Abstract
Virus-induced gene silencing was used to assess the function of random Nicotiana benthamiana cDNAs in disease resistance. Out of 4992 cDNAs tested from a normalized library, there were 79 that suppressed a hypersensitive response (HR) associated with Pto-mediated resistance against Pseudomonas syringae. However, only six of these clones blocked the Pto-mediated suppression of P.syringae growth. The three clones giving the strongest loss of Pto resistance had inserts corresponding to HSP90 and also caused loss of Rx-mediated resistance against potato virus X and N-mediated tobacco mosaic virus resistance. The role of HSP90 as a cofactor of disease resistance is associated with stabilization of Rx protein levels and could be accounted for in part by SGT1 and other cofactors of disease resistance acting as co-chaperones. This approach illustrates the potential benefits and limitations of RNA silencing in forward screens of gene function in plants.
Keywords: gene silencing/HSP90/plant disease/resistance
Introduction
Plant viruses are activators of RNA silencing through a process that involves 21–25 nucleotide short interfer ing RNA (siRNA) (Hamilton and Baulcombe, 1999; Waterhouse et al., 2001). By analogy with findings from animals, it seems that the siRNA is processed from a double-stranded RNA (dsRNA) precursor by a DICER RNase III homologue and that it is incorporated as the specificity determinant into an RNA interference specificity complex (RISC) (Hannon, 2002). In virus-induced silencing, the dsRNA either is a viral replication intermediate or is produced by a host-encoded RNA polymerase using a viral RNA template (Mourrain et al., 2000). The viral RNA in this process is thus both an initiator and a target of the silencing mechanism. At the beginning of the infection cycle, the levels of dsRNA and siRNA would be correspondingly low and the viral RNA replication would be unchecked by silencing. However, at later stages of the cycle when viral dsRNA is more abundant, the higher levels would target RISC to the viral RNA and the rate of viral replication would slow down. Eventually the accumulation of viral RNA would stop or even decline.
Virus-induced RNA silencing, also referred to as virus-induced gene silencing (VIGS), is normally specific for viral rather than host RNA. However, if the viral genome is modified to include host sequences, the corresponding host RNAs are targeted and the symptoms in the infected plant resemble the phenotype of a null or reduced function mutant (Kumagai et al., 1995; Ruiz et al., 1998). This approach to suppression of host gene expression is useful in the assignment of gene function because it is fast (Baulcombe, 1999). The virus vector constructs can be assembled using standard DNA manipulations and, as symptoms develop on the infected plant by 7–21 days post-inoculation (d.p.i.), a loss-of-function phenotype can be produced within a month of having identified a gene of interest.
The use of VIGS for analysis of gene function has been validated by the use of virus vector constructs targeted against genes involved in primary and secondary metabolism, development and disease resistance (Atkinson et al., 1998; Kjemtrup et al., 1998; Jones et al., 1999; Burton et al., 2000; Ratcliff et al., 2001; Jin et al., 2002; Liu et al., 2002a,c; Peart et al., 2002a,b; Slaymaker et al., 2002). Here we describe an application of VIGS in a forward screen of genes required for disease resistance. We produced a normalized library of Nicotiana benthamiana cDNA in a vector derived from potato virus X (PVX). This vector was in an Agrobacterium Ti plasmid so that Agrobacterium inoculation could be used as a high throughput means of initiating virus infection without isolation and in vitro transcription of the PVX cDNA (Turpen et al., 1993). Plants were infected with individual PVX constructs from the library, and the resulting symptoms were used as a guide to the function of cloned cDNA. To identify cDNAs of genes involved in disease resistance, the infected plants were assayed for Pto-mediated resistance against Pseudomonas syringae. We anticipated that silencing would lead to susceptibility to P.syringae if the target of VIGS encoded a protein that is, or is similar to, a cofactor of disease resistance.
The primary screen for Pto resistance was based on a hypersensitive cell death response that is elicited by the bacterial AvrPto protein in the presence of Pto (Rommens et al., 1995). Out of 4992 cDNAs that were silenced, there were 79 that affected the Pto-mediated hypersensitive response (HR), of which there were six that silenced genes required both for the HR and for the Pto-mediated suppression of bacterial growth. Three of these PVX clones with the most pronounced effect on the Pto phenotype carried inserts corresponding to HSP90-like chaperonins encoded by a small multigene family. Silencing of these genes also compromised virus resistance mediated by the Rx and N proteins conferring resistance against PVX and tobacco mosaic virus (TMV), respectively. Thus, VIGS is an effective means of screening for novel gene function and is informative about multigene families that may not be readily accessible by mutagenesis because of functional redundancy. The HSP90 proteins may play a role either in stabilization of proteins involved in disease resistance or in assembly of multisubunit complexes required for activation of R proteins and intracellular signalling.
Results
A cDNA library in PVX
A cDNA library for insertion into a PVX agroinfection vector (Figure 1, pGR106) was synthesized using polyadenylated RNA isolated from fully developed leaves of young N.benthamiana. The cDNA was normalized through three cycles of PCR to ensure that cDNAs of abundant mRNAs were not over-represented in the library. The abundance of clones representing rubisco, assessed by colony hybridization of aliquots of the library, was ∼100-fold lower (1/600) than in an equivalent library of non-normalized cDNA (1/6). In contrast, rare mRNAs [phytoene desaturase (PDS) and PRF] were similarly abundant (∼1/10 000) before and after normalization (data not shown).
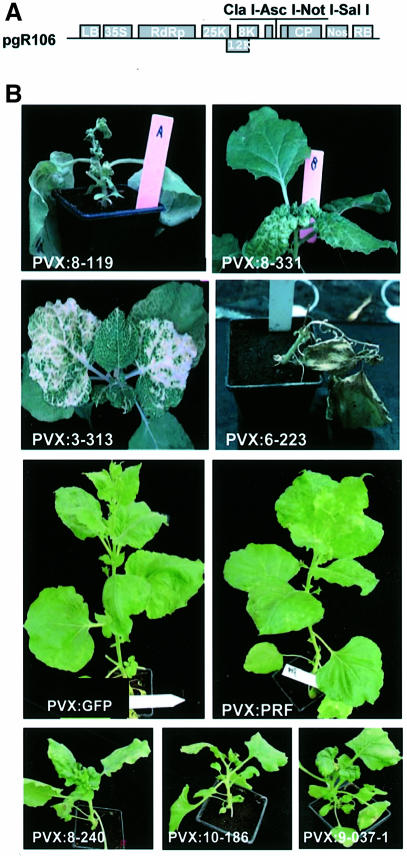
Fig. 1. PVX VIGS vectors. (A) Diagrammatic representation of a PVX vector in the pGreen0000 binary plasmid: 35S = 35S promoter of cauliflower mosaic virus; RdRp = PVX RNA-dependent RNA polymerase gene; 25K, 12K and 8K = PVX movement protein genes encoding proteins of the indicated molecular weight; CP = viral coat protein gene; nos = nopaline synthase transcriptional terminator; LB and RB are the left and right T-DNA border sequences. Unique restriction sites for insertion of sequence into the viral vector are indicated. (B) Symptoms due to VIGS of histone H4 (PVX:8-119), 16S ribosomal protein L30 (PVX:8-331), chlorophyll a/b-binding protein (PVX:3-213), receptor-like protein kinase (PVX:6-223) and HSP90 (PVX:9-037-1, PVX10-186hsp and PVX:8-240). The PVX:GFP- and PVX: PRF-infected plants did not show symptoms due to VIGS. The plants were photographed 4 weeks after inoculation with the indicated viral constructs.
