Abstract
Recent advances in molecular cloning have led to the identification of a large number of mammalian zinc finger-containing transcription factors that exhibit homology to the Drosophila melanogaster protein, Krüppel. Although the amino acid sequences in the zinc finger domains of these Krüppel-like factors (KLFs) are closely related to one another, the regions outside the zinc fingers of the proteins are usually unique. KLFs display seemingly different and broad biological properties with each functioning as an activator of transcription, a repressor or both. This review article provides a current phylogenetic classification of the identified KLFs to date. More importantly, the currently known biological activities of the KLFs in regulating transcription, cell proliferation, differentiation and development are summarized and compared. Further characterization of this interesting protein family should provide additional insights into the their respective regulatory role in various important biological processes. © 2000 Elsevier Science Ltd. All rights reserved.
Keywords: Activation, Gene expression, Krüppel-like factors, Repression, Transcription factors, Zinc fingers
1. Introduction
The regulation of tissue- and development-specific expression of eukaryotic genes is a fundamentally important process that represents the focus of research for numerous molecular biologists. A great deal of attention has been paid to the study of transcription factors, which directly participate in the regulation of gene transcription by interacting with cis-regulatory DNA elements in specific genes. Most transcription factors are classified based on the structural motifs that they use to bind to DNA (reviewed in Ref. [1]). The zinc finger motif is an important example of a DNA-binding motif. As the name implies, a zinc finger contains a single zinc atom, which serves as a critical structural component of the finger motif. The zinc atom is tetrahedrally coordinated by amino acids, such as cysteine and histidine. A frequently encountered zinc finger protein contains two cysteine and two histidine residues per finger and is referred to as a C2H2 zinc finger. Each finger is a simple structure consisting of 25–30 amino acid residues that includes two β-pleaded sheets in the amino terminal half and an α-helix in the carboxyl terminal half, held together at the base by the zinc atom. Many zinc fingertranscription factors contain multiple fingers that are continuously aligned with one another, which fit in the major groove of DNA [1].
The C2H2 zinc finger motif has a remarkably conserved primary sequence. In the completely sequenced Caenorhabditis elegans genome, 138 proteins (0.7% of all proteins) contain the C2H2 motif [2,3]. In comparison, there are 352 (2.6%) C2H2 zinc finger proteins in the recently completed genome of Drosophila melanogaster [3,4]. It has been estimated that ≈1% of the human genome consists of genes encoding C2H2 type of zinc finger protein [5]. This would correspond to between 700 and 1000 genes encoding distinct zinc finger proteins [6]. It is therefore, not surprising that 434 matches of human zinc finger proteins were found in a recent search in the GenBank protein database for protein sequences related to the three C2H2 zinc fingers of the ubiquitous transcription factor, Sp1 [7].
A subset of C2H2 zinc finger proteins contains amino acid sequences that resemble those of the segmentation gene product of D. melanogaster, Krüppel [8]. In addition to the conserved amino acid sequence in the zinc finger, these proteins share a highly conserved seven-amino acid inter-finger spacer, TGEKP(Y/F)X, often referred to as a H/C link. Sp1, for example, exhibits homology to Krüppel. Many other transcription factors containing the Krüppel motif have been identified. Studies indicate that they play key roles in regulating a diverse range of biological processes, including cell growth, differentiation, embryogenesis and tumorigenesis. More recently, a family of Krüppel-like factors (KLFs) that are highly related to the Krüppel protein erythroid Krüppel-like factor (EKLF) [9] has been described [10,11]. This family is expanding rapidly such that since EKLF was first cloned in 1993, a total of 12 KLFs have been identified. The KLFs have been given numerical designations by the Human Gene Nomenclature Committee (HGNC) [12]. EKLF, for instance, is now KLF1. Despite the relatively short span of time since their discovery, this family of proteins has been shown to exhibit important tissue- or organ-specific regulatory functions. The present article will review the known biological activities of the KLFs identified thus far.
2. Phylogenetic classification of the human Krüppel-like factor family
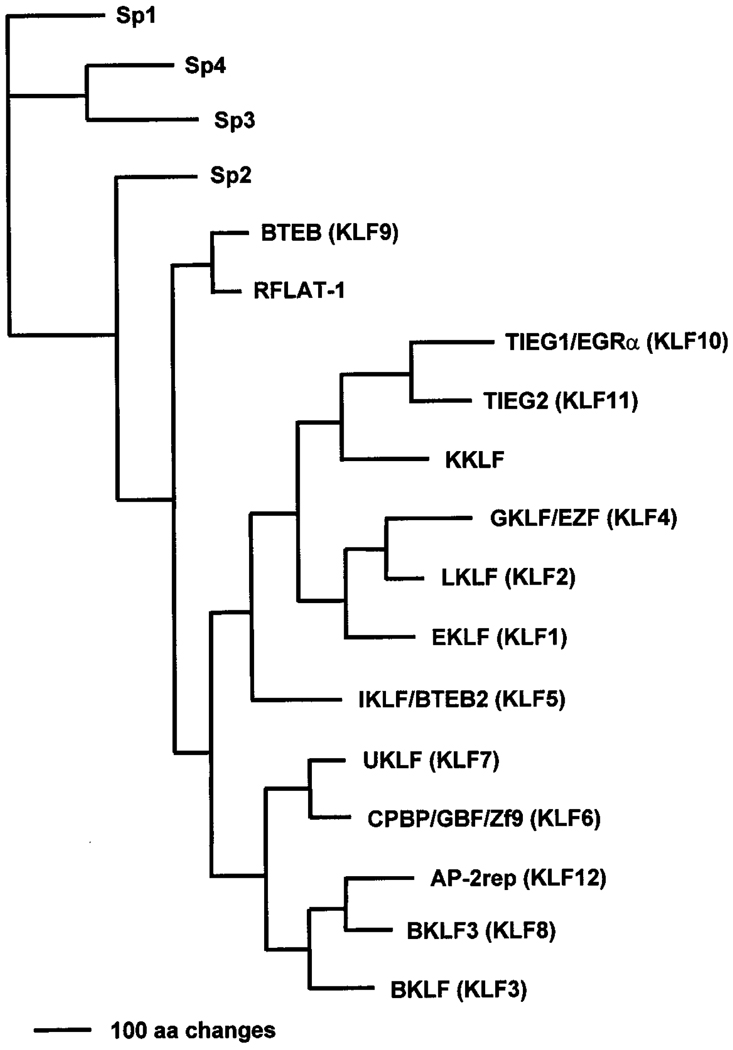
Fig. 1 illustrates the phylogenetic relationship among the 12 human KLFs along with an additional six other Krüppel-like proteins that have not received a HGNC numerical designation. Table 1 provides information on the GenBank accession number, the Unigene number, the length of the polypeptide, and the chromosome localization, when known, for each of the 12 KLFs. All proteins in Fig. 1 contain three zinc fingers. With the exception of KLFs 9–11, the zinc fingers of the remaining nine KLFs are localized to the extreme carboxyl terminus and are followed by an additional one to three amino acid residues before the termination codon. It is also clear from the diagram that subfamilies, or clusters, of related proteins could be defined due to close homology. Thus, EKLF (KLF1), LKLF (KLF2), and GKLF (KLF4) are more closely related to one another than to other members of the Krüppel protein family. In fact, this close relationship has previously been documented based on a functional analysis of the motif required for nuclear localization of the different Krüppel proteins [13]. Another example is provided by the concurrent regulation of two closely related Krüppel-like factors, TIEG1 (KLF10) and TIEG2 (KLF11), by the transforming growth factor-β1 (TGF-(β1) [14,15]. The latter example suggests that there are possibly functional, in addition to sequence, conservation within subfamilies of highly related Krüppel proteins.
Fig. 1.
Classification of related human Krüppel-like factors. Human KLF1 and related proteins were identified by BLAST searching, then multiple aligned using the PileUp program of GCG (Genetics Computer Group version 9.0, Madison, WI). The relationship of 18 Krüppel-like proteins was visualized as an unrooted phylogram using a heuristic search with Phylogeny Analysis Using Parsimony (PAUP version 4.0b3a; Sinauer Associates, Sunderland, MA). The horizontal bar in the left lower corner of the figure indicates 100 amino acid changes. Branch lengths are proportional to amino acid changes. For keys to abbreviations, see Table 1. RFLAT-1 and KKLF have not received a HNGC nomenclature. RFLAT-1 is RANTES (regulated upon activation, normal T cells expressed and secreted) factor of late activated T lymphocytes-1 [117]. Its gene is expressed in T cells 3 days after activation and coincides with RANTES expression. KKLF represents kidney-enriched Krüppel-like factor whose identification was only recently described [118]. It functions to repress transcription of two kidney-specific chloride channel genes by blocking the activating effect of another transcription factor, MA2 [118].
Table 1.
The 12 mammalian Krüppel-like factorsa
| HGNCb name | Gene | # AAc | Accession No. (protein) | Unigene No. | Chromosome localization | References |
|---|---|---|---|---|---|---|
| KLF1 | EKLF | 362 | Q13351 | Hs.37860 | 19p13.12– p13.13 | [9] |
| KLF2 | LKLF | 355 | NP_006066 | Hs.107740 | 19p13.11–p13.13 | [37] |
| KLF3 | BKLF | 345 | BAA92271 | Hs. 129026 | NDd | [44] |
| KLF4 | GKLF/EZF | 470 | NP_04226 | Hs.182965 | 9q31 | [50,52,54] |
| KLF5 | BTEB2/IKLF | 457 | AAF18307 | Hs.84728 | ND | [69,71,72] |
| KLF6 | CPBP/Zf9/GBF | 283 | AAC39929 | Hs.4055 | 10p15 | [75,76] |
| KLF7 | UKLF | 302 | NP_003700 | Hs.21599 | 2q32 | [78] |
| KLF8 | BKLF3 | 359 | NP_009181 | Hs.25442 | Xp11.21 | [84] |
| KLF9 | BTEB | 244 | NP_001197 | Hs.150557 | ND | [59] |
| KLF10 | TIEG1/EGRαTF | 480 | NP_005646 | Hs.82173 | 8 | [14,93] |
| KLF11 | TIEG2 | 512 | NP_003588 | Hs.193776 | ND | [15] |
| KLF12 | AP–2rep | 402 | NP_009180 | Hs.23510 | 13q22 | [108] |
EKLF, erythroid Krüppel-like factor; LKLF, lung Krüppel-like factor; BKLF, basic Krüppel-like factor; GKLF, gut-enriched Krüppel-like factor; EZF, epithelial zinc finger; BTEB2, basic transcription element binding protein 2; IKLF, intestinal-enriched Krüppel-like factor; CPBP, core promoter-binding protein; Zf9, Zinc finger 9; GBF, GC-rich sites binding factor; UKLF, ubiquitous Krüppel-like factor; BTEB, basic transcription element binding protein; TIEG1, TGF-β-inducible early gene 1; EGRαTF, early growth response α transcription factor; TIEG2, TGF-β-inducible early gene 2; AP-2rep, AP-2 repressor.
