Abstract
During atrial fibrillation (AF), rapid stimulation causes atrial remodeling that increases arrhythmia susceptibility. Using an established atrial (HL-1) myocyte model, we investigated the transcriptional profile associated with early atrial myocyte remodeling. Spontaneously contracting HL-1 cells were cultured in the absence and presence of rapid stimulation for 24 hrs and RNA harvested for microarray analysis. We identified 758 genes that were significantly altered with rapid stimulation (626 up- and 132 down-regulated). Results were confirmed using real-time quantitative RT-PCR for selected genes based on physiological relevance in human AF and/or experimental atrial tachycardia (AT), and regulation in the microarray results. In some cases, transcriptional changes were rapid, occurring within 3 hours. For a selected group of genes, results were validated for the expressed protein, with findings that correlated with observed transcriptional changes. Significantly regulated genes were classified using the Gene Ontology Database to permit direct comparison of our findings with previously published myocardial transcriptional profiles. For broad functional categories, there was strong concordance between rapidly stimulated HL-1 myocytes and human AF, but not for other remodeling paradigms (cardiomyopathy and exercise). Many individual gene changes were conserved with AF/AT, with marked up-regulation of genes encoding brain and atrial natriuretic peptide precursors, and heat shock proteins. For the conserved genes, both a cellular stress and survival response was evident. Our results demonstrate similarities with human AF/experimental AT with respect to large-scale patterns of transcriptional remodeling, as well as regulation of specific individual genes. Importantly, we identified novel pathways and molecules that were concordantly regulated in vivo.
Keywords: Microarray, HL-1 cells, atrial myocytes, atrial fibrillation, remodeling, western, PCR, enzyme immunoassay
Introduction
Atrial fibrillation (AF) is the most common sustained cardiac arrhythmia in the Western World, and it remains a major cause of stroke and death. Nevertheless, currently available therapies are inadequate. The clinical course of AF is usually progressive, due to rapid atrial activation that increases arrhythmia susceptibility as a result of electrical and structural remodeling. The molecular events that initiate the remodeling process are not currently known, although circulating hormones and/or non-cardiomyocyte elements (e.g., the renin-angiotensin system, inflammation and oxidative stress) have been proposed.
A hallmark feature of experimental atrial tachycardia (AT) and human AF is abbreviation of atrial action potential duration (APD) and refractoriness, due at least in part to reduced ICa,L. Changes in other ionic currents have been less consistent across studies.[1] A possible explanation is the variability of in vivo parameters such as structural heart disease, heart failure and altered hemodynamics[2–4] that can promote atrial remodeling in a manner independent of rapid activation. Prominent histological features of AT remodeling are myolysis,[5] cellular hypertrophy, degeneration of the sarcoplasmic reticulum, dedifferentiation (e.g., reappearance of a fetal gene program)[6] and neurogenesis.[7] Patients with AF have increased levels of atrial fibrosis compared to those in sinus rhythm, although this has not been a consistent finding in animal models.[8] The structural and hemodynamic-based remodeling that occurs in vivo complicates identification of the early molecular pathways that promote AF. An experimental approach that is free of these confounding issues would facilitate investigation of the initial cellular events that mediate AT remodeling.
Our laboratory has previously shown that rapid stimulation of an atrial cell line (HL-1 cells) for 24 hrs in culture results in electrical remodeling resembling that seen AF/AT.[9] The utility of this in vitro model system has been confirmed in other studies, with experimental results that correlate with findings in both human AF[10,11] and in vivo AT models.[12] These results demonstrate that HL-1 atrial cells themselves contain the essential components necessary to produce AF/AT remodeling. Despite the inherent limitations of an in vitro model system, rapid stimulation of atrial cells in culture provides an opportunity to investigate the initial, myocyte-specific events during rapid stimulation. Therefore, we investigated the initial transcriptional regulation occurring in rapidly stimulated HL-1 atrial cells using microarray analysis and real time quantitative RT-PCR. Our results demonstrate similarities between these myocytes and human AF and experimental AT with respect to large-scale patterns of transcriptional remodeling, as well as specific single gene events. Importantly, we identified novel pathways and molecules that were regulated in both the in vitro model and in vivo.
Methods
Experimental preparation: HL-1 atrial myocytes
Nearly confluent HL-1 atrial myocytes were cultured in the absence (control) or presence of rapid stimulation, as described previously.[9] Cells were stimulated at 300 bpm, a rate that exceeded the spontaneous beat rate by 1.5 – 2 fold, but did not cause prohibitive cell death. Electrical capture in the stimulated cells was confirmed using qualitative fluorescence microscopy to monitor intracellular Ca2+ release after cells were loaded with the calcium sensitive dye Xrod-1 (5μM, Invitrogen). Complete details are presented in the Online Methods Section.
mRNA preparation
mRNA samples were extracted from control or stimulated cells using the RNAeasy Mini Kit with DNAse treatment (Qiagen). To ensure rigorous quality control, all samples were analyzed by the Vanderbilt Microarray Shared Resource for concentration, purity, and integrity (degradation) using spectrophotometry (NanoDrop ND-1000 UV-Vis Spectrophotometer) and a microfluidic assay (Agilent 2100 Bioanalyzer, Agilent Technologies). Synthesis of cDNA for real-time quantitative RT-PCR (q-PCR) was performed using random hexamers and Taqman reverse transcription reagents (Applied Biosystems).
Microarray experiments: Hybridization to GeneChip Mouse Genome 430 2.0 Array
Near-confluent HL-1 cells were split into 2 dishes, with one subjected to rapid pacing at 300 bpm for 24 hr, and the other containing control, spontaneously-beating cells that were cultured in parallel. mRNA was harvested from each dish. A total of 5 pairs of control and rapidly-paced cells were generated, with the mRNA extracted from each group (control vs. paced) combined to generate pooled mRNA samples for microarray analysis. This process was replicated using 5 time-independent pairs to generate an additional pooled sample for control and rapidly-paced cells (n=4 arrays). The Vanderbilt Microarray Shared Resource completed the synthesis of ds cDNA and biotinylated cRNA, as well as subsequent hybridization steps. Samples were hybridized to the GeneChip Mouse Genome 430 2.0 Array. Gene symbols and titles were based on Affymetrix Mu 430 2.0 annotation.
Real-time quantitative RT-PCR microarray validation
To validate the microarray analysis, the expression pattern of 24 genes was evaluated by q-PCR (ABI Prism 7900HT Sequence Detection System, Applied Biosystems) using Sybr Green PCR Master Mix (Applied Biosystems). These genes were selected by their physiological relevance in human AF, and their regulation status based on microarray analysis (up-, down-, or non-regulated). Primers (Online Supplement 1) were designed such that all amplified products spanned an exon-exon boundary within the coding regions (with the exception of Apba3) and produced a single melting curve after the q-PCR reactions. All primer sets amplified their specific gene sequences with no cross-reaction before 35 cycles. The reference gene for normalization of amplification results was 18S ribosomal RNA (18S). Equivalent reaction efficiency was confirmed by performing q-PCR at various cDNA concentrations for 18S and each gene of interest (GOI). For each gene, 3 independent pairs of control and stimulated HL-1 atrial cell mRNA were analyzed. All samples were measured in triplicate for both the amplification efficiency curves (GOI vs. 18S) and for q-PCR amplification.
Validation of results for the expressed proteins
Whole-cell lysate was generated from 5 sets of paired dishes of control and rapidly stimulated HL-1 atrial cells (24 hrs). Heat shock protein 70 (Hsp70) was quantitated using Western analysis, with lysate from heat-shocked HL-1 atrial cells as the positive control. For atrial and brain natriuretic peptides (ANP and BNP), protein release into the cell culture medium was quantitated using enzyme immunoassay methods according to the manufacturer’s instructions. These procedures are presented in detail in the Online Supplemental Expanded Methods Section.