After ligation of the normalized cDNA into the Ti plasmid PVX vectors and transformation into Agrobacterium, we confirmed that >95% of the colonies contained inserts of between 400 and 1000 bp in length. Each of 4992 PVX clones in Agrobacterium was then inoculated to a single 3 cm high N.benthamiana plant at the five-leaf stage. Nearly all (∼95%) of the plants developed symptoms of virus infection that, in some instances, were similar to the normal mosaic associated with PVX. However, there were also plants with chlorosis, necrosis, leaf distortion or stunting (Figure 1B). These symptoms were present in many of the plants to some extent but, in ∼15% of the plants, they were pronounced. Figure 1B illustrates examples of the most frequently observed symptoms. Presumably these symptoms were due to VIGS of endogenous genes.
Screening of genes required for Pto-mediated resistance
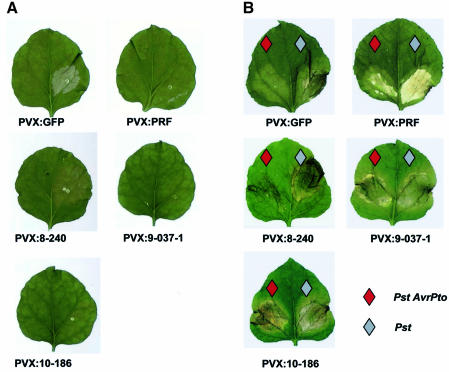
To find out whether genes required for Pto-mediated resistance were represented in the PVX library, we used an HR assay. AvrPto and Pto (Scofield et al., 1996; Tang et al., 1996) were transiently expressed in leaves of the PVX vector-infected plant in the expectation that an HR would develop only in plants that were fully competent to carry out Pto-mediated resistance. Consistent with this expectation, there was an HR in the infiltrated regions of non-infected or PVX:green fluorescent protein (GFP)-infected plants (Figure 2A). Thus PVX did not interfere with the interaction of AvrPto and Pto or with the signalling pathway leading to the HR. In contrast, the HR did not develop if the plants were infected with a virus vector construct (PVX:PRF) that would silence the Prf cofactor of Pto-mediated resistance (Figure 2A) (Peart et al., 2002a). Therefore, the loss of the HR could be used as an indicator of genes that are required for Pto-mediated resistance.
Fig. 2. Pto responses are compromised by VIGS of Prf. (A) Nicotiana benthamiana plants were inoculated with the indicated PVX constructs and, after 3 weeks, the upper leaves were infiltrated with a mixture of Agrobacterium cultures carrying 35S:Pto and 35S:AvrPto transgenes. In the non-PVX-infected or PVX:GFP-infected leaves, an HR developed after 48 h. This HR was suppressed in plants infected with PVX:PRF or with the indicated clones from the PVX cDNA library. The suppression of HR was weaker with 8-240 than with the other HSP90 constructs. (B) Pto-transgenic N.benthamiana plants (line 38-12) were inoculated with the indicated viral constructs. After 4 weeks, the upper leaves of these plants were challenge-inoculated with dilute cultures of Pst that either carried (red symbol) or did not (black symbol) carry AvrPto. The Pst chlorotic and necrotic symptoms, shown here at 4 days post-challenge inoculation, developed with the virulent Pst on all plants or with avirulent Pst(AvrPto) inoculated to plants infected with PVX:PRF or with the indicated HSP90 clones from the PVX cDNA library. The symptoms were weaker with PVX:8-240 than with the other HSP90 constructs as with PVX:8-320, PVX:11-9 and PVX:13-21 (not shown).
Out of the 4992 PVX vector clones tested, there were 79 for which the silencing phenotype resulted in compromised AvrPto-induced HR. For each of these 79 clones, the loss of the HR was consistent in at least two replicate tests involving a minimum of three plants each. In some instances, as with PVX:PRF, the loss of HR was complete (e.g. PVX:10-186 and PVX:9-037 shown in Figure 2A). However, there were other clones (e.g. PVX:8-240 shown in Figure 2A) that caused only partial suppression or delay of the HR. The GenBank accession numbers of the insert sequences are presented as Supplementary table 1 available at The EMBO Journal Online and on our website (www.sainsbury-laboratory.ac.uk).
To find out whether loss of the AvrPto-induced HR was accompanied by loss of disease resistance, Pto transgenic plants (line 38-12) (Rommens et al., 1995) were infiltrated with a dilute suspension (5 × 104 c.f.u./ml) of P.syringae pv tabaci either without (Pst) or with AvrPto [Pst(AvrPto)]. In plants inoculated with Pst, the disease symptoms were manifested at 3 days post-inoculation as a water-soaked lesion in the infiltrated patch of the leaf. After a further 3 days, this lesion became necrotic and surrounded by a chlorotic halo. These symptoms developed irrespective of whether the plants were previously infected with PVX:GFP or PVX:PRF (Figure 2B). However, in the Pto transgenic plants infected with PVX:GFP, Pst(AvrPto) did not cause necrotic symptoms and the inoculated regions developed only a slight green chlorosis (Figure 2B). Presumably the lack of symptoms in these plants was due to the elicitation of the Pto-mediated resistance by AvrPto.
In the Pto transgenic plants that had been previously infected with PVX:PRF, the water-soaked lesions and subsequent necrosis due to Pst(AvrPto) were as severe as with Pst (Figure 2B). Similarly, with six out of the 79 PVX clones that suppressed the Pto-AvrPto-mediated HR, the Pto-mediated resistance was suppressed. With two of these clones (PVX:10-186 and PVX:9-037), the suppression of resistance was as effective as with PVX:PRF. The severe Pst symptoms developed within 4 d.p.i. (Figure 2B). However, with the other clones (PVX:8-240, PVX:8-320, PVX:11-9 and PVX:13-21), the Pst symptoms did not develop until 6 days or later or were milder than on PVX:PRF-infected plants (Figure 2B). We conclude that these six PVX clones contained inserts representing genes that are required for Pto-mediated disease resistance.
Two of the six PVX clones (PVX:10-186 and PVX:9-037) contained chimeric inserts separated by AscI or NotI sites used in the original cDNA cloning process. For each of these chimeras, we produced derivative clones (PVX:10-186hsp and PVX:9-037-1) with single inserts that suppressed Pto-mediated resistance in exactly the same way as the progenitor. Further studies described below used these single insert clones.
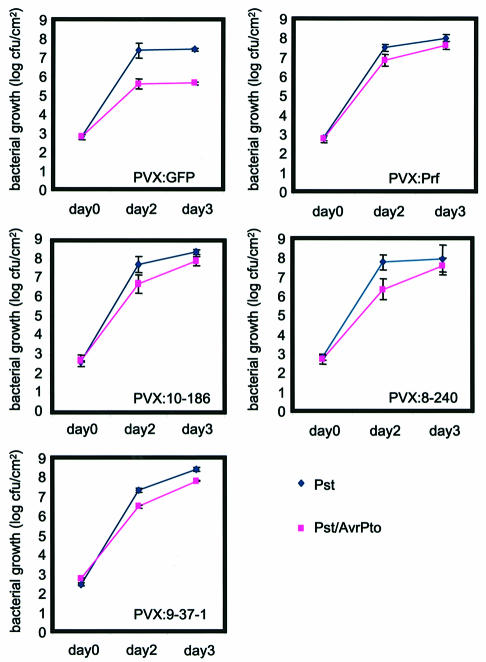
To investigate the effect of these six clones on Pto resistance further, we measured bacterial growth in the virus-infected plants. In plants infected with PVX:GFP, the manifestation of Pto resistance was differential growth of Pst and Pst(AvrPto): Pst(AvrPto) grew more slowly than Pst and, by 3 d.p.i., was 30-fold less abundant than Pst (Figure 3). As expected, in Pto plants infected with PVX:PRF, the loss of Pto resistance reduced the differential in growth of these strains. Similarly, following infection with PVX:10-186hsp, PVX:9-037-1 and PVX:8-240, the growth of Pst(AvrPto) was accelerated and both strains grew at similar rates (Figure 3). However, with PVX:8-320, PVX:11-9 and PVX:13-21, despite the delayed symptoms, there was differential growth of Pst and Pst(AvrPto) as in the PVX:GFP-infected plants (not shown).
