Abstract
Microorganisms that live in fluctuating environments must constantly adapt their behavior to survive. The host constitutes an important microenvironment in opportunistic and primary fungal pathogens like Cryptococcus neoformans (C. neoformans) and Cryptococcus gattii (C. gattii). In clonal populations, adaptation may be achieved through the generation of diversity. For fungi phenotype switching constitutes a mechanism that allows them to change rapidly. Both C. neoformans and C. gattii undergo phenotypic switching, which allows them to be successful pathogens and cause persistent disease. Similar to other encapsulated microbes that exhibit phenotypic variation, phenotypic switching in Cryptococcus changes the polysaccharide capsule. Most importantly, in animal models phenotypic switching affects virulence and can change the outcome of infection. Virulence changes because C. neoformans and C. gattii switch variants elicit different inflammatory responses in the host. This altered host response can also affect the response to antifungal therapy and in some cases may even promote the selection of switch variants. This review highlights the similarity and difference between phenotypic switching in C. neoformans and C. gattii, the two dominant species that cause cryptococcosis in humans.
Keywords: Cryptococcus, Switching
Introduction
Microorganisms that live in fluctuating environments must constantly adapt their behavior to survive. In clonal populations, this may be achieved through the generation of diversity by phenotypic switching [1]. Phenotypic switching has been observed both in prokaryotic as well as eukaryotic microbes. For fungi phenotypic switching is defined as the reversible change manifested as altered colony morphology at a rate higher than the somatic mutation rate. Phenotypic switching has been reported first in Candida over 20 years ago [2, 3] and the molecular mechanism has been studied extensively [4, 5]. In Candida phenotypic switching controls mating and has also been proposed to contribute to virulence [6].
Classification of Cryptococcus neoformans and C. gattii
The genus Cryptococcus includes around 37 species. Among these, C. neoformans (var. neoformans and var. grubii) and C. gattii are the etiologic agents of cryptococcosis and thus the predominant pathogenic species although rarely others are described [7]. Four serotypes (A–D) are identified by sero-typing, a method that distinguishes the encapsulated yeasts based on capsular agglutination reactions. However serotypes are not necessarily stable and thus should not be the sole basis of species assignment. The heterogeneity of the two species became clear when two distinct sexual forms (telemorphs) were found. The telemorph, Filobasidiella neoformans was found to be produced only by strains of serotypes A and D, whereas Filobasidiella bacillispora was found to be produced by strains of serotypes B and C. Ensuing studies revealed many more differences between the anamorphs of the two Filobasidiella species with regard to their epidemiology, ecology, biochemistry, pathobiology, and genetics. Hence, at present the serotypes are assigned to two species, C. neoformans (serotypes A and D) and C. gattii (serotypes B and C). Until recently, serotypes A and D were included in var. neoformans while serotypes B and C were included in var. gattii. However, itwas proposed that a new variety, var. grubii, be created to contain serotype A because of observed phenotypic differences, and significant genetic variations between serotypes A and D [8]. This leaves serotype D as the sole serotype in var. neoformans. A separate assignment of a third species to serotype A has been debated and rejected [9]. The need for recognition of the two species within C. neoformans is warranted not only because of their biologic and ecological differences, but also because they tend to infect different hosts. C. gattii infections have been documented more commonly in immunocompetent individuals, whereas a majority of the patients infected with C. neoformans are immunocompromised, often due to HIV infection [10, 11]. Furthermore, some reports have suggested that infections due to C. gattii carry a worse prognosis [12].
C. neoformans var. grubii is the most prevalent agent causing cryptococcosis worldwide [13, 14]. C. neoformans var. neoformans has a comparable environmental and clinical distribution worldwide and in some areas such as sub-Saharan Africa up to 20% of the isolates are serotype D strains [15, 16]. Differences in the immunological status of the host infected by C. neoformans var. neoformans and var. grubii, however, have not been documented. C. gattii (serotypes B and C) was originally predominantly found in subtropical regions but a newly recombined serotype B clone has emerged over the past 10 years as a cause of cryptococcal infections in animals and otherwise healthy humans on Vancouver Island [17]. More recently, this epidemic is spreading to the Pacific Northwest of the United States [18, 19]. The outbreak clone appears to have descended from two alpha mating-type parents. Cryptic same-sex reproduction enabled expansion to a new geographical niche and contributed to the ongoing production of infectious spores [20]. Serotype C strains only rarely cause human infection [21, 22].
