Abstract
Nucleotide substitutions are found in recombined Ig switch (S) regions and also in unrecombined (germline, GL) Sµ segments in activated splenic B cells. Herein we examine whether mutations are also introduced into the downstream acceptor S regions prior to switch recombination, but find very few mutations in GL Sγ3 and Sγ1 regions in activated B cells. These data suggest that switch recombination initiates in the Sµ segment and secondarily involves the downstream acceptor S region. Furthermore, the pattern and specificity of mutations in GL and recombined Sµ segments differ, suggesting different repair mechanisms. Mutations in recombined Sµ regions show a strong bias toward G/C base pairs and WRCY/RGYW hotspots, whereas mutations introduced into the GL Sµ do not. Additionally, induction conditions affect mutation specificity within the GL Sµ segment. Mutations are most frequent near the S–S junctions and decrease rapidly with distance from the junction. Finally, we find that mice expressing a transgene for terminal deoxynucleotidyl transferase (TdT) have nucleotide insertions at S–S junctions, indicating that the recombining DNA ends are accessible to end-processing enzyme activities.
Keywords: class switch recombination/mismatch repair/Msh2/mutations/TdT
Introduction
Antibody class switching occurs in B cells after activation by antigen. Switching results in a change from IgM and IgD expression to IgG, IgE or IgA expression, thereby diversifying antibody effector functions, while maintaining the identical antigen specificity. Class switching occurs by an intrachromosomal deletional recombination within tandemly repeated switch (S) region sequences located upstream of each Ig heavy chain constant region gene. Recombination seems to occur anywhere within each S region segment, which differ in sequence from each other and also vary in length from 2 to 10 kb.
Frequent nucleotide mutations (substitutions, insertions and deletions), reminiscent of those found in hypermutated Ig variable regions, occur near S recombination junctions (Dunnick et al., 1993). By sequencing several products from one class switch recombination (CSR) event in a B-cell line (I.29µ) and also the donor and acceptor Sµ and Sα regions from IgM+ cells of that cell line, it was determined that the mutations were introduced during CSR rather than during subsequent cell divisions after CSR (Dunnick et al., 1989; Dunnick and Stavnezer, 1990). Since the products of one switch event were analyzed, it could not be determined whether mutations were also introduced prior to CSR. Several investigators subsequently have observed mutations in segments near S junctions in normal mouse and human B cells (Du et al., 1997; Nagaoka et al., 2002; Schrader et al., 2002, 2003; Pan-Hammarstrom et al., 2003).
Although the mechanism of CSR is unclear, it has been shown that activation-induced cytidine deaminase (AID) is required for CSR and for somatic hypermutation (SHM) of antibody variable region genes (Muramatsu et al., 1999, 2000; Revy et al., 2000). Recent data suggest that AID may directly deaminate genomic DNA and that enzymes of the base excision repair (BER) pathway convert the resulting dU residues to DNA breaks that initiate CSR (Di Noia and Neuberger, 2002; Petersen-Mahrt et al., 2002; Rada et al., 2002; Bransteitter et al., 2003; Chaudhuri et al., 2003). This would involve excision of the dU base by uracil DNA glycosylase, followed by creation of a single-strand nick at the abasic site by AP endonuclease (Lindahl, 2000). Consistent with this model, mice that are deficient in the uracil DNA glycosylase UNG have greatly impaired CSR (Rada et al., 2002; reviewed in Storb and Stavnezer, 2002).
In addition to DNA breaks, the AID–BER pathway could lead to mutations in S regions. If replication occurs prior to DNA break formation, mutations would be generated at the dU residues (Petersen-Mahrt et al., 2002; Storb and Stavnezer, 2002). A dU residue could serve as a template for incorporation of an opposing dA residue, resulting in a transition mutation. If, instead, DNA polymerase encounters an abasic site, it will pause and a translesion error-prone polymerase, e.g. Pol ι, η or ζ, will replace the high fidelity polymerase and insert any nucleotide opposite the abasic site and sometimes in adjacent regions (Haracska et al., 2001; Storb and Stavnezer, 2002; McDonald et al., 2003). It is also possible that multiple DNA breaks could lead to deletions within the S region, without resulting in CSR (Dudley et al., 2002).
To obtain evidence for DNA lesions within S regions undergoing CSR, two groups recently have examined the nucleotide sequences of the unrearranged (germline, GL) Sµ alleles in splenic B cells that have been treated with inducers of IgG1 CSR: lipopolysaccharide (LPS) and interleukin-4 (IL-4) (Petersen et al., 2001; Nagaoka et al., 2002). Although many of the Sµ segments in these cells have recombined with Sγ1 segments, a significant portion have not. Interestingly, the investigators found nucleotide substitutions in the GL Sµ regions in activated B cells, but not in the Sµ regions of resting cells. As predicted by the deamination model for AID function, the mutations were not detected in AID-deficient B cells, although these cells proliferated normally in response to the activators. These data suggest that AID, which is induced by LPS + IL-4 (Muramatsu et al., 1999), deaminates dC residues, leading to mutations in the Sµ region in activated B cells even in cells that do not complete the process of CSR.
The mismatch repair (MMR) proteins Msh2, Mlh1 and Pms2 each contribute to CSR, although the mechanism of their action is not clear (Ehrenstein and Neuberger, 1999; Ehrenstein et al., 2001; Schrader et al., 1999, 2002, 2003). Splenic B cells from mice deficient in Msh2, Mlh1 and Pms2 show 2- to 7-fold reductions in switching, and also show altered switch recombination junctions. The available data suggest that Msh2 may be involved in processing of the DNA ends prior to their recombination (Ehrenstein and Neuberger, 1999; Schrader et al., 2002). To explore the end-processing hypothesis further, we now examine whether the location of mutations surrounding S–S junctions differs between wild-type and msh2–/– B cells. In addition, we also examine whether Msh2 affects the sequence specificity of S region mutations, as it does during the process of SHM of Ig variable region segments (Rada et al., 1998)
In order to learn more about the mechanism of CSR, we have investigated the frequency, distribution and nucleotide specificity of mutations introduced into Ig S regions during CSR and obtained several unanticipated results. First, we have observed that although mutations are introduced into the GL Sµ segments in splenic B cells treated with inducers of CSR, they are absent or rare in the GL Sγ3 and Sγ1 acceptor regions in these same cell populations. These data suggest that switching initiates asymmetrically in the Sµ segment. Furthermore, although the frequencies of mutations in the GL Sµ and recombined Sµ segments are similar, the sequence specificity of the mutations in these segments differs, suggesting that although AID most probably initiates the process (Petersen et al., 2001; Nagaoka et al., 2002), the repair mechanisms that determine the final pattern of mutations differ. Additionally, the distribution of mutations in the GL Sµ and recombined Sµ–Sγ3 segments differs. Although mutations appear to extend from the 5′ to 3′ ends of the GL Sµ segments, nearly all mutations in recombined Sµ–Sγ3 junctions are within 150 bp of the junction. Junctions from msh2–/– B cells have an even greater clustering of mutations near the S–S junctions. Finally, we provide evidence that the DNA ends formed during CSR are accessible to DNA-modifying enzymes by demonstrating that they are accessible to terminal deoxynucleotidyl transferase (TdT).
Results
Nucleotide substitutions are found in the GL Sµ segment in activated wild-type and Msh2-deficient B cells, but rarely in GL Sγ3 or Sγ1 segments
To determine the nucleotide sequences of GL Sµ segments in activated normal B cells, T-depleted spleen cells were activated to switch to IgG3 by treatment for 4 days with LPS ± anti-δ dextran. No difference in mutation frequency or sequence specificity was obtained by induction with LPS or with LPS + anti-δ dextran (not shown). DNA was isolated and S regions were amplified and cloned, using primers complementary to sequences at the 5′ and 3′ ends of both the Sµ and Sγ3 segments. These primers do not amplify the switch regions that have undergone S–S recombination. Figure 1 presents a schematic of the primer positions, and their locations are described further in Materials and methods. The GL 5′Sµ segment sequenced has the identical 5′ end to the segment analyzed by Petersen et al. (2001) and is closely located to the segment analyzed by Nagaoka et al. (2002).
