Abstract
The Tn7 transposon avoids inserting into a target DNA that contains a pre-existing copy of Tn7. This phenomenon, known as ‘target immunity’, is established when TnsB, a Tn7 transposase subunit, binds to Tn7 sequences in the target DNA and mediates displacement of TnsC, a critical transposase activator, from the DNA. Paradoxically, TnsB–TnsC interactions are also required to promote transposon insertion. We have probed Tn7 target immunity by isolating TnsB mutants that mediate more frequent insertions into a potentially immune target DNA because they fail to provoke dissociation of TnsC from the DNA. We show that a single region of TnsB mediates the TnsB–TnsC interaction that underlies both target immunity and transposition, but that TnsA, the other transposase subunit, channels the TnsB–TnsC interaction toward transposition.
Keywords: DNA transposition/enzyme activation/target immunity/Tn7/transposase
Introduction
Transposable elements use many different strategies to select their target DNAs (Craig, 1997; Craig et al., 2002). For example, insertion into nearby DNA is common for IS10/Tn10 elements. Insertion into DNA flanking the transposon can lead to the evolution of new composite IS10/Tn10 elements; however, insertion within the transposon itself can lead to element destruction (Kleckner et al., 1996). Other elements, such as Tn7, Mu and Tn3, preferentially insert into target sites that are distally located and do not already contain a copy of the transposable element (Robinson et al., 1977; Hauer and Shapiro, 1984; Reyes et al., 1987; Arciszewska et al., 1989). This bias in selecting a distal target site is known as ‘target immunity’ because the presence of a transposable element in a target DNA renders the DNA ‘immune’ to future insertions. The immunity signal is provided by the sequences at the ends of the transposon DNA that are bound by the transposase. In the case of Tn7 (Stellwagen and Craig, 1997a,b) and Mu (Maxwell et al., 1987; Adzuma and Mizuuchi, 1988), target immunity has been shown to depend on the interaction of the transposase with an element-encoded target DNA binding transposase activator protein; the interaction of these proteins results in the dissociation of the transposase activator from the target DNA. In this report, we explore the molecular mechanism by which TnsB, the component of the Tn7 transposase that binds to the ends of the transposon, interacts with TnsC, the target DNA binding transposase activator, to establish target immunity.
Tn7 target immunity is effective over large distances in vivo (Arciszewska et al., 1989; DeBoy and Craig, 1996). For example, the presence of Tn7 transposon end sequences in the Escherischia coli chromosome decreases insertions into a target site 190 kb away; however, insertion into a target site 1900 kb away is not affected (DeBoy and Craig, 1996). Thus Tn7 target immunity is a distance-dependent cis-acting phenomenon.
While Tn7 avoids inserting into DNA containing a ‘negative’ signal, i.e. Tn7 transposon end sequences, it can also be attracted to a target DNA by ‘positive’ signals. The Tn7 target selection proteins TnsD or TnsE are responsible for recognizing ‘positive’ target signals (Peters and Craig, 2001; Craig, 2002). The target selection proteins collaborate with the TnsAB transposase and the TnsC transposase activator to promote transposition into two types of target DNAs: (i) TnsABC + D direct insertions into DNA containing attTn7, a unique site in the E.coli chromosome (Bainton et al., 1993), and (ii) TnsABC + E direct insertions into DNA that is undergoing lagging strand DNA synthesis, such as conjugating DNA (Wolkow et al., 1996; Peters and Craig, 2001). However, both the TnsABC + D and the TnsABC + E transposition pathways are inhibited by target immunity, i.e. the presence of a ‘negative’ target signal is dominant to a ‘positive’ signal present on the same DNA (Arciszewska et al., 1989; DeBoy and Craig, 1996).
Insight into the molecular interactions that promote Tn7 insertion into target DNA containing a ‘positive’ signal and inhibit insertion into DNA containing a ‘negative’ signal has come from the in vitro reconstituted TnsABC + D transposition system (Bainton et al., 1993; Craig, 2002). The attTn7 target DNA is recognized in a sequence-specific manner by TnsD, which then recruits the ATP-dependent sequence-independent DNA binding protein TnsC (Gamas and Craig, 1992; Bainton et al., 1993; Kuduvalli et al., 2001). TnsC, in turn, interacts with the TnsAB transposase and the transposon DNA resulting in the assembly of an essential transposition intermediate, the TnsABC + D transposon–target nucleoprotein complex (Skelding et al., 2002). Thus, the presence of TnsC on a target DNA is key to attracting Tn7.
What feature of Tn7 provides a negative signal? That is, how does a pre-existing Tn7 element block insertions into the DNA in which it resides? The presence of TnsB binding sites on an attTn7 target DNA render it immune to insertions by increasing the local concentration of TnsB on the target DNA, which enables a TnsB–TnsC interaction that provokes displacement of TnsC from the DNA via a mechanism dependent on ATP hydrolysis by TnsC (Stellwagen and Craig, 1997a). Therefore ‘positive’ and ‘negative’ target signals determine whether or not TnsC binds stably to a DNA, which, in turn, determines whether that target DNA will receive a Tn7 insertion.
Tn7 transposition and target immunity both depend on interactions between TnsB and TnsC. These interactions are essential for the assembly of a stable transposon–target complex (Skelding et al., 2002), and for the displacement of TnsC from DNA during target immunity (Stellwagen and Craig, 1997a). Are the TnsB–TnsC interactions that mediate the assembly and disassembly processes the same or different?
To examine this question, we have probed the role of TnsB in establishing target immunity by isolating and analyzing TnsB mutants that are impaired in their ability to impose immunity on a target DNA that contains Tn7 ends close to attTn7. The mutations map to the carboxyl terminus of TnsB, a region shown previously to be essential for the TnsB–TnsC interaction that promotes transposition (Skelding et al., 2002). We show that these TnsB mutants are defective for imposing target immunity owing to a decrease in their ability to interact with TnsC and promote displacement of TnsC from target DNA. We have also found that TnsA, the other component of the Tn7 transposase, plays a critical role in influencing this TnsB–TnsC interaction: TnsA decreases the TnsB-mediated displacement of TnsC from target DNA, and thus promotes transposition. We discuss how a similar TnsB–TnsC interaction is able both to build nucleoprotein complexes for transposition in conjunction with TnsA and to displace TnsC from target DNA containing transposon ends to establish target immunity.
Results
Isolation of TnsB mutants that bypass target immunity
Central to Tn7 target immunity is the interaction of TnsB, the component of the Tn7 transposase that binds specifically to the ends of Tn7, with TnsC, the target binding protein (Stellwagen and Craig, 1997a). A DNA containing a copy of Tn7 is made immune to further insertions because the TnsB–TnsC interaction results in the displacement of TnsC from the target DNA (Stellwagen and Craig, 1997a). To understand better the role of TnsB in Tn7 target immunity, we isolated TnsB mutants that are defective in establishing immunity but still maintain their ability to promote transposition.
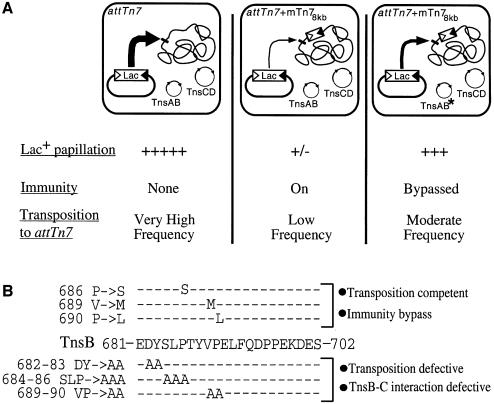
To isolate these mutants, we exploited the ability of Tn7 to insert at high frequency into attTn7, a specific chromosomal site 3′ of the bacterial glmS gene. Due to target immunity, insertion of a second element into attTn7 is much reduced in a strain containing a non-mobilizable mini-Tn7 (mTn7) element 8 kb from attTn7 (attTn7 + mTn78kb) (DeBoy and Craig, 1996). We used a visual assay to screen for TnsB mutants that promote transposition at a higher frequency into the attTn7 + mTn78kb site than TnsBwt (see Materials and methods) (Figure 1A).