Although C. neoformans and C. gattii exhibit some differences in the host response they elicit, and in their response to antifungal therapy, both can cause chronic meningoencephalitis that can be difficult to treat, especially in immunocompromised hosts. Of note is that treatment failure cannot be attributed to antifungal resistance; however, there is evidence that these fungi can undergo phenotypic switching [23] which may affect the outcome of chronic infection.
Evidence of Microevolution and Phenotypic Switching in C. neoformans
Microevolution is the process of change in traits of a pathogen population in a brief time. These changes are selected, stable, and inherited. They can be due to many different mechanisms including mutations, transfer of genes from one population to another, phenotypic switching, and other epigenetic mechanisms. Several lines of evidence indicate that both serotype D as well serotype A strains of C. neoformans undergoe microevolution. First, relapse of cryptococcal meningitis results during persistent infection with a single infecting strain rather than re-infection with a new strain [24]. Second, serial Cryptococcus isolates from AIDS patients exhibit minor electrophoretic karyotype and they can also differ in growth rates, capsule size, or virulence in mice [23, 25]. Additional changes that occur during chronic infection include stable alterations in cell membrane sterol composition and differences in the glucuronoxylomannan (GXM) structure of the capsule [26]. Third, analysis of a standard strain maintained in various laboratories reveals significant differences in capsule size, melanin production, growth rates, and virulence in mice [27]. Fourth, phenotypic switching that enables microorganisms to undergo rapid microevolution and to adapt to different microenvironments. Phenotypic switching is defined as the emergence of reversible colony morphology at a rate higher than somatic mutation rate. Reversible switching between various colony morphologies (smooth, wrinkled, mucoid, and pseudohyphal) has been observed in standard strains (SB4, J32, RC2) of serotypes A and D [28, 29]. This colony-type switching is associated with changes in virulence and in host inflammatory and antibody responses in murine and rat model. Most serotypes D and A strains manifest smooth colony morphology. One study reported that 96% of Indian var. neoformans and var. grubii isolates were smooth in colony morphology whereas 100% of C. gattii were mucoid [25]. Switching in a smooth (SM) parent strain (RC2, serotype D) that switches to mucoid (MC) variants and reversibly in vitro and in vivo has been thoroughly investigated [30]. However, other colony morphologies in switch variants such as serrated, wrinkled, and pseudohyphal are also described but these rare colony phenotypes are not usually seen in clinical isolates.
Much less is known about microevolution in C. gattii strains. Several clinical case reports and epidemiologic studies have examined C. gattii strains from clinical isolates [31]. Clinical C. gattii strains were found to be less diverse in different studies [25, 32] and often are typed as serotype B, MATalpha strains with a typical VGI molecular type [14]. C. gattii is predominantly clonal, and except for the newly emerged RG strain in Canada, the majority to C. gattii strains are not fertile and cannot generate haploid spores by monokaryotic fruiting [33]. Recently investigations from our laboratory have demonstrated that a clinical C. gattii strain can also undergo phenotypic switching [34]. Differences compared to switching in the C. neoformans var. neoformans and var. grubii strains were noted and are discussed.
Phenotypic Switching in C. neoformans var. neoformans and C. gattii
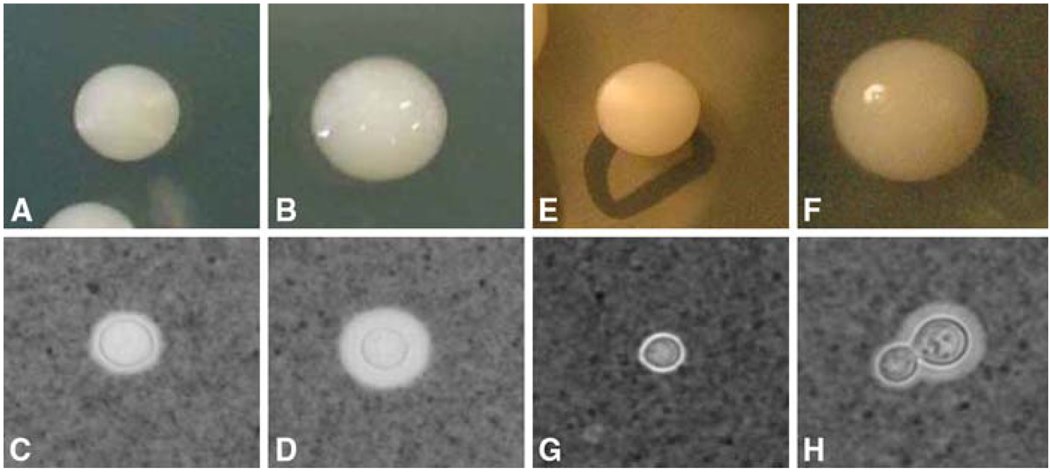
Phenotypic switching in var. neoformans has been described in both serotypes A and D strains. ATCC 24067 is a serotype D strain, and was isolated from the cerebrospinal fluid of a leukemic patient and placed into the ATCC bank in 1968 [35]. This strain has a high predisposition to undergo microevolution and thus several variants have been described [27]. One of the variant named RC2 was originated from the Cherniak Laboratory and presumably emerged after spontaneous microevolution [27]. Fries et al. demonstrated that RC2 has a tendency to switch from a smooth parent to mucoid colony morphology (Fig. 1: a, b), which are both naturally occurring morphologies in clinical isolates. In addition the phenotypes are very stable and thus this strain has been a model strain to investigate phenotypic switching. However, other switching strains are described and include serotypes A and other D strains and hence highlight the notion that phenotypic switching is a general phenomenon [28, 29].