Human Gene Nomenclature Committee.
Amino acid.
Not determined.
3. The biology of mammalian Krüppel-like factors
3.1. Erythroid Krüppel-like factor (EKLF) (KLF1)
As the earliest identified KLF, EKLF represents a ‘prototype’ of mammalian Krüppel-like factors. Since its initial identification [9], a large body of literature has accumulated that describes the biochemical and physiological functions of EKLF (reviewed in Ref. [16]). EKLF was originally isolated by subtraction cloning of a cDNA library enriched for genes expressed in a mouse erythroleukemia cell line [9]. Expression of EKLF is predominantly restricted to cells of the erythroid lineage [9]. Moreover, EKLF binds to and activates a CACCC element in the promoter of the β-globin gene in humans and mice [9]. Because CACCC element is a functionally important and evolutionarily conserved cis-acting element of the β-globin gene as well as other erythroid cell-specific promoters and enhancers [17], EKLF is considered an important regulator of globin and other erythroid cell-specific gene expression. Indeed, EKLF has been shown to be critical for the cell-specific inducibility of the β-globin gene promoter and this activity is localized to the proline-rich transactivation domain of EKLF [18].
The biochemical mechanisms by which EKLF regulates transcription are well-established [16]. Human EKLF contains 362 amino acid (aa) residues [19], including three C2H2 zinc finger motifs near the very carboxyl (C) terminus. A short peptide sequence in the region immediately amino (N) terminal to the zinc fingers is very rich in basic aa residues. A similar basic region is found in the mouse GKLF, which was shown to be crucial for the nuclear localization of the protein [13]. The N-terminal half of EKLF, similar to GKLF and LKLF, is rich in proline (~ 15%), serine, theronine and acidic residues, all of which have been implicated in activation or repression of transcription [20]. EKLF binds strongly to the CACCC element in the β-globin promoter (5′-CCACACCCT) but to a much less extent to a similar element in the γ-globin promoter (5′-CTCCACCCA) [21]. One potential explanation for this difference is the presence of other neighboring cis-elements in the γ-, but not in the β-globin promoter that may prevent the recruitment of EKLF to the γ-globin promoter [22]. Added to the fact that EKLF is expressed at a higher level in adult erythroid tissue compared to the fetal tissue [21], it has been suggested that EKLF is involved in the human γ- to β-globin gene switch during development.
EKLF is an activator of transcription of the β-globin gene promoter [9]. The transactivation domain of EKLF has been localized to the N-terminal portion of the protein between aa residues 20 and 291 [18]. Further analyses of the mechanisms of transactivation by EKLF, however, indicate that they are far more complex than previously thought. Thus, the transactivation domain of EKLF contains distinct stimulatory and inhibitory subdomains [23]. The inhibitory subdomain resides between aa residues 196–291 and exerts its effect in cis by interfering with DNA binding [23]. In contrast, the stimulatory subdomain resides between aa residues 20–124, which coincides with an acidic aa-rich region of the protein that is also involved in interaction with other proteins [23]. Additional experiments suggest that either the conformation or phosphorylation status of this subdomain may be critical for such interactions [23]. Indeed, a casein kinase II (CKII) [24] site is present in the interaction domain of EKLF and can be phosphorylated by CKII [25]. Moreover, the ability of EKLF to transactivate gene expression has been shown to critically depend on the phosphorylation status of this CKII site [25].
An exciting recent advance in understanding EKLF’s biochemical properties is in the area of chromatin remodeling. A clue to the overall contribution of chromatin structure to EKLF function came from the observation that the nucleosome structure in the mouse β-globin gene promoter is altered (remains in a closed configuration) in mice that are null for the EKLF allele when compared to their wild type littermates [26]. Using an in vitro chromatin assembly system, a protein complex named E-RC1 (for EKLF coactivator-remodeling complex 1) has been identified and shown to induce transcription of an assembled chromatin template in collaboration with EKLF [27]. E-RC1 exhibits homology to the yeast SWI/SNF family of chromatin remodeling complexes, which are involved in the activation of transcription of a subset of genes [28]. The SWI/ SNF complex functions in order to disrupt chromatin structure and facilitate binding of transcription factors to the remodeled chromatin [29,30]. Although the ultimate nature of E-RC1 remains to be defined, it is possible that EKLF’s principal function as a transcriptional activator is to attract E-RC1 to the β-globin promoter.
Another important component of the EKLF transcription complex is a family of transcription co-activators that exhibits histone acetyltransferase (HAT) activity and modifies chromatin structures (reviewed in Ref. [31]). Thus, EKLF was shown to physically associate with HATs such as CBP, p300, and P/CAF [32]. Moreover, CBP and p300 acetylate at least two lysine residues in the inhibitory subdomain of EKLF immediately on the N-terminal side of the zinc fingers [23,32]. The interaction between EKLF and CBP/p300 leads to an enhancement of transcriptional activation of the β-globin promoter in erythroid cells [32]. Taken together, these observations indicate that EKLF is a tissue-specific transcription factor that undergoes post-translational modifications, including phosphorylation and acetylation and assists in the remodeling of chromatin structure by recruiting co-activator complexes to the β-globin gene cluster.
The in vivo function of EKLF can partially be elucidated by its pattern of expression during development [33]. The earliest detectable sign of EKLF expression in a developing mouse embryo is on embryonic day 7.5 (E7.5) in the primitive erythroid cells at the very beginning of blood island formation in the yolk sac. By E9, EKLF is expressed in the hepatic primordia and remains high in the developing liver, which becomes the sole source of EKLF mRNA in an E14.5 fetus. In the adult spleen, EKLF expression is strictly localized to the red pulp. These studies demonstrate that EKLF is a specific, early marker of erythroid differentiation and its expression in the adult is consistent with its requirement for later stage β-globin gene expression. More definitive proofs of the physiological functions of EKLF came from studies involving gene knockout. EKLF−/− mice die in utero at E15 due to severe ineffective erythropoiesis [34,35]. The levels of β-globin mRNA in the fetal livers of EKLF−/− mice are reduced 10-fold compared to heterozygous (EKLF+/−) littermates, whereas the levels of α-globin mRNA are normal. In addition, the embryonic stage of hematopoiesis in the EKLF−/− mice appears to be normal, including expression of the fetal and embryonic globin genes, ε and ξ [34,35]. It is not until hematopoiesis has been switched to the adult phenotype before the fatal anemia sets in. This stage-specific and β-globin-gene-specific requirement indicates that EKLF is necessary to facilitate the fetal-to-adult hemoglobin switch in mammals. The contribution of EKLF to red blood cell physiology is further documented by the inability of a human fetal Aγ-globin transgene to rescue the abnormal red blood cells, therefore prenatal lethality, in EKLF−/− embryos despite an apparent restoration of globin chain imbalance [36]. This last study suggests that other non-globin genes regulated by EKLF are essential for the normal progression of erythropoiesis.
3.2. Lung Krüppel-like factor (LKLF) (KLF2)
Using the zinc finger region of EKLF as a hybridization probe to screen a mouse genomic library, Lingrel et al. isolated a cDNA clone encoding a 354-aa polypeptide that they named LKLF [37]. Expression of the mouse LKLF gene is primarily found in the lung, heart and spleen [37]. The tissue distribution of the human LKLF transcript is similar to that of the mouse with the exception that the heart has a higher level of transcripts than the other organs [38]. Expression of LKLF is also developmentally regulated with expression first detected at E7 of the mouse embryo followed by a down-regulation at E11 and subsequent reactivation at E15 [37]. Similar to EKLF, the region of the protein outside its three zinc fingers has a high percentage of proline residues. Lastly, LKLF is able to transactivate a human β-globin gene promoter containing a CACCC element [37].
To better understand the in vivo function of LKLF, gene knockout experiments have been performed. These studies yielded interesting and somewhat surprising results. LKLF-deficient (−/−) mice die in utero from severe intra-embryonic and intra-amniotic hemorrhaging between E12.5 and E14.5 of development [39,40]. This bleeding disorder is associated with specific defects in the morphology of blood vessels. Umbilical veins and arteries in the LKLF−/− embryos display an abnormally thin tunica media and aneurysmal dilatation before rupturing into the amniotic cavity. Similarly, vascular smooth muscle cells in the aorta from LKLF−/− animals fail to organize into a compact tunica media. These results indicate that LKLF is a necessary component in the assembly of the vascular tunica media and blood vessel stabilization during embryogenesis.
To circumvent the problem of embryonic lethality in LKLF-null mice, Lingrel et al. generated mouse aggregation chimeras by injecting LKLF−/− embryonic stem (ES) cells into blastocysts of wild type mice, and analyzed the contribution of LKLF-deficient cells to the formation of various internal organs [41]. In chimeric mice that survive after birth, LKLF−/− ES cells contribute significantly to all of the major organs except the lungs. Although some highly chimeric animals die at birth, histopathological examination of their lungs suggests an abnormality in lung development [41]. It is concluded that LKLF plays an important role in normal lung development.
Additional insights into the physiological functions of LKLF are provided by an independent study, also involving mouse aggregation chimeras. In addition to the aforementioned organs, LKLF expression is also found in lymphoid tissues [42]. Specifically, LKLF mRNA is present in both CD4+ and CD8+ single-positive (SP) quiescent T lymphocytes in the thymus and spleen, but absent in double-positive (DP) CD4+CD8+ lymphocytes. Upon T cell receptor (TCR)-mediated activation of the quiescent splenic T cells, LKLF mRNA and protein are rapidly degraded, suggesting that LKLF play an important role in regulating the function of resting SP T cells in vivo. To address this issue and to circumvent the embryonic lethality in LKLF-null mice, Leiden et al. generated a model in which LKLF−/− ES cells are injected into recombinase activating gene 2-deficient (RAG2−/−) blastocysts to produce LKLF−/−RAG2−/− chimeric mice [42]. Because mature B and T cells cannot develop in the absence of RAG2, all B and T cells in the chimeric mice are derived from the LKLF−/− ES cells. In control experiments, injection of LKLF+/− ES cells into RAG2−/− blastocysts fully rescued the defects in B and T cell development in the RAG2−/− animals. In contrast, chimeric mice produced by injection of RAG2−/− blastocysts with LKLF−/− ES cells display profound peripheral T cell defects. Total splenic and lymph node T cell numbers are reduced by > 90%. In addition, the few LKLF−/− splenic and lymph node T cells display a constellation of cell surface markers that is characteristic of an activated phenotype, which leads to death from Fas ligand-induced apoptosis [42]. These results, therefore, indicate that LKLF is required to program the quiescent state of SP T lymphocytes and to maintain their viability in the peripheral lymph organs and blood. A more recent study showed that LKLF is reexpressed following culture of activated CD8+ T cells in certain cytokines (IL-2, IL-7) that are known to influence T cell development [43]. Evidence was presented that supports a role for the induction of LKLF reexpression in determining long-term T cell survival and development of memory T cells [43].