Fig. 3. Growth of Pst(AvrPto) on Pto-transgenic N.benthamiana after VIGS of HSP90 and PRF. Pto-transgenic N.benthamiana (line 38-12) were inoculated with PVX vector constructs as indicated and challenge-inoculated with Pst or Pst(AvrPto) after 4 weeks. The Pst inocula were a 10 000-fold dilution of bacterial suspensions (OD600 = 1.0 in 10 mM MgCl2), and the in planta level of both Pst (blue) and Pst (AvrPto)(pink) was monitored at 0, 48 and 72 h post-bacterial inoculation. The values shown (c.f.u./cm2) with standard errors were derived from three batches of three plants.
To rule out that the loss of Pto resistance was an artefact associated with PVX, the 10-186hsp, 9-037-1 and 8-240 cDNA inserts were transferred into vectors based on tobacco rattle virus (TRV). These vectors were inoculated to N.benthamiana plants and, 3 weeks later, the plants were tested for the Pto-dependent HR and resistance against Pst (AvrPto). In each instance, the results were the same as with the PVX vector: resistance and HR were compromised (R.Lu, unpublished data).
Characterization of genes required for Pto-mediated resistance
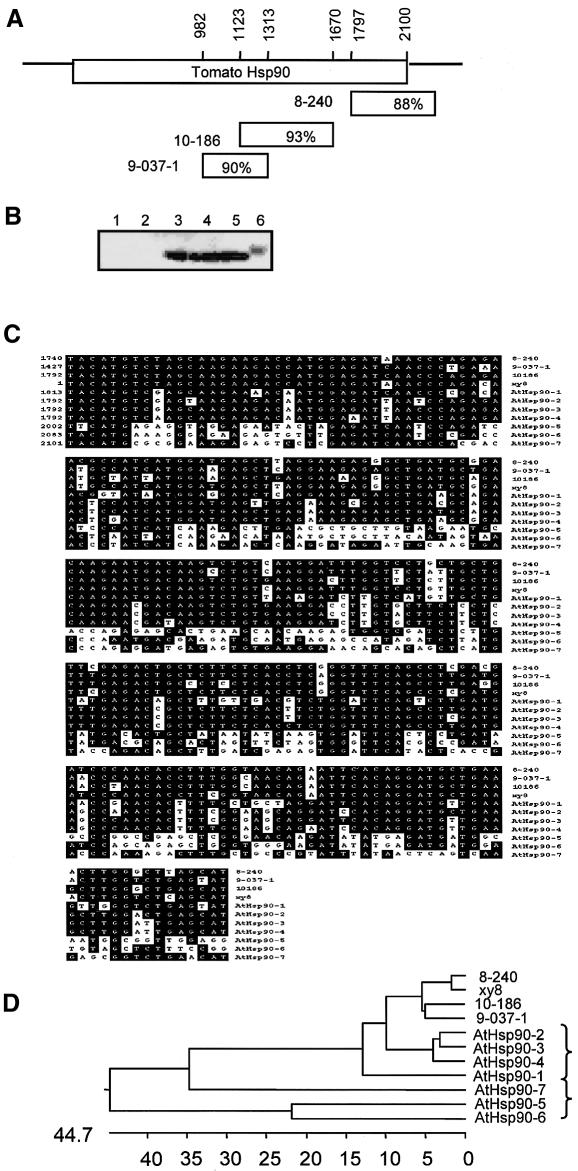
The PVX clones causing pronounced loss of Pto disease resistance (10-186hsp, 8-240 and 9-037-1) carried inserts of 549, 499 and 332 bp, respectively, that are similar to an HSP90 cDNA from tomato (GenBank accession No. M86549) (Figure 4A and C). Correspondingly, the plants infected with PVX:10-186hsp and PVX:9-037-1 contained less HSP90 protein than the equivalent control plants (Figure 4B). From these results, it seems likely that the loss of disease resistance phenotype was due to VIGS of HSP90. The other three PVX clones causing loss of Pto disease resistance carried inserts corresponding to snRNA-associated proteins (8-320 and 11-9) or to proteins without known function (13-21) (Supplementary table 1). These clones have not yet been analysed in detail because there was weaker loss of disease resistance than with the HSP90 clones.
Fig. 4. PVX clones that compromise Pto-mediated resistance caused reduced levels of HSP90. (A) Sequence alignments. The amino acid sequence corresponding to the ORFs of the 8-240, 9-037-1 and 10-186hsp inserts showed the strongest match to an HSP90 homologue from several organisms. The diagram illustrates the alignment of these sequences with a tomato HSP90 (accession No. M96549) (see C) indicating the region and percentage of homology. The arrow corresponds to the RT–PCR-amplified region of N.benthamiana HSP90 cDNA that generated the sequences shown in (C). (B) Western analysis of N.benthamiana extracts probed with HSP90-specific antibody. The extracts were taken from plants that had been inoculated 4 weeks previously with TRV constructs carrying the HSP90-specific inserts from the PVX clones 9-037-1 (lane 1) and 10-186hsp (lane 2) or from non-inoculated plants (lanes 3–5); a molecular weight marker was loaded on lane 6. Equal loading of the different lanes was confirmed by Ponceau staining of the western blot membrane (not shown). The samples were from Pto transgenic (lanes 1–4) or non-transgenic (lane 5) N.benthamiana. (C) Sequence alignments. The nucleotide sequences of HSP90 from N.benthamiana and A.thaliana were aligned using the program Clustal W (Thompson et al., 1994). The aligned sequences correspond to a conserved region in the 3′ end of the coding sequence from the seven different HSP90s of A.thaliana: four cytosolic type Athsp90-1, (AB025606), Athsp90-2 (AB011476), AtHsp90-3 (AB011476), AtHsp90-4 (AB011476); mitochondrial AtHsp90-5 (AC007167); chloroplastic AtHsp90-6 (AC009176); and ER-localized AtHsp90-7 (AL078637) and the extended cDNAs of the N.benthamiana 8-240 (2280 nucleotides, accession No. AY310776), 9-037-1 (1893 nucleotides, accession No. AY310781) and 10-186hsp (full length coding sequence of 2301 nucleotides, accession No. AY310789). An additional N.benthamiana sequence (xy8) was amplified from a N.benthamiana cDNA library with gene-specific primers designed from a conserved region in the cytoplasmic Hsp90 sequences. Numbers on the left side of the sequence correspond to the starting position of the sequence used for the alignment for each gene. (D) Dendrogram derived from the sequence alignment described in (C) generated with the program ClustalW (Thompson et al., 1994). The brackets indicate the groups of cytoplasmic (upper) and organellar (lower) forms of HSP90 (Milioni and Hatzopoulos, 1997; Krishna and Gloor, 2001).