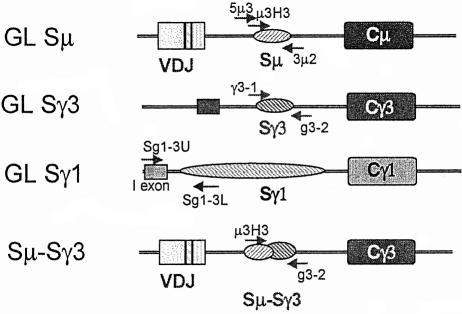
Fig. 1. Diagrams (not to scale) of the DNA segments analyzed in this study. The arrows indicate the positions of the primers used for amplification of the segments analyzed by nucleotide sequencing. See Materials and methods for the precise locations of the primers and the segments sequenced. The primer names are indicated above or below the arrows. To amplify recombined Sµ–Sγ3 segments, primers located at the 5′ end of Sµ and 3′ end of Sγ3 (µ3H3 and g3-2) were used. The 5′ primer used for GL Sµ amplification (5µ3) was located 640 nucleotides 5′ to the primer used to amplify Sµ–Sγ3 junctions. The more 5′ location of the former primer allowed us to compare our data with previously published data (Petersen et al., 2001; Nagaoka et al., 2002). The use of this primer also increased the length of the GL Sµ segment that could be sequenced from the 5′ end prior to reaching the region of Sµ that has tandemly repeated pentamers and frequently undergoes deletions. Both the 5′ and 3′ ends of the GL Sγ3 segments were sequenced to determine their mutation frequency. Both ends gave similar results, and the average is shown in Table I. Since the Sγ1 segment is too large to be amplified in its entirety, primers were designed to amplify a 2 kb segment located near the 5′ end of the Sγ1 segment, which would be deleted in cells having undergone Sµ–Sγ1 recombination at any of the sites that have been reported (Dunnick et al., 1993; W.Dunnick, personal communication). The mutation frequency was determined by sequencing the 3′ end of such fragments, which is located downstream of the Iγ1 exon, near the 5′ end of the tandem repeats. Nested primers were not used for any of the amplifications.
Nucleotide substitutions were detected in the GL 5′Sµ segments from splenic B cells induced to undergo CSR (Table I, row 1), similar to previous reports (Petersen et al., 2001; Nagaoka et al., 2002). Surprisingly, we found that the mutation frequency of the GL 5′Sµ segment is as high as in the recombined Sµ segments from Sµ–Sγ3 recombination junctions amplified from the same cell populations (Table I, rows 1 and 2). Even more surprising was the finding of very few mutations in the GL Sγ3 segments from the activated splenic B cells (row 3). In fact, the mutation frequency of the GL Sγ3 segment was no higher than the PCR error frequency in our experiments (row 12). About 20% of B cells undergo switching in these conditions (Schrader et al., 1999; and data not shown) and, although all switching involves the Sµ segment, only half of this switching is to IgG3, as switching to IgG2b also occurs. Thus, it was possible that the lack of mutations might simply be because a lower proportion of Sγ3 segments than Sµ segments are acted upon by AID. To examine this possibility, we determined the nucleotide sequences of the GL Sγ1 segment in B cells induced to switch to IgG1 with LPS + IL-4. Under these conditions, 30–40% of B cells switch and 90% of the switching is from IgM to IgG1 (Schrader et al., 1999; and data not shown). We found that, like the GL Sγ3 segment, the GL Sγ1 segment also has a very low mutation frequency (Table I, row 7), not significantly different from PCR error frequency. The fact that there are mutations in GL 5′Sµ segments in activated B cells, while the GL Sγ3 and Sγ1 segments are not or are rarely mutated, suggests that switch recombination initiates asymmetrically. CSR may initiate in the Sµ region and only subsequently involve the downstream S region.
Table I. Mutation frequency in unrecombined and recombined switch region segments from splenic B cells induced to undergo CSR in culture for 4 days.
| Mutation frequency (×10–4) | Total nucleotides sequenced | No. of mutations | P-valuea | |
|---|---|---|---|---|
| Wild-type B cells | ||||
| LPS + anti-δ-dextran treated | ||||
| 1. GL 5′Sµ | 28.0 | 11423 | 32 | <0.001 |
| 2. Recombined Sµ | 22.7 | 9711 | 22 | 0.003 |
| 3. GL Sγ3 (5′ + 3′) | 8.0 | 16245 | 13 | 1.0 |
| 4. Recombined Sγ3 | 18.9 | 9011 | 17 | 0.027 |
| 5. GL Sγ3 from ex vivo cells (no stimulation) | 7.4 | 20250 | 15 | 1.0 |
| LPS + IL-4 treated: | ||||
| 6. GL Sµ | 21.8 | 14232 | 31 | 0.002 |
| 7. GL Sγ1 |
10.9 |
17442 |
19 |
0.443 |
|
msh2–/– B cells | ||||
| LPS + anti-δ-dextran treated | ||||
| 8. GL 5′Sµ | 24.7 | 9713 | 24 | 0.001 |
| 9. Recombined Sµ | 31.6 | 6019 | 19 | <0.001 |
| 10. GL Sγ3 (5′ + 3′) | 8.4 | 11871 | 10 | 0.822 |
| 11. Recombined Sγ3 | 32.9 | 6074 | 20 | <0.001 |
| 12. PCR error frequencyb | 7.4 | 12182 | 9 | |
aSignificance of difference from PCR error frequency; Fisher’s exact t-test.
bDetermined from eight independent PCR amplifications of a recombined Sµ–Sγ3 segment from the Sµ segment in the TIB114 myeloma cell line (see Materials and methods).
We have shown recently that recombined Sµ–Sγ3 junctions from Msh2-deficient B cells have a higher mutation frequency than junctions from wild-type cells (Schrader et al., 2003) (also Table I, compare rows 2 and 4 with rows 9 and 11), suggesting that Msh2 is involved in repairing mutations introduced during CSR by error-prone repair. To learn more about the mechanism of introduction of mutations into GL Sµ segments, we examined whether Msh2 also has a role in reducing mutations in GL S regions in B cells stimulated with LPS or with LPS + anti-δ dextran, as above. Similar to the results with wild-type B cells, the GL Sγ3 segments were unmutated (row 10), whereas the GL 5′Sµ segments were mutated (row 8). The frequency of mutation of the GL 5′Sµ segment in Msh2-deficient cells was similar to the mutation frequency of the wild-type GL 5′Sµ segment, suggesting that Msh2 is not involved in repairing mutations that accumulate in the GL 5′Sµ segment.
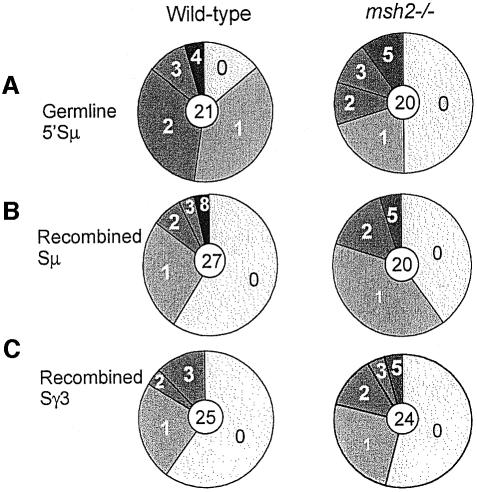
Mutations are found in a large fraction of the GL 5′Sµ and recombined S regions, rather than being restricted to rare subsets. This is demonstrated by a series of pie charts, in which the number of sequences analyzed is indicated by the number in the center, and the proportion of sequences with the indicated numbers of mutations is indicated by the size of the slices (Figure 2). Additionally, the portion of recombined Sµ segments that have mutations is not greater than that of the GL 5′Sµ segments, consistent with the finding that the frequency of mutations in recombined Sµ segments is not greater than in the GL 5′Sµ segments.