3.3. Basic Krüppel-like factor (BKLF) (KLF3)
In addition to EKLF, the CACCC element in the β-globin gene promoter interacts with a number of other proteins when incubated with nuclear extracts isolated from the murine erythroleukemia cell line, MEL [44]. One of these proteins is Sp1. To identify the remaining protein(s) that binds to CACCC, Crossley et al. screened a MEL cDNA library with a mixed probe containing sequences in the zinc finger regions of EKLF and Sp1 [44]. Several of the identified positive clones encode a novel cDNA sequence, which is designated BKLF. The open reading frame of BKLF contains a protein of 344 aa that is subsequently shown to bind to the CACCC sequence in the β-globin promoter [44]. A recent report indicates that BKLF also binds to a CACCC element present in the promoter of the human C4 complement gene [45]. Similar to other KLFs, the region of BKLF outside the zinc fingers is rich in proline residues. However, a notable difference between BKLF and EKLF is the unusually high isoelectric point of 10.2 of BKLF, thus the name basic KLF, as compared to 7.0 for EKLF. The tissue distribution of BKLF in the adult mouse is also wider than that of EKLF and includes liver, lung, muscle and brain, in addition to hematopoietic tissues [44]. It is of interest to note that the level of BKLF is selectively reduced in fetal liver erythrocytes in mice lacking the EKLF gene due to knockout [44]. This suggests that BKLF may participate in the regulation of erythropoiesis in conjunction with EKLF. A preliminary report indicating that mice with BKLF knockout display abnormalities in hematopoiesis, albeit less severe than the EKLF−/− mice, supports the notion that BKLF is involved in erythropoiesis [46].
BKLF can activate a reporter gene containing a CACCC element although the degree of activation is less than that caused by EKLF [44]. However, when the CACCC sequence is linked to a glucocorticoid response element (GRE), BKLF becomes a potent repressor of the reporter gene while EKLF remains a strong activator [47]. The repression domain is mapped to the N-terminal 74 aa of BKLF [47]. Using this region of BKLF as a bait in a yeast two-hybrid screen, Turner and Crossley isolated a cDNA clone encoding a co-repressor called C-terminal-binding protein 2 (CtBP2) [47]. A model of the mechanism of repression by BKLF and CtBP2 has recently been proposed [48]. Given that EKLF is highly expressed in late stages of erythroid development and that EKLF may be an activator of the BKLF gene, it is not surprising that BKLF is also highly expressed in similar stages. Relative to EKLF, BKLF binds to the CACCC element in the fetal γ-globin promoter with a much higher affinity [44]. It is, therefore, possible that one of the primary physiological functions of BKLF is the suppression of fetal and embryonic globin gene expression in adult tissues. This hypothesis is supported by the observation that there is an excess of fetal globin chains in the embryos of EKLF−/− mice when the BKLF level is concomitantly decreased [34,35].
3.4. Gut-enriched Krüppel-like factor (GKLF) (KLF4)
In an effort to identify novel zinc finger-containing transcription factors with a role in regulating cell growth, Yang et al. screened a NIH 3T3 cDNA library at reduced stringency with the zinc finger portion of an immediate early transcription factor zif268 [49]. One of the positive clones contained a novel sequence, which was subsequently named gut-enriched Krüppel-like factor (GKLF) [50]. GKLF encodes a polypeptide of 483 aa and, similar to KLFs 1–3, contains three Krüppel-type zinc fingers in the very C-terminal end. The region immediately N terminal to the three zinc fingers is a 20-aa peptide containing a cluster of basic aa residues, which is essential for the nuclear localization of the protein [13]. Only two other KLFs, EKLF and LKLF, exhibit any degree of sequence homology to this basic region of GKLF, indicating that the three KLFs belong to a subfamily of closely related Krüppel proteins [13] (see Fig. 1).
The tissue distribution of GKLF is enriched for the gastrointestinal tract; hence the name gut-enriched KLF [50,51]. Expression of GKLF can also be found in a select number of other organs including the lung [50], testis [50], skin [52] and thymus [53], and in vascular endothelial cells [51]. In situ hybridization experiments indicate that expression of GKLF primarily occurs in the epithelial cells of the gut [50,52] and skin [52]. In these epithelial tissues, the GKLF transcript is primarily localized to the post-mitotic cells of the gut [50] and skin epithelium [52]. Similarly, GKLF expression has been correlated with maturation of thymus epithelial cells [53]. Collectively, these studies establish that expression of GKLF is primarily associated with a terminally differentiated state of epithelial cells. This is supported by the observation that fetal GKLF expression is highest around E17 [52,53,55], a time during fetal development in which the gut, skin and thymus epithelial tissues undergo dramatic changes prior to maturation.
The biochemical properties of GKLF are well established. GKLF is able to interact with the CACCC element in the β-globin promoter [52,54]. Using an empirical approach, Shields and Yang identified a consensus DNA sequence to which GKLF binds [56]. This sequence is GC-rich and is similar but not identical to the binding site for Sp1. One of the naturally occurring cis-elements called the basic transcription element (BTE), found in the promoter and essential for the expression of a family of cytochrome P-450 drug-metabolizing genes [57], is highly related to the consensus binding sequence of GKLF and shown to interact with GKLF [58]. Indeed, GKLF can inhibit the promoter activity of one of the cytochrome P-450 genes, CYP1A1, in a BTE-dependent fashion [58]. This inhibition is due primarily to the ability of GKLF to physically compete with Sp1, a strong activator of the CYP1A1 promoter [59], for binding to the BTE [58]. In contrast, other studies have shown that GKLF can act as an activator of transcription either by itself [51,56] or in conjunction with other Krüppel-like factors [60,61]. GKLF is therefore a pleiotropic transcription factor with a context-dependent transcriptional activating or suppressing activity. A detailed characterization of the mechanism of transactivation by GKLF has recently been performed [62]. This study identified a number of acidic aa residues near the N-terminus that are crucial for the transactivating effect of GKLF [62]. In addition, GKLF physically interacts with the p300/CBP transcription co-activators through the same acidic residues and this interaction is necessary for the transactivating ability of GKLF [62]. In this regard, the mechanism of activation by GKLF is similar to EKLF [32].
The association of GKLF expression with a growth-arrested state in vivo has been demonstrated in vitro as well. Thus, in cultured NIH 3T3 cells, expression of GKLF is found in a growth-arrest state that is either induced by serum deprivation or contact inhibition [50]. In contrast, there is little or no GKLF expression in cells that are actively proliferating [50]. In cells rendered quiescent by serum deprivation then stimulated to reenter the cell cycle by serum addition, there is a temporal down-regulation of GKLF expression during a critical phase of DNA synthesis [50]. Conversely, forced expression of GKLF by transfection inhibits DNA synthesis [50]. These results suggest that GKLF is a potent negative regulator of cell growth. This conclusion is consistent with the finding that expression of GKLF is decreased in the intestine of a murine tumor model, the Min mouse, in which large numbers of intestinal adenomas form as a result of mutation of the tumor suppressor gene, APC [55]. More recently, it was shown that expression of GKLF is reduced in the intestinal adenomas of Min mice when compared to the surrounding normal mucosa, as well as in the colonic adenomas of patients with familial adenomatous polyposis, a hereditary colon cancer syndrome resulted from mutation in the APC gene [63]. Combined together, these observations strongly support a role of GKLF in regulating proliferation of the intestinal epithelial cells.
The involvement of GKLF in the control of the cell cycle is further supported by recent studies showing that GKLF is induced during cell cycle arrest due to DNA damage [64]. This induction is caused by the activation of the tumor suppressor, p53, and a consequence of this induction is the transcriptional activation of the gene encoding the cyclin-dependent kinase inhibitor, p21WAF/Cip1, through a direct binding of GKLF to the p21WAF1/Cip1 promoter [64]. Overexpression of GKLF in the human colonic carcinoma cells, HT-29, also results in a reduction of cyclin D1 expression and that this reduction is due to a repressive effect of GKLF on the cyclin D1 promoter [65]. Moreover, treatment of HT-29 cells with interferon-γ inhibits proliferation and induces apoptosis, which is accompanied by a rise in the levels of GKLF mRNA [66]. It is of interest to note that the inductive effect of interferon-γ on GKLF expression, unlike that resulted from DNA damage, is independent of p53 [66]. Be that as it may, it is clear that there appears to be two distinct classes of ‘target’ genes regulated by GKLF. One includes genes with functions primarily related to terminal differentiation such as CYP1A1 [58], keratin 4 [60] and keratin 19 [61]. The other group belongs to genes that are directly involved in the regulation of cell growth including p21WAF1/Cip1 [64], cyclin D1 [65], and even GKLF itself [67]. These studies suggest that GKLF may have a dually important function in regulating differentiation- and growth-specific gene expression in epithelial cells.
A hint of the physiological function of GKLF is provided by a recent study in knockout mice [68]. Mice null for the GKLF alleles are born normal but die within 24 h after birth due to a loss of skin barrier function [68]. This loss of barrier function occurs without significant morphological or biochemical alterations to the well-known structural features of epidermis that are essential for mechanical integrity. Instead, late stage differentiation structures are selectively perturbed, including the cornified envelope [68]. Although the guts of the GKLF−/− mice are described to be normal, no histological or physiological information is provided to allow evaluation whether gastrointestinal function is perturbed due to the loss of GKLF.
3.5. Intestinal-enriched Krüppel-like factor (IKLF) (KLF5)
In an attempt to identify novel members of the Krüppel-like family of transcription factors, Lingrel et al. conducted a search of the expressed sequence tag database (dbEST) using the zinc finger region of LKLF [69]. Four overlapping cDNA clones were obtained that encode a 446-aa polypeptide, subsequently named IKLF [69]. Like GKLF, IKLF is primarily expressed in the gasatrointestinal epithelium although its site of expression is toward the proliferating zone of crypt cells [69,70], as opposed to the more luminal expression of GKLF [50]. Thus, expression of GKLF and IKLF in the intestinal epithelium appears to be complementary, rather than redundant, to each other. The developmental expression of the two genes also seems to contrast each other. While expression of GKLF is higher in later stages of gestation (E16-E17), that of IKLF is higher in earlier stages (E7). Like GKLF, IKLF is able to bind to the CACCC element of the [β-globin promoter and activates a reporter gene linked to two copies of the CACCC sequence [69]. A human homologue of the mouse IKLF gene was recently identified based on its ability to bind to the epidermal growth factor response element (EGFRE) of the lactoferrin gene and found to be 93% identical [71]. Interestingly, this human IKLF represses expression of a reporter driven by the lactoferrin EGFRE [71].
