 6I).
Figure 6.
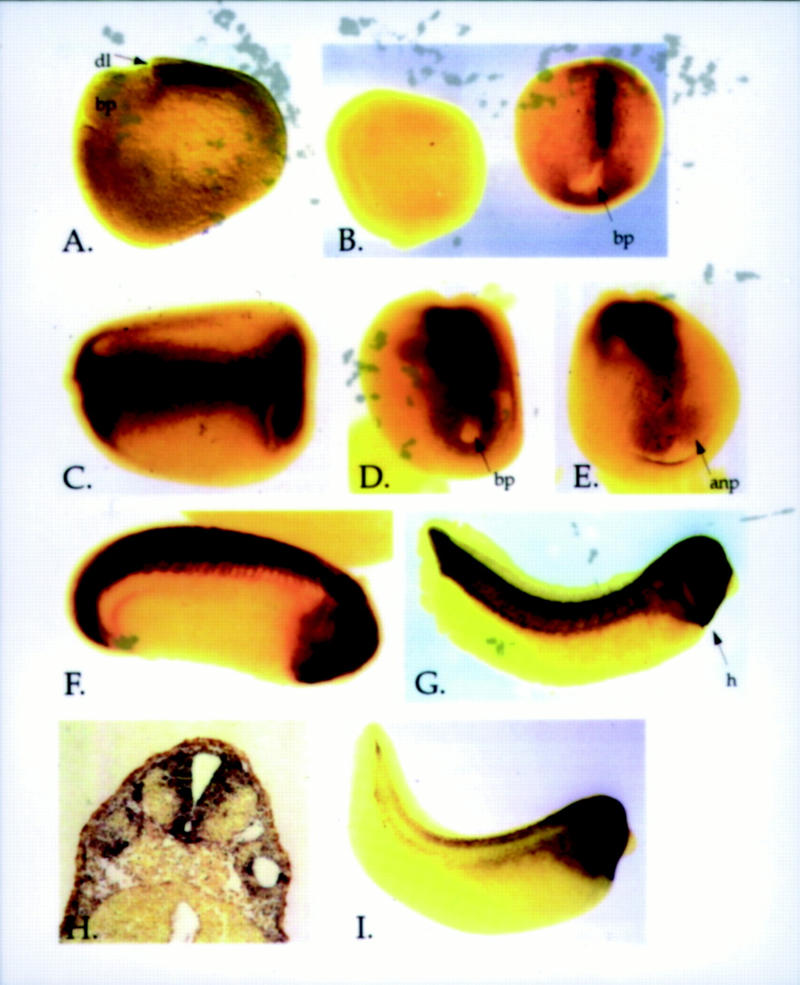
Embryonic expression of Xqua mRNA. Whole-mount in situ hybridization using an RNA probe specific to the 5-kb embryonic form of Xqua. (A) Lateral view of a stage 11 gastrula embryo shows that transcripts are first detected in the chordamesoderm of the dorsal blastopore lip. (B) By late gastrula, Xqua is strongly expressed in the notochord with weak expression in the circumblastoporal region (B, embryo on the right, dorsal view). Embryos hybridized with sense strand control probes (B, embryo on the left) show no staining. In neurula embryos (stage 18) (C, dorsal view; D, posterior view; E, anterior view), the expression domain has expanded to include the notochord, paraxial mesoderm, circumblastoporal region, and the neural plate. In tailbud embryos (stages 24–30) (F,G), Xqua is strongly expressed in the brain and neural tube, the developing heart, tail blastema, and branchial arches. (G) By late tailbud (stage 30), the expression in the notochord and somites is declining. (H) Cross sections through the head of the stage 30 whole-mount embryo show that expression in the brain is largely restricted to the ventricular and marginal zones and the floor plate. (I) Later in development at hatched tadpole stage, Xqua expression is almost exclusively in the head and heart. (anp) Anterior neural plate; (bp) blastopore; (dl) dorsal lip; (h) heart; (n) notochord.
Overexpression of wild-type and mutant Xqua mRNA disrupts anterior/axial development
Our expression studies show that Xqua is expressed in a variety of tissues of mesodermal and neural origin. To investigate the role of Xqua during development of these tissues, we have overexpressed pXqua357 in the embryo by microinjection of synthetic mRNA and then examined embryos for developmental effects at the late tailbud stage. Microinjection of mRNA into two ventral blastomeres (2.5 ng/blastomere) targeted ectopic Xqua overexpression primarily to regions of the embryo that do not normally express Xqua, such as the non-neural ectoderm, the ventral mesoderm, or the endoderm. The distribution of the injected mRNA was monitored by coinjection of mRNA encoding green fluorescent protein (GFP) (Zernicka-Goetz et al. 1996) or β-galactosidase as a lineage marker (data not shown). Ectopic Xqua overexpression in the ventral region had no obvious effects on the development of the embryo (Table 1). However, targeting overexpression to regions of the embryo where Xqua is normally expressed, by microinjection into dorsal blastomeres (1.25–2.5 ng/blastomere), resulted in abnormal anterior development. At the highest dose of Xqua mRNA (2.5 ng/blastomere) over half of the embryos exhibit features of exaggerated dorsal-anterior development, including expanded hindbrains, reduced cement glands, enlarged eyes, and occasionally duplicated head structures (Fig. 7B; Table 1). Control injections of an equivalent amount of rRNA into dorsal blastomeres produced no observable phenotype (Fig. 7A; Table 1). These data indicate that when pXqua357 is expressed in tissues where it is not normally present, it has no obvious effects on development. In contrast, overexpression of pXqua357 in tissues where it is normally expressed produces developmental defects.
Table 1.
Summary of phenotypes resulting from Xqua and ΔD mRNA microinjections
| RNA injected
|
Location (ng/blastomere)a
|
Number of embryos
|
Exaggerated anterior development (%)
|
Microcephalic (%)
|
Acephalic (%)
|
Severe anterior truncation (%)
|
Tail defects (%)
|
Normal development (%)
|
|---|---|---|---|---|---|---|---|---|
| rRNA | dorsal (2.5 ng) | 90 | 0 | 1 | 0 | 0 | 0 | 99 |
| rRNA | ventral (2.5 ng) | 30 | 0 | 0 | 0 | 0 | 3 | 97 |
| Xqua | dorsal (1.25 ng) | 18 | 11 | 0 | 0 | 0 | 0 | 89 |
| Xqua | dorsal (2.5 ng) | 50 | 54 | 0 | 0 | 0 | 0 | 46 |
| Xqua | ventral (2.5 ng) | 25 | 0 | 0 | 0 | 0 | 12 | 88 |
| ΔD | dorsal (0.5 ng) | 31 | 0 | 35 | 0 | 0 | 0 | 65 |
| ΔD | dorsal (1.25 ng) | 20 | 0 | 32 | 56 | 2 | 0 | 2 |
| ΔD | dorsal (2.5 ng) | 66 | 0 | 27 | 24 | 49 | 0 | 0 |
| ΔD | ventral (2.5 ng) | 51 | 0 | 8 | 4 | 0 | 57b | 31 |
mRNA was injected into the equatorial region of either the two dorsal or the two ventral blastomeres at the four-cell stage.
Tail defects occurred only when the majority of injected mRNA was located in the circumblastoporal region as monitored by the coinjection of GFP or β-galactosidase mRNA as a lineage marker (see Fig. 71).
Figure 7.