Fig. 2. Pie charts indicate that the mutations are quite widely distributed. The fraction of S segments that have the indicated number of mutations are indicated by the size of the pie slice. (A) GL 5′Sµ segments from wild-type and msh2–/– cells. (B) Recombined Sµ segments. (C) Recombined Sγ3 segments. The average lengths of the GL 5′Sµ segments analyzed were 544 for wild-type and 486 for msh2–/–; recombined Sµ segments, 365 for wild-type and 301 for msh2–/–; and recombined Sγ3 segments, 360 for wild-type and 253 for msh2–/–. Only Sµ and Sγ3 segments sequenced in their entirety are included.
Mutations in recombined S regions show a preference for G/C base pairs and a striking preference for WRCY/RGYW hotspots
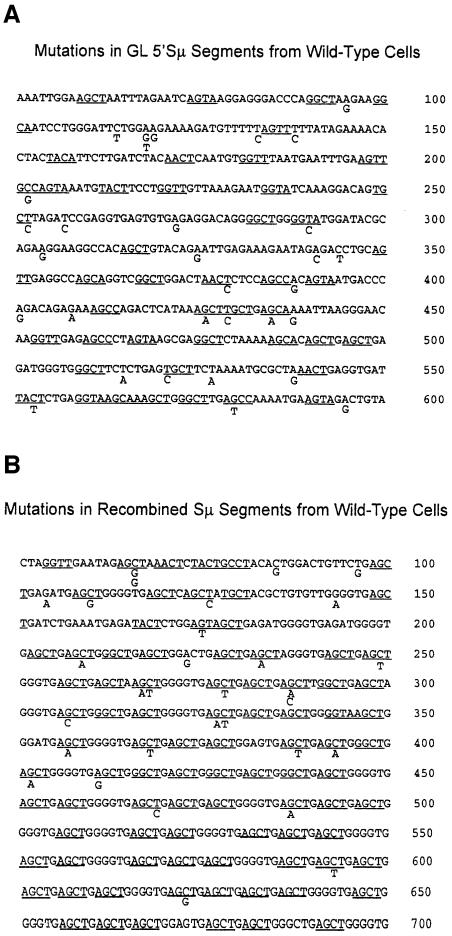
To learn more about the mechanism of introduction of mutations into S regions, we analyzed their nucleotide specificity. Mutations introduced by AID activity should target G/C base pairs. In addition, evidence suggests that the known SHM hotspot, WRCY, and its complement, RGYW, are favored AID targets (Bransteitter et al., 2003; Pham et al., 2003), although this conclusion is controversial (Dickerson et al., 2003; Sohail et al., 2003). Nonetheless, AGCT, the major motif of the Sµ tandem repeats, is the hottest of the mutation hotspots (Milstein et al., 1998). Figure 3A presents the mutations observed in the wild-type GL 5′Sµ segments, and Figure 3B the mutations in recombined Sµ segments in wild-type cells. The WRCY/RGYW hotspots are underlined. Analysis of these data for wild-type and msh2–/– cells is shown in Table II. Over 80% of the mutations in recombined Sµ segments and ≥75% in recombined Sγ3 segments occur preferentially at G/C base pairs from both wild-type and msh2–/– cells. As the percentage of G/C base pairs in these segments is ∼60%, there is a preference (1.2- to 1.4-fold) for mutating G/C base pairs relative to A/T base pairs (see Table II). The mutation frequency at WRCY and RGYW in recombined Sµ segments showed a striking and highly significant preference for mutation at hotspots in recombined Sµ segments, although there was no such bias in recombined Sγ3 segments from wild-type cells. However, in msh2–/– cells, the mutations were significantly targeted to hotspots. In fact, both G/C base pair and hotspot targeting within Sγ3 were increased in Msh2-deficient cells, suggesting that Msh2 may preferentially repair mutations introduced by AID at G/C base pairs in hotspots. Although such an increase was not observed in recombined Sµ regions, the level of targeting was already very high in wild-type cells. All of the sequences analyzed in Table II are shown in Supplementary figures S1–S9 available at The EMBO Journal Online. These data appear consistent with the hypothesis that AID initiates CSR by targeting G/C base pairs within WRCY/RGYW motifs.
Fig. 3. Nucleotide sequences and the mutations observed in (A) GL Sµ segments and (B) recombined Sµ segments from activated wild-type splenic B cells. Somatic hypermutation hotspots RCYW/RGYW are underlined. There were no mutations in the first 50 bp of each sequence, so these sequences are not shown. An additional mutation is located in the GL Sµ segment at nucleotide 672, which is not shown here but is shown in Supplementary figure S1 which presents all of the GL Sµ sequences. Likewise, an additional mutation is located in the recombined Sµ segment at nucleotide 702, which is shown in Supplementary figure S4.
Table II. Sequence specificity of mutations in Sµ and Sγ3 segments from activated splenic B cellsa.
| Recombined Sµb | P-valuec | Recombined Sγ3b | P-value | GL 5′ Sµd | P-value | |
|---|---|---|---|---|---|---|
| Percentage mutations at G/C nucleotides | ||||||
| Wild-type | 84% (32 mutations)e | 0.045 | 75% (24 mutations) | 0.205 | 25% (32 mutations) | 0.045 |
| msh2–/– | 82% (22 mutations) | 0.056 | 87% (23 mutations) | 0.015 | 12% (25 mutations) | 0.002 |
| TdT transgenic | 79% (24 mutations) | 0.056 | 63% (19 mutations) | 1.0 | 21% (52 mutations) | 0.002 |
| Sequence | 59% G/C | 61% G/C | 43% G/C | |||
| Percentage mutations at (RGYW/WRCY) hotspots | ||||||
| Wild-type | 84% (32 mutations) | 0.009 | 50% (24 mutations) | 0.647 | 28% (32 mutations) | 0.358 |
| msh2–/– | 86% (22 mutations) | 0.002 | 83% (23 mutations) | <0.001 | 32% (25 mutations) | 0.834 |
| TdT transgenic | 88% (24 mutations) | 0.018 | 68% (19 mutations) | 0.102 | 24% (52 mutations) | 0.053 |
| Sequence | 54% hotspots | 43% hotspots | 35% hotspots | |||
aActivated 4 days with LPS or with LPS + anti-δ dextran.
bSegments amplified using the µ3H3 and g3-2 primers.
cDifference from random by Fisher’s exact test.
dSegments amplified using the 5µ3 and 3µ2 primers.
eThe number of nucleotides analyzed in Table II for recombined segments is larger than in Table I, because mutation frequency shown in Table I is determined only from Sµ or Sγ3 segments which were sequenced in their entirety.
The sequences analyzed here are shown in the Supplementary figures.
The sequence specificity of the mutations in GL Sµ segments differs from recombined Sµ segments
We also analyzed the G/C and hotspot preferences of mutations in GL 5′Sµ segments. Unlike the mutations in recombined Sµ–Sγ3 segments, mutations in the GL 5′Sµ segment in both wild-type and msh2–/– cells tended to avoid G/C base pairs and showed no preference for hotspots (Table II; Figure 3A; Supplementary figures S1 and S2). The 5′Sµ segment sequenced in this study was located 640 nucleotides upstream of the segment sequenced for the recombined Sµ sequences (Figure 1 legend). Although this segment still contains abundant hotspot pentamers, they are not as dense as within the region sequenced in the recombined Sµ segments, and the percentage of G/C base pairs is also lower (see Table II). Nonetheless, it is clear that mutations in the GL 5′Sµ segment in B cells activated with LPS ± anti-δ dextran occur preferentially at A/T base pairs and show no preference for WRCY/RGYW hotspots. The difference between mutation specificity in recombined Sµ–Sγ3 segments and GL Sµ segments was also found in segments cloned from mice transgenic for the TdT gene (Table II; Supplementary figure S3; see below). Similar results were obtained in an analysis of 18 mutations at the 3′ end of the GL Sµ segments from wild-type B cells. These mutations also disfavored hotspots (22% at hotspots, whereas the sequence contains 36% hotspots) and showed no preference for G/C base pairs (28% at G/C, whereas the sequence is 29% G/C). These data suggest that some aspect of the error-prone repair mechanism that resolves DNA lesions/breaks occurring during successful CSR differs from the mechanism involved in repairing Sµ lesions that do not lead to productive CSR.