It should be noted that IKLF is identical to a previously isolated protein called BTEB2 (for basic transcription element binding protein 2) with the exception that BTEB2 is only 219 aa in length [72]. It appears that the open reading frame of BTEB2 represents only a part of the IKLF as shown by Ohnishi et al.[70]. A cDNA clone encoding a rabbit homologue of BTEB2 has also been recently identified from a rabbit aorta smooth muscle cell line [73]. In that model, expression of the BTEB2 gene is highly induced in the neointimal smooth muscle cell of the rat aorta after balloon injury [73]. In cultured cells, the levels of BTEB2 mRNA are rapidly and persistently induced by treatment with phorbol ester and basic fibroblast growth factor [74]. This induction is mediated by the immediate early gene product Egr-1 through its binding to an Egr-1-binding site in the promoter of BTEB2 [74]. It therefore appears that BTEB2 may be a mediator of the cellular responses to mitogenic stimulation.
3.6. Core promoter binding protein (CPBP)/GC-rich sites binding factor (GBF)/Zf9 (KLF 6)
Within the span of a year, three groups independently isolated cDNA clones encoding KLF6, which was given such different names as CPBP [75], GBF [76] and Zf9 [77]. Phylogenetically, KLF6 is most related to KLF7 [78] (Fig. 1). Human KLF6 contains 283 aa and is present in many tissues, including the placenta, heart, lung, liver and pancreas [75]. As an activator of transcription, KLF6 interacts with the core promoter element present in both TATA-less [75] or TATA box-containing promoters [76]. The cellular ‘target’ genes of KLF6 identified to date include those encoding pregnancy-specific glycoprotein 5 (PSG5) [75], collagen α1(I) [76], leukotriene C4 synthase (LTC4S) [79], urokinase plasminogen activator (uPA) [80], transforming growth factor (TGF)-β1 [81], and types I and II TGF-β receptors [81]. In addition, KLF6 physically interacts with GKLF and the two coactivate the human keratin 4 promoter [60].
Although initially thought to be constitutively expressed in a diverse array of tissues, KLF6 has recently been shown to be an inducible gene with characteristics resembling an immediate-early gene [80,82]. For example, the level of KLF6 protein increases significantly in liver stellate cells within 3 h of liver injury caused by CCl4 [77]. Expression of KLF6 is also transiently induced in 3T3-L1 preadipocytes stimulated to differentiate by various adipogenic hormones including serum, insulin, phorbol ester, and agents that elevate intracellular cyclic AMP levels [82]. Similarly, KLF6 is induced by phorbol ester in bovine aortic endothelial cells [80]. These observations suggest that KLF6 expression is regulated in a number of physiological processes and the induced production of KLF6 may mediate some of the subsequent physiological responses to these stimuli.
Studies by Friedman et al. have shed additional light on the function of KLF6. Activation of liver stellate cells is a physiological response to many forms of liver injury [83]. The activated stellate cells play a crucial role in the formation of extracellular matrix, which leads to subsequent liver fibrosis [83]. Induction of KLF6 expression occurs rapidly during stellate cell activation and precedes induction of structural and cytokine genes, including collagens, platelet-derived growth factor (PDGF) receptor, and transforming growth factor-β1 and its receptors [77]. Induction of KLF6 occurs in distinct models of liver injury in vivo and in vitro, suggesting that the upregulation of KLF6 is a general feature of stellate cell activation. The significance of this induction is further enhanced by the observation that KLF6 functions to activate the promoters of collagen α1(I) [77], TGF-β1 [81], and types I and II TGF-β receptors [81]. The combined expression of these down-stream genes may contribute to the process of hepatic fibrogenesis. Additional evidence supporting a role for KLF6 in mediating extracellular matrix homeostasis is provided by a recent study demonstrating the induction of KLF6 in balloon-injured vascular endothelium [80]. A consequence of this induction is the KLF6-mediated transcriptional activation of the gene encoding urokinase plasminogen activator (uPA), which in turn causes an increased level of bioactive TGF-β via enhancement of proteolytic activation of latent TGF-β [80]. These studies suggest that KLF6 may have a wide range of physiological functions in mediating fibrogenic or fibrinolytic responses of various tissues to injury.
3.7. Ubiquitous Krüppel-like factor (UKLF) (KLF7)
Only one publication to date describes the identification and isolation of human ubiquitous Krüppel-like factor (UKLF) [78]. UKLF was originally identified during a search for novel Krüppel-like proteins expressed in endothelial cells using a PCR strategy designed to amplify EKLF-related sequences. After its cloning, it becomes clear that UKLF is present in almost all tissues surveyed, hence the name ubiquitous [78]. Like other KLFs, UKLF binds strongly to the CACCC element but weakly to the Sp1 element. In a manner similar to the mechanism of transactivation by GKLF [62], the activation domain of UKLF is localized to the 72 aa in the N-terminal portion of the protein in a region that is rich in acidic residues [78]. Among all the KLFs, UKLF is most related to KLF6 although the two genes are localized to different chromosomes [78].
3.8. Basic Krüppel-like factor 3 (BKLF3)/ZNF741 (KLF8)
Using the sequence for an expressed sequence tag, ZNF741, a cDNA clone named BKLF3 or KLF8 was isolated from the K562 cell line [84]. Like many KLFs, KLF8 is a CACCC-binding protein that exerts a repressive effect on a CACCC-dependent promoter [84]. At least part of this repressive effect can be attributed to the association of KLF8 with the co-repressor C-terminal binding protein (CtBP) [84]. It is interesting to note that all three closely related KLFs, KLF3, KLF8 and KLF12 (Fig. 1), contain a conserved motif that is used to contact CtBP [84]. This group of KLFs, therefore, is functionally conserved and behaves mostly as transcriptional repressors.
3.9. Basic transcription element-binding protein (BTEB) (KLF9)
Of the 12 KLFs, BTEB was the first identified [59]. Along with Sp1, BTEB was cloned based on its ability to bind to the basic transcription element (BTE), which is a single GC-box sequence in the promoter and necessary for the constitutive expression of the cytochrome P-450IA1 (CYP1A1) gene [85]. Human BTEB is 244 aa in length [86], the zinc fingers of which bind to BTE with an affinity equal to Sp1 [87]. In contrast to some of the other KLFs, such as EKLF and GKLF, the transcriptional activation domain of BTEB is localized to two regions that are rich in hydrophobic aa residues [88]. Interestingly, like GKLF, BTEB is a bifunctional transcription factor, capable of activating the expression of genes containing multiple GC box sequences in promoters such as the simian virus 40 early promoter, but repressing a BTE-containing promoter present in the CYP1A1 gene [59].
Although a BTEB-knockout model is not available, several recent studies have partially revealed its potential physiological functions. These studies suggest that BTEB has a regulatory role in diverse biological processes. For example, a recent study implicates BTEB in mediating collagen α1(I) gene expression which is induced by acetaldehyde, the major active metabolite of alcohol, in rat liver stellate cells [89]. In this regard, BTEB is similar to Zf9, which also activates collagen α1(I) gene expression in stellate cells during liver injury [77]. Since an excessive production of collagen matrix proteins is central to the pathogenesis of hepatic cirrhosis, which results from various forms of liver injury including those caused by alcohol, both BTEB and Zf9 appear to have fundamentally important roles in mediating the fibrotic responses in the liver. A different group provides evidence that BTEB may have a regulatory function in endometrial epithelial gene expression associated with pregnancy. First, BTEB is specifically localized to the porcine endometrial epithelium during different stages of pregnancy [90]. Second, in conjunction with progesterone receptor, BTEB is able to activate expression of the uteroferrin gene, which encodes a uterine en-dometrial secretory protein [91]. Although these studies are suggestive that BTEB may be involved in mediating hormone-regulated gene expression, they do not present direct evidence. In contrast, another recent study showed that BTEB expression is regulated by the thyroid hormone, T3, and that this regulation is specific to neuronal cells in the developing central nervous system [92]. Overexpression of BTEB in neuronal cells increases the number and length of neurites in a dose-dependent manner, suggesting that BTEB play a critical role in neural development.
3.10. Early growth response α (EGRα)/transforming growth factor-β-inducible early gene 1(TIEG1) (KLF10) and transforming growth factor-β-inducible early gene 2(TIEG2) (KLF11)
TIEG1/EGRα (KLF10) and TIEG2 (KLF11) are two highly related Krüppel-like factors (Fig. 1) (for a detailed recent review, see Ref. [93]). Human TIEG1 encodes a 480-aa ubiquitously expressed polypeptide. It was initially identified by differential display-polymerase chain reaction in a human osteoblastic cell line that had been treated with transforming growth factor-β1 (TGF-β1) [14]. In the same study, a select number of growth-stimulating factors other than TGF-β1, including bone morphogenetic factor 2 (BMP2) and epidermal growth factor (EGF), have been shown to induce TIEG1 gene expression [14]. A second group concomitantly identified TIEG1 as early growth response α (EGRα) gene in several prostate cancer cell lines and showed that its expression is regulated by serum, phorbol ester, and EGF 8594. TIEG1 and EGRα subsequently were confirmed to be encoded by the same gene [95]. In addition to being subjected to regulation by various growth factors, TIEG1/EGRα expression is also influenced by various hormones. For example, the estrogen 17β-estradiol (E2) causes a rapid increase in expression of TIEG1/EGRα in estrogen receptor (ER)-positive human fetal osteoblastic cells [96]. In contrast, androgens such as 5α-dihydroxytestosterone (DHT) inhibit expression of TIEG1/EGRα in prostate cancer cell lines [94,97].
Consistent with its pattern of expression in response to growth factor and hormonal treatments, the presumed physiological function of TIEG1/EGRα is its involvement in the regulation of cell growth. In synchronized prostate cells, for example, EGRα mRNA is highly expressed in the G1 phase of the cell cycle [94]. Similarly, the rapid induction in TIEG1/EGRα expression in human fetal osteoblastic cells by estrogen is correlated with an estrogen-induced inhibition of DNA synthesis in the same cells [96]. A converse relationship between TIEG1/EGRα expression and the state of cell growth is also illustrated by a decreased TIEG1/ EGRα mRNA level in breast cancer cells as compared to normal breast epithelium [98]. The expression of TIEG1/EGRα has also been shown to be similarly decreased in prostate cancers compared to normal prostate [99]. Taken together, these studies are highly suggestive of an anti-proliferative nature of TIEG1/ EGRα activity.