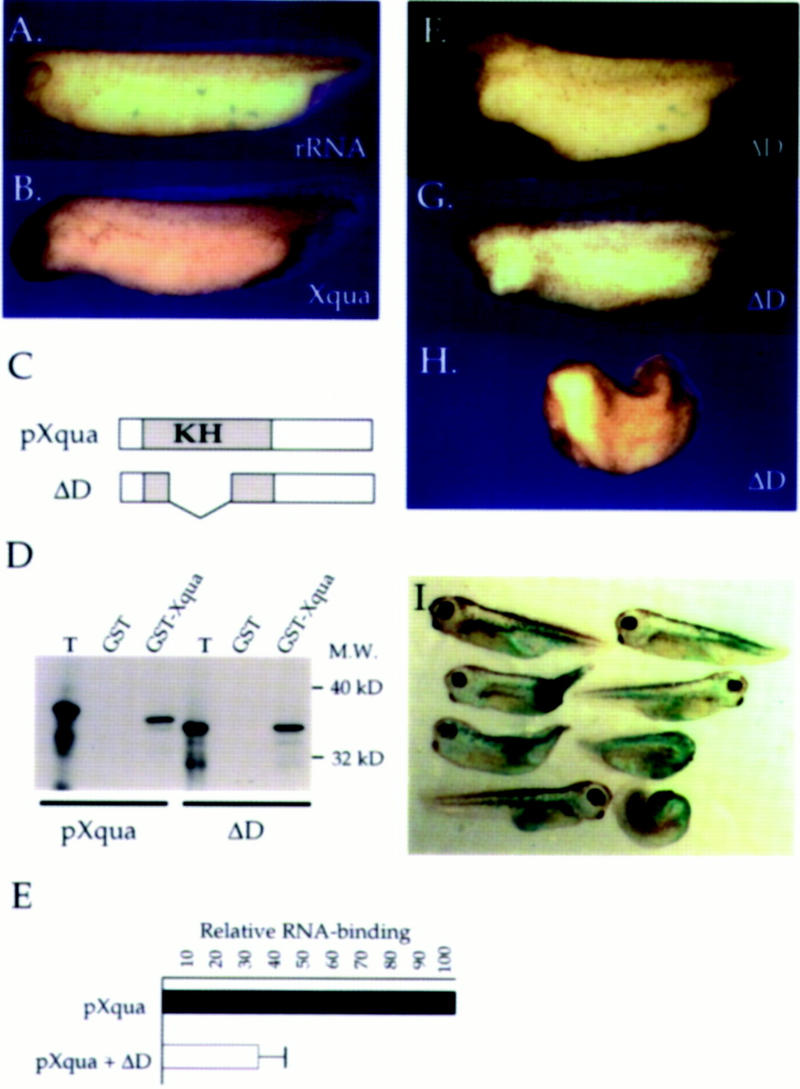
Overexpression of Xqua and ΔD causes defects in anterior development. Synthetic mRNA encoding either pXqua357, the truncated pXqua protein ΔD (which lacks the KH motif), or control rRNA was overexpressed in embryos by microinjection into the equatorial region of each of the two dorsal blastomeres at the four-cell stage. (A) Overexpression of rRNA (2.5 ng/blastomere) has no effect on development. (B) Dorsal injection of Xqua mRNA (2.5 ng/blastomere) resulted in exaggerated anterior development including elongation of the hindbrain region and reduced cement glands. (C) Schematic representation of pXqua357 and ΔD proteins. Representative embryos illustrating the phenotypes are shown. (D) The ΔD protein is able to form dimers with wild-type pXqua. 35S-Labeled pXqua357 or ΔD was incubated with either 2 μg of GST alone or a GST–Xqua fusion protein coupled to glutathione–Sepharose. The bound proteins were resolved on 12% SDS-PAGE and visualized by fluorography. The lane marked T represents 20% of the input translation. (E) ΔD/pXqua heterodimers have reduced RNA-binding activity. 35S-Labeled pXqua was synthesized in vitro, either alone or by cotranslation with a fivefold excess of ΔD mRNA. The RNA-binding activity of the wild-type pXqua, or the ΔD/pXqua mixture, was assayed by binding to poly(G)–agarose. Bound proteins were resolved on 12.5% SDS-PAGE and quantitated by PhosphorImager analysis. The results, averaged from three separate experiments, are presented as a histograph that indicates that ΔD/pXqua heterodimers exhibit impaired RNA binding. Dorsal overexpression of ΔD resulted in a dose-dependent deletion of anterior structures ranging from (F) microcephalic and (G) acephalic embryos at low mRNA levels (0.5–1.25 ng/blastomere) to (H) severe anterior-half truncations at higher doses (2.5 ng/blastomere). (I) Lineage tracing of embryos coinjected with a high dose of ΔD and β-galactosidase mRNA shows that the phenotypic effects of ΔD are specific to dorsal tissues. The tissue expressing the injected mRNA is stained blue.
To further elucidate the role of pXqua in development, we wished to disrupt its function in the embryo. We chose to overexpress the ΔD deletion mutant (Fig. 7C) that lacks the KH domain and exhibits severely impaired RNA-binding activity (Fig. 2). Because pXqua is able to form homodimers, the ΔD mutant should interact with endogenous pXqua and inhibit the normal biochemical function. To validate the use of ΔD as a dominant interfering agent, we have carried out two sets of experiments: the first demonstrating that ΔD is able to dimerize with wild-type pXqua (Fig. 7D) and the second demonstrating that ΔD/pXqua heterodimers exhibit impaired RNA-binding activity (Fig. 7E). The protein binding experiments presented in Figure 7D show that ΔD is able to bind to GST–Xqua at levels equivalent to wild-type pXqua, strongly suggesting that ΔD will form heterodimers with endogenous pXqua in the embryo. The in vitro RNA-binding assays presented in Figure 7E show that a fivefold excess of ΔD over wild-type pXqua357 reduces RNA binding to just 35% of control levels. We estimate that our overexpression protocol will result in at least a 100-fold excess of ΔD over endogenous pXqua (based on relative RNA levels). Thus, overexpression of ΔD in the embryo should effectively inhibit endogenous pXqua function, by interfering with the protein’s ability to bind to its in vivo target RNA.
Synthetic mRNA encoding ΔD was microinjected into either the dorsal or ventral equatorial region at the four-cell stage, and embryos were allowed to develop until the late tailbud stage. Dorsal overexpression of ΔD resulted in a dose-dependent deletion of anterior/axial structures. The phenotype ranged from microcephaly and acephaly at lower doses (0.5–1.25 ng/blastomere) to loss of the entire anterior half of the embryo at the highest dose (2.5 ng/blastomere) (Fig. 7F–I; Table 1). Spina bifida was also frequently observed in cases showing severe anterior truncation. Overexpression of ΔD in ventral regions resulted in about half of the embryos having tail defects, but otherwise the morphology was normal. Lineage tracing, using coinjected GFP of β-galactosidase mRNA, indicates that tail defects only occur when the majority of injected mRNA is localized to the posterior region including the ventral circumblastoporal tissue and the tail blastema (Fig. 7I), both of which normally express Xqua (Fig. 6B,D,F,G). In these cases we observe a disruption of blastopore closure. When the majority of the injected ΔD mRNA is localized to regions of the embryo that do not normally express Xqua, such as the gut or ectoderm, the embryos develop normally. We therefore conclude that the ΔD effects are specific to cells that normally express Xqua.
We have examined embryos overexpressing pXqua357 and ΔD by histology and by whole-mount immunostaining using antibody markers to detect neural tissue, notochord, and somites. Histological sections and immunostaining show that in control embryos, the anterior end of the notochord usually terminates in a hook structure just behind the forebrain (Fig. 8A,G). Dorsal overexpression of Xqua results in an anterior extension of the notochord, in many cases until it contacts the ventral pharyngeal endoderm (Fig. 8B). This enhanced development of the anterior chordamesoderm is often accompanied by a disruption in the overlying neural tissue with the loss of forebrain features and a single expanded midbrain/hindbrain vesicle (Fig. 8B,H). In cases where Xqua overexpression caused a duplication of anterior head structures such as cement glands or eyes, immunostaining shows a duplication or forking of the notochord. In a few cases, as many as three notochords can be seen lying side by side (Fig. 8J). In contrast, dorsal overexpression of the ΔD mutant has the opposite effect to overexpression of wild-type Xqua and caused a deletion of anterior structures. In microcephalic and acephalic embryos resulting from overexpression of ΔD at moderate levels (0.5–1.25 ng/blastomere), the anterior notochord is truncated, and this is accompanied by a loss of forebrain and facial structures (Fig. 8C). Histology and immunostaining of the more severe anterior truncations resulting from high-level ΔD overexpression (2.5 ng/blastomere) show a complete loss of all head structures and a total absence of the anterior portion of the notochord as well as a dramatic reduction in neural and somitic tissue (Fig. 8D, K–M). In most cases a small amount of posterior notochord tissue is observed in the tip of the tail, which is split in two as a result of spina bifida (Fig. 8M). Lineage tracing confirmed that the residual notochord developed in the posterior region of the axial mesoderm that received a relatively low dose of ΔD mRNA (data not shown).
Figure 8.