Fig. 1. Isolation of TnsB immunity bypass mutants (A) A mini-Tn7lac (mTn7lac) element, consisting of a promoterless lacZY gene cassette flanked by the Tn7 left (Tn7L) and right (Tn7R) sequences required for its mobilization (triangles), resides in a transcriptionally silent location on a plasmid such that, in the absence of transposition, cells containing the plasmid have a Lac– phenotype. However, when the mTn7lac element inserts into attTn7, it is transcribed by the glmUS promoter upstream of attTn7 (Bainton et al., 1993), generating a cell with a Lac+ phenotype, i.e. formation of Lac+ papillae on a MacConkey lactose indicator plate. Thus, in a strain containing a non-mobilizable mTn7 element 8 kb from attTn7 (attTn7 + mTn78kb) (DeBoy and Craig, 1996), the frequency of mTn7lac insertion into attTn7 is very low, and this strain produces very few Lac+ papillae when grown on a MacConkey lactose indicator plate (second column). TnsB mutants that bypass immunity will promote higher-frequency transposition to attTn7 + mTn78kb, which increases the number of Lac+ papillae per colony (third column). (B) TnsB mutations that affect target immunity and transposition overlap. The TnsB immunity bypass mutations (top) were localized to the C-terminus of TnsB (middle). Alanine scanning mutagenesis was shown previously to block TnsB–TnsC interaction and transposition (bottom) (Skelding et al., 2002).
Four TnsB ‘immunity bypass’ mutants were recovered by screening ∼42 000 colonies that were transformed with mutagenized TnsB plasmids. Although the entire TnsB gene was subjected to mutagenesis, DNA sequencing revealed that the mutations were tightly clustered at the carboxyl terminus of the TnsB 702 aa protein: TnsBP686S, TnsBV689M (two isolates) and TnsBP690L (Figure 1B).
The location of the mutations is particularly striking because, as reported elsewhere (Skelding et al., 2002), other mutations within the same region prevent TnsB from interacting with TnsC-bound target DNA, which profoundly decreases TnsABC + D transposition (Figure 1B). Therefore mutation of a single region within TnsB can affect two processes that have been shown to depend upon contact between TnsB and TnsC: target immunity and TnsABC + D transposition. Indeed, the experiments presented in this work provide strong support for the idea that the same TnsB–TnsC interaction is being utilized for both processes.
The ability of TnsBwt and the TnsB immunity bypass mutants to catalyze TnsABC + D transposition in vivo was examined using a quantitative assay that measures the translocation of a mTn7 element from a lambda phage to the bacterial chromosome (Materials and methods). Consistent with previous work (DeBoy and Craig, 1996), the presence of transposon ends 8 kb from attTn7 decreased TnsABC + D transposition into this site >60-fold (Table I, column 5), reflecting the target immunity effect.
Table I. The TnsB mutants promote increased insertion into an attTn7 + mTn78kb chromosome.
| TnsB allele | Transposition frequency into attTn7 + mTn78kba | TnsBmut attTn7 + mTn78kbb |
Transposition frequency into attTn7 | Tpn. attTn7c |
|---|---|---|---|---|
| TnsBwt attTn7 + mTn78kb | Tpn. attTn7 + mTn78kb | |||
| TnsABwtC + D | 0.4 (± 0.13) | 1d | 25 (± 5.4) | 62.5 |
| TnsABP686SC + D | 1.7 (± 0.50) | 4.2 | 5.0 (± 1.2) | 2.9 |
| TnsABV689MC + D | 1.9 (± 0.53) | 4.8 | 7.8 (± 0.71) | 4.1 |
| TnsABP690LC + D | 1.1 (± 0.22) | 2.8 | 3.5 (± 0.29) | 3.2 |
aTransposition frequency is expressed as the ratio of chloramphenicol-resistant colonies to particle forming units of lambda phage (see Materials and methods). Each value represents the average of ≥3 trials where the final value is multiplied by 104.
bTnsABmutC + D transposition frequency in a strain containing attTn7 + mTn78kb divided by the TnsABWTC + D transposition frequency in a strain containing attTn7 + mTn78kb.
cTnsABC + D transposition frequency in a strain containing attTn7 divided by the TnsABC + D transposition frequency in a strain containing attTn7 + mTn78kb.
dThe numerator and denominator are equivalent: TnsABWTC + D transposition frequency in a strain containing attTn7 + mTn78kb.
The TnsB immunity bypass mutants promoted 2.8–4.8 times more frequent transposition into attTn7 + mTn78kb than TnsBwt (Table I, column 3). Sequence analysis verified that 25/25 TnsABV689MC + D reactions contained a mTn7 element inserted at the appropriate position and with the appropriate orientation within attTn7. Thus the TnsB immunity bypass mutants do indeed promote increased transposition into attTn7 + mTn78kb, a site normally refractory to insertion because of target immunity.
When the TnsB immunity bypass mutants were examined for their ability to carry out transposition into an attTn7 site not flanked by Tn7 ends, they all promoted less transposition than TnsABwtC + D (Table I, column 4). Thus, while able to promote increased transposition into a target site that lies close to ‘immunizing’ TnsB binding sites, these TnsB mutants were also slightly defective for transposition in general.
The TnsBV689M mutant was further characterized because it increases transposition to attTn7 + mTn78kb more than the other mutants, while also promoting the most transposition into an attTn7 site lacking nearby Tn7 ends. We purified TnsBV689M and analyzed its properties in vitro.
TnsBV689M is defective in establishing transposition immunity in vitro as evaluated by a redistribution assay
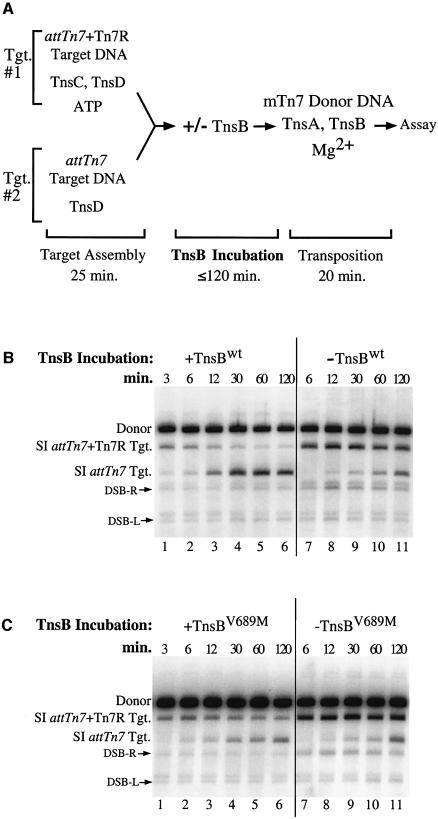
In a previous study, target immunity was reconstituted in an in vitro system in which TnsB was able to provoke the ‘redistribution’ of TnsC from a target DNA containing the right end of Tn7 (attTn7 + Tn7R) to a target DNA lacking TnsB binding sites (attTn7). This reaction depends upon ATP hydrolysis (Stellwagen and Craig, 1997a). We examined the ability of TnsBV689M to provoke TnsC redistribution with a similar assay (Figure 2A). TnsD and a limited quantity of TnsC were bound to an attTn7 + Tn7R target DNA (Figure 2A, Tgt. #1). In a separate reaction, TnsD was bound to an attTn7 target DNA, which was present in equimolar concentration to the attTn7 + Tn7R target DNA (Figure 2A, Tgt. #2). The protein-bound target DNAs were combined in one tube and then incubated with TnsB for various times. After the TnsB incubation period, the distribution of TnsC among the target DNAs was evaluated by adding mTn7 donor DNA, TnsA, additional TnsB and Mg2+ to determine which target DNAs received Tn7 insertions.
Fig. 2. TnsBV689M is defective for promoting TnsC ‘redistribution’ (A) Reaction scheme. (B) In lanes 1–6, half of the TnsBwt was added during the ‘TnsB incubation’ and the remainder was added during the ‘transposition’ phase. In lanes 7–11, all the TnsBwt was added during the ‘transposition’ phase. The DNA products of the reaction were linearized and detected with a mTn7-specific probe. The slowest mobility species is the unreacted mTn7 donor DNA (Donor). The next two bands arise from simple insertion (SI) of the mTn7 element into either the attTn7 + Tn7R target DNA or the attTn7 target DNA. A double-strand break (DSB) at the Tn7R or the Tn7L end in the mTn7 donor DNA produces DSB-R and DSB-L, respectively. The band above DSB-L is due to cross-hybridization of the mTn7-specific probe to the unreacted attTn7 + Tn7R target DNA. (C) Reactions in lanes 1–11 were identical to the reactions in (B) except that TnsBV689M was substituted for TnsBwt.