Mutation specificity in the GL 5′Sµ segment is altered by different B-cell activation conditions
Two other groups have also examined the nucleotide specificity of mutations within the identical GL 5′Sµ segment in wild-type B cells induced to switch with LPS + IL-4, rather than the LPS ± anti-δ dextran we used here (Petersen et al., 2001; Nagaoka et al., 2002). They both found that the mutations occurred preferentially at G/C base pairs (58–71%) and also preferentially targeted WRCY/RGYW hotspots (60–63%). Although their G/C and hotspot mutation specificity is less than we found for recombined Sµ segments, it is considerably higher than we found for the same GL 5′Sµ segment. To determine if this difference might be due to the different methods used to activate the splenic B cells, we amplified and sequenced the identical GL 5′Sµ segments from cells treated with LPS + IL-4 for 4 days. Interestingly, we found that under these conditions, the mutation specificity was similar to the findings of these two groups (Table III; Supplementary figure S10). These data indicate that LPS + IL-4 treatment results in a mutation spectrum different from LPS ± anti-δ dextran. Both the G/C and hotspot targeting differ significantly from the spectra found in sequences from cells treated with LPS ± anti-δ dextran (P = 0.021 and 0.011, respectively).
Table III. Sequence specificity of mutations in Sµ and Sγ3 segments from wild-type splenic B cells activated with LPS + IL-4.
| GL 5′ Sµa | P-valueb |
|---|---|
| Percentage mutations at G/C nucleotides | |
| 55% (31 mutations)c | 0.138 |
| 43% G/C content | |
| Percentage mutations at hotspots | |
| 61% (31 mutations) | 0.004 |
| 35% hotspots in sequence | |
aSegments amplified using the 5µ3 and 3µ2 primers.
bDifference from random by Fisher’s exact test.
cSequences shown in Supplementary figure S10.
Mutation frequency decreases with nucleotide distance from the Sµ–Sγ3 junction
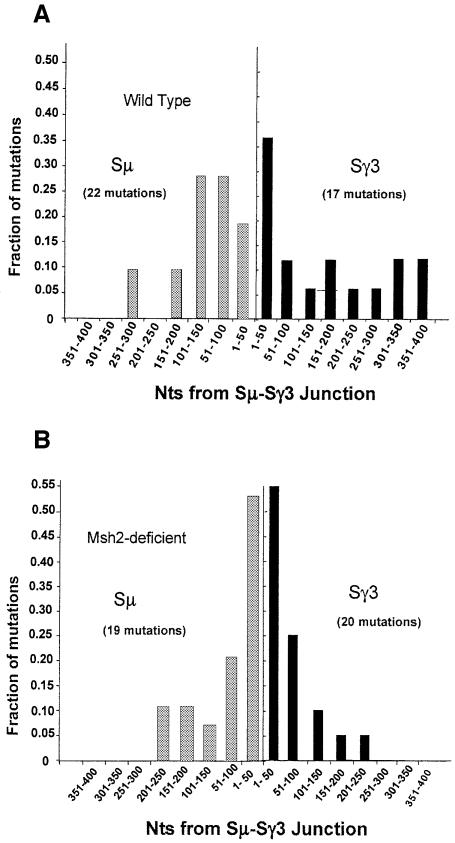
To obtain information about the size of the AID-targeted region and/or the extent of end processing during CSR, we examined the distance that mutations extend from the S–S junctions. Due to the presence of lesions introduced by AID, the repair synthesis involved in end processing is likely to be error prone. Although mutations have been shown to extend at least 200 bp from the junctions (Dunnick et al., 1993), the extent of the mutated region has not been studied systematically. The finding that GL Sµ segments have mutations near their 5′ ends (Figure 3A) and 3′ ends (data not shown) suggests that mutations are initially introduced over the entire Sµ segment. Figure 4A shows the fraction of mutations found relative to the distance in base pairs from the Sµ–Sγ3 junctions in wild-type B cells. The fraction of mutations per 50 bp segment is plotted. Mutation frequency is highest near the junctions on both the Sµ and Sγ3 sides and most mutations are within 150 bp of the junction. Thus, although mutations in GL Sµ segments extend considerably 5′ to the Sµ segments sequenced in the recombined Sµ–Sγ3 junctions, mutations in the Sµ–Sγ3 recombinant molecules are found to be focused near the S–S junctions. In agreement with the difference in nucleotide sequence specificity, these data are consistent with the hypothesis that the repair mechanisms leading to mutations in recombined Sµ–Sγ3 segments differ in some aspect from those leading to mutations in GL Sµ segments.
Fig 4. Mutations are most frequent near the Sµ–Sγ3 junctions from (A) wild-type and (B) msh2–/– B cells. The proportion of the mutations in each 50 bp segment is plotted. Only segments in which the entire sequence was available are analyzed here.
Although the role of Msh2 in CSR is unknown, the available data suggest that it may be involved in processing of DNA ends prior to their joining (Ehrenstein and Neuberger, 1999; Schrader et al., 2002, 2003; Min et al., 2003), similar to its role in double-strand break repair in yeast (Paques and Haber, 1997). Sµ–Sγ3 junctions in Msh2-deficient B cells show an increase in the occurrence of inserted nucleotides at the junction that do not appear to originate from the Sµ or Sγ3 parental sequences (Schrader et al., 2002). These results may be explained by problems in forming neat end-to-end junctions due to improper end processing without Msh2. If end processing is reduced during CSR in Msh2-deficient cells, one might expect mutations to be specifically increased near the S–S junctions. In fact, that is what we found. Figure 4B shows that ∼50% of the mutations found in msh2–/– S–S junctions are located within 50 bp of the junction, whereas only 26% are located this close to wild-type junctions. These data further support the hypothesis that Msh2 is important for end processing during CSR.
DNA breaks at Sµ–Sγ3 junctions are accessible to nucleotide addition by TdT
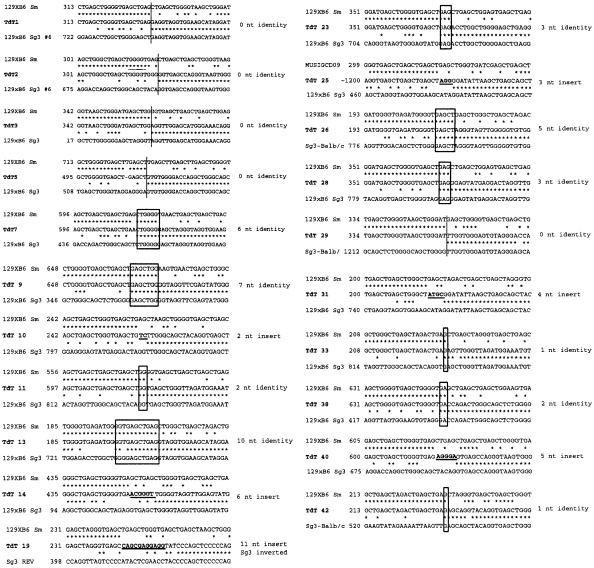
Although AID and BER should initially produce single-strand DNA breaks in S regions, blunt double-strand breaks have been detected by linker-mediated PCR analysis in cells undergoing CSR (Wuerffel et al., 1997). Blunt double-strand breaks might be produced by end processing of staggered breaks by endonuclease, exonuclease and/or by fill-in DNA synthesis. If the DNA ends undergoing CSR are indeed processed prior to recombination, they should be accessible to enzymes with end-processing activities. We tested this hypothesis by asking whether the DNA ends that form S–S junctions can be modified by the enzyme TdT. Splenic B cells from mice bearing a ubiquitously expressed TdT transgene (Marshall et al., 1998) were induced to switch to IgG3 with LPS + anti-δ dextran. The B cells could be induced to switch in culture to levels similar to wild-type cells (data not shown). On day 4 of culture, genomic DNA was isolated, Sµ–Sγ3 junctions were cloned and their sequences determined (Figure 5). Thirty percent of the sequences showed 2–11 untemplated nucleotides inserted at the Sµ–Sγ3 junctions, whereas only 3% of sequences from wild-type B cells showed inserts, using an identical approach (Schrader et al., 2002). These data indicate that the DNA ends involved in forming Sµ–Sγ3 junctions are accessible to end-processing activities. We did not detect an increased frequency of insertions at sites of mutations or small deletions within recombined and GL 5′ Sµ segments (Supplementary figures S3, S8 and S9).
