Abstract
Macroautophagy has been implicated as a mechanism of cell death. However, the relationship between this degradative pathway and cell death is unclear as macroautophagy has been shown recently to protect against apoptosis. To better define the inter-play between these two critical cellular processes, we determined whether inhibition of macroautophagy could have both pro-apoptotic and anti-apoptotic effects in the same cell. Embryonic fibroblasts from mice with a knock-out of the essential macroautophagy gene atg5 were treated with activators of the extrinsic and intrinsic death pathways. Loss of macroautophagy sensitized these cells to caspase-dependent apoptosis from the death receptor ligands Fas and tumor necrosis factor-α (TNF-α). Atg5−/− mouse embryonic fibroblasts had increased activation of the mitochondrial death pathway in response to Fas/TNF-α in concert with decreased ATP levels. Fas/TNF-α treatment failed to up-regulate macroautophagy, and in fact, decreased activity at late time points. In contrast to their sensitization to Fas/TNF-α, Atg5−/− cells were resistant to death from menadione and UV light. In the absence of macroautophagy, an up-regulation of chaperone-mediated autophagy induced resistance to these stressors. These results demonstrate that inhibition of macroautophagy can promote or prevent apoptosis in the same cell and that the response is governed by the nature of the death stimulus and compensatory changes in other forms of autophagy. Experimental findings that an inhibition of macroautophagy blocks apoptosis do not prove that autophagy mediates cell death as this effect may result from the protective up-regulation of other autophagic pathways such as chaperone-mediated autophagy.
The relationship between the cellular processes of macroautophagy, quantitatively the most important form of autophagy in mammalian cells, and cell death remains unclear (1). Experimental evidence from several model systems suggests that autophagy is a mechanism of cell death. These findings have led to the concept that programmed cell death can result from one of two distinct pathways, apoptosis or autophagy (2). These two forms of cell death can be distinguished by their morphological differences. Apoptosis is marked by cytoskeletal collapse that leads to cellular condensation and caspase-dependent DNA fragmentation but the relative preservation of organelles. In contrast, the critical initiating feature of autophagic cell death is not cytoskeletal breakdown but organelle degradation. Macro-autophagy has been implicated as a mechanism of cell death from a variety of factors including radiation, tumor necrosis factor-α (TNF-α),3 viruses, and low potassium (3–6). This conclusion is supported by two findings: 1) the presence of increased numbers of autophagosomes in cells responding to these death stimuli, indicative of an up-regulation of macroautophagy that may kill the cell; and 2) the prevention of cell death by inhibition of macroautophagy with 3-methyladenine (3-MA) or RNA interference against autophagy-related genes (ATG) (3–6).
Complicating the classification of macroautophagy as a mechanism of cell death are recent studies indicating that this degradative pathway acts as a cellular survival mechanism (1). Macroautophagy protects against cell death induced by nutrient deprivation, DNA damage, ischemia/reperfusion injury, and endoplasmic reticulum stress (7–11). In all of these examples, macroautophagy was up-regulated in response to the death-inducing stimulus (7–11). The conclusion that the induction of macroautophagy under these conditions is a protective response that inhibits cell death casts doubt on the concept that autophagy promotes cell death.
There are several reasons to question findings that macroautophagy is a mechanism of cell death. First, the fact that autophagy is induced in these models may alternatively represent the activation of a cellular protective mechanism that is ineffective or partially effective in blocking death. Second, the mechanistic investigations implicating autophagy as a form of cell death have often relied on nonspecific pharmacological inhibition of autophagy. In particular, the widely used inhibitor 3-MA inhibits phosphatidylinositol 3-kinase (12), which itself regulates cell death through its downstream effector Akt (13, 14). Investigations with RNA interference-induced genetic knockdowns of autophagic genes have also indicated that autophagy is a mechanism of cell death (15, 16). However, these studies were conducted in cells in which apoptosis had been blocked, raising the possibility that autophagy may be a mechanism of death only in cells in which apoptosis is inhibited and not in normal cells (1). Thus, these studies may not be proof of a normal function of autophagy in cell death.
A simple explanation for the discrepant findings may be that cell type specificity determines whether macroautophagy exerts a pro-or anti-apoptotic effect. Macroautophagy has been reported recently to promote or protect against death from endoplasmic reticulum stress depending on the cell type (9). However, the nature of the stress may also determine the cellular response. It is therefore necessary to examine whether the inhibition of macroautophagy can have different effects in a single cell depending on the type of death pathway that is activated.
An additional reason for the disparate results may be that studies have focused only on macroautophagy without examining the contributions of other autophagic pathways. Chaper-one-mediated autophagy (CMA) is a second stress-induced form of autophagy that is selective for certain soluble cytosolic proteins (17). Recently we also implicated CMA in the regulation of cell death by demonstrating that an impairment in CMA sensitized cells to apoptosis from oxidants and UV light even in the presence of normal macroautophagic function (18). In addition, cross-talk was shown to occur between these two autophagic pathways as the absence of CMA led to a compensatory up-regulation of macroautophagy. Inhibition of this increase in macroautophagy in cells with a blockage in CMA further increased levels of apoptosis, demonstrating that activation of macroautophagy was protective in the absence of CMA. These findings suggest the possibility that the consequences of an inhibition of macroautophagy could vary depending on the ability of the cell to compensate by up-regulating CMA and possibly other lysosomal and nonlysosomal pathways.
One way to reconcile the opposing findings on the function of autophagy in cell death is that there may be no simple, constant relationship between the two. Rather, the interactions may be complex and vary both with cell type and with the nature of the environmental stress, as well as with the ability of the cell to elicit compensatory changes in other forms of autophagy. To test this hypothesis, we examined the effects of activators of the extrinsic and intrinsic death pathways in a well established genetic knock-out of macroautophagy, Atg5−/− mouse embryonic fibroblasts (MEFs) (19, 20). The absence of macroautophagy in Atg5−/− cells sensitized them to apoptosis from the Fas- or TNF-α-induced extrinsic death pathway. In contrast, the loss of macroautophagy was protective against apoptosis from activation of the intrinsic death pathway by menadione-generated oxidative stress and UV light. However, these findings may be explained not by a pro-apoptotic effect of macroautophagy but by a compensatory up-regulation of CMA that led to resistance to oxidative stress and UV light. These findings demonstrate that inhibition of macroautophagy can either promote or protect against cell death in the same cell type depending on the context of the death stimulus and the compensatory changes in other forms of autophagy.
EXPERIMENTAL PROCEDURES
Cells and Culture Conditions
The studies were performed in MEFs derived from wild-type and Atg5−/− mouse embryos (kind gift of Noboru Mizushima, Tokyo Medical and Dental University, Japan) (19, 20), mouse fibroblasts (NIH3T3) from the American Type Culture Collection (Manassas, VA), and NIH3T3 LAMP-2A RNA interference-expressing cells, which were generated as described previously (18). The cells were cultured in Dulbecco’s modified Eagle’s medium (Mediatech Inc., Herndon, VA) supplemented with 15% fetal bovine serum (Atlanta Biologicals, Lawrenceville, GA) and antibiotics (Invitrogen). Experiments were performed in confluent cultures 48 h after cell plating in fresh medium unless otherwise indicated. Cells were treated with Jo2 antibody (0.67µg/ml; BD Biosciences), mouse recombinant TNF-α (10 ng/ml; R&D Systems, Minneapolis, MN), menadione, staurosporine, or tunicamycin (Sigma). Some cells were cotreated with actinomycin D (75 ng/ml; Sigma), the caspase inhibitors Q-Val-Asp-OPh (Q-VD-OPh; MP Biomedicals, Aurora, OH), or IDN-1529 (Pfizer), 3-MA (Sigma), rapamycin (EMD Bioscience, San Diego, CA), leupeptin, or ammonium chloride (Fisher). UV light exposure was performed in a Stratalinker UV cross-linker (Stratagene, La Jolla, CA).
3-(4,5-Dimethylthiazol-2-yl)-2,5-diphenyltetrazolium Bromide (MTT) Assay
Levels of cell death were quantitated by MTT assay (21). After 24 or 48 h of treatment, the cell culture medium was removed, and an equal volume of a 1 mg/ml MTT solution, pH 7.4, in Dulbecco’s modified Eagle’s medium was added to the cells. After incubation at 37 °C for 1 h, the MTT solution was removed, and N-propyl alcohol was added to solubilize the formazan product whose absorbance was measured in a spectrophotometer at a wavelength of 560 nm. The percentage of cell death was calculated by dividing the optical density of a treatment group by the optical density for untreated, control cells, multiplying by 100, and subtracting that number from 100.
Fluorescence Microscopy
The numbers of apoptotic and necrotic cells were quantified by fluorescence microscopy after costaining with acridine orange and ethidium bromide (22), as described previously (23). Cells with shrunken cytoplasm and condensed or fragmented nuclei as determined by acridine orange staining were scored as apoptotic. Necrotic cells were detected by positive staining with ethidium bromide. A minimum of 400 cells/dish were examined, and the numbers of apoptotic and necrotic cells are expressed as a percentage of the total number of cells counted.
Immunofluorescence studies of cultured cells were per-formed following conventional procedures (24). Cells grown on coverslips were treated for the required times and then fixed in 3% formaldehyde. After blocking, the coverslips were incubated with mouse anti-light chain 3 (LC3; MBL International Corp., Woburn, MA), mouse anti-hsc70 (AbCam, Cambridge, MA), or rabbit anti-LAMP-2A (25) antibodies and corresponding fluorescein isothiocyanate-or Cy5-conjugated secondary antibodies. Mito Tracker (Invitrogen) was added directly to the culture medium, and after 15 min at 37 °C, the cells were fixed and mounted. Mounting medium contained 4′,6-diamidino-2-phenylindole (Fisher) to stain and highlight the cell nucleus. Images were acquired with an Axiovert 200 fluorescence microscope (Carl Zeiss, Thornwood, NY) and subjected to deconvolution with the manufacturer’s software. Morphometric analysis was performed with the ImageJ software (National Institutes of Health, Bethesda, MD). The mean distance of lysosomes from the nucleus was calculated in 20 different cells for each condition by tracing lines (six in each cell) from the nuclear membrane to the most distal fluorescent vesicle (lysosome). The distribution of fluorescent vesicles along each line was calculated with the distribution function of the ImageJ program. Distances and numbers of vesicles at each distance for each cell were averaged, and mean values were calculated from the individual distributions in 20 cells. The numbers of mitochondria were calculated after thresholding using the measure function of the ImageJ program, and the percentage of colocalization for LAMP-2A and hsc70 signals was determined using the JACoP plugin. All digital microscopic images were prepared using Adobe PhotoShop 6.0 software (Adobe Systems, Mountain View, CA).