TGF-β1 is a potent inhibitor of cell growth in many different cell types, in part by causing apoptosis [100–102]. The fact that TIEG1 expression is induced by TGF-β1 suggests that TIEG1 may be involved in the regulation of apoptosis. Indeed, overexpression of TIEG1 in a TGF-β1-sensitive pancreatic cell line, PANC1, led to the induction of apoptosis [103]. A similar apoptotic-promoting effect by TIEG1 has been documented in another epithelial cell line of lung origin, Mv1Lu [104]. More recently, the mechanisms by which TIEG1 causes apoptosis was defined in further detail in the liver cell line, Hep 3B, that has been rendered to undergo apoptosis by TGF-β1 [105]. In these cells, the induction in TIEG1 expression by TGF β1 precedes that of any morphological features of apoptosis. The expression of genes encoding some of the classical apoptotic proteins, such as Bax and Bcl-XL, does not change after treatment with TGF-β1. Instead, TGF-β1- and TIEG1-induced cell death are accompanied by an increase in the generation of reactive oxygen species and a loss of mitochondrial membrane potential preceding the morphological changes of apoptosis [105]. This mechanism by which TIEG1 induces oxidative stress is reminiscent of the mechanism of action by p53 to generate reactive oxygen species. p53-Induced apoptosis is accompanied by the transcriptional induction of redox-related genes [106]. It is proposed that these gene products form reactive oxygen species, which lead to oxidative mitochondrial damage. Due to the similarities between TIEG1- and p53-dependent apoptosis, it is possible that TIEG1 expression may also cause the transcriptional induction of redox-related genes.
A closely related cDNA to TIEG1, called TIEG2 (KLF11), was recently identified based on the homology to TIEG1 [15]. TIEG2 shares 79% amino acid identity with TIEG1 in the zinc finger region and 40% identity outside the zinc fingers. Similar to TIEG1, the tissue expression of TIEG2 is ubiquitous but with an enrichment in pancreas and muscle [15]. Moreover, expression of TIEG2 is similarly induced by TGF-β1 and that its overexpression inhibits cell growth [15]. It is of interest to note that both TIEG1 and TIEG2 are repressors of transcription [15] and that their repressive activities are localized to three conserved repressor domains in the two proteins [107]. Combined together, these findings highly suggest that TIEG1 and TIEG2 are two closely related Krüppel-like proteins with similar physiological roles in mediating apoptosis caused by TGF-β1.
3.11. AP-2rep (KLF12)
AP-2rep (KLF12) was first identified by its ability to bind to a crucial cis-element in the promoter of the gene encoding AP-2α, an important mammalian transcription factor [108]. This element confers autoregulatory activity with a core consensus binding site for AP-2 and was found to interact with three different transcription factors, AP-2rep, BTEB (KLF9), and AP-2 [108]. Whereas BTEB and AP-2 are both activators of the AP-2α promoter, AP-2rep is a potent repressor of the promoter activity [108]. There is also an excellent correlation between induction of AP-2rep mRNA expression and downregulation of AP-2α mRNA during development of the kidney [108]. Moreover, AP-2rep is able to suppress the endogenous AP-2α gene expression and is inversely negatively regulated by AP-2α [109]. These findings therefore point to a role for AP-2rep as a transcriptional silencer and reveal reciprocal regulation of AP-2α and AP-2rep.
4. Why so many related Krüppel-like factors?
It was merely 15 years ago when the zinc finger was first proposed to represent a stably folded structural domain that involves the binding of zinc to conserved cysteine and histidine residues in the repeated sequences of the Xenopus transcription factor TFIIIA [110,111]. At the time,Klug et al. noted that “it would not be surprising if the same 30 residue units were found to occur in varying numbers in other related gene control proteins” [110]. Indeed, the zinc finger motif proves to be one of the most prevalent structures in biology. As stated in Section 1, it has been estimated that human may have close to 1000 distinct C2H2 zinc finger protein-encoding genes, accounting for ≈ 1% of the genome [6]. A significant portion of these proteins contains the Krüppel motif, making the 18 KLFs in Fig. 1 only a small fraction of all zinc finger proteins. An obvious question is why so many zinc finger proteins are in existence in the human genome. One possible answer may rest on the remarkably stable nature of the zinc finger structure, as demonstrated by numerous crystallographic studies (see for example Ref. [112]). The lack of redox activity of zinc and its characteristic binding kinetics make it an especially suitable metal ion to stabilize a peptide structure [113]. The tetrahedral coordination of zinc then facilitates the interactions between proteins and DNA. Moreover, since the zinc finger is modular in nature, the specificity of DNA sequence recognition can be modified by the specific residues in the finger used to contact DNA and by the number of repeats each protein may contain.
It is clear that the Krüppel-like factors described in this articles share a high degree of similarity in the DNA sequences to which they recognize. However, there appears to be two loosely divided subgroups of sequences, based on the DNA binding affinity for each KLF. The Sp family of proteins favor the classical GC-rich Sp1-binding box [114], whereas the remaining KLFs favor the CACCC-, or GT-, element. As both sequences are found in a relatively high frequency in the regulatory regions of various mammalian genes, it is not unreasonable to assume that the KLFs play an important role in the regulation of expression of myriad genes. The significance of these cis-sequences in modulating gene expression is underscored by their frequent presence in promoters containing CpG-islands that are subject to DNA methylation, an important mechanism for regulating cell growth and development [115]. The versatility of the DNA-protein interactions is further enhanced by the presence of certain cis-elements in the promoters of some genes that appear to have evolved to exhibit strong affinities for both subgroups of KLFs. For example, while GKLF binds strongly to a GC-rich, empirically determined consensus sequence, it binds much weaker to a classical GC-rich Sp1-binding sequence [56]. However, both GKLF and Sp1 bind with high affinity to the basic transcription element (BTE) [55], a single GC-box sequence present in the promoters of a family of cytochrome P450 genes, including CYP1A1 [85]. It is of interest to note that the BTE resembles a composite sequence between the empirically determined GKLF-binding site and the classical Sp1-binding site. Perhaps this is one mechanism by which regulatory sequences evolve in which to attract the participation of DNA-binding proteins with different sequence preferences.
Another reason for the existence of multiple KLFs with similar recognition sequences is the diverse biochemical mechanisms by which these proteins function. While some KLFs are potent activators of transcription, such as Sp1 [116], some are repressors of transcription, like KLF12 [108]. Some KLFs are bifunctional, acting as activators or repressors, depending on the circumstances, as exemplified by GKLF [51,58,62]. The mechanisms by which the KLFs modulate transcription are also diverse and may depend on the presence of intrinsic activation or repression domains, competition or cooperation with other KLFs, and interactions with co-activators or co-repressors. It is, therefore, not unreasonable to assume that the purpose of such a large repertoire has important biological implications, and may allow the fine-tuning of gene expression under various physiological conditions.
5. Conclusion
This article summarizes the biology of 12 mammalian Krüppel-like factors, the majority of which were discovered within the last 5 years. There is significant conservation in several aspects among the members of this family. For example, they share a high degree of sequence homology in the zinc finger regions. In addition, the zinc fingers of most of the KLFs are located to the extreme carboxyl terminal end of the proteins, which suggests that they may have originated from the same ancestral gene. All KLFs bind to a similar DNA sequence that has a CACCC homology or is rich in GC-content. As a result, there is a certain degree of overlap in the target genes that the KLFs regulate. In fact, it is not unusual for several KLFs to interact with the same cis-element in the same gene and perhaps with each other. Lastly, each of the KLFs appears to exert important regulatory functions on many biological processes, such as growth, development, differentiation and apoptosis. Despite these similarities, the majority of KLFs seem to have unique tissue-specific roles in an in vivo setting. Some of these roles have begun to be elucidated primarily by in vivo experiments involving gene knockout. It is anticipated that within the next few years, there will be further experimentation on these proteins that is likely to provide additional insights into the mechanisms of action of the KLFs and their physiological relevancy.
Acknowledgements
This work was in part supported by Grants DK10020, DK52230 and CA84197, from the National Institutes of Health, USA.