Fig 5. Sequences of Sµ–Sγ3 junctions (middle sequence in each group) from in vitro activated splenic B cells from TdT transgenic mice demonstrate the presence of nucleotide insertions at the junctions. The parental Sµ sequences (top sequence) and parental Sγ3 sequences are aligned above and below the recombinant sequences. Boxes surround nucleotides at the Sµ–Sγ3 junctions that are identical in the cloned recombination product and both parental sequences (Sµ and Sγ3). To the right of each sequence are indicated the number of identical or inserted junctional nucleotides.
Discussion
Mutations occur in the Ig Sµ region but rarely in the Sγ3 and Sγ1 regions prior to class switch
One of the most interesting findings of this study is that mutations are introduced into the GL 5′Sµ segment in B cells activated to switch, but rarely into the GL acceptor Sγ3 or Sγ1 regions in these same cultures. This difference leads us to suggest the model that switch recombination may be initiated by AID attacking the Sµ region. If the downstream S region is also attacked, the allele would usually undergo CSR, whereas the Sµ region could be attacked and subsequently repaired during unsuccessful attempts to switch. The downstream Sγ3 and Sγ1 regions appear rarely to undergo the reciprocal event of being attacked and repaired without undergoing CSR. The finding that the GL Sγ1 segments in cells treated with LPS + IL-4 did not show an appreciable mutation frequency, even under conditions in which nearly all isotype switching is to IgG1, suggests that the difference in mutation frequency of the donor and acceptor S regions is not simply due to a greater frequency of switch events involving the Sµ region compared with each of the two different acceptor S regions, since under these conditions nearly all switching is to IgG1.
These data are consistent with the recent report by Reina-San-Martin et al. (2003) in which genomic Southern blotting experiments were used to examine whether internal deletions occur in GL Sµ and Sγ1 segments in hybridomas prepared from splenic B cells treated to induce switching with LPS + IL-4. Their data showed that Sµ segments that had not undergone switch recombination frequently had sustained internal deletions, whereas GL Sγ1 segments remained intact in these cells. In addition, they examined mutations in splenic B cells that had divided five times after addition of LPS + IL-4 and found more mutations in the GL 5′Sµ segment than in the GL 5′Sγ1 segment, although a small portion of the GL Sγ1 segments had sustained some mutations. Together, these data suggest that DNA lesions might be introduced first into the Sµ region and only subsequently into the downstream acceptor S region, and that introduction of lesions into the downstream S region usually does not occur without S–S recombination. Perhaps the lesions within Sµ are important for activation of factors that result in subsequent recruitment of the downstream S region. Interestingly, activated splenic B cells from mice deficient in γ-H2AX have been shown to accrue normal levels of mutations in the GL 5′Sµ segment, but they do not undergo CSR (Reina-San-Martin et al., 2003). It is possible that γ-H2AX is required for recruitment of and/or synapsis with the downstream S region.
One could hypothesize further that the reason CSR initiates at the GL Sµ segment rather than the downstream GL S region is because the GL Sµ segment is much more transcriptionally active. The VDJ promoter, with its strong octamer element and close proximity to the µ intron enhancer, drives much more transcription than the weak downstream GL promoters. Additional transcription is driven across the Sµ region by GL Sµ promoters located within and near to the µ intron enhancer (Lennon and Perry, 1985). This should make the GL Sµ segment a much better target for AID, since the substrate for AID appears to be single-stranded DNA, transiently created by active transcription (Chaudhuri et al., 2003; Ramiro et al., 2003) and also by formation of RNA–DNA hybrids (R loops) with S regions (Yu et al., 2003). Furthermore, the frequency of CSR in an integrated switch plasmid substrate has been shown to correlate with the rate of transcription of the recombining S regions (Lee et al., 2001).
The nucleotide specificity of the mutations in GL Sµ and recombined Sµ–Sγ3 segments differs
Surprisingly, the sequence specificity of the mutations differs between GL Sµ (both 5′ and 3′ segments) and recombined Sµ, suggesting that the processes creating these mutations differ. The lack of specificity of the GL Sµ mutations for G/C base pairs and for WRCY hotspots indicates that most of the mutations in GL Sµ regions cannot simply be caused by deamination of dC residues in hotspots by AID. However, since these mutations are known to be AID dependent (Petersen et al., 2001; Nagaoka et al., 2002), it is likely that the lesions created after deamination of dC residues activate error-prone mechanisms that introduce mutations at additional sites, e.g., error-prone translesion DNA polymerases (Diaz et al., 2001; Zeng et al., 2001; Faili et al., 2002a; Storb and Stavnezer, 2002; McDonald et al., 2003). To explain the lack of G/C and hotspot targeting in the GL Sµ segments, perhaps BER correctly repairs the initiating lesion, but during this process error-prone translesion polymerases introduce mutations at the surrounding nucleotides. Alternatively, it is possible that the GL Sµ regions we sequenced are located 5′ and 3′ to the main segment targeted by AID, and thus the mutations observed might be due mostly to repair processes that occur over extensive regions in response to AID-induced lesions occurring in a central region of Sµ.
McDonald et al. (2003) recently showed that pol ι and η together, or to a lesser extent individually, can perform displacement DNA synthesis, and thus these polymerases could introduce mutations at sites away from the initial dU residue. These investigators also demonstrated that the 129 strain of mice lack pol ι, although their levels of and specificity of SHM are similar to those of pol ι+/+ C57BL/6 mice. To investigate whether pol ι could be affecting the mutation frequencies and spectra in our mice, we determined their pol ι genotype. We found that some of the wild-type splenic B cells used in our experiments, which were always littermate controls, were from mice heterozygous for pol ι and some were from pol ι-deficient mice. As expected from the findings of McDonald et al. (2003), the frequency of mutations in recombined Sµ and Sγ3 segments from pol ι+/– and pol ι–/– mice was identical (23.3 × 10–4 and 20.0 × 10–4, respectively). In addition, the G/C base pair and hotspot targeting were similar (P = 0.765). Therefore, differences in the pol ι background do not appear to affect our results.
The mutation frequency in GL 5′Sµ segments from wild-type cells is as high as found after Sµ–Sγ3 recombination (Table I). This seems surprising because it has been demonstrated that mutations are introduced into S regions during CSR, and thus one would expect more mutations in recombined Sµ segments (Dunnick et al., 1989, 1993). However, it is likely that GL Sµ segments cloned from the day 4 cultures have undergone several rounds of mutations and repair that did not lead to successful CSR, whereas it is likely that after CSR, introduction of mutations may cease (Dunnick et al., 1989; Dunnick and Stavnezer, 1990). This could result in an accumulation of more mutations in GL Sµ segments than in recombined Sµ segments.