When the TnsBwt incubation period was short, the attTn7 + Tn7R target received all the insertions and target immunity was not observed, i.e. TnsC was not redistributed to the attTn7 target plasmid that lacked TnsB binding sites (Figure 2B, lane 1). However, after a 30 min TnsBwt incubation period, the majority of insertions were directed to the attTn7 target DNA (Figure 2B, lane 4), reflecting a redistribution of TnsC from the attTn7 + Tn7R target to the attTn7 target. When the combined target DNAs were incubated for 2 h in the absence of TnsB, the attTn7 + Tn7R target DNA still received most of the insertions, indicating that the spontaneous redistribution of TnsC is a slow process (Figure 2B, lane 11). Thus TnsBwt incubation is required to impose immunity on the attTn7 + Tn7R target DNA.
In contrast, when we examined the ability of TnsBV689M to promote TnsC redistribution, we found that, even after a 60 min incubation, insertions occurred equally into both target DNAs (Figure 2C, lane 5), revealing that TnsBV689M does not effectively displace TnsC from the attTn7 + Tn7R target DNA. Thus, as was true in vivo, TnsBV689M is less able than TnsBwt to discourage transposition into a target DNA containing TnsB binding sites.
Incubation with TnsBwt ‘inactivates’ TnsCD–attTn7 target DNA
In the ‘redistribution’ experiment described above, the attTn7 + Tn7R target DNA became immune to insertion after incubation with TnsBwt because TnsC was displaced from attTn7 + Tn7R, leading to an accumulation of TnsC on the attTn7 target DNA. However, the mechanism by which TnsB interacts with TnsC in vitro to promote its displacement from a target DNA does not strictly depend upon TnsB and TnsC being bound to the same target DNA. For example, when Tn7R and attTn7 are on separate, but catenated plasmids, incubation with TnsB still decreases transposition into the tethered attTn7 plasmid (Stellwagen and Craig, 1997a). This finding suggests that binding of TnsB to the same DNA as TnsC is important only to increase the local concentration of TnsB around TnsC, as opposed to a model where the DNA itself plays an essential role in enabling TnsB to interact with TnsC (Adzuma and Mizuuchi, 1989). Consistent with this view, incubation of a TnsCD-bound attTn7 target DNA with TnsBwt can promote moderate redistribution of TnsC from its original attTn7 target DNA to another attTn7 target DNA, even when the original attTn7 target DNA lacks TnsB binding sites (Stellwagen and Craig, 1997a).
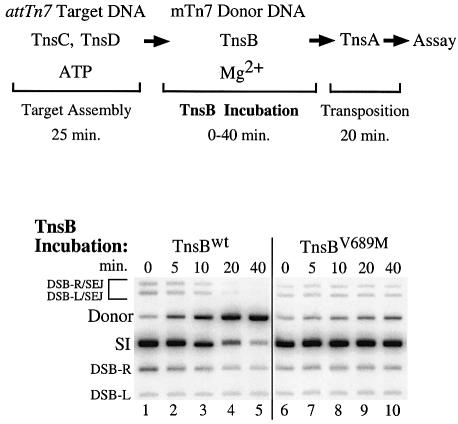
Building on these observations, we evaluated the effect of TnsB incubation on TnsC DNA binding using an assay that does not depend upon the redistribution of TnsC from one target DNA to another. We bound TnsC and TnsD to an attTn7 target DNA, and then added TnsB, mTn7 donor DNA and Mg2+ and incubated for various times (Figure 3). The effect of TnsB incubation on the TnsCD-bound attTn7 target DNA was assessed by determining how much transposition occurred into the target DNA upon addition of TnsA.
Fig. 3. Incubation of TnsCD-bound attTn7 target DNA with TnsB decreases subsequent transposition. TnsBwt was added to the reactions in lanes 1–5 and TnsBV689M was added to the reactions in lanes 6–10. Bands are labeled as in Figure 2B. The bands above the donor result from a double-strand break at either Tn7R or Tn7L that was ‘single end joined’ to the target DNA (DSB-R/SEJ and DSB-L/SEJ) (Bainton et al., 1993).
In the absence of TnsBwt incubation, transposition was robust; most of the mTn7 donor DNA was converted to the simple insertion product (Figure 3, lane 1). However, as the TnsBwt incubation period was increased, the amount of simple insertion product decreased (Figure 3, lanes 2–5). Thus incubation with TnsBwt can decrease the ability of the TnsCD-bound attTn7 target DNA to participate in transposition, even when that target DNA lacks specific TnsB binding sites.
We found that TnsBV689M, like TnsBwt, promoted substantial transposition into the TnsCD–attTn7 target (Figure 3, lane 6 versus lane 1). In striking contrast, TnsBV689M incubation did not significantly reduce the amount of simple insertion product (Figure 3, lanes 6–10). Thus, while TnsBV689M can promote efficient transposition, it has lost considerable ability to ‘inactivate’ the TnsCD-bound attTn7 target DNA for transposition. The finding that incubation of the target complex with TnsBwt results in a decrease in target activity, whereas incubation with TnsBV689M does not, supports the view that ‘inactivation’ of the attTn7 target DNA occurs by a mechanism similar to the mechanism used to establish immunity on a target DNA that contains TnsB binding sites.
TnsBV689M interacts poorly with TnsC
The TnsC ‘redistribution’ experiment and the target ‘inactivation’ experiment described above demonstrate that the V689M mutation alters the ability of TnsB to impose target immunity. How might TnsBV689M fail to ‘inactivate’ the transposition potential of a TnsC-bound target DNA?
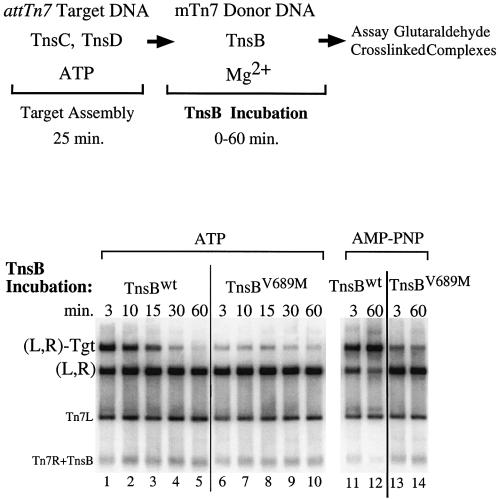
One hypothesis is that TnsBV689M interacts poorly with TnsC and thus is incapable of displacing TnsC from a target DNA. Alternatively, TnsBV689M may interact with TnsC but be unable to provoke some downstream event required for displacement of TnsC from the target DNA, for example TnsC-ATP hydrolysis (Stellwagen and Craig, 1997a, 1998). To explore these possibilities, we directly examined TnsB–TnsC interaction by using a glutaraldehyde cross-linking method to assay for the assembly of a Tns-mediated transposon–target DNA nucleoprotein complex that depends upon TnsB–TnsC interaction (Figure 4)(Skelding et al., 2002).
Fig. 4. TnsBwt and TnsBV689M interact differently with TnsCD-bound attTn7 target DNA. The reactions were similar to Figure 3 except that a cross-linker was added instead of TnsA. The cross-linked products were digested with PflMI which cuts the mTn7donor DNA into two fragments containing either Tn7L or Tn7R. In lanes 1–10 TnsC and TnsD were bound to attTn7 target DNA in the presence of ATP, while in lanes 11–14 AMP–PNP was substituted for ATP.
All the reactions produced an abundant amount of complex in which the Tn7 left and right ends were paired, designated (L,R) (Skelding et al., 2002); thus TnsBV689M is normal for Tn7 end binding and pairing (Figure 4, lanes 1–10). However, TnsBwt-bound transposon ends and TnsBV689M-bound transposon ends interact very differently with TnsCD-bound target DNA. Brief incubation of TnsBwt and the mTn7 donor DNA with the TnsCD-bound target DNA produced an abundant amount of nucleoprotein complex containing the Tn7 left and right ends and the target DNA designated (L,R)-Tgt (Figure 4, lane 1). In marked contrast, reactions containing TnsBV689M produced only a small amount of the (L,R)-Tgt complex (Figure 4, lane 6 versus lane 1). Thus TnsBV689M bound to the transposon ends interacts poorly with TnsC-bound target DNA.
We also examined the effect of extended TnsB incubation on (L,R)-Tgt complex formation. As the TnsBwt incubation period was increased, the amount of (L,R)-Tgt complex decreased in an ATP-dependent manner (Figure 4, lanes 1 and 5 versus lanes 11 and 12). Taken together, the ATP dependence of the (L,R)-Tgt complex instability and the TnsB-provoked displacement of TnsC from DNA during target immunity suggests that (L,R)-Tgt complex dissociation is a consequence of TnsB-provoked TnsC displacement. The amount of TnsBV689M (L,R)-Tgt complex appears to decrease over time as well, but because such a small amount of complex is formed, it is difficult to compare dissociation of this complex with the TnsBwt (L,R)-Tgt complex (Figure 4, lanes 6–10).