Data analysis
In brief, microarray data were normalized using Robust Multichip Analysis (RMA) and absent calls removed. Significantly regulated genes were identified using the Significance Analysis of Microarrays approach (false discovery rate or FDR of q<0.06). The original data files have been deposited in Gene Expression Omnibus (GEO, http://www.ncbi.nlm.nih.gov/geo) and are accessible through GEO series accession number GSE10598. Genes were functionally classified using the Gene Ontology (GO) Database via the web-based program FatiGO+ to permit comparison of the rapidly stimulated HL-1 gene profile to other myocardial transcriptional profiles after conversion to species-consistent homologues. For time course q-PCR and protein measurement, paired t-tests were used to determine statistical significance (P≤0.05 or 0.01) and expressed as mean±SEM.
Results
Effects of rapid stimulation on gene expression in HL-1 cells at 24 hrs
We used microarray gene expression profiling to investigate early transcriptional regulation in rapidly stimulated HL-1 atrial myocytes. These experiments enabled us to identify 919 Affymetrix probes, representing 758 genes (626 up-regulated and 132 down-regulated) whose expression was significantly altered by rapid stimulation of HL-1 cells for 24 hrs. A complete list of significantly regulated transcripts with probe ID, numeric fold-change, and q-value is provided in Online Supplement 2. Significantly regulated genes were classified using the Gene Ontology Database and Panther Classification System.[13] Up-regulated transcripts were found predominantly in functional classes that included signaling, transcription, and metabolism (Table 1), as well as those involved in the cellular response to stress. A summary of specific genes involved in the stress response as defined by relevant subcategories (e.g., apoptosis, immunity and defense response, inflammation and oxidative stress) is presented in Online Supplement 3, with the entire list of significantly regulated genes, separated into their major functional categories and subclassifications shown in Online Supplement 4. Transcription factors were generally up-regulated, a result that is consistent with the early time point in the remodeling process (24 hrs) that we investigated. In contrast, down-regulation of transcription factors was observed in permanent AF,[14] likely resulting from the end-stage nature of the remodeled substrate.
TABLE 1.
Highly regulated gene clusters in rapidly stimulated HL-1 atrial myocytes
| Category | Total | Up-regulated | Down-regulated |
|---|---|---|---|
| Signaling | 118 | 92 | 26 |
| Transcription | 80 | 59 | 21 |
| Response to Stress | 74 | 63 | 11 |
| Metabolism | 66 | 55 | 11 |
| Protein Handling | 49 | 41 | 8 |
| Structural Components | 47 | 32 | 15 |
| Neurogenesis | 26 | 20 | 6 |
| Proteolysis | 17 | 11 | 6 |
| Muscle Development & Function | 13 | 11 | 2 |
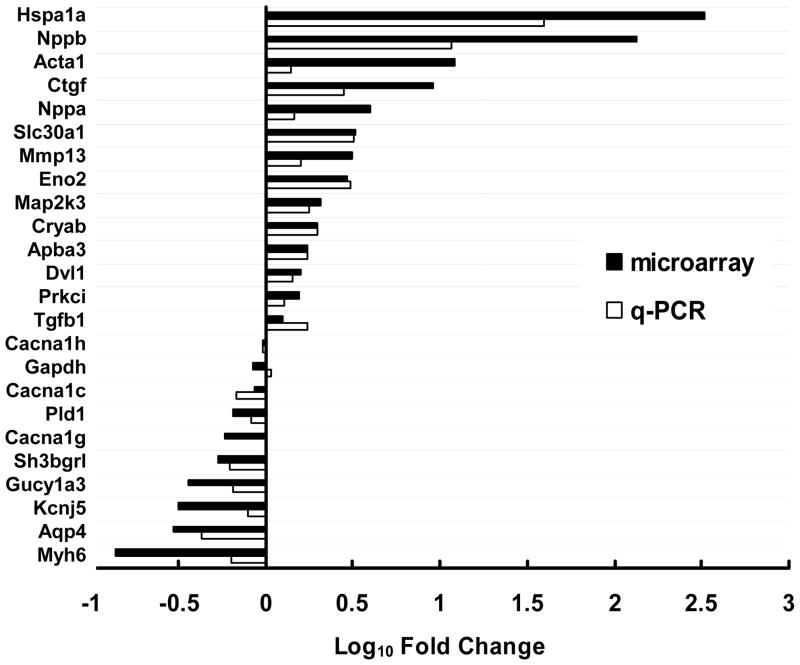
To validate the microarray analysis, q-PCR was performed for 24 genes with physiological relevance in human AF that displayed differential regulation in the microarray analysis (up-, down- or non-regulated). In 23 of 24 genes tested, microarray and q-PCR methods produced concordant results with respect to the direction of regulation (Figure 1), although variability was observed in the degree of fold change (e.g., Hspa1a, Nppa and Acta1).
Figure 1. Comparison of real-time quantitative RT-PCR (q-PCR) and microarray results.
Data are expressed as log fold change (black bars represent microarray results and unfilled bars represent q-PCR results). Genes were selected by virtue of significant up-, down- or non-regulation in the microarray and/or physiological relevance in AF.
Time course of transcriptional remodeling
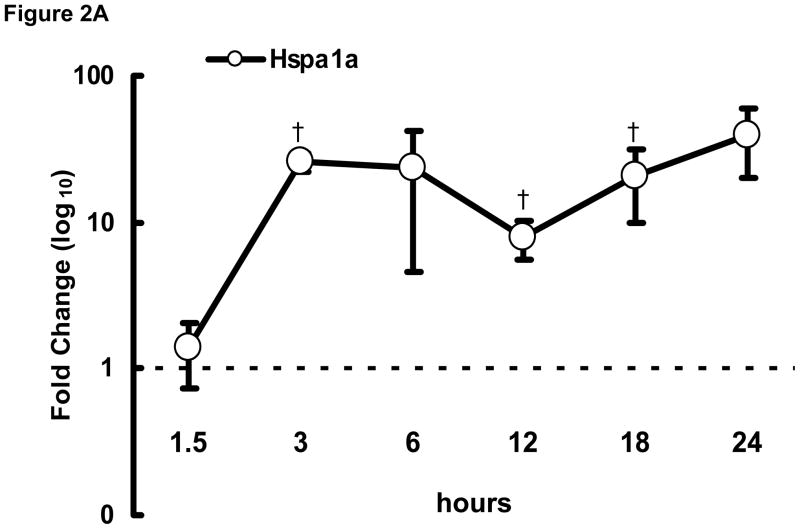
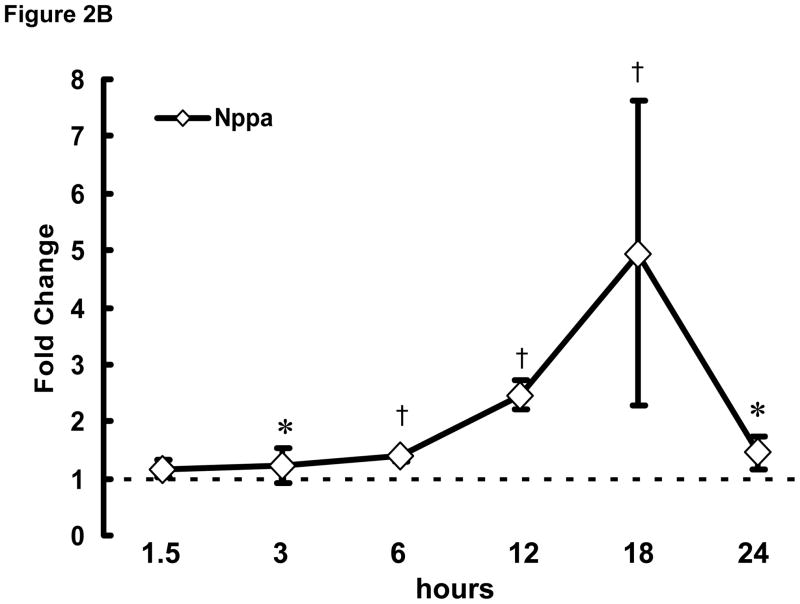
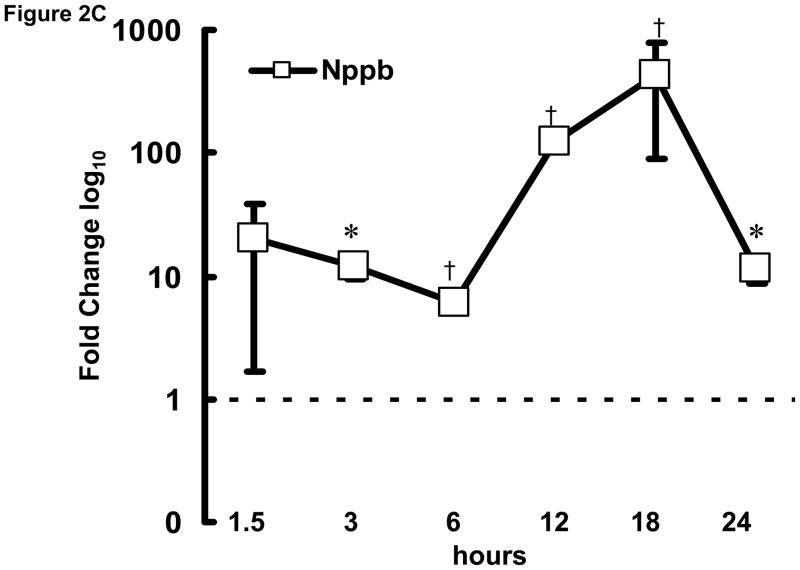
To examine the initial phase of transcriptional remodeling in rapidly stimulated HL-1 atrial cells in greater detail, q-PCR was performed at various time points (1.5–24 hrs) following the onset of stimulation for a selected group of genes, including Hspa1a, Nppa, and Nppb (encoding heat shock protein 70 [Hsp70], atrial and brain natriuretic peptides [ANP and BNP], respectively), which are involved in the early stress response. As shown in Figure 2, significant changes were detected as early as 3 hr after initiation of rapid stimulation. Thus, the onset of transcriptomic changes in response to the stimulus is rapid in these cells for at least some genes.
