Abstract
DNA damage repair is crucial for the maintenance of genome integrity and cancer suppression. We found that loss of the mouse transcription factor YY1 resulted in polyploidy and chromatid aberrations, which are signatures of defects in homologous recombination. Further biochemical analyses identified a YY1 complex comprising components of the evolutionarily conserved INO80 chromatin-remodeling complex. Notably, RNA interference–mediated knockdown of YY1 and INO80 increased cellular sensitivity toward DNA-damaging agents. Functional assays revealed that both YY1 and INO80 are essential in homologous recombination–based DNA repair (HRR), which was further supported by the finding that YY1 preferentially bound a recombination-intermediate structure in vitro. Collectively, these observations reveal a link between YY1 and INO80 and roles for both in HRR, providing new insight into mechanisms that control the cellular response to genotoxic stress.
Genomic instability is a hallmark of cancer development and progression. Defects in DNA double-strand break (DSB) repair are believed to be responsible for chromosomal rearrangements and can be rectified by homologous recombination, which is particularly active in the S and G2 phases of the cell cycle, or by nonhomologous end joining (NHEJ), active in G1 phase1,2. Homologous recombination is an important mechanism for the repair of damaged chromosomes and damaged replication forks, and for several other aspects of chromosome maintenance3. Furthermore, impairment of homologous recombination is probably one of the underlying causes of breast, ovarian and other cancers4,5. INO80, a member of the Snf2p family of DNA-dependent ATPases, has been shown to positively regulate homologous recombination in a number of organisms6–8. The Ino80 complex is evolutionarily conserved, and components of the complex have been identified in mammalian cells9. DNA damage induces phosphorylation of histone H2A, and the yeast INO80 complex is known to be recruited directly to DSBs through an interaction with the phosphorylated histone H2A7,8,10,11; however, the precise function of INO80 in DSB repair and its recruitment to the damage site are not completely understood12.
Yin Yang-1 (YY1) is a zinc finger–containing Polycomb group (PcG) transcription factor that is essential in development13–16. Recently, a number of studies have shown that loss of YY1 increases the p53 protein level17,18, raising the possibility that YY1 may contribute to the regulation of genomic integrity. To investigate this possibility, we used genetic, biochemical and proteomic approaches. We discovered essential roles for YY1 in the cellular response to genotoxic stress and in the maintenance of chromosomal stability. Furthermore, we found multiple lines of evidence that link YY1 and the ATP-dependent chromatin-remodeling complex INO80 in DNA repair. We identified a YY1 complex containing components of the INO80 complex and confirmed this interaction of YY1 with components of the INO80 complex by additional co-immunoprecipitation and in vitro protein-interaction assays. Notably, both YY1 and INO80 were found to be necessary for the cellular DNA damage response, and both were crucial for HRR. In vitro, recombinant YY1 and the YY1 complex preferentially bound the Holliday-junction structure, an important homologous recombination intermediate. Together, these results suggest that the PcG protein YY1 and the ATP-dependent chromatin-remodeling complex INO80 may function together to mediate the cellular response to DNA damage. These findings shed new light not only on YY1’s function in the DNA repair process but also on that of mammalian INO80.
RESULTS
Loss of YY1 induces polyploidy and chromosomal aberrations
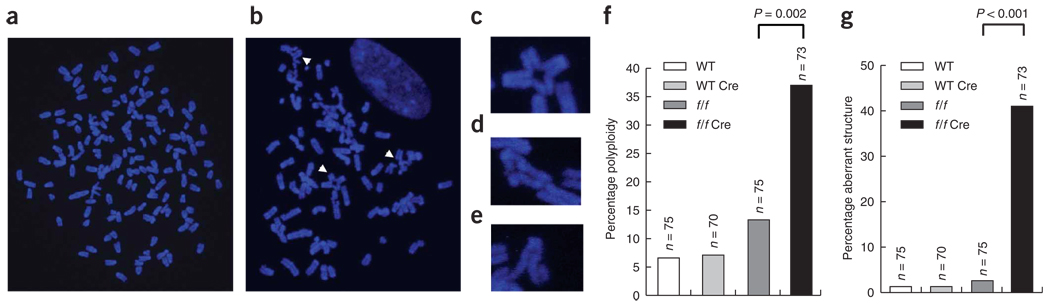
We investigated the possibility that YY1 may regulate genomic stability by analyzing the karyotypes of mouse embryonic fibroblast (MEF) cells isolated from Yy1–conditional knockout (YY1f/f) mice recently developed in our lab19. The wild-type and YY1f/f MEF cells were infected with Ad-Cre, and depletion of YY1 was analyzed by western blotting, which revealed essentially undetectable levels of YY1 in the YY1f/f/Cre cells (Supplementary Fig. 1a online). For each karyotyping experiment, metaphase spreads from MEF cells prepared from two independently isolated embryos were inspected for chromosome abnormalities. We found an increase in chromosome number as well as abnormal chromosome structures in YY1f/f MEFs transduced with Ad-Cre, compared with the control MEFs (Fig. 1a,b). In contrast to the three controls, in which less than 3% of cells had chromosomal abnormalities, 35.6% of YY1f/f MEF cells treated with Ad-Cre showed aneuploidy or polyploidy, and 41% contained chromosomal abnormalities, including chromatid and chromosome breaks as well as triradial and quadriradial chromosomes (Fig. 1c–g). These abnormal chromosomal structures are characteristic of cells with defects in DNA repair, such as those of individuals with Fanconi anemia20, Bloom’s syndrome21,22 or the hereditary breast and ovarian cancer syndromes involving BRCA1 and BRCA2 (refs. 23,24). These findings suggest a role for YY1 as a genomic caretaker.
Figure 1.
Loss of YY1 results in polyploidy and chromosome structural aberrations. (a) Metaphase spread from YY1f/f MEFs plus Cre, showing polyploidy. Wild-type and YY1f/f MEFs were transduced with Ad-Cre. (b) Metaphase spread from the YY1f/f MEFs plus Cre with multiple structural abnormalities (arrowheads). (c–e) Enlargements of the typical quadriradial structure (c), triradial structure (d) and chromatid break (e). (f) Percentages of aneuploid and polyploid chromosomes, which differ significantly between YY1f/f and YY1f/f plus Cre cultures. Difference between wild-type (WT) and YY1f/f plus Cre cultures was also statistically significant but is not shown. n indicates the number of metaphase spreads scored. (g) Percentages of abnormal chromosome structures, which differ significantly between YY1f/f and YY1f/f plus Cre cultures.
Accordingly, the YY1-knockout MEFs showed greater sensitivity to UV-C and camptothecin (a topoisomerase I inhibitor) treatments than the control cells (Supplementary Fig. 1b,c). In human U2OS osteosarcoma cells, small hairpin RNA (shRNA)-mediated knockdown of YY1 (Supplementary Fig. 1h) also increased sensitivity toward various DNA-damaging agents (Supplementary Fig. 1d–g). These findings suggest that YY1 is crucial in the cellular response to many genotoxins.
