Abstract
It is generally believed that significant ribosomal frameshifting during translation does not occur without a functional purpose. The distribution of two frameshift-prone sequences, A_AAA_AAG and CCC_TGA, in coding regions of Escherichia coli has been analyzed. Although a moderate level of selection against the first sequence is evident, 68 genes contain A_AAA_AAG and 19 contain CCC_TGA. The majority of those tested in their genomic context showed >1% frameshifting. Comparative sequence analysis was employed to assess a potential biological role for frameshifting in decoding these genes. Two new candidates, in pheL and ydaY, for utilized frameshifting have been identified in addition to those previously known in dnaX and nine insertion sequence elements. For the majority of the shift-prone sequences no functional role can be attributed to them, and the frameshifting is likely erroneous. However, none of frameshift sequences is in the 306 most highly expressed genes. The unexpected conclusion is that moderate frameshifting during expression of at least some other genes is not sufficiently harmful for cells to trigger strong negative evolutionary pressure.
Keywords: frameshifting/genomics/proline/translational errors
Introduction
During readout of genetic information into proteins, translation is the last and probably the least accurate process, although considerable accuracy of protein synthesis is crucial for cell survival. Errors in translation are divided into two types: missense errors and processivity errors. Missense errors occur when ribosomes accept a non-cognate AA-tRNA or an aminoacyl tRNA synthetase mischarges a tRNA with a wrong amino acid. Missense errors are the most benign of possible errors, since the mistake is limited to a particular amino acid and does not necessarily inactivate the protein product. Processivity errors include frameshift errors, false recognition of a sense codon by a release factor and drop-off (also termed ribosomal editing—dissociation of a nascent polypeptidyl-tRNA from an mRNA-programmed ribosome). Mistakes in processivity often result in truncated products, and in the case of frameshifting, the sequence of amino acids incorporated after the shift is gibberish. Therefore, unless these errors occur near the end of an open reading frame (ORF), the product is likely to be inactive. As a result, it is believed that selection has resulted in processivity errors being significantly less frequent than missense errors (Kurland et al., 1996). Earlier studies support this idea. The frequency of missense errors was estimated to be between 10–3 and 10–4 (Donner and Kurland, 1972; Loftfield and Vanderjagt, 1972; Edelmann and Gallant, 1977; Parker et al., 1983; Kurland and Gallant, 1986), while processivity errors were estimated to be in the range 10–4–10–7 (Kurland, 1979; Jørgenesen et al., 1993). At the same time it has been noted that processivity errors occur in a sequence-dependent manner and are likely to be more efficient in particular places than in others (Atkins et al., 1972; Manley, 1978; Atkins et al., 1983). Later, a substantial number of relatively simple sequence motifs that can cause significantly high levels of frameshifting in E.coli were characterized (Weiss et al., 1990; Curran, 1993).
The discovery of genes whose expression requires non-standard processivity, such as programmed frameshifting, was one of the threads that lead to the term ‘recoding’ (Gesteland et al., 1992), which describes the phenomenon where non-standard translational events are used for gene expression purposes. In the majority of recoding cases in which frameshifting is involved, the efficiency of frameshifting on a specific mRNA site is much higher than the above estimates for error frameshifting due to the presence of stimulatory sequences in the mRNA. Frameshifting programmed in this manner is sometimes even more efficient than standard translation at the same site. The general assumption is that unless they have a functional role, sequences prone to high levels of frameshifting are subject to negative selection. In this scenario frameshift events fall into a low-efficiency frameshift error category or a highly efficient programmed frameshifting class (although it is possible that low-level frameshifting is utilized for gene expression purposes). Despite the studies cited above, it is hard to know what the efficiency of frameshifting errors can be before selection is triggered against them. Here we have analyzed the frequency of occurrence in E.coli genes of two sequences prone to either +1 (CCC_UGA) or –1 (A_AAA_AAG) frameshifting (the triplets indicate the zero frame codons) and measured frameshift efficiency on these sequences in their native contexts.
Results
A_AAA_AAG
The sequence A_AAA_AAG supports efficient –1 ribosomal frameshifting in E.coli. This sequence alone causes ∼2% frameshifting (Weiss et al., 1989) and the efficiency can be greatly increased by the presence of stimulatory signals. Frameshifting at A_AAA_AAG is used for expression of E.coli dnaX, which encodes two subunits of DNA polymerase III: τ and γ. While τ is synthesized by standard translation, synthesis of γ is dependent on a –1 frameshift event on the sequence A_AAA_AAG (Blinkowa and Walker, 1990; Flower and McHenry, 1990; Tsuchihashi and Kornberg, 1990). This frameshifting is 50% efficient, so that τ and γ subunits are synthesized in equal amounts. There are two stimulatory elements in E.coli dnaX mRNA, and both are conserved in related species: an internal Shine–Dalgarno sequence 10 bases upstream (Larsen et al., 1994) and a stem–loop downstream of the frameshift site (Larsen et al., 1997). These stimulators are essential to elevate frameshifting to such a high level. The same sequence (A_AAA_AAG) is also used for programmed frameshifting in bacterial insertion elements in E.coli (Chandler and Fayet, 1993; Hu et al., 1996) and related species (Polard et al., 1991; Rettberg et al., 1999). However, it is likely that this frameshifting is limited to only those bacteria that lack a tRNALys with the anticodon 3′-UUC-5′ (Tsuchihashi and Brown, 1992; Baranov et al., 2002).
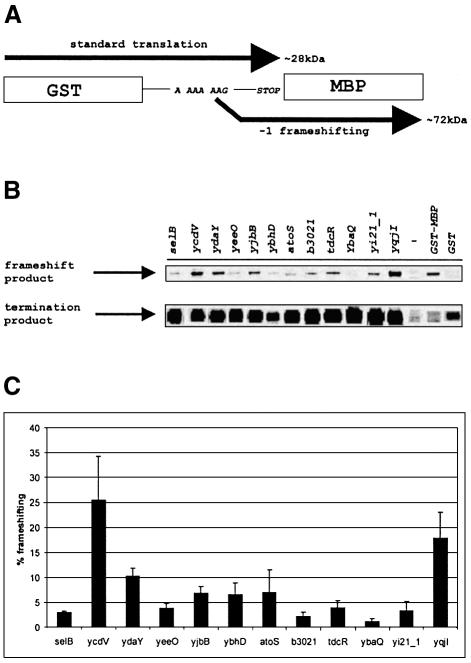
It is reasonable to expect that the sequence A_AAA_AAG is avoided in E.coli genes that do not utilize frameshifting for their expression. In E.coli K12, there are 70 instances of A_AAA_AAG in 68 genes (two genes have this sequence twice). These genes are listed in Supplementary Table I available at The EMBO Journal Online. Out of the 68 genes, 12 were selected to check whether frameshifting does in fact take place during translation of their mRNAs (Table I). We cloned gene sequences including ∼10 codons upstream and downstream of the shift site into the pGHM57 vector between the glutathione S-transferase (GST) and maltose-binding protein (MBP) genes (see Materials and methods). In those cases where nearby potential 3′ secondary structures were identified their sequences was fully included. Selection of the 12 chosen genes was somewhat biased as they had the first stop codon in the –1 frame at least 10 codons downstream of the shift site, so that the stop codon is not included in the cloned sequence. The sequence in the ‘zero’ frame was placed in-frame with GST and the sequence in the –1 frame was placed in-frame with MBP. Ribosomes that translate through A_AAA_AAG in a standard manner will terminate either at a stop codon in the cloned insert or just after the insert. The resulting products have approximately the same mass as GST (∼28 kDa). Shifting into the –1 frame on A_AAA_AAG yields products of roughly the same mass as the GST–MBP fusion (∼72 kDa) (Figure 1A). Frameshifting was assayed by pulse–chase experiments with [35S]Met as a label and the products were separated by SDS–PAGE (Figure 1B). All tested sequences support –1 frameshifting at levels ranging from 1.2 to 25.5% (Figure 1C). Distant sequences are unlikely to affect frameshifting when transcription and translation are tightly coupled. However, one further A_AAA_AAG sequence was tested, but in the context of the entire gene sequence. Gene ycdB was used (as both its frameshift and termination products are readily distinguishable), and showed a frameshifting efficiency of 8% (Figure 2 and Table I).