Caspase 8 Activity
Caspase 8 activity was determined in total cellular lysates with the BD ApoAlert™ caspase 8 colorimetric assay kit (BD Biosciences), according to the manufacturer’s instructions.
Protein Isolation and Western Blotting
Total cellular protein was isolated from cells harvested in phosphate-buffered saline, centrifuged, and resuspended in cell lysis buffer containing 20 mm Tris, pH 7.5, 1% Triton X-100, 1 mm EDTA, 1 mm EGTA, and protease and phosphatase inhibitors as described previously (26). The cells were then sonicated, and the lysate was used for Western blotting. Protein concentrations were determined using the Bio-Rad protein assay according to the manufacturer’s instructions.
Western blotting was performed by denaturing 50µg of protein at 100 °C for 5 min in Laemmli sample buffer containing 62.5mm Tris, pH 6.8, 2% SDS, 25% glycerol, 0.01% bromphenol blue, and 5% β-mercaptoethanol. Samples were applied to 12% SDS-polyacrylamide gels and resolved at 100 V over 3 h. Proteins were transferred to nitrocellulose membrane (Schleicher & Schuell) in transfer buffer containing 25mm Tris, pH 8.3, 192 mm glycine, 0.01% SDS, and 15% methanol using a Bio-Rad Trans-blot SD semidry transfer cell to which 150 mA were applied for 90 min. Membranes were blocked in 5% nonfat dry milk, 20 mm Tris, pH 7.5, 500 mm sodium chloride, and 0.5% Tween 20 (TBS-T) for 1 h. Membranes were incubated for 18 h at 4 °C with the following primary antibodies at 1:1,000–1:2,000 dilutions in 5% bovine serum albumin or non-fat milk: rabbit anti-caspase 3, rabbit anti-Atg7, rabbit anti-mammalian target of rapamycin (mTOR), rabbit anti-phospho-mTOR (Cell Signaling, Beverly, MA), rabbit anti-poly(ADP-ribose) polymerase (PARP), rabbit anti-cIAP2, rabbit anti-TRAF2 (Santa Cruz Biotechnology, Santa Cruz, CA), rabbit anti-Beclin 1, rabbit anti-Atg5 (Novus Biologicals, Littleton, CO), and mouse anti-light chain 3 (LC3). Membranes were exposed to goat anti-rabbit or anti-mouse (Kirkegaard & Perry Laboratories, Gaithersburg, MD) secondary antibodies conjugated with horseradish peroxidase at a dilution of 1:10,000 in 5% nonfat milk in TBS-T for 1 h at room temperature. Signals were detected with a chemiluminescence detection system (Western Lightning chemiluminescence Plus, PerkinElmer Life Sciences) and exposure to x-ray film. To ensure that equivalent amounts of protein were loaded among samples, membranes were stripped and immunoblotted with a mouse antibody for β-actin (AbCam). Carbonyl groups on oxidized proteins were detected using the OxyBlot™ oxidized protein detection kit from Chemicon International (Temecula, CA) according to the manufacturer’s instructions.
Mitochondrial and cytosolic protein fractions were obtained by homogenization of cells in buffer A (20mm Hepes, pH 7.5, 250 mm sucrose, 10 mm KCl, 1.5 mm MgCl2, 0.5 mm EDTA, 0.5 mm EGTA, 1 mm 1,4-dithiotheitol, 1 mm phenylmethyl-sulfonyl fluoride) with a Dounce homogenizer and centrifugation of the homogenate at 4 °C for 10 min at 600 × g. The resultant supernatant was centrifuged a second time under the same conditions. This supernatant was centrifuged at 4 °C for 15 min at 10,000 × g. The pellet was redissolved in buffer A and formed the mitochondrial fraction. The supernatant was further centrifuged at 4 °C for 10 min at 10,000 × g. This last supernatant comprised the cytosolic protein fraction. Western blotting was performed as above with mouse anti-cytochrome c (BD Biosciences), mouse anti-Bax, rabbit anti-Bcl-XL (Santa Cruz Biotechnology), rabbit anti-Bad (Cell Signaling), rabbit anti-Bid (kind gift of Xiao-Ming Yin, University of Pittsburgh, PA), and mouse anti-cytochrome oxidase (MitoSciences, Eugene, OR) antibodies.
Adenovirus Preparation and Infection
To inhibit the mitochondrial death pathway, cells were infected with adenoviruses that express Bcl-2 or Bcl-XL (27, 28). Ad5LacZ, which expresses the Escherichia coli β-galactosidase gene (29), served as a control for the nonspecific effects of adenoviral infection. Viruses were amplified in 293 cells, purified by banding twice on CsCl gradients as described previously (30), and titered by plaque assay. Infections were performed as described previously (23), at a multiplicity of infection of 20.
ATP Assay
Intracellular ATP concentrations were determined by the Roche Applied Science ATP bioluminescence assay kit HS II using the manufacturer’s instructions. Briefly, the reaction was initiated by the addition of 50µl of the reagent containing luciferase and luciferin to an equal volume of the blank, standard, or sample by automated injection. The ATP-dependent luciferase-catalyzed bioluminescence was determined in a 96-well plate luminometer using a light integration time of 3 s following a delay of 1 s. ATP levels were determined from a standard curve prepared from the manufacturer’s standard.
Intracellular Protein Degradation
To measure the degradation of long-lived proteins, cells were labeled with [3H]leucine (2 µCi/ml) for 48 h at 37 °C. The cells were then extensively washed and maintained in complete medium containing an excess of unlabeled leucine (2.8 mm) to prevent reutilization of radiolabeled leucine (31). Aliquots of medium were removed at different times, proteins in the medium were precipitated with trichloroacetic acid, and the amount of proteolysis was expressed as the percentage of the initial acid-insoluble radioactivity (protein) transformed into acid-soluble radioactivity (amino acids and small peptides) at different times. Total radioactivity incorporated into cellular proteins was determined as the amount of acid-precipitable radioactivity in labeled cells after washing. Where indicated, 20 mm ammonium chloride and 100µm leupeptin or 10mm3-MA were added to the culture medium during the chase. Lysosomal degradation was calculated as the percentage of protein degradation sensitive to ammonium chloride/leupeptin. The lysosomal protein degradation inhibited by 3-MA was attributed to macroautophagy.
Statistical Analysis
All numerical results are reported as mean ± S.E. and represent data from a minimum of 3 independent experiments unless otherwise stated. Groups were compared by the Student’s t test. Statistical significance was defined as p < 0.05. Calculations were made with SigmaPlot (Jandel Scientific, San Rafael, CA).
RESULTS
Atg5−/− MEFs Are Sensitized to Death Receptor Ligand-mediated Apoptosis
Apoptosis is divided mechanistically into the extrinsic and intrinsic death pathways with the primary extrinsic forms being Fas and TNF-α death receptor-mediated. To determine the function of macroautophagy in death receptor-induced apoptosis, the toxicity of both the Fas agonist antibody Jo2 and TNF-α was examined in wild-type and Atg5−/− MEFs. Jo2 treatment induced a low level of cell death in wild-type MEFs as determined by a 24-h MTT assay (Fig. 1A). In contrast, Atg5−/− MEFs were highly sensitive to Jo2 killing with a 6-fold increase in cell death over that in wild-type cells at 24 h (Fig. 1A). Wild-type MEFs were completely resistant to TNF-α toxicity (Fig. 1A). However, TNF-α induced a significant amount of cell death in Atg5−/− cells at 24 h (Fig. 1A). To ensure that wild-type MEFs had an intact TNF-α death pathway, the cells were sensitized to TNF-α toxicity by cotreatment with the RNA synthesis inhibitor actinomycin D. Wild-type and Atg5−/− MEFs both underwent cell death from actinomycin D/TNF-α cotreatment (data not shown).
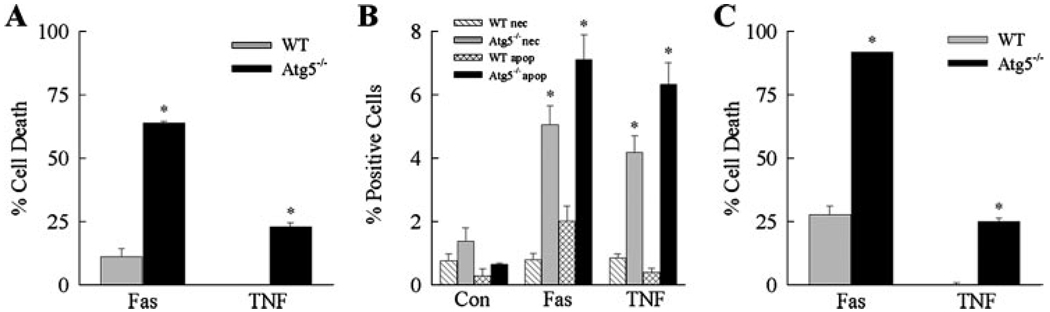
FIGURE 1. Loss of Atg5 sensitizes MEFs to death from Jo2 antibody and TNF-α.
Wild-type (WT) and Atg5−/− MEFs were treated with Jo2 antibody (Fas) or TNF-α for various times. A, the percentage cell death was determined by MTT assay at 24 h. Data are from 6 independent experiments (*, p < 0.0001 as compared with wild-type cells). B, cells that were untreated (Con) or treated with Jo2 or TNF-α for 12 h were costained with acridine orange and ethidium bromide, and the percentage of necrotic (nec) and apoptotic (apop) cells was determined by fluorescence microscopy as described under “Experimental Procedures.” Results are from 3 independent experiments performed in duplicate (*, p<0.0001 as compared with the same type of cell death in wild-type cells). C, the percentage cell death was determined by MTT assay after 48 h of treatment. Data are from 4 experiments performed in duplicate (*, p < 0.0001 as compared with wild-type cells).
To confirm this differential death receptor response in wild-type and Atg5−/− cells by a second cell death assay, the steady-state numbers of apoptotic and necrotic cells were determined by acridine orange/ethidium bromide costaining and fluorescence microscopy. When untreated, both cell types had low levels of apoptosis and necrosis (Fig. 1B). Wild-type cells had a small increase in the number of apoptotic cells at 12 h after Jo2 treatment and no change with TNF-α treatment (Fig. 1B), findings that paralleled those of the MTT assay. Atg5−/− cells had markedly increased steady-state levels of both apoptotic and necrotic cells at 12 h after Jo2 or TNF-α treatment (Fig. 1B). The nuclei of most of the ethidium bromide-positive cells were condensed, suggesting that they were actually apoptotic cells that had undergone secondary necrosis. These data together with those of the MTT assays support the conclusion that macroautophagy is critical for resistance to death receptor ligand-induced cell death in MEFs.