The increase in chromosome abnormalities may result from either a defect in cell cycle checkpoint control or a defect in DNA repair per se. When exposed to DNA-damaging agents such as UV-C radiation, cells can be arrested at two cell-cycle checkpoints. The G1/S checkpoint involves p53 and p21 and prevents replication of damaged DNA templates, whereas the G2/M checkpoint prevents segregation of the damaged chromosomes25. To determine whether loss of YY1 affects cell cycle checkpoint controls in cells exposed to damaging agents, we used FACS analysis to monitor cell-cycle progression after UV irradiation of the control and YY1 shRNA–transfected U2OS cells26. The YY1-knockdown cells had essentially intact G1/S and G2/M checkpoint responses similar to those seen in control cells (Supplementary Fig. 2 online). Together, these findings suggest that loss of YY1 does not affect cell-cycle checkpoints; thus, YY1 probably functions directly in DNA repair.
Biochemical identification of a YY1–INO80 complex
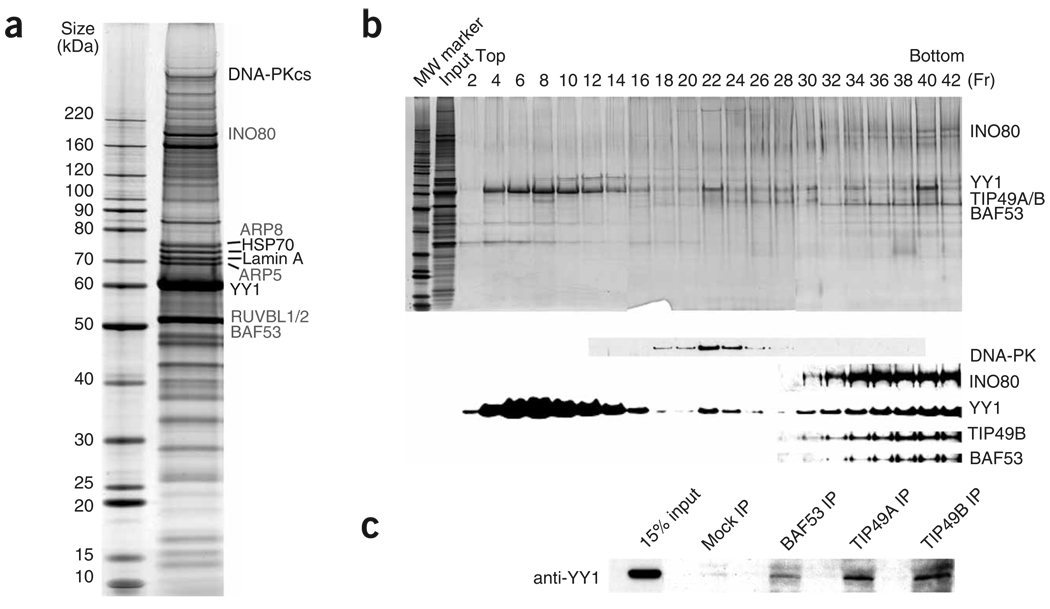
To address the possibility that YY1 may participate in DNA repair and to understand the underlying mechanism, we purified YY1 from HeLaS cells using a double–affinity tag purification technique described previously27,28. Briefly, HeLaS cells were transduced with either empty or recombinant retroviruses expressing YY1 tagged with Flag and hemagglutinin (HA) epitope at its N terminus (Flag-HA-YY1). Nuclear extracts were prepared from the mock-infected and Flag-HA-YY1 retrovirus–infected cells and then subjected to two successive rounds of affinity chromatography using affinity matrices with immobilized Flag antibody and HA antibody, respectively. Many proteins were found to copurify with YY1 (Fig. 2a), whereas essentially no proteins were found from purification using the mock-infected cells (data not shown). Proteins copurified with YY1 were then identified by tandem mass spectrometry (Fig. 2a and Supplementary Table 1 online). Notably, we identified mammalian homologs of seven subunits of the yeast Ino80 complex, which has been shown to be crucial for DSB repair in yeast10. These seven human INO80 complex components were TIP49A and TIP49B (previously identified as ‘RuvB-like’ proteins, and labeled RUVBL1 and RUVBL2 in Fig. 2a)10; KIAA1259, the human homolog of yeast Ino80, a SWI/SNF-like helicase; and human ARP4 (BAF53), ARP5, ARP8 and β-actin. Similar observations have been reported recently29. We also identified DNA-PKcs and Ku86, components of the machinery that mediates NHEJ30–32. In this study, we focused our analysis on the association of YY1 with the INO80 complex.
Figure 2.
YY1 associates with mammalian Ino80 complex. (a) Silver staining of the YY1 complex separated on 4%–12% SDS-PAGE gel. The YY1 complex was immunopurified from nuclear extract prepared from HeLaS cells stably expressing Flag-HA-YY1. Subunits of INO80 complex are labeled in gray. (b) The double-purified YY1 complex in a was further fractionated by 10%–40% glycerol-gradient sedimentation. Fractions were visualized by silver staining (top) and immunoblotting with indicated antibodies (bottom). Input (3%) and molecular weight (MW) marker were also included on the silver-staining gel. (c) YY1 interacts with subunits of INO80 complex in vivo. Total HeLa cell lysates were immunoprecipitated with BAF53, TIP49A and TIP49B antibodies. YY1 levels in immunoprecipitates and 15% input were compared by western blotting with the YY1 antibody.
Western blotting readily identified the INO80 components in the purified YY1 complex (Supplementary Fig. 3a online), confirming the mass spectrometry result. To further investigate whether YY1 is in a stable complex with INO80, we carried out glycerol-gradient sedimentation and analyzed the protein fractions by both silver staining and immunoblotting. The affinity-purified YY1 complex was separated into three peaks (Fig. 2b). Given their molecular-weight distribution and the fact that recombinant YY1 can form multimers (Supplementary Fig. 4c,d online), fractions 4–16 probably represent free and oligomerized YY1. YY1 cofractionated with DNA-PKcs (Fig. 2b, lanes 20–26), a protein that is involved in NHEJ, but not with Ku proteins, which are also necessary for NHEJ. YY1 also cofractionated with many subunits of the INO80 complex, including TIP49B, INO80 and BAF53, in the high–molecular weight fractions (Fig. 2b, lanes 30–42). Thus, we have identified distinct subcomplexes in the affinity-purified YY1 mixture, including the YY1–DNA-PK and YY1–INO80 complexes, that are separable by glycerol-gradient sedimentation.