Table I. Analyzed genes containing the A_AAA_AAG sequence.
| Gene name (Accession no.) | Position of A_AAA_AAG nucleotides (no. of codons after the shift in 0/–1 frame) | Sequence around shift site | Frameshift level (%) |
|---|---|---|---|
| atoS (16130156) | 1647–1653 (107/24) | CTC TCG CTG CAA AAA AAG ATC TTC GAT | 7.0 ± 4.6 |
| b3021 (16130917) | 234–240 (51/41) | GTG AAG GTT CGA AAA AAG CTC TCT CTT | 2.3 ± 0.8 |
| selB (16131461) | 93–99 (581/28) | CTG CCG GAA GAA AAA AAG CGC GGC ATG | 2.9 ± 0.3 |
| tdcR (16131012) | 57–63 (93/11) | GTG GTT AAT ACA AAA AAG GGG CTG AGA | 4.0 ± 1.5 |
| ybaQ (33347458) | 381–387 (2/11) | GAA GAG CGT GCA AAA AAG GTC GCG TAA | 1.2 ± 0.5 |
| ybhD (33347481) | 177–183 (277/10) | ACG CGA AGA ATA AAA AAG ATG GAG GAA | 6.6 ± 2.3 |
| ycdB (16128983) | 462–468 (267/19) | CCA CAG ATG CCA AAA AAG CTG CAG AAG | 8.3 ± 3.2 |
| ycdV (1787269) | 198–204 (69/9) | CAA TAC ACG AAA AAA AAG CCC GTA CTT | 25.5 ± 8.8 |
| ydaY (16129327) | 348–354 (1/102) | CAG GAT ACG ATA AAA AAG CCA TAG CTG | 10.3 ± 1.6 |
| yeeO (16129928) | 1620–1626 (5/42) | CAA AAG TGT GAA AAA AAG CCA GTT GTG | 3.9 ± 1.0 |
| yi21_1 (1786557) | 363–369 (13/114) | TAT GGA CGG GCA AAA AAG TGG ATA GCG | 3.4 ± 1.9 |
| yjbB (16131846) | 126–132 (499/17) | CGG AGC GTC GAA AAA AAG CCG CTC GCC | 7.0 ± 1.2 |
Fig. 1. Measurement of frameshifting efficiency on the A_AAA_AAG sequences. (A) Schematic representation of constructs used to assay frameshifting. (B) Pulse–chase analysis of the products expressed from cassettes with the A_AAA_AAG contexts from different genes. The areas from the gels corresponding to the termination and frameshifting products are shown. The GST lane shows the corresponding products from the parental vector in which the stop codon is located after GST; the GST–MBP lane shows products from the parental vector in which the GST and MBP genes are in-frame. The (–) lane contains labeled proteins from the uninduced control (Materials and methods). (C) Quantitation of the efficiency of frameshifting. Average frameshifting in three independent pulse–chase experiments was calculated for each construct and is represented by black bars. Error bars show standard deviations.
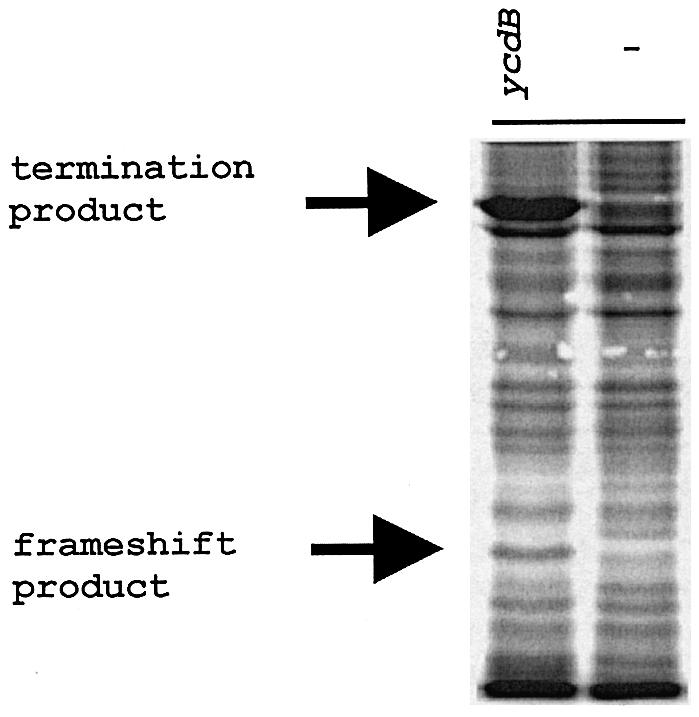
Fig. 2. Pulse–chase analysis of the products expressed from the construct with the entire sequence of the ycdB gene. The (–) lane contains labeled proteins from the uninduced control.
Since A_AAA_AAG alone supports efficient frameshifting without any stimulatory signals, it may be under-represented. To assess possible bias in its representation, codon usage for AAA (3.36%), AAG (1.03%) and occurrence of A in the wobble position (17.79%) were taken into account (see Materials and methods). Then on an unselected basis, in 1 365 282 codons of annotated E.coli K12 ORFs, this sequence should occur 1 365 282 × 0.0103 × 0.0336 × 0.17 ≈ 84 times (though this estimate does not take into account that A_AAA_AAG cannot occur in the first and in the last position of the gene). Therefore, the sequence A_AAA_AAG is somewhat under-represented (∼83% of the expected value). However, this estimate does not take into account how frequently two adjacent lysine residues occur in E.coli proteins. It is possible that the frequencies of tandem lysines are biased thereby influencing the occurrence of A_AAA_AAG. A control for this is the ‘non-shifty’ sequence A_AAG_AAA, in which two lysine codons are retained but their positions swapped. This sequence occurs 132 times, almost twice as frequently as A_AAA_AAG. Thus, both estimates show that the sequence A_AAA_AAG is moderately under-represented in the coding regions of the E.coli K12 genome.
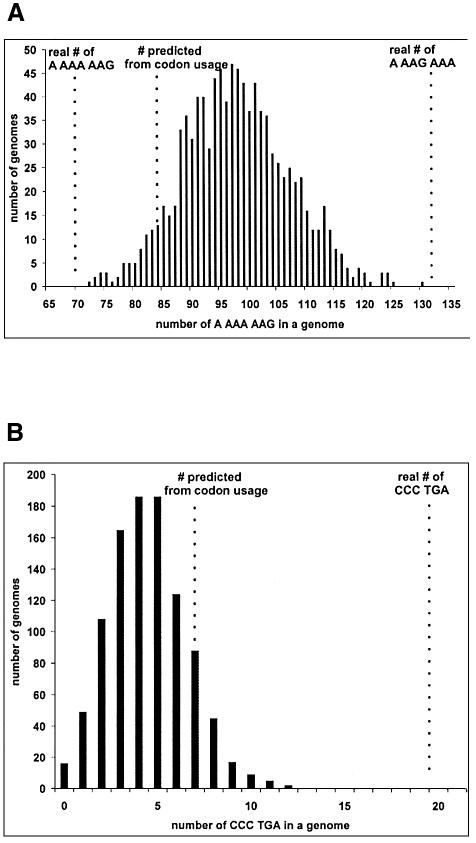
In a more rigorous test, 1000 random genomes were generated using the following rules: protein sequences from the original E.coli K12 genome were preserved, but the codons encoding the amino acids were randomized taking into account codon usage. Such random genomes are relieved of selective pressure to avoid slippery sequences. The distribution of A_AAA_AAG occurrences in the genomes generated is shown in Figure 3A. The mean occurrence of A_AAA_AAG is 97.6 per genome. The standard deviation is 9.3 and the standard error of mean is 0.3. None of the 1000 genomes had 70 A_AAA_AAG (the number of A_AAA_AAG in the real E.coli genome) and only one had 72, the lowest count of A_AAA_AAG in the 1000 genomes. One sample t-test was carried out using 70 as a hypothetical mean. The t-value was 98 and the p-value is <0.0001. This p-value suggests that the difference between the mean occurrence count from the randomized genomes and the occurrence in the actual genome is highly statistically significant. Therefore, A_AAA_AAG is indeed under-represented; however, it is not avoided since 68 genes constitute 1.7% of all genes.