Figure 1.
Colony morphology and capsule phenotype of RC2 and NP1 variants. The colonies are grown on sabouraud dextrose agar plates. For capsular phenotype, cells grown overnight in sabouraud dextrose broth were observed microscopically in India ink preparation. (a) RC2-SM colony; (b) RC2-MC colony; (c) RC2-SM capsule; (d) RC2-MC capsule; (e) NP1-SM colony; (f) NP12-MC colony; (g) NP1-SM capsule; (h) NP1-MC capsule
NP1 is a serotype B of C. gattii strain that exhibits a VG1 RAPD pattern similar to other C. gattii strains. This strain was isolated from an immunocompetent 34-year-old male patient with meningitis in India. Upon plating the CSF two distinct colonies morphologies, namely smooth and mucoid were noted and further subcultured (Fig. 1e, f). Both the colonies were found to be the same strain by standard molecular typing methods. Switching experiments with this C. gattii strain NP1 [34] were performed and documented the presence of reversible switching from a mucoid colony (NP1-MC) to a smooth colony (NP1-SM) morphology. Mostly serotypes A and D strains manifest a smooth colony morphology and C. gattii strains exhibit a mucoid colony morphology. Thus, we proposed that the smooth variant, RC2-SM constitutes the parent colony morphology in RC2 whereas, NP1-MC the mucoid variant constitutes the parent morphology in NP1. In NP1 rates of switching and reversion differ and switching from NP1-MC to NP1-SM occurred at 1 in 5 × 10−3 colonies which was higher than the reversion rate from NP1-SM to NP1-MC, which occurred 1 in 7 × 10−5. In contrast, the two rates of switching and reversion in the RC2 were comparable and occur at about 1 in 10−4, when 5 × 10−4 colonies plated.
Comparison of Phenotypic Switching in C. neoformans and C. gattii (Table 1)
Table 1.
Comparison of phenotypic characteristics of RC2 and NP1
| Characteristic parameters |
C. neoformans |
C. gattii |
||
|---|---|---|---|---|
| RC2 SM | RC2 MC | NP1 SM | NP1 MC | |
| Capsule size at 37°C | 1.7 ± 0.6 | 2.9 ± 0.54 | 2.4 ± 0.4 | 5.3 ± 0.2 |
| Capsule size in 5% CO2 | 6.2 ± 09 | 6.9 ± 05 | 2.7 ± 0.09 | 9.8 ± 0.3 |
| Cell size | 5. 9 ± 3 | 7.1 ± 0.79 | 4.7 ± 0.2 | 5.4 ± 0.4 |
| Doubling time at 30°C | 2.5 h | 2.8 h | 11.1 h | 6.4 h |
| Doubling time at 37°C | 2.6 h | 2.7 h | 13.9 h | 6.6 h |
| Doubling time at 37°C in DMEM | - | - | 7.3 h | 6.3 h |
| Phagocytosis index | 39.16 ± 8.6 | 8.9 ± 2 | 14.5 ± 3.6 | 15 ± 8.3 |
| Melanization | Yes | Yes | Yes | Yes |
| Osmotic resistance (Colony morphology on different plates) | SM ≫ MC | SM ≫ MC | SM ≫ MC | SM ≫ MC |
| 1 M NaCl | SM | SM | SM | SM |
| 1M Sorbitol | SM | SM | SM | SM |
| 10 mM Glycerol | SM | SM | SM | SM |
| Lysing enzyme resistance | SM ≫ MC | SM ≫ MC | SM ≫ MC | SM ≫ MC |
| Concentration of lysing enzyme required for complete lysis (µg/ml) | 48 | 12 | 48 | 12 |
| Amphotericin B MIC, (µg/ml) | 0.25 | 0.25 | 0.25 | 0.25 |
| Fluconazole MIC, (µg/ml) | 16 | 16 | 16 | 16 |
| Viscosity of GXM | MC ≫ SM | MC ≫ SM | MC ≫ SM | MC ≫ SM |
| GXM triad structure, as determined by NMR | M1 | M1 | M3, M1, M6 | M3, M1, M6 |
Cellular Characteristics
The doubling time of RC2 is shorter compared to NP1 but in both switching systems the switch variant (RC2-MC and NP1-SM) grows slower when compared to the parent strain (RC2-SM and NP1-MC). Both strains manifest differences in capsule size in their switch variants as the mucoid variant exhibits a significantly larger capsule in both strains (Fig. 1). The capsule induces significantly in both the parent and switch variant of RC2 in the presence of CO2, whereas in the NP1 strain background significant capsule induction was observed only in NP1-MC. There was no difference in MICs of amphotericin B and fluconazole for the switch variants in both strains. RC2-MC and NP1-MC exhibited increased sensitivity to lysing enzyme. Whereas the RC2-SM and NP1-SM were more resistant to osmotic stress. Cell charge and sugar assimilation profile were not affected by phenotypic switching in the two switch systems.