B-cell activation conditions alter mutation specificity
Surprisingly, both the G/C and hotspot targeting are much higher in GL 5′Sµ segments amplified from cells induced with LPS + IL-4 than with LPS ± anti-δ dextran (Tables II and III). It is known that IL-4 treatment can increase CSR without increasing GL transcript levels (Shockett and Stavnezer, 1991; McIntyre et al., 1995). Perhaps IL-4 increases the size of the region targeted by AID, resulting in introduction of mutations at G/C base pairs and hotspots over a greater region, although the frequency of mutation in the GL 5′Sµ region was not increased in cells treated with LPS + IL-4 (Table I, row 6). Alternatively, IL-4 might alter subsequent processes that affect how the mutations are processed and repaired.
S region mutation frequency is increased in MMR-deficient B cells
MMR appears to partially correct mutations introduced into S regions during CSR. B cells deficient in Msh2, Mlh1 or doubly deficient for both Msh2 and Mlh1 have increased mutation frequencies relative to junctions from wild-type B cells (Table I and Schrader et al., 2003). Data presented herein indicate that Msh2 may specifically correct mutations created during CSR at G/C base pairs within hotspots, because there is an increase in hotspot and G/C focusing of mutations in recombined Sγ3 segments from msh2–/– cells relative to wild-type cells (Supplementary figure S7 and Table II), although this was not found for the recombined or GL Sµ segments. Msh2 deficiency also specifically increases hotspot and G/C targeting of V gene SHM, although, unlike CSR, Msh2 appears to do this by increasing mutations at sites other than G/C base pairs and hotspots (Phung et al., 1998; Rada et al., 1998).
Mutation frequency decreases with nucleotide positions distal to S–S junctions and a role for Msh2 in end processing
The high level of mutations within 150 bp of the switch junction may be due to end processing and error-prone repair of S region DNA that has sustained lesions due to AID–BER activity. Although AID may broadly target the Sµ region prior to CSR, the focusing of mutations observed in recombined Sµ–Sγ3 segments to regions near the S junctions indicates that DNA lesions, end processing and repair DNA synthesis occurring during CSR are localized to segments near the recombination junctions. In msh2–/– cells, mutations are specifically increased at the junction regions, consistent with data indicating that Msh2 may contribute to end processing during resolution of DNA breaks (Ehrenstein and Neuberger, 1999; Schrader et al., 2002). These results suggest that Msh2 may have similar roles in end processing in CSR and in double-strand break repair in Saccharomyces cerevisiae, in which it has been shown to be required for removal of heterologous sequences that are >30 nucleotides long (Paques and Haber, 1997). In addition, the recent finding that Msh2 is required for CSR in mice lacking the Sµ tandem repeats, where RGYW hotspots occur further apart than in wild-type Sµ (Min et al., 2003), is also consistent with the model that Msh2 is required for removal of the single-strand tails produced when the breaks on opposite strands are separated from each other.
TdT has access to DNA ends during CSR
The finding that TdT expressed from a transgene is able to modify 30% of the recombining S–S junctions indicates that some of the DNA ends at the S junctions are not sequestered into a complex that blocks them, nor are they so rapidly recombined that they are rendered inaccessible. These data are reminiscent of the finding that 30% of the VH genes in TdT-expressing Ramos B-cell lines undergoing constitutive SHM have insertions of 2–11 nucleotides (Sale and Neuberger, 1998). These insertions occurred at or near to SHM hotspots. It is likely that TdT can insert nucleotides at both single-strand and double-strand breaks, and SHM can occur at single-strand breaks (Faili et al., 2002b). Although we detected insertions at Sµ–Sγ3 junctions from TdT transgenic B cells, we did not detect any increase in nucleotide insertions within GL and recombined Sµ–Sγ3 segments at sites away from the junctions. These data suggest that the mutation sites within S region segments are inaccessible to TdT, unlike sites of SHM in VH genes. Perhaps S region mutations occur independently of the formation of DNA breaks or perhaps single-strand breaks within the S region segments are rapidly repaired. However, it is possible that TdT could insert nucleotides at sites undergoing internal deletions within Sµ, as these insertions would be difficult to detect.
Altogether, our data support a model positing that CSR is initiated by AID attacking the Sµ region, and that only in cells undergoing successful CSR is the downstream S region also attacked. The apparent preference for Sµ may be due to differential rates of transcription of the expressed VDJ-Cµ gene and the downstream I–S–CH gene segments. Perhaps the initial lesions in the GL Sµ segment activate and recruit repair factors that are necessary to obtain synapsis with a downstream S region. Furthermore, if the initiating lesions in the Sµ region do not lead to successful recombination with an acceptor S region, they are repaired. This process may occur a few times over several cell divisions, resulting in accumulation of mutations in the GL Sµ region. Several aspects of our data indicate that processes that result in nucleotide mutations in the GL Sµ segments differ from processes resulting in mutations in recombined Sµ–Sγ3 segments. The most compelling data supporting this conclusion are the findings that mutations in the recombined Sµ segments show a much greater preference for G/C base pairs and for hotspot sequences than mutations within the GL Sµ segments. Furthermore, the mutations are localized to the S–S junctions in recombined molecules but are more evenly distributed in the GL Sµ sequences. In addition, Msh2 is involved in repair of mutations in recombined Sµ–Sγ3 segments, but does not appear to be involved in repairing mutations within the GL 5′Sµ segments.
Since it has been shown that mice deficient in γ-H2AX sustain GL 5′Sµ mutations but do not undergo CSR (Reina-San-Martin et al., 2003), we further hypothesize that Sµ lesions activate γ-H2AX and that this is necessary for recruitment or synapsis of a downstream S region with Sµ. Perhaps γ-H2AX recruits and organizes factors that are necessary for proper recombination. In cells that fail to undergo CSR, DNA lesions generated within the GL Sµ segment are repaired by error-prone processes that introduce mutations preferentially at A/T base pairs. However, during resolution of DNA breaks that result in successful CSR, mutations are introduced mostly within G/C base pairs near the S–S junction, suggesting that DNA breaks at the S–S junction recruit repair factors different from those recruited by DNA lesions within GL Sµ regions that do not undergo successful CSR.
Materials and methods
Mice
Msh2-deficient mice were obtained from W.Edelmann (Wei et al., 2002) and from T.Mak (Reitmair et al., 1996). Mice transgenic for TdT were obtained from G.Wu (Marshall et al., 1998).
B-cell isolation and cultures
Splenic B cells were isolated and cultured as described (Schrader et al., 2002). To induce switch recombination to IgG3, LPS ± anti-δ-dextran (0.3 ng/ml; gift from C.Snapper, Uniformed Services University of the Health Sciences, Bethesda MD) was added at the initiation of culture. To induce switch recombination to IgG1, LPS and IL-4 (800 U/ml, from Dr W.Paul, NIH) were added.
PCR amplification of Sµ–Sγ3 junctions and germline Sµ, Sγ3 and Sγ1 segments
Genomic DNA was isolated from purified splenic B cells ex vivo or cultured as indicated (Schrader et al., 2002). Sµ–Sγ3 junctions were amplified from genomic DNA by PCR using the Expand Long Template Taq and Pfu polymerase mix (Roche, Piscataway NJ) and the primers µ3-H3 (5′-AACAAGCTTGGCTTAACCGAGATGAGCC-3′) (AC073553.5 at nucleotides 137 284–137 303; first eight nucleotides were added for cloning) and g3-2 (5′-TACCCTGACCCAGGAGCTGCATAAC-3′) (MUSIGHANA nucleotides 2601–2626) (Schrader et al., 2002). Different primers were used for GL 5′Sµ sequences, 5µ3 (5′-AATGG ATACCTCAGTGGTTTTTAATGGTGGGTTTA-3′) and 3µ2 (5′-AGA GGCCTAGATCCTGGCTTCTCAAGTAG-3′) (AC073443.5 at nucleotides 136 645–136 679 and 139 861–139 889, respectively) (Petersen et al., 2001). Sequences located at the 5′ end of this segment are analyzed in Tables I and II, Figure 3A and Supplementary figures S1–S3. The primers used to amplify the GL Sγ3 fragment were γ3-1 (5′-CAGGCTAAGATGGATGCTACAGGGA-3′) (Wuerffel et al., 1997) and g3-2, located in MUSIGHANA at nucleotides 404–428 and 2601–2626, respectively. Sequences from both ends of the GL Sγ3 segment were analyzed for the data in Table I. The primers used for amplification of the 5′ segment of the GL Sγ1 segments were Sg1-3U (5′-CACTC TGGCCTTTTTGGTCCCTTACGC-3′) and Sg1-3L (5′-TTCCTCTAC TTGTCTTTCCCTCCTTCA-3′), located in MUSIGHANB (D78344) at nucleotides 113–140 and 2133–2159, respectively.