Figure 2. Time course of transcriptional remodeling.
q-PCR was performed on HL-1 atrial cells subjected to rapid stimulation for 1.5–24 hrs. Transcription of (A) Hspa1a, (B) Nppa, and (C) Nppb increased over time. Mean ± SEM (n=3), *:P≤0.05, †:P≤0.01.
Validation of results for the expressed proteins
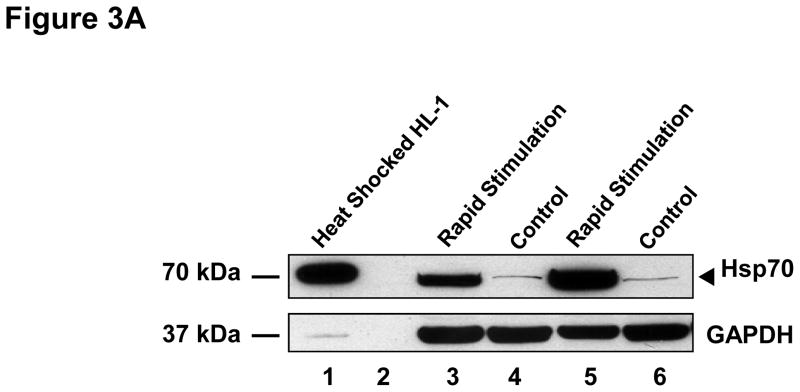
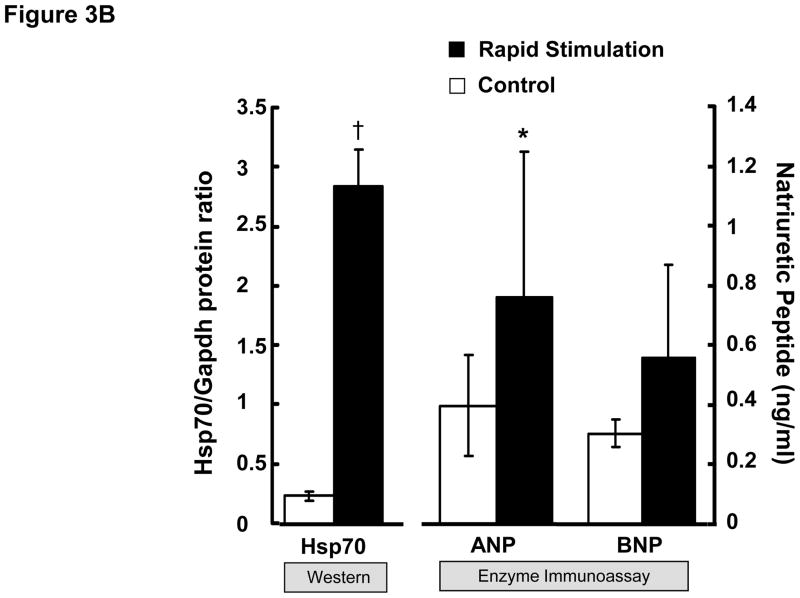
To further confirm the results of the microarray analysis, we investigated the effects of 24 hrs of rapid stimulation on protein levels for Hsp70, ANP and BNP in these cells. The production of Hsp70 by HL-1 atrial cells was dramatically increased (P=0.013) by rapid stimulation (Figure 3). ANP and BNP are hormones secreted into the blood by atrial cells during AF. Indeed, we could not detect ANP in either control or rapidly stimulated HL-1 atrial cells by Western blotting (n=14 pairs, data not shown). However, as shown in Figure 3B, rapid stimulation of HL-1 atrial cells led to an increase in both ANP and BNP protein concentrations in the culture medium, which was statistically significant for ANP (p=0.02).
Figure 3. Effects of rapid stimulation on protein expression.
(A) HL-1 atrial cells were cultured in the absence (control) and presence of rapid stimulation for 24 hrs. Cell lysate was harvested and Western analysis was performed using an antibody directed against heat shock protein 70 (Hsp70, 90 μg in lanes 3–6) or GAPDH. Lysate (8 μg) from heat shocked HL-1 cells was loaded as a positive control (lane 1; lane 2 is blank). (B) Summary data are shown for Western analysis of Hsp70 normalized to GAPDH (n=3) on the left panel. ANP (n=5) and BNP (n=3) were quantitated in the culture medium from HL-1 atrial cells in the absence and presence of 24 hrs of rapid stimulation, with summary data shown in the right panel. Mean ± SEM, *:P≤0.05, †:P≤0.01
Transcriptomic signature of rapidly stimulated HL-1 cells: Concordance with human AF
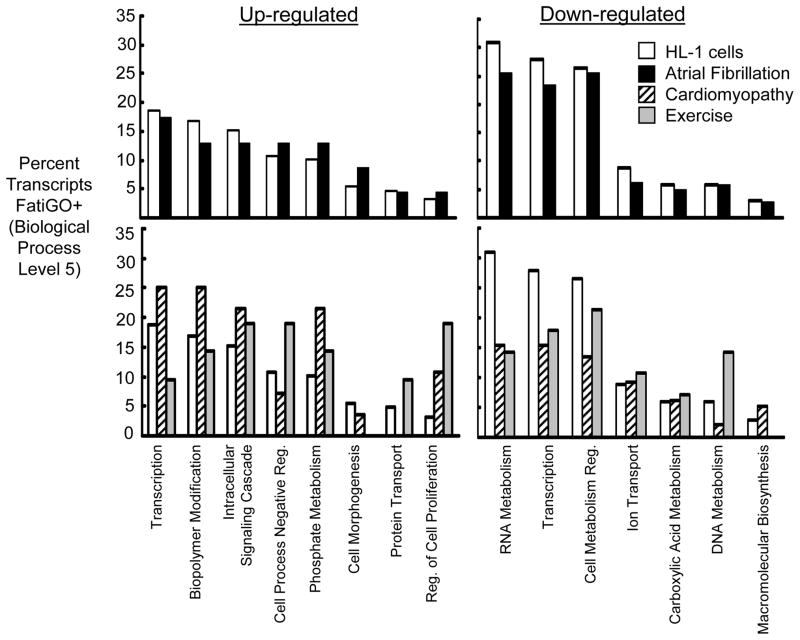
To investigate whether atrial cells subjected to rapid stimulation produced broad transcriptional changes that were similar to human AF, significantly regulated transcripts were compared to a permanent human AF transcriptome[14] using FatiGO+ (a web-based tool for Gene Ontology classification). The top panels of Figure 4 show functional classification of the most significantly regulated HL-1 transcripts compared to permanent human AF based on the percentage of transcripts up- or down-regulated (using Biological Process categories at a complexity level of 5). The categories shown represent those most dominantly regulated in the HL-1 data set, that were also present in the permanent human AF data set. The results of this comparison provide evidence for strong concordance between the two data sets.