Differences in GXM of RC2 and NP1
An understanding of the basic composition of the cryptococcal capsular structure is important to appreciate the specific capsule alterations observed in phenotypic variants. Switch variants of serotypes D and A strains (RC2, SB4, 24067A) as well as the serotype B strain exhibit changes in polysaccharide capsule that affect virulence. GXM is the predominant capsular polysaccharide and is composed of (1 → 3)-linked linear α-D-mannopyranan with β-D-xylopyranosyl (Xylp) and β-D-glucupyranosyluronic acid (GlcpA) residues added to the mannose at various positions. Six (M1-M6) structural reporter groups (SRG) are defined based on the amount of 2–0-linked, 4–0-linked Xylp residues, and 2–0-linked Glcp A residues [36]. In C. neoformans strain SB4 and in strain 24067A phenotypic switching resulted in significant changes of the biochemical composition of GXM [28]. The GXMs of the C colony type of SB4 are composed of mixtures of SRGs (M2 and M3 for C), whereas SB4-SM exhibits predominately SRG M2. In a similar fashion the PH and WR colonies of 24067A exhibit a mix of SRGs (M1 and M5) whereas the 24067A-SM parents are predominately M1 and M2. The addition of a Xylp group at the 4–0 position in M3 and M5 most likely requires a different enzyme than linkage to the 2–0 position. Interestingly, M3 SRGs are traditionally thought to be present only in the GXM of C. gattii isolates (serotypes B and C), and not in GXM of C. neoformans var. neoformans isolates (serotypes A and D). In RC2, phenotypic switching alters the biophysical and biochemical properties of GXM [37]. Viscosity data in solutions of different ionic strength, suggest that the spacing of GlcpA along the Mannose backbone differs between the GXM of the RC2-SM and that of the RC2-MC strain. Because NMR measures an average repeat unit, no differences between RC2-SM and RC2-MC were detected. NMR analysis of GXMs from the NP1 strain’s switch variants yielded similar serotype B specific reporter group structure for both NP1-SM and NP1-MC GXM. Similar to the RC2 strain, the GXM of the NP1-MC was more viscous compared to the NP1-SM variant.
Most importantly, as a result of altered capsular polysaccharide both antibody and complement mediated phagocytosis of the RC2-MC variant was significantly reduced compared with RC2-SM cells both in vitro and in vivo. Although antibody mediated phagocytosis was comparable for NP1-SM and NP1- MC cells, we found that intracellular survival was significantly enhanced for NP1-MC cells when compared to NP1-SM cells. Hence, the NP1-MC parent phenotype was more resistant to intracellular killing by macrophages.