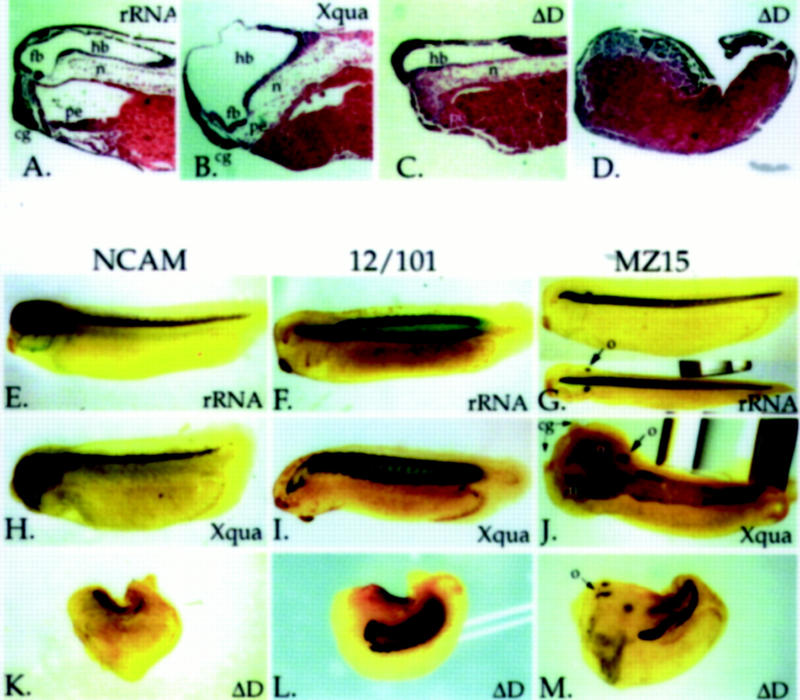
Histological and immunostaining analysis of abnormal development resulting from dorsal Xqua and ΔD overexpression. Control rRNA, Xqua, or ΔD was microinjected into the equatorial region of each of the two dorsal blastomeres at the four-cell stage (2.5 ng/blastomere, except embryo in C that was 1.25 ng/blastomere). Representative embryos were examined by (A–D) histological sectioning and hemotoxylin/eosin staining or by whole-mount immunostaining with the monoclonal antibodies (E,H,K) Anti-NCAM for neural tissue, (F,I,L) 12/101 for somites, and (G,J,M) MZ15 for notochord. Control embryos injected with rRNA are shown in A, E, F, and G; embryos overexpressing Xqua are shown in B, H, I, and J; and embryos overexpressing ΔD are shown in C, D, K, L, and M. In all panels, anterior is left and a lateral view is shown, except the bottom image in G and J, which are dorsal views. (A,G) In control embryos the notochord ends anteriorly behind the forebrain. (B) Dorsal injection of Xqua results in an elongation of the notochord into the head until it reaches the ventral pharyngeal endoderm and (B,H) a reduction in forebrain and anterior facial structures with an enlarged hindbrain vesicle. (J, dorsal view) Xqua overexpression occasionally results in duplications of the notochord and facial structures; notice two notochords lying side by side and two cement glands. (I) The somites are relatively normal in Xqua overexpressing embryos. (C) Histological section of embryos with a low-level overexpression of ΔD (1.25 ng/blastomere) shows a truncation of the anterior notochord with a loss of forebrain and anterior head structures. (D,K,L,M) High-level overexpression of ΔD (2.5 ng/blastomere) causes a complete truncation of the anterior half of the embryos. (K) These embryos exhibit little neural tissue, (L) reduced somites, and (M) only small amounts of posterior notochord tissue. ΔD embryos show a high frequency of spina bifida resulting in a splitting of the residual notochord tissue; notice the duplicated notochord in the tail. (cg) Cement gland; (fb) forebrain; (hb) hindbrain; (n) notochord; (o) otic vesicle; (pe) pharyngeal endoderm.
Xqua is essential for notochord differentiation
Because Xqua is normally expressed in a number of neural and mesodermal tissues, it is likely that overexpression of Xqua or ΔD influences the development of each of these tissues directly. However, many of the phenotypic effects that we observe are likely to result from disruption of the normal development of the chordamesoderm and its derivative, the notochord. In this case, impaired chordamesoderm development could influence its neural inducing and embryonic patterning properties, thereby causing defects in neural tissue and other anterior structures. In practice, we expect chordamesoderm and notochord tissues to be particularly vulnerable in these overexpression studies because they are the earliest domains of Xqua expression in the embryo and levels of most injected mRNA decline rapidly after gastrulation (Vize et al. 1991). To simplify analysis, subsequent experiments have focused on the role of Xqua in notochord development.
To avoid possible secondary effects that may occur in whole-embryo studies, the effects of Xqua and ΔD overexpression on notochord differentiation have been examined in dorsal mesoderm explants. In these experiments, Xqua or ΔD mRNA was microinjected into dorsal blastomeres at the four-cell stage (2.5 ng/blastomere). At early gastrula, the dorsal half of the mesoderm, which contains the prospective notochord and somites, was dissected from the embryo, and two of these dorsal mesoderm pieces were combined and cultured in a saline solution. The results of a typical experiment are presented in Figure 9. Because chordamesoderm undergoes convergent extension cell movements, the external morphology of the explants provides some indication of the amount of notochord that has differentiated (Fig. 9C,E,G). Dorsal marginal zone explants from embryos overexpressing Xqua (Fig. 9E) consistently produce more extended structures than control explants injected with rRNA (Fig. 9C). This result is consistent with the extended notochords observed in whole embryos overexpressing Xqua. Conversely, explants from embryos injected with ΔD mRNA (Fig. 9G) showed less elongation compared with control explants. When sibling control embryos had reached tailbud stage, explants were fixed, sectioned, and immunostained for the presence of notochord (black) and somite (red). Twelve independent explants from two separate injection experiments produced consistent results, and a representative section through a typical explant of each type is shown (Fig. 9D,F,H). Explants overexpressing Xqua were more elongated but showed only a slight increase in the total amount of notochord tissue, ∼10% more notochord tissue than in control explants (Fig. 9F). It is possible that the apparent increase in notochord tissue may be owing to enhanced morphogenic movements. In contrast, in all 12 cases, much less notochord tissue differentiated in explants overexpressing ΔD mRNA, relative to rRNA-injected controls (Fig. 9, cf. D and H). ΔD overexpressing explants had ∼10% of the amount of notochord found in control explants. In general, somite differentiation was unaffected in these experiments. To demonstrate that ΔD overexpression is inhibiting notochord development by specifically interfering with Xqua function, we have performed rescue experiments. In every case (six explants), notochord development was significantly rescued in explants coexpressing Xqua and ΔD (at a 1:1 mRNA ratio) relative to explants overexpressing ΔD alone (Fig. 9, cf. D, H, and I). Rescue explants had on average 50% of the amount of notochord found in uninjected control explants, up from only 10% found in ΔD explants. Similar rescue experiments using whole embryos were inconclusive owing to the large amount of mRNA required (at least 10 ng) that caused nonspecific gastrulation defects. As a result of the Xqua and ΔD overexpression experiments, we conclude that normal Xqua function is essential for notochord differentiation.
Figure 9.