References
- 1.Yang VW. Eukaryotic transcription factors: identification, characterization and functions. J. Nutr. 1998;128:2045–2051. doi: 10.1093/jn/128.11.2045. [DOI] [PubMed] [Google Scholar]
- 2.Clarke ND, Berg JM. Zinc fingers in Caenorhabditis elegans: finding families and probing pathways. Science. 1998;282:2018–2022. doi: 10.1126/science.282.5396.2018. [DOI] [PubMed] [Google Scholar]
- 3.Rubin GM, Yandell MD, Wortman JR, et al. Comparative genomics of the eukaryotes. Science. 2000;287:2204–2215. doi: 10.1126/science.287.5461.2204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Adams MD, Celniker SE, Holt RA, et al. The genome sequence of Drosophila melanogaster. Science. 2000;287:2185–2195. doi: 10.1126/science.287.5461.2185. [DOI] [PubMed] [Google Scholar]
- 5.Hoovers JM, Mannens M, John R, et al. High-resolution localization of 69 potential human zinc finger protein genes: a number are clustered. Genomics. 1992;12:254–263. doi: 10.1016/0888-7543(92)90372-y. [DOI] [PubMed] [Google Scholar]
- 6.Klug A, Schwabe JWR. Protein motifs 5. Zinc fingers. FASEB J. 1995;9:597–604. [PubMed] [Google Scholar]
- 7.Kadonaga JT, Carner KR, Masiarz FR, Tjian R. Isolation of cDNA encoding transcription factor Sp1 and functional analysis of the DNA binding domain. Cell. 1987;51:1079–1090. doi: 10.1016/0092-8674(87)90594-0. [DOI] [PubMed] [Google Scholar]
- 8.Schuh R, Aicher W, Gaul U, et al. A conserved family of nuclear proteins containing structural elements of the finger protein encoded by Krüppel, a Drosophila segmentation gene. Cell. 1986;47:1025–1032. doi: 10.1016/0092-8674(86)90817-2. [DOI] [PubMed] [Google Scholar]
- 9.Miller IJ, Bieker JJ. A novel, erythroid cell-specific murine transcription factor that binds to the CACCC element and is related to the Krüppel family of nuclear proteins. Mol. Cell. Biol. 1993;13:2776–2786. doi: 10.1128/mcb.13.5.2776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Turner J, Crossley M. Mammalian Krüppel-like transcription factors: more than just a pretty finger. Trends Biochem. Sci. 1999;24:236–240. doi: 10.1016/s0968-0004(99)01406-1. [DOI] [PubMed] [Google Scholar]
- 11.Philipsen S, Suske G. A tale of three fingers: the family of mammalian Sp/XKLF transcription factors. Nucl. Acids Res. 1999;27:2991–3000. doi: 10.1093/nar/27.15.2991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.White JA, McAlpine PJ, Antonarakis S, et al. Guidelines for human gene nomenclature . HUGO Nomenclature Committee. Genomics. 1997;45:468–471. doi: 10.1006/geno.1997.4979. [DOI] [PubMed] [Google Scholar]
- 13.Shields JM, Yang VW. Two potent nuclear localization signals in the gut-enriched Krüppel-like factor define a subfamily of closely related Krüppel proteins. J. Biol. Chem. 1997;272:18504–18507. doi: 10.1074/jbc.272.29.18504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Subramaniam M, Harris SA, Oursler MJ, et al. Identification of a novel TGF-β-regulated gene encoding a putative zinc finger protein in human osteoblasts. Nucl. Acids Res. 1995;23:4907–4912. doi: 10.1093/nar/23.23.4907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cook T, Gebelein B, Mesa K, Mladek A, Urrutia R. Molecular cloning and characterization of TIEG2 reveals a new subfamily of transforming growth factor-β-inducible Sp1-like zinc finger-encoding genes involved in the regulation of cell growth. J. Biol. Chem. 1998;273:25929–25936. doi: 10.1074/jbc.273.40.25929. [DOI] [PubMed] [Google Scholar]
- 16.Perkins A. Erythroid Krüppel like factor: from fishing expedition to gourmet meal. Int. J. Biochem. Cell Biol. 1999;31:1175–1192. doi: 10.1016/s1357-2725(99)00083-7. [DOI] [PubMed] [Google Scholar]
- 17.Hartzog GA, Myers RM. Discrimination among potential activators of the β-globin CACCC element by correlation of binding and transcriptional properties. Mol. Cell. Biol. 1993;13:44–56. doi: 10.1128/mcb.13.1.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bieker JJ, Southwood CM. The erythroid Krüppel-like factor transactivation domain is a critical component for cell-specific inducibility of a β-globin promoter. Mol. Cell. Biol. 1995;15:852–860. doi: 10.1128/mcb.15.2.852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bieker JJ. Isolation, genomic structure, and expression of human erythroid Krüppel-like factor (EKLF) DNA Cell Biol. 1996;15:347–352. doi: 10.1089/dna.1996.15.347. [DOI] [PubMed] [Google Scholar]
- 20.Mitchell PJ, Tjian R. Transcriptional regulation in mammalian cells by sequence-specific DNA binding proteins. Science. 1989;245:371–378. doi: 10.1126/science.2667136. [DOI] [PubMed] [Google Scholar]
- 21.Donze D, Townes TM, Bieker JJ. Role of erythroid Krüppel-like factor in human γ- to β-globin gene switching. J. Biol. Chem. 1995;270:1955–1959. doi: 10.1074/jbc.270.4.1955. [DOI] [PubMed] [Google Scholar]
- 22.Lee JS, Ngo H, Kim D, Chung JH. Erythroid Krüppel-like factor is recruited to the CACCC box in the β-globin promoter but not to the CACCC box in the γ-globin promoter: The role of the neighboring promoter elements. Proc. Natl. Acad. Sci. USA. 2000;97:2468–2473. doi: 10.1073/pnas.040476297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen X, Bieker JJ. Erythroid Krüppel-like factor (EKLF) contains a multifunctional transcriptional activation domain important for inter-and intramolecular interactions. EMBO J. 1996;15:5888–5896. [PMC free article] [PubMed] [Google Scholar]
- 24.Meisner H, Czech MP. Phosphorylation of transcriptional factors and cell-cycle-dependent proteins by casein kinase II. Curr. Opin. Cell Biol. 1991;3:474–483. doi: 10.1016/0955-0674(91)90076-b. [DOI] [PubMed] [Google Scholar]
- 25.Ouyang L, Chen X, Bieker JJ. Regulation of erythroid Krüppel-like factor (EKLF) transcriptional activity by phosphorylation of a protein kinase casein kinase II site within its interaction domain. J. Biol. Chem. 1998;273:23019–23025. doi: 10.1074/jbc.273.36.23019. [DOI] [PubMed] [Google Scholar]
- 26.Wijgerde M, Gribnau J, Trimborn T, et al. The role of EKLF in human β-globin gene competition. Genes Dev. 1996;10:2894–2902. doi: 10.1101/gad.10.22.2894. [DOI] [PubMed] [Google Scholar]
- 27.Armstrong JA, Bieker JJ, Emerson BM. A SWI/ SNF-related chromatin remodeling complex, E-RC1, is required for tissue-specific transcriptional regulation by EKLF in vitro. Cell. 1998;95:93–104. doi: 10.1016/s0092-8674(00)81785-7. [DOI] [PubMed] [Google Scholar]
- 28.Winston F, Carlson M. Yeast SNF/SWI transcriptional activators and the SPT/SIN chromatin connection. Trends Genet. 1992;8:387–391. doi: 10.1016/0168-9525(92)90300-s. [DOI] [PubMed] [Google Scholar]
- 29.Cote J, Quinn J, Workman JL, Peterson CL. Stimulation of GAL4 derivative binding to nucleosomal DNA by the yeast SWI/SNF complex. Science. 1994;265:53–60. doi: 10.1126/science.8016655. [DOI] [PubMed] [Google Scholar]
- 30.Kwon H, Imbalzano AN, Khavari PA, Kingston RE, Green MR. Nucleosome disruption and enhancement of activator binding by a human SW1/SNF complex. Nature. 1994;370:477–481. doi: 10.1038/370477a0. [DOI] [PubMed] [Google Scholar]
- 31.Kouzarides T. Histone acetylases and deacetylases in cell proliferation. Curr. Opin. Genet. Dev. 1999;9:40–48. doi: 10.1016/s0959-437x(99)80006-9. [DOI] [PubMed] [Google Scholar]
- 32.Zhang W, Bieker JJ. Acetylation and modulation of erythroid Krüppel-like factor (EKLF) activity by interaction with histone acetyltransferases. Proc. Natl. Acad. Sci. USA. 1998;95:9855–9860. doi: 10.1073/pnas.95.17.9855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Southwood CM, Downs KM, Bieker JJ. Erythroid Krüppel-like factor exhibits an early and sequentially localized pattern of expression during mammalian erythroid ontogeny. Dev. Dynam. 1996;206:248–259. doi: 10.1002/(SICI)1097-0177(199607)206:3<248::AID-AJA3>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- 34.Nuez B, Michalovich D, Bygrave A, Ploemacher R, Grosveld F. Defective haematopoiesis in fetal liver resulting from inactivation of the EKLF gene. Nature. 1995;375:316–318. doi: 10.1038/375316a0. [DOI] [PubMed] [Google Scholar]
- 35.Perkins AC, Sharpe AH, Orkin SH. Lethal β-thalas-saemia in mice lacking the erythroid CACCC-transcription factor EKLF. Nature. 1995;375:318–322. doi: 10.1038/375318a0. [DOI] [PubMed] [Google Scholar]
- 36.Perkins AC, Peterson KR, Stamatoyannopoulos G, Witkowska HE, Orkin SH. Fetal expression of a human Aγ globin transgene rescues globin chain imbalance but not hemolysis in EKLF null mouse embryos. Blood. 2000;95:1827–1833. [PubMed] [Google Scholar]
- 37.Anderson KP, Kern CB, Crable SC, Lingrel JB. Isolation of a gene encoding a functional zinc finger protein homologous to erythroid Krüppel-like factor: Identification of a new multigene family. Mol. Cell. Biol. 1995;15:5957–5965. doi: 10.1128/mcb.15.11.5957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wani MA, Conkright MD, Jeffries S, Hughes MJ, Lingrel JB. cDNA isolation, genomic structure, regulation, and chromosomal localization of human lung Krüppel-like factor. Genomics. 1999;60:78–86. doi: 10.1006/geno.1999.5888. [DOI] [PubMed] [Google Scholar]
- 39.Kuo CT, Veselits ML, Barton KP, et al. The LKLF transcription factor is required for normal tunica media formation and blood vessel stabilization during murine embryogenesis. Genes Dev. 1997;11:2996–3006. doi: 10.1101/gad.11.22.2996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wani MA, Means RT, Lingrel JB., Jr Loss of LKLF function results in embryonic lethality in mice. Transgen. Res. 1998;7:229–238. doi: 10.1023/a:1008809809843. [DOI] [PubMed] [Google Scholar]
- 41.Wani MA, Wert SE, Lingrel JB. Lung Krüppel-like factor, a zinc finger transcription factor, is essential for normal lung development. J. Biol. Chem. 1999;274:21180–21185. doi: 10.1074/jbc.274.30.21180. [DOI] [PubMed] [Google Scholar]
- 42.Kuo CT, Veselits ML, Leiden JM. LKLF: A transcriptional regulator of single-positive T cell quiescence and survival. Science. 1997;277:1986–1990. doi: 10.1126/science.277.5334.1986. [DOI] [PubMed] [Google Scholar]