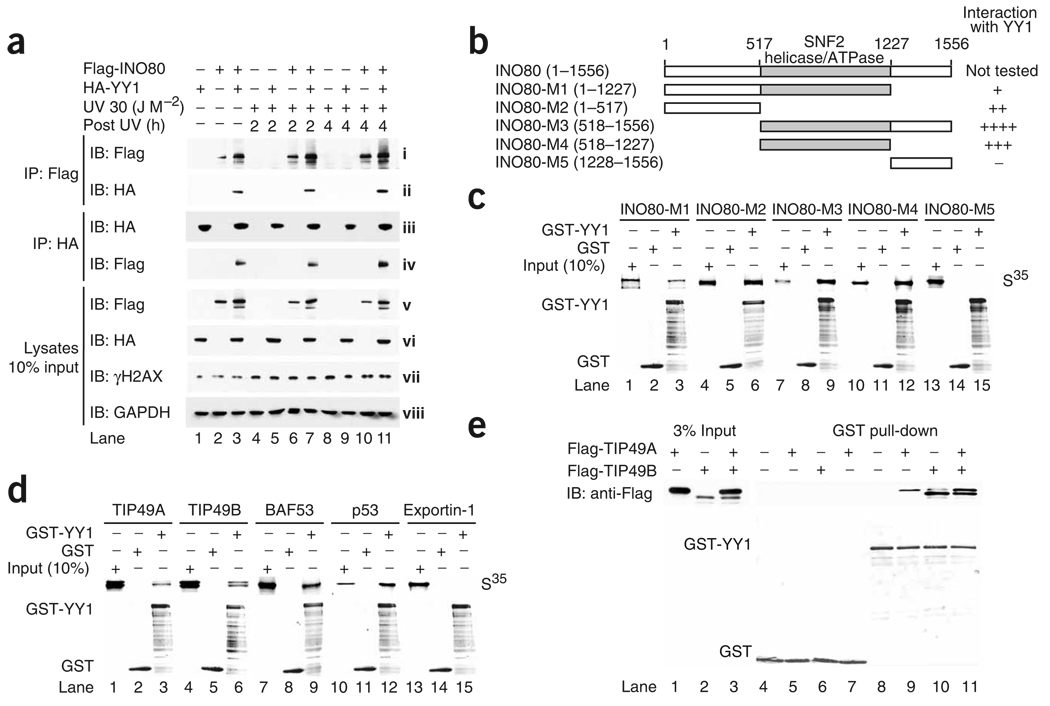
To analyze the YY1–INO80 association further, we carried out co-immunoprecipitation experiments. YY1 was co-immunoprecipitated by BAF53, TIP49A and TIP49B antibodies (Fig. 2c). We also analyzed YY1’s interaction with INO80 in HeLa cells transfected with HA-tagged YY1 and Flag-tagged INO80. The Flag antibody that recognized Flag-INO80 also precipitated HA-YY1 only when both proteins were expressed together in HeLa cells (Fig. 3a, ii, compare lane 3 with lanes 1 and 2). Similarly, HA antibody precipitated HA-YY1 as well as Flag-INO80 (Fig. 3a, iv, lane 3). In mammalian cells, RNA interference (RNAi) knockdown of YY1 and INO80 led to increased UV sensitivity (see below). We therefore investigated whether DNA damage influences the interaction between YY1 and INO80. Treatment of cells with UV had no overt effects on the interaction between ectopically expressed YY1 and INO80 4 h after radiation (Fig. 3a, ii,iv, compare lanes 3, 7 and 11). Together, these findings suggest that YY1 interacts with the INO80 complex components in vivo, and the interaction seems to be stable, changing little in response to UV stress. Accordingly, the YY1 complex purified from UV-stressed HeLa cells showed no substantial difference in composition from the YY1 complex purified without UV treatment (Supplementary Fig. 3b).
Figure 3.
YY1 interacts with INO80 subunits in vitro and in vivo. (a) Co-immunoprecipitation between YY1 and INO80 upon DNA damage. HeLa cells transfected with Flag-INO80, HA-YY1 or both were radiated with UV (30 J M−2) and lysed 2 or 4 h after stress. Lysates were immunoprecipitated with Flag or HA affinity beads, probed with anti-Flag and reprobed with anti-HA. Lysate western blots show the protein levels of Flag-INO80 (v), HA-YY1 (vi), γH2AX (vii) and GAPDH (viii) in 10% input. IP, immunoprecipitation; IB, immunoblot. (b) Diagrams of human INO80 full-length protein and fragments used to map the YY1-binding domain. Abilities of indicated INO80 fragments to bind GST-YY1 protein are summarized on the right. (c,d) In vitro binding of indicated proteins to YY1 protein. Upper gels, amounts of 35S-labeled, in vitro–translated proteins retained on glutathione-agarose beads after pull-down assay, compared to the corresponding input (10%). Lower gels, Coomassie blue staining of GST and GST-YY1 proteins used for this experiment. In d, p53 and exportin 1 were used as positive and negative binding controls, respectively. (e) GST-YY1 pull-down assay with bacterially purified Flag-TIP49A and Flag-TIP49B. Top gel, pulled down samples analyzed by western blotting with the Flag antibody. Bottom gel, GST fusion proteins visualized by Coomassie blue staining.
YY1 interacts with INO80, BAF53, TIP49A and TIP49B in vitro
We next carried out glutathione S-transferase (GST) pull-down experiments to identify components of the INO80 complex that may interact with YY1 in vitro. Figure 3b shows a schematic representation of the full-length human INO80 (residues 1–1556) and of the five deletion mutants (INO80 M1 to M5). A potential physical interaction between the full-length human INO80 and YY1 could not be tested because we were not able to express full-length recombinant INO80. In GST pull-down assays using in vitro–translated INO80 truncation mutants, both the N-terminal segment (1–517) and the middle segment (518–1227) of INO80 interacted with GST-YY1 but not GST alone (Fig. 3c,d), suggesting that YY1 makes multiple contacts with INO80. We also found interactions of YY1 with TIP49A, TIP49B and BAF53 (Fig. 3c,d). Notably, GST-YY1 pulled down TIP49A and TIP49B in a 1:1 stoichiometric ratio (Fig. 3e, top blot, lane 11). Together, these findings suggest that YY1 may physically contact several components of the INO80 complex.
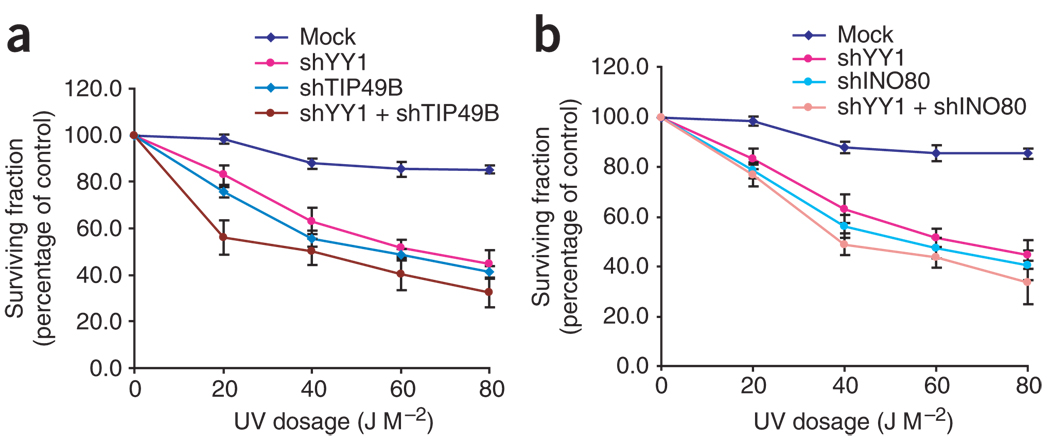
Knockdown of TIP49B and INO80 leads to UV hypersensitivity
The biochemical results suggest that YY1 and the INO80 complex might function together to regulate DNA repair. To address this possibility, we first examined whether cells in which the INO80 components were knocked down by RNAi showed increased sensitivity to DNA-damaging agents (Supplementary Methods online), as did the YY1-deficient MEFs described above. U2OS cells subjected to RNAi knockdown of INO80, YY1 and TIP49B had very similar responses to UV irradiation, as determined by survival assays (Fig. 4). Knockdown of YY1 simultaneously with either INO80 or TIP49B did not further decrease cell survival in response to DNA damage, supporting the hypothesis that YY1, TIP49B and INO80 function in the same pathway, and probably at the same step, in the cellular response to UV damage. The RNAi knockdown efficiencies of the various shRNAs used were monitored by western blotting (Supplementary Fig. 3c).