To ensure that the absence of Atg5 in fact sensitized MEFs to death receptor-mediated cell death and did not merely accelerate an existing death pathway, levels of cell death were also examined at 48 h after Jo2 or TNF-α treatment. Over the second 24-h period after Jo2 treatment, the amount of cell death increased moderately in wild-type and Atg5−/− cells but remained significantly increased in the knock-out cells as compared with wild-type MEFs (Fig. 1C). Wild-type cells were completely resistant to TNF-α toxicity even after 48 h of treatment, whereas the amount of death in Atg5−/− cells remained unchanged (Fig. 1C). Thus, the absence of Atg5 and macroautophagy did not merely accelerate death from Fas and TNF-α but acted to sensitize MEFs to these death stimuli.
Loss of Atg5 Sensitizes MEFs to Caspase-dependent Fas- and TNF-α-induced Apoptosis
The death receptor ligands Fas and TNF-α trigger activation of the initiator caspase 8 that induces cleavage and activation of the downstream effector caspases 3 and 7, which then mediate apoptosis (32, 33). To determine whether Atg5−/− cell death from Fas and TNF-α involved this classical death pathway, caspase activation was examined in Jo2- and TNF-α-treated cells. Caspase 8 activation, as determined by activity assay, was similarly increased in wild-type and Atg5−/− cells 8 h after Fas treatment (Fig. 2A). However, by 12 h, the level of activation decreased in wild-type cells, whereas caspase 8 activity remained significantly increased in knock-out cells (Fig. 2A). Consistent with this result was the finding that activation of downstream caspase 3 was detectable in Atg5−/− but not wild-type cells, as indicated by the presence of cleaved caspase 3 on immunoblots (Fig. 2B). Supportive of increased activation of downstream caspases in Atg5−/− MEFs was that PARP cleavage was only detectable in these cells (Fig. 2B). Identical to the findings with Jo2 administration, caspase 3 and PARP cleavage were detected in TNF-α-treated Atg5−/− cells but not in wild-type cells (Fig. 2C).
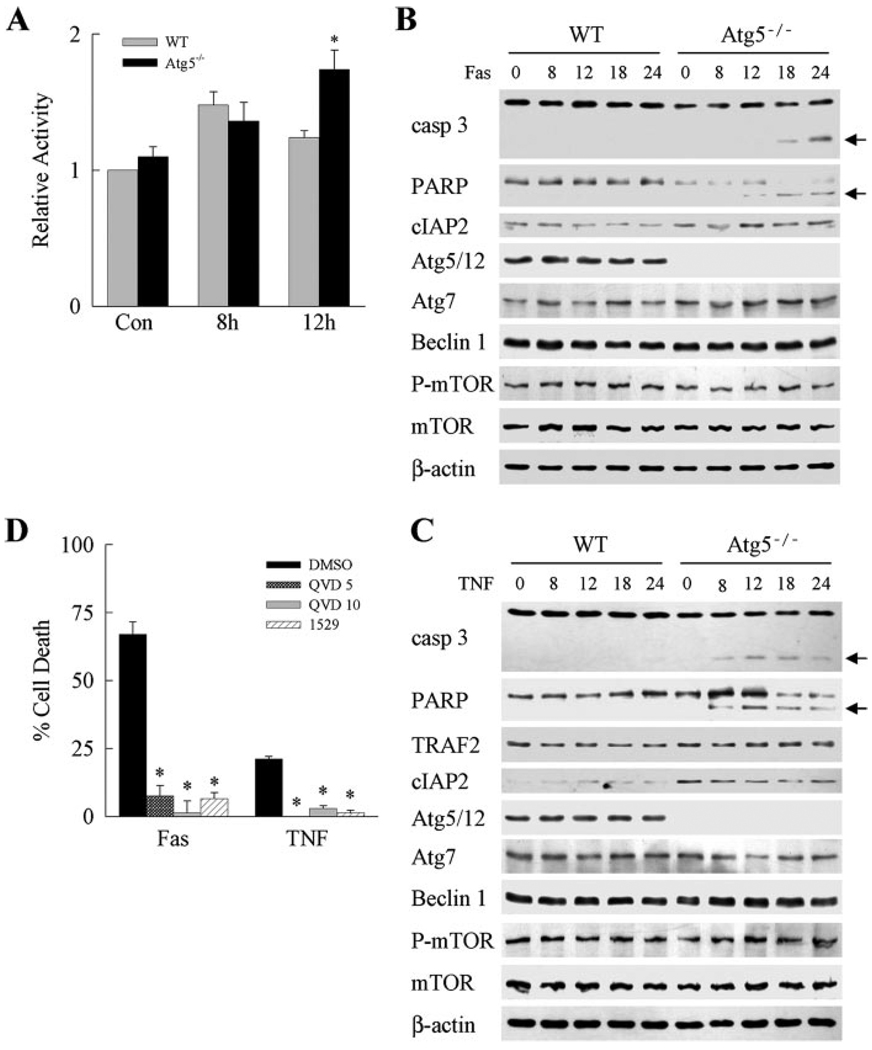
FIGURE 2. Death receptor-induced Atg5−/− cell death results from caspase-dependent apoptosis.
A, caspase 8 activity was assayed in wild-type (WT) and Atg5−/− MEFs treated with Jo2 antibody (Fas) for 8 or 12 h. Results are from 4 experiments (*, p < 0.01 as compared with wild-type cells). Con, untreated. B and C, protein was isolated from untreated wild-type and Atg5−/−cells and cells treated with Jo2 (Fas) (B) or TNF-α (C) for the indicated hours. Aliquots of protein were immunoblotted with antibodies for caspase 3 (casp 3), PARP, cIAP2, TRAF2, Atg5, Atg7, Beclin 1, phospho-mTOR (P-mTOR), and mTOR. For Atg5, the band corresponding to the Atg5/12 conjugate protein is shown. Stripped membranes were reprobed with β-actin as a measure of equivalent protein loading among samples. The caspase 3 and PARP cleavage products are indicated by arrows. Immunoblots are representative of 3 independent experiments. D, Atg5−/− cells were pretreated with Me2SO (DMSO) vehicle, 5 (QVD 5) or 10 (QVD 10) µm Q-VD-OPh, or 25 µm IDN-1529 (1529). Cells were then treated with Jo2 antibody (Fas) or TNF-α, and the percentage of cell death was determined by MTT assay at 24 h. Results are from 3 independent experiments (*, p < 0.00001 as compared with Me2SO-reated cells).
The effect of caspase inhibition on Jo2- and TNF-α-induced death was examined to determine the function of caspases in the sensitization of Atg5−/− cells to death receptor-mediated apoptosis. Caspase inhibitors decreased both Jo2-induced and TNF-α-induced cell death in Atg5−/− cells by 90–100% (Fig. 2D). These data indicate that Atg5−/− MEFs were sensitized to the classical, caspase-dependent pathway of death receptor-induced apoptosis.
In Response to Jo2 or TNF-α, Atg5−/− MEFs Have Increased Activation of the Mitochondrial Death Pathway
An important mechanism of Fas- and TNF-α-induced caspase-dependent cell death is activation of the mitochondrial death pathway (34, 35). Critical to mitochondrial death pathway activation is the cleavage and mitochondrial translocation of the pro-apoptotic Bcl-2 family members Bid and Bad (36, 37). This event induces mitochondrial changes that promote cytochrome c release and trigger the activation of downstream effector caspases. The possibility that increased caspase 3 cleavage in Jo2- and TNF-α-treated Atg5−/− MEFs resulted from overactivation of the mitochondrial death pathway was investigated. After Jo2 antibody treatment, mitochondrial cytochrome c release occurred in Atg5−/− cells but not wild-type cells, as indicated by the cytosolic presence of cytochrome c in conjunction with decreased mitochondrial levels of this protein on immunoblots (Fig. 3A). Mitochondrial translocation of cleaved Bid was also detected in Jo2-treated Atg5−/− cells but not in wild-type MEFs (Fig. 3A). No change was detected in cytosolic or mitochondrial levels of Bad, Bax, or the anti-apoptotic protein Bcl-XL (Fig. 3A). The purity of the fractions was demonstrated by the exclusive presence of cytochrome oxidase in the mitochondrial isolates (Fig. 3A). Similar findings of differential mitochondrial death pathway activation, as indicated by cytosolic cytochrome c release and mitochondrial translocation of truncated Bid, were found in TNF-α-treated Atg5−/− but not wild-type MEFs (Fig. 3B). Thus, the increased cleavage of caspase 8 that occurred in Atg5−/− cells was associated with augmented Bid cleavage and activation of the mitochondrial death pathway.
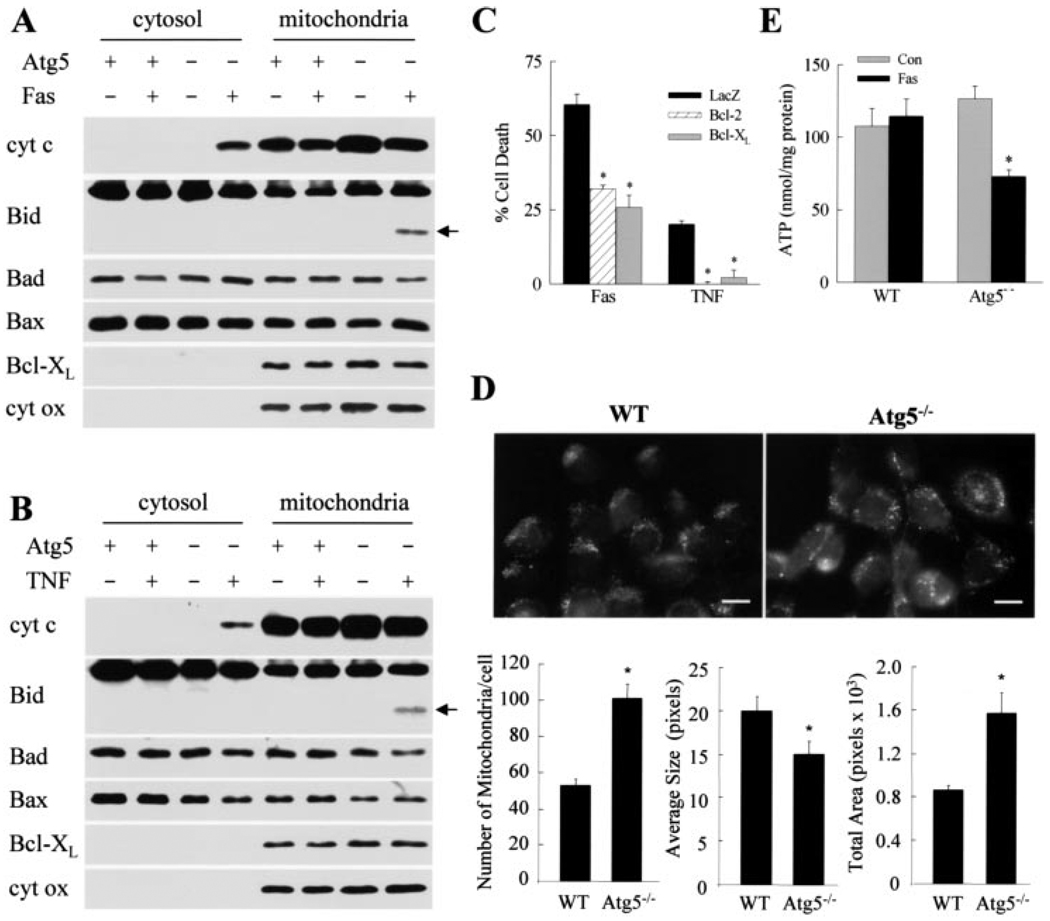
FIGURE 3. Mitochondrial death pathway activation is increased in Atg5−/− cells after death receptor stimulation.
