Abstract
The elongation phase of transcription by RNA polymerase II (RNAP II) is tightly controlled by a large number of transcription elongation factors. Here we describe experimental approaches for the isolation of RNAPII elongation complexes in vitro and the use of these complexes in the examination of the function of a variety of factors. The methods start with formation of elongation complexes on DNA templates immobilized to paramagnetic beads. Elongation is halted by removing the nucleotides and the ternary elongation complexes are then stripped of factors by a high salt wash. The effect of any factor or mixture of factors on elongation is determined by adding the factor(s) along with nucleotides and observing the change in the pattern of RNAs generated. Association of a factor with elongation complexes can be examined using an elongation complex electrophoretic mobility shift assay (EC-EMSA) in which elongation complexes that have been liberated from the beads are analyzed on a native gel. Besides being used to dissect the mechanisms of elongation control, these experimental systems are useful for analyzing the function of termination factors and mRNA processing factors. Together these experimental systems permit detailed characterization of the molecular mechanisms of elongation, termination, and mRNA processing factors by providing information concerning both physical interactions with and functional consequences of the factors on RNAPII elongation complexes.
1. Introduction
Transcription of RNAPII proceeds through multiple stages designated preinitiation, initiation, elongation, and termination. Historically, studies of the elongation stage of eukaryotic mRNA synthesis lagged behind studies of the preinitiation and initiation stages because many researchers believed that initiation was the rate limiting step in expression of most genes. However, in recent years, with the discovery of a diverse collection of transcription elongation factors that regulate the activity of elongating RNAPII, the importance of RNAPII elongation control in regulation of gene expression is being gradually revealed [1-5]. Very recently, global analyses of RNAPII occupancy across the Drosophila and human genomes have provided strong evidence that regulation of elongation is very prevalent and may direct the patterns of gene expression during development [6-9].
RNAPII elongation is an elaborate biochemical process that requires the concerted action of a large number of transcription elongation factors. It can be divided into two distinct stages: promoter proximal pausing or abortive elongation, and productive elongation [1, 10]. Early in elongation RNAPII is subject to the control of negative elongation factors including DSIF and NELF, which leave polymerases paused in promoter proximal regions [2, 4, 11]. These polymerases are also termed “poised polymerases” [12, 13]. During this process, only short transcripts are synthesized that ultimately terminate prematurely. The length of time that human polymerases spend in the engaged state has not been measured, but a wide range of poising times, from several minutes to only once or twice per cell division, fit the available data. To make full length mRNA, P-TEFb is required to allow the poised polymerase to enter the productive elongation mode, in part, by reversing the negative effect of NELF and simultaneously facilitating the action of positive elongation factors including TFIIF [2-4, 11, 14, 15]. In addition to the factors that directly affect and control elongation, the mRNA processing machinery has been found to interact with and act on the elongation complexes [16-19].
Functional biochemical assays have played an essential role in the discovery of many factors that directly affect the elongation activity of RNAPII. These assays have allowed the examination of the process of RNAPII elongation in detail [20-23] and the detection of factor dependent changes in elongation properties of RNAPII [1, 20, 24-26]. Such assays have not only allowed the identification of elongation factor activity, but have also provided a means to follow the activity of specific factors during purification leading to the identification of the proteins responsible for the activity [2, 4, 5, 27, 28]. We have found that in vitro transcription systems using isolated elongation complexes that are initially formed on an immobilized DNA template with a natural promoter have a lot of advantages for identification and functional examination of factors that affect elongation. Following sections describe methods that we have developed for isolating active RNAPII elongation complexes and for analyzing them in functional transcription assay and factor binding assay systems (Fig. 1). These systems have been and continue to be invaluable for examining the molecular mechanisms of RNAPII elongation and associated events.
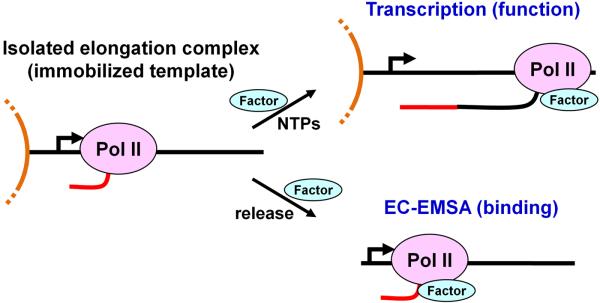
Figure 1. Overview of methods covered.