The sequences of the 10-186hsp, 8-240 and 9-037-1 HSP90 cDNAs were extended by RT–PCR (GenBank accession Nos AY310789, AY310781 and AY310776). These sequences clearly represent a gene family because they were non-identical in overlapping regions. To investigate the number of HSP90 genes, we carried out RT–PCR of total cDNA with a primer from a conserved region that would have amplified cDNA of other HSP90 homologues in addition to 10-186, 9-037-1 and 8-240. These RT–PCR cDNAs corresponded to the C-terminal 103 amino acids and the 3′-non-coding region (see arrow in Figure 4A).
Most of the amplified cDNAs corresponded to the extended sequence of 10-186hsp or 8-240. However, there was an additional sequence (xy8) that is 2.9, 9.7 and 6.8% mismatched with the equivalent region of the 8-240, 9-037-1 and 10-186 cDNAs, respectively (Figure 4D). These four sequences were more similar to the cytoplasmic HSP90s (Milioni and Hatzopoulos, 1997; Krishna and Gloor, 2001) of Arabidopsis than the organellar forms (Krishna and Gloor, 2001). However, these four N.benthamiana genes were more similar to each other than to any of the Arabidopsis HSP90s. It is likely, therefore, that the families of genes coding for cytoplasmic HSP90 proteins in these two species evolved from a common gene ancestor through separate gene duplications. The functional difference of cytoplasmic and organellar HSP90 is reinforced by the finding that a PVX clone targeted at an organellar HSP90 (5-328) caused loss of Pto-mediated HR but had no effect on the Pto disease resistance (Supplementary table 1).
In addition to a role in disease resistance, it is likely that HSP90 proteins have functions in plant growth and development. Consequently, with both the PVX and TRV vectors, there was a VIGS phenotype of stunting and leaf deformation (Figure 1D) that, as with the disease resistance phenotype, was more severe with 10-186 and 9-037-1 than 8-240. In some experiments, these symptoms eventually were so severe that the plants died.
HSP90 is not specific for the Pto disease resistance pathway
From genetic and biochemical analyses, it can be inferred that there are overlapping signal transduction pathways associated with different disease resistance genes (Innes, 1998). It was of interest, therefore, to find out whether HSP90 silencing affected disease resistance associated with proteins with nucleotide-binding site and leucine-rich repeat domains (NBS-LRRs) encoded by the N and Rx genes. These proteins confer resistance against PVX (Rx) or TMV (N), and the experiments were carried out with N.benthamiana carrying either N or Rx transgenes (Bendahmane et al., 1999; Peart et al., 2002a). These plants were first infected with TRV:HSP90 and later challenge-inoculated with either PVX or TMV constructs that were modified to carry GFP reporter genes.
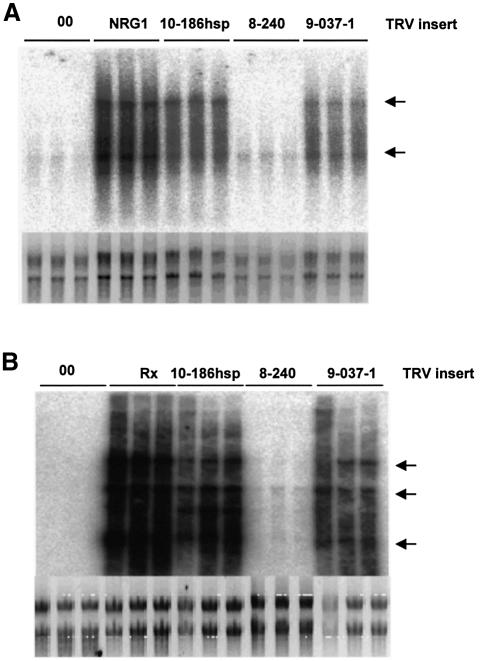
In N genotype plants infected with the TRV empty vector or with TRV:8-240, there was no detectable accumulation of TMV:GFP RNA (Figure 5A). However, following infection with TRV:10-186hsp or TRV:9-037-1, there was marked accumulation of TMV:GFP that could be monitored by northern analysis (Figure 5A). The level of TMV accumulation was similar to that in plants infected with TRV:NRG1 in which the target of VIGS is required for N-mediated resistance (J.Peart and D.C.Baulcombe, unpublished). However, the loss of N-mediated resistance in TRV:HSP90-infected plants was incomplete because we could not detect systemic spread of the TMV:GFP. This partial loss of resistance is not intrinsic to the methods used because VIGS of SGT1 or N leads to loss of both local and systemic N-mediated resistance against TMV:GFP (Peart et al., 2002a,b).
Fig. 5. VIGS of HSP90-compromised N- and Rx-mediated disease resistance. (A) N-mediated resistance. VIGS was established on N transgenic N.benthamiana (line 310A) using TRV vectors carrying the inserts from the indicated PVX clones (Peart et al., 2002a). TRV:NRG is a positive control; the insert corresponds to a gene required for N-mediated disease resistance that was identified in a separate VIGS screen and will be described in detail elsewhere (J.Peart and D.C.Baulcombe, in preparation). Four weeks post-TRV inoculation, these plants were challenged with TMV:GFP. Accumulation of the TMV:GFP was monitored after a further 7 days by northern analysis. The northern analysis is shown for three independent total RNA preparations for each VIGS construct probed with a GFP-specific probe. The lower panel shows an rRNA loading control. The arrows indicate the major species of viral genomic and subgenomic RNA. (B) Rx-mediated resistance. VIGS was established on Rx transgenic N.benthamiana (line Rx18) (Bendahmane et al., 1999) using TRV vectors carrying the inserts from the indicated PVX clones or with an Rx-specific insert (Peart et al., 2002a). Four weeks post-TRV inoculation, these plants were challenged with PVX:GFP. Accumulation of the PVX:GFP was monitored after a further 5 days by northern analysis. The northern analysis is shown for three independent total RNA preparations for each VIGS construct probed with a PVX coat protein-specific probe. The lower panel shows an rRNA loading control.
Rx-mediated resistance against PVX was compromised in plants infected with TRV:9-037-1 and TRV:10-186hsp but not with TRV:8-240 (Figure 5B). This loss of Rx resistance was indicated by GFP fluorescence in the leaf inoculated with PVX:GFP or by northern analysis (Figure 5B) on Rx genotype N.benthamiana inoculated with PVX:GFP. However, the loss of Rx resistance was not as strong as in plants infected with TRV:Rx (Figure 5B) and did not allow systemic spread of PVX.
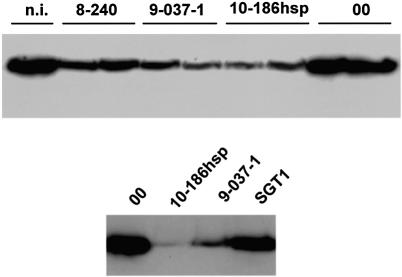
From these results, it seems that HSP90 is required for a process in disease resistance that is common to the three different disease resistance genes Pto, N and Rx. These three examples of disease resistance all involve NBS-LRR proteins: N and Rx both encode proteins in this class (Whitham et al., 1994; Bendahmane et al., 1999) and Pto resistance is dependent on the NBS-LRR product of Prf (Salmeron et al., 1996). It was therefore of interest to monitor directly the effect of HSP90 VIGS on an NBS-LRR protein. To carry out this analysis, we used N.benthamiana carrying an Rx-4HA transgene. The haemagglutinin (HA) epitope-tagged protein conferred resistance against PVX as with the wild-type Rx (P.Moffett, unpublished data) and, despite its low level expression from the endogenous Rx promoter, was readily detectable by western blotting (Figure 6). After VIGS with the TRV:HSP90 constructs, the level of Rx-4HA was reduced (Figure 6) in parallel with the effects of these recombinant viruses on Rx disease resistance (Figure 5). Thus the 10-186 and 9-037-1 constructs caused a greater reduction of Rx-4HA than did TRV:8-240. In contrast, in plants infected with TRV:SGT1 in which Rx-mediated resistance was also suppressed, there was no detectable effect on the steady-state level of Rx-4HA (Figure 6). It seems, therefore, that the effect of HSP90 in disease resistance could be through direct or indirect effects on the accumulation on the NBS-LRR protein products of disease resistance genes. It also seems that HSP90 and SGT1 influence different stages of the Rx resistance mechanism.