This analysis of donor–target nucleoprotein complex formation reveals directly that the ability of TnsBV689M-bound transposon ends to interact with TnsC-bound target DNA is much reduced compared with the interaction of TnsBwt with TnsC. The decreased ability of TnsBV689M to interact with TnsC likely prevents TnsBV689M from provoking displacement of TnsC from DNA, allowing the TnsC-bound target DNA to attract Tn7 insertions.
TnsBV689M-bound transposon DNA forms a stable complex with TnsCD-bound target DNA in the presence of TnsA
In the experiment described above, TnsBV689M bound to the transposon ends interacted poorly with TnsC-bound target DNA. This TnsB–TnsC interaction is also required for the assembly of a critical transposition intermediate, the TnsABC + D (L,R)-Tgt complex (Skelding et al., 2002). We tested for TnsABV689MC + D (L,R)-Tgt complex formation by setting up a reaction just like our ‘standard’ in vitro transposition reaction except that we substituted Ca2+ for Mg2+, a change which supports complex assembly but not the cleavage activity of the TnsAB transposase.
We found both TnsABwtC + D and TnsABV689MC + D formed abundant (L,R)-Tgt complex (Supplementary figure 1, lanes 1–4, available at The EMBO Journal Online). We also found that TnsABV689MC + D and TnsABwtC + D formed equal amounts of (L,R)-Tgtsc complex in the presence of Mg2+ (Supplementary figure 1, lanes 5–8). Thus TnsA enables TnsBV689M-bound transposon DNA to interact effectively with the TnsC-bound target DNA, consistent with the ability of TnsBV689M to promote high-frequency TnsABC + D transposition.
TnsB binding sites adjacent to attTn7 impose target immunity on attTn7
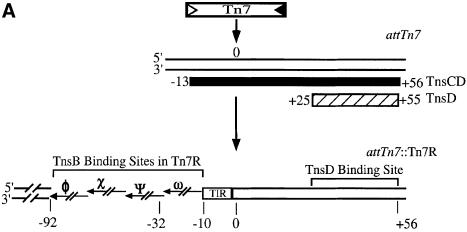
We have argued above and elsewhere (Stellwagen and Craig, 1997a) that the greater the local concentration of TnsB around a TnsC-bound target DNA, the greater the possibility that TnsB will impose immunity on that target DNA. One potential target DNA which contains TnsB binding sites positioned very close to DNA bound by TnsC is the simple insertion product of the TnsABC + D transposition reaction (McKown et al., 1988; Kuduvalli et al., 2001). Thus we created a DNA fragment containing Tn7R immediately adjacent to attTn7 (attTn7::Tn7R), as occurs following TnsABC + D transposition into attTn7 (Figure 5A). We evaluated the binding of the TnsC and TnsD proteins to this fragment with a band-shift assay (Figure 5B). We found that just as much TnsCD complex formed on the attTn7::Tn7R DNA fragment as on the attTn7 DNA fragment (compare lanes 1 in Figure 5C and D); thus the TnsB binding sites do not affect assembly of the TnsCD complex on attTn7::Tn7R. It should be noted that TnsD–attTn7 complex cannot be detected using this assay because TnsD alone cannot remain bound to attTn7 in the presence of 15 mM Mg2+.
Fig. 5. A direct assay for TnsB provoked dissociation of a TnsCD–attTn7::Tn7R complex. (A) The hatched bar beneath the attTn7 target DNA represents the hydroxyl radical footprint of the TnsD–attTn7 complex, while the solid bar represents the hydroxyl radical footprint of the TnsCD–attTn7 complex which extends beyond the position of Tn7 insertion centered around bp 0 (Kuduvalli et al., 2001). Insertion of Tn7 into attTn7 places the four contiguous TnsB binding sites of Tn7R adjacent to the region of DNA bound by the TnsCD complex. As the concentration of TnsB is increased, the sites in Tn7R are bound in the following order: χ; χ + ψ and χ + ω; χ + ψ + ω; χ + ψ + ω + Φ (Arciszewska and Craig, 1991). The outermost 8 bp of Tn7R is conserved with the outermost 8 bp of Tn7L; this sequence is referred to as the ‘terminal inverted repeat’ (TIR). (B) Reaction scheme. (C) Reactions in which different TnsBs were incubated with TnsCD–attTn7::Tn7R. The various TnsB–attTn7::Tn7R complexes are indicated using the χ, ψ and ω symbols. (D) Reactions in which different TnsBs were incubated with TnsCD–attTn7.
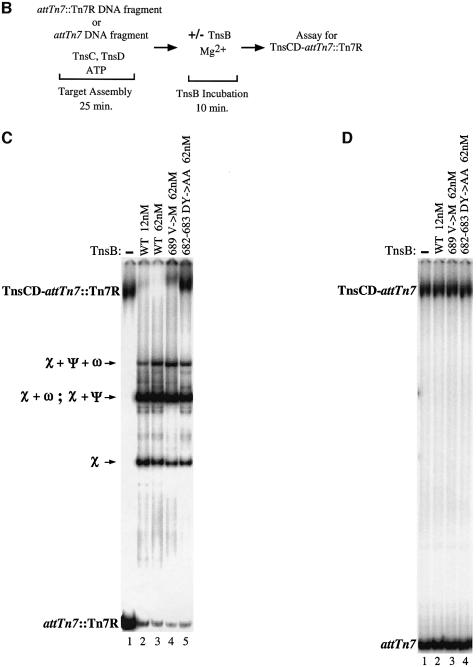
Incubation of the assembled TnsCD–attTn7::Tn7R complex with a low concentration of TnsBwt (12 nM), dramatically decreased the amount of TnsCD–attTn7 complex and produced several new bands (Figure 5C, lane 1 versus lane 2). These new bands are the result of TnsB binding to different combinations of its binding sites in Tn7R (Figure 5A) (Arciszewska and Craig, 1991). In contrast, a TnsCD complex bound to an attTn7 fragment lacking TnsB binding sites was not affected by incubation with TnsBwt (Figure 5D, lane 1 versus lane 2). Thus the presence of TnsB binding sites adjacent to the TnsD binding site greatly facilitates the TnsB-provoked dissociation of the TnsCD–attTn7 complex, likely by increasing the local concentration of TnsB around TnsCD.
We also incubated the TnsCD–attTn7::Tn7R complex with a high concentration of TnsBV689M (62 nM), which we showed above has some residual ability to interact with TnsC-bound target DNA. TnsBV689M provoked limited dissociation of the TnsCD–attTn7::Tn7R complex (Figure 5C, lane 4), consistent with the reduced ability of TnsBV689M to bypass immunity in vivo. In contrast, TnsBDY682-683AA, a mutant unable to assemble a donor–target complex with TnsC-bound target DNA or to promote transposition (Skelding et al., 2002), had no effect on the amount of TnsCD–attTn7::Tn7R complex (Figure 5C, lane 5). Thus TnsB mutants that interact defectively with TnsC in trans, as measured by TnsBC + D donor–target complex formation, are also less able to provoke dissociation of TnsCD–attTn7::Tn7R in cis.
It is notable that the mobility of the TnsCD–attTn7::Tn7R complex decreased in the reactions containing TnsBV689M and TnsBDY682-683AA (Figure 5C, lane 1 versus lanes 4 and 5). To determine whether this supershift reflected the binding of TnsB to the target DNA, we incubated the TnsCD–attTn7::Tn7R complex with TnsB C-terminal truncations that bind normally to Tn7R but are unable to interact with TnsC (R.Sarnovsky and N.Craig, unpublished results). We found that the TnsB C-terminal truncations also supershifted the target complex by an amount consistent with their decreased mass (Supplementary figure 2, lanes 2–5), confirming that TnsB can be incorporated into the TnsCD–attTn7::Tn7R complex.
Taken together, these results suggest that TnsBwt provokes dissociation of TnsC from TnsCD–attTn7::Tn7R by a target immunity mechanism, rather than by occluding the binding of TnsC or TnsD to attTn7::Tn7R. Thus we believe target immunity is likely essential for preventing Tn7 from inserting more than once into attTn7 (Hauer and Shapiro, 1984; Arciszewska et al., 1989; Bainton et al., 1993).