Figure 4. Comparison of transcripts regulated in rapidly stimulated HL-1 atrial cells with human AF and other experimental models.
FatiGO+ analysis classifications containing the highest percentage of significantly regulated transcripts in rapidly stimulated HL-1 cells are shown. Categories are quantitated as a percentage of total transcripts (Biological Process, complexity level 5). Results are shown for rapidly stimulated HL-1 cells (unfilled bars), permanent AF14 (black bars), cardiomyopathy16 (striped bars) and chronic endurance exercise17 (grey bars).
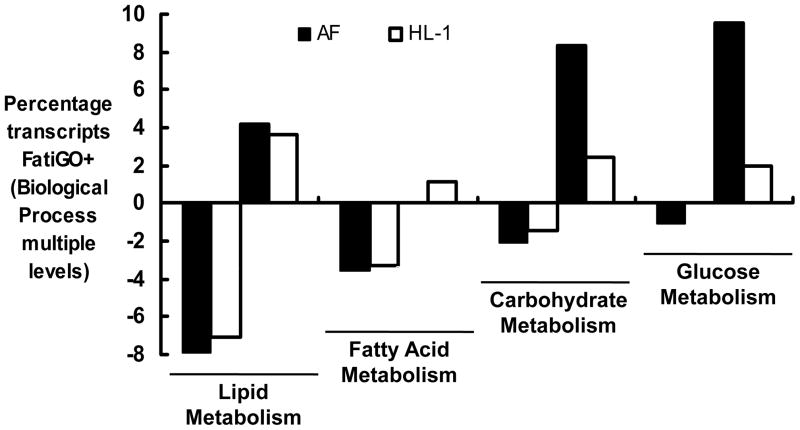
In human AF, the pattern of adaptation that occurs in metabolic transcripts resembles a fetal energy program,[14,15] with a shift from fatty acid oxidation to greater reliance on glucose utilization. The fatty acid/glucose metabolic profile of the two gene sets was also compared (Figure 5). As in the data set for human AF,[14] there was evidence of coordinated transcriptional down-regulation of fatty acid metabolism in rapidly stimulated HL-1 cells, with parallel up-regulation of genes involved in glucose utilization. The percentage of transcripts up-regulated in carbohydrate/glucose metabolism categories was more robust in the permanent human AF data set, possibly related to the longer duration of the stimulus.
Figure 5. Metabolic substrate utilization: Comparison of rapidly stimulated HL-1 atrial cells to human AF.
Classifications are quantitated as in Figure 4 with Biological Process, complexity level 4 for lipid and carbohydrate metabolism, level 7 for fatty acid metabolism, and level 8 for glucose metabolism. Data are shown for HL-1 cells (unfilled bars) and permanent human AF14 (black bars).
Comparison to other cardiac remodeling paradigms
To determine whether the transcriptional regulation observed for rapidly stimulated HL-1 cells was relatively specific to AF, additional comparisons were performed between the stimulated atrial cell data set and atrial regulation found in cardiomyopathy[16] or physiologic hypertrophy induced by endurance exercise.[17] The results of these comparisons are shown in the bottom panels of Figure 4 (using the same Biological Process categories and complexity level as for the top panels). In both cases, the percentage of transcripts up or down-regulated is markedly different from that observed with rapidly stimulated HL-1 cells. Taken together, these comparisons suggest that the similarities observed in the transcriptional signatures for permanent human AF and rapidly stimulated HL-1 cells do not represent a nonspecific cellular response the experimental paradigm or to stress.
Conservation of specific genes and processes known to be altered in AF/AT
To further test the relevance of our results to those observed in vivo, we investigated whether transcriptional conservation was evident between rapidly stimulated HL-1 atrial cells and AF/AT at the level of individual transcripts. The transcriptional profile of the stimulated HL-1 atrial cells was compared to recent AF/AT reports on a gene by gene basis after converting the transcripts to their species-specific homologues. This comparison identified 30 transcripts that were significantly regulated in a highly concordant, isoform-specific manner in both stimulated HL-1 atrial cells and AF/AT (Table 2). Importantly, for 16 of the 30 genes, findings were confirmed by q-PCR. In Table 2 the results are grouped into three categories: specific genes/proteins previously reported to be regulated in AF/AT in vivo (top); those that participate in cellular pathways implicated in AF/AT, although the specific transcript had not been previously identified (middle); and conserved genes/proteins for which involvement in AF/AT remodeling is entirely novel (bottom).
Table 2.
Significantly regulated genes in rapidly stimulated HL-1 atrial cells conserved with AF/AT
| Genes/proteins reported in vivo | HL-1 (fold change) | AF/AT (citation #) |
|---|---|---|
| Natriuretic peptide precursors | ||
| Natriuretic peptide precursor type A (Nppa) | ↑ (4.0) | ↑ (14,21,22) |
| Natriuretic peptide precursor type B (Nppb) | ↑ (134.4) | ↑ (14,21) |
| Heat shock proteins | ||
| Heat shock protein 1A (Hspa1a) | ↑ (329.5) | ↑ (14) |
| Heat shock protein 1 (chaperone 10) (Hspe1) | ↑ (2.0) | ↑ (24) |
| Heat shock protein 1 (Hspb1) | ↑ (2.1) | ↑ (12) |
| Crystallin, alpha B (Cryab) | ↑ (2.0) | ↑ (23) |
| Signaling | ||
| Mitogen activated protein kinase kinase 3 (Map2k3) | ↑ (2.1) | ↑ (14) |
| Contractile function | ||
| Myosin, heavy polypeptide 6, cardiac muscle, alpha (Myh6) | ↓(0.1) | ↓(19) |
| Ion channel | ||
| Potassium inwardly-rectifying channel, subfamily J, memb 5 (Kcnj5) | ↓(0.3) | ↓ (14) |
|
Cellular pathways reported previously | ||
| Signaling | ||
| Guanylate cyclase 1, soluble, alpha 3 (Gucy1a3) | ↓(0.4) | ↓ (14) |
| Phosphatidylinositol 3-kinase, regulatory subunit, polypeptide 1(Pik3r1) | ↓ (0.5) | ↓ (14) |
| Phospholipase D1 (Pld1) | ↓ (0.7) | ↓ (14) |
| Oxidative stress | ||
| SH3-binding domain glutamic acid-rich protein like (Sh3bgrl) | ↓ (0.5) | ↓ (14) |
| Contractile function | ||
| Luc7 homolog (S. cerevisiae)-like (Luc7l) | ↑ (2.2) | ↑ (14) |
| Calcium handling | ||
| Calmodulin 3 (Calm3) | ↓(0.5) | ↓↑ (3,14) |
| Solute carrier family 30 (zinc transporter), member 1 (Slc30a1) | ↑ (3.3) | ↑ (38,39) |
| Growth Factors | ||
| Connective tissue growth factor (Ctgf) | ↑ (9.1) | ↑ (23) |
| WNT1 inducible signaling pathway protein 1 (Wisp1) | ↑ (2.1) | ↑ (45) |
|
Novel | ||
| Structure/cytoskeleton | ||
| Erythrocyte protein band 4.1-like 2 (Epb4.1l2) | ↓ (0.5) | ↓(14) |
| Neurogenesis | ||
| Fasciculation and elongation protein zeta 2 (zygin II) (Fez2) | ↑ (1.8) | ↑ (19) |
| Enolase 2, gamma neuronal (Eno2) | ↑ (3.0) | ↑ (14) |
| Transcription factors | ||
| Homeobox A2 (Hoxa2) | ↓ (0.5) | ↓(23) |
| Energy handling | ||
| Transferrin Receptor (Tfrc) | ↓(0.4) | ↓(23) |
| Protein processing | ||
| Mannosidase 1, alpha (Man1a) | ↓ (0.6) | ↓ (14) |
| Membrane-bound transcription factor peptidase, site 1(Mbtps1) | ↓ (0.5) | ↓(14) |
| Protein Trafficking | ||
| Clathrin, light polypeptide (Lcb) (Cltb) | ↑ (2.0) | ↑ (23) |
| Amyloid beta (A4) precursor protein-binding, family A, memb 3 (Apba3) | ↑ (1.7) | ↑ (23) |
| WNT signaling | ||
| Dishevelled, dsh homolog 1 (Drosophila) (Dvl1) | ↑ (1.6) | ↑ (45) |
| Membrane transport | ||
| Solute carrier family 25 (mito/phosphate carrier), memb 25 (Slc25a25) | ↑ (1.6) | ↑ (14) |
| Aquaporin 4 (Aqp4) | ↓ (0.3) | ↓ (14,23) |
Some of the most well described consequences of AF/AT in this conserved group were also the most significantly regulated transcripts (i.e., up-regulation of natriuretic peptide precursors and heat shock proteins). Additionally, the majority of the conserved transcripts (18/30) are associated with pathways and cascades known to be relevant to AF/AT remodeling, such as cellular responses to stress, contractile remodeling and dysfunction, fibrosis and hypertrophic cascades.