Figure 4.
Knockdown of TIP49B and INO80 leads to UV hypersensitivity. (a) U2OS cells were transfected with plasmids expressing shRNAs (sh) that target mGFP (negative control) (Supplementary Fig. 1g), YY1 or TIP49B, or both YY1 and TIP49B, for 48 h and treated with indicated doses of UV light. Survival fraction is the number of live cells after irradiation divided by the number of live cells after mock treatment and is expressed as a percentage. About 4,000 cells were counted in at least three independent experiments. Data are presented as means ± s.e.m. (b) As in a, cells were transfected with plasmids expressing shRNAs that target YY1, INO80 or both.
YY1 and INO80 promote homology-directed DNA repair
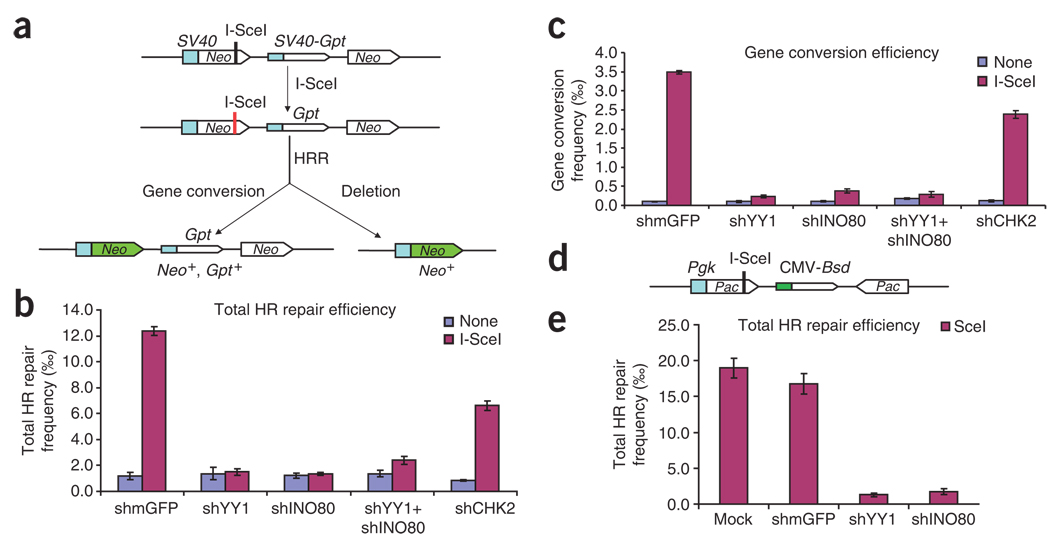
Next, to investigate whether YY1 is involved in homology-directed repair, we used a reporter-based functional assay in which the Neo gene can be expressed only upon repair of DSBs produced by the endonuclease I-SceI33 (Fig. 5a). HR-293T cells were transfected with plasmids that express YY1 and INO80 shRNAs either singly or together, as well as with a plasmid that expresses I-SceI. After I-SceI induction, the number of Neo-positive clones was counted. Cells transfected with YY1 shRNA, INO80 shRNA or both produced approximately eight-fold fewer I-SceI–induced Neo-positive colonies (Fig. 5b) than did cells transfected with control shRNA. These data were normalized to plating efficiencies of shRNA-treated HR-293T cells (see Methods). RNAi knockdown of YY1 did not affect the expression of HA-tagged I-SceI, as shown by western blotting (data not shown). Because both YY1 and INO80 also regulate transcription, the deficiency in HRR of I-SceI–induced DSBs observed in cells expressing little YY1 and/or INO80 could reflect a reduction in transcription of the Neo gene. However, the frequency of Neo-positive colonies arising in the absence of I-SceI expression was unaffected by reductions in YY1 or INO80 expression (Fig. 5b, light gray bars). Thus, the frequency of Neo-positive cells arising by spontaneous homologous recombination in these populations was not affected by the RNAi treatment, indicating that YY1 and INO80 levels do not affect transcription of Neo. In HR-293T cells, gene conversion gives rise to Neo-positive cells that retain the Gpt gene and are resistant to both G418 and mycophenolic acid. We found that cells transfected with YY1 shRNA, INO80 shRNA or both showed markedly less gene conversion, ranging from 10- to 14-fold less (Fig. 5c). The large reduction in total homologous recombination and gene conversion suggests that YY1 and INO80 may function together as regulators of HRR.
Figure 5.
Impaired homology-directed repair of a chromosomal DSB in YY1- and INO80-deficient 293T and HT-1080 cell lines. (a) Schematic showing how, in the Neo direct-repeat substrate, I-SceI–induced DSBs can result in gene-conversion or deletion products. Neo direct repeats of 1.4 kilobases (open boxes) flank the Gpt gene. Transcription of the upstream Neo is driven by the TEAD1 (here called SV40) promoter and an I-SceI recognition site interrupts the reading frame, whereas the downstream Neo lacks a promoter. Shaded box, SV40 promoter; green arrow, functional Neo gene after repair. (b) Plasmid expressing shRNA constructs was cotransfected with I-SceI plasmid or control plasmid (SceI− plasmid; ‘None’ in key) into HR-293T cells. Total HRR frequencies were calculated as the number of G418-resistant colonies per viable cell plated in selective medium. Blue bars, spontaneous homologous recombination events; purple bars, I-SceI–induced events. Error bars indicate s.e.m. (c) Gene-conversion frequencies, calculated as the number of colonies resistant to both G418 and mycophenolic acid per viable cell plated. (d) Structure of inverted puromycin (Pac) repeats flanking the cytomegalovirus (CMV) Bsd promoter in the HT1080–1885 cell line. The upstream Pac is driven by the mouse Pgk promoter and inactivated by insertion of an I-SceI site in the coding sequence; the downstream donor Pac lacks a promoter. (e) Total HRR frequencies in Pac inverted-repeat HT1080–1885 strain. shRNA constructs were cotransfected with I-SceI plasmid or control plasmid into HT1080–1885 cells. Total HRR frequencies were calculated as the number of puromycin-resistant colonies per viable cell plated in selective medium. Error bars indicate s.e.m.
To ensure that the observed role for YY1 and INO80 in HRR is not limited to just 293T cells, which express SV40 large T antigen and therefore are defective in p53 function34, we measured HRR in a derivative of the HT1080 strain that is wild-type with respect to p53 function35. HT1080–1885 cells carry inverted puromycin repeats36 but are otherwise analogous to HR-293T cells (Fig. 5d). Consistent with the HR-293T results, HT1080–1885 cells transfected with YY1 shRNA, INO80 shRNA or both showed about 13-fold less HRR of I-SceI– induced DSBs than cells transfected with control shRNA (Fig. 5e). Together, these findings support a model in which YY1 and INO80 both function in homologous recombination.
YY1 binds recombination-intermediate structure in vitro
Several findings discussed above suggest that YY1 may have a role in HRR37. Given that YY1 directly interacts with TIP49A and TIP49B (Fig. 2c), which are potential mammalian homologs of RuvB, we wished to test whether YY1 functions similarly to RuvA, although YY1 is not related to RuvA at the protein sequence level.