TnsB can provoke dissociation of TnsCD–attTn7 in the absence of TnsB binding sites
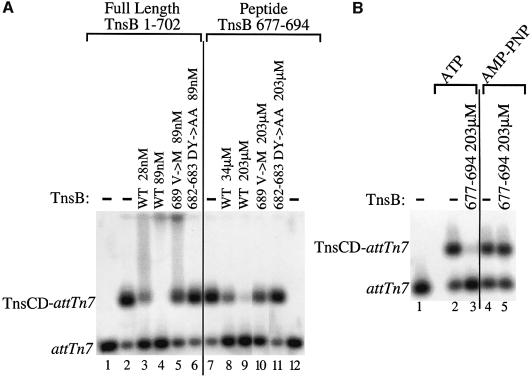
In the experiment described above, a low concentration of TnsBwt (12 nM) provoked dissociation of TnsCD from attTn7::Tn7R, but not from an attTn7 target DNA, i.e. a target DNA lacking TnsB binding sites. However, the effective concentration of TnsB can be increased by decreasing the concentration of non-specific competitor DNA. Under these conditions, and using a slightly different gel system, we found that 28nM TnsBwt can provoke dissociation of the TnsCD–attTn7 complex (Figure 6A, lane 2 versus lanes 3 and 4), while TnsCD–attTn7 complex assembled with AMP–PNP was not affected by incubation with TnsBwt (data not shown). Thus, even in the absence of TnsB binding sites, TnsBwt can provoke dissociation of the TnsCD–attTn7 complex in an ATP-dependent reaction consistent with target immunity being a function of the local concentration of TnsB around TnsC (Stellwagen and Craig, 1997a).
Fig. 6. Full-length TnsB and a TnsB peptide can provoke dissociation of a TnsCD–attTn7 complex by an ATP-dependent reaction. (A) Lanes 1 and 12 contain no Tns proteins. In lanes 2–11 TnsCD–attTn7 was incubated with TnsB protein or peptide as indicated, followed by the addition of glutaraldehyde cross-linker and separation by agarose gel electrophoresis. (B) In lane 1, no Tns proteins were added. Lanes 2 and 3 contained TnsCD–attTn7 complexes assembled in ATP, whereas lanes 4 and 5 were assembled in AMP–PNP. Lanes 3 and 5 also included the TnsB peptide.
In contrast, a high concentration of TnsBDY682-683AA (89 nM) had no effect on the amount of TnsCD–attTn7 complex (Figure 6A, lane 6 versus lane 2), while a small reduction in the amount of TnsCD–attTn7 complex was observed in the presence of 89 nM TnsBV689M immunity bypass mutant (Figure 6A, lane 5). Thus TnsB C-terminal mutations that disrupt TnsB–TnsC interaction commensurately block the ability of TnsB to provoke dissociation of TnsCD–attTn7, regardless of whether the TnsB mutant is bound adjacent to the TnsCD–attTn7 complex or acting in the absence of explicit TnsB binding sites.
A TnsB peptide is sufficient to provoke TnsCD–attTn7 complex disassembly
All the TnsB mutants isolated in this study and elsewhere (Skelding et al., 2002) that affect TnsB–TnsC interaction lie within a small region of TnsB, extending from aa682 to aa690. We asked whether a TnsB peptide containing this region (aa677–694) could alter the stability of the TnsCD–attTn7 complex. Incubation of the TnsCD–attTn7 complex with the TnsBwt peptide provoked complex dissociation. The concentration of peptide required for this effect was ∼1200-fold greater than the concentration of TnsBwt protein required for a similar effect (Figure 6A, lane 8 versus lane 3). The peptide was also unable to provoke dissociation of a TnsCD–attTn7 complex in the presence of AMP–PNP (Figure 6B, lanes 4 and 5 versus lanes 2 and 3). Thus, like full-length TnsB, the TnsB peptide provokes dissociation of the TnsCD–attTn7 complex in an ATP-dependent reaction.
We also examined how the DY682-683AA and V689M mutations affect the ability of the TnsB peptide to provoke dissociation of the TnsCD–attTn7 complex. The effect of the mutant peptides on the TnsCD–attTn7 complex mirrored the effect of the cognate full-length TnsB mutants on the TnsCD–attTn7 complex (Figure 6A, compare lanes 10 and 11 with lanes 5 and 6). This similarity suggests that residues within the aa677–694 region of TnsB are the primary determinants of the TnsB–TnsC interaction.
TnsA protects TnsC-bound target DNA from transposition ‘inactivation’
The preceding sections have focused on the TnsB–TnsC interaction that provokes displacement of TnsC from the target DNA to establish target immunity. However, a TnsB–TnsC interaction mediated by the same region of TnsB is required for Tn7 transposition (Skelding et al., 2002). How can the same interaction be involved with both target immunity and transposition? In addition to TnsB, Tn7 transposition requires the other subunit of the Tn7 transposase, TnsA. We have demonstrated elsewhere that TnsA stabilizes the interaction of TnsB-bound transposon DNA with TnsC-bound target DNA to form an (L,R)-Tgt complex (Skelding et al., 2002), and that TnsA interacts with TnsC (Lu and Craig, 2000; Stellwagen and Craig, 2001). Does TnsA also affect the TnsB–TnsC interaction that provokes displacement of TnsC from target DNA?
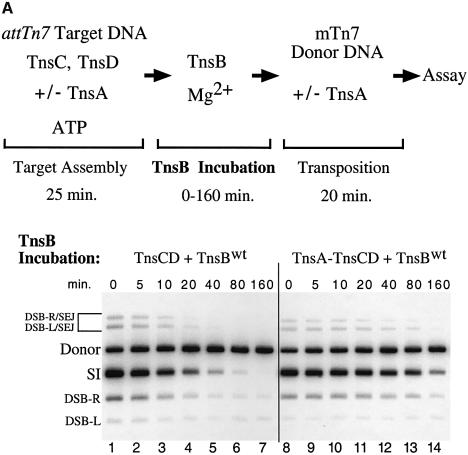
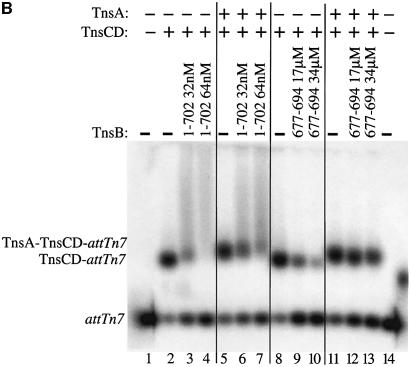
We compared the effect of TnsB incubation on TnsCD–attTn7 and TnsA–TnsCD–attTn7 target DNAs using transposition as a read-out of target ‘inactivation’ (Figure 7A). As demonstrated in Figure 3, addition of TnsB to the TnsCD-bound target DNA in the absence of TnsA caused the amount of simple insertion product to decrease as the time of TnsB incubation increased (Figure 7A, lanes 1–7). However, when TnsA was added at the start of target assembly, significant simple insertion product was seen even after a lengthy incubation with TnsB (Figure 7A, lanes 8–14). Therefore assembly of target DNA with TnsC and TnsD in the presence of TnsA protected the target DNA from the ‘inactivating’ effect of TnsB such that the target DNA received more insertions when transposition was initiated.
Fig. 7. TnsA inhibits TnsB-provoked dissociation of TnsCD–attTn7. (A) In lanes 1–7, TnsC and TnsD were assembled on the target DNA and TnsA was present only during the ‘transposition’ phase. In lanes 8–14, TnsA, TnsC and TnsD were assembled on the target DNA and no additional TnsA was added to the reaction. (B) TnsCD–attTn7 complex was assembled in lanes 2–4 and 8–10 while a TnsA + TnsCD–attTn7 complex was assembled in lanes 5–7 and 11–13.
TnsA protects the TnsCD–attTn7 complex from TnsB peptide provoked dissociation
In the experiment above, the presence of TnsA decreased the TnsB-provoked ‘inactivation’ of the TnsCD-bound target DNA. We directly examined the effect of TnsA on TnsCD–attTn7 complex stability during TnsB incubation using the band-shift assay. Incubation of the attTn7 target DNA with TnsA, TnsC and TnsD produced a TnsA–TnsCD–attTn7 complex with a distinct mobility compared with the TnsCD–attTn7 complex (Figure 7B, lane 5 versus lane 2); this complex is the result of TnsA–TnsC interaction (Lu and Craig, 2000).
When the TnsA–TnsCD–attTn7 complex was incubated with TnsB or TnsB peptide, we found that it was more resistant to TnsB-provoked dissociation than the TnsCD–attTn7 complex (Figure 7B, lanes 5–7 versus lanes 2–4 and lanes 11–13 versus lanes 8–10). We are attracted to the hypothesis that TnsA, likely through interaction with TnsC, stabilizes the TnsCD–attTn7 complex against TnsB-provoked dissociation, thus channeling the complexes resulting from TnsB–TnsC interaction toward transposition (see below).