Cytosolic and mitochondrial proteins were isolated from wild-type (Atg5+) and Atg5−/− (Atg5−) cells that were untreated or treated with Jo2 (Fas) (A) or TNF-α (B) and immunoblotted with antibodies for cytochrome c (cyt c), Bid, Bad, Bax, or Bcl-XL. Stripped membranes were reprobed for cytochrome oxidase. Bid cleavage product is indicated by an arrow. C, Atg5−/− cells were infected with adenoviruses that express β-galactosidase (LacZ), Bcl-2, or Bcl-XL. Cells were then treated with Jo2 (Fas) or TNF-α, and the percentage of cell death was determined at 24 h by MTT assay. Data are from 4 independent experiments (*, p < 0.0002). Untreated, wild-type (WT) and knock-out MEFs were stained with Mito Tracker to highlight mitochondria. Top, representative field (bar: 10µm). Bottom, number of mitochondria per cell, average mitochondrial size, and the total cellular area occupied by mitochondria were quantitated. Values are the means of 6–8 fields such as the ones shown (*, p < 0.01 as compared with wild-type cells). E, ATP levels in wild-type and knock-out MEFs untreated (Con) and treated with Jo2 (Fas) for 24 h. Results are from 4 independent experiments (*, p < 0.001 as compared with control cells).
To examine whether mitochondrial death pathway activation led to Atg5−/− cell death from Fas and TNF-α, the effects of adenoviral Bcl-2 and Bcl-XL overexpression on cell death were investigated. Death from Jo2 was inhibited ~50% by Bcl-2 or Bcl-XL overexpression, whereas TNF-α-induced cell death was completely blocked by adenoviral expression of either protein (Fig. 3C). Overactivation of the mitochondrial death pathway therefore played a mechanistic role in the sensitization of Atg5−/− MEFs to death from Fas and TNF-α.
Steady-state levels of mitochondrial cytochrome c were increased in untreated Atg5−/− cells as compared with wild-type cells by immunoblotting (Fig. 3, A and B), suggesting that the knock-out cells may have more mitochondria. This finding was confirmed by fluorescence microscopy with the mitochondrial fluorophore MitoTracker, which demonstrated significantly increased numbers of mitochondria per cell in untreated Atg5−/− cells and a net increase in the percentage of cellular area occupied by mitochondria despite a smaller average mitochondrial size (Fig. 3D). These data are consistent with the fact that removal of excessive or dysfunctional mitochondria is impaired in Atg5−/− cells that lack macroautophagy (38). To assess whether defective removal resulted in Atg5−/− cell mitochondrial dysfunction in response to death receptor stimulation, cellular ATP levels were examined after Jo2 treatment. ATP levels 24 h after Jo2 administration were unchanged in wild-type cells but decreased 45% in Atg5−/− cells (Fig. 3E). Mitochondrial function was therefore significantly impaired in Atg5−/− cells in response to stimulation of the Fas death receptor pathway.
Levels of Pro- and Anti-apoptotic Proteins Are Equivalent in Wild-type and Atg5−/− MEFs
The differential death effects of Jo2 and TNF-α in wild-type and Atg5−/− cells could reflect altered expression of a pro- or anti-apoptotic factor critical to both death pathways. However, levels of the pro-apoptotic proteins TRAF2, Bid, Bad, and Bax and the anti-apoptotic proteins cIAP2 and Bcl-XL were equivalent in wild-type and Atg5−/− cells (Fig. 2, B and C, and Fig. 3, A and B). Thus, changes in the levels of apoptotic pathway proteins did not mediate the altered death responses in Atg5−/− cells.
Death Receptor Ligands Do Not Increase Levels of Autophagy
Although autophagy was initially considered a stress-induced cellular pathway, recent evidence supports a constitutive function for this process as well (18, 38–40). Basal levels of autophagy may be sufficient to maintain MEF resistance to death receptor-induced apoptosis. Alternatively, active up-regulation of autophagy by death receptor ligands may be required for protection from cell death, and stimuli of both the extrinsic and the intrinsic death pathways have been reported to increase levels of autophagy (5, 41, 42). To distinguish between these two possibilities, we examined whether Jo2 or TNF-α increased levels of autophagy in wild-type MEFs. As a measure of autophagy, rates of degradation of long-lived proteins that are the typical substrates for lysosomal degradation were compared in untreated and Jo2- or TNF-α-treated cells. Instead of an increase in protein degradation that would be expected with an up-regulation of autophagy, the rate of long-lived protein degradation was unchanged until 24 h after Jo2 or TNF-α treatment, at which time a slight decrease occurred (supplemental Fig. 1, A and B). This small but significant decrease in protein degradation was mostly due to reduced rates of lysosomal degradation (as defined by sensitivity to inhibition by ammonium chloride/leupeptin) (supplemental Fig. 1, C and D). Interestingly, although the changes in the absolute amounts of protein degradation were small, death receptor stimulation induced a switch in the type of autophagy responsible for this degradation by 24 h. In untreated cells, 25–30% of total lysosomal degradation was mediated by macroautophagy (as determined by the percentage that was inhibited by 3-MA). In contrast, at 24 h after treatment with Jo2 or TNF-α, a significant reduction in 3-MA-sensitive proteolysis was observed (supplemental Fig. 1, E and F). These treatments did not affect the total rates of protein degradation in Atg5−/− cells that lack macro-autophagy (supplemental Fig. 2), further confirming that the inhibitory effect of both death stimuli was preferentially on macroautophagy. The failure of MEFs to up-regulate macroautophagy in response to Jo2/TNF-α treatment supports the conclusion that basal levels of macroautophagy mediate resistance to death receptor-induced apoptosis. In addition, the switch in the form of autophagy suggests that up-regulation of alternative types of autophagy such as CMA may also affect the cellular response to these death stimuli.
For additional evidence of the failure of Jo2 and TNF-α to up-regulate macroautophagy, the effects of these treatments on critical regulators and effectors of macroautophagy were determined by immunoblotting. Atg5 was not detected in the knock-out cells, and levels of the Atg5/12 conjugate, required for the formation of autophagosomes, were unchanged in wild-type cells by either Jo2 or TNF-α treatment (Fig. 2, B and C). Protein levels of Atg7, a limiting ligase for macroautophagy, the macro-autophagy activator Beclin 1, and inhibitor of autophagy phospho-mTOR, were equivalent in the two cell types and unaffected by death receptor ligand stimulation (Fig. 2, B and C).
As a final confirmation that resistance to Jo2/TNF-α in wild-type MEFs occurred in the absence of any increase in macroautophagy, cellular levels of LC3 after Jo2 and TNF-α treatment were examined by immunoblotting. A critical event in macroautophagy is the conversion of microtubule-associated protein 1 LC3 from the free LC3-I form to the membrane-bound LC3-II form (43). Intracellular levels of LC3-II there-fore serve as an index of the steady-state level of autophagosomes in the cell, whereas turnover of LC3-II can be used as index of autophagosome clearance rates (44). Immunoblot analysis of the amount of cellular conversion of LC3 from the I to II form was employed to determine whether death receptor ligands induced macroautophagy in MEFs. Levels of LC3-I and LC3-II were unchanged in wild-type MEFs at 4 and 24 h after Jo2 or TNF-α treatment by immunoblotting (Fig. 4, A and B). Despite the observed decrease in macroautophagy-dependent protein degradation, conjugated LC3-II was still detected at 24 h after the Jo2/TNF-α treatment, demonstrating that the ability to form autophagosomes was preserved in these cells. Turnover of LC3-II, measured as the increase in LC3-II when lysosomal proteolysis was inhibited by ammonium chloride and leupeptin, was comparable in control and treated cells (Fig. 4, A and B). Interestingly, as reported previously for TNF-α in other cells (45), the number of autophagosomes was higher in Jo2/TNF-α-treated cells as determined by immunofluorescence microscopy with an anti-LC3 antibody (Fig. 4, C and D). The fact that the LC3-positive vesicles were slightly smaller in size in the treated cells as compared with untreated controls (Fig. 4D) may explain the finding that LC3 levels were comparable in untreated and treated cells despite the slight increase in auto-phagosome number. This could also be the reason why LC3 turnover remains constant but the net degradation of long-lived proteins does eventually decrease. In addition, treatment with rapamycin, which up-regulates macroautophagy in MEFs by inhibiting activity of the macroautophagy inhibitor mTOR (46), failed to induce macroautophagy in TNF-α-treated cells (supplemental Fig. 3). These data indicate that apoptotic stimuli eventually impair both basal and inducible levels of macroautophagy.