Elongation complexes containing labeled nascent transcripts are generated on templates that are immobilized to paramagnetic beads and isolated by high salt wash. A functional assay is used to demonstrate the effect of the factor of interest during transcription elongation. This is accomplished by addition of the factor to the isolated elongation complexes followed by the re-addition of NTPs. To directly analyze factor binding, elongation complexes are released from the beads using a restriction enzyme and resolved in a native gel before or after incubation with the factor. Factor binding causes an electrophoretic mobility shift of the elongation complexes (EC-EMSA).
2. Description of Methods
2.1. Materials
2.1.1. Preparation of Nuclear Extract
High-quality nuclear extract provides RNA polymerase II and the transcription factors which are essential for initiating transcription on a DNA template in vitro. Detailed protocols for preparing nuclear extracts from HeLa cells or Drosophila Kc cells can be found in previous publications [29, 30]. We routinely start with 20 liters of HeLa S3 cells harvested in mid-log phase growth by the National Cell Culture Center (Minneapolis, MN) that are washed with PBS and shipped on wet ice. After the preparation which includes 360 mM ammonium sulfate extraction of the nuclei, ammonium sulfate precipitation and dialysis, we normally obtain about 10 milliliters of the final nuclear extract in 150 mM HGKEDP (20 mM HEPES (pH 7.6), 15% glycerol, 150 mM KCl, 0.1 mM EDTA, 1 mM DTT, 0.1% PMSF saturated in isopropanol). These extracts are significantly more concentrated (25-100 mg/ml protein) and have a somewhat different mix of proteins than those that are prepared without ammonium sulfate precipitation. Aliquots of the extract can be stably stored at -80°C for many years without any significant reduction of activity.
2.1.2. Preparation of Immobilized DNA Templates
The DNA template for transcription assays should contain a robust promoter known to be efficient for transcription initiation in vitro and a downstream transcribed fragment. In our assays, we routinely use a 1-2 kb template that contains the full cytomegalovirus (CMV) promoter (~ 0.8 kb) driving the production of a run-off transcript (0.2 to 1 kb). The transcribed DNA fragment can be designed for particular experimental purposes. For example, we used a template containing the sequences of HIV-LTR to study transcription activation by the viral protein Tat [29].
Immobilization of the DNA template facilitates isolation of elongation complexes. In addition, as you will see in the examples described in later sections, it greatly increases the possible means of reaction manipulations, which exponentially amplifies the potential usefulness of our experimental systems. The DNA template is generated using polymerase chain reaction (PCR) in which the upstream primer is 5′-biotinylated. Products from about 20 PCR reactions are pooled, ethanol precipitated, and dissolved into TE (10 mM Tris (pH 8.0), 1 mM EDTA). The concentrated PCR products are purified to remove the unincorporated primers using the Wizard SV Gel and PCR Clean-up kit (Promega, Madison, WI) and finally eluted with water. The resulting biotinylated DNA is bound to streptavidin-coated paramagnetic Dynabeads M-280 (Invitrogen, Carlsbad, CA) through the streptavidin-biotin linkage. The capacity of the beads varies inversely with the length of the DNA and usually we use 0.5 ml of bead slurry (5 mg of beads) for binding of 60 μg of DNA that is about 1 kb in length. Binding takes place most efficiently in the presence of high salt (1 M NaCl) which reduces repulsion between the long negatively charged template molecules. The beads are washed 2-3 times with TE after one hour incubation with the purified DNA and then stored in 0.5 ml TE at 4°C for use over the next few months. The 5′-biotinylated end of the template should be at least 200-300 bp upstream of the transcription start site since we found that the minimal CMV promoter (less than 100 bp) did not support efficient transcription initiation once being attached to the beads.
2.2. Functional analysis of RNAPII elongation using pulse-chase assays
To specifically examine the elongation phase of transcription in vitro, the basic strategy is to generate and isolate early elongation complexes associated with radiolabeled nascent transcripts (pulse) and then follow the progress of the subsequent elongation of these transcripts upon the addition of cold nucleotides (chase). The short pulse facilitates the synchronization of transcription initiation and because there is no more label incorporation during the chase, new initiation or reinitiation events are rendered undetectable by autoradiography. Therefore, only the elongation of the pulse-labeled transcripts found in elongation complexes is analyzed.
2.2.1. Generation and isolation of early elongation complexes (EECs) with labeled nascent RNA
Transcription is allowed to initiate in the presence of a crude nuclear extract and radiolabeled ribonucleotides, and proceed for a short time (usually 30 seconds) to allow the nascent transcripts be pulse labeled. Then transcription is halted by the addition of EDTA and the resulting EECs are isolated. For consistency, the production and isolation of a whole experiment worth of EECs is typically done in bulk (multiple reactions in one tube) and after isolation the EECs are divided into individual reaction tubes. All steps are carried out at room temperature as detailed below.