Fig. 6. VIGS of HSP90 caused reduced accumulation of Rx-4HA. Rx-4HA-2 N.benthamiana were infected with TRV vectors that were either empty (TRV:00), with an SGT1 insert (Peart et al., 2002b) or with HSP90 inserts from the PVX clones 9-037-1, 10-186hsp and 8-240, as indicated. Protein samples were taken at 18 days post-inoculation from the upper leaves. The samples were processed for western blotting and probed with anti-HA antibody. The two panels represent separate experiments. Equal loading was confirmed by Coomassie blue staining of the same samples (not shown). n.i. = non-infected leaf sample.
Discussion
VIGS was developed as an alternative to insertional mutagenesis for analysis of gene function. It was proposed as an approach that would be informative about genes that could not be easily investigated by mutagenesis because they have a lethal phenotype (Baulcombe, 1999). In addition, it was intended to be more suitable than transgenic approaches with antisense or inverted repeat constructs for high throughput applications because the viral vector symptoms could be induced without the time-consuming and labour-intensive procedures for plant transformation. Here, by carrying out a survey of nearly 5000 cDNAs, we have validated the suitability of VIGS for high throughput analysis. Moreover, from the high frequency and nature of the extreme symptoms (Figure 1), we have indications that VIGS is informative about genes that are required for growth. Many of the VIGS targets, for example those producing stunted plants, would most probably have a lethal phenotype in a null insertion mutant. However, because the VIGS symptoms are established gradually on a fully developed plant, it is possible to differentiate ways of growth arrest that would not be straightforward in mutants with an embryo lethal phenotype. An additional hypothetical advantage of VIGS over insertional mutagenesis is the possibility of investigating genes in functionally redundant families. The discussion of VIGS and multigene families is included below in the assessment of the HSP90 gene family in disease resistance.
At present, VIGS has limitations. For example, it may not be a suitable approach for analysis of embryo and seedling characteristics or of genes expressed in fruit and seeds and, for reasons that we do not fully understand, it is more effective in N.benthamiana and related species than in other hosts. A further limitation of VIGS is the need for large-scale facilities for growing many plants under physical containment. However, it may be possible to miniaturize the procedures so that seedlings are agroinoculated as they germinate and the VIGS symptoms are assayed in seedlings. Such developments will reduce the time course, scale and expense of forward screens with VIGS. The use of vectors adapted for other hosts may also extend the applicability of VIGS forward screens (Holzberg et al., 2002; Liu et al., 2002b; Turnage et al., 2002).
A challenge for forward screens of gene function based on VIGS or other silencing approaches will be to ensure complete genome coverage. In the screen described here with 4992 cDNAs, if we conservatively assume 50% redundancy in the library and a genome size equivalent to a typical 25 000 genes, we would have covered ∼10% of the transcriptome in our survey. To extend this coverage, we can simply test more clones. However, at some point, there would be a diminishing return and it would be necessary to target known expressed sequence tags (ESTs).
The role of the HR in disease resistance
Previous analyses of disease resistance in plants have concluded that the HR is secondary and not necessary for the suppression of the pathogen. For example, in Rx-mediated resistance against PVX, the HR was only induced if expression of the elicitor coat protein was uncoupled from virus replication (Bendahmane et al., 1999). It was concluded that the HR is a secondary mechanism that is activated only when the, as yet unidentified, primary mechanism failed to arrest the pathogen completely. The analyses of other resistance systems have come to the same conclusion. These systems involve the dnd1 mutant in Arabidopsis (Yu et al., 1998), the Sw-5-mediated resistance against Tospoviruses (Brommonschenkel et al., 2000), Rsv1-mediated virus resistance in soybean (Hajimorad and Hill, 2001) and resistance against cauliflower mosaic virus (Cole et al., 2001) in Nicotiana species.
If the HR is a secondary feature of disease resistance, it would be expected, as we found, that VIGS of some targets would not affect the mechanisms leading to suppression of the pathogen but would interfere with the secondary cell death response. However, there was an unexpected high proportion of genes (73/79) with an HR-specific VIGS phenotype and no effect on pathogen growth. An explanation for this finding could be related to the number of components in the signal transduction pathways. If the part of the pathway affecting the growth of the pathogen involves fewer proteins than the HR branch, there would be correspondingly fewer VIGS targets. Alternatively, the HR part of the pathway may be affected by diverse VIGS targets that, perhaps including those that affect the general metabolic status of the cell, are not directly involved in disease resistance.
However, whatever the explanation, the discrepant results with the HR and disease resistance assays emphasize how these are very different tests. The HR test involves expression of the elicitor at a high level from the 35S promoter, whereas in the pathogen it would normally accumulate only at very low levels. The lesson for future VIGS screens is that direct tests of disease resistance are more useful than assays based on an HR.
The role of HSP90 proteins in disease resistance
HSP90 proteins facilitate protein folding, and it could be that NBS-LRR proteins or other components of the disease resistance pathways are dependent on HSP90 for correct folding, as in other systems (Nathan et al., 1997). The misfolded proteins could be either degraded or not biologically active. We cannot rule out this explanation. However, there are indications that HSP90 may be more specifically involved in disease resistance than is implied by a general role in protein folding. For example, there are mutations in HSP90 genes of Arabidopsis resulting in specific loss of disease resistance that do not lead to severe morphological phenotypes. One HSP90 mutation affects RPS2 (Takahashi et al., 2003), whereas another affects RPM1 (J.Dangl, personal communication). The findings that HSP90 interacts directly with proteins involved in disease resistance including SGT1 (Takahashi et al., 2003; J.Dangl, personal communication) are also an indication that this chaperonin has a specific role in disease resistance.
The VIGS phenotype of HSP90 is similar to that of SGT1 in that there is partial or complete loss of many disease resistance responses including those associated with Rx, N and Pto (Figures 3 and 6) (Peart et al., 2002b). Silencings of SGT1 and HSP90 were also similar in causing loss of non-host resistance against P.syringae pv. maculicola and in having no effect on non-host resistance against Xanthomonas campestris pv campestris (I.Malcuit, unpublished data). These similarities may be significant because there are structural motifs that are common to SGT1 and HSP90-associated proteins known as co-chaperones. One co-chaperone of HSP90, p23, has sequence characteristics of the CS domain of SGT1 (Garcia-Ranea et al., 2002) that binds directly to the RAR1 disease resistance signalling protein. Other HSP90 co-chaperones including Hop have a tandem arrangement of three degenerate 34 amino acid repeats referred to as the tetratricopeptide repeats (TPRs) that are also present in Sgt1 (Azevedo et al., 2002). TPR domains bind specifically to the conserved C-terminal motif MEEVD of HSP90 (Chen et al., 1998; Young et al., 1998).