Importantly, highly conserved transcriptional regulation was evident for multiple pathways and molecules not previously reported to be altered in AF/AT, as shown at the bottom of Table 2. While novel, many of the transcriptional changes observed are consistent with the known consequences of atrial cell remodeling that occurs in response to rapid stimulation (e.g., altered structural components, energy handling and protein processing/trafficking). However, other processes (e.g., neurogenesis and Wnt signaling) and genes (e.g., aquaporin 4) were identified for which regulation in human AF or experimental AT has not been described. However, their concordant regulation in AF/AT strongly suggests a potential role in the remodeling process in AF/AT.
Discussion
In this study, we used microarray technology and an established cellular model to investigate the early transcriptional changes that occur in atrial myocytes with rapid stimulation. In a global comparison based on functional classification, the transcriptional signature was remarkably similar to that observed with human AF, but discordant with other cardiac remodeling paradigms (Figure 4). We found evidence for robust transcriptional changes in a wide variety of genes known to be regulated in AF/AT (e.g., natriuretic peptide precursors, heat shock proteins, myosin isoforms and growth factors). Importantly, a number of novel, unanticipated genes were identified that were similarly regulated in previously published AT and/or human AF transcriptomes. Such concordant regulation implies that these protein/processes likely play an important role in the remodeling process in vivo.
While it is virtually impossible to quantitatively compare transcriptional profiles derived from different microarray studies (i.e., having variable conditions and platforms), broad qualitative comparisons are feasible based on the functional classification of significantly regulated transcripts.[18] Using this approach, we observed striking concordance between our data set and that derived from permanent human AF.[14] Interestingly, this was also largely the case for a data set taken from a 24 hr canine atrial tachycardia model. As illustrated in Online Supplement 5, important similarities were identified for multiple categories of genes when classified based on their function.[19] An important distinction between our results and the human AF study is that significantly regulated transcripts (including transcription factors) were largely up-regulated for rapidly stimulated HL-1 atrial cells, and down-regulated in permanent AF. Considering the different experimental paradigms, these results are not surprising. For HL-1 atrial cells, transcriptional profiling was conducted early in the remodeling process, whereas permanent AF represents an end-stage substrate during which compensatory changes have already occurred.[20] In this respect, the similarities that remain between the data sets are particularly noteworthy.
A dominant theme in our results was transcriptional regulation consistent with a cardiomyocyte stress response, such as that observed with mechanical stretch. The rapid and striking up-regulation of genes encoding natriuretic peptides, as well as multiple heat shock proteins, provide strong support for phenotypic conservation for the atrial myocyte stress that occurs during AF/AT. Both atrial and brain natriuretic peptides are rapidly released by the fibrillating atria, with elevated circulating levels in patients with AF[14,21] that return to normal when sinus rhythm is restored.[21,22] It is likely that up-regulation of natriuretic peptides, which increase cGMP production, is responsible for the apparent compensatory down-regulation in guanylate cyclase (encoded by Gucy1a3, Table 2). In our data set, heat shock proteins were the most highly regulated of the conserved genes known to be activated by myocytes during mechanical stress.[12,14,23,24] These cytoprotective proteins are produced during a variety of cellular stressors and appear to be cardioprotective in AF/AT. Pharmacological induction of heat shock proteins protects against tachycardia-induced remodeling in experimental AT[12] and may limit progression to persistent AF.[11] The robust up-regulation of transcripts encoding heat shock proteins (Table 2 and Figure 2A) and heat shock protein 70 itself (Figure 3) likely represents an early survival response of atrial myocytes subjected to rapid activation. Heat shock proteins also act to directly suppress proapoptotic signaling events to promote cellular survival.[25,26] Consistent with this, we observed down-regulation of the gene encoding caspase-3 (Casp3, Online Supplements 2, 3 and 4), which is required for DNA fragmentation and morphological changes that occur during the apoptotic process.[27]
Additional evidence for a myocyte stress response was apparent in the deregulation of genes encoding mitogen activated protein kinase kinase 3, phosphatidylinositol 3-kinase, phospholipase D1 and SH3-binding domain glutamic acid-rich protein like (Table 2). Moreover, functional analysis revealed that many of the significantly regulated genes are components of cellular cascades that have been implicated in mechanical stress-induced hypertrophy[28] and pressure overload,[29] as well as ischemia/reperfusion injury,[30] protection against cardiocyte apoptosis,[31] oxidative stress,[32] and arrhythmogenic atrial structural remodeling[33] (Table 1, Online Supplements 2 and 3). Indeed, in response to mechanical stress, multiple hypertrophic and survival cascades are activated in cardiomyocytes, along with contractile remodeling. Our results for both individual genes (e.g., natriuretic peptides, MAP kinases, tyrosine kinases, c-jun and the skeletal isoform of α-actin) and global pathway analysis (Online Supplements 2 and 4) indicate transcriptional regulation for multiple hypertrophic and survival pathways. Nonetheless, the transcriptional profile that we observed in this in vitro model system was most specific for AF, compared to other types of remodeling (Figure 4).
Contractile remodeling is a recognized consequence of human AF and is often accompanied by adaptive changes in myosin isoform expression[34] consistent with activation of the fetal gene program. The myosin isoform shift from the high velocity, atrial isoform, alpha myosin heavy chain (αMHC, encoded by Myh6), to a lower velocity, ventricular isoform, β-myosin heavy chain (βMHC), is theorized to be a compensatory response to reduced ATP production and/or ADP accumulation. In both experimental AT[19,34] and stimulated HL-1 cells, Myh6 was dramatically down-regulated (Figure 1 and Table 2), indicative of an isoform shift. βMHC up-regulation was observed in permanent AF,[34] but not in HL-1 cells stimulated in culture. This discrepancy may result from the shorter duration of rapid stimulation in our in vitro system. We also observed up-regulation of genes encoding immature isoforms in other contractile proteins (Ryr1 and Acta1; Figure 1 and Online Supplements 2 and 4).