Fig. 3. Distribution of occurrences of slippery sequences in 1000 randomized genomes. (A) A_AAA_AAG. (B) CCC_TGA.
Interestingly, the value of mean occurrence of A_AAA_AAG (97.6) is greater than the value predicted from codon usage (84). The probable explanation for this discrepancy lies in the fact that tandem lysines appear in the E.coli genome more often (3044 times) than if their distribution was random (2643). Our results also demonstrate that A_AAG_AAA is over-represented (see Figure 3A). Perhaps part of the reason for over-representation of A_AAG_AAA is compensation for under-representation of the slippery A_AAA_AAG sequence.
Another factor that can influence occurrence of A_AAA_AAG is the dinucleotide frequency of As in the third position of the upstream codon and the first position of the downstream codon (XXA_AYY). We have estimated this bias using a random genomes approach and found that such dinucleotides are slightly over-represented in the real genome (∼44 000 in the real genome versus ∼41 000 average in random). Although consideration of this bias may improve the accuracy of our analysis, it is unlikely to affect the general conclusion that there is a moderate selection against A_AAA_AAG sequences.
CCC_UGA
+1 frameshifting on the sequence CCC_UGA has been reported in several artificial constructs expressed in E.coli (de Smit et al., 1994; Vilbois et al., 1994; O’Connor, 2002). The observed efficiency of frameshifting ranged between 2 and 4%. Frameshifting on this sequence is utilized for expression of antizyme of some eukaryotes (Ivanov et al., 2000) and of tsh gene of Listeria monocytogenes phage PSA (Zimmer et al., 2003). However, it has not so far been found to be used for gene expression in E.coli.
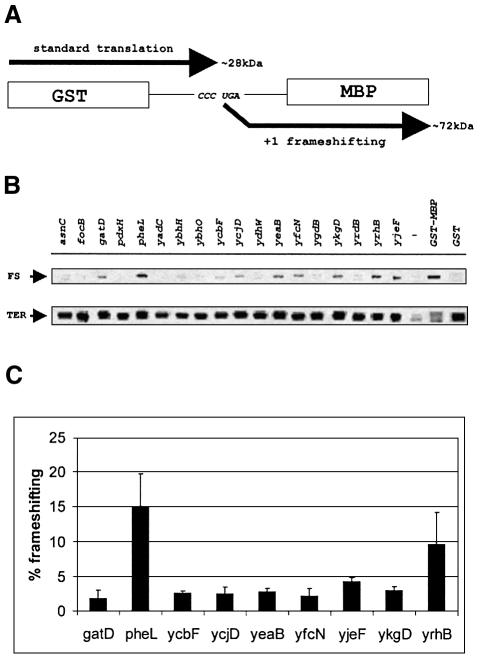
To identify coding sequences ending with CCC_TGA, we searched through the annotated E.coli K12 ORFs (Blattner et al., 1997) using the Colibri database (Medigue et al., 1993) and found 18 genes. GenBank has 20 in the nucleotide sequence file, but one of the genes was recently excluded from the annotation. Therefore we consider that there are 19 genes that end with CCC_TGA in E.coli. These genes and the nucleotide sequences surrounding their corresponding termination sites are listed in Table II. We examined the level of frameshifting on the 18 CCC_UGA sites (originally identified using the Colibri database) in their natural context. Sequences from each of the 18 genes including ∼10 codons upstream of the CCC_UGA and 10 codons downstream (or as far as, but not including, the stop codon in +1 frame) were cloned into the pGHM57 vector between the GST and MBP genes. The sequence upstream of the shift site was placed in-frame with GST and the sequence in the +1 frame downstream of shift site was placed in-frame with the MBP gene. Termination at CCC_UGA results in a protein similar in mass to GST, while the product of +1 frameshifting is approximately the same mass as the GST–MBP fusion protein (Figure 4A). The efficiency of frameshifting was assayed as before (Figure 4B). Of the 18 gene sequences, nine support +1 frameshifting at levels higher than 1% (Figure 4C and Table II). Frameshifting at levels lower than 1% is difficult to distinguish from the background in pulse–chase experiments. In two cases frameshifting is very efficient: 15% for pheL and 9.7% for yrhB.
Table II. All known E.coli K12 coding sequences terminating with CCC_UGA.
| Gene name (Accession no.) | Sequence around termination/frameshift site | Frameshift level (%) | Number of sense codons after a frameshift |
|---|---|---|---|
| asnC (16131611) | ACC ATC AAG CCC TGAT CGG CTT TTT | <1 | 3 |
| focB (16130417) | CGT CAG GAA CCC TGAA AAA TCA GCC | <1 | 10 |
| gatD (16130029) | TTG CTC ATT CCC TGAA ACC GCG GGC | 1.8 ± 1.2 | 25 |
| pdxH (16129596) | CGT CTT GCA CCC TGAA AAG ATG CAA | <1 | 12 |
| pheL (16130519) | TTT ACC TTC CCC TGAA TGG GAG GCG | 15 ± 4.7 | 50 |
| yadC (16128128) | GTA ACC TAT CCC TGAT AAC GTA GCA | <1 | 21 |
| ybhH (16128737) | GTT TAT CTT CCC TGAA AAA ATT CGT | <1 | 10 |
| ybhO (16128757) | GGG GTA AAA CCC TGAT GAG TAA ATC | <1 | 1 |
| ycbF (33347497) | CAA AAT CTG CCC TGAA ACA GGT TCG | 2.6 ± 0.3 | 43 |
| ycjD (16129250) | TCA CCC TCT CCC TGAA AGA GCG AGG | 2.5 ± 1.0 | 71 |
| ydhW (16129628) | TTT CAG AAC CCC TGAA ATT TCA GGG | <1 | 7 |
| yeaB (16129767) | GGT GTG AAA CCC TGAC TAT ACT TAT | 2.8 ± 0.5 | 32 |
| yfcN (16130266) | CCG GAG TTG CCC TGAG GAG TTG AGC | 2.2 ± 1.1 | 21 |
| ygdB (33347702) | TGT CAG CTT CCC TGAA GAA TCA ACA | <1 | 5 |
| yjeF (16131989) | AAT TCC GCT CCC TGAT GAG CAG GCA | 4.3 ± 0.6 | 144 |
| ykgD (16128290) | CAG CTT GCA CCC TGAA TAA AAC CGC | 3.0 ± 0.6 | 0 |
| yrdB (16131161) | GTC TGG TTA CCC TGAT CCA GAT ATT | <1 | 29 |
| yrhB (16131318) | TTC GGC TTG CCC TGAC AAA ATA GCC | 9.7 ± 4.5 | 17 |
| yzgL (33347755) | GCG GTA ATT CCC TGAA TTA AAA AGT | Not assayed | 8 |
Fig. 4. Measurement of the frameshifting efficiency in cassettes with sequences from genes ending with CCC_UGA. (A) Schematic representation of expression constructs for analysis of frameshifting efficiency. (B) Pulse–chase analysis of the products expressed from vectors containing inserts of different genes ending with CCC_UGA. The areas from the gels corresponding to the termination and frameshifting products are shown. FS indicates frameshift product; TER indicates termination product. The GST lane shows the corresponding products from the parental vector in which the stop codon is located after GST; the GST–MBP lane shows products from the parental vector in which the GST and MBP genes are in-frame. The (–) lane contains labeled proteins from uninduced control. (C) Quantitation of the frameshifting efficiency. Average frameshifting efficiency of three independent pulse–chase experiments was calculated for each construct and is represented by black bars. Sequences in which frameshifting is <1% are omitted. Error bars show standard deviations.