To address this possibility, we carried out gel-shift assays to determine whether YY1, like RuvA, can bind a synthetic Holliday junction, an important intermediate in the DSB repair model38,39. We estimated the amount of YY1 in the YY1 complex by western blotting, using recombinant YY1 of known concentrations for comparison (Supplementary Fig. 4a). The purity of the recombinant YY1 is shown in Supplementary Figure 4b. When the YY1 complex and the C-terminal binding protein (CtBP) complex were incubated with the Holliday-junction probe, several band-shift complexes formed, but only with the YY1 and not the CtBP complex (Fig. 6a,b). This result suggests binding specificity. In competition experiments, unlabeled Holliday-junction probe competed with a double-stranded DNA (dsDNA) having a sequence identical one of the ‘arms’ of the four-way junction, and the YY1 complex preferentially bound the Holliday junction (Fig. 6b, compare lanes 5 and 6). YY1 antibody supershift assays demonstrated that essentially all the band-shift complexes contained YY1 (Fig. 6b, compare lane 3 with lanes 2 and 4). We were unable to supershift the complexes with TIP49B and INO80 antibodies, probably owing to antibody quality, epitope masking or both (data not shown). However, the DNA–protein complexes formed with the YY1 complex are different from those formed with purified recombinant YY1 (see below). This observation is consistent with the idea that the shifted bands (Fig. 6a, band-shifted complexes I–V) contain additional proteins that may be components of the INO80 complex.
Figure 6.
YY1 binds a recombination-intermediate structure in vitro. (a) Purified YY1 complex was incubated with biotinylated Holliday-junction DNA (probe), and the binding reactions were resolved on 4% nondenaturing PAGE gels. Reactions contained 10 nM probe and YY1 complex at 0, 10, 20, 40, 60, 80, 100 and 200 nM per reaction in lanes 1–8, respectively. The positions of unbound probe, gel origin and YY1 band-shift complexes I–V are indicated. (b) Specificity determination using YY1 supershift and unlabeled competitor DNA binding assays. Lane 1, probe only; lane 2, 40 nM YY1 complex; lane 3, supershift with YY1 antibody; lane 4, supershift with control antibody; lane 5, excess unlabeled Holliday junction (50 nM); lane 6, excess unlabeled dsDNA (50 nM); lane 7, CtBP complex. Reactions contained 10 nM probe. (c) Purified His-YY1 was incubated with biotinylated Holliday junction DNA (probe) or dsDNA probes, and the binding reactions were resolved on 4% nondenaturing PAGE gels. Reactions contained 1 nM probe and His-YY1 at 0, 100, 200, 400, 500, 600, 700, 800 and 900 nM in lanes 1–9, respectively. The positions of unbound probe and His-YY1 band-shift complexes I–V are indicated. (d) Specificity determination. Unlabeled DNA structures were titrated against biotinylated Holliday junction probe (1 nM). Lanes 1, 6 and 11 contain 600 nM His-YY1. The type of unlabeled competitor DNA is indicated above each of the three titration series (lanes 2–5, 7–10 and 12–15); concentrations of competitor DNA in each series were 5-fold, 10-fold, 25-fold and 50-fold molar excess.
We next used purified recombinant YY1 to determine whether YY1 alone is sufficient to mediate binding to a Holliday-junction structure. Titration of YY1 against a fixed molar concentration of probes (1 nM) resulted in concentration-dependent binding of YY1 to the Holliday-junction structure (Fig. 6c). YY1 seems to bind preferentially to Holliday junctions and form slowly migrating complexes, which may represent Holliday junctions bound by YY1 multimers. Recombinant YY1 also bound the dsDNA probe, though less efficiently. However, the pattern for the Holliday junction–YY1 complexes is clearly distinct from that of the dsDNA–YY1 complexes. In contrast, purified recombinant TIP49A and TIP49B did not bind the Holliday-junction probe (data not shown). To determine whether recombinant YY1 binding to Holliday junctions is specific, we carried out competition assays with molar excesses of unlabeled DNA representing a Holliday junction, a ‘Y’ structure and dsDNA. The formation of YY1–Holliday junction complexes I and II was inhibited by the unlabeled Holliday-junction probe in a dose-responsive manner, whereas the effect of the linear duplex DNA was ten-fold less (Fig. 6). Excess Y-structure DNA also specifically reduced the formation of complexes I and II (Fig. 6d, lanes 11 and 12), suggesting that YY1 may also bind this structure, a putative intermediate in models of both synthesis-dependent strand annealing and break-induced replication. Together, these results suggest binding of YY1 to the Holliday-junction structure.
DISCUSSION
In this report, we have provided multiple lines of evidence suggesting a role for YY1 in HRR, the most important evidence being the association of YY1 loss with the chromatid and chromosome aberrations that are characteristic of HRR defects. This model is further supported by findings from reporter assays that analyze the effects of YY1 on HRR. The identification of YY1 associating with the INO80 complex suggests that YY1 may function together with this ATP-dependent chromatin-remodeling complex to regulate DNA repair. Notably, the association of YY1 with Ino80 is conserved evolutionarily in Drosophila melanogaster40, consistent with the idea that the YY1-Ino80 interaction is functionally important. A recent study has also shown that mammalian INO80 associates with YY1 and implicated this interaction in transcriptional activation29.
Studies have shown that YY1 depletion causes a proliferation defect in many known cell types. One common phenotype of YY1 depletion is that replicative ability is progressively lost, culminating in proliferative arrest or apoptosis. Notably, we found frequent quadriradial and triradial structures in the YY1-deficient MEFs, which may be the basis for their observed retarded cell growth19.
Biochemical and molecular studies point to the similarities between YY1 and RuvA, including the ability to form tetramers (Supplementary Fig. 4c,d) and to preferentially bind Holliday junctions. Recently, RAD54, a SWI2/SNF2 protein, has been shown to bind Holliday junction–like structures and promote their bidirectional branch migration in an ATPase-dependent manner41. Our findings raise the possibility that YY1 may function similarly to RAD54, regulating HRR by binding the homologous recombination intermediates. The binding affinity of recombinant YY1 for the Holliday junction (estimated Km of 500 nM, Supplementary Fig. 4e) is lower than that of bacterial RuvA (Km of 10–200 nM)39,42. However, it is possible that when YY1 is in complex with other proteins or post-translationally modified in vivo, its binding affinity may be higher (Fig. 6b). Determining whether YY1 binding to Holliday junctions is functionally significant in a physiological setting will require further studies.
A common feature of many DNA repair proteins is that they relocalize to distinct nuclear foci in response to DNA damage43. These foci have been proposed to be DNA repair centers containing gigadalton-sized protein assemblies44. We also investigated nuclear localization of YY1 and components of the INO80 complex in response to DNA damage in human cells. Notably, although we found no distinct YY1 and INO80 foci after ion irradiation, we observed formation of YY1 and INO80 nuclear foci in response to replication stress (S.W. and Yang Shi, unpublished data). However, we saw no evidence of colocalization of YY1 with γH2AX, 53BP1, NBS-1, DNA-PK and Ku80 in response to UV or camptothecin treatment. Recent studies have reported that HRR proteins show nuclear-localization patterns different from those of the DNA damage sensors45. We are currently investigating the functional significance of the YY1– INO80 foci; in preliminary studies (S.W. and Yang Shi, unpublished data), RNAi inhibition of YY1 seemed to compromise INO80 focus formation, suggesting that YY1 may have a recruitment role.