Assembly of preinitiation complexes
Preinitiation complexes are formed during a 10 minute preincubation of a reaction mixture containing the immobilized DNA template and a nuclear extract. Each individual reaction (8 μl) in the master preincubation mixture contains 1 μl of HeLa nuclear extract (HNE), ~ 0.5 pmol of immobilized DNA template (usually concentrated from 2-4 μl of bead slurry), 20 mM HEPES (pH 7.6), 7 mM MgCl2, 60 mM KCl and 10 units of RNaseOUT (Invitrogen, Carlsbad, CA). In our experiments, we typically have 1 μM flavopiridol present in the preincubation mixture to inhibit the function of P-TEFb during the pulse labeling. The storage buffer is removed from the beads (after concentrating the beads using a magnet) prior to addition of the rest of preincubation mixture. Please note that the amounts of extract and DNA template listed here can only be taken as guidelines since each preparation of nuclear extract has a different activity that also affects the optima of template DNA. In practice, it is important to initially set up an experiment to assay from 1 to 4 μl of HNE and 1 to 8 μl of the immobilized template bead slurry using the standard pulse-chase assay described here to determine the amounts of extract and template that produce the optimum amount of transcription.
Transcription initiation and pulse labeling
Transcription is initiated upon the addition of a pulse mix (2 μl per transcription reaction) containing physiological concentrations of ATP, GTP, UTP (500 μM), and 5 μCi of [α-32P]CTP, 20 mM HEPES (pH 7.6), 7 mM MgCl2 and 60 mM KCl. The exact concentration of CTP is not known because the bulk of CTP comes as a contaminant of other NTPs. Unpublished experiments suggest that the final concentration is between 1 and 5 μM. We have found that initiation take place most efficiently in the presence of 7 mM MgCl2. After 30 seconds of pulse, elongation is halted with the addition of EDTA to 20 mM. The resulting EECs are associated with labeled nascent RNA predominantly less than 25 nt in length. Most functional preinitiation complexes initiate within 10-20 seconds, but longer or shorter pulse times will produce slightly longer or shorter RNA with longer RNA having a higher specific activity than shorter RNA.
Isolation of EECs
The beads with EECs associated are collected using a magnetic concentrator to remove the reaction solution and washed three times (5 minute each) in a high salt EEC isolation buffer (20 mM HEPES (pH 7.6), and 1.6 M KCl) to strip all the factors (bound specifically or non-specifically) from the polymerase and template. Tubes should be left in the magnetic field during the removal of the supernatants and then removed quickly to facilitate resuspension of the beads in the next solution. In our early studies, we also added 1% Sarkosyl in the high salt EEC isolation buffer [29]. Recently, we found that elimination of Sarkosyl resulted in the isolated EECs that responded more efficiently to various elongation factors including DSIF, NELF, and TFIIF [15]. Other detergents, such as 1% NP-40 or 1% Triton X-100 were also found to negatively influence the subsequent function of elongation factors in the add-back transcription assays (data not shown). After the stringent high salt washes, the beads are washed twice in a low salt EEC isolation buffer (20 mM HEPES, 60 mM KCl, and 200 μg/ml bovine serum albumin), and resuspended in the low salt EEC isolation buffer.
2.2.2. Chase reactions (transcript extension)
The nascent transcripts in isolated EECs can be further extended upon the addition of cold nucleotides. Isolated EECs generated in bulk are aliquotted into each individual reaction and a chase reaction is carried out by adding 500 μM of cold NTP to isolated EECs in a chase buffer containing 20 mM HEPES, 60 mM KCl, 200 μg/ml bovine serum albumin, 3 mM MgCl2 and 10 units of RNaseOUT. We have found that lowering the MgCl2 from 7 mM (optimum for initiation) to 3 mM facilitates elongation factor function. Using 5 mM MgCl2 for both steps is also acceptable. After a chosen chase time, each reaction is terminated by the addition of 200 μl of stop solution (100 mM Tris (pH 8.0), 100 mM NaCl, 10 mM EDTA, 1% Sarkosyl with 200 μg/ml low molecular weight Torula yeast RNA [Sigma, St. Louis, MO]). Then the RNA is isolated using a phenol extraction followed by an ethanol precipitation as previously described [29]. The pellet of RNA in each tube is dissolved in 10 μl RNA loading buffer (8 M urea, 0.25x TBE containing bromophenol blue and xylene cyanol) by heating at 75°C for five minutes. The RNA is analyzed on a 6% polyacrylamide gel with 1x TBE buffer and 6 M urea in the gel and 1x TBE running buffer. Before drying, the gel can be soaked in water containing ethidium bromide (EtBr) for ten minutes to stain the nucleic acids. Although the radiolabeled transcripts cannot be detected upon EtBr staining due to the small amount generated, the intensity of the bands for the DNA template and any RNAs that might be present in the extract or factor preparations, as well as those added during extraction, will reveal if the recovery of the samples is good and uniform. Autoradiography of the dried gels provides images of the results, and quantitation is accomplished with a Packard InstantImager (Perkin Elmer, Woodbridge, Ontario) or other phosphoimager.