Cloning, identification and sequence analysis of PCR products
PCR products were cloned into the vector pGEM®-T Easy (Promega, Madison, WI) or into pCR4-TOPO (Invitrogen, Carlsbad, CA). Segments with deletions were not used for calculation of mutation frequencies although, depending on the clarity of the alignment, they could be used for determination of mutation specificity. The unmutated Sγ3 and the GL Sµ sequences were determined previously (Schrader et al., 2002). The GL Sγ1 sequence in the 129xB6 mice we used was determined by consensus among our sequences, and it differed in a few positions from MUSIGHANB. The PCR error frequency in our experiments was determined by sequencing several independent amplifications of the single Sµ–Sγ3 junction from the plasmacytoma TIB114, using procedures identical to those used for splenic B-cell junctions (Table I). TIB114, like other plasmacytomas, does not have AID mRNA and does not undergo SHM or CSR (Ma et al., 2002; Martin et al., 2002).
Supplementary data
Supplementary data are available at The EMBO Journal Online.
Acknowledgments
Acknowledgements
We thank Dr Stephen P.Baker for help with statistical analysis, Michael Twarog for technical assistance, Harold Hills and Phyllis Spatrick for DNA sequencing, Drs Amy L.Kenter, Pan Qiang and Denise A.Kaminski for helpful criticisms, and Dr Wesley A.Dunnick for information about the sites of Sµ–Sγ3 recombination. The research was supported by grants from the NIH AI23283 and AI42108.
References
- Bransteitter R., Pham,P., Scharff,M.D. and Goodman,M.F. (2003) Activation-induced cytidine deaminase deaminates deoxycytidine on single-stranded DNA but requires the action of RNase. Proc. Natl Acad. Sci. USA, 100, 4102–4107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhuri J., Tian,M., Khuong,C., Chua,K., Pinaud,E. and Alt,F.W. (2003) Transcription-targeted DNA deamination by the AID antibody diversification enzyme. Nature, 422, 726–730. [DOI] [PubMed] [Google Scholar]
- Di Noia J. and Neuberger,M.S. (2002) Altering the pathway of immunoglobulin hypermutation by inhibiting uracil-DNA glycosylase. Nature, 419, 43–48. [DOI] [PubMed] [Google Scholar]
- Diaz M., Verkoczy,L.K., Flajnik,M.F. and Klinman,N.R. (2001) Decreased frequency of somatic hypermutation and impaired affinity maturation but intact germinal center formation in mice expressing antisense RNA to DNA polymerase ζ. J. Immunol., 167, 327–335. [DOI] [PubMed] [Google Scholar]
- Dickerson S.K., Market,E., Besmer,E. and Papavasiliou,F.N. (2003) AID mediates hypermutation by deaminating single stranded DNA. J. Exp. Med., 197, 1291–1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du J., Shanmugam,A. and Kenter,A.L. (1997) Analysis of immunoglobulin Sγ3 recombination breakpoints by PCR: implications for the mechanism of isotype switching. Nucleic Acids Res., 25, 3066–3073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudley D.D., Manis,J.P., Zarrin,A.A., Kaylor,L., Tian,M. and Alt,F.W. (2002) Internal IgH class switch region deletions are position-independent and enhanced by AID expression. Proc. Natl Acad. Sci. USA, 99, 9984–9989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunnick W. and Stavnezer,J. (1990) Copy choice mechanism of immunoglobulin heavy chain switch recombination. Mol. Cell. Biol., 10, 397–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunnick W., Wilson,M. and Stavnezer,J. (1989) Mutations, duplication and deletion of recombined switch regions suggest a role for DNA replication in the immunoglobulin heavy-chain switch. Mol. Cell. Biol., 9, 1850–1856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunnick W., Hertz,G.Z., Scappino,L. and Gritzmacher,C. (1993) DNA sequences at immunoglobulin switch region recombination sites. Nucleic Acids Res., 21, 365–372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrenstein M.R. and Neuberger,M.S. (1999) Deficiency in Msh2 affects the efficiency and local sequence specificity of immunoglobulin class-switch recombination: parallels with somatic hypermutation. EMBO J., 18, 3484–3490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrenstein M.R., Rada,C., Jones,A.M., Milstein,C. and Neuberger,M.S. (2001) Switch junction sequences in PMS2-deficient mice reveal a microhomology-mediated mechanism of Ig class switch recombination. Proc. Natl Acad. Sci. USA, 98, 14553–14558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faili A., Aoufouchi,S., Flatter,E., Gueranger,Q., Reynaud,C.A. and Weill,J.C. (2002a) Induction of somatic hypermutation in immunoglobulin genes is dependent on DNA polymerase ι. Nature, 419, 944–947. [DOI] [PubMed] [Google Scholar]
- Faili A., Aoufouchi,S., Gueranger,Q., Zober,C., Leon,A., Bertocci,B., Weill,J.C. and Reynaud,C.A. (2002b) AID-dependent somatic hypermutation occurs as a DNA single-strand event in the BL2 cell line. Nature Immunol., 3, 815–821. [DOI] [PubMed] [Google Scholar]
- Haracska L., Unk,I., Johnson,R.E., Johansson,E., Burgers,P.M., Prakash,S. and Prakash,L. (2001) Roles of yeast DNA polymerases δ and ζ and of Rev1 in the bypass of abasic sites. Genes Dev., 15, 945–954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C.G., Kinoshita,K., Arudchandran,A., Cerritelli,S.M., Crouch,R.J. and Honjo,T. (2001) Quantitative regulation of class switch recombination by switch region transcription. J. Exp. Med., 194, 365–374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lennon G.G. and Perry,R.P. (1985) Cµ-containing transcripts initiate heterogeneously within the IgH enhancer region and contain a novel 5′-nontranslatable exon. Nature, 318, 475–478. [DOI] [PubMed] [Google Scholar]
- Lindahl T. (2000) Suppession of spontaneous mutagenesis in human cells by base excision-repair. Mutat. Repair, 462, 129–135. [DOI] [PubMed] [Google Scholar]
- Ma L., Wortis,H.H. and Kenter,A.L. (2002) Two new isotype-specific switching activities detected for Ig class switching. J. Immunol., 168, 2835–2846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall A.J., Doyen,N., Bentolila,L.A., Paige,C.J. and Wu,G.E. (1998) Terminal deoxynucleotidyl transferase expression during neonatal life alters D(H) reading frame usage and Ig-receptor-dependent selection of V regions. J. Immunol., 161, 6657–6663. [PubMed] [Google Scholar]
- Martin A., Bardwell,P.D., Woo,C.J., Fan,M., Shulman,M.J. and Scharff,M.D. (2002) Activation-induced cytidine deaminase turns on somatic hypermutation in hybridomas. Nature, 415, 802–806. [DOI] [PubMed] [Google Scholar]
- McDonald J.P., Frank,E.G., Plosky,B.S., Rogozin,I.B., Masutani,C., Hanaoka,F., Woodgate,R. and Gearhart,P.J. (2003) 129-derived strains of mice are deficient in DNA polymerase ι and have normal immunoglobulin hypermutation. J. Exp. Med., 198, 635–643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McIntyre T.M., Kehry,M.R. and Snapper,C.M. (1995) Novel in vitro model for high-rate IgA class switching. J. Immunol., 154, 3156–3161. [PubMed] [Google Scholar]