Discussion
Transposon Tn7 is distinguished by its target site selectivity (Craig et al., 2002). The results presented here provide insight into how the TnsABC + D proteins can both encourage transposition into a target DNA containing a ‘positive’ signal, e.g. attTn7, and discourage transposition into a target DNA containing a ‘negative’ signal, i.e. binding sites for TnsB, resulting in target immunity. In this work, we have explored several issues: Is the same TnsB–TnsC interaction involved in both transposition and target immunity? How does TnsA, the other transposase subunit which also interacts with TnsC, promote transposition in favor of target immunity?
To probe the role of TnsB in target immunity, we isolated and analyzed TnsB mutants that can promote transposition into target DNAs containing TnsB binding sites, i.e. mutants that can bypass target immunity. All the immunity bypass mutations occurred near the C-terminus of TnsB. Notably, substitution of some of the same C-terminal residues with alanines has been shown to block TnsB–TnsC interaction such that the TnsABC + D transposon–target nucleoprotein complex, a key intermediate in Tn7 transposition, does not form (Skelding et al., 2002). Thus interaction between the same region of TnsB and TnsC is involved in both transposition and target immunity.
Using a band-shift assay, we have shown directly that TnsBwt provokes dissociation of a TnsCD–attTn7 target complex in an ATP-dependent fashion, demonstrating the ability of TnsB to affect the removal of the TnsC from a target DNA and preclude that DNA from acting as a target for Tn7 insertion. Moreover, a TnsB immunity bypass mutant provokes much less dissociation of a TnsCD–attTn7 complex than TnsBwt. This difference in TnsC dissociation can account for the immunity bypass phenotype: a TnsCD–attTn7 target complex that lies 8 kb from another Tn7 element is able to persist in the presence of the TnsB immunity mutant, allowing this target to recruit insertions effectively.
It may seem paradoxical to suggest that the same interaction between TnsB and TnsC can mediate transposition and target immunity. We suggest the following working model to accommodate the involvement of the TnsB–TnsC interaction in both processes.
TnsC is an ATP-dependent DNA binding protein that must be present on a DNA for that DNA to be a transposition target. When TnsB-bound transposon ends interact with TnsC-bound target DNA, multiple outcomes are possible. TnsB can provoke TnsC to hydrolyze ATP and dissociate from the target DNA, such that the DNA is no longer attractive to Tn7, i.e. the DNA has become immune (Bainton et al., 1993; Stellwagen and Craig, 1997a). However, we also suggest that before TnsC hydrolyzes its ATP, the TnsB–TnsC transposon–target nucleoprotein complex could serve as a transposition intermediate. We describe below how the presence of the other transposase subunit, TnsA, can determine the fate of the TnsB–TnsC interaction.
TnsA and the choice between transposition and target immunity
The ability of TnsA to stabilize TnsB–TnsC interaction was examined in previous work which showed that a TnsABC + D transposon–target complex was more stable than a TnsBC + D transposon–target complex (Skelding et al., 2002). In this work, we have extended our understanding of the ability of TnsA to stabilize TnsB–TnsC interaction by demonstrating that TnsA can inhibit the TnsB-provoked dissociation of TnsC from a potential target DNA. We have also shown that TnsA is capable of rescuing the weakened TnsBV689M–TnsC interaction enabling the immunity bypass mutant to promote efficient transposition in vitro.
What is the molecular mechanism that underlies the ability of TnsA to channel the TnsB–TnsC interaction toward transposition in vitro? Our finding that TnsA inhibits the dissociation of TnsC from a target DNA in response to either full-length TnsB or the TnsB peptide is most consistent with TnsA acting on TnsC to change the response of TnsC to TnsB. Perhaps TnsA stabilizes the binding of TnsC to the target DNA in the presence of TnsB by altering ATP hydrolysis by TnsC (Stellwagen and Craig, 1998).
TnsA may also channel the TnsB–TnsC interaction toward transposition by other means. In addition to interacting with TnsC (Lu and Craig, 2000; Stellwagen and Craig, 2001), TnsA also likely interacts with TnsB (Biery et al., 2000; Lu and Craig, 2000) and with the DNA at the 5′ end of Tn7 that it cuts during transposition (Bainton et al., 1991; Sarnovsky et al., 1996; Hickman et al., 2000). These other interactions may also stabilize the transposon–target complex, thus favoring transposition.
Comparison of Tn7 and bacteriophage Mu
The Mu transposition proteins share some similarities with TnsB and TnsC (Chaconas and Harshey, 2002). The MuA transposase, like TnsB, binds to multiple sites at the ends of the transposon and mediates transposon 3′ end cleavage and joining to the target DNA. MuB, like TnsC, is an ATP-dependent DNA binding protein that binds to the target DNA and activates the MuA transposase (Maxwell et al., 1987; Adzuma and Mizuuchi, 1988).
There are also parallels between the mechanisms of Tn7 and Mu target immunity. A target DNA that contains a copy of Mu will be bound by MuA, which increases the probability of MuA–MuB interactions that provoke MuB to hydrolyze its bound ATP and dissociate from the DNA, resulting in Mu target immunity (Adzuma and Mizuuchi, 1988, 1989; Greene and Mizuuchi, 2002b). Interestingly, TnsB and MuA both interact with their cognate ATP-dependent, DNA binding proteins through residues in their C-terminal domain and, in both cases, a peptide corresponding to this region of TnsB or MuA retains the ability to displace TnsC or MuB from DNA in vitro (Wu and Chaconas, 1994; Levchenko et al., 1997). Additionally, both Tn7 and Mu rely on interactions of the C-terminal regions of TnsB with TnsC or MuA with MuB to mediate assembly of a key transposition intermediate, the transposon–target complex (Mizuuchi et al., 1992; Naigamwalla and Chaconas, 1997; Skelding et al., 2002).
However, in contrast with these similarities, Tn7 and Mu modify their core TnsB–TnsC or MuA–MuB interactions in strikingly different ways to promote transposition into suitable target DNAs and avoid transposition into immune DNA. In the case of Tn7 transposition to attTn7, TnsD-bound attTn7 recruits TnsC to the DNA and a discrete TnsCD nucleoprotein complex is formed (Bainton et al., 1993; Kuduvalli et al., 2001). The TnsCD complex promotes Tn7 insertion at a specific position within the region of DNA bound by TnsC (Kuduvalli et al., 2001).
In contrast, Mu inserts into DNA with much lower specificity, although some regional preferences are observed (Mizuuchi and Mizuuchi, 1993; Manna et al., 2001). Regional specificity is dependent on the activity of MuB, which forms heterogeneous polymers at the particular regions of DNA that are preferred sites of Mu insertion (Greene and Mizuuchi, 2002a,b). Thus the different ways in which TnsC + D and MuB associate with the target DNA enables these proteins to direct insertion into a single site or into many different sites, respectively.
Tn7 and Mu also use different strategies to decide whether a TnsB–TnsC or MuA–MuB interaction will lead to transposition or target immunity. Mu seems to rely on different interactions between two proteins, the MuA transposase and the MuB target-binding transposase activator, to determine whether insertion will occur into a particular target DNA or immunity will be imposed on that DNA. The molecular interactions that determine whether a given MuA–MuB interaction will lead to Mu transposition or dissociation of MuB from the target DNA, and hence target immunity, have yet to be determined.
Tn7, in comparison, has divided its transposase activity between two polypeptides, TnsA and TnsB, each of which depends on the other for its own transposon end-cleavage activity, as well as upon the TnsC + D or TnsC + E target binding proteins. Using its two transposase subunits, Tn7 has the ability to separate the regulation of target immunity from transposition: TnsA + TnsB–TnsC interaction leading to transposition, and TnsB–TnsC interaction leading to target immunity. It will be interesting to explore how TnsA, TnsB and TnsC interactions are balanced to allow Tn7 to decide whether a TnsB–TnsC interaction will activate insertion of Tn7 into a TnsC-bound DNA or impose immunity on that DNA by provoking TnsC dissociation.
Materials and methods
Strains and plasmids
All strains were derived from E.coli NLC28, a valine-resistant derivative of MC4100 F-araD139 Δ(argF-lac)U169 rpsL150 relA1 flb5301 deoC1 ptsF25 rbsR (McKown et al., 1987). NLC51 is a recA56 derivative of NLC28 (McKown et al., 1987). ZS410 is NLC51 bglS-bglB::mTn7kan. The BglS-BglB genes are 8 kb from attTn7 and the mTn7kan has been immobilized by mutation of its terminal dinucleotides (DeBoy and Craig, 1996). ZS410 was created by using P1 transduction to make a recA56 derivative of BD288 (DeBoy, 1997).