- 43.Schober SL, Kuo CT, Schluns KS, et al. Expression of the transcription factor lung Krüppel-like factor is regulated by cytokines and correlates with survival of memory T cells in vitro and in vivo. J. Immunol. 1999;163:3662–3667. [PubMed] [Google Scholar]
- 44.Crossley M, Whitelaw E, Perkins A, et al. Isolation and characterization of the cDNA encoding BKLF/ TEF-2, a major CACCC-box-binding protein in erythroid cells and selected other cells. Mol. Cell. Biol. 1996;16:1695–1705. doi: 10.1128/mcb.16.4.1695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ulgiati D, Subrata LS, Abraham LJ. The role of Sp family members, basic Krüppel-like factor, and E box factors in the basal and IFN-γ regulated expression of the human complement C4 promoter. J. Immunol. 2000;164:300–307. doi: 10.4049/jimmunol.164.1.300. [DOI] [PubMed] [Google Scholar]
- 46.Perkins AC, Yang H, Crossley M, Fujiwara Y, Orkin SH. Blood. 1997;90(Suppl 1):575. [Google Scholar]
- 47.Turner J, Crossley M. Cloning and characterization of mCtBP2, a co-repressor that associates with basic Krüppel-like factor and other mammalian transcriptional regulators. EMBO J. 1998;17:5129–5140. doi: 10.1093/emboj/17.17.5129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Turner J, Crossley M. Basic Krüppel-like factor functions within a network of interacting haematopoietic transcription factors. Int. J. Biochem. Cell Biol. 1999;31:1169–1174. doi: 10.1016/s1357-2725(99)00067-9. [DOI] [PubMed] [Google Scholar]
- 49.Christy BA, Lau LF, Nathans D. A gene activated in mouse 3T3 cells by serum growth factors encodes a protein with ‘zinc finger’ sequences. Proc. Natl. Acad. Sci. USA. 1988;85:7857–7861. doi: 10.1073/pnas.85.21.7857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shields JM, Christy RJ, Yang VW. Identification and characterization of a gene encoding a gut-enriched Krüppel-like factor expressed during growth arrest. J. Biol. Chem. 1996;271:20009–20017. doi: 10.1074/jbc.271.33.20009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jenkins TD, Opitz OG, Okano J, Rustgi AK. Transactivation of the human keratin 4 and Epstein–Barr virus ED-L2 promoters by gut-enriched Krüppel-like factor. J. Biol. Chem. 1998;273:10747–10754. doi: 10.1074/jbc.273.17.10747. [DOI] [PubMed] [Google Scholar]
- 52.Garrett-Sinha LA, Eberspaecher H, Seldin MF, de Crombrugghe B. A gene for a novel zinc-finger protein expressed in differentiated epithelial cells and transiently in certain mesenchymal cells. J. Biol. Chem. 1996;271:31384–31390. doi: 10.1074/jbc.271.49.31384. [DOI] [PubMed] [Google Scholar]
- 53.Panigada M, Porcellini S, Sutti F, Doneda L, Pozzoli O, Consalez GG, Guttinger M, Grassi F. GKLF in thymus epithelium as a developmentally regulated element of thymocyte-stroma cross-talk. Mech. Dev. 1999;81:103–113. doi: 10.1016/s0925-4773(98)00237-8. [DOI] [PubMed] [Google Scholar]
- 54.Yet SF, McA’Nulty MM, Folta SC, et al. Human EZF, a Krüppel-like zinc finger protein, is expressed in vascular endothelial cells and contains transcriptional activation and repression domains. J. Biol. Chem. 1998;273:1026–1031. doi: 10.1074/jbc.273.2.1026. [DOI] [PubMed] [Google Scholar]
- 55.Ton-That H, Kaestner KH, Shields JM, Mahatanankoon CS, Yang WW. Expression of the gut-enriched Krüppel-like factor gene during development and intestinal tumorigenesis. FEBS Letts. 1997;419:239–243. doi: 10.1016/s0014-5793(97)01465-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Shields JM, Yang VW. Identification of the DNA sequence that interacts with the gut-enriched Krüppel-like factor. Nucl. Acids Res. 1998;26:796–802. doi: 10.1093/nar/26.3.796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Fujii-Kuriyama Y, Imataka H, Sogawa K, Yasumoto K, Kikuchi Y. Regulation of CYP1A1 expression. FASEB J. 1992;6:706–710. doi: 10.1096/fasebj.6.2.1537460. [DOI] [PubMed] [Google Scholar]
- 58.Zhang W, Shields JM, Sogawa K, Fujii-Kuriyama Y, Yang VW. The gut-enriched Krüppel-like factor suppresses the activity of the CYP1A1 promoter in an Sp1-dependent fashion. J. Biol. Chem. 1998;273:17917–17925. doi: 10.1074/jbc.273.28.17917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Imataka H, Sogawa K, Yasumoto K, Kikuchi Y, Sasano K, Kobayashi A, Hayami M, Fujii-Kuriyama Y. Two regulatory proteins that bind to the basic transcription element (BTE), a GC box sequence in the promoter region of the rat P-4501A1 gene. EMBO J. 1992;11:3663–3671. doi: 10.1002/j.1460-2075.1992.tb05451.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Okano J, Opitz OG, Nakagawa H, et al. The Krüppel-like transcriptional factors Zf9 and GKLF coactivate the human keratin 4 promoter and physically interact. FEBS Letts. 2000;473:95–100. doi: 10.1016/s0014-5793(00)01468-x. [DOI] [PubMed] [Google Scholar]
- 61.Brembeck FH, Rustgi AK. The tissue-dependent Keratin 19 gene transcription is regulated by GKLF/KLF4 and Sp1. J. Biol. Chem. 2000;275:28230–28239. doi: 10.1074/jbc.M004013200. [DOI] [PubMed] [Google Scholar]
- 62.Geiman DE, Ton-That H, Johnson JM, Yang VW. Transactivation and growth suppression by the gut-enriched Krüppel-like factor (Krüppel-like factor 4) are dependent on acidic amino acid residues and protein–protein interaction. Nucl. Acids. Res. 2000;28:1106–1113. doi: 10.1093/nar/28.5.1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Dang DT, Bachman KE, Mahatan CS, et al. Decreased expression of the gut-enriched Krüppel-like factor gene in intestinal adenomas of multiple intestinal neoplasia mice and in colonic adenomas of familial adenomatous polyposis patients. FEBS. Letts. 2000;476:203–207. doi: 10.1016/s0014-5793(00)01727-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhang W, Geiman DE, Shields JM, et al. The gut-enriched Krüppel-like factor (Krüppel-like factor 4) mediates the transactivating effect of p53 on the p21WAF1/Cip1 promoter. J. Biol. Chem. 2000;275:18391–18398. doi: 10.1074/jbc.C000062200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Shie JL, Pestell RG, Tseng CC. Gut-enriched Krüppel-like factor represses cyclin D1 promoter activity through Sp1 motif. Nucl. Acids Res. 2000;28:2969–2976. doi: 10.1093/nar/28.15.2969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Chen ZY, Shie J, Tseng C. Up-regulation of gut-enriched Krüppel-like factor by interferon-γ in human colon carcinoma cells. FEBS Letts. 2000;477:67–72. doi: 10.1016/s0014-5793(00)01764-6. [DOI] [PubMed] [Google Scholar]
- 67.Mahatan CS, Kaestner KH, Geiman DE, Yang VW. Characterization of the structure and regulation of the murine gene encoding gut-enriched Krüppel-like factor (Krüppel-like factor 4) Nucl. Acids Res. 1999;27:4562–4569. doi: 10.1093/nar/27.23.4562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Segre JA, Bauer C, Fuchs E. Klf4 is a transcription factor required for establishing the barrier function of the skin. Nature Gen. 1999;22:356–360. doi: 10.1038/11926. [DOI] [PubMed] [Google Scholar]
- 69.Conkright MD, Wani MA, Anderson KP, Lingrel JB. A gene encoding an intestinal-enriched member of the Krüppel-like factor family expressed in intestinal epithelial cells. Nucl. Acids Res. 1999;27:1263–1270. doi: 10.1093/nar/27.5.1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ohnishi S, Laub F, Matsumoto N, et al. Developmental expression of the mouse gene coding for the Krüppel-like transcription factor KLF5. Dev. Dynam. 2000;217:421–429. doi: 10.1002/(SICI)1097-0177(200004)217:4<421::AID-DVDY9>3.0.CO;2-1. [DOI] [PubMed] [Google Scholar]
- 71.Shi H, Zhang Z, Wang X, Liu S, Teng CT. Isolation and characterization of a gene encoding human Krüppel-like factor 5 (IKLF): binding to the CAAT/GT box of the mouse lactoferrin gene promoter. Nucl. Acids Res. 1999;27:4807–4815. doi: 10.1093/nar/27.24.4807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sogawa K, Imataka H, Yamasaki Y, et al. cDNA cloning and transcriptional properties of a novel GC box-binding protein, BTEB2. Nucl. Acids Res. 1993;21:1527–1532. doi: 10.1093/nar/21.7.1527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Watanabe N, Kurabayashi M, Shimomura Y, et al. BTEB2, a Krüppel-like transcription factor, regulates expression of the SMemb/Nonmuscle myosin heavy chain B Memb/NMHC-B gene. Circulation Res. 1999;85:182–191. doi: 10.1161/01.res.85.2.182. [DOI] [PubMed] [Google Scholar]
- 74.Kawai-Kowase K, Kurabayashi M, Hoshino Y, Ohyama Y, Nagai R. Transcriptional activation of the zinc finger transcription factor BTEB2 gene by Egr-1 through mitogen-activated protein kinase pathways in vascular smooth muscle cells. Circulation Res. 1999;85:787–795. doi: 10.1161/01.res.85.9.787. [DOI] [PubMed] [Google Scholar]
- 75.Koritschoner NP, Bocco JL, Panzetta-Dutari GM, et al. A novel human zinc finger protein that interacts with the core promoter element of a TATA box-less gene. J. Biol. Chem. 1997;272:9573–9580. doi: 10.1074/jbc.272.14.9573. [DOI] [PubMed] [Google Scholar]
- 76.Suzuki T, Yamamoto T, Kurabayashi M, et al. Isolation and initial characterization of GBF, a novel DNA-binding zinc finger protein that binds to the GC-rich binding sites of the HIV-1 promoter. J. Biochem. (Tokyo) 1998;124:389–395. doi: 10.1093/oxfordjournals.jbchem.a022124. [DOI] [PubMed] [Google Scholar]
- 77.Ratziu V, Lalazar A, Wong L, et al. Zf9, a Krüppel-like transcription factor up-regulated in vivo during early hepatic fibrosis. Proc. Natl. Acad. Sci. USA. 1998;95:9500–9505. doi: 10.1073/pnas.95.16.9500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Matsumoto N, Laub F, Aldabe R, et al. Cloning the cDNA for a new human zinc finger protein defines a group of closely related Krüppel-like transcription factors. J. Biol. Chem. 1998;273:28229–28237. doi: 10.1074/jbc.273.43.28229. [DOI] [PubMed] [Google Scholar]
- 79.Zhao JI, Austen KF, Lam BK. Cell-specific transcription of leukotriene C(4) synthase involves a Krüppel-like transcription factor and Sp1. J. Biol. Chem. 2000;275:8903–8910. doi: 10.1074/jbc.275.12.8903. [DOI] [PubMed] [Google Scholar]