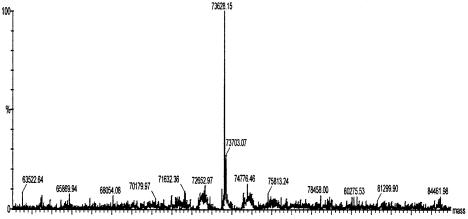
To verify that the +1 frameshifting indeed occurs at CCC_UGA, we used affinity tag purification, via GST and MBP, of the fusion protein translated from the construct with yjeF sequence (frameshifting efficiency 4.3%). The mass of the purified protein, as determined by mass spectrometry, is 73 628.15 Da, which is within 2 Da of the predicted mass of the fusion protein, 73 629.9 Da, that would result from shifting from CCC to CCU at the sequence CCC_UGA (Figure 5).
Fig. 5. Mass spectrum of the GST–MBP fusion protein synthesized from a cassette containing the yjeF sequence. The major peak at 73 628.15 Da corresponds to the predicted mass of the fusion protein (73 629.91 Da). The satellite peak at 73 703.07 Da corresponds to the β-mercaptoethenol adduct of the fusion protein.
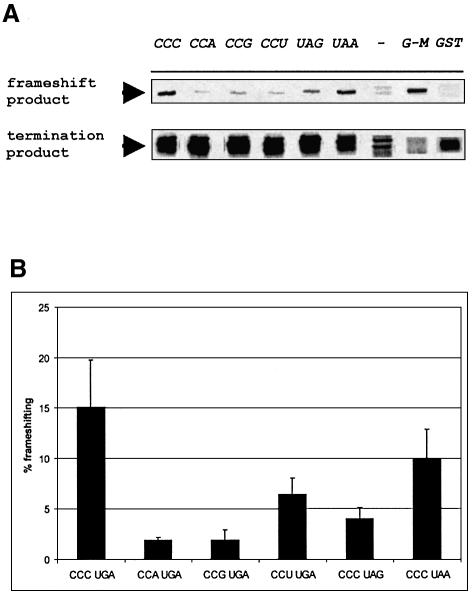
Does identity of either the stop or proline codons influence the efficiency of frameshifting? Changing the CCC codon to either CCA or CCG in the construct containing pheL decreases frameshifting from 15 to ∼2%, while changing it to CCU decreases frameshifting to 6%. Changing UGA to either UAG or UAA decreases frameshifting to 4 and 10%, respectively (Figure 6).
Fig. 6. Analysis of the frameshifting efficiency on different CCN-Stop combinations. (A) Pulse–chase experiments with expression vectors containing mutations in pheL. CCC denotes the wild-type pheL context. Other abbreviations indicate the mutation of either a Pro or a Stop codon. The areas from the gels corresponding to the termination and frameshifting products are shown. The GST lane shows the corresponding products from the parental vector in which the stop codon is located after GST; the G–M lane shows products from the parental vector in which the GST and MBP genes are in-frame. The (–) lane contains labeled proteins from uninduced cultures. (B) Quantitation of the pulse–chase results with mutated pheL constructs. Average frameshifting in three independent pulse–chase experiments was calculated for each construct and is represented by black bars. Error bars show standard deviations.
Since the sequence CCC_UGA is prone to relatively high efficiency frameshifting, its occurrence is expected to be under-represented in the E.coli K12 genome. The theoretical frequency of CCC_UGA can be calculated by multiplication of the absolute values for CCC and UGA codon usage (7506 and 1252, respectively) divided by the total number of codons in E.coli K12 (1 365 282). This gives a value 6.9, which is significantly less than the observed number of 19. The random genome approach was also applied to analyze the distribution of CCC_TGA (Figure 3B). On average only 4.5 genes end with CCC_TGA in 1000 random genomes. The lower value of the mean (4.5) than the one predicted based on codon usage (6.9) probably reflects the fact that proline codons are under-represented in the last position of ORFs. At the same time, none of the 1000 genomes contains more than 12 genes ending with CCC_TGA. This analysis clearly demonstrates that CCC_TGA is over-represented in E.coli K12 genome, even though it can support efficient frameshifting. This is surprising since a simple change of either the CCC codon to another proline codon or UGA to another stop codon can eliminate significant propensity for frameshifting.
Discussion
Assessment of the numerous occurrences of the two shift-prone sequences identified requires distinguishing those where there is a selective advantage for specific ribosomal frameshifting from those where it is simply an error, which wastes the cells resources. While some cases of utilized frameshifting may be organism specific, many will be evolutionarily conserved in related species. Comparative analysis with orthologs, juxtaposition of ORFs and features relevant to possible regulatory frameshifting help distinguish between the two categories. The following analysis deals with the two shift-prone sequences separately, and later with common features of the distribution of all members of the erroneous frameshifting category. Similar analysis was also performed on some previously published cases of frameshifting for which no functional role is evident.
A_AAA_AAG
Statistical analysis of the occurrence of A_AAA_AAG shows that this shift-prone sequence is somewhat under-represented in E.coli. Nevertheless, the total number of such sequences in E.coli, 70, is substantial. Interestingly, similar observations were made for the two other related bacteria, which also lack tRNALys with the anticodon 3′-UUC-5′. In Salmonella typhimurium A_AAA_AAG is slightly under-represented, while in Shigella flexneri 2a it is slightly over-represented, showing that there is also no major avoidance of this slippery sequence in these bacteria. In the 13 E.coli sequences tested (Table I), the frameshifting levels varied from 1 to 25%. With this limited set of sequences, a correlation was not evident between frameshifting efficiency and the presence of 3′ nucleotides with stacking potential (Bertrand et al., 2002). Most probably the cumulative effect of other sequence context surrounding the shift site in these particular cases is more important than the effect of the single 3′ adjacent nucleotide.
In several cases the frameshifting is expected not to have significant negative consequences. In the genes ybaQ, yeeO, atoS, b3021 and yqjI, A_AAA_AAG occurs near the end of the ORF. In these cases there is a termination codon in the –1 frame within the next 42 codons. As a result, the product of frameshifting contains almost the same information as the product of standard decoding. In others, however, frameshifting should result in the production of truncated dysfunctional proteins. In selB, ybhD, tdcR, yjbB, ycdV and ycdB, A_AAA_AAG occurs in the early or middle parts of their coding sequences. Ribosomes that frameshift at A_AAA_AAG will encounter a stop codon and terminate. Theoretically such frameshifting could be used for down-regulation of expression, but there is no experimental evidence that frameshifting on A_AAA_AAG can be specifically regulated. Alternatively, the short protein can have a separate function. If the frameshifting is used for gene expression, conservation of the shift site is expected in homologous genes from related species. The frameshift cassette, in genes other than selB and ybhD, is limited amongst sequenced genomes to E.coli species and S.flexneri. For selB and ybhD, it appears that the conservation of tandem lysines is important, rather then the frameshift cassette itself. In the Haemophilus influenzae selB gene, the corresponding sequence is A_AAA_AAA, and in Vibrio cholerae ybhD gene it is A_AAG_AAA.