2.2.3. Add-back assays
Isolated EECs are extremely useful in add-back assays to examine the effects of the add-back component(s) on elongation (evaluated by the resultant changes in the pattern/lengths of transcripts generated during chase). Normally, isolated EECs are left untreated (as a control) or supplemented with either a crude extract or purified factors and then the chase reactions are performed under the same conditions as described above. To facilitate the following descriptions, transcription assays with a whole extract added back or with only purified factors added back will be referred to as the “crude system” or “defined system,” respectively. The crude system can utilize a whole nuclear extract, a factor-depleted nuclear extract, or a particular crude fraction resulting from a fractionation of an extract. The defined system is named not because every component is known to be 100% pure, but because the components are highly purified and have defined methods of isolation. Please refer to our early articles for the detailed protocols of the expression and purification of human elongation factors and mRNA processing factors, including DSIF and NELF [11], P-TEFb [31], TFIIF [32], TFIIS [33], TTF2 [34], and HCE and HCM [19].
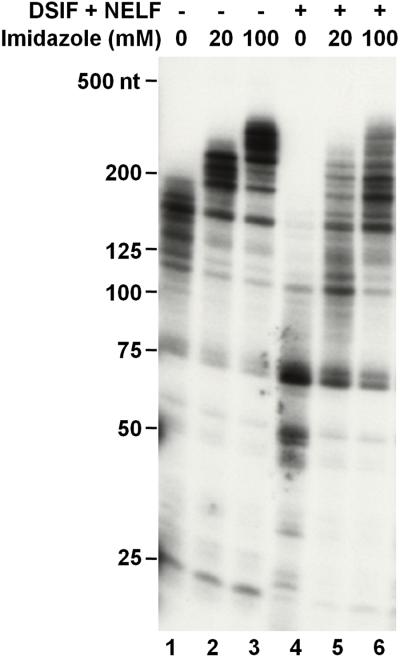
One consideration for the factor add-back assay is that some of the reagents in the protein storage buffer may have direct influence on transcription. Because the buffer for eluting His-tagged recombinant proteins from a Ni-column routinely contains imidazole, we tested the effect of imidazole on elongation in a standard pulse-chase assay. Isolated EECs were allowed to elongate for six minutes in the absence or presence of increasing amounts of imidazole. As illustrated in Figure 2, inclusion of imidazole stimulated the elongation rate in a dose dependent manner evidenced by the increase of the lengths of RNA (lanes 1-3). In addition, this stimulatory effect of imidazole may impact the observation of the effects of factors added back. DSIF and NELF together showed a strong negative effect on elongation (Fig. 2, compare lanes 4 and 1) while this effect became less detectable in the presence of imidazole (Fig. 2, lanes 5 and 6). Ammonium ions have been shown to have a positive effect on elongation [20] and the levels of NaCl or KCl normally found in purified protein fractions can negatively influence factor function [24]. Therefore, dialysis or further purification is needed to eliminate imidazole or ammonium ions from the protein storage buffer prior to their add-back to elongation reactions. We routinely use HGKEDP as a protein storage buffer, which does not impact elongation. Also care must be taken to precisely maintain identical salt concentrations across a set of reactions so that effects seen can be ascribed to the factors added.
Figure 2. Influences of immidazol on transcription of isolated elongation complexes.
Isolated EECs were allowed to elongate transcripts for six minutes in the absence or presence of indicated concentrations of immidazol and/or purified factors (0.45 pmol of DSIF and 0.03 pmol of NELF). The synthesized transcripts were extracted and analyzed on a 6% denaturing RNA gel followed by autoradiography of the dried gel.