A simple prediction, by extrapolation from these results, is that HSP90 would bind directly to SGT1 via the TPR domain. The bound SGT1 would then serve as a co-chaperone that could influence downstream signalling proteins through interactions of the CS domain. However, from the Rx-4HA accumulation data (Figure 6), we infer that HSP90 cannot only act in association with SGT1. HSP90 silencing results in reduced levels of Rx-4HA, whereas SGT1 silencing has no effect. Thus, there must also be an HSP90 effect at a level that is SGT1 independent and that affects the stability of Rx. This process may influence stability of other similar proteins because an HSP90 mutation in Arabidopsis causes destabilization of a disease resistance protein that, like Rx, has a nucleotide-binding site and leucine-rich repeat (J.Dangl, personal communication).
Does VIGS silence one or more members of the HSP90 multigene family?
VIGS can be effective with as few as 28 nucleotides of sequence identity between the insert in the virus vector and the target RNA (Thomas et al., 2001). It is therefore quite likely that the 9-037-1 and 10-186hsp inserts from the highly conserved coding sequence region would have silenced all HSP90 mRNAs. Consistent with this expectation, the western analysis showed almost complete absence of HSP90 in the tissues infected with the PVX vectors carrying these inserts (Figure 4B).
In contrast, the VIGS clones targeted at less divergent regions of the HSP90 mRNA had either weak or no suppression of disease resistance. For example, the 8-240 clone that corresponds to the 3′ region coding sequence and the untranslated region had only small and inconsistent effects on HSP90 protein levels and weaker effects than 9-037-1 or 10-186 on the various types of disease resistance (Figures 3 and 5) or on Rx levels (Figure 6). Similarly, PVX constructs with inserts from the 3′-untranslated regions of HSP90 mRNAs did not cause loss of disease resistance (9-295; Supplementary table 1, and R.Lu, unpublished information). One interpretation of these results is that there is functional redundancy in the HSP90 gene family. However, we could not assess how efficiently these 3′-specific constructs had silenced the corresponding mRNA and formally we cannot rule out the possibility that there is functional differentiation of HSP90 isoforms.
HSP90 VIGS affected plant growth (Figure 1B) as well as disease resistance. The growth effects correlated with the effects on disease resistance in that 9-037-1 and 10-186hsp had more pronounced effects than 8-240. From this result, it could be concluded that HSP90 is involved in similar signalling pathways affecting normal plant growth and disease resistance. However, it could also be that HSP90 in plants, as in other eukaryotes, is required to chaperone correct folding of a subset of essential proteins (Nathan et al., 1997). Alternatively, as suggested recently, it could be that HSP90 is required to buffer cells against the epigenetic effects of misfolded proteins (Queitsch et al., 2002). If that is the case, the growth phenotypes observed here (Figure 1D) could reflect the damage caused by the accumulation of misfolded proteins.
Materials and methods
Plant material
The plants were either non-transgenic N.benthamiana or transgenic derivatives carrying Rx (line Rx18) and N (line 310A) and Pto (lines 38-12) (Rommens et al., 1995; Bendahmane et al., 1999; Peart et al., 2002a) as indicated in the text. The line Rx4HA-2 carrying an Rx-4HA construct was assembled by transformation with a modified version of pB1-RxHA (Bendahmane et al., 1999). The modification was the addition of three HA epitope tags (data not shown) to the 5′ end of the Rx opening reading frame in pB1-RxHA (Bendahmane et al., 1999). The resulting construct was used for Agrobacterium-mediated transformation of N.benthamiana and stable transformants were selected on kanamycin. The experiments shown were carried out in plants of line Rx4HA-2 that are homozygous for the Rx-4HA transgene and in which the PVX resistance was as strong as in lines transformed with wild-type Rx (P.Moffett unpublished data).
Viral constructs, virus inoculation and cDNA library construction
The TRV vector construct for agroinoculation (pTV00) and inoculation procedures have been described previously (Ratcliff et al., 2001). The PVX vector construct (pgR106) is a derivative of previously described clones (Chapman et al., 1992) (GenBank accession No. AY297843). A similar vector with ClaI, SmaI and SalI sites for insertion of foreign sequence is also available (pgR107: GenBank accession No. AY297842). Inoculation procedures for the TRV vector were as described (Ratcliff et al., 2001). With PVX constructs, we used a toothpick that was touched to an Agrobacterium colony and then used to pierce the leaf or petiole of a 2–3 cm high seedling. Systemic PVX infection developed after 5–10 days presumably because wounded cells were transformed with the Ti plasmid and were a source of infection for the rest of the plant.
Construction of the cDNA library in the PVX vector is described below. The PRF insert in PVX:PRF and TRV:PRF was generated by PCR with primers Prf2a (5′-GTTGGCATGCCAGGATTGGGC-3′) and Prf1s (5′-ACAAGGCTTAAG ATAGTGTGG-3′); the PCR products correspond to nucleotides 3361–4035 in the ORF of the tomato Prf gene (Peart et al., 2002a) (accession No. AF479624).
Total RNA for cDNA library construction was treated with RNase-free DNase (Promega), and recovered by ethanol precipitation. Reverse transcription used standard procedures, and cDNA normalization has been described previously (Ko, 1990; Kohchi et al., 1995). To facilitate cDNA amplification and subsequent cloning, a ‘lone’ linker, which is an annealing product of two primers AscI A (5′-GAGATATTAGGCGCGCCTACTC) and AscI B (5′-GAGTAGGCGCGCCTAATAT), was added to both ends of all random cDNAs. Three rounds of normalization were performed consecutively. The normalized cDNAs were digested with AscI and NotI, and cDNA fragments of 400 bp to 1 kb were purified, ligated into the PVX vector construct pGR106 and transformed into Agrobacterium GV3101 competent cells. To assess the efficiency of normalization, an aliquot of the ligation reaction was transformed into Escherichia coli and the transformants were assayed by colony hybridization with probes specific for rubisco, PDS and Prf.
PCR of HSP 90 sequences
Hsp90 cDNA sequences from N.benthamiana were amplified using the SMART™ cDNA RACE amplification kit (BD Bioscience-Clontech, Palo Alto, CA) with HSP90 gene-specific primer X (5′-GATATTTACTACATTACTGGTGAGAGCAAGAAGGC-3′) and the library primer AP1. Nested PCR was performed with an HSP-specific nested primer Y (5′-TCTAGCATGGCTGGATACATGTCTAGCAAGA-3′) and nested library primer AP2 according to the manufacturer’s instructions (BD Bioscience-Clontech). Both X and Y primers were designed from a region in the HSP90 sequence that is conserved in N.benthamiana, tomato and Arabidopsis cytosolic Hsp90 sequences.
Monitoring of HR and bacterial and viral disease on plants
The HR test was carried out by infiltration of N.benthamiana upper leaves with a mixture of Agrobacterium cultures carrying 35S:Pto and 35S:AvrPto transgenes (Scofield et al., 1996). Pseudomonas syringae pv tabaci (Pst) used for Pto-mediated resistance assay was Pst 11528 which is virulent on resistant tomato cultivars expressing Pto on Pto-transgenic N.benthamiana plants (Rommens et al., 1995). Pst 11528 (AvrPto) is a AvrPto-expressing Pst transconjugant strain which is avirulent on resistant tomato cultivars expressing Pto and on Pto-transgenic N.benthamiana plants (Rommens et al., 1995). All Pst strains were grown in L medium only supplemented with 25 µg/ml kanamycin to maintain the transconjugation vector pDSK519 and 100 µg/ml rifampicin for the selection of Pst strains. Bacterial suspensions were inoculated onto leaves using a needleless syringe. Inoculation of diluted inocula [10 000-fold dilution from original cell suspension (OD600 ± 1.0)] was used for disease symptom and bacterial growth assays. To measure in planta growth of Pst strains, three replicate samples, each containing leaf discs from three plants, were collected from inoculated leaves. Leaf discs were ground in 10 mM MgCl2, serially diluted and plated onto L medium containing kanamycin (25 µg/ml) and rifampicin (100 µg/ml). Colonies were counted after 24 h growth at 28°C.