Consistent with a shift to a fetal phenotype,[35] we observed evidence for reduced fatty acid oxidation and increased dependence on glucose metabolism. In both permanent AF and the stimulated HL-1 atrial cells, genes in fatty acid oxidation pathways were generally down-regulated, while those involved in glucose utilization were up-regulated (Figure 5). During the fetal/perinatal period, the heart uses glycolysis for nearly half of its ATP production, shifting predominantly to fatty acid utilization in the adult myocardium.[36] This return to a fetal metabolic profile is likely an adaptive response, allowing atrial myocytes to survive for prolonged periods of increased wall stress, a relative shortage of oxygen and higher energy demands.[35]
As in the ventricle, fibrillation in the atrium likely creates intracellular Ca2+ overload, and the expression of proteins involved in Ca2+ handling is altered during AF/AT. Indeed, recent work demonstrates transient elevation in intracellular Ca2+ in rapidly stimulated atrial cells, which promotes transcriptional downregulation of ICa,L.[37] Our results demonstrated conserved transcriptional up-regulation of the solute carrier protein, Slc30a1 or Znt-1 (Figure 1 and Table 2), and down-regulation of the Ca2+ binding protein, calmodulin 3.[3,14] Znt-1 was recently shown to inhibit current through the L-type Ca2+ channels without altering α1C expression.[38] Znt-1 expression is up-regulated in patients with AF,[39] and it may be at least partially responsible for the reduced ICa,L observed in vivo. Transcriptional deregulation of Znt-1 and calmodulin in both AF/AT and rapidly stimulated HL-1 atrial cells is consistent with intracellular Ca2+ overload.
Electrical remodeling is an early consequence of rapid activation, with changes that occur within 24 hrs. Abbreviation of atrial APD and reduced ICa,L, are found in human AF, canine AT and rapidly stimulated HL-1 atrial cells.[9] In longstanding AF, many studies report down-regulation of mRNA (Cacna1c) and protein (Cav1.2) for the L-type Ca2+ channel α1C-subunit.[2,10] However, in our data and the permanent human AF study[14], Cacna1c transcription was not significantly regulated (Figure 1). Previous studies have revealed that L-type calcium channel α-subunit proteolysis,[10] as well as posttranslational modifications of channel function (i.e., up-regulation of Znt-1),[38,39] likely play a role in reducing ICa,L.
Histologic studies have typically shown evidence of atrial fibrosis in patients with AF.[8] By increasing conduction heterogeneity, fibrosis is related to the development of the AF/AT substrate in some experimental models.[40] The results shown in Table 2, indicate transcriptional up-regulation of connective tissue growth factor (Ctgf) and the Wnt1 inducible signaling pathway protein 1 (Wisp1). In the heart, Ctgf plays a role in interstitial fibrotic remodeling[41] and Wisp1 is a prohypertrophic, profibrotic growth factor.[42] Interestingly, a related transcript, Cyr61, and other genes involved in extracellular matrix remodeling (e.g., Mmp13, Timp3, and Gja5; Online Supplements 2 and 4) were also deregulated in the stimulated HL-1 cells. Taken together, these findings provide evidence for early, myocyte-specific signals in response to rapid stimulation that promote structural remodeling. To further support this concept, it was recently shown that rapidly paced atrial cells release substances into the culture medium that activate cardiac fibroblasts, to promote expression of extracellular matrix genes.[43]
An important advantage of microarray technology is the ability to screen for transcriptional regulation in an unbiased manner. We detected a number of novel, myocyte-specific transcriptional changes that were conserved in vivo. Given that we studied a cardiomyocyte autonomous preparation, deregulation of neurogenesis pathways (Table 1, Online Supplements 2 and 4) that was confirmed by conserved individual genes (Table 2), was an unanticipated finding. As shown in Table 1, there was evidence of global transcriptional regulation of genes involved in neurogenesis, with 20 genes up-regulated and 6 down-regulated. In the canine AT model, nerve sprouting and sympathetic hyperinnervation is significantly increased with rapid atrial stimulation,[7] resulting in heterogeneous changes in atrial sympathetic innervation and ultimately, atrial refractoriness.[44] Our evidence of myocyte-specific up-regulation of genes involved in neurogenesis offers the intriguing possibility that sympathetic hyperinnervation in response to rapid atrial stimulation may be mediated, at least in part, by the atrial myocytes themselves.
Another unexpected result from our study was the concordant up-regulation of the gene encoding disheveled1, Dvl1, with human AF (Figure 1 and Table 2).[45] Disheveled 1 is a critical component of canonical Wnt signaling. Up-regulation of 9 genes encoding other proteins involved in Wnt signaling further supports deregulation of this pathway with rapid atrial stimulation (Online Supplements 3 and 4). In the heart, Wnt signaling has been primarily implicated in development, and suppression of Wnt signaling may play a role in maintaining atrial chamber specificity.[46] It is known that permanent AF causes expression of a genomic signature in the atrium that is very similar to the ventricle. It is possible that deregulation of Wnt signaling plays a role in this ventricularization. Recent studies have also elucidated its participation in remodeling processes that include cardiac hypertrophy.[47,48] Although the role of Wnt signaling in AF/AT has not been investigated, the transcriptional profile presented here implies that this pathway is deregulated during initial atrial remodeling. These results highlight the strength of using microarray analysis to reveal previously unexplored pathways and key molecules.
Potential Limitations
Because we sought to investigate transcriptional regulation in response to rapid stimulation at the level of the atrial myocyte, we chose a myocyte autonomous preparation. While this preparation is appropriate to address our scientific question, the results must be interpreted with caution. A limitation of this model system relates to its cellular nature, with an absence of in vivo parameters that can influence transcription. Nevertheless, we and others have observed remodeling in this rapidly stimulated preparation that resembles AF/AT, including transcriptional concordance. These findings provide further evidence that important components of the remodeling process may be mediated by the myocytes themselves.
In conclusion, our results demonstrate that rapid stimulation of HL-1 atrial cells in culture causes early transcriptional regulation, with many features concordant with in vivo changes observed in AF/AT, as well as conserved, novel findings that merit further investigation. This in vitro preparation provides a complementary experimental paradigm to investigate tachycardia-induced remodeling, and it is uniquely suited to elucidate the cardiomyocyte-specific signals in this process.