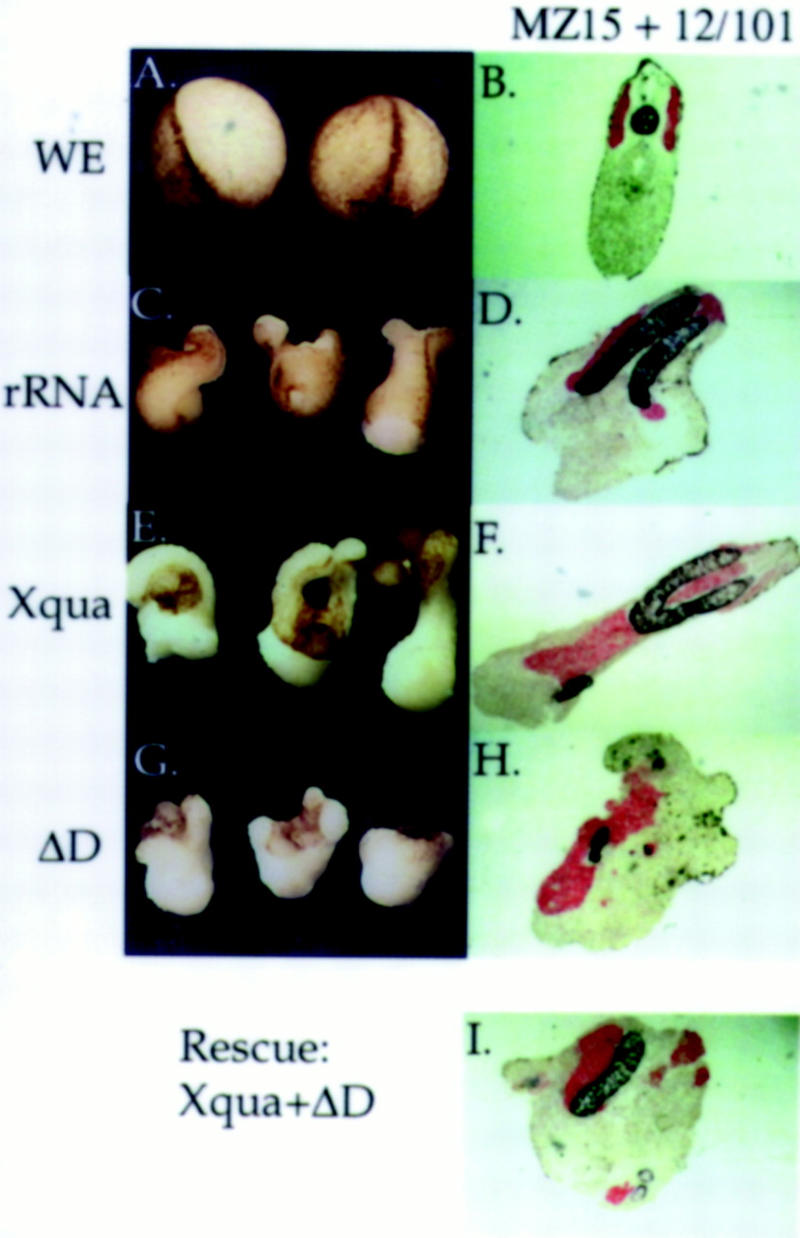
Overexpression of the pXqua mutant ΔD inhibits notochord differentiation. rRNA (2.5 ng), Xqua, or ΔD mRNA was injected into each of the two dorsal blastomeres at the four-cell stage. Dorsal mesoderm explants containing presumptive notochord and somite tissue were dissected from injected embryos at early gastrula (stage 10–10.5) and cultured in isolation to examine notochord differentiation directly. (A,C,E,G) External morphology of the explants is shown when sibling embryos (WE) reached early neurula stage. (B,D,F,H,I). At later tailbud stage, explants were fixed, sectioned, and immunostained with MZ15 for notochord (black) and 12/101 for somite (red). From a total of 12 explant experiments, a section through a typical explant is presented. (C) Control rRNA explants exhibit considerable elongation resulting from convergent extention movements of the developing notochord, and (D) immunostaining of control explants shows a substantial amount of notochord and somite tissue. (E) Xqua overexpressing explants exhibit more dramatic morphological elongation and (F) contain moderately more notochord and somite tissue than controls (cf. amount of unstained tissue to that in D). (G) Explants overexpressing ΔD exhibit very little morphological elongation compared with controls (cf. to C) and (H) show dramatically less notochord tissue but little change in the amount of somite tissue. (I) Inhibition of notochord differentiation owing to ΔD overexpression is rescued by coinjection of an equal amount of synthetic Xqua mRNA (5 ng of ΔD + 5 ng of Xqua) (cf. amount of notochord in I to H and D), indicating that the biological effects of ΔD overexpression are specific to the Xqua pathway.
Xqua is involved in early stages of notochord specification
Our explant experiments show that Xqua function is essential for notochord development. To learn more about this effect, we have investigated whether Xqua is required for terminal differentiation of the notochord or whether it is functioning during specification. Because Xqua is expressed early in the prenotochord tissue during gastrulation, we have looked at the effect of ΔD overexpression on early chordamesoderm and notochord specific markers. Embryos were injected in the two dorsal blastomeres at the four-cell stage with 2.5 ng/blastomere mRNA encoding the dominant inhibitor ΔD or with ΔD plus β-galactosidase mRNA as a lineage tracer. Previous experiments have demonstrated that this dose is sufficient to strongly inhibit notochord differentiation (Figs. 7H, 8M, and 9H). At the gastrula stage, injected embryos were assayed for the following transcripts: (1) Xnot-2 (Gont et al. 1993), which marks the early notochord; (2) Xbra (Smith et al. 1991), Xenopus bracyhury, which is expressed in the notochord in late gastrula and is panmesodermal in early gastrula; and (3) gsc (Cho et al. 1991), which marks the dorsal lip and the presumptive prechordal plate mesoderm. The first morphological effects of ΔD overexpression are detected at the late gastrula stage when the embryos exhibit disrupted gastrulation and the blastopore does not close properly (Fig. 10, cf. A and D, morphology of control embryos, with B, C, and E, experimental embryos). Analyis using in situ hybridization indicates that transcript levels for all three markers are severely down-regulated by ΔD overexpression. In particular, expression of Xnot-2 and Xbra is completely absent from the dorsal midline where the notochord would normally form (Fig. 10A–F). Residual Xnot-2 expression is often seen in two lateral patches. These apparently correlate with the two remnant notochords that are often observed at later stages in the tail of ΔD-injected embryos (Fig. 8M). Lineage tracing using β-galactosidase activity (Fig. 10C,F,I,L) confirms that the effect is restricted to cells that express the dominant-negative ΔD. These results demonstrate that Xqua is acting very early during development of the chordamesoderm and that pXqua function is required for the accumulation of several essential mRNAs. Given the down-regulation of Xnot-2, Xbra, and gsc transcripts in embryos in which Xqua function has been disrupted, it is not surprsing that notochord development is impaired in these embryos.
Figure 10.
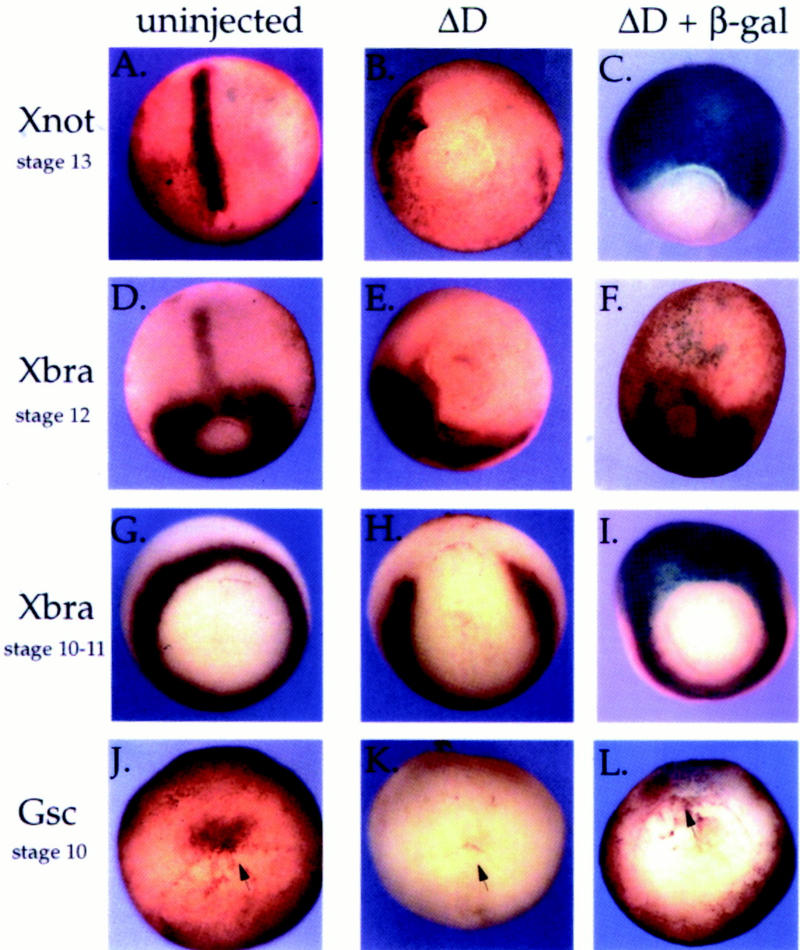
Inhibition of Xqua function results in down-regulation of early notochord marker mRNAs. mRNA (2.5 ng) encoding the dominant-negative pXqua protein ΔD or ΔD plus β-galactosidase was microinjected into the equatorial region of each of the two dorsal blastomeres at the four-cell stage. The resulting embryos, together with uninjected control embryos, were fixed at gastrula stages and analyzed by whole-mount in situ hybridization for expression of early dorsal mesoderm and notochord mRNA markers: Xnot-2 (A–C), Xbra (D–I), and gsc (J–L). Arrows indicate the location of the dorsal lip in early stage embryos. In C, F, I, and L, embryos were stained with X-gal (blue) prior to in situ hybridization (purple/brown), to indicate the cells that received the ΔD mutant. Overexpression of ΔD in the dorsal mesoderm results in disruption of gastrulation and a loss of detectable Xnot-2, Xbra, and gsc transcripts in recipient cells.