PVX:GFP and TMV:GFP derivatives were inoculated by infiltration or toothpick of Agrobacterium strains carrying the viral cDNA in Ti plasmid vectors, and RNA analysis was as described previously (Peart et al., 2002a). The probe used to detect TMV is the full-length GFP sequence. The full-length PVX coat protein sequence was used to make a probe for detection of PVX:GFP in the resistance assay test.
Protein analysis
For the analysis of the HSP90 protein, leaf samples were extracted in 50 mM Tris–HCl pH 7.5, 150 mM NaCl, 5 mM EDTA, 5 mM dithiothreitol (DTT), 1% Triton X-100, 1.5% polyvinylpyrrolidone supplemented with protease inhibitors (protease inhibitor cocktail for plant and tissue extracts, Sigma). A 50 µg aliquot of total protein was loaded onto a 6% SDS–polyacrylamide gel and, after electrophoresis, the gels were electroblotted onto PVDF membrane. Immunological reactions were carried with a primary antibody HSP90α (aE-17), (Santa Cruz Biotechnology Inc., Santa Cruz, CA) which is an affinity-purified goat polyclonal antibody against a conserved peptide mapping near the N-terminus of HSP90α of Arabidopsis thaliana. For analysis of Rx-4HA levels, the western blots were prepared and probed with anti-HA antibody (3F10, Roche) as described (Bendahmane et al., 2002).
Supplementary data
Supplementary data are available at The EMBO Journal Online.
Acknowledgments
Acknowledgements
Ken Shirasu and Jeff Dangl have communicated unpublished work indicating that HSP90 is implicated in disease resistance, and we are grateful for their openness. Recombinant viruses and transgenic plants were contained in greenhouses under DEFRA license PHL 161/4080. We are grateful to The Gatsby Charitable Foundation and the BBSRC for support of this work and to EMBO for a long-term fellowship to P.M.
Note added in proof
A recent paper [Kanzaki,H., Saitoh,H., Ito,A., Fujisawa,S., Kamoun,S., Katou,S., Yoshioka,H. and Terauchi,R. (2003) Cytosolic HSP90 and HSP70 are essential components of IFN1-mediated hypersensitive response and non-host resistance to pseudomonas cichorii in Nicotiana benthamiana. Mol. Plant Pathol., 1, 383–391] also describes the use of VIGS to reveal a role of HSP90 in disease resistance.
References
- Atkinson R.G., Bieleski,L.R.F., Gleave,A.P., Jannsen,B.J. and Morris,B.A.M. (1998) Post-transcriptional silencing of chalcone synthase in petunia using a geminivirus-based episomal vector. Plant J., 15, 593–604. [DOI] [PubMed] [Google Scholar]
- Azevedo C., Sadanandom,A., Kitagawa,K., Freialdenhoven,A., Shirasu,K. and Schulze-Lefert,P. (2002) The RAR1 interactor SGT1, an essential component of R gene-triggered disease resistance. Science, 295, 2073–2076. [DOI] [PubMed] [Google Scholar]
- Baulcombe D.C. (1999) Fast forward genetics based on virus-induced gene silencing. Curr. Opin. Plant Biol., 2, 109–113. [DOI] [PubMed] [Google Scholar]
- Bendahmane A., Kanyuka,K. and Baulcombe,D.C. (1999) The Rx gene from potato controls separate virus resistance and cell death responses. Plant Cell, 11, 781–791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bendahmane A., Farnham,G., Moffett,P. and Baulcombe,D.C. (2002) Constitutive gain-of-function mutants in a nucleotide binding site–leucine rich repeat protein encoded at the Rx locus of potato. Plant J., 32, 195–204. [DOI] [PubMed] [Google Scholar]
- Brommonschenkel S.H., Frary,A., Frary,A. and Tanksley,S.D. (2000) The broad-spectrum tospovirus resistance gene Sw-5 of tomato is a homolog of root-knot nematode resistance gene Mi. Mol. Plant-Microbe Interact., 13, 1130–1138. [DOI] [PubMed] [Google Scholar]
- Burton R.A., Gibeaut,D.M., Bacic,A., Findlay,K., Roberts,K., Hamilton,A., Baulcombe,D.C. and Fincher,G.B. (2000) Virus-induced silencing of a plant cellulose synthase gene. Plant Cell, 12, 691–705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman S.N., Kavanagh,T.A. and Baulcombe,D.C. (1992) Potato virus X as a vector for gene expression in plants. Plant J., 2, 549–557. [DOI] [PubMed] [Google Scholar]
- Chen S., Sullivan,W.P., Toft,D.O. and Smith,D.F. (1998) Differential interactions of p23 and the TPR-containing proteins Hop, Cyp40, FKBP52 and FKB51 with Hsp90 mutants. Cell Stress Chaperones, 3, 118–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole A.B., Kiraly,L., Ross,H. and Schoelz,J.E. (2001) Uncoupling resistance from cell death in the hypersensitive response of Nicotiana species to cauliflower mosaic virus infection. Mol. Plant-Microbe Interact., 14, 31–41. [DOI] [PubMed] [Google Scholar]
- Garcia-Ranea J., Mirey,G., Camonis,J. and Valencia,A. (2002) p23 and HSP20/α-crystallin proteins define a conserved sequence domain present in other eukaryotic protein families. FEBS Lett., 529, 162. [DOI] [PubMed] [Google Scholar]
- Hajimorad M.R. and Hill,J.H. (2001) Rsv1-mediated resistance against soybean mosaic virus-N is hypersensitive response-independent at inoculation site, but has the potential to initiate a hypersensitive response-like mechanism. Mol. Plant-Microbe Interact., 14, 587. [DOI] [PubMed] [Google Scholar]
- Hamilton A.J. and Baulcombe,D.C. (1999) A species of small antisense RNA in post-transcriptional gene silencing in plants. Science, 286, 950–952. [DOI] [PubMed] [Google Scholar]
- Hannon G.J. (2002) RNA interference. Nature, 418, 244–251. [DOI] [PubMed] [Google Scholar]
- Holzberg S., Brosio,P., Gross,C. and Pogue,G.P. (2002) Barley stripe mosaic virus-induced gene silencing in a monocot plant. Plant J., 30, 315–327. [DOI] [PubMed] [Google Scholar]
- Innes R.W. (1998) Genetic dissection of R gene signal transduction pathways. Curr. Opin. Plant Biol., 1, 299–304. [DOI] [PubMed] [Google Scholar]
- Jin H.L., Axtell,M.J., Dahlbeck,D., Ekwenna,O., Zhang,S.Q., Staskawicz,B. and Baker,B. (2002) NPK1, an MEKK1-like mitogen-activated protein kinase kinase kinase, regulates innate immunity and development in plants. Dev. Cell, 3, 291–297. [DOI] [PubMed] [Google Scholar]
- Jones L., Hamilton,A.J., Voinnet,O., Thomas,C.L., Maule,A.J. and Baulcombe,D.C. (1999) RNA–DNA interactions and DNA methylation in post-transcriptional gene silencing. Plant Cell, 11, 2291–2302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kjemtrup S., Sampson,K.S., Peele,C.G., Nguyen,L.V., Conkling,M.A., Thompson,W.F. and Robertson,D. (1998) Gene silencing from plant DNA carried by a geminivirus. Plant J., 14, 91–100. [DOI] [PubMed] [Google Scholar]