Inflammatory Response and Virulence of Switch Variants
In all the switching strains of C. neoformans, phenotypic switching affects virulence and pathogenesis. RC2-MC variant was significantly more virulent in all murine and rat animal models. In a murine pulmonary infection model histological analysis demonstrated significant differences in the inflammatory tissue response elicited by RC2-SM and RC2-MC. At day 14, lungs of RC2-SM mice exhibited moderate inflammatory changes with cellular infiltrates composed primarily of lymphocytes and only a few macrophages. By day 28, the cellular infiltrates progressed to orderly granuloma formation with little concomitant lung damage. In contrast, at day 14, RC2-MC-infected lungs exhibited extensive cellular infiltrates beyond the peri-bronchial regions, which were predominantly composed of macrophages and neutrophils with only a few lymphocytes. Near the time of death the inflammatory response increased and resulted in extensive destruction of alveolar membranes. In addition, the RC2-MC variant was able to promote increased intra-cranial pressure in a rat model of cryptococcal meningitis [38]. This finding was important because in human infection increased intra-cranial pressure is the leading cause of high morbidity and mortality [39]. When the virulence of NP1-SM and NP1-MC variant were compared in pulmonary and intra-venous (i.v.) murine animal models, NP1-SM-infected mice survived significantly longer than NP1-MC-infected mice in (i.t.) (P = 0.021) as well as in i.v. (P = 0.008) infection model [34]. Consistent with this, the CFU in the lung of NP1-SM-infected mice after 14 days was significantly lower (P ≤ 0.03) than NP1-MC infected. The inflammatory response also differed for the NP1-SM and NP1-MC. However, in this switching system we did not observe a damage promoting over-stimulated inflammatory response. Histological analysis of the lung sections demonstrated an appropriate and effective inflammatory response in lung tissue infected with NP1-SM. The mononuclear inflammation was composed of lymphocytes and macrophages. In contrast the lung of NP1-MC-infected Balb/c mice exhibited minimal inflammatory response and consistent with the failure to elicit an inflammatory response, a large accumulation of yeast cells in lakes of polysaccharide consistent with cryptococcomas was seen on lung tissue sections. Histological analysis of the brain of NP1-SM and NP1-MC-infected mice demonstrated multiple cryptococcomas but the cryptococcomas of NP1-SM infected were smaller and elicited more inflammation in brain tissue [34].
Phenotypic Switching In vivo, True for Both Switching Systems
For RC2 the proof that phenotypic switching truly occurs in vivo was confirmed by Poisson calculations in mice that were infected with low inocula. In mice that were infected with RC2-SM we recovered RC2-MC colonies with increasing frequency in chronic infection. In mice infected with RC2-MC, we did not recover RC2-SM colonies although; the in vitro switching rate was comparable for RC2-SM to RC2-MC and RC2-MC to RC2-SM. Hence the switching was only one directional in vivo for this strain. In vitro MICs of amphotericin B (AMB) did not differ for the RC2-SM and RC2-MC variants (Table 1), in vivo experiments has demonstrated a more pronounced CFU reduction after AMB treatment in RC2-SM infected than in RC2-MC-infected mice. Treatment with AMB or anti-capsular MAb also promoted the selection of RC2-MC variants in mice [40]. In contrast, in mice infected with either with NP1-SM or NP1-MC, we recovered both phenotypes in the lungs, similar to the CSF of the patient from which the strain was originally grown. Interestingly, from the brains of mice-infected i.v. or i.t., only the smooth phenotype was recovered regardless whether the mouse was infected with the NP1-SM or NP1-MC. Thus in contrast to RC2, phenotypic switching occurs in both directions in vivo in NP1 and appears to be necessary for the NP1-MC variant to cross the blood brain barrier.
Conclusion
The phenomenon of phenotypic switching in C. neoformans has been well studied in serotypes A, D strains and recently also in a C. gattii strain. C. neoformans is an excellent model organism for studying phenotypic switching and its role in the pathogenesis and in progression of chronic infection. Both C. neoformans and C. gattii undergo phenotypic switching during experimental infection and enhance virulence. The switch variants of RC2 and NP1 exhibit changes in the polysaccharide capsule and elicit qualitatively different inflammatory responses in the host. This is reminiscent of phase variation in encapsulated bacteria. The molecular mechanism of phenotypic switching in C. neoformans is still unknown. The results of differential display on RC2-SM and RC2-MC mRNA demonstrated that phenotypic switching is associated with upregulation of genes in MC switch variant relative to the SM switch variant. The function of most of the genes in unknown; however, some encode proteins containing immunogenic epitopes [41]. For C. gattii these studies have not been undertaken.
List of Abbreviations
- SM
Smooth colony
- MC
Mucoid colony
- GXM
Glucuronoxylomannan
- ICP
Intra-cranial pressure
- CNS
Central nervous system
Contributor Information
Neena Jain, Email: njain@aecom.yu.edu, Department of Medicine, Albert Einstein College of Medicine, 1300 Morris Park Avenue, UL 1223, Bronx, NY, USA.