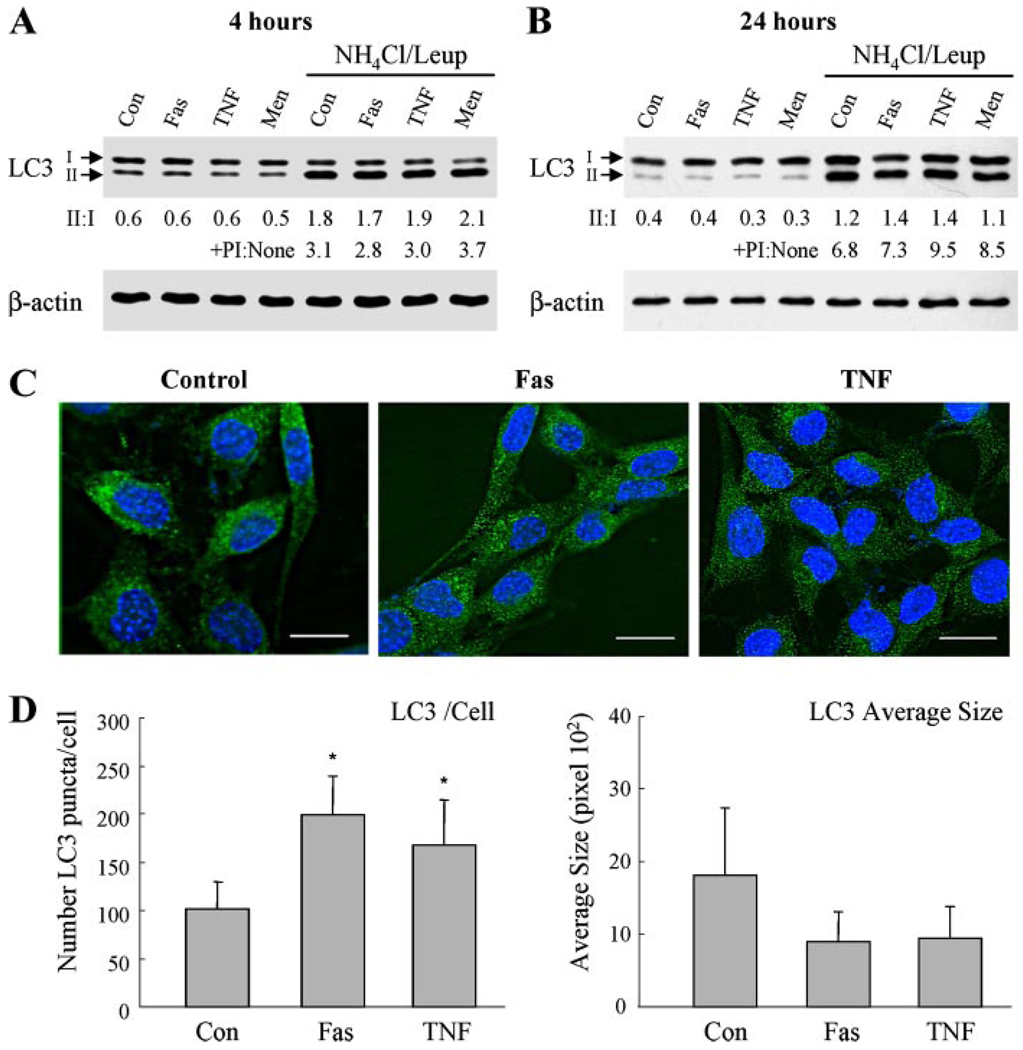
FIGURE 4. Jo2 and TNF-α induce changes in the macroautophagic compartment.
A and B, MEFs treated with Jo2 (Fas), TNF-α, or menadione (Men) for 4 (A) or 24 h (B) were subjected to immunoblotting with an antibody for LC3. Where indicated, 20mm ammonium chloride and 100µm leupeptin (NH4Cl/Leup) were added to the incubation medium. Stripped membranes were reprobed for β-actin. The unconjugated (I) and conjugated (II) LC3 forms are indicated. The LC3-II to LC3-I ratio (II:I) and the -fold increase in LC3-II in the presence of the protease inhibitors (+PI:None) are shown. Values are the means from densitometric scans of immunoblots from 3 independent experiments. Con, untreated. C and D, immunofluorescence staining for LC3 in the same cells. C, representative fields are shown (bar: 5µm). D, the numbers of fluorescent punctate structures per cell (left) and the average size of the LC3-positive puncta (right) were determined in 20 cells/experiment. Data are from 3 independent experiments (*, p < 0.05 between control and Fas or TNF-α treatment).
Inhibition of Macroautophagy Both Protects against and Sensitizes to Stimuli of the Intrinsic Death Pathway
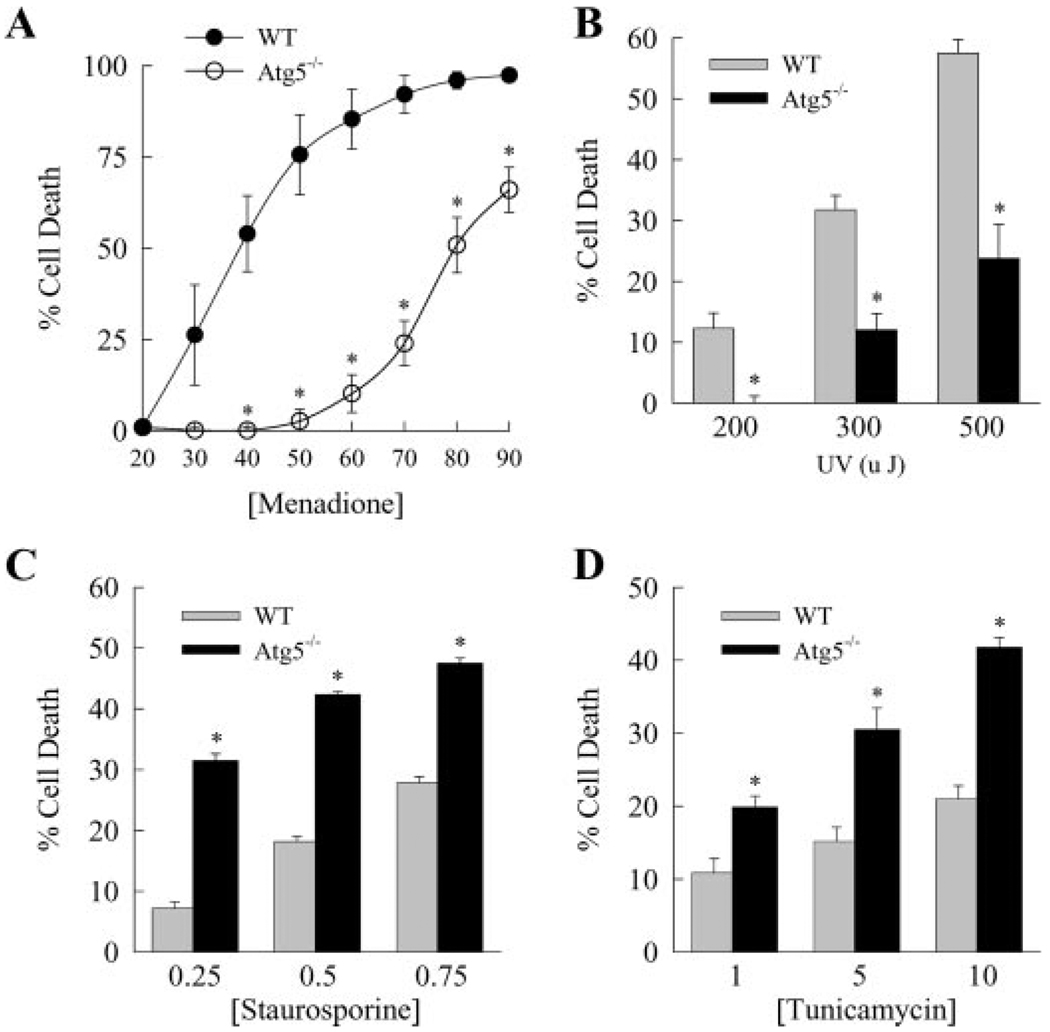
The engulfment of cytosolic components by macroautophagy may act as a generalized protective response against any form of apoptosis in MEFs or specifically mediate resistance against death receptor ligand-induced apoptosis. To distinguish between these possibilities, the effects of Atg5 loss on death from specific stimuli of the intrinsic death pathway were examined. First, increasing levels of oxidative stress were induced in wild-type and Atg5−/− cells by the superoxide generator menadione (47, 48). In direct opposition to the findings for death receptor ligand-induced apoptosis, Atg5−/− cells were significantly more resistant to death from menadione-induced oxidative stress (Fig. 5A). Atg5−/− cells were completely resistant to concentrations of menadione that induced death in 75% of wild-type cells (Fig. 5A). Similarly, the loss of Atg5 sensitized the cells to death from various doses of UV light (Fig. 5B). A possible mechanism for resistance to these two death stimuli could be an inability of the knock-out cells to generate an Atg5 cleavage product that has been reported to directly promote apoptosis through an autophagy-independent mechanism (49). However, no Atg5 cleavage fragment was detected in menadione-treated wild-type MEFs (data not shown), indicating that loss of this direct pro-apoptotic effect of Atg5 could not explain the resistance of Atg5−/− cells to death from menadione.
FIGURE 5. Loss of Atg5 protects against toxicity from menadione and UV light but not from staurosporine- and endoplasmic reticulum stress-induced death.
Wild-type (WT) and Atg5−/− MEFs were treated with the indicated µm concentrations of menadione (A), energy levels of UV light (B), and µm concentrations of staurosporine (C) or tunicamycin (D). The percentage cell death was determined by MTT assay at 6 h for staurosporine and at 24 h for the other treatments. The data are from 3–4 independent experiments (*, p < 0.001 as compared with wild-type cells).
In contrast to the findings for menadione and UV light, Atg5−/− MEFs were sensitized to death from the intrinsic death pathway activator staurosporine. The percentages of cell death over a range of concentrations of the protein kinase inhibitor staurosporine were significantly greater in knock-out MEFs as compared with wild-type cells (Fig. 5C). In addition, Atg5 ablation sensitized the cells to death from the endoplasmic reticulum stress inducer tunicamycin (50). Tunicamycin induced a dose-dependent cell death in wild-type cells that was significantly increased in Atg5−/− cells at every concentration of tunicamycin (Fig. 5D). These findings are consistent with prior reports in other cell types that macroautophagy protects against death from endoplasmic reticulum stress (9, 11). Thus, macroautophagy regulates MEF death pathways other than those mediated by death receptor ligands, but a loss of macroautophagy can either increase or decrease the amount of cell death depending on the apoptotic stimulus.