Given that YY1 is a transcriptional regulator, we also considered the possibility that YY1 might affect DNA repair indirectly by regulating genes important for repair. Our previous microarray analyses of MEF cells with or without YY1 revealed no change in the transcription of many genes associated with DNA repair, including RAD51, BRCA1 and NBS1 (ref. 19). Thus, although a transcriptional role cannot be formally excluded, several lines of evidence described here, including YY1 binding to Holliday junctions and YY1 regulation of homologous recombination reporters, suggest that YY1 directly participates in DNA repair. It should be noted, however, that the potential direct and indirect effects of YY1 are not mutually exclusive.
In summary, we have identified a crucial role for YY1 in regulating genomic stability and in the cellular response to DNA damage. Our findings further uncover a link between YY1 and the mammalian INO80 complex, suggesting that YY1 and INO80 work together to regulate HRR. These observations not only provide important insights into YY1 function but also shed new light on mechanisms that control genomic stability and DNA damage repair in mammals.
METHODS
Plasmid construction and antibodies
To construct retrovirus expression vectors for Flag and HA epitope–tagged YY1 proteins, we PCR-amplified YY1 coding sequences and subcloned them into pOZ-N vector using the XhoI and NotI sites. Full-length human INO80 cDNA was cloned into pCMV vector (Sigma) encoding an N-terminal Flag tag and into pEGFP-C1 (Clontech). pET-Flag-TIP49A, pET-Flag-TIP49B and pCMV-Flag-BAF53 were gifts (see Acknowledgments). pBS/U6 vectors encoding INO80 and TIP49B shRNA were constructed as described46.
Antibodies were as follows: anti-TIP49B rabbit polyclonal 1:1,000, anti-TIP49A rabbit polyclonal 1:1,000, anti-INO80 rabbit polyclonal and, anti-BAF53 rabbit polyclonal 1:1,000 were gifts (see Acknowledgments); anti-YY1 H10 mouse monoclonal 1:100 was from Santa Cruz; anti-HA 1:1000 was from Covance; anti-Flag 1:1,000 was from Sigma.
Cytogenetics
Cell preparations started 96 h after viral infection. Yy1-knockout or wild-type MEFs were treated with colcemid (0.1 µg ml−1) for 4 h, trypsinized and pelleted at 800 r.p.m. for 10 min in an Eppendorf model 5415D centrifuge. After hypotonic swelling in 0.075 M KCl for 30 min at 37 °C, cells were fixed in 3:1 methanol/glacial acetic acid at room temperature for 10 min. Cells resuspended in a drop of fixative were dropped onto humidified, clean, chilled glass slides. After adding 50 µl of the DAPI/anti-fade medium to the metaphase slide, we visualized chromosome structure by fluorescence microscopy. Chromosomes during metaphase spreading were counterstained with DAPI and examined with a Zeiss LSM 510 Meta UV upright confocal microscope and associated Zeiss LSM 510 software. Typically, 400–500 mitotic cells were selected randomly and scanned, with at least 100 cells being fully analyzed with the routine cytogenetic procedure. The number of chromosomes in each mitotic cell was counted, and each chromosome was examined closely for any of the structural changes described in the literature47. The percentage of metaphase spreads with one or more aberrations was calculated for each culture. χ2 analysis (SPSS) was used to test the significance of differences between all possible pairs.
Tandem affinity purification of YY1 protein complex
A recombinant retrovirus expressing a bicistronic messenger RNA containing open reading frames of Flag-HA-tagged human YY1 and interleukin-2 receptor-α (IL-2R-α) was constructed and transduced into HeLaS cells. The infected HeLaS cells were sorted using a monoclonal antibody to IL-2R conjugated with magnetic beads, and the resulting Flag-HA-YY1–expressing stable cell lines were propagated as suspension cells. Nuclear extracts of YY1-expressing stable cells were prepared as described27. YY1 and associated components were isolated from nuclear extracts by immunoprecipitation with anti-Flag and then with anti-HA according to established procedures27,48. YY1-associated protein bands were excised from SDS-PAGE gels and digested with trypsin, and their peptide sequences were determined by MALDI-TOF mass spectrometry at the DanaFarber Cancer Institute Molecular Biology Core Facility of Harvard University.
Protein interactions
Protein interactions were examined in 293T cells 36 h after Lipofectamine 2000–mediated transfection of the indicated plasmids. Cells were lysed in 0.1% (v/v) Nonidet P40 buffer (50 mM Tris-HCl (pH 7.5), 150 mM NaCl, 1 mM EDTA and protease inhibitors (Roche)) and cleared by centrifugation (14,000 r.p.m. for 15 min) before immunoprecipitation. For in vitro and in vivo binding, immobilized GST fusion proteins (made in BL21/DE3 cells) were incubated with bacterially purified Flag-TIP49A and Flag-TIP49B or with protein expressed in mammalian cells. Beads were washed with high-salt washing buffer (50 mM Tris (pH 7.3), 500 mM NaCl, 0.1% (v/v) Nonidet P-40, 200 µg ml−1 BSA, 0.5 mM DTT, 1 mM PMSF and Roche protease inhibitor cocktail), then subjected to SDS-PAGE and western blotting.
Electrophoretic mobility shift assay
The synthetic 50-mer four-way junction X12 comprised four annealed oligonucleotides, X12–1 (5′-GACGCTGCC GAATTCTGGCTTGCTAGGACATCTTTGCCCACGTTGACCCG-3′), X12–2 (5′-CGGGTCAACGTGGGCAAAGATGTCCTAGCAATGTAATCGTCTATGAC GTC-3′), X12–3 (5′-GACGTCATAGACGATTACATTGCTAGGACATGCT GTCTAGAGACTATCGC-3′) and X12–4 (5′-GCGATAGTCTCTAGACAGCAT GTCCTAGCAAGCCAGAATTCGGCAGCGTC-3′)49, and was purified by PAGE. We prepared a linear duplex by annealing X12–2 with its complement. The Y structure was prepared with X12–1, X12–4, FlapX12–1 (5′-CGG GTCAACGTGGGCAAAGA-3′) and Flap-X12–4 (5′-TGCTGTCTAGAGACTA TCGC-3′). Assays were done as described . Briefly, His-YY1, YY1 complex, Flag -TIP49A, Flag -TIP49B or CtBP complex were incubated with 1 nM biotinylated Holliday-junction probe in 10-µl reactions for 30 min at room temperature. The reaction buffer contained 20 mM Tris-HCl (pH 7.5), 10 mM magnesium acetate, 20% (v/v) glycerol, 100 µM EDTA, 40 µg ml−1 BSA and 10 mM 2-mercaptoethanol. Reactions were initiated upon probe addition. For supershift assays, 2 µg of YY1 H-414 antibody (Santa Cruz) or control G9a antibody (Upstate) were added immediately before probe addition. Unlabeled competitor DNA was added at the same time as probe. For gel analysis, 5 µl of 15% (v/v) Ficoll (type 400; Amersham Pharmacia) and 0.25% (w/v) xylene cyanol FF were added and reactions were resolved at room temperature on 4% nondenaturing PAGE gels containing 0.5 × Tris Boric buffer (TBE). Gels were transferred to Hybond-N+ membranes (Amersham Pharmacia), cross-linked with UV (120 mJ cm−2) and probed using the Light Shift electrophoretic mobility shift assay kit (Pierce).