Bettina C. Fries, Department of Microbiology & Immunology, Albert Einstein College of Medicine, Bronx, NY, USA e-mail: njain@aecom.yu.edu
References
- 1.Kussell E, Leibler S. Phenotypic diversity, population growth, and information in fluctuating environments. Science. 2005;309(5743):2075–2078. doi: 10.1126/science.1114383. doi: 10.1126/science.1114383. [DOI] [PubMed] [Google Scholar]
- 2.Slutsky B, Buffo J, Soll DR. High-frequency switching of colony morphology in Candida albicans. Science. 1985;230(4726):666–669. doi: 10.1126/science.3901258. doi: 10.1126/science.3901258. [DOI] [PubMed] [Google Scholar]
- 3.Slutsky B, Staebell M, Anderson J, Risen L, Pfaller M, Soll DR. “White-opaque transition”: a second high-frequency switching system in Candida albicans. J Bacteriol. 1987;169(1):189–197. doi: 10.1128/jb.169.1.189-197.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Miller MG, Johnson AD. White-opaque switching in Candida albicans is controlled by mating-type locus homeodomain proteins and allows efficient mating. Cell. 2002;110(3):293–302. doi: 10.1016/s0092-8674(02)00837-1. doi: 10.1016/S0092-8674(02)00837-1. [DOI] [PubMed] [Google Scholar]
- 5.Perez-Martin J, Uria JA, Johnson AD. Phenotypic switching in Candida albicans is controlled by a SIR2 gene. Embo J. 1999;18(9):2580–2592. doi: 10.1093/emboj/18.9.2580. doi: 10.1093/emboj/18.9.2580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nikawa H, Mikihira S, Egusa H, Fukushima H, Kawabata R, Hamada T, et al. Candida adherence and biofilm formation on oral surfaces. Nippon Ishinkin Gakkai Zasshi. 2005;46(4):233–242. doi: 10.3314/jjmm.46.233. [DOI] [PubMed] [Google Scholar]
- 7.Kantarcioglu AS, Boekhout T, De Hoog GS, Theelen B, Yucel A, Ekmekci TR, et al. Subcutaneous cryptococcosis due to Cryptococcus diffluens in a patient with sporotrichoid lesions case report, features of the case isolate and in vitro antifungal susceptibilities. Med Mycol. 2007;45(2):173–181. doi: 10.1080/13693780601045166. doi: 10.1080/13693780601045166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Franzot SP, Salkin IF, Casadevall A. Cryptococcus neoformans var. grubii: separate varietal status for Cryptococcus neoformans serotype A isolates. J Clin Microbiol. 1999;37(3):838–840. doi: 10.1128/jcm.37.3.838-840.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kwon-Chung KJ, Varma A. Do major species concepts support one, two or more species within Cryptococcus neoformans? FEMS Yeast Res. 2006;6(4):574–587. doi: 10.1111/j.1567-1364.2006.00088.x. doi: 10.1111/j.1567-1364.2006.00088.x. [DOI] [PubMed] [Google Scholar]
- 10.Speed B, Dunt D. Clinical and host differences between infections with the two varieties of Cryptococcus neoformans. Clin Infect Dis. 1995;21(1):28–34. doi: 10.1093/clinids/21.1.28. discussion 35-6. [DOI] [PubMed] [Google Scholar]
- 11.Chen S, Sorrell T, Nimmo G, Speed B, Currie B, Ellis D, et al. Epidemiology and host-and variety-dependent characteristics of infection due to Cryptococcus neoformans in Australia and New Zealand. Australasian Cryptococcal Study Group. Clin Infect Dis. 2000;31(2):499–508. doi: 10.1086/313992. doi: 10.1086/313992. [DOI] [PubMed] [Google Scholar]
- 12.Mitchell DH, Sorrell TC, Allworth AM, Heath CH, McGregor AR, Papanaoum K, et al. Cryptococcal disease of the CNS in immunocompetent hosts: influence of cryptococcal variety on clinical manifestations and outcome. Clin Infect Dis. 1995;20(3):611–616. doi: 10.1093/clinids/20.3.611. [DOI] [PubMed] [Google Scholar]
- 13.Banerjee U, Datta K, Casadevall A. Serotype distribution of Cryptococcus neoformans in patients in a tertiary care center in India. Med Mycol. 2004;42(2):181–186. doi: 10.1080/13693780310001615376. doi: 10.1080/13693780310001615376. [DOI] [PubMed] [Google Scholar]