The mTn7 reporter element used in the TnsB mutant screen was constructed by inserting the promoterless lacZY BamHI fragment from pOHO (Hughes, 1993) into the SalI site within a mTn7::kan element residing on pBD14 (DeBoy, 1997). The resulting mTn7lac element was inserted via TnsABC + E transposition into pOX-G, a conjugal derivative of the F plasmid, and a Lac– derivative, pZS97, was isolated by a mating-out procedure (Stellwagen and Craig, 1997b).
The transposition functions for the TnsB mutant screen and the lambda hop assay were expressed from a pSC101-based TnsAB plasmid, pZS564 (Skelding et al., 2002), and a pACYC184-based TnsCD plasmid, pZS568 (Skelding et al., 2002). The TnsB immunity bypass mutants were cloned into pZS564 by site-directed mutagenesis creating pZS681 (TnsABP686S), pZS680 (TnsABV689M) and pZS594E (TnsABP690L). A TnsBV689M-intein-chitin-binding-domain (intein-CBD) expression plasmid, pZS627B (New England Biolabs), was derived from pRS513 (Skelding et al., 2002) by site-directed mutagenesis.
The donor plasmid for in vitro transposition was pEMΔ, which contains a kan resistance gene flanked by Tn7L and Tn7R (Bainton et al., 1993). The attTn7-containing target plasmids were pKAO4-3 (McKown et al., 1988), pRM2 (McKown et al., 1988) and pPK13 (Kuduvalli et al., 2001). pZS811 and pLA11 (Bainton et al., 1991) contain Tn7R positioned adjacent to and ∼1 kb from attTn7, respectively. pZS811 was constructed by TnsABC + D transposition of the mTn7 element from pBD14 into the attTn7 site of pPK13, followed by deletion of the SalI–HindIII fragment.
Tns proteins and peptides
TnsA (May and Craig, 1996) and TnsC (Gamas and Craig, 1992) were purified as described, and TnsBwt, TnsBDY682-683AA and TnsBV689M were purified as intein-CBD fusions. Native TnsB WT or mutant was isolated as described elsewhere (Skelding et al., 2002). Native TnsD was isolated by the intein-CBD protocol (New England Biolabs) (R.Mitra and N.Craig, unpublished results). The TnsB peptides (purity ≥ 95%) were obtained from the Johns Hopkins Medical Institute Synthesis facility.
Isolation of TnsB immunity bypass mutants and the lambda hop transposition assay
To generate TnsB immunity bypass mutants we mutagenized pZS564 with 1 M hydroxylamine as described elsewhere (Stellwagen and Craig, 1997b), and then subcloned the SacI–SalI fragment containing TnsB and the portion of TnsA encoding aa190–273 into untreated pZS564. The ligated DNAs were electroporated into ZS410 containing pZS97 and pZS568, and transformants were screened on MacConkey lactose plates.
The lambda hop transposition assay has been described elsewhere (McKown et al., 1988). In this work, transposition was measured in strains NLC51 and ZS410 containing TnsABC + D expressed from various plasmids; all strains contained pZS568 and one of pZS564, pZS681, pZS680 or pZS594E.
TnsB-provoked target ‘inactivation’ and TnsBC + D donor–target complex formation
Each 100 µl reaction presented in Figures 3B and 7A has a composition that mirrors a ‘standard’ TnsABC + D transposition reaction (Skelding et al., 2002). In brief the reaction mixture contained 3.3 nM pRM2, 0.21 nM pEMΔ, 25.7 mM HEPES pH 7.5, 2.01 mM ATP, 1.2% glycerol, 15 mM MgAc, 12 nM TnsA, 6.2 nM TnsB, 4.8 nM TnsC, 3.7 nM TnsD and various salts. TnsC and TnsD were preassembled on attTn7 because these proteins cannot bind to attTn7 in the presence of the Mg2+ required for transposition (Bainton et al., 1993).
We examined the TnsBC + D donor–target complex assembly by slightly modifying the target ‘inactivation’ experiment. In some reactions, 2 mM AMP–PNP was substituted for ATP. After the TnsB incubation, nucleoprotein complexes were captured by adding glutaraldehyde (Sigma) to 0.05% and incubating for 2.5 min at 30°C. Then lysine and Tris pH 8.0 were added to 50 mM each, and the reaction mixture was incubated for 10 min at 22°C. The reaction mixtures were digested with 16 units of PflMI and assayed as described elsewhere (Skelding et al., 2002).
TnsB-provoked TnsC ‘redistribution’
The TnsB-provoked redistribution of TnsC from an attTn7 + Tn7R target to an attTn7 target DNA was nearly identical to a previously described assay (Stellwagen and Craig, 1997a). The only differences were as follows: (i) the ‘immune’ target DNA, pLA11, contained Tn7R, while pRM2L, a plasmid containing Tn7L, was used in the previous work (Stellwagen and Craig, 1997a); (ii) TnsC was redistributed from TnsCD–attTn7 + Tn7R to a TnsD-bound pKAO4-3, while in the earlier work (Stellwagen and Craig, 1997a) TnsCA225V, a mutant that binds to DNA in the absence of TnsD and is able to catalyze transposition as well as respond to target immunity, was redistributed from pRM2L to pKAO4-3.
attTn7 and attTn7::R90 band-shift experiments
The attTn7 and attTn7::R90 target fragments for the band-shift assay were generated by cutting pPK13 and pZS811 with HindIII–XbaI to generate fragments of 205 bp and 222 bp, respectively, which were 3′ end labeled with Klenow DNA polymerase (New England Biolabs). The labeled fragments were isolated by 6% polyacrylamide gel electrophoresis and crush-and-soak extraction (Sambrook et al., 1989).
For the reactions in Figure 5C and D, 1 µg of sheared salmon sperm DNA and ∼0.03 pmol of labeled attTn7::Tn7R or attTn7 target fragment were incubated with 36 nM TnsD and 38 nM TnsC at 30°C in a reaction volume of 18 µl containing 29 mM HEPES, 4.7 mM dithiothreitol, 2.3 mM ATP, 0.08 mM EDTA, 48 mM NaCl, 36 mM KCl, 0.5 mM MgCl, 0.5 mM CHAPS and 5% glycerol. After target complex assembly, MgAc was added to 10 mM and TnsB was added as indicated in the figure, bringing the reaction volume to 20 µl. The products were examined by electrophoresis on a 5% (29:1) (acrylamide:bisacrylamide) 0.5× TBE gel run at 4°C. The gel was dried, and the radioactive species were detected with a phosphoimager and quantified with ImageQuant software.
For the other band-shift reactions, 250 ng (Figure 6A and B) or 750 ng (Figure 7B) of sheared salmon sperm DNA and ∼0.03 pmol of labeled attTn7 DNA were incubated with 31 nM TnsD, 32 nM TnsC and 31 nM TnsA (Figure 7B only) at 30°C in a 21 µl reaction volume containing the buffer components listed above with the following changes: 0.26 mM EDTA, 65 mM NaCl and 71 mM KCl. After target complex assembly, TnsB and MgAc were added as described above, increasing the reaction volume to 25 µl. Tns nucleoprotein complexes were stabilized with 0.008% glutaraldehyde and visualized by 1.2% agarose gel electrophoresis in 1× TBE followed by gel processing as described above.
Analysis of the products of the transposition reactions
Following transposition, the mTn7 donor and target DNAs from the reactions shown in Figures 2, 3 and 7A were purified via phenol/chloroform extraction and then linearized with NdeI. The cut DNAs were visualized by agarose gel electrophoresis and Southern blotting with a mTn7-specific probe followed by quantitative analysis as described elsewhere (Skelding et al., 2002).
Supplementary data
Supplementary data are available at The EMBO Journal Online.
Acknowledgments
Acknowledgements
We thank Dr Prasad Kuduvalli and the Craig laboratory for their criticism of the manuscript. This work was supported by grant P01 CA16519–28 (N.L.C) from the National Institutes of Health. N.L.C. is an investigator with the Howard Hughes Medical Institute.