Recombination assays
The human 293T cell line with a homologous recombination substrate (HR-293T) has been described33. I-SceI–induced homologous recombination was assayed as follows. For each experiment, 5 × 106 cells were seeded into one 10-cm-diameter plate, incubated for 24 h and lipofected with 20 µg of shRNA and 10 µg of either pCMV(3 × NLS)I-SceI to induce DSBs or the negative control vector pCMV(I-SceI−). Forty-eight hours after transfection, 105 cells were seeded into 10-cm dish as triplicates. After an additional 24 h, G418 was added to a final concentration of 750 µg ml−1 and the cells were incubated for 14 d. We measured plating efficiency by seeding 1,000 transfected cells into nonselective medium and scoring colony formation after 12–14 d. HRR frequencies were calculated as the ratio of the total number of G418-resistant (G418r) colonies over the plating efficiency. To measure gene-conversion frequencies, 106 cells were seeded into each 10-cm dish in triplicate. After an additional 24 h, both G418 and GPT Selection Reagent (Chemicon) were added and the cells were incubated for 16–21 d to form colonies. Data shown represent the averages from two independent experiments, each performed in triplicate.
Supplementary Material
ACKNOWLEDGMENTS
We thank members of the Shi laboratory for suggestions and helpful discussions; R. Scully and A. Xie (Harvard Institute of Medicine) for reagents and discussion; M.D. Cole (Dartmouth University), Y. Nakatani (Dana Farber Cancer Institute), W Harper (Harvard Medical School) and J. Lieberman (Harvard Medical School) for antibodies and constructs; and G. Sui (Wake Forest University) and Y. Li (Dana Farber Cancer Institute) for technical assistance. pET-Flag-TIP49A and pET-Flag-TIP49B were from T.-a. Tamura (Chiba University); pCMV-Flag-BAF53 and anti-BAF53 were from M.D. Cole; anti-TIP49B was from Y Nakatani; anti-TIP49A was from A. Dutta (Dana Farber Cancer Institute); anti-INO80 was from C. Wu (US National Cancer Institute). This project was supported by a grant from the US National Institutes of Health (GM53874) to Yang Shi.
Footnotes
Note: Supplementary information is available on the Nature Structural & Molecular Biology website.
Reprints and permissions information is available online at http://npg.nature.com/reprintsandpermissions
References
- 1.Khanna KK, Jackson SP. DNA double-strand breaks: signaling, repair and the cancer connection. Nat. Genet. 2001;27:247–254. doi: 10.1038/85798. [DOI] [PubMed] [Google Scholar]
- 2.van Gent DC, Hoeijmakers JH, Kanaar R. Chromosomal stability and the DNA double-stranded break connection. Nat. Rev. Genet. 2001;2:196–206. doi: 10.1038/35056049. [DOI] [PubMed] [Google Scholar]
- 3.Sung P, Klein H. Mechanism of homologous recombination: mediators and helicases take on regulatory functions. Nat. Rev. Mol. Cell Biol. 2006;7:739–750. doi: 10.1038/nrm2008. [DOI] [PubMed] [Google Scholar]
- 4.Jasin M. Homologous repair of DNA damage and tumorigenesis: the BRCA connection. Oncogene. 2002;21:8981–8993. doi: 10.1038/sj.onc.1206176. [DOI] [PubMed] [Google Scholar]
- 5.Moynahan ME. The cancer connection: BRCA1 and BRCA2 tumor suppression in mice and humans. Oncogene. 2002;21:8994–9007. doi: 10.1038/sj.onc.1206177. [DOI] [PubMed] [Google Scholar]
- 6.Fritsch O, Benvenuto G, Bowler C, Molinier J, Hohn B. The INO80 protein controls homologous recombination in Arabidopsis thaliana. Mol. Cell. 2004;16:479–485. doi: 10.1016/j.molcel.2004.09.034. [DOI] [PubMed] [Google Scholar]
- 7.Tsukuda T, Fleming AB, Nickoloff JA, Osley MA. Chromatin remodelling at a DNA double-strand break site in Saccharomyces cerevisiae. Nature. 2005;438:379–383. doi: 10.1038/nature04148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Morrison AJ, et al. INO80 and gamma-H2AX interaction links ATP-dependent chromatin remodeling to DNA damage repair. Cell. 2004;119:767–775. doi: 10.1016/j.cell.2004.11.037. [DOI] [PubMed] [Google Scholar]
- 9.Jin J, et al. A mammalian chromatin remodeling complex with similarities to the yeast INO80 complex. J. Biol. Chem. 2005;280:41207–41212. doi: 10.1074/jbc.M509128200. [DOI] [PubMed] [Google Scholar]
- 10.Shen X, Mizuguchi G, Hamiche A, Wu C. A chromatin remodelling complex involved in transcription and DNA processing. Nature. 2000;406:541–544. doi: 10.1038/35020123. [DOI] [PubMed] [Google Scholar]
- 11.van Attikum H, Fritsch O, Hohn B, Gasser SM. Recruitment of the INO80 complex by H2A phosphorylation links ATP-dependent chromatin remodeling with DNA double-strand break repair. Cell. 2004;119:777–788. doi: 10.1016/j.cell.2004.11.033. [DOI] [PubMed] [Google Scholar]
- 12.van Attikum H, Gasser SM. The histone code at DNA breaks: a guide to repair? Nat. Rev. Mol. Cell Biol. 2005;6:757–765. doi: 10.1038/nrm1737. [DOI] [PubMed] [Google Scholar]
- 13.Donohoe ME, et al. Targeted disruption of mouse Yin Yang 1 transcription factor results in peri-implantation lethality. Mol. Cell. Biol. 1999;19:7237–7244. doi: 10.1128/mcb.19.10.7237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brown JL, Fritsch C, Mueller J, Kassis JA. The Drosophila pho-like gene encodes a YY1-related DNA binding protein that is redundant with pleiohomeotic in homeotic gene silencing. Development. 2003;130:285–294. doi: 10.1242/dev.00204. [DOI] [PubMed] [Google Scholar]
- 15.Atchison L, Ghias A, Wilkinson F, Bonini N, Atchison ML. Transcription factor YY1 functions as a PcG protein in vivo. EMBO J. 2003;22:1347–1358. doi: 10.1093/emboj/cdg124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brown JL, Mucci D, Whiteley M, Dirksen ML, Kassis JA. The Drosophila Polycomb group gene pleiohomeotic encodes a DNA binding protein with homology to the transcription factor YY1. Mol. Cell. 1998;1:1057–1064. doi: 10.1016/s1097-2765(00)80106-9. [DOI] [PubMed] [Google Scholar]
- 17.Sui G, et al. Yin Yang 1 is a negative regulator of p53. Cell. 2004;117:859–872. doi: 10.1016/j.cell.2004.06.004. [DOI] [PubMed] [Google Scholar]
- 18.Gronroos E, Terentiev AA, Punga T, Ericsson J. YY1 inhibits the activation of the p53 tumor suppressor in response to genotoxic stress. Proc. Natl. Acad. Sci. USA. 2004;101:12165–12170. doi: 10.1073/pnas.0402283101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Affar el B, et al. Essential dosage-dependent functions of the transcription factor yin yang 1 in late embryonic development and cell cycle progression. Mol. Cell. Biol. 2006;26:3565–3581. doi: 10.1128/MCB.26.9.3565-3581.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.D’Andrea AD, Grompe M. The Fanconi anaemia/BRCA pathway. Nat. Rev. Cancer. 2003;3:23–34. doi: 10.1038/nrc970. [DOI] [PubMed] [Google Scholar]