Discussion
RNA-binding properties of pXqua
Our experiments show that pXqua, which contains a single KH motif, can bind RNA in vitro. This is also true for S3 (Urlaub et al. 1995) and the GSG protein SAM68 (Wong et al. 1992; Taylor and Shalloway 1994; Wang et al. 1995), which also contain single KH motifs. However, most other KH proteins require the cooperation of multiple RNA-binding motifs for efficient RNA binding (Siomi et al. 1994). Therefore, it was not entirely clear how proteins with a single KH motif could bind RNA. In pXqua the KH domain is essential but not sufficient for full RNA-binding activity, because the carboxyl terminus is also required. There are several possibilities to explain the necessity of the carboxy-terminal region. The carboxyl terminus may contain an RNA-binding motif that cooperates directly with the KH domain for RNA binding. This seems unlikely, however, because extensive sequence comparisons of the carboxy-terminal region have detected no known RNA-binding motifs. On the other hand, we have demonstrated that pXqua can homodimerize, and it is possible that this is an essential precondition for RNA binding. This argument is supported by the observation that addition of ΔD to pXqua inhibits RNA binding, presumably owing to the formation of ΔD/pXqua heterodimers. Deletion of the carboxyl terminus of pXqua may remove or disrupt sequences essential for dimerization thereby inhibiting RNA-binding activity.
The proteins FMR1, FXR1, and FXR2, all of which have two KH motifs, can homo- and heterodimerize with each other (Zhang et al. 1995), although it is not known whether this interaction is required for RNA binding. It has been proposed that GSG proteins may be involved in protein–protein interactions (Jones and Schedl 1995; Musco et al. 1996), and our pXqua studies provide the first demonstration of homodimerization in this class of protein. It is possible that homodimerization is a general feature of GSG proteins, explaining how they can effectively bind RNA with only one KH domain. If dimerization-dependent RNA binding is a general feature of GSG proteins, then, in addition to elements in the carboxy-terminal region of pXqua, it is reasonable to suggest that sequences within the conserved GSG domain might also be involved in protein interactions. Structural analysis of the GSG-type KH domain predicts an extended loop region that is absent in typical KH motifs, and it is speculated that this loop may be involved in protein–protein interactions (Musco et al. 1996). The ΔD mutant, in which the first part of the KH core domain is deleted, still dimerizes with pXqua, indicating that this region is not essential for protein–protein interactions. The loop region that is conserved between GSG family members is left intact in the ΔD mutant and therefore remains a prime candidate for a dimerization motif. We are currently mapping the protein interaction domains of pXqua and investigating the relationship between dimerization and RNA binding.
Subcellular distribution of quaking
We have shown that pXqua is distributed between the nuclear and cytoplasmic compartments of the cell and that the nuclear localization signal (NLS) is contained within the last 60 amino acids of the protein. Examination of the carboxyl terminus does not reveal a recognizable NLS although the last 60 amino acids contain a concentration of basic residues that are often found in the NLS. In mouse, the 5-, 6-, and 7-kb qkI mRNAs generate the QK1-5, QK1-6 and QK1-7 proteins, respectively, which are identical except for the extreme carboxy-terminal sequences (Ebersole et al. 1996). Recent immunolocalization studies in mouse brain tissue using antibodies specific for each of the three distinct QK1 carboxy-terminal regions show that QK1-5 (equivalent to pXqua357) is detected in the nucleus and cytoplasm, whereas QK1-6 and QK1-7 are almost exclusively cytoplasmic (Hardy et al. 1996). These observations are consistent with our results; furthermore, it appears that the alternative carboxy-terminal regions regulate the subcellular location of the QK1 proteins. Because the last 60 amino acids of the mouse and Xenopus 5-kb quaking sequences are identical (Zorn et al. 1997), the NLS of pXqua357 (QK1-5) is likely to reside in the last 30 amino acid residues that are alternatively spliced. It will be interesting to determine whether the different isoforms can heterodimerize and if so, what effect this has on their subcellular distribution and hence their function.
Xqua is essential for notochord development
The earliest embryonic expression of Xqua occurs in the chordamesoderm of the dorsal blastopore lip. This region of tissue, which is known as the organizer, is particularly interesting because it differentiates into the notochord, has neural-inducing activity, has a major role in embryonic patterning, and is responsible for the morphogenic movements of gastrulation. To our knowledge, this is the first example of an RNA-binding protein that is expressed in the vertebrate organizer region and its major derivative, the notochord. To date, expression of qkI mRNA in the chordamesoderm of the mouse embryo has not been reported, but expression studies on early embryos have been limited. Our experiments show that ectopic expression of Xqua does not cause notochord differentiation at ectopic locations, but it can enhance the development of notochord tissue, suggesting that Xqua is probably involved in the maintenance of the differentiation process rather than determination of notochord cell fate. Similar phenotypic effects, that is, increased notochord development and subsequent enlargement of the hindbrain, have been observed from overexpression of the Xnot-2 homeobox sequence, which is also normally expressed in the organizer and notochord (Gont et al. 1996). In our experiments, blocking endogenous Xqua function by overexpression of the dominant interfering Xqua protein, ΔD, inhibits the differentiation of the notochord, demonstrating that Xqua is essential for this process. Furthermore, the neural inducing properties of the chordamesoderm/organizer are inhibited in these embryos, resulting in little or no anterior neural tissue. Phenotypically similar aneural embryos are observed when formation of the chordamesoderm/organizer is inhibited by UV irradiation of fertilized Xenopus eggs (for review, see Kao and Danilchik 1991). Analysis of early gene transcripts suggests that the phenotypic effects of ΔD overexpression can be explained by a down-regulation of essential mRNAs such as Xnot-2, Xbra, and gsc and possibly many others. Based on comparisons to the mouse bracyhury mutation (Herrmann et al. 1990) and the zebrafish floating head (zebrafish Xnot) mutations (Talbot et al. 1995), down-regulation of Xbra and Xnot mRNAs could largely account for the observed phenotype. Interestingly, some of the qke embryonic lethal mutant mice have axial defects and failure of head development (Justice and Bode 1988), with a phenotype reminiscent of the Xenopus embryos resulting from overexpression of mutant ΔD protein. These qke phenotypes could be explained, by analogy with our results, if quaking also functions in mammals to maintain chordamesoderm development and its embryonic organizer activity.
The role of quaking in vertebrate development
Our overexpression experiments have focused on the role of pXqua in the development of the chordamesoderm and notochord. These tissues are the sites of earliest Xqua expression in the embryo and are therefore the most easily disrupted in overexpression studies. Whereas our experiments show that Xqua function is critical for notochord development, quaking is expressed in a number of different embryonic and adult tissues and is therefore likely to be required for the differentiation or development of many cell types. The later embryonic expression of Xqua in the head, heart, and somites is similar to the pattern of qkI expression observed in mouse embryos (Ebersole et al. 1996) and correlates well with the diminished head folds, disorganized somites, and malformed hearts observed in qke mutant embryos (Justice and Bode 1988). The high levels of Xqua expression in the developing central nervous system, particularly in the proliferating and actively differentiating layers of the developing brain where the neuronal and oligodendrocytes originate, are exactly where qkI expression is detected in the mouse neonatal brain (Ebersole et al. 1996) and are consistent with the myelination defects observed in qkv mice (Sidman et al. 1964; Samorajski et al. 1970). Recent immunostaining studies show a dramatic reduction of QK1 (equivalent to pXqua357) protein levels in oligodendrocytes and Schwann cells of qkv mutant mice, compared with wild-type mice (Hardy et al. 1996). The reduction of QK1 is directly correlated with the dismyelination defect and suggests that lack of the QK1 protein, and hence lack of QK1 function, prevents proper differentiation of myelinating cells. This observation parallels our experimental results where inhibition of pXqua357 function by overexpression of pXqua mutants results in an inhibition of notochord differentiation.