- 14.Chen J, Varma A, Diaz MR, Litvintseva AP, Wollenberg KK, Kwon-Chung KJ. Cryptococcus neoformans strains and infection in apparently immunocompetent patients, China. Emerg Infect Dis. 2008;14(5):755–762. doi: 10.3201/eid1405.071312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dromer F, Mathoulin-Pelissier S, Launay O, Lortholary O. Determinants of disease presentation and outcome during cryptococcosis: the CryptoA/D study. PLoS Med. 2007;4(2):e21. doi: 10.1371/journal.pmed.0040021. doi: 10.1371/journal.pmed.0040021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Irokanulo EO, Makinde AA, Akuesgi CO, Ekwonu M. Cryptococcus neoformans var neoformans isolated from droppings of captive birds in Nigeria. J Wildl Dis. 1997;33(2):343–345. doi: 10.7589/0090-3558-33.2.343. [DOI] [PubMed] [Google Scholar]
- 17.Kidd SE, Hagen F, Tscharke RL, Huynh M, Bartlett KH, Fyfe M, et al. A rare genotype of Cryptococcus gattii caused the cryptococcosis outbreak on Vancouver Island (British Columbia, Canada) Proc Natl Acad Sci USA. 2004;101(49):17258–17263. doi: 10.1073/pnas.0402981101. doi: 10.1073/pnas.0402981101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hoang LM, Maguire JA, Doyle P, Fyfe M, Roscoe DL. Cryptococcus neoformans infections at Vancouver Hospital and Health Sciences Centre (1997–2002): epidemiology, microbiology and histopathology. J Med Microbiol. 2004;53(Pt 9):935–940. doi: 10.1099/jmm.0.05427-0. doi: 10.1099/jmm.0.05427-0. [DOI] [PubMed] [Google Scholar]
- 19.Upton A, Fraser JA, Kidd SE, Bretz C, Bartlett KH, Heitman J, et al. First contemporary case of human infection with Cryptococcus gattii in Puget Sound: evidence for spread of the Vancouver Island outbreak. J Clin Microbiol. 2007;45(9):3086–3088. doi: 10.1128/JCM.00593-07. doi: 10.1128/JCM.00593-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Heitman J. Sexual reproduction and the evolution of microbial pathogens. Curr Biol. 2006;16(17):R711–R725. doi: 10.1016/j.cub.2006.07.064. doi: 10.1016/j.cub.2006.07.064. [DOI] [PubMed] [Google Scholar]
- 21.Lizarazo J, Linares M, de Bedout C, Restrepo A, Agudelo CI, Castaneda E. Results of nine years of the clinical and epidemiological survey on cryptococcosis in Colombia, 1997–2005. Biomedica. 2007;27(1):94–109. [PubMed] [Google Scholar]
- 22.Escandon P, Sanchez A, Martinez M, Meyer W, Castaneda E. Molecular epidemiology of clinical and environmental isolates of the Cryptococcus neoformans species complex reveals a high genetic diversity and the presence of the molecular type VGII mating type a in Colombia. FEMS Yeast Res. 2006;6(4):625–635. doi: 10.1111/j.1567-1364.2006.00055.x. doi: 10.1111/j.1567-1364. 2006.00055.x. [DOI] [PubMed] [Google Scholar]
- 23.Fries BC, Casadevall A. Serial isolates of Cryptococcus neoformans from patients with AIDS differ in virulence for mice. J Infect Dis. 1998;178(6):1761–1766. doi: 10.1086/314521. doi: 10.1086/314521. [DOI] [PubMed] [Google Scholar]
- 24.Klepser ME, Pfaller MA. Variation in electrophoretic karyotype and antifungal susceptibility of clinical isolates of Cryptococcus neoformans at a university-affiliated teaching hospital from 1987 to 1994. J Clin Microbiol. 1998;36(12):3653–3656. doi: 10.1128/jcm.36.12.3653-3656.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jain N, Wickes BL, Keller SM, Fu J, Casadevall A, Jain P, et al. Molecular epidemiology of clinical Cryptococcus neoformans strains from India. J Clin Microbiol. 2005;43(11):5733–5742. doi: 10.1128/JCM.43.11.5733-5742.2005. doi: 10.1128/JCM.43.11.5733-5742.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.San-Blas G, Travassos LR, Fries BC, Goldman DL, Casadevall A, Carmona AK, et al. Fungal morphogenesis and virulence. Med Mycol. 2000;38(suppl1):79–86. [PubMed] [Google Scholar]
- 27.Franzot SP, Mukherjee J, Cherniak R, Chen LC, Hamdan JS, Casadevall A. Microevolution of a standard strain of Cryptococcus neoformans resulting in differences in virulence and other phenotypes. Infect Immun. 1998;66(1):89–97. doi: 10.1128/iai.66.1.89-97.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fries BC, Goldman DL, Cherniak R, Ju R, Casadevall A. Phenotypic switching in Cryptococcus neoformans results in changes in cellular morphology and glucuronoxylomannan structure. Infect Immun. 1999;67(11):6076–6083. doi: 10.1128/iai.67.11.6076-6083.