Supplementary Material
Acknowledgments
This work was supported by US Public Health Service (HL71002) and a postdoctoral fellowship grant from the American Heart Association, Southeast Affiliate (0625270B). All microarray experiments were performed in the Vanderbilt Microarray Shared Resource (VMSR). The VMSR is supported by the Vanderbilt Ingram Cancer Center (P30 CA68485), the Vanderbilt Diabetes Research and Training Center (P60 DK20593), the Vanderbilt Digestive Disease Center (P30 DK58404) the Genomics of Inflammation Program Project Grant (1 P01 HL6744-01), and the Vanderbilt Vision Center (P30 EY08126). The authors thank Dr. Al George and Jennifer Kunic for advice and guidance with performing quantitative gene expression assays.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Nattel S, Maguy A, Le Bouter S, Yeh YH. Arrhythmogenic Ion-Channel Remodeling in the Heart: Heart Failure, Myocardial Infarction, and Atrial Fibrillation. Physiol Rev. 2007;87(2):425–56. doi: 10.1152/physrev.00014.2006. [DOI] [PubMed] [Google Scholar]
- 2.Brundel BJJM, van Gelder IC, Henning RH, Tieleman RG, Tuinenburg AE, Wietses M, et al. Ion channel remodeling is related to intraoperative atrial effective refractory periods in patients with paroxysmal and persistent atrial fibrillation. Circulation. 2001;103(5):684–90. doi: 10.1161/01.cir.103.5.684. [DOI] [PubMed] [Google Scholar]
- 3.Gaborit N, Steenman M, Lamirault G, Le Meur N, Le Bouter S, Lande G, et al. Human atrial ion channel and transporter subunit gene-expression remodeling associated with valvular heart disease and atrial fibrillation. Circulation. 2005;112(4):471–81. doi: 10.1161/CIRCULATIONAHA.104.506857. [DOI] [PubMed] [Google Scholar]
- 4.Li D, Fareh S, Leung TK, Nattel S. Promotion of atrial fibrillation by heart failure in dogs: Atrial remodeling of a different sort. Circulation. 1999;100(1):87–95. doi: 10.1161/01.cir.100.1.87. [DOI] [PubMed] [Google Scholar]
- 5.Ausma J, Wijffels M, Thone F, Wouters L, Allessie M, Borgers M. Structural changes of atrial myocardium due to sustained atrial fibrillation in the goat. Circulation. 1997;96(9):3157–63. doi: 10.1161/01.cir.96.9.3157. [DOI] [PubMed] [Google Scholar]
- 6.Ausma J, Wijffels M, van Eys GJJM, Koide M, Ramaekers FCS, Allessie M, et al. Dedifferentiation of atrial cardiomyocytes as a result of chronic atrial fibrillation. Am J Pathol. 1997;151:985–97. [PMC free article] [PubMed] [Google Scholar]
- 7.Chang CM, Wu TJ, Zhou S, Doshi RN, Lee MH, Ohara T, et al. Nerve sprouting and sympathetic hyperinnervation in a canine model of atrial fibrillation produced by prolonged right atrial pacing. Circulation. 2001;103(1):22–5. doi: 10.1161/01.cir.103.1.22. [DOI] [PubMed] [Google Scholar]
- 8.Everett TH, IV, Olgin JE. Atrial fibrosis and the mechanisms of atrial fibrillation. Heart Rhythm. 2007;4(3 Supplement 1):S24–S27. doi: 10.1016/j.hrthm.2006.12.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yang Z, Shen W, Rottman JN, Wikswo JP, Murray KT. Rapid stimulation causes electrical remodeling in cultured atrial myocytes. J Mol Cell Cardiol. 2005;38(2):299–308. doi: 10.1016/j.yjmcc.2004.11.015. [DOI] [PubMed] [Google Scholar]
- 10.Brundel BJJM, Kampinga HH, Henning RH. Calpain inhibition prevents pacing-induced cellular remodeling in a HL-1 myocyte model for atrial fibrillation. Cardiovasc Res. 2004;62(3):521–8. doi: 10.1016/j.cardiores.2004.02.007. [DOI] [PubMed] [Google Scholar]
- 11.Brundel BJJM, Henning RH, Ke L, van Gelder IC, Crijns HJGM, Kampinga HH. Heat shock protein upregulation protects against pacing-induced myolysis in HL-1 atrial myocytes and in human atrial fibrillation. J Mol Cell Cardiol. 2006;41(3):555–62. doi: 10.1016/j.yjmcc.2006.06.068. [DOI] [PubMed] [Google Scholar]
- 12.Brundel BJJM, Shiroshita-Takeshita A, Qi X, Yeh YH, Chartier D, van Gelder IC, et al. Induction of heat shock response protects the heart against atrial fibrillation. Circ Res. 2006;99(12):1394–402. doi: 10.1161/01.RES.0000252323.83137.fe. [DOI] [PubMed] [Google Scholar]
- 13.Mi H, Guo N, Kejariwal A, Thomas PD. PANTHER version 6: protein sequence and function evolution data with expanded representation of biological pathways. Nucl Acids Res. 2007;35(Supplement 1):D247–D252. doi: 10.1093/nar/gkl869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Barth AS, Merk S, Arnoldi E, Zwermann L, Kloos P, Gebauer M, et al. Reprogramming of the human atrial transcriptome in permanent atrial fibrillation: Expression of a ventricular-like genomic signature. Circ Res. 2005;96(9):1022–9. doi: 10.1161/01.RES.0000165480.82737.33. [DOI] [PubMed] [Google Scholar]
- 15.Ingwall JS, Weiss RG. Is the Failing Heart Energy Starved?: On Using Chemical Energy to Support Cardiac Function. Circ Res. 2004;95(2):135–45. doi: 10.1161/01.RES.0000137170.41939.d9. [DOI] [PubMed] [Google Scholar]
- 16.Kaab S, Barth A, Margerie D, Dugas M, Gebauer M, Zwermann L, et al. Global gene expression in human myocardium-oligonucleotide microarray analysis of regional diversity and transcriptional regulation in heart failure. J Mol Med. 2004;82(5):308–16. doi: 10.1007/s00109-004-0527-2. [DOI] [PubMed] [Google Scholar]
- 17.Iemitsu M, Maeda S, Miyauchi T, Matsuda M, Tanaka H. Gene expression profiling of exercise-induced cardiac hypertrophy in rats. Acta Physiol Scand. 2005;185(4):259–70. doi: 10.1111/j.1365-201X.2005.01494.x. [DOI] [PubMed] [Google Scholar]
- 18.Bammler T, Beyer R, Bhattacharya S, Boorman G, Boyles A, Bradford B, et al. Standardizing global gene expression analysis between laboratories and across platforms. Nature Methods. 2005;2(5):351–6. doi: 10.1038/nmeth754. [DOI] [PubMed] [Google Scholar]
- 19.Cardin S, Libby E, Pelletier P, Le Bouter S, Shiroshita-Takeshita A, Le Meur N, et al. Contrasting gene expression profiles in two canine models of atrial fibrillation. Circ Res. 2007;100(3):425–33. doi: 10.1161/01.RES.0000258428.09589.1a. [DOI] [PubMed] [Google Scholar]
- 20.Tomaselli GF. Ventricularization of atrial gene expression in the fibrillating heart? Circ Res. 2005;96(9):923–4. doi: 10.1161/01.RES.0000168041.46134.8e. [DOI] [PubMed] [Google Scholar]
- 21.Kurosaki K, Tada H, Hashimoto T, Ito S, Miyaji K, Naito S, et al. Plasma natriuretic peptide concentrations as a predictor for successful catheter ablation in patients with drug-refractory atrial fibrillation. Circ J. 2007;71(3):313–20. doi: 10.1253/circj.71.313. [DOI] [PubMed] [Google Scholar]
- 22.Wozakowska-Kaplon B, Opolski G. Effects of sinus rhythm restoration in patients with persistent atrial fibrillation: a clinical, echocardiographic and hormonal study. Int J Cardiol. 2004;96(2):171–6. doi: 10.1016/j.ijcard.2003.05.033. [DOI] [PubMed] [Google Scholar]
- 23.Lai LP, Lin JL, Lin C-S, Yeh H-M, Tsay Y-G, Lee C-F, et al. Functional genomic study on atrial fibrillation using cDNA microarray and two-dimensional protein electrophoresis techniques and identification of the myosin regulatory light chain isoform reprogramming in atrial fibrillation. J Cardiovasc Electrophysiol. 2004;15(2):214–23. doi: 10.1046/j.1540-8167.2004.03423.x. [DOI] [PubMed] [Google Scholar]