Although more information is needed to define the biochemical function of quaking, available evidence suggests that it regulates expression of transcripts essential for differentiation of some cells. KH proteins have been implicated in a diversity of processes at all levels of RNA metabolism although in most cases their precise biochemical functions are unclear. Because pXqua is present in both cytoplasmic and nuclear components, we should consider a role for the protein in each compartment. Interestingly, some abnormalities in RNA metabolism have been observed in the neural tissue of qkv mutant mice. Proportions of particular alternatively spliced mRNA isoforms of myelin-associated glycoprotein, MAG (Fujita et al. 1988; Braun et al. 1990; Bartoszewicz et al. 1995), and myelin basic protein, MBP (Carnow et al. 1984), are altered in qk mice, whereas other mRNAs are unaffected. These observations are consistent with the experimental results indicating that blocking Xqua function prevents the accumulation of Xnot-2, Xbra, and gsc transcripts. The fact that levels of all three tested marker transcripts were reduced suggests that pXqua somehow regulates a number of different mRNAs. It is also possible that quaking may act more generally by regulating global mRNA levels in mesodermal and neural cells. Although the mechanism of action is unknown, available evidence suggests that quaking regulates mRNA stability, processing, or both and that blocking quaking function prevents specific mRNAs from accumulating to detectable levels.
Materials and methods
Production of synthetic mRNA and proteins
PCR with Vent polymerase (NEB) was used to generate deletions of the Xqua357 coding sequence, starting with pSKXqua8c cDNA as a template (Zorn et al. 1997). Fragments were cloned into the expression vector pT7TS (a modified version of pSP64T; Krieg and Melton 1984) or pT7TS–HA, which provides an amino-terminal HA epitope tag. Templates pT7TS–Xqua357, pT7TS–Xqua365, pT7TS–HAXqua357, and the Xqua deletion constructions were linearized with BamHI and transcribed with T7 RNA polymerase. pSPXMax4 (Tonissen and Krieg 1994) was linearized with BamHI and transcribed with SP6 RNA polymerase. pXef1 (Krieg et al. 1989) was linearized with EcoRI and transcribed with T7 RNA polymerase. pFR, encoding the Xenopus fibroblast growth factor (FGF) receptor (a gift from Dr. E. Amaya, Wellcome/CRC Institute, Cambridge, UK), was linearized with PstI and transcribed with SP6 RNA polymerase. pXK1 encoded the Xenopus hnRNP K (Siomi et al. 1993a). Synthetic capped mRNA was synthesized as described (Krieg and Melton 1984) or with the Mega-Script kit (Ambion) using cap analog (NEB). 35S-labeled proteins were translated in vitro in reticulocyte lysates (BRL or Promega).
RNA binding assays
RNA binding of in vitro translated proteins was performed essentially as described by Swanson and Dreyfuss (1988). Binding reactions used 105 cpm of labeled protein and 30 μl of RNA–agarose resin (10–60 μg of RNA) in 0.5 ml of binding buffer (10 mm Tris-HCl at pH 7.5, 100 mm NaCl, 2.5 mm MgCl2, 0.5% Triton X-100, and 0.1 mg/ml of BSA) for 10 min with rocking at 4°C. The beads were washed five times with ice-cold binding buffer. Proteins bound to the RNA–agarose were released by boiling in SDS–protein sample buffer, resolved on SDS-PAGE, and visualized by fluorography. Homopolymer RNA–agarose was purchased from Sigma. Total Xenopus embryonic–agarose was generated as follows: RNA at 1 mg/ml in water was biotinylated with photoactivated biotin–acetate (Vector Labs) according to manufacturers instructions, and RNA–biotin was bound to streptavidin–agarose resin (BRL) at a final concentration of 0.1–2 μg of RNA per 10 μl of agarose resin.
Immunoprecipitation
Approximately one-fifth of the labeled proteins generated in a translation reaction were incubated with 5 μg/ml of monoclonal antibody 12CA5 (Boehringer Mannheim) and 20 μl of protein A–agarose (Sigma) in IP buffer (50 mm Tris-HCl at pH 7.5, 150 mm NaCl, 5 mm MgCl2, 1 mm EDTA, 0.25% NP-40, 1 mm PMSF, and 1 mm DTT) for 2 hr at 4°C with rocking. After precipitation, pellets were washed three times in ice-cold IP buffer and specifically bound proteins were eluted by incubation in 50 μl of 1 mm HA peptide (YPYDVPYA) (a gift from Dr. T. Kouzarides, Wellcome/CRC Institute, Cambridge, UK) at room temperature for 15 min. Precipitated proteins were resolved on 10% SDS-PAGE and visualized by fluorography.
GST fusion protein binding assays
The Xqua357 coding sequence was cloned in-frame with GST in the vector pGEX-3X. GST–Xqua or GST alone (a gift from Dr. P. Lavender, Wellcome/CRC, Cambridge, UK) was expressed and purified on glutathione–Sepharose. Approximately one-fifth of the labeled proteins generated in a translation reaction were incubated with 2 μg of GST–Xqua or GST alone coupled to glutathione–Sepharose in 500 μl of binding buffer (25 mm HEPES at pH 7.5, 12.5 mm MgCl2, 150 mm KCl, 20% glycerol, 0.1% NP-40, 1 mm DTT, and 1 mg/ml of BSA). Following 1-hr incubation at room temperature, the Sepharose was pelleted and washed four times in NETN (20 mm Tris at pH 8, 100 mm NaCl, 1 mm EDTA, and 0.5% NP-40). The bound proteins were resolved on 12.5% SDS-PAGE and visualized by fluorography.
In situ hybridization, immunocytochemistry, and histology
Embryos and explants were fixed in MEMFA (Hemmati-Brivanlou and Harland 1989) for 2–3 hr and then stored in methanol at −20°C. Whole-mount in situ hybridization was performed essentially as described by Harland (1991) with modifications detailed by Drysdale et al. (1994). Antisense DIG RNA probes, specific to the embryonic 5-kb Xqua mRNA, were transcribed from NotI linearized pSKXqua11c cDNA template (Zorn et al. 1997). Other probe templates were prepared as described previously. pBSK Xnot-2 (Gont et al. 1993), pXbra (Smith et al. 1991), and pGsc (Cho et al. 1991). Whole-mount immunocytochemistry was performed using the following monoclonal antibodies. Anti-NCAM (Sigma, NCAM-OB11) for neural tissue, 12/101 (Kintner and Brockes 1984) for somite, and MZ15 (Smith and Watt 1985) for notochord. Embryos were rehydrated in MAB (100 mm maleic acid at pH 7.5 and 150 mm NaCl), blocked for 2 hr in BR-MAB [2% blocking reagent (Boehringer Mannheim) in MAB], incubated with the appropriate antibody (1/200 dilution in each case) overnight at 4°C in BR-MAB followed by washing, 12 × 30 min in MAB. These steps were repeated sequentially with rabbit anti-mouse Ig antibody (ICN, 1/200 dilution) followed by alkaline phosphatase–anti-alkaline phosphatase (APAAP; Boehringer Mannheim, 5 U/ml). Color reactions were performed either by NBT/BCIP (black) or BM Purple (Boehringer Mannheim). Stained embryos were fixed in Bouin’s solution and photographed in benzyl alcohol/benzyl benzoate (1:2). Sections (10 μm) were cut from tissue embedded in Histoplast/beeswax (98:2) followed by either staining with hematoxylin/eosin or double-immunostained with MZ15 and 12/101 as described in Carnac et al. (1996).
Xenopus oocytes and embryo manipulations
Oocytes were surgically removed from anesthetized female Xenopus, manually defolliculated, and cultured in 1× MBS (Gurdon 1977) at 18°C. Oocytes were microinjected with ∼30 ng of synthetic capped mRNA in a volume of ∼30 nl. After 16–24 hr of incubation in the presence of 1 mCi/ml of [35S]methionine, the nucleus was manually removed from the oocyte and both nuclear and cytoplasmic fractions were homogenized in IP buffer. Protein extracts were resolved on SDS-PAGE and visualized by fluorography. Embryos were cultured as described (Gurdon 1977) and staged according to Nieuwkoop and Faber (1967). Embryos, in 4% Ficoll/MBS, were microinjected with synthetic mRNA (4.9 nl/blasotomere) and subsequently transferred to 1/10 MBS. Dorsal–ventral polarity was predicted by differential pigmentation and verified by coinjection of GFP mRNA as a lineage marker (Zernicka-Goetz et al. 1996). Dorsal marginal zone tissue was dissected at stage 10–10.5, and two pieces of tissue were placed in opposition and cultured in 1× MBS.