Another gene with A_AAA_AAG is yi21_1, which belongs to the IS2 family of bacterial insertion sequences. –1 frameshifting results in fusion of yi21_1 and yi22_1. Frameshifting in the IS2 element on the sequence A_AAA_AAG was previously reported by Hu et al. (1996). Thus, in this case frameshifting is functional, and it is a true case of recoding. In fact, we found that there are five more IS2 related sequences with A_AAA_AAG sequence in the E.coli K12 genome (see Supplementary table I).
Frameshifting during decoding of the ydaY gene on A_AAA_AAG is also likely to be a recoding event. Standard translation of ydaY terminates one codon after the A_AAA_AAG site. However, –1 frameshifting on A_AAA_AAG yields a fusion of the ydaY product with that of the downstream ORF b1367. Additional evidence that A_AAA_AAG in ydaY is purposeful comes from the fact that there are putative stimulatory signals: an internal Shine–Dalgarno sequence AGAAG, 11 bases upstream of the A_AAA_AAG (with the nearest 3′ start codon 95 nt downstream) and a potential RNA secondary structure appropriately positioned downstream of it. Unfortunately, the functions of both ydaY and b1367 are currently unknown. None of the completed bacterial genomes contain homologs. However, homologous sequences occur in Klebsiella pneumoniae whose sequencing is currently under way. Both ORFs are present in the same orientation as in E.coli K12, and the frameshift site as well as the tentative stimulator signals are also preserved.
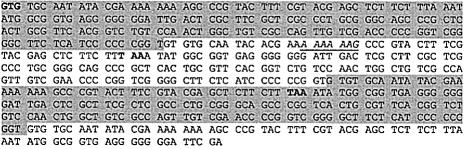
The gene whose decoding exhibits the highest level of frameshifting, 25.5%, is ycdV. This high level prompted a closer examination of this gene. However, the analysis raised very serious doubt that it is in fact a protein-encoding gene. The annotated gene consists of three exact nucleotide repeats that occur in all three translational frames (Figure 7). Thus, repeats occur at the nucleotide level and not at the protein level. No protein homologs were found in other bacteria including other sequenced strains of E.coli. At the same time, we found similar nucleotide repeats in several bacteria. Nevertheless, the fact that extremely efficient frameshifting occurs on the misannotated gene sequence indicates that in real genes there might be selection for sequences surrounding A_AAA_AAG to lower frameshifting efficiency.
Fig. 7. Sequence of the ycdV gene. Annotated initiation codon, termination codon and termination codon in the –1 frame are in bold. The A_AAA_AAG sequence in the ‘0’ frame is underlined. Repeated sequences are differently highlighted.
Overall, we have found that –1 frameshifting on a A_AAA_AAG sequences is employed for gene expression purposes in addition to the previously characterized cases, e.g. in decoding dnaX. However, in the majority of cases, although frameshifting is evident, any functional role is unclear. Moreover, lack of sequence conservation in the corresponding genes among related species suggests that there is no functional role for these frameshifting events (unless its role is specific to E.coli).
CCC_UGA
The sequence CCC_UGA which supports +1 frameshifting at the end of a gene is surprisingly over-represented in E.coli (and in the two other bacteria examined, S.flexneri 2a and S.typhimurium LT2). The most efficient frameshifting (15%) occurs in decoding pheL—the leader peptide in the pheA biosynthetic operon. The sole role of this gene is to provide regulation of downstream gene expression via transcriptional attenuation. It is known that there is a certain level of basal transcriptional readthrough in the pheA operon even in the presence of an adequate concentration of tRNAPhe. The level of basal expression depends on the efficiency of ribosome release from the UGA stop codon at the end of pheL (Gavini and Pulakat, 1991). Thus, it is likely that +1 frameshifting at the end of pheL provides an additional path for the ribosome to escape the UGA stop codon and possibly fine-tunes the basal level of expression of the pheA operon. Even though other effects of CCC_UGA may also be relevant (Hayes et al, 2002), the presence of CCC_UGA at the same position in S.typhimurium LT2 and Salmonella typhi provides additional support for a functional role for this particular sequence and probably for frameshifting.
Decoding the cassette with yjeF gene sequences flanking CCC_UGA yields 4.3% +1 frameshifting. In the native context, frameshifting results in fusion of the yjeF and yjeE proteins, since the ORF for the gene yjeE overlaps with the yjeF ORF. The initiation codon for yjeE is located 28 nt upstream of the termination site for yjeF. It could be that all three products, from the two ORFs separately and the frameshift product, have special functions. If so, this organization of genes and the frameshift site should likely be conserved in other bacteria. In fact, conservation is limited to E.coli O157:H7 and CFT073 strains and S.flexneri. In S.typhimurium LT2, the two genes overlap, but the termination site is ACC UGA, which is unlikely to promote +1 frameshifting. In Yersinia pestis the termination codon for the yjeF gene occurs prior to the initiation codon of yjeE. Therefore, it is unlikely that this frameshifting has a functional role.
In the majority of the remaining genes, frameshifting on CCC_UGA results in the addition of a few amino acids at the C-terminus of the protein. Such extensions are unlikely to be deleterious and the resulting protein probably retains its function. In a few cases, the frameshift products are significantly longer (71 amino acids in ycjD). Nevertheless the absence of conservation of the shift site in related species suggests that the frameshifting does not play a significant functional role.
What determines whether a ribosome frameshifts at CCC_UGA? There are several known stimulators of +1 frameshifting at termination codons. An internal Shine–Dalgarno sequence stimulates the required +1 frameshifting event in decoding the RF2 gene when it is located 3 nt upstream of the shift site (CUU UGA). Other major contributors are the base following the stop codon and the two last amino acids in the nascent peptide, as their identity influences efficiency of the competing reaction—termination (Mottagui-Tabar et al., 1994; Poole et al., 1995). In a few cases the observed frameshifting can be explained by the presence of a Shine–Dalgarno-like sequence (yfcN) or suboptimal termination context (yrhB and yeaB). However, in the majority of cases it is hard to draw any conclusions from the obtained results, since we observed that frameshifting in genes with similar contexts occurs at very different levels (see Table II). Another factor that influences frameshifting on CCC_UGA is the availability of release factor 2. The concentration of release factor 2 and its ability to compete with recoding events for UGA changes at different growth rates (Adamski et al., 1993; Mansell et al., 2001). Therefore, it is likely that certain growth conditions favor frameshifting at CCC_UGA more than the others.
Other cases of efficient non-programmed ribosomal frameshifting
Relatively simple sequences (in addition to the ones used in this study) can trigger ribosomes to shift frames. However, the efficiency of frameshifting can depend on the sequence surrounding the frameshift site. Sequence elements that affect frameshifting can be located far away from the frameshift site in eukaryotes (Barry and Miller, 2002). In such a situation finding a simple shift-prone sequence in the gene does not necessarily imply that ribosomes efficiently shift frame at this sequence in vivo as the surrounding sequence may have evolved to suppress this phenomenon. The consequent difficulty of shiftiness predictability is exemplified by the present CCC_UGA analysis, where some simple frameshift-prone sequences support efficient frameshifting and others do not.