CMA Mediates Cellular Resistance to Death from Menadione
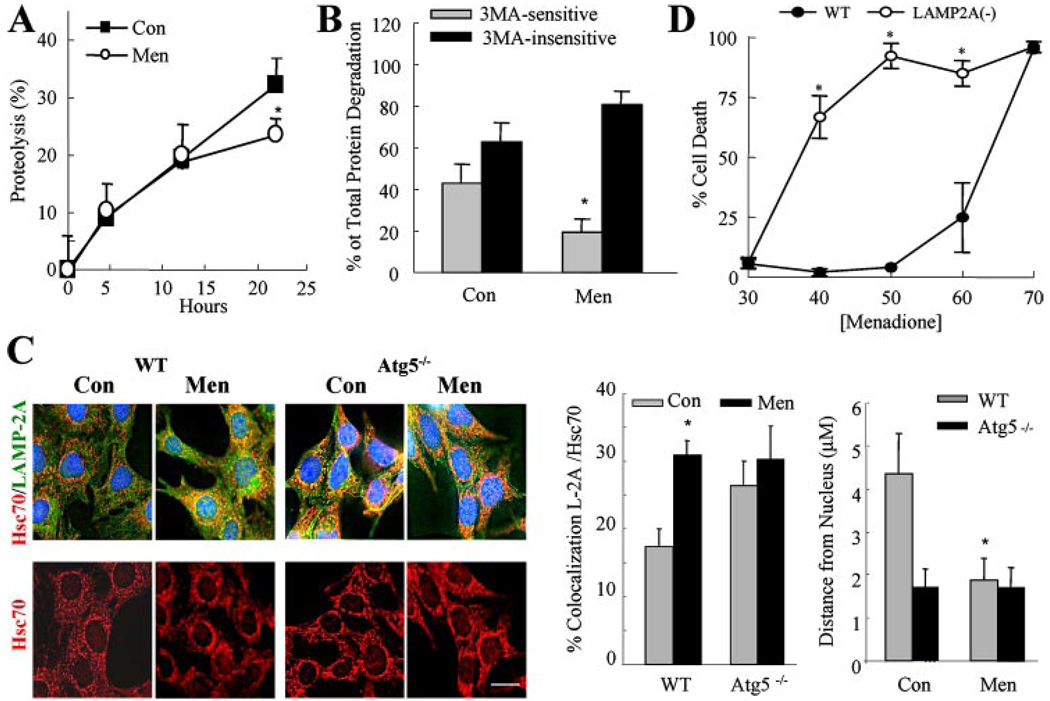
The increased resistance of Atg5−/− cells to death from menadione and UV light despite their increased sensitivity to death receptor-mediated apoptosis could be explained by the ability of macroautophagy to both promote and inhibit distinct cell death pathways. Alternatively, the explanation may be more complex with the cellular death response dependent on the interplay between the various forms of autophagy. The later possibility was suggested by two findings: 1) data in the present study that the contributions of the different types of autophagy to protein degradation change in response to death stimuli (supplemental Fig. 1, E and F); and 2) that the absence of macroautophagy in Atg5−/− MEFs induces an up-regulation of CMA.4 We have previously demonstrated that CMA contributes to the removal of oxidized proteins (24) and that the integrity of this pathway is critical for the preservation of cell viability during mild oxidative stress (18). Menadione-induced oxidative stress did not significantly alter the rate of total protein degradation until 24 h, at which time menadione had an inhibitory effect on macroautophagy similar to that described for Jo2 and TNF-α (Fig. 6, A and B). We confirmed that the menadione-induced switch from macroautophagy to 3-MA-insensitive forms of lysosomal protein degradation reflected an up-regulation of CMA activity. Menadione-treated wild-type MEFs had increased levels of CMA-active lysosomes as demonstrated by increased colocalization of LAMP-2A and hsc70 by immunofluorescence (Fig. 6C, top panels). In addition, with menadione treatment, CMA-active lysosomes were mobilized from the cell periphery to the perinuclear area (Fig. 6C, bottom panels), a process known to occur during maximal CMA activation (18, 51). As we have recently demonstrated,4 the numbers of CMA-positive lysosomes and those with a perinuclear location were increased in Atg5−/− MEFs as compared with wild-type cells under basal conditions and remained elevated after treatment with menadione (Fig. 6C).
FIGURE 6. CMA up-regulation protects against menadione toxicity.
A, total rates of protein degradation in wild-type MEFs untreated (Con) or treated with menadione (Men) were calculated as described under “Experimental Procedures.” Data are from 3 independent experiments with triplicate wells for each experiment (*, p< 0.05 as compared with control). B, macroautophagic activity in wild-type MEFs was calculated as the percentage of lysosomal degradation sensitive to inhibition by 3-MA in the same experiments (*, p<0.01 as compared with control). C, intracellular distribution of LAMP-2A/hsc70 enriched lysosomes in wild-type (WT) and Atg5−/− MEFs untreated or treated with menadione. Top, representative image of double labeled cells. Bottom, representative image of the distribution of hsc70 puncta (bar: 5 µm). Right, the percentage of LAMP-2A and hsc70 colocalization and the distance of hsc70-containing lysosomes from the nucleus. Data are from the quantification of six different fields (average 20 cells total) in 2 independent experiments (*, p<0.01 as compared with untreated wild-type cells). D, LAMP-2A RNA interference NIH3T3 cells were treated with menadione, and the percentage of cell death was determined by MTT assay at 24 h. Results are from 4 independent experiments (*, p < 0.001 as compared with wild-type cells).
Activation of CMA to compensate for the lack of macroautophagy in Atg5−/− MEFs suggested a mechanism for the resistance of these cells to menadione-induced oxidative stress. Increased levels of CMA in these cells did inhibit the effects of oxidative stress as reflected in a reduction in the content of oxidized proteins in Atg5−/− MEFs as compared with wild-type cells (supplemental Fig. 4). To confirm that CMA up-regulation could explain the resistance of Atg5−/− cells to toxicity from menadione, the effect of the inhibition of CMA on cellular sensitivity to menadione killing was examined. CMA blockage was attained through RNA interference against LAMP-2A in NIH3T3 cells, as described previously (18).
Inhibition of CMA by LAMP-2A knockdown significantly increased cell sensitivity to menadione toxicity (Fig. 6D). Similarly, we have previously reported that inhibition of CMA sensitizes to death from UV light (17). Cross-talk between the two forms of autophagy such that down-regulation of macroautophagy results in increased CMA could therefore explain the ability of Atg5−/− cells to be both sensitized to death receptor-mediated apoptosis and resistant to death from menadione and UV light.
DISCUSSION
The results demonstrate for the first time that a knock-out of macroautophagy can both promote and block cell death in the same cell type challenged with different stressors. Loss of macroautophagy sensitized MEFs to death via the extrinsic pathway but made them resistant to intrinsic pathway-mediated death from oxidative stress and a DNA-damaging agent. Thus, the findings of opposing effects of a loss of macroautophagy on cell death in different cell types (3–11, 15, 16) likely do not represent inherent cell type-specific functions of autophagy. That is, an inhibition of macroautophagy does not always promote death in one cell type and prevent death in a second, different cell type. Rather, a decrease in macroautophagy can serve to promote death or survival in the same cell depending on the nature of the death stimulus and the ability of the cell to compensate for the loss of macroautophagy.
Prior studies examining the relationship of macroautophagy to death receptor ligand-induced apoptosis have demonstrated an increase in TNF-α-induced death in muscle and leukemia cell lines (5, 52). However, MEFs that were resistant to death from Fas/TNF-α became sensitized to death receptor ligand cytotoxicity with a loss of macroautophagy. Death was apoptotic as indicated by activation of caspase 8 and the mitochondrial death pathway, the induction of effector caspase cleavage, and the blockage of death by overexpression of anti-apoptotic Bcl-2 family members or treatment with caspase inhibitors. Caspase 8 activation occurred in wild-type and knock-out cells in response to Jo2 antibody, but the period of activation was more prolonged in Atg5−/− cells. As a result, cleavage of the downstream effector caspase 3 and the caspase substrate PARP occurred in Atg5−/− cells but not wild-type cells after Jo2 antibody or TNF-α treatment. Thus, the loss of macroautophagy sensitized MEFs to death by the classical, caspase-dependent apoptotic pathway. A calpain-generated fragment of Atg5 has been shown to have pro-apoptotic properties in certain cell types and in response to particular death stimuli (49, 53). Our results indicate that an Atg5 fragment does not mediate Jo2- and TNF-α-induced cell death in MEFs as death occurred in cells lacking this protein.
Macroautophagy mediates the degradation of cellular organelles including mitochondria (38, 54). Death receptor ligands induce apoptosis via the mitochondrial death receptor pathway in which changes in the mitochondrial membrane, termed the mitochondrial permeability transition, lead to the release of pro-apoptotic factors such as cytochrome c from mitochondria (55). In rat hepatocytes in which macroautophagy was induced by serum deprivation and glucagon, the mitochondrial permeability transition led to mitochondrial depolarization and the subsequent movement of these mitochondria into autophagosomes (56). These findings suggest the possibility that macroautophagy may limit cellular injury from Fas/TNF-α by sequestering and degrading mitochondria that have undergone the mitochondrial permeability transition and preventing their release of pro-apoptotic factors (41, 56). Suggestive that an impairment in this process is the mechanism for Fas/TNF-α-induced apoptosis in Atg5−/− MEFs is that increased cytochrome c release occurred in these cells with Jo2 or TNF-α treatment in concert with mitochondrial dysfunction as indicated by decreased ATP levels. Increased activation of the initiator caspase 8 also occurred, suggesting that a loss of autophagy may have had an effect upstream of mitochondrial activation. However, it is well recognized that death receptor-induced caspase 8 activation requires the actions of effector caspases. Caspase 8 activation sufficient to induce cell death does not occur until effector caspases are activated by the mitochondrial death pathway and act to promote further caspase 8 cleavage via the caspase feedback amplification loop (57–59). Consistent with the conclusion that in the absence of autophagy pro-apoptotic mitochondrial factors augmented caspase 8 activation in our model are data that increased caspase 8 activation did not occur until a late time point when caspase 3 and PARP cleavage had already resulted from activation of the mitochondrial death pathway. The present studies therefore suggest that macroautophagy performs the critical function of limiting activation of the mitochondrial death pathway in response to death receptor stimulation and thus provides resistance to the cytotoxic effects of these factors. However, the current studies cannot rule out the alternative possibility that in the setting of death receptor stimulation, the loss of autophagy in some way leads directly to increased caspase 8 activation.
One finding critical to the concept that macroautophagy is a mechanism of cell death is the demonstrated up-regulation of macroautophagy in association with specific forms of cell death (3, 5, 6, 60). Less consideration has been given to the physiological effects of lower, constitutive levels of macroautophagy. Recent investigations have demonstrated the importance of basal levels of macroautophagy in the maintenance of cellular homeostasis in brain and liver cells (38–40). Our findings indicate that constitutive levels of macroautophagy are critical to protect against acute cellular stresses such as death receptor stimulation as well. If, as our data suggest, basal sequestration of cytosolic components by macroautophagy has a protective effect against Jo2- or TNF-α-mediated cell death, why is macroautophagy not up-regulated under these conditions? It is possible that these death stimuli induce intracellular changes that directly prevent the ability of cells to up-regulate this pathway. The fact that Jo2 and TNF-α treatment abolished the stimulatory effect of rapamycin on macroautophagy supports this possibility. Very little is known about the regulation of basal macroautophagy, but recent descriptions of mTOR-independent forms of macroautophagy (61, 62) raise the possibility that different regulatory mechanisms exist for the basal and inducible forms of macroautophagy. The anti-apoptotic effect described in other systems with rapamycin-induced up-regulation of macroautophagy (63) therefore did not occur in our model. Although further studies are required, one possible reason for this discrepancy is that our studies were performed in a normal cellular background in contrast to other studies in which death was induced in an already pro-apoptotic context (cells expressing a mutant or altered protein).