- Ko M.S. (1990) An ‘equalized cDNA library’ by the reassociation of short double-stranded cDNAs. Nucleic Acids Res., 18, 5705–5711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohchi T., Fujishige,K. and Ohyama,K. (1995) Construction of an equalized cDNA library from Arabidopsis thaliana. Plant J., 8, 771–776. [DOI] [PubMed] [Google Scholar]
- Krishna P. and Gloor,G. (2001) The hsp90 family of proteins in Arabidopsis thaliana. Cell Stress Chaperones, 6, 238–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumagai M.H., Donson,J., Della-Cioppa,G., Harvey,D., Hanley,K. and Grill,L.K. (1995) Cytoplasmic inhibition of carotenoid biosynthesis with virus-derived RNA. Proc. Natl Acad. Sci. USA, 92, 1679–1683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Schiff,M., Serino,G., Deng,X.W. and Dinesh-Kumar,S.P. (2002a) Role of SCF ubiquitin-ligase and the COP9 signalosome in the N gene-mediated resistance response to tobacco mosaic virus. Plant Cell, 14, 1483–1496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y.L., Schiff,M. and Dinesh-Kumar,S.P. (2002b) Virus-induced gene silencing in tomato. Plant J., 31, 777–786. [DOI] [PubMed] [Google Scholar]
- Liu Y.L., Schiff,M., Marathe,R. and Dinesh-Kumar,S.P. (2002c) Tobacco Rar1, EDS1 and NPR1/NIM1 like genes are required for N-mediated resistance to tobacco mosaic virus. Plant J., 30, 415–429. [DOI] [PubMed] [Google Scholar]
- Milioni D. and Hatzopoulos,P. (1997) Genomic organization of hsp90 gene family in Arabidopsis. Plant Mol. Biol., 35, 955–961. [DOI] [PubMed] [Google Scholar]
- Mourrain P. et al. (2000) Arabidopsis SGS2 and SGS3 genes are required for posttranscriptional gene silencing and natural virus resistance. Cell, 101, 533–542. [DOI] [PubMed] [Google Scholar]
- Nathan D.F., Vos,M.H. and Lindquist,S. (1997) In vivo functions of the Saccharomyces cerevisiae Hsp90 chaperone. Proc. Natl Acad. Sci. USA, 94, 12949–12956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peart J.R., Cook,G., Feys,B.J., Parker,J.E. and Baulcombe,D.C. (2002a) An EDS1 orthologue is required for N-mediated resistance against tobacco mosaic virus. Plant J., 29, 569–579. [DOI] [PubMed] [Google Scholar]
- Peart J.R. et al. (2002b) Ubiquitin ligase-associated protein SGT1 is required for host and nonhost disease resistance in plants. Proc. Natl Acad. Sci. USA, 99, 10865–10869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Queitsch C., Sangster,T.A. and Lindquist,S. (2002) Hsp90 as a capacitor of phenotypic variation. Nature, 417, 618–624. [DOI] [PubMed] [Google Scholar]
- Ratcliff F., Martin-Hernandez,A.M. and Baulcombe,D.C. (2001) Tobacco rattle virus as a vector for analysis of gene function by silencing. Plant J., 25, 237–245. [DOI] [PubMed] [Google Scholar]
- Rommens C.M.T., Salmeron,J.M., Oldroyd,G.E.D. and Staskawicz,B.J. (1995) Intergeneric transfer and functional expression of the tomato disease resistance gene Pto. Plant Cell, 7, 1537–1544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz M.T., Voinnet,O. and Baulcombe,D.C. (1998) Initiation and maintenance of virus-induced gene silencing. Plant Cell, 10, 937–946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salmeron J.M., Oldroyd,G.E.D., Rommens,C.M.T., Scofield,S.R., Kim,H.-S., Lavelle,D.T., Dahlbeck,D. and Staskawicz,B.J. (1996) Tomato Prf is a member of the leucine-rich repeat class of plant disease resistance genes and lies embedded within the Pto kinase gene cluster. Cell, 86, 123–133. [DOI] [PubMed] [Google Scholar]
- Scofield S.R., Tobias,C.M., Rathjen,J.P., Chang,J.H., Lavelle,D.T., Michelmore,R.W. and Staskawicz,B.J. (1996) Molecular basis of gene-for-gene specificity in bacterial speck disease of tomato. Science, 274, 2063–2065. [DOI] [PubMed] [Google Scholar]
- Slaymaker D.H., Navarre,D.A., Clark,D., del Pozo,O., Martin,G.B. and Klessig,D.F. (2002) The tobacco salicylic acid-binding protein 3 (SABP3) is the chloroplast carbonic anhydrase, which exhibits antioxidant activity and plays a role in the hypersensitive defence response. Proc. Natl Acad. Sci. USA, 99, 11640–11645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi A.A., Casais,C., Ichimura,K. and Shirasu,K. (2003) HSP90 interacts with RAR1 and SGT1 and is essential for RPS2-mediated disease resistance in Arabidopsis. Proc. Natl Acad. Sci. USA, 100, 11777–11782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang X.Y., Frederick,R.D., Zhou,J.M., Halterman,D.A., Jia,Y.L. and Martin,G.B. (1996) Initiation of plant disease resistance by physical interaction of AvrPto and Pto kinase. Science, 274, 2060–2063. [DOI] [PubMed] [Google Scholar]
- Thomas C.L., Jones,L., Baulcombe,D.C. and Maule,A.J. (2001) Size constraints for targeting post-transcriptional gene silencing and for RNA-directed methylation in Nicotiana benthamiana using a potato virus X vector. Plant J., 25, 417–425. [DOI] [PubMed] [Google Scholar]
- Thompson J.D., Higgins,D.G. and Gibson,T.J. (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position specific gap penalties and weight matrix choice. Nucleic Acids Res., 22, 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turnage M.A., Muangsan,N., Peele,C.G. and Robertson,D. (2002) Geminivirus-based vectors for gene silencing in Arabidopsis. Plant J., 30, 107–114. [DOI] [PubMed] [Google Scholar]
- Turpen T.H., Turpen,A.M., Weinzettl,N., Kumagai,M.H. and Dawson,W.O. (1993) Transfection of whole plants from wounds inoculated with Agrobacterium tumefaciens containing cDNA of tobacco mosaic virus. J. Virol. Methods, 42, 227–240. [DOI] [PubMed] [Google Scholar]
- Waterhouse P.M., Wang,M.B. and Lough,T. (2001) Gene silencing as an adaptive defence against viruses. Nature, 411, 834–842. [DOI] [PubMed] [Google Scholar]
- Whitham S., Dinesh-Kumar,S.P., Choi,D., Hehl,R., Corr,C. and Baker,B.J. (1994) The product of the tobacco mosaic virus resistance gene N: similarity to Toll and the interleukin-1 receptor. Cell, 78, 1101–1115. [DOI] [PubMed] [Google Scholar]
- Young J.C., Obermann,W.M. and Hartl,F.U. (1998) Specific binding of tetratricopeptide repeat proteins to the C-terminal 12-kDa domain of hsp90. J. Biol. Chem., 273, 18007–18010. [DOI] [PubMed] [Google Scholar]
- Yu I.C., Parker,J. and Bent,A.F. (1998) Gene-for-gene disease resistance without the hypersensitive response in Arabidopsis dnd1 mutant. Proc. Natl Acad. Sci. USA, 95, 7819–7824. [DOI] [PMC free article] [PubMed] [Google Scholar]