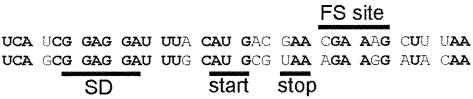
A few other cases of non-programmed ribosomal frameshifting have been reported when expression of particular genes were analyzed. Fu and Parker (1994) have demonstrated that ribosomal frameshifting occurs at a frequency of between 3 and 16% during translation of E.coli argI mRNA at the sequence UUU_U/C. Frameshifting results in a truncated protein and it is not obvious whether this has any functional role. Mejlhede et al. (1999) have reported 16% efficient –1 frameshifting on the sequence CGA_AAG in the cdd gene of Bacillus subtilis. The product of frameshifting has the same enzymatic activity as the product of standard translation. Frameshifting is stimulated by the presence of a Shine–Dalgarno sequence upstream of the frameshift site. The authors speculated about the possible involvement of this frameshifting in regulation of expression of the gene located downstream of cdd. However, comparative sequence analysis of the cdd gene from B.subtilis and Bacillus firmus (Figure 8) shows that while there is a clear sequence similarity, including the stimulatory signals between these genes, there is no identical frameshift site in B.firmus. Moreover, a stop codon is located before the site corresponding to the frameshift site in B.subtilis. The Shine–Dalgarno sequence that stimulates frameshifting in B.subtilis exists in both species as it is required for the initiation of translation of the gene downstream of cdd, not for the frameshifting. Therefore the frameshifting in B.subtilis cdd is likely accidental.
Fig. 8. Comparative sequence analysis of cdd genes from B.subtilis (upper sequence) and B.firmus (lower sequence). FS site, position of frameshifting site in cdd gene from B.subtilis; SD, Shine–Dalgarno sequence facilitating initiation on initator AUG (marked as start) and known to stimulate frameshifting. stop, stop codon in B.firmus cdd.
Taking into account that only a small proportion of sequences known to be prone to ribosomal frameshifting were analyzed, we can conclude that in the decoding of quite a number of genes significant ribosomal frameshifting may occur without a clear functional role. It is likely that this conclusion is not limited to prokaryotic systems. Recent genome-scale analysis of yeast mRNAs suggests that the average processing error is 0.6% per step of elongation (Arava et al., 2003). Considering the variety of frameshifting errors, it is likely that a substantial number of shift-prone sequences exist in the coding regions of diverse organisms.
However, frequency of frameshift-prone sequences in a genome is very unlikely to simply correlate with the total load of aberrant frameshift products synthesized. None of the genes containing frameshift-prone sequences described in this study belongs to the pool of the 306 most highly expressed genes in E.coli (Karlin et al., 2001). It is likely that these genes adapted their codon usage against shift-prone sequences much more successfully than genes expressed at lower levels. A correlation between gene length and occurrence of shift-prone sequences may exist, as exceptionally long genes such as that for human titin (38 138 codons; Bang et al., 2001) will not be successfully decoded if shift-prone sequences are not, at least, very rare in its sequence. It is also possible that shift-prone sequences are not equally distributed throughout coding sequences. Frameshift errors near the 3′ end of genes should generally be less harmful, since they affect only protein C-terminal heterogeneity. This idea is in part supported by the difference in the genomic distribution of sequences considered here. While selection against A_AAA_AAG is evident, CCC_TGA sequences are even over-represented in E.coli genome. Although there may be an unknown functional reason for positive selection of such sequences, it is clear that selection based on shift-prone characteristics of these sequences is unable to compensate. It is also possible that shift-prone sequences occur more frequently early rather than in the middle of genes as in this case a cell would spend fewer resources for the synthesis of aberrant products compared to the situation with shift-prone sequence located in the middle of a gene.
Selection against shift-prone sequences most probably correlates with the efficiency of the frameshifting they mediate. Shah et al., (2002) explored the idea that heptameric sequences are under-represented in the reading frame in which they would mediate high levels of frameshifting. It has been found that translation of heptameric sequences, which are extremely under-represented in the Saccharomyces cerevisiae genome, indeed results in highly efficient frameshifting. It is likely that selection against less efficient shift-prone sequences such as A_AAA_AAG is less strong. Not surprisingly with the smaller genomes of bacteria particular heptameric sequences are absent. Whether the reason for such avoidance is shift-prone properties of such sequences needs to be elucidated in a separate study.
The efficiency of erroneous frameshifting can be influenced by a number of different factors. Frameshift errors are known to increase on certain ‘hungry’ codons under starvation conditions (Lindsley and Gallant, 1993). In this case, what could be considered as erroneous frameshifting may actually facilitate recycling and enhance the effect of the stringent response. Frame maintenance is known to be dependent on tRNA modifications (Urbonavièius et al., 2001) and consequently may be influenced by tRNA modifications as well as changes in the relative concentrations of tRNAs.
As biology enters the proteomic era, it is important to understand that certain proteins may be expressed in heterogeneous forms from a single mRNA and that those forms may, or may not, differ in their function. Recent attempts to detect the complete proteome of Deionococcus radiodurans indicate that there are a considerable number of proteins produced from ORFs in different translational phases (Lipton et al., 2002). It appears that synthesis of a small amount of dysfunctional protein product as a result of frameshift errors is not significantly harmful for the cell to drive strong selection against frameshift-prone sequences at least in moderately expressed genes.
Materials and methods
Plasmids and bacterial strains
GenBank accession numbers of the bacterial strains discussed in this study are as follows: E.coli K12, U00096; E.coli O157:H7, BA000007; E.coli CFT073, AE014075; S.flexneri 2a, AE005674; S.typhimurium LT2, AE006468; S.typhi, AL513382; Y.pestis KIM, AE009952.
Escherichia coli strains DH5α and SU1675 were used throughout the experiments. The GST–MBP fusion expression vector (GHM57), containing BamHI and EcoRI restriction sites between the coding sequences of GST and MBP has been described previously (Herr et al., 2001). Gene sequences were either amplified by PCR from E.coli genomic DNA or made from complementary oligonucleotides and cloned between BamHI and EcoRI sites. Mutations in the CCC_TGA frameshift site in the construct containing the pheL were introduced by two-step PCR using oligonucleotides complimentary to the frameshift site and carrying appropriate mutations. pSKAGS vector was described previously (Wills et al, 1997). The entire sequence of the ycdB gene was amplified by PCR and cloned into pSKAGS between the XbaI and HindIII sites. All plasmid constructions were confirmed by DNA sequencing on automated sequencing machines (ABI-100).
Frameshifting assay
Overnight cultures of strains expressing the appropriate construct were grown in MOPS–glucose (Neidhardt et al., 1974) containing 100 µg/ml ampicillin and all amino acids (150 µg/ml each) except methionine and tyrosine and diluted 1:50 in 300 µl of the same. After 2 h incubation at 37°C, cultures were induced with 2 mM IPTG for 10 min [except for the (–) control]. The cells were pulsed for 2 min by addition of 7.5 µCi [35S]Met in 30 µl media, chased for 2 min by addition of 30 µl 50 mg/ml cold methionine, chilled on ice and harvested by centrifugation. The pellet was resuspended in 50 µl cracking buffer (6 M urea, 1% SDS, 50 mM Tris–HCl pH 7.2) and heated at 95°C for 5 min. 10 µl aliquots were loaded on 4–12% NuPAGE Gels (Invitrogen Inc.) and electrophoresed according to the manufacturer’s instructions in MOPS–SDS buffer. Gels were exposed overnight and visualized with a Molecular Dynamics PhosporImager.
Protein analysis
Overnight cultures of the strain expressing the yjeF-containing construct were diluted 1:50 in terrific broth, grown at 37°C to mid-log phase, and induced with IPTG (final concentration 1 mM) for an additional 4 h at 37°C. Harvested cells were lysed using Novagen’s BugBuster reagent. The GST–yjeF–MBP fusion protein was purified by sequential passages over glutathionine–Sepharose (AP Biotec) and Amylose Resin (New England BioLabs). Purified protein was concentrated and washed extensively with Nanopure H2O using a Centricon 30 (Millipore) filtration unit. Final clean-up and mass measurements were performed as described (Herr et al., 2001) except only C4 P10 ZipTip (Millipore) were used for clean-ups and proteins were eluted with 56% (v/v) methanol + 1.5% formic acid with three aliquots of 2 ml, which were then pooled and introduced into the mass spectrometer by infusion at 3 ml/min.