In contrast to prior reports of an induction of macroautophagy during cell death, the long term effect of death receptor stimulation in MEFs was to decrease levels of macroautophagy. Interestingly, despite this blockage in macroautophagy progression, wild-type cells still had significantly higher survival after Jo2/TNF-α treatment than cells unable to form autophagosomes, suggesting that sequestration of cytosolic contents rather than their degradation was sufficient for cellular resistance to death. The mechanism(s) responsible for this reduction in macroautophagy is unclear. Activation of both the extrinsic and the intrinsic death pathways slowed the clearance of auto-phagic vacuoles. As this effect appeared late after treatment, it is possible that in the advanced stages of both apoptotic pathways, lysosomal alterations occur that render them unable to fuse with autophagic vacuoles or degrade their contents. This failure of autophagosome clearance may occur by a common mechanism or distinct mechanisms specific for each pathway or death stimulus. The sequestration of dysfunctional mitochondria into autophagosomes resulting from activation of the extrinsic apoptotic pathway could directly alter the ability of autophagosomes to fuse with lysosomes (64). In contrast, although mild oxidative stress stimulates lysosomal function, more severe oxidant stress with menadione as in our study could promote lysosomal membrane instability and leakage that interferes with normal lysosomal function (65).
The studies also examined for the first time the interplay between macroautophagy and CMA in the response of a cell with impaired macroautophagy to death stimuli. We have previously shown that CMA is activated in response to prolonged nutritional stress, probably as a late source of essential amino acids, and under conditions resulting in protein damage such as oxidative stress and exposure to toxic compounds (17). The finding that death from menadione-induced oxidative stress and UV-mediated DNA damage was decreased in MEFs lacking macroautophagy could be interpreted as evidence to support the conclusion that macroautophagy functions to promote cell death. However, the finding that menadione up-regulated protein degradation that was not mediated by macroautophagy, along with the proven protective role of CMA in oxidative stress, suggested that an up-regulation of CMA in the absence of macroautophagy may mediate this effect. Supportive of this fact is that cells with higher basal CMA activity (Atg5−/− cells) were resistant to menadione toxicity and that a knockdown of CMA sensitized cells to death from menadione. Thus, the finding that an inhibition of macroautophagy blocks death from a particular stimulus cannot be taken as definitive proof that death is mediated by this autophagic pathway. Cellular compensation for the loss of macroautophagy in the form of up-regulation of CMA, or even other proteolytic pathways, may be the true mechanism for this effect and lead to the false interpretation that macroautophagy mediated the death response. The conclusion that autophagy promotes cell death must now be reconsidered in light of this ability of compensatory CMA up-regulation to alter the outcome from a cell death stimulus.
The fact that up-regulation of CMA had a protective function against menadione- and UV-induced cell death but not against the extrinsic apoptotic dead pathway may be related to the type of cellular damage caused by each of these stimuli. The early involvement of mitochondrial dysfunction in Fas/TNF-α-induced apoptosis could explain the lack of a protective effect of CMA against these stimuli as this autophagic pathway can only remove soluble cytosolic proteins and not whole organelles. In contrast, the ability of CMA to clear oxidized proteins can protect against death from menadione-induced oxidative damage. CMA may similarly protect against UV light, which also induces an oxidative stress (66, 67). Although mitochondria and other organelles also get damaged eventually, the efficient removal of oxidized proteins by CMA, at least in the early stages of menadione- and UV-induced cell dead, improves cell survival.
These studies demonstrate the complexity of the role of autophagy in modulating the cellular death response. In particular, conclusions regarding the function of one autophagic pathway cannot be made unless the compensatory effects of the other pathways are taken into account. Studies examining the cross-talk between macroautophagy and CMA in different cell types challenged with different death stimuli are necessary to better delineate what protective and possibly harmful roles autophagy plays in apoptosis.
Supplementary Material
Acknowledgments
The wild-type and Atg5−/− MEFs were kindly provided by Noboru Mizushima, the caspase inhibitor IDN-1529 was provided by Pfizer, the anti-Bid antibody was provided by Xiao-Ming Yin, and the adenoviruses were provided by David Brenner.
Footnotes
This work was supported by National Institutes of Health Grants AG021904 and DK041918 (to A. M. C.), DK044234 (to M. J. C.), and T32AG023475 (to A. C. M.) and an American Liver Foundation Postdoctoral Research Fellowship Award (to R. S.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
This article was selected as a Paper of the Week.
The on-line version of this article (available at http://www.jbc.org) contains four supplemental figures.
The abbreviations used are: TNF-α, tumor necrosis factor-α; ATG, autophagy-related gene; hsc70, heat shock cognate protein of 70 kDa; LAMP-2A, lysosome-associated membrane protein type 2A; LC3, light chain 3; 3-MA, 3-methyladenine; MEF, mouse embryonic fibroblast; MTT, 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide; PARP, poly (ADP-ribose) polymerase; Q-VD-OPh, Q-Val-Asp-OPh; CMA, chaperone-mediated autophagy; mTOR, mammalian target of rapamycin.
S. Kaushik, A. C. Massey, N. Mizushima, and A. M. Cuervo, submitted for publication.
REFERENCES
- 1.Levine B, Yuan J. J. Clin. Investig. 2005;115:2679–2688. doi: 10.1172/JCI26390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gozuacik D, Kimchi A. Oncogene. 2004;23:2891–2906. doi: 10.1038/sj.onc.1207521. [DOI] [PubMed] [Google Scholar]
- 3.Canu N, Tufi R, Serafino AL, Amadoro G, Ciotti MT, Calissano P. J. Neurochem. 2005;92:1228–1242. doi: 10.1111/j.1471-4159.2004.02956.x. [DOI] [PubMed] [Google Scholar]
- 4.Espert L, Denizot M, Grimaldi M, Robert-Hebmann V, Gay B, Varbanov M, Codogno P, Biard-Piechaczyk M. J. Clin. Investig. 2006;116:2161–2172. doi: 10.1172/JCI26185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jia L, Dourmashkin RR, Allen PD, Gray AB, Newland AC, Kelsey SM. Br. J. Haematol. 1997;98:673–685. doi: 10.1046/j.1365-2141.1997.2623081.x. [DOI] [PubMed] [Google Scholar]
- 6.Paglin S, Hollister T, Delohery T, Hackett N, McMahill M, Sphicas E, Domingo D, Yahalom J. Cancer Res. 2001;61:439–444. [PubMed] [Google Scholar]
- 7.Abedin MJ, Wang D, McDonnell MA, Lehmann U, Kelekar A. Cell Death Differ. 2006;14:500–510. doi: 10.1038/sj.cdd.4402039. [DOI] [PubMed] [Google Scholar]
- 8.Boya P, Gonzalez-Polo RA, Casares N, Perfettini JL, Dessen P, Larochette N, Metivier D, Meley D, Souquere S, Yoshimori T, Pierron G, Codogno P, Kroemer G. Mol. Cell. Biol. 2005;25:1025–1040. doi: 10.1128/MCB.25.3.1025-1040.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ding WX, Ni HM, Gao W, Hou YF, Melan MA, Chen X, Stolz DB, Shao ZM, Yin XM. J. Biol. Chem. 2007;282:4702–4710. doi: 10.1074/jbc.M609267200. [DOI] [PubMed] [Google Scholar]
- 10.Hamacher-Brady A, Brady NR, Gottlieb RA. J. Biol. Chem. 2006;281:29776–29787. doi: 10.1074/jbc.M603783200. [DOI] [PubMed] [Google Scholar]
- 11.Ogata M, Hino S, Saito A, Morikawa K, Kondo S, Kanemoto S, Murakami T, Taniguchi M, Tanii I, Yoshinaga K, Shiosaka S, Hammarback JA, Urano F, Imaizumi K. Mol. Cell. Biol. 2006;26:9220–9231. doi: 10.1128/MCB.01453-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Blommaart EF, Krause U, Schellens JP, Vreeling-Sindelarova H, Meijer AJ. Eur. J. Biochem. 1997;243:240–246. doi: 10.1111/j.1432-1033.1997.0240a.x. [DOI] [PubMed] [Google Scholar]
- 13.Madrid LV, Wang CY, Guttridge DC, Schottelius AJ, Baldwin AS, Jr, Mayo MW. Mol. Cell. Biol. 2000;20:1626–1638. doi: 10.1128/mcb.20.5.1626-1638.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Suhara T, Kim HS, Kirshenbaum LA, Walsh K. Mol. Cell. Biol. 2002;22:680–691. doi: 10.1128/MCB.22.2.680-691.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shimizu S, Kanaseki T, Mizushima N, Mizuta T, Arakawa-Kobayashi S, Thompson CB, Tsujimoto Y. Nat. Cell Biol. 2004;6:1221–1228. doi: 10.1038/ncb1192. [DOI] [PubMed] [Google Scholar]