- 21.German J. Bloom syndrome: a mendelian prototype of somatic mutational disease. Medicine (Baltimore) 1993;72:393–406. [PubMed] [Google Scholar]
- 22.Auerbach AD, Wolman SR. Susceptibility of Fanconi’s anaemia fibroblasts to chromosome damage by carcinogens. Nature. 1976;261:494–496. doi: 10.1038/261494a0. [DOI] [PubMed] [Google Scholar]
- 23.Scully R, Livingston DM. In search of the tumour-suppressor functions of BRCA1 and BRCA2. Nature. 2000;408:429–432. doi: 10.1038/35044000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Patel KJ, et al. Involvement of Brca2 in DNA repair. Mol. Cell. 1998;1:347–357. doi: 10.1016/s1097-2765(00)80035-0. [DOI] [PubMed] [Google Scholar]
- 25.Elledge SJ. Cell cycle checkpoints: preventing an identity crisis. Science. 1996;274:1664–1672. doi: 10.1126/science.274.5293.1664. [DOI] [PubMed] [Google Scholar]
- 26.Manke IA, et al. MAPKAP kinase-2 is a cell cycle checkpoint kinase that regulates the G2/M transition and S phase progression in response to UV irradiation. Mol. Cell. 2005;17:37–48. doi: 10.1016/j.molcel.2004.11.021. [DOI] [PubMed] [Google Scholar]
- 27.Groisman R, et al. The ubiquitin ligase activity in the DDB2 and CSA complexes is differentially regulated by the COP9 signalosome in response to DNA damage. Cell. 2003;113:357–367. doi: 10.1016/s0092-8674(03)00316-7. [DOI] [PubMed] [Google Scholar]
- 28.Shi Y, et al. Coordinated histone modifications mediated by a CtBP co-repressor complex. Nature. 2003;422:735–738. doi: 10.1038/nature01550. [DOI] [PubMed] [Google Scholar]
- 29.Cai Y, et al. YY1 functions with INO80 to activate transcription. Nat. Struct. Mol. Biol. 2007;14:872–874. doi: 10.1038/nsmb1276. [DOI] [PubMed] [Google Scholar]
- 30.Taccioli GE, et al. Ku80: product of the XRCC5 gene and its role in DNA repair and V(D)J recombination. Science. 1994;265:1442–1445. doi: 10.1126/science.8073286. [DOI] [PubMed] [Google Scholar]
- 31.Allen C, Kurimasa A, Brenneman MA, Chen DJ, Nickoloff JA. DNA-dependent protein kinase suppresses double-strand break-induced and spontaneous homologous recombination. Proc. Natl. Acad. Sci. USA. 2002;99:3758–3763. doi: 10.1073/pnas.052545899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Blunt T, et al. Defective DNA-dependent protein kinase activity is linked to V(D)J recombination and DNA repair defects associated with the murine scid mutation. Cell. 1995;80:813–823. doi: 10.1016/0092-8674(95)90360-7. [DOI] [PubMed] [Google Scholar]
- 33.Schildkraut E, Miller CA, Nickoloff JA. Gene conversion and deletion frequencies during double-strand break repair in human cells are controlled by the distance between direct repeats. Nucleic Acids Res. 2005;33:1574–1580. doi: 10.1093/nar/gki295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gaffney EV, et al. Established lines of SV40-transformed human amnion cells. Cancer Res. 1970;30:1668–1676. [PubMed] [Google Scholar]
- 35.Labrecque S, Matlashewski GJ. Viability of wild type p53-containing and p53-deficient tumor cells following anticancer treatment: the use of human papillomavirus E6 to target p53. Oncogene. 1995;11:387–392. [PubMed] [Google Scholar]
- 36.Allen C, Halbrook J, Nickoloff JA. Interactive competition between homologous recombination and non-homologous end joining. Mol. Cancer Res. 2003;1:913–920. [PubMed] [Google Scholar]
- 37.Tsaneva IR, Muller B, West SC. ATP-dependent branch migration of Holliday junctions promoted by the RuvA and RuvB proteins of E. coli. Cell. 1992;69:1171–1180. doi: 10.1016/0092-8674(92)90638-s. [DOI] [PubMed] [Google Scholar]
- 38.Parsons CA, Tsaneva I, Lloyd RG, West SC. Interaction of Escherichia coli RuvA and RuvB proteins with synthetic Holliday junctions. Proc. Natl. Acad. Sci. USA. 1992;89:5452–5456. doi: 10.1073/pnas.89.12.5452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Iwasaki H, Takahagi M, Nakata A, Shinagawa H. Escherichia coli RuvA and RuvB proteins specifically interact with Holliday junctions and promote branch migration. Genes Dev. 1992;6:2214–2220. doi: 10.1101/gad.6.11.2214. [DOI] [PubMed] [Google Scholar]
- 40.Klymenko T, et al. A Polycomb group protein complex with sequence-specific DNA-binding and selective methyl-lysine-binding activities. Genes Dev. 2006;20:1110–1122. doi: 10.1101/gad.377406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bugreev DV, Mazina OM, Mazin AV. Rad54 protein promotes branch migration of Holliday junctions. Nature. 2006;442:590–593. doi: 10.1038/nature04889. [DOI] [PubMed] [Google Scholar]
- 42.Ingleston SM, et al. Holliday junction binding and processing by the RuvA protein of Mycoplasma pneumoniae. Eur. J. Biochem. 2002;269:1525–1533. doi: 10.1046/j.1432-1033.2002.02805.x. [DOI] [PubMed] [Google Scholar]
- 43.Lisby M, Rothstein R. DNA damage checkpoint and repair centers. Curr. Opin. Cell Biol. 2004;16:328–334. doi: 10.1016/j.ceb.2004.03.011. [DOI] [PubMed] [Google Scholar]
- 44.Lisby M, Rothstein R, Mortensen UH. Rad52 forms DNA repair and recombination centers during S phase. Proc. Natl. Acad. Sci. USA. 2001;98:8276–8282. doi: 10.1073/pnas.121006298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gao H, Chen XB, McGowan CH. Mus81 endonuclease localizes to nucleoli and to regions of DNA damage in human S-phase cells. Mol. Biol. Cell. 2003;14:4826–4834. doi: 10.1091/mbc.E03-05-0276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sui G, Shi Y. Gene silencing by a DNA vector-based RNAi technology. Methods Mol. Biol. 2005;309:205–218. doi: 10.1385/1-59259-935-4:205. [DOI] [PubMed] [Google Scholar]
- 47.Savage JR. Classification and relationships of induced chromosomal structual changes. J. Med. Genet. 1976;13:103–122. doi: 10.1136/jmg.13.2.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ikura T, et al. Involvement of the TIP60 histone acetylase complex in DNA repair and apoptosis. Cell. 2000;102:463–473. doi: 10.1016/s0092-8674(00)00051-9. [DOI] [PubMed] [Google Scholar]
- 49.Karow JK, Constantinou A, Li JL, West SC, Hickson ID. The Bloom’s syndrome gene product promotes branch migration of holliday junctions. Proc. Natl. Acad. Sci. USA. 2000;97:6504–6508. doi: 10.1073/pnas.100448097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kaplan DL, O’Donnell M. RuvA is a sliding collar that protects Holliday junctions from unwinding while promoting branch migration. J. Mol. Biol. 2006;355:473–490. doi: 10.1016/j.jmb.2005.10.075. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.