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Goldman DL, Fries BC, Franzot SP, Montella L, Casadevall A. Phenotypic switching in the human pathogenic fungus Cryptococcus neoformans is associated with changes in virulence and pulmonary inflammatory response in rodents. Proc Natl Acad Sci U S A. 1998;95(25):14967–14972. doi: 10.1073/pnas.95.25.14967. doi: 10.1073/pnas.95.25.14967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fries BC, Taborda CP, Serfass E, Casadevall A. Phenotypic switching of Cryptococcus neoformans occurs in vivo and influences the outcome of infection. J Clin Invest. 2001;108(11):1639–1648. doi: 10.1172/JCI13407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tay ST, Lim HC, Tajuddin TH, Rohani MY, Hamimah H, Thong KL. Determination of molecular types and genetic heterogeneity of Cryptococcus neoformans and C. gattii in Malaysia. Med Mycol. 2006;44(7):617–622. doi: 10.1080/13693780600857330. doi: 10.1080/13693780600857330. [DOI] [PubMed] [Google Scholar]
- 32.Almeida AM, Matsumoto MT, Baeza LC, de Oliveira ESRB, Kleiner AA, Melhem Mde S, et al. Molecular typing and antifungal susceptibility of clinical sequential isolates of Cryptococcus neoformans from Sao Paulo State, Brazil. FEMS Yeast Res. 2007;7(1):152–164. doi: 10.1111/j.1567-1364.2006.00128.x. doi: 10.1111/j.1567-1364.2006.00128.x. [DOI] [PubMed] [Google Scholar]
- 33.Wickes BL, Mayorga ME, Edman U, Edman JC. Dimorphism and haploid fruiting in Cryptococcus neoformans: association with the alpha-mating type. Proc Natl Acad Sci U S A. 1996;93(14):7327–7331. doi: 10.1073/pnas.93.14.7327. doi: 10.1073/pnas.93.14.7327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jain N, Li L, McFadden DC, Banarjee U, Wang X, Cook E, et al. Phenotypic switching in a Cryptococcus neoformans variety gattii strain is associated with changes in virulence and promotes dissemination to the central nervous system. Infect Immun. 2006;74(2):896–903. doi: 10.1128/IAI.74.2.896-903.2006. doi: 10.1128/IAI.74.2.896-903.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wilson DE, Bennett JE, Bailey JW. Serologic grouping of Cryptococcus neoformans. Proc Soc Exp Biol Med. 1968;127(3):820–823. doi: 10.3181/00379727-127-32812. [DOI] [PubMed] [Google Scholar]
- 36.Cherniak R, O’Neill EB, Sheng S. Assimilation of xylose, mannose, and mannitol for synthesis of glucuronoxylomannan of Cryptococcus neoformans determined by 13C nuclear magnetic resonance spectroscopy. Infect Immun. 1998;66(6):2996–2998. doi: 10.1128/iai.66.6.2996-2998.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McFadden DC, Fries BC, Wang F, Casadevall A. Capsule structural heterogeneity and antigenic variation in Cryptococcus neoformans. Eukaryot Cell. 2007;6(8):1464–1473. doi: 10.1128/EC.00162-07. doi:10.1128/EC.00162-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fries BC, Lee SC, Kennan R, Zhao W, Casadevall A, Goldman DL. Phenotypic switching of Cryptococcus neoformans can produce variants that elicit increased intracranial pressure in a rat model of cryptococcal meningoencephalitis. Infect Immun. 2005;73(3):1779–1787. doi: 10.1128/IAI.73.3.1779-1787.2005. doi: 10.1128/IAI.73.3.1779-1787.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Graybill JR, Sobel J, Saag M, Van Der Horst C, Powderly W, Cloud G, et al. Diagnosis and management of increased intracranial pressure in patients with AIDS and cryptococcal meningitis. The NIAID mycoses study group and AIDS cooperative treatment groups. Clin Infect Dis. 2000;30(1):47–54. doi: 10.1086/313603. doi: 10.1086/313603. [DOI] [PubMed] [Google Scholar]
- 40.Fries BC, Cook E, Wang X, Casadevall A. Effects of antifungal interventions on the outcome of experimental infections with phenotypic switch variants of Cryptococcus neoformans. Antimicrob Agents Chemother. 2005;49(1):350–357. doi: 10.1128/AAC.49.1.350-357.2005. doi: 10.1128/AAC.49.1.350-357.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Guerrero A, Jain N, Cook E, Casadevall A, Fries B. Phenotypic switch variant of Cryptococcus differ in gene expression profile. 103rd ASM General Meeting, F-039; Washington, DC. 2003. [Google Scholar]