Statistical analysis
Three programs were written to analyze the E.coli K12 genome. The programs are CodonUsageTable, ColiGenerator and MotifCounter. All three programs were written with Java2 (http://java.sun.com/) and BioJava (http://www.biojava.org/). Two files were downloaded from the National Center for Biotechnology Information’s website (http://www.ncbi.nlm.nih.gov/): NC_000913.ffn, which contains the nucleotide sequences of all E.coli K12 protein coding genes, and NC_000913.faa, which contains the corresponding protein sequences.
The CodonUsageTable program read and counted each codon from the nucleotide file NC_000913.ffn. These counts were then totaled and output as a codon usage table. Then, the probability with which particular codon encodes a particular amino acid was calculated by dividing its codon usage by the sum of usages of all synonymous codons.
The second program, ColiGenerator creates a new nucleotide sequences file based on protein sequences and the above codon usage table. The ColiGenerator program reads each amino acid from the E.coli protein file NC_000913.faa. As each amino acid is read, it is replaced in a new file by its corresponding codon with the probability counted by the CodonUsageTable program. The sequences in the new file are the same length as in the original nucleotide file and encode exactly the same proteins. The only difference is that the codons may, or may not, match. 1000 such nucleotide files were generated.
The final program, MotifCounter searched through the 1000 new files and counted how many times a motif (A_AAA_AAG or CCC_TGA) occurred in each genome.
Supplementary data
Supplementary data are available at The EMBO Journal Online.
Acknowledgments
Acknowledgements
We thank Dr Chad Nelson for his valuable help and experience with mass-spectrometry, and an anonymous referee for the valuable suggestion of comparison with random genome sequences. This work was supported by a grant from NIH (GM48152) to J.F.A. and from DOE (DE-FG03-01ER63132) to R.F.G.
References
- Adamski F.M., Donly,B.C. and Tate,W.P. (1993) Competition between frameshifting, termination and suppression at the frameshift site in the Escherichia coli release factor-2 mRNA. Nucleic Acids Res., 21, 5074–5078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arava Y., Wang,Y., Storey,J.D., Liu,C.L., Brown,P.O. and Herschlag,D. (2003) Genome-wide analysis of mRNA translation profiles in Saccharomyces cerevisiae. Proc. Natl Acad. Sci. USA, 100, 3889–3894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atkins J.F., Elseviers,D. and Gorini,L. (1972) Low activity of β-galactosidase in frameshift mutants of Escherichia coli. Proc. Natl Acad. Sci. USA, 69, 1192–1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atkins J.F., Nichols,B.P. and Thompson,S. (1983) The nucleotide sequence of the first externally suppressible –1 frameshift mutant, and of some nearby leaky frameshift mutants. EMBO J., 2, 1345–1350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bang M.L., Centner,T., Fornoff,F., Geach,A.J., Gotthardt,M., McNabb,M., Witt,C.C., Labeit,D., Gregorio,C.C., Granzier,H. and Labeit,S. (2001) The complete gene sequence of titin, expression of an unusual approximately 700-kDa titin isoform, and its interaction with obscurin identify a novel Z-line to I-band linking system. Circ. Res., 89, 1065–1072. [DOI] [PubMed] [Google Scholar]
- Baranov P.V., Gesteland,R.F. and Atkins,J.F. (2002) Recoding: translational bifurcations in gene expression. Gene, 286, 187–201. [DOI] [PubMed] [Google Scholar]
- Barry J.K. and Miller,W.A. (2002) A –1 ribosomal frameshift element that requires base pairing across four kilobases suggests a mechanism of regulating ribosome and replicase traffic on a viral RNA. Proc. Natl Acad. Sci. USA. 99, 11133–11138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertrand C., Prère,M.F., Gesteland,R.F., Atkins,J.F. and Fayet,O. (2002) Influence of the stacking potential of the base 3′ of tandem shift codons on –1 ribosomal frameshifting used for gene expression. RNA, 8, 16–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Björnsson A., Mottagui-Tabar,S. and Isaksson,L.A. (1996) Structure of the C-terminal end of the nascent peptide influences translation termination. EMBO J., 15, 1696–1704. [PMC free article] [PubMed] [Google Scholar]
- Blattner F.R. et al. (1997) The complete genome sequence of Escherichia coli K-12. Science, 277, 1453–1474. [DOI] [PubMed] [Google Scholar]
- Blinkowa A.L. and Walker,J.R. (1990) Programmed ribosomal frameshifting generates the Escherichia coli DNA polymerase III γ subunit from within the τ subunit reading frame. Nucleic Acids Res., 18, 1725–1729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandler M. and Fayet,O. (1993) Translational frameshifting in the control of transposition in bacteria. Mol. Microbiol., 7, 497–503. [DOI] [PubMed] [Google Scholar]
- Curran J.F. (1993) Analysis of effects of tRNA:message stability on frameshift frequency at the Escherichia coli RF2 programmed frameshift site. Nucleic Acids Res., 21, 1837–1843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Smit M.H., van Duin,J., van Knippenberg,P.H. and van Eijk,H.G. (1994) CCC.UGA: a new site of ribosomal frameshifting in Escherichia coli. Gene, 143, 43–47. [DOI] [PubMed] [Google Scholar]
- Donner D. and Kurland,C.G. (1972) Changes in the primary structure of a mutationally altered ribosomal protein S4 of Escherichia coli. Mol. Gen. Genet., 115, 49–53. [DOI] [PubMed] [Google Scholar]
- Edelmann P. and Gallant,J. (1977) Mistranslation in E. coli. Cell, 10, 131–137. [DOI] [PubMed] [Google Scholar]
- Flower A.M. and McHenry,C.S. (1990) The γ subunit of DNA polymerase III holoenzyme of Escherichia coli is produced by ribosomal frameshifting. Proc. Natl Acad. Sci. USA, 87, 3713–3717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu C. and Parker,J. (1994) A ribosomal frameshifting error during translation of the argI mRNA of Escherichia coli. Mol. Gen. Genet., 243, 434–441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gesteland R.F., Weiss,R.B. and Atkins,J.F. (1992) Recoding: reprogrammed genetic decoding. Science, 257, 1640–1641. [DOI] [PubMed] [Google Scholar]
- Gavini N. and Pulakat,L. (1991) Role of ribosome release in the basal level of expression of the Escherichia coli gene pheA. J. Gen. Microbiol., 137, 679–684. [DOI] [PubMed] [Google Scholar]
- Hayes C.S., Bose,B. and Sauer,R.T. (2002) Proline residues at the C terminus of nascent chains induce SsrA tagging during translation termination. J. Biol. Chem., 277, 33825–33832. [DOI] [PubMed] [Google Scholar]
- Herr A.J., Nelson,C.C., Wills,N.M., Gesteland,R.F. and Atkins,J.F. (2001) Analysis of the roles of tRNA structure, ribosomal protein L9, and the bacteriophage T4 gene 60 bypassing signals during ribosome slippage on mRNA. J. Mol. Biol., 309, 1029–1048. [DOI] [PubMed] [Google Scholar]
- Hu S.T., Lee,L.C. and Lei,G.S. (1996) Detection of an IS2-encoded 46-kilodalton protein capable of binding terminal repeats of IS2. J. Bacteriol., 178, 5652–5659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov I.P., Gesteland,R.F. and Atkins,J.F. (2000) Antizyme expression: a subversion of triplet decoding, which is remarkably conserved by evolution, is a sensor for an autoregulatory circuit. Nucleic Acids Res., 28, 3185–3196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jørgensen F., Adamski,F.M., Tate,W.P. and Kurland,C.G. (1993) Release factor-dependent false stops are infrequent in Escherichia coli. J. Mol. Biol., 230, 41–50. [DOI] [PubMed] [Google Scholar]