References
- Adzuma K. and Mizuuchi,K. (1988) Target immunity of Mu transposition reflects a differential distribution of MuB protein. Cell, 53, 257–266. [DOI] [PubMed] [Google Scholar]
- Adzuma K. and Mizuuchi,K. (1989) Interaction of proteins located at a distance along DNA: Mechanism of target immunity in the Mu DNA strand-transfer reaction. Cell, 57, 41–47. [DOI] [PubMed] [Google Scholar]
- Arciszewska L.K. and Craig,N.L. (1991) Interaction of the Tn7-encoded transposition protein TnsB with the ends of the transposon. Nucleic Acids Res., 19, 5021–5029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arciszewska L.K., Drake,D. and Craig,N.L. (1989) Transposon Tn7 cis-acting sequences in transposition and transposition immunity. J. Mol. Biol., 207, 35–52. [DOI] [PubMed] [Google Scholar]
- Bainton R., Gamas,P. and Craig,N.L. (1991) Tn7 transposition in vitro proceeds through an excised transposon intermediate generated by staggered breaks in DNA. Cell, 65, 805–816. [DOI] [PubMed] [Google Scholar]
- Bainton R.J., Kubo,K.M., Feng,J.-N. and Craig,N.L. (1993) Tn7 transposition: target DNA recognition is mediated by multiple Tn7-encoded proteins in a purified in vitro system. Cell, 72, 931–943. [DOI] [PubMed] [Google Scholar]
- Biery M., Lopata,M. and Craig,N.L. (2000) A minimal system for Tn7 transposition: the transposon-encoded proteins TnsA and TnsB can execute DNA breakage and joining reactions that generate circularized Tn7 species. J. Mol. Biol., 297, 25–37. [DOI] [PubMed] [Google Scholar]
- Chaconas G. and Harshey,R.M. (2002) Transposition of phage Mu DNA. In Craig,N.L., Craigie,R., Gellert,M. and Lambowitz,A. (eds), Mobile DNA II. ASM Press, Washington, DC, pp. 384–402. [Google Scholar]
- Craig N.L. (1997) Target site selection in transposition. Annu. Rev. Biochem., 66, 437–474. [DOI] [PubMed] [Google Scholar]
- Craig N.L. (2002) Tn7. In Craig,N.L., Craigie,R., Gellert,M. and Lambowitz,A. (eds), Mobile DNA II. ASM Press, Washington, DC, pp. 423–456. [Google Scholar]
- DeBoy R.T. (1997) Transposon Tn7: target DNAs can modulate Tn7 insertion. In Molecular Biology and Genetics. Johns Hopkins University, Baltimore, MD, p. 250. [Google Scholar]
- DeBoy R. and Craig,N.L. (1996) Tn7 transposition as a probe of cis interactions between widely separated (190 kilobases apart) DNA sites in the Escherichia coli chromosome. J. Bacteriol., 178, 6184–6191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamas P. and Craig,N.L. (1992) Purification and characterization of TnsC, a Tn7 transposition protein that binds ATP and DNA. Nucleic Acids Res., 20, 2525–2532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greene E.C. and Mizuuchi,K. (2002a) Direct observation of single MuB polymers: evidence for a DNA-dependent conformational change for generating an active target complex. Mol. Cell, 9, 1079–1089. [DOI] [PubMed] [Google Scholar]
- Greene E.C. and Mizuuchi,K. (2002b) Dynamics of a protein polymer: the assembly and disassembly pathways of the MuB transposition target complex. EMBO J., 21, 1477–1486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauer B. and Shapiro,J.A. (1984) Control of Tn7 transposition. Mol. Gen. Genet., 194, 149–158. [DOI] [PubMed] [Google Scholar]
- Hickman A.B., Li,L., Mathew,S.V., May,E.W., Craig,N.L. and Dyda,F. (2000) Unexpected structural diversity in DNA recombination: the restriction endonuclease connection. Mol. Cell, 5, 1025–1034. [DOI] [PubMed] [Google Scholar]
- Hughes O. (1993) Host Components of Tn7 Transposition. University of California, San Francisco, CA. [Google Scholar]
- Kleckner N., Chalmers,R., Kwon,D., Sakai,J. and Bolland,S. (1996) Tn10 and IS10 transposition and chromosome rearrangements: mechanism and regulation in vivo and in vitro. Curr. Top. Microbiol. Immunol., 204, 49–82. [DOI] [PubMed] [Google Scholar]
- Kuduvalli P., Rao,J.E. and Craig,N.L. (2001) Target DNA structure plays a critical role in Tn7 transposition. EMBO J., 20, 924–932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levchenko I., Yamauchi,M. and Baker,T.A. (1997) ClpX and MuB interact with overlapping regions of Mu transposase: implications for control of the transposition pathway. Genes Dev., 11, 1561–1572. [DOI] [PubMed] [Google Scholar]
- Lu F. and Craig,N.L. (2000) Isolation and characterization of Tn7 transposase gain-of-function mutants: a model for transposase activation. EMBO J., 19, 3446–3457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manna D., Wang,X. and Higgins,N.P. (2001) Mu and IS1 transpositions exhibit strong orientation bias at the Escherichia coli bgl locus. J. Bacteriol., 183, 3328–3335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxwell A., Craigie,R. and Mizuuchi,K. (1987) B protein of bacteriophage Mu is an ATPase that preferentially stimulates intermolecular DNA strand transfer. Proc. Natl Acad. Sci. USA, 84, 699–703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- May E.W. and Craig,N.L. (1996) Switching from cut-and-paste to replicative Tn7 transposition. Science, 272, 401–404. [DOI] [PubMed] [Google Scholar]
- McKown R.L., Waddell,C.S., Arciszewska,L.A. and Craig,N.L. (1987) Identification of a transposon Tn7-dependent DNA-binding activity that recognizes the ends of Tn7. Proc. Natl Acad. Sci. USA, 84, 7807–7811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKown R.L., Orle,K.A., Chen,T. and Craig,N.L. (1988) Sequence requirements of Escherichia coli attTn7, a specific site of transposon Tn7 insertion. J. Bacteriol., 170, 352–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuuchi M. and Mizuuchi,K. (1993) Target site selection in transposition of phage Mu. Cold Spring Harbor Symp. Quant. Biol., 58, 515–523. [DOI] [PubMed] [Google Scholar]
- Mizuuchi M., Baker,T.A. and Mizuuchi,K. (1992) Assembly of the active form of the transposase-Mu DNA complex: a critical control point in Mu transposition. Cell, 70, 303–311. [DOI] [PubMed] [Google Scholar]
- Naigamwalla D.Z. and Chaconas,G. (1997) A new set of Mu DNA transposition intermediates: alternate pathways of target capture preceding strand transfer. EMBO J., 16, 5227–5234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters J.E. and Craig,N.L. (2001) Tn7 recognizes target structures associated with DNA replication using the DNA binding protein TnsE. Genes Dev., 15, 737–747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyes I., Beyou,A., Mignotte-Vieux,C. and Richaud,F. (1987) Mini-mu transduction: cis-inhibition of the insertion of Mud transposons. Plasmid, 18, 183–192. [DOI] [PubMed] [Google Scholar]
- Robinson M.K., Bennett,P.M. and Richmond,M.H. (1977) Inhibition of TnA translocation by TnA. J. Bacteriol., 129, 407–414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J., Fritsch,E.F. and Maniatis,T. (1989) Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Press, Cold Spring Harbor, NY. [Google Scholar]
- Sarnovsky R., May,E.W. and Craig,N.L. (1996) The Tn7 transposase is a heteromeric complex in which DNA breakage and joining activities are distributed between different gene products. EMBO J., 15, 6348–6361. [PMC free article] [PubMed] [Google Scholar]
- Skelding Z., Sarnovsky,R. and Craig,N.L. (2002) Formation of a nucleoprotein complex containing Tn7 and its target DNA regulates transposition initiation. EMBO J., 21, 3494–3504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stellwagen A. and Craig,N.L. (1997a) Avoiding Self: Two Tn7-encoded proteins mediate target immunity in Tn7 transposition. EMBO J., 16, 6823–6834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stellwagen A. and Craig,N.L. (1997b) Gain-of-function mutations in TnsC, an ATP-dependent transposition protein which activates the bacterial transposon Tn7. Genetics, 145, 573–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stellwagen A. and Craig,N.L. (1998) Mobile DNA elements: controlling transposition with ATP-dependent molecular switches. Trends Biochem. Sci., 23, 486–490. [DOI] [PubMed] [Google Scholar]
- Stellwagen A.E. and Craig,N.L. (2001) Analysis of gain of function mutants of an ATP-dependent regulator of Tn7 transposition. J. Mol. Biol., 305, 633–642. [DOI] [PubMed] [Google Scholar]
- Wolkow C.A., DeBoy,R.T. and Craig,N.L. (1996) Conjugating plasmids are preferred targets for Tn7. Genes Dev., 10, 2145–2157. [DOI] [PubMed] [Google Scholar]
- Wu Z. and Chaconas,G. (1994) Characterization of a region in phage Mu transposase that is involved in interaction with the MuB protein. J. Biol. Chem., 269, 28829–28833. [PubMed] [Google Scholar]