Acknowledgments
We are grateful to Prof. J. B. Gurdon in whose lab the overexpression experiments were performed. We thank Gilles Carnac for advice and encouragement and E. Bellefroid, F. Stennard, and K. Ryan for critical reading of the manuscript. We thank Andy Mitchell for help with histology and P. Lavander and C. Le Chalony for advice on the GST-fusion protein experiments. We especially thank T. Ebersole, Q. Chen, and K. Artzt for providing the original mouse quaking probe. A.M.Z. was supported by a Natural Sciences and Engineering Research Council of Canada postdoctoral fellowship and the Cancer Research Campaign. This work was supported by National Institutes of Health grant HD25179 to P.A.K.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL amz@mole.bio.cam.ac.uk; FAX 01223-334089.
References
- Bartoszewicz ZP, Noronha AB, Fujita N, Sato S, Bo L, Trapp BD, Quarles RH. Abnormal expression and glycosolation of the large and small isoforms of myelin-associated glycoprotein in dysmyelinating quaking mutants. J Neurosci Res. 1995;41:27–38. doi: 10.1002/jnr.490410105. [DOI] [PubMed] [Google Scholar]
- Bode VC. Ethylnitrosourea mutagenesis and the isolation of mutant alleles for specific genes located in the t region of the mouse chromosome 17. Genetics. 1984;108:457–470. doi: 10.1093/genetics/108.2.457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun PE, Horvath E, Edwards AM. Two isoforms of myelin-associated glycoprotein accumulate in quaking mice: Only the large polypeptide is phosphorylated. Dev Neurosci. 1990;12:286–292. doi: 10.1159/000111857. [DOI] [PubMed] [Google Scholar]
- Carnac G, Kodjabachia L, Gurdon JB, Lemaire P. The homeobox gene Siamois is a target of the Wnt dorsalisation pathway and triggers organiser activity in the absence of mesoderm. Development. 1996;122:3055–3065. doi: 10.1242/dev.122.10.3055. [DOI] [PubMed] [Google Scholar]
- Carnow TB, Carson JH, Brostoff SW, Hogan EL. Myelin basic protein gene expression in quaking, jimpy and myelin synthesis-deficient mice. Dev Biol. 1984;106:38–44. doi: 10.1016/0012-1606(84)90058-7. [DOI] [PubMed] [Google Scholar]
- Cho KWY, Blumberg B, Steinbeisser H, DeRobertis EM. Molecular nature of Spemann’s organizer: The role of the Xenopus homeobox gene goosecoid. Cell. 1991;67:1111–1120. doi: 10.1016/0092-8674(91)90288-a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colman A. Translation of eukaryotic messenger RNA in Xenopus oocytes. In: Hames B, Higgins S, editors. Transcription and translation. Oxford, UK: IRL; 1984. pp. 271–302. [Google Scholar]
- Cruz-Alvarez M, Pellicer A. Cloning of a full-length complementary DNA for an Artemia salina glycine-rich protein. J Biol Chem. 1987;262:13377–13380. [PubMed] [Google Scholar]
- De Boulle K, Verkerk AJ, Reyniers E, Vits L, Hendrickx J, VanRoy B, VanDen Bos F, deGraaff E, Oostra BA, Willems PJ. A point mutation in the FMR-1 gene associated with fragile X mental retardation. Nature Genet. 1993;3:31–35. doi: 10.1038/ng0193-31. [DOI] [PubMed] [Google Scholar]
- Draper BW, Mello CC, Bowerman B, Hardin J, Priess JR. MEX-3 is a KH domain protein that regulates blastomere identity in early C. elegans embryos. Cell. 1996;87:205–216. doi: 10.1016/s0092-8674(00)81339-2. [DOI] [PubMed] [Google Scholar]
- Drysdale TA, Tonissen KF, Patterson KD, Crawford MJ, Krieg PA. Cardiac Troponin is a heart-specific marker in the Xenopus embryo: Expression during abnormal heart development. Dev Biol. 1994;165:432–441. doi: 10.1006/dbio.1994.1265. [DOI] [PubMed] [Google Scholar]
- Ebersole T, Chen Q, Justice MJ, Artzt K. The quaking gene product necessary in embryogenesis and myelination combines features of RNA binding and signal transduction proteins. Nature Genet. 1996;12:260–265. doi: 10.1038/ng0396-260. [DOI] [PubMed] [Google Scholar]
- Fujita N, Sato S, Kurihara T, Inuzuka T, Takahashi Y, Miyatake T. Developmentally regulated alternative splicing of the brain myelin-associated glycoprotein mRNA is lacking in the quaking mouse. FEBS Lett. 1988;232:323–327. doi: 10.1016/0014-5793(88)80762-2. [DOI] [PubMed] [Google Scholar]
- Fumagalli S, Totty NF, Hsuan JJ, Courtneidge SA. A target for Src in mitosis. Nature. 1994;368:871–874. doi: 10.1038/368871a0. [DOI] [PubMed] [Google Scholar]
- Gibson TJ, Thompson JD, Heringa J. The KH domain occurs in a diverse set of RNA-binding proteins that include the anti terminator NusA and is probably involved in binding to nucleic acid. FEBS Lett. 1993;324:361–366. doi: 10.1016/0014-5793(93)80152-k. [DOI] [PubMed] [Google Scholar]
- Golbfarb D, Michaud N. Pathways for the nuclear transport of proteins and RNAs. Trends Cell Biol. 1991;1:20–24. doi: 10.1016/0962-8924(91)90065-h. [DOI] [PubMed] [Google Scholar]
- Gont LK, Steinbeisser H, Blumberg B, De Robertis EM. Tail formation as a continuation of gastrulation: The multiple cell populations of the Xenopus tail bud derived from the late blastopore lip. Development. 1993;119:991–1004. doi: 10.1242/dev.119.4.991. [DOI] [PubMed] [Google Scholar]
- Gont LK, Fainsod A, Kim S-H, De Robertis EM. Overexpression of the Homeobox gene Xnot-2 leads to notochord formation in Xenopus. Dev Biol. 1996;174:174–178. doi: 10.1006/dbio.1996.0061. [DOI] [PubMed] [Google Scholar]
- Gurdon JB. Methods for nuclear transplantation in Amphibian. Methods Cell Biol. 1977;16:125–139. doi: 10.1016/s0091-679x(08)60096-5. [DOI] [PubMed] [Google Scholar]
- Hamburger V. The heritage of experimental embryology. Oxford, UK: Oxford University Press; 1988. Hanns Spemann and the Organiser. [Google Scholar]
- Hardy RJ, Loushin CL, Friedrich VL, Chen Q, Ebersole TA, Lazzarini RA, Artzt K. Neural cell type-specific expression of QK1 proteins is altered in quaking mutant mice. J Neuro Sci. 1996;16:7941–7949. doi: 10.1523/JNEUROSCI.16-24-07941.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harland RM. In situ hybridisation: An improved whole-mount method for Xenopus embryos. Methods Cell Biol. 1991;36:685–695. doi: 10.1016/s0091-679x(08)60307-6. [DOI] [PubMed] [Google Scholar]
- Hemmati-Brivanlou A, Harland RM. Expression of an Engrailed-related protein is induced in the anterior neural ectoderm of early Xenopus embryos. Development. 1989;106:611–617. doi: 10.1242/dev.106.3.611. [DOI] [PubMed] [Google Scholar]
- Herrmann BG, Labeit S, Poustka A, King TR, Lehrach H. Cloning of the T gene required in mesoderm formation in the mouse. Nature. 1990;343:617–622. doi: 10.1038/343617a0. [DOI] [PubMed] [Google Scholar]
- Hogan EL, Greenfield S. Animal models of genetic disorders of myelin. In: Morrell P, editor. Myelin. New York, NY: Plenum Press; 1984. pp. 489–534. [Google Scholar]
- Jones AR, Schedl T. Mutations in gld-1, a female germ cell-specific tumor suppressor gene in C. elegans, affect a conserved domain also found in Src-associated protein Sam68. Genes & Dev. 1995;9:1491–1504. doi: 10.1101/gad.9.12.1491. [DOI] [PubMed] [Google Scholar]