The effects of factor(s) or extracts on RNAPII elongation can be examined by carrying out time-course reactions and/or titration reactions. Time-course experiments are particularly useful for estimation and comparison of the elongation rates of the polymerases in the absence or presence of the factor(s) or extract added back. For consistency, master reactions are first set up that contain isolated EECs, and factor(s) of interest or extract and protein storage buffer (control) respectively, in the chase buffer (without NTP added) described above. In addition, prior to the start of chase reactions, a set of tubes is prepared with each of them having 200 μl of stop solution added. The chase reaction starts upon the addition of NTPs to 500 μM and at each indicated time point, an individual reaction worth of reaction mixture is taken out and immediately added into a prepared tube containing stop solution. Titration experiments are routinely used to optimize the amount(s) of factor(s) or the relative ratio of factors needed to exhibit the maximal effect on elongation. To set up a titration experiment, a master reaction that contains isolated EECs in the chase buffer is assembled and aliquotted into each individual reaction. A series of dilutions of the factor(s) or extract are prepared in a buffer identical to the buffer the factor or extract is in except that 200 μg/ml BSA is added and then an equal volume of each dilution is added to each individual reaction. The BSA is included to avoid losses of the factors on the walls of the microfuge tubes during dilution. A reaction with only buffer added back serves as a negative control. Then a mixture of all four NTPs is added to start the chase reaction. Both types of the add-back assays have been used to study the effect of negative elongation factors DSIF and NELF and positive elongation factor TFIIF in a defined system [11, 29] and should be the starting point for examination of any potential elongation factor. An example of a detailed protocol actually used for a published figure that showed the effect of NELF and DSIF on kinetics of elongation is provided as an Excel file (Supplementary File 1). This file demonstrates an efficient way to set up a specific experiment and indicates what type of information is important to document.
2.2.4. Identification and characterization of new activities by comparing the defined and crude systems
In the crude system, a whole nuclear extract is supposed to recapitulate the intracellular distribution of a variety of transcription regulatory factors such that meaningful information about controlling RNAPII transcription can be extracted. On the other hand, establishing a defined system makes it possible to dissect the effect of a particular factor or a group of factors on RNAPII elongation. Activities missing in a defined system reconstituted with all factors in hand added back will lead to differences in elongation properties of polymerases compared to those in the crude system. Therefore, a detailed comparison of the results from two systems can help identify unknown activities and ultimately provide us a more comprehensive picture about the process and control of RNAPII elongation.
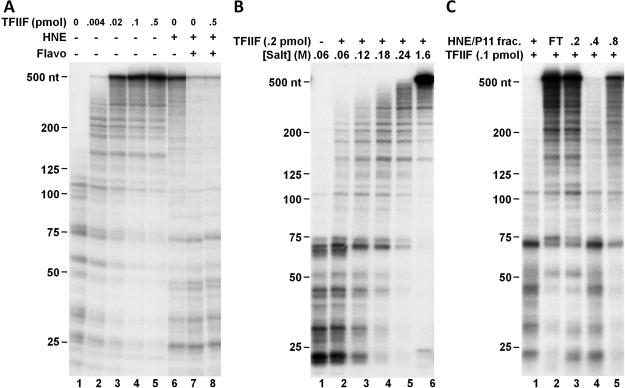
Here we show an example of how to make comparisons of the two transcription systems and to further characterize a newly identified activity. The function of positive elongation factor TFIIF was examined in both defined and crude systems. Isolated EECs were allowed to elongate for three minutes with no addition or with indicated component(s). Addition of recombinant TFIIF in the defined system dramatically stimulated elongation as evidenced by the generation of much longer transcripts than in the control reaction containing only EECs (Fig. 3A, compare lanes 2-5 to lane 1). The stimulatory effect of TFIIF was virtually saturated when 0.1 pmol/reaction of TFIIF was used, as evidenced by the accumulation of most transcripts at the 548-nt run-off site. Similarly, when whole HNE was added back to isolated EECs, most transcripts reached the run-off site within three minutes of chase (Fig. 3A, lane 6), suggesting that TFIIF in the extract was allowed to function under these conditions. Addition of 1 μM flavopiridol, a potent P-TEFb inhibitor, blocked the transition into productive elongation such that the generation of run-off transcripts was effectively inhibited (Fig. 3A, lane 7). Different from what was seen in the defined system, when P-TEFb was inhibited, addition of extra amounts of recombinant TFIIF to HNE did not stimulate the generation of run-off transcripts (Fig. 3A, lanes 8). This suggests that before P-TEFb functions, the polymerases are dominantly controlled by negative elongation factors and resistant to the function of positive factors including TFIIF. Although it was known that the negative elongation factors, DSIF and NELF, could functionally compete with TFIIF, we found that they were not sufficient to