- 80.Kojima S, Hayashi S, Shimokado K, et al. Transcriptional activation of urokinase by the Krüppel-like factor Zf9/COPEB activates latent TGF-β1 in vascular endothelial cells. Blood. 2000;95:1309–1316. [PubMed] [Google Scholar]
- 81.Kim Y, Ratziu V, Choi SG, et al. Transcriptional activation of transforming growth factor beta1 and its receptors by the Krüppel-like factor Zf9/core promoter-binding protein and Sp1: Potential mechanisms for autocrine fibrogenesis in response to injury. J. Biol. Chem. 1998;273:33750–33758. doi: 10.1074/jbc.273.50.33750. [DOI] [PubMed] [Google Scholar]
- 82.Inuzuka H, Wakao H, Masuho Y, et al. cDNA cloning and expression analysis of mouse zf9, a Krüppel-like transcription factor gene that is induced by adipogenic hormonal stimulation in 3T3-L1 cells. Biochim. Biophys. Acta. 1999;1447:199–207. doi: 10.1016/s0167-4781(99)00161-x. [DOI] [PubMed] [Google Scholar]
- 83.Friedman SL. Hepatic stellate cells. Prog. Liver Dis. 1996;14:101–130. [PubMed] [Google Scholar]
- 84.van Vliet J, Turner J, Crossley M. Human Krüppel-like factor 8: a CACCC-box binding protein that associates with CtBP and represses transcription. Nucl. Acids Res. 2000;28:1955–1962. doi: 10.1093/nar/28.9.1955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Yanagida A, Sogawa K, Yasumoto KI, Fujii-Kuriyama Y. A novel cis-acting DNA element required for a high level of inducible expression of the rat P-450c gene. Mol. Cell. Biol. 1990;10:1470–1475. doi: 10.1128/mcb.10.4.1470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Ohe N, Yamasaki Y, Sogawa K, et al. Chromosomal localization and cDNA sequence of human BTEB, a GC box binding protein. Som. Cell Mol. Genet. 1993;19:499–503. doi: 10.1007/BF01233255. [DOI] [PubMed] [Google Scholar]
- 87.Sogawa K, Kikuchi Y, Imataka H, Fujii-Kuriyama Y. Comparison of DNA-binding properties between BTEB and Sp1. J. Biochem. (Tokyo) 1993;114:605–609. doi: 10.1093/oxfordjournals.jbchem.a124224. [DOI] [PubMed] [Google Scholar]
- 88.Kobayashi A, Sogawa K, Imataka H, Fujii-Kuriyama Y. Analysis of functional domains of a GC box-binding protein, BTEB. J. Biochem. (Tokyo) 1995;117:91–95. doi: 10.1093/oxfordjournals.jbchem.a124727. [DOI] [PubMed] [Google Scholar]
- 89.Chen A, Davis BH. The DNA binding protein BTEB mediates acetaldehyde-induced, jun N-terminal kinase-dependent αI(I) collagen gene expression in rat hepatic stellate cells. Mol. Cell. Biol. 2000;20:2818–2826. doi: 10.1128/mcb.20.8.2818-2826.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Wang Y, Michel FJ, Wing A, Simmen FA, Simmen RC. Cell-type expression, immunolocalization, and deoxyribonucleic acid-binding activity of basic transcription element binding transcription factor, an Sp-related family member, in porcine endometrium of pregnancy. Biol. Reprod. 1997;57:707–714. doi: 10.1095/biolreprod57.4.707. [DOI] [PubMed] [Google Scholar]
- 91.Simmen RC, Chung TE, Imataka H, et al. Trans-activation functions of the Sp-related nuclear factor, basic transcription element-binding protein, and progesterone receptor in endometrial epithelial cells. Endocrinology. 1999;140:2517–2525. doi: 10.1210/endo.140.6.6625. [DOI] [PubMed] [Google Scholar]
- 92.Denver RJ, Ouellet L, Furling D, Kobayashi A, Fujii-Kuriyama Y, Puymirat J. Basic transcription element-binding protein (BTEB) is a thyroid hormone-regulated gene in the developing central nervous system. Evidence for a role in neurite outgrowth. J. Biol. Chem. 1999;274:23128–23134. doi: 10.1074/jbc.274.33.23128. [DOI] [PubMed] [Google Scholar]
- 93.Cook T, Urrutia R. TIEG proteins join the Smads as TGF-β-regulated transcription factors that control pancreatic cell growth. Am. J. Physiol: Gastrointes. Liver Physiol. 2000;278:G513–G521. doi: 10.1152/ajpgi.2000.278.4.G513. [DOI] [PubMed] [Google Scholar]
- 94.Blok LJ, Grossmann ME, Perry JE, Tindall DJ. Characterization of an early growth response gene, which encodes a zinc finger transcription factor, potentially involved in cell cycle regulation. Mol. Endocrinol. 1995;9:1610–1620. doi: 10.1210/mend.9.11.8584037. [DOI] [PubMed] [Google Scholar]
- 95.Fautsch MP, Vrabel A, Rickard D, et al. Characterization of the mouse TGFβ-inducible early gene (TIEG): conservation of exon and transcriptional regulatory sequences with evidence of additional transcripts. Mammal. Genome. 1998;9:838–842. doi: 10.1007/s003359900878. [DOI] [PubMed] [Google Scholar]
- 96.Tau KR, Hefferan TE, Waters KM, et al. Estrogen regulation of a transforming growth factor-beta inducible early gene that inhibits deoxyribonucleic acid synthesis in human osteoblasts. Endocrinology. 1998;139:1346–1353. doi: 10.1210/endo.139.3.5830. [DOI] [PubMed] [Google Scholar]
- 97.Hofbauer LC, Hicok KC, Khosla SJ. Effects of gonadal and adrenal androgens in a novel androgen-responsive human osteoblastic cell line. J. Cell. Biochem. 1998;71:96–108. [PubMed] [Google Scholar]
- 98.Subramaniam M, Hefferan TE, Tau K, et al. Tissue, cell type, and breast cancer stage-specific expression of a TGF-β inducible early transcription factor gene. J. Cell. Biochem. 1998;68:226–236. [PubMed] [Google Scholar]
- 99.Eid MA, Kumar MV, Iczkowski KA, Bost-wick DG, Tindall DJ. Expression of early growth response genes in human prostate cancer. Cancer Res. 1998;58:2461–2468. [PubMed] [Google Scholar]
- 100.Oberhammer FA, Pavelka M, Sharma S, et al. Induction of apoptosis in cultured hepatocytes and in regressing liver by transforming growth factor β1. Proc. Natl. Acad. Sc. USA. 1992;89:5408–5412. doi: 10.1073/pnas.89.12.5408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Lomo J, Blomhoff HK, Beiske K, Stokke T, Smeland EB. TGF-ゲ1 and cyclic AMP promote apoptosis in resting human B lymphocytes. J. Immunol. 1995;154:1634–1643. [PubMed] [Google Scholar]
- 102.Hsing AY, Kadomatsu K, Bonham MJ, Danielpour D. Regulation of apoptosis induced by transforming growth factor-beta1 in nontumorigenic rat prostatic epithelial cell lines. Cancer Res. 1996;56:5146–5149. [PubMed] [Google Scholar]
- 103.Tachibana I, Imoto M, Adjei PN, et al. Overexpression of the TGFβ-regulated zinc finger encoding gene, TIEG, induces apoptosis in pancreatic epithelial cells. J. Clin. Investig. 1997;99:2365–2374. doi: 10.1172/JCI119418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Chalaux E, Lopez-Rovira T, Rosa JL, et al. A zinc-finger transcription factor induced by TGF-β promotes apoptotic cell death in epithelial Mv1Lu cells. FEBS. Letts. 1999;457:478–482. doi: 10.1016/s0014-5793(99)01051-0. [DOI] [PubMed] [Google Scholar]
- 105.Ribeiro A, Bronk SF, Roberts PJ, Urrutia R, Gores GJ. The transforming growth factor β(1)-inducible transcription factor TIEG1, mediates apoptosis through oxidative stress. Hpatology. 1999;30:1490–1497. doi: 10.1002/hep.510300620. [DOI] [PubMed] [Google Scholar]
- 106.Polyak K, Xia Y, Zweier JL, Kinzler KW, Vogelstein B. A model for p53-induced apoptosis. Nature. 1997;389:300–305. doi: 10.1038/38525. [DOI] [PubMed] [Google Scholar]
- 107.Cook T, Gebelein B, Belal M, Mesa K, Urrutia R. Three conserved transcriptional repressor domains are a defining feature of the TIEG subfamily of Sp1-like zinc finger proteins. J. Biol. Chem. 1999;274:29500–29504. doi: 10.1074/jbc.274.41.29500. [DOI] [PubMed] [Google Scholar]
- 108.Imhof A, Schuierer M, Werner O, et al. Transcriptional regulation of the AP-2α promoter by BTEB-1 and AP-2rep, a novel wt-1/egr-related zinc finger repressor. Mol. Cell. Biol. 1999;19:194–204. doi: 10.1128/mcb.19.1.194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Roth C, Schuierer M, Gunther K, Buettner R. Genomic structure and DNA binding properties of the human zinc finger transcriptional repressor AP-2rep (KLF12) Genomics. 2000;63:384–390. doi: 10.1006/geno.1999.6084. [DOI] [PubMed] [Google Scholar]
- 110.Miller J, McLachlan AD, Klug A. Repetitive zinc-binding domains in the protein transcription factor IIIA from Xenopus oocytes. EMBO J. 1985;4:1609–1614. doi: 10.1002/j.1460-2075.1985.tb03825.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Brown RS, Sander C, Argos P. The primary structure of transcription factor TFIIIA has 12 consecutive repeats. FEBS. Letts. 1985;186:271–274. doi: 10.1016/0014-5793(85)80723-7. [DOI] [PubMed] [Google Scholar]
- 112.Elrod-Erickson M, Rould MA, Nekludova L, Pabo CO. Zif268 protein-DNA complex refined at 1.6 A: a model system for understanding zinc finger-DNA interactions. Structure. 1996;4:1171–1180. doi: 10.1016/s0969-2126(96)00125-6. [DOI] [PubMed] [Google Scholar]
- 113.Berg JM, Shi Y. The galvanization of biology: a growing appreciation for the roles of zinc. Science. 1996;271:1081–1085. doi: 10.1126/science.271.5252.1081. [DOI] [PubMed] [Google Scholar]
- 114.Thiesen HJ, Bach C. Target detection assay (TDA): a versatile procedure to determine DNA binding sites as demonstrated on Sp1 protein. Nucl. Acids Res. 1990;18:3203–3209. doi: 10.1093/nar/18.11.3203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Robertson KD, Jones PA. DNA methylation: past, present and future directions. Carcinogenesis. 2000;21:461–467. doi: 10.1093/carcin/21.3.461. [DOI] [PubMed] [Google Scholar]
- 116.Suske G. The Sp-family of transcription factors. Gene. 1999;238:291–300. doi: 10.1016/s0378-1119(99)00357-1. [DOI] [PubMed] [Google Scholar]
- 117.Song A, Chen YF, Thamatrakoln K, Storm TA, Krensky AM. RFLAT-1: a new zinc finger transcription factor that activates RANTES gene expression in T lymphocytes. Immunity. 1999;10:93–103. doi: 10.1016/s1074-7613(00)80010-2. [DOI] [PubMed] [Google Scholar]
- 118.Uchida S, Tanaka Y, Ito H, Saitoh-Ohara F, Inazawa J, Yokoyama KK, Sasaki S, Marumo F. Transcriptional regulation of the CLC-K1 promoter by myc-associated zinc finger protein and kidney-enriched Krüppel-like factor, a novel zinc finger repressor. Mol. Cell. Biol. 2000;20:1319–1331. doi: 10.1128/mcb.20.19.7319-7331.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]