- Justice MJ, Bode VC. Three ENU-induced alleles of the murine quaking locus are recessive embryonic lethal mutations. Genet Res Camb. 1988;51:95–102. doi: 10.1017/s0016672300024101. [DOI] [PubMed] [Google Scholar]
- Kao K, Danilchik M. Generation of body plan phenotypes in early embryogenesis. Methods Cell Biol. 1991;36:271–284. doi: 10.1016/s0091-679x(08)60282-4. [DOI] [PubMed] [Google Scholar]
- Kintner CR, Brockes JP. Monoclonal antibodies identify blastema cells derived from dedifferentiating muscle in newt limb regeneration. Nature. 1984;308:67–69. doi: 10.1038/308067a0. [DOI] [PubMed] [Google Scholar]
- Krieg PA, Melton DA. Functional messenger RNAs produced in vitro by SP6 in vitro transcription of cloned cDNAs. Nucleic Acids Res. 1984;12:7057–7071. doi: 10.1093/nar/12.18.7057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krieg PA, Varnum SM, Wormington WM, Melton DA. The mRNA encoding elongation factor 1-α (EF-1α) is a major transcript at the midblastula transition in Xenopus. Dev Biol. 1989;133:93–100. doi: 10.1016/0012-1606(89)90300-x. [DOI] [PubMed] [Google Scholar]
- Lock P, Fumagalli S, Polakis P, McCormick F, Courtneidge SA. The human p62 cDNA encodes Sam68 and not the RasGap-associated p62 protein. Cell. 1996;84:23–24. doi: 10.1016/s0092-8674(00)80989-7. [DOI] [PubMed] [Google Scholar]
- Mahone M, Saffman EE, Lasko PL. Localized Bicaudal-C RNA encodes a protein containing a KH domain, the RNA binding motif of FMR1. EMBO J. 1995;14:2043–2055. doi: 10.1002/j.1460-2075.1995.tb07196.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musco G, Stier G, Joseph C, Antonietta M, Morelli C, Nilges M, Gibson TJ, Pastore A. Three-dimensional structure and stability of the KH domain: Molecular insights into the Fragile X Syndrome. Cell. 1996;85:237–245. doi: 10.1016/s0092-8674(00)81100-9. [DOI] [PubMed] [Google Scholar]
- Nieuwkoop PD, Faber J. In: Normal table of Xenopus laevis. 2nd ed. Daudin, editor. Amsterdam, Netherlands: North-Holland; 1967. [Google Scholar]
- Piñol-Roma S, Choi YD, Matunis MJ, Dreyfuss G. Immunopurification of heterogeneous nuclear ribonucleoprotein particles reveals an assortment of RNA-binding proteins. Genes & Dev. 1987;2:215–227. doi: 10.1101/gad.2.2.215. [DOI] [PubMed] [Google Scholar]
- Powers MA, Forbes DJ. Cytosolic factors in nuclear transport: What’s importin? Cell. 1994;79:931–943. doi: 10.1016/0092-8674(94)90024-8. [DOI] [PubMed] [Google Scholar]
- Samorajski T, Friede RL, Reiner PR. Hypomyelination in the quaking mouse. A model for the analysis of disturbed myelin synthesis. J Neuropathol Exp Neurol. 1970;29:507–523. doi: 10.1097/00005072-197010000-00001. [DOI] [PubMed] [Google Scholar]
- Shedlovsky A, King TR, Dove WF. Saturation germ line mutagenesis of the murine t region including a lethal allele of the quaking locus. Proc Natl Acad Sci. 1988;85:180–184. doi: 10.1073/pnas.85.1.180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sidman RL, Dickie MM, Appel SH. Mutant mice (quaking and jimpy) with deficient myelination in the central nervous system. Science. 1964;144:309–311. doi: 10.1126/science.144.3616.309. [DOI] [PubMed] [Google Scholar]
- Siomi H, Matunis MJ, Matthew W, Dreyfuss G. The pre-mRNA binding K protein contains a novel evolutionary conserved motif. Nucleic Acids Res. 1993a;21:1193–1198. doi: 10.1093/nar/21.5.1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siomi H, Siomi MC, Nussabaum RL, Dreyfuss G. The protein product of the fragile X gene, FMR1, has characteristics of an RNA-binding protein. Cell. 1993b;74:291–298. doi: 10.1016/0092-8674(93)90420-u. [DOI] [PubMed] [Google Scholar]
- Siomi H, Choi M, Siomi MC, Nussbaum RL, Dreyfuss G. Essential role for KH domains in RNA binding: Impaired RNA binding by a mutation in the KH domain of FMR1 that causes Fragile X Syndrome. Cell. 1994;77:33–39. doi: 10.1016/0092-8674(94)90232-1. [DOI] [PubMed] [Google Scholar]
- Smith JC, Watt FM. Biochemical specificity of Xenopus notochord differentiation. Development. 1985;29:109–115. doi: 10.1111/j.1432-0436.1985.tb00302.x. [DOI] [PubMed] [Google Scholar]
- Smith JC, Price BMJ, Green JBA, Weigel D, Herrman BG. Expression of a Xenopus homologue of Brachyury (T) is an immediate-early response to mesoderm induction. Cell. 1991;67:79–87. doi: 10.1016/0092-8674(91)90573-h. [DOI] [PubMed] [Google Scholar]
- Swanson MS, Dreyfuss G. Classification and purification of proteins of heterogeneous nuclear ribonucleoprotein particles by RNA-binding specificities. Mol Cell Biol. 1988;8:2237–2241. doi: 10.1128/mcb.8.5.2237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talbot WS, Trevarrow B, Halpern ME, Melby AE, Farr G, Postlethwait JH, Jowett T, Kimmel CB, Kimelman D. The organizer-specific homeobox gene floating head is essential for notochord development in Zebrafish. Nature. 1995;378:150–157. doi: 10.1038/378150a0. [DOI] [PubMed] [Google Scholar]
- Taylor SJ, Shalloway D. An RNA-binding protein associated with Src through its SH2 and SH3 domains in mitosis. Nature. 1994;368:867–871. doi: 10.1038/368867a0. [DOI] [PubMed] [Google Scholar]
- Tonissen KD, Krieg PA. Analysis of a variant Max sequence expressed in Xenopus laevis. Oncogene. 1994;9:33–38. [PubMed] [Google Scholar]
- Urlaub H, Kruft V, Bischof O, Muller E-C, Wittmann-Liebold B. Protein-rRNA binding features and their structural and functional implications in ribosomes as determined by cross-linking studies. EMBO J. 1995;14:4578–4588. doi: 10.1002/j.1460-2075.1995.tb00137.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vize PD, Melton DA, Hemmati-Brivanlou A, Harland RM. Assays for gene function in developing Xenopus embryos. Methods Cell Biol. 1991;36:367–387. doi: 10.1016/s0091-679x(08)60288-5. [DOI] [PubMed] [Google Scholar]
- Wang LL, Richard S, Shaw AS. p62 Association with RNA is regulated by tyrosine phosphorylation. J Biol Chem. 1995;270:2010–2013. doi: 10.1074/jbc.270.5.2010. [DOI] [PubMed] [Google Scholar]
- Wong G, Muller O, Clark R, Conroy L, Moran MF, Polakis P, McCormick F. Molecular cloning and nucleic acid binding properties of the GAP-associated tyrosine phosphoprotein p62. Cell. 1992;69:551–558. doi: 10.1016/0092-8674(92)90455-l. [DOI] [PubMed] [Google Scholar]
- Zernicka-Goetz M, Pines J, Ryan K, Siemering KR, Haseloff J, Evans MJ, Gurdon JB. An indelible marker for Xenopus using a mutated green fluorescent protein. Development. 1996;122:3719–3724. doi: 10.1242/dev.122.12.3719. [DOI] [PubMed] [Google Scholar]
- Zhang Y, O’Conner JP, Siomi MC, Srinivasan S, Dutra A, Nussbaum RL, Dreyfuss G. The Fragile X Mental Retardation Syndrome protein interacts with novel homologs FXR1 and FXR2. EMBO J. 1995;14:5358–5366. doi: 10.1002/j.1460-2075.1995.tb00220.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zorn AM, Grow M, Patterson KD, Ebersole TA, Chen Q, Artzt K, Krieg PA. Remarkable sequence conservation of transcripts encoding amphibian and mammalian homologues of quaking, a KH-domain RNA-binding protein. Gene. 1997;188:199–206. doi: 10.1016/s0378-1119(96)00795-0. [DOI] [PubMed] [Google Scholar]