- Milstein C., Neuberger,M.S. and Staden,R. (1998) Both DNA strands of antibody genes are hypermutation targets. Proc. Natl Acad. Sci. USA, 95, 8791–8794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Min I., Schrader,C., Vardo,J., D’Avirro,N., Luby,T., Stavnezer,J. and Selsing,E. (2003) Sµ tandem repeat region is required for isotype switching in the absence of Msh2. Immunity, 19(4), 515–524. [DOI] [PubMed] [Google Scholar]
- Muramatsu M., Sankaranand,V.S., Anant,S., Sugai,M., Kinoshita,K., Davidson,N.O. and Honjo,T. (1999) Specific expression of activation-induced cytidine deaminase (AID), a novel member of the RNA-editing deaminase family in germinal center B cells. J. Biol. Chem., 274, 18470–18476. [DOI] [PubMed] [Google Scholar]
- Muramatsu M., Kinoshita,K., Fagarasan,S., Yamada,S., Shinkai,Y. and Honjo,T. (2000) Class switch recombination and hypermutation require activation-induced cytidine deaminase (AID), a potential RNA editing enzyme. Cell, 102, 553–563. [DOI] [PubMed] [Google Scholar]
- Nagaoka H., Muramatsu,M., Yamamura,N., Kinoshita,K. and Honjo,T. (2002) Activation-induced deaminase (AID)-directed hypermutation in the immunoglobulin Sµ region: implication of AID involvement in a common step of class switch recombination and somatic hypermutation. J. Exp. Med., 195, 529–534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan-Hammarstrom Q., Dai,S., Zhao,Y., van Dijk-Hard,I.F., Gatti,R.A., Borresen-Dale,A.L. and Hammarstrom,L. (2003) ATM is not required in somatic hypermutation of V(H), but is involved in the introduction of mutations in the switch micro region. J. Immunol., 170, 3707–3716. [DOI] [PubMed] [Google Scholar]
- Paques F. and Haber,J.E. (1997) Two pathways for removal of nonhomologous DNA ends during double-strand break repair in Saccharomyces cerevisiae. Mol. Cell. Biol., 17, 6765–6771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen S. et al. (2001) AID is required to initiate Nbs1/γ-H2AX focus formation and mutations at sites of class switching. Nature, 414, 660–665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen-Mahrt S.K., Harris,R.S. and Neuberger,M.S. (2002) AID mutates E.coli suggesting a DNA deamination mechanism for antibody diversification. Nature, 418, 99–104. [DOI] [PubMed] [Google Scholar]
- Pham P., Bransteitter,R., Petruska,J. and Goodman,M.F. (2003) Processive AID-catalysed cytosine deamination on single-stranded DNA simulates somatic hypermutation. Nature, 424, 103–107. [DOI] [PubMed] [Google Scholar]
- Phung Q.H., Winter,D.B., Cranston,A., Tarone,R.E., Bohr,V.A., Fishel,R. and Gearhart,P.J. (1998) Increased hypermutation at G and C nucleotides in immunoglobulin variable genes from mice deficient in the MSH2 mismatch repair protein. J. Exp. Med., 187, 1745–1751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rada C., Ehrenstein,M.R., Neuberger,M.S. and Milstein,C. (1998) Hot spot focusing of somatic hypermutation in MSH2-deficient mice suggests two stages of mutational targeting. Immunity, 9, 135–141. [DOI] [PubMed] [Google Scholar]
- Rada C., Williams,G.T., Nilsen,H., Barnes,D.E., Lindahl,T. and Neuberger,M.S. (2002) Immunoglobulin isotype switching is inhibited and somatic hypermutation perturbed in UNG-deficient mice. Curr. Biol., 12, 1748–1755. [DOI] [PubMed] [Google Scholar]
- Ramiro A.R., Stavropoulos,P., Jankovic,M. and Nussenzweig,M.C. (2003) Transcription enhances AID-mediated cytidine deamination by exposing single-stranded DNA on the nontemplate strand. Nature Immunol., 4, 452–456. [DOI] [PubMed] [Google Scholar]
- Reina-San-Martin B., Difilippantonio,S., Hanitsch,L., Masilamani,R.F., Nussenzweig,A. and Nussenzweig,M.C. (2003) H2AX is required for recombination between immunoglobulin switch regions but not for intra-switch region recombination or somatic hypermutation. J. Exp. Med., 197, 1767–1778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reitmair A.H. et al. (1996) MSH2 deficiency contributes to accelerated APC-mediated intestinal tumorigenesis. Cancer Res., 56, 2922–2926. [PubMed] [Google Scholar]
- Revy P., et al. (2000) Activation-induced cytidine deaminase (AID) deficiency causes the autosomal recessive form of the hyper-IgM syndrome (HIGM2). Cell, 102, 565–575. [DOI] [PubMed] [Google Scholar]
- Sale J.E. and Neuberger,M.S. (1998) TdT-accessible breaks are scattered over the immunoglobulin V domain in a constitutively hypermutating B cell line. Immunity, 9, 859–869. [DOI] [PubMed] [Google Scholar]
- Schrader C.E., Edelmann,W., Kucherlapati,R. and Stavnezer,J. (1999) Reduced isotype switching in splenic B cells from mice deficient in mismatch repair enzymes. J. Exp. Med., 190, 323–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrader C.E., Vardo,J. and Stavnezer,J. (2002) Role for mismatch repair proteins Msh2, Mlh1 and Pms2 in immunoglobulin class switching shown by sequence analysis of recombination junctions. J. Exp. Med., 195, 367–373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrader C., Vardo,J. and Stavnezer,J. (2003) Mlh1 can function in antibody class switch recombination independently of Msh2. J. Exp. Med., 197, 1377–1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shockett P. and Stavnezer,J. (1991) Effect of cytokines on switching to IgA and α germline transcripts in the B lymphoma I.29µ: transforming growth factor-β activates transcription of the unrearranged Cα gene. J. Immunol., 147, 4374–4383. [PubMed] [Google Scholar]
- Sohail A., Klapacz,J., Samaranayake,M., Ullah,A. and Bhagwat,A.S. (2003) Human activation-induced cytidine deaminase causes transcription-dependent, strand-biased C to U deaminations. Nucleic Acids Res., 31, 2990–2994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storb U. and Stavnezer,J. (2002) Immunoglobulin genes: generating diversity with AID and UNG. Curr. Biol., 12, R725–R727. [DOI] [PubMed] [Google Scholar]
- Wei K., Kucherlapati,R. and Edelmann,W. (2002) Mouse models for human DNA mismatch-repair gene defects. Trends Mol. Med., 8, 346–353. [DOI] [PubMed] [Google Scholar]
- Wuerffel R.A., Du,J., Thompson,R.J. and Kenter,A.L. (1997) Ig Sγ3 DNA-specific double strand breaks are induced in mitogen-activated B cells and are implicated in switch recombination. J. Immunol., 159, 4139–4144. [PubMed] [Google Scholar]
- Yu K., Chedin,F., Hsieh,C.L., Wilson,T.E. and Lieber,M.R. (2003) R-loops at immunoglobulin class switch regions in the chromosomes of stimulated B cells. Nature Immunol., 4, 442–451. [DOI] [PubMed] [Google Scholar]
- Zeng X., Winter,D.B., Kasmer,C., Kraemer,K.H., Lehmann,A.R. and Gearhart,P.J. (2001) DNA polymerase η is an A–T mutator in somatic hypermutation of immunoglobulin variable genes. Nature Immunol., 2, 537–541. [DOI] [PubMed] [Google Scholar]