- 16.Yu L, Alva A, Su H, Dutt P, Freundt E, Welsh S, Baehrecke EH, Lenardo MJ. Science. 2004;304:1500–1502. doi: 10.1126/science.1096645. [DOI] [PubMed] [Google Scholar]
- 17.Massey AC, Zhang C, Cuervo AM. Curr. Top. Dev. Biol. 2006;73:205–235. doi: 10.1016/S0070-2153(05)73007-6. [DOI] [PubMed] [Google Scholar]
- 18.Massey AC, Kaushik S, Sovak G, Kiffin R, Cuervo AM. Proc. Natl. Acad. Sci. U. S. A. 2006;103:5805–5810. doi: 10.1073/pnas.0507436103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kuma A, Hatano M, Matsui M, Yamamoto A, Nakaya H, Yoshimori T, Ohsumi Y, Tokuhisa T, Mizushima N. Nature. 2004;432:1032–1036. doi: 10.1038/nature03029. [DOI] [PubMed] [Google Scholar]
- 20.Mizushima N, Yamamoto A, Hatano M, Kobayashi Y, Kabeya Y, Suzuki K, Tokuhisa T, Ohsumi Y, Yoshimori T. J. Cell Biol. 2001;152:657–668. doi: 10.1083/jcb.152.4.657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mosmann T. J. Immunol. Methods. 1983;65:55–63. doi: 10.1016/0022-1759(83)90303-4. [DOI] [PubMed] [Google Scholar]
- 22.Duke RC, Cohen JJ. In: Current Protocols in Immunology. Coligan JE, Kruisbeek AM, Marguiles DH, Shevack EM, Strober W, editors. New York: John Wiley & Sons, Inc.; 1992. pp. 1–16. [Google Scholar]
- 23.Liu H, Lo CR, Jones BE, Pradhan Z, Srinivasan A, Valentino KL, Stockert RJ, Czaja MJ. J. Biol. Chem. 2000;275:40155–40162. doi: 10.1074/jbc.M001565200. [DOI] [PubMed] [Google Scholar]
- 24.Kiffin R, Christian C, Knecht E, Cuervo AM. Mol. Biol. Cell. 2004;15:4829–4840. doi: 10.1091/mbc.E04-06-0477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cuervo AM, Dice JF. Science. 1996;273:501–503. doi: 10.1126/science.273.5274.501. [DOI] [PubMed] [Google Scholar]
- 26.Schattenberg JM, Wang Y, Singh R, Rigoli RM, Czaja MJ. J. Biol. Chem. 2005;280:9887–9894. doi: 10.1074/jbc.M410310200. [DOI] [PubMed] [Google Scholar]
- 27.Liedtke C, Plumpe J, Kubicka S, Bradham CA, Manns MP, Brenner DA, Trautwein C. Hepatology. 2002;36:315–325. doi: 10.1053/jhep.2002.34615. [DOI] [PubMed] [Google Scholar]
- 28.Shinoura N, Koike H, Furitu T, Hashimoto M, Asai A, Kirino T, Hamada H. Hum. Gene Ther. 2000;11:1123–1137. doi: 10.1089/10430340050015185. [DOI] [PubMed] [Google Scholar]
- 29.Iimuro Y, Nishiura T, Hellerbrand C, Behrns KE, Schoonhoven R, Grisham JW, Brenner DA. J. Clin. Investig. 1998;101:802–811. doi: 10.1172/JCI483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xu Y, Bialik S, Jones BE, Iimuro Y, Kitsis RN, Srinivasan A, Brenner DA, Czaja MJ. Am. J. Physiol. 1998;275:C1058–C1066. doi: 10.1152/ajpcell.1998.275.4.C1058. [DOI] [PubMed] [Google Scholar]
- 31.Auteri JS, Okada A, Bochaki V, Dice JF. J. Cell. Physiol. 1983;115:167–174. doi: 10.1002/jcp.1041150210. [DOI] [PubMed] [Google Scholar]
- 32.Pinkoski MJ, Brunner T, Green DR, Lin T. Am. J. Physiol. 2000;278:G354–G366. doi: 10.1152/ajpgi.2000.278.3.G354. [DOI] [PubMed] [Google Scholar]
- 33.Wajant H, Pfizenmaier K, Scheurich P. Cell Death Differ. 2003;10:45–65. doi: 10.1038/sj.cdd.4401189. [DOI] [PubMed] [Google Scholar]
- 34.Hatano E, Bradham CA, Stark A, Iimuro Y, Lemasters JJ, Brenner DA. J. Biol. Chem. 2000;275:11814–11823. doi: 10.1074/jbc.275.16.11814. [DOI] [PubMed] [Google Scholar]
- 35.Jones BE, Lo CR, Liu H, Srinivasan A, Streetz K, Valentino KL, Czaja MJ. J. Biol. Chem. 2000;275:705–712. doi: 10.1074/jbc.275.1.705. [DOI] [PubMed] [Google Scholar]
- 36.Condorelli F, Salomoni P, Cotteret S, Cesi V, Srinivasula SM, Alnemri ES, Calabretta B. Mol. Cell. Biol. 2001;21:3025–3036. doi: 10.1128/MCB.21.9.3025-3036.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhao Y, Li S, Childs EE, Kuharsky DK, Yin XM. J. Biol. Chem. 2001;276:27432–27440. doi: 10.1074/jbc.M102465200. [DOI] [PubMed] [Google Scholar]
- 38.Komatsu M, Waguri S, Ueno T, Iwata J, Murata S, Tanida I, Ezaki J, Mizushima N, Ohsumi Y, Uchiyama Y, Kominami E, Tanaka K, Chiba T. J. Cell Biol. 2005;169:425–434. doi: 10.1083/jcb.200412022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hara T, Nakamura K, Matsui M, Yamamoto A, Nakahara Y, Suzuki-Migishima R, Yokoyama M, Mishima K, Saito I, Okano H, Mizushima N. Nature. 2006;441:885–889. doi: 10.1038/nature04724. [DOI] [PubMed] [Google Scholar]
- 40.Komatsu M, Waguri S, Chiba T, Murata S, Iwata J, Tanida I, Ueno T, Koike M, Uchiyama Y, Kominami E, Tanaka K. Nature. 2006;441:880–884. doi: 10.1038/nature04723. [DOI] [PubMed] [Google Scholar]
- 41.Lemasters JJ, Nieminen AL, Qian T, Trost LC, Elmore SP, Nishimura Y, Crowe RA, Cascio WE, Bradham CA, Brenner DA, Herman B. Biochim. Biophys. Acta. 1998;1366:177–196. doi: 10.1016/s0005-2728(98)00112-1. [DOI] [PubMed] [Google Scholar]
- 42.Mills KR, Reginato M, Debnath J, Queenan B, Brugge JS. Proc. Natl. Acad. Sci. U. S. A. 2004;101:3438–3443. doi: 10.1073/pnas.0400443101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kabeya Y, Mizushima N, Ueno T, Yamamoto A, Kirisako T, Noda T, Kominami E, Ohsumi Y, Yoshimori T. EMBO J. 2000;19:5720–5728. doi: 10.1093/emboj/19.21.5720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tanida I, Minematsu-Ikeguchi N, Ueno T, Kominami E. Autophagy. 2005;1:84–91. doi: 10.4161/auto.1.2.1697. [DOI] [PubMed] [Google Scholar]
- 45.Jia G, Cheng G, Gangahar DM, Agrawal DK. Immunol. Cell Biol. 2006;84:448–454. doi: 10.1111/j.1440-1711.2006.01454.x. [DOI] [PubMed] [Google Scholar]
- 46.Noda T, Ohsumi Y. J. Biol. Chem. 1998;273:3963–3966. doi: 10.1074/jbc.273.7.3963. [DOI] [PubMed] [Google Scholar]
- 47.Monks TJ, Hanzlik RP, Cohen GM, Ross D, Graham DG. Toxicol. Appl. Pharmacol. 1992;112:2–16. doi: 10.1016/0041-008x(92)90273-u. [DOI] [PubMed] [Google Scholar]
- 48.Thor H, Smith MT, Hartzell P, Bellomo G, Jewell SA, Orrenius S. J. Biol. Chem. 1982;257:12419–12425. [PubMed] [Google Scholar]
- 49.Yousefi S, Perozzo R, Schmid I, Ziemiecki A, Schaffner T, Scapozza L, Brunner T, Simon HU. Nat. Cell Biol. 2006;8:1124–1132. doi: 10.1038/ncb1482. [DOI] [PubMed] [Google Scholar]
- 50.Boyce M, Yuan J. Cell Death Differ. 2006;13:363–373. doi: 10.1038/sj.cdd.4401817. [DOI] [PubMed] [Google Scholar]
- 51.Cuervo AM, Dice JF. J. Cell Sci. 2000;113:4441–4450. doi: 10.1242/jcs.113.24.4441. [DOI] [PubMed] [Google Scholar]
- 52.Martinet W, De Meyer GR, Herman AG, Kockx MM. Biotechnol. Lett. 2005;27:1157–1163. doi: 10.1007/s10529-005-0007-y. [DOI] [PubMed] [Google Scholar]
- 53.Pyo JO, Jang MH, Kwon YK, Lee HJ, Jun JI, Woo HN, Cho DH, Choi B, Lee H, Kim JH, Mizushima N, Oshumi Y, Jung YK. J. Biol. Chem. 2005;280:20722–20729. doi: 10.1074/jbc.M413934200. [DOI] [PubMed] [Google Scholar]
- 54.Kim I, Rodriguez-Enriquez S, Lemasters JJ. Arch. Biochem. Biophys. 2007;462:245–253. doi: 10.1016/j.abb.2007.03.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bradham CA, Qian T, Streetz K, Trautwein C, Brenner DA, Lemasters JJ. Mol. Cell. Biol. 1998;18:6353–6364. doi: 10.1128/mcb.18.11.6353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Elmore SP, Qian T, Grissom SF, Lemasters JJ. FASEB J. 2001;15:2286–2287. doi: 10.1096/fj.01-0206fje. [DOI] [PubMed] [Google Scholar]
- 57.Nieminen AI, Partanen JI, Klefstrom J. Cell Cycle. 2007;6:2464–2472. doi: 10.4161/cc.6.20.4917. [DOI] [PubMed] [Google Scholar]
- 58.Slee EA, Keogh SA, Martin SJ. Cell Death Differ. 2000;7:556–565. doi: 10.1038/sj.cdd.4400689. [DOI] [PubMed] [Google Scholar]
- 59.Sohn D, Schulze-Osthoff K, Janicke RU. J. Biol. Chem. 2005;280:5267–5273. doi: 10.1074/jbc.M408585200. [DOI] [PubMed] [Google Scholar]
- 60.Kanzawa T, Germano IM, Komata T, Ito H, Kondo Y, Kondo S. Cell Death Differ. 2004;11:448–457. doi: 10.1038/sj.cdd.4401359. [DOI] [PubMed] [Google Scholar]
- 61.Sarkar S, Floto RA, Berger Z, Imarisio S, Cordenier A, Pasco M, Cook LJ, Rubinsztein DC. J. Cell Biol. 2005;170:1101–1111. doi: 10.1083/jcb.200504035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yamamoto A, Cremona ML, Rothman JE. J. Cell Biol. 2006;172:719–731. doi: 10.1083/jcb.200510065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ravikumar B, Berger Z, Vacher C, O’Kane CJ, Rubinsztein DC. Hum. Mol. Genet. 2006;15:1209–1216. doi: 10.1093/hmg/ddl036. [DOI] [PubMed] [Google Scholar]
- 64.Terman A, Dalen H, Brunk UT. Exp. Gerontol. 1999;34:943–957. doi: 10.1016/s0531-5565(99)00070-4. [DOI] [PubMed] [Google Scholar]
- 65.Terman A, Brunk UT. Cardiovasc. Res. 2005;68:355–365. doi: 10.1016/j.cardiores.2005.08.014. [DOI] [PubMed] [Google Scholar]
- 66.Morliere P, Moysan A, Tirache I. Free Radic. Biol. Med. 1995;19:365–371. doi: 10.1016/0891-5849(95)00043-w. [DOI] [PubMed] [Google Scholar]
- 67.Przybyszewski WM, Widel M, Palyvoda O. Teratog. Carcinog. Mutagen. 2002;22:93–102. doi: 10.1002/tcm.10006. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.