- Karlin S., Mrazek,J., Campbell,A. and Kaiser,D. (2001) Characterizations of highly expressed genes of four fast-growing bacteria. J. Bacteriol., 183, 5025–5040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurland C.G. (1979) Reading frame errors on ribosomes. In Celis,J.E. and Smith,J.D. (eds), Nonsense Mutations and tRNA Suppressors. Academic Press, London, UK, pp. 97–108. [Google Scholar]
- Kurland C.G. and Gallant,J.A. (1986) The secret life of the ribosome. In Kirkwood,T.B.L., Rosenberger,R.F. and Galas,D.J. (eds), Accuracy in Molecular Processes. Chapman and Hall, New York, NY, pp. 127–158. [Google Scholar]
- Kurland C.G., Hughes,D. and Ehrenberg,M. (1996) Limitations of translational accuracy. In Neidhart,F.C. et al. (eds), Escherichia coli and Salmonella Cellular and Molecular Biology. ASM Press, Washington, DC, pp. 979–1004. [Google Scholar]
- Larsen B., Wills,N.M., Gesteland,R.F. and Atkins,J.F. (1994) rRNA–mRNA base pairing stimulates a programmed –1 ribosomal frameshift. J. Bacteriol., 176, 6842–6851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsen B., Gesteland,R.F. and Atkins,J.F. (1997) Structural probing and mutagenic analysis of the stem–loop required for Escherichia coli dnaX ribosomal frameshifting: programmed efficiency of 50%. J. Mol. Biol., 271, 47–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindsley D. and Gallant,J. (1993) On the directional specificity of ribosome frameshifting at a “hungry” codon. Proc. Natl Acad. Sci. USA, 90, 5469–5473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipton M.S. et al. (2002) Global analysis of the Deinococcus radiodurans proteome by using accurate mass tags. Proc. Natl Acad. Sci. USA, 99, 11049–11054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loftfield R.B. and Vanderjagt,D. (1972) The frequency of errors in protein biosynthesis. Biochem. J. 128, 1353–1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manley J.L. (1978) Synthesis and degradation of termination and premature-termination fragments of β-galactosidase in vitro and in vivo. J. Mol. Biol., 125, 407–432. [DOI] [PubMed] [Google Scholar]
- Mansell J.B., Guevremont,D., Poole,E.S. and Tate,W.P. (2001) A dynamic competition between release factor 2 and the tRNA(Sec) decoding UGA at the recoding site of Escherichia coli formate dehydrogenase H. EMBO J., 20, 7284–7293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medigue C., Viari,A., Henaut,A. and Danchin,A. (1993) Colibri: a functional data base for the Escherichia coli genome. Microbiol. Rev., 57, 623–654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mejlhede N., Atkins,J.F. and Neuhard,J. (1999) Ribosomal –1 frameshifting during decoding of Bacillus subtilis cdd occurs at the sequence CGA AAG. J. Bacteriol., 181, 2930–2937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mottagui-Tabar S., Björnsson,A. and Isaksson,L.A. (1994) The second to last amino acid in the nascent peptide as a codon context determinant. EMBO J., 13, 249–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mottagui-Tabar S. and Isaksson,L.A. (1997) Only the last amino acids in the nascent peptide influence translation termination in Escherichia coli genes. FEBS Lett., 414, 165–170. [DOI] [PubMed] [Google Scholar]
- Neidhardt F.C., Bloch,P.L. and Smith,D.F. (1974) Culture medium for enterobacteria. J. Bacteriol., 119, 736–747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Connor M. (2002) Imbalance of tRNA(Pro) isoacceptors induces +1 frameshifting at near-cognate codons. Nucleic Acids Res., 30, 759–765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker J., Johnston,T.C., Borgia,P.T., Holtz,G., Remaut,E. and Fiers,W. (1983) Codon usage and mistranslation. In vivo basal level misreading of the MS2 coat protein message. J. Biol. Chem., 258, 10007–10012. [PubMed] [Google Scholar]
- Polard P., Prère,M.F., Chandler,M. and Fayet,O. (1991) Programmed translational frameshifting and initiation at an AUU codon in gene expression of bacterial insertion sequence IS911. J. Mol. Biol., 222, 465–477. [DOI] [PubMed] [Google Scholar]
- Poole E.S., Brown,C.M. and Tate,W.P. (1995) The identity of the base following the stop codon determines the efficiency of in vivo translational termination in Escherichia coli. EMBO J., 14, 151–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rettberg C.C., Prère,M.F., Gesteland,R.F., Atkins,J.F. and Fayet,O. (1999) A three-way junction and constituent stem–loops as the stimulator for programmed –1 frameshifting in bacterial insertion sequence IS911. J. Mol. Biol., 286, 1365–1378. [DOI] [PubMed] [Google Scholar]
- Shah A.A., Giddings,M.C., Parvaz,J.B., Gesteland,R.F., Atkins,J.F. and Ivanov,I.P. (2002) Computational identification of putative programmed translational frameshift sites. Bioinformatics, 18, 1046–1053. [DOI] [PubMed] [Google Scholar]
- Tsuchihashi Z. and Kornberg,A. (1990) Translational frameshifting generates the γ subunit of DNA polymerase III holoenzyme. Proc. Natl Acad. Sci. USA, 87, 2516–2520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuchihashi Z. and Brown,P.O. (1992) Sequence requirements for efficient translational frameshifting in the Escherichia coli dnaX gene and the role of an unstable interaction between tRNA(Lys) and an AAG lysine codon. Genes Dev., 6, 511–519. [DOI] [PubMed] [Google Scholar]
- Urbonavièius J., Qian,Q., Durand,J.M., Hagervall,T.G. and Björk,G.R. (2001) Improvement of reading frame maintenance is a common function for several tRNA modifications. EMBO J., 20, 4863–4873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vilbois F., Caspers,P., da Prada,M., Lang,G., Karrer,C., Lahm,H.W. and Cesura,A.M. (1994) Mass spectrometric analysis of human soluble catechol O-methyltransferase expressed in Escherichia coli. Identification of a product of ribosomal frameshifting and of reactive cysteines involved in S-adenosyl-l-methionine binding. Eur. J. Biochem., 222, 377–386. [DOI] [PubMed] [Google Scholar]
- Weiss R.B., Dunn,D.M., Shuh,M., Atkins,J.F. and Gesteland,R.F. (1989) E. coli ribosomes re-phase on retroviral frameshift signals at rates ranging from 2 to 50 percent. New Biol., 1, 159–169. [PubMed] [Google Scholar]
- Weiss R.B., Dunn,D.M., Atkins,J.F. and Gesteland,R.F. (1990) Ribosomal frameshifting from –2 to +50 nucleotides. Prog. Nucleic Acid Res. Mol. Biol., 39, 159–183. [DOI] [PubMed] [Google Scholar]
- Wills N.M., Ingram,J.A., Gesteland,R.F. and Atkins,J.F. (1997) Reported translational bypass in a trp′-lacZ′ fusion is accounted for by unusual initiation and +1 frameshifting. J. Mol. Biol., 271, 491–498. [DOI] [PubMed] [Google Scholar]
- Zimmer M., Sattelberger,E., Inman,R.B., Calendar,R. and Loessner,M.J. (2003) Genome and proteome of Listeria monocytogenes phage PSA: an unusual case for programmed +1 translational frameshifting in structural protein synthesis. Mol. Microbiol., 50, 303–317. [DOI] [PubMed] [Google Scholar]