- 24.Schafler AE, Kirmanoglou K, Pecher P, Hannekum A, Schumacher B. Overexpression of heat shock protein 60/10 in myocardium of patients with chronic atrial fibrillation. Ann Thorac Surg. 2002;74(3):767–70. doi: 10.1016/s0003-4975(02)03830-4. [DOI] [PubMed] [Google Scholar]
- 25.Beere HM. Death versus survival: functional interaction between the apoptotic and stress-inducible heat shock protein pathways. J Clin Invest. 2005;115(10):2633–9. doi: 10.1172/JCI26471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gill C, Mestril R, Samali A. Losing heart: The role of apoptosis in heart disease-a novel therapeutic target? FASEB J. 2002;16(2):135–46. doi: 10.1096/fj.01-0629com. [DOI] [PubMed] [Google Scholar]
- 27.Janicke RU, Sprengart ML, Wati MR, Porter AG. Caspase-3 is required for DNA fragmentation and morphological changes associated with apoptosis. J Biol Chem. 1998;273(16):9357–60. doi: 10.1074/jbc.273.16.9357. [DOI] [PubMed] [Google Scholar]
- 28.Ruwhof C, van der Laarse A. Mechanical stress-induced cardiac hypertrophy: Mechanisms and signal transduction pathways. Cardiovasc Res. 2000;47(1):23–37. doi: 10.1016/s0008-6363(00)00076-6. [DOI] [PubMed] [Google Scholar]
- 29.Luo J, McMullen JR, Sobkiw CL, Zhang L, Dorfman AL, Sherwood MC, et al. Class IA phosphinositide 3-kinase regulates heart size and physiological cardiac hypertrophy. Mol Cell Biol. 2005;25(21):9491–502. doi: 10.1128/MCB.25.21.9491-9502.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tsang A, Hausenloy DJ, Mocanu MM, Yellon DM. Postconditioning: A form of “modified reperfusion” protects the myocardium by activating the phosphatidylinositol 3-kinase-Akt pathway. Circ Res. 2004;95(3):230–2. doi: 10.1161/01.RES.0000138303.76488.fe. [DOI] [PubMed] [Google Scholar]
- 31.Nishida K, Yamaguchi O, Hirotani S, Hikoso S, Higuchi Y, Watanabe T, et al. p38alpha mitogen-activated protein kinase plays a critical role in cardiomyocyte survival but not in cardiac hypertrophic growth in response to pressure overload. Mol Cell Biol. 2004;24(24):10611–20. doi: 10.1128/MCB.24.24.10611-10620.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mazzocco M, Maffei M, Egeo A, Vergano A, Arrigo P, Di Lisi R, et al. The identification of a novel human homologue of the SH3 binding glutamic acid-rich (SH3BGR) gene establishes a new family of highly conserved small proteins related to Thioredoxin Superfamily. Gene. 2002;291(1–2):233–9. doi: 10.1016/s0378-1119(02)00602-9. [DOI] [PubMed] [Google Scholar]
- 33.Li D, Shinagawa K, Pang L, Leung TK, Cardin S, Wang Z, et al. Effects of angiotensin-converting enzyme inhibition on the development of the atrial fibrillation substrate in dogs with ventricular tachypacing-induced congestive heart failure. Circulation. 2001;104(21):2608–14. doi: 10.1161/hc4601.099402. [DOI] [PubMed] [Google Scholar]
- 34.Mihm MJ, Yu F, Carnes CA, Reiser PJ, McCarthy PM, Van Wagoner DR, et al. Impaired myofibrillar energetics and oxidative injury during human atrial fibrillation. Circulation. 2001;104(2):174–80. doi: 10.1161/01.cir.104.2.174. [DOI] [PubMed] [Google Scholar]
- 35.Thijssen VL, Ausma J, Borgers M. Structural remodelling during chronic atrial fibrillation: act of programmed cell survival. Cardiovasc Res. 2001;52(1):14–24. doi: 10.1016/s0008-6363(01)00367-4. [DOI] [PubMed] [Google Scholar]
- 36.Stanley WC, Recchia FA, Lopaschuk GD. Myocardial substrate metabolism in the normal and failing heart. Physiol Rev. 2005;85(3):1093–129. doi: 10.1152/physrev.00006.2004. [DOI] [PubMed] [Google Scholar]
- 37.Qi XY, Yeh YH, Xiao L, Burstein B, Maguy A, Chartier D, et al. Cellular Signaling Underlying Atrial Tachycardia Remodeling of L-type Calcium Current. Circ Res. 2008;103(8):845–54. doi: 10.1161/CIRCRESAHA.108.175463. [DOI] [PubMed] [Google Scholar]
- 38.Beharier O, Etzion Y, Katz A, Friedman H, Tenbosh N, Zacharish S, et al. Crosstalk between L-type calcium channels and ZnT-1, a new player in rate-dependent cardiac electrical remodeling. Cell Calcium. 2007;42(1):71–82. doi: 10.1016/j.ceca.2006.11.007. [DOI] [PubMed] [Google Scholar]
- 39.Etzion Y, Ganiel A, Beharier O, Shalev A, Novack V, Volvich L, et al. Correlation between atrial ZnT-1 expression and atrial fibrillation in humans: A pilot study. J Cardiovasc Electrophysiol. 2007;19(2):157–64. doi: 10.1111/j.1540-8167.2007.01008.x. [DOI] [PubMed] [Google Scholar]
- 40.Cha TJ, Ehrlich JR, Zhang L, Shi YF, Tardif JC, Leung TK, et al. Dissociation between ionic remodeling and ability to sustain atrial fibrillation during recovery from experimental congestive heart failure. Circulation. 2004;109(3):412–8. doi: 10.1161/01.CIR.0000109501.47603.0C. [DOI] [PubMed] [Google Scholar]
- 41.Gabrielsen A, Lawler PR, Yongzhong W, Steinbruchel D, Blagoja D, Paulsson-Berne G, et al. Gene expression signals involved in ischemic injury, extracellular matrix composition and fibrosis defined by global mRNA profiling of the human left ventricular myocardium. J Mol Cell Cardiol. 2007;42(4):870–83. doi: 10.1016/j.yjmcc.2006.12.016. [DOI] [PubMed] [Google Scholar]
- 42.Colston JT, de la Rosa SD, Koehler M, Gonzales K, Mestril R, Freeman GL, et al. Wnt-induced secreted protein-1 is a prohypertrophic and profibrotic growth factor. Am J Physiol Heart Circ Physiol. 2007;293(3):H1839–H1846. doi: 10.1152/ajpheart.00428.2007. [DOI] [PubMed] [Google Scholar]
- 43.Burstein B, Qi XY, Yeh YH, Calderone A, Nattel S. Atrial cardiomyocyte tachycardia alters cardiac fibroblast function: A novel consideration in atrial remodeling. Cardiovasc Res. 2007;76(3):442–52. doi: 10.1016/j.cardiores.2007.07.013. [DOI] [PubMed] [Google Scholar]
- 44.Jayachandran JV, Sih HJ, Winkle W, Zipes DP, Hutchins GD, Olgin JE. Atrial fibrillation produced by prolonged rapid atrial pacing is associated with heterogeneous changes in atrial sympathetic innervation. Circulation. 2000;101(10):1185–91. doi: 10.1161/01.cir.101.10.1185. [DOI] [PubMed] [Google Scholar]
- 45.Kim NH, Ahn Y, Oh SK, Cho JK, Park HW, Kim YS, et al. Altered Patterns of Gene Expression in Response to Chronic Atrial Fibrillation. Int Heart J. 2005;46(3):383–95. doi: 10.1536/ihj.46.383. [DOI] [PubMed] [Google Scholar]
- 46.Tabibiazar R, Wagner RA, Liao A, Quertermous T. Transcriptional profiling of the heart reveals chamber-specific gene expression patterns. Circ Res. 2003;93:1193–201. doi: 10.1161/01.RES.0000103171.42654.DD. [DOI] [PubMed] [Google Scholar]
- 47.Brade T, Manner J, Kuhl M. The role of Wnt signalling in cardiac development and tissue remodelling in the mature heart. Cardiovasc Res. 2006;72(2):198–209. doi: 10.1016/j.cardiores.2006.06.025. [DOI] [PubMed] [Google Scholar]
- 48.MacRae CA, Birchmeier W, Thierfelder L. Arrhythmogenic right ventricular cardiomyopathy: moving toward mechanism. J Clin Invest. 2006;116(7):1825–8. doi: 10.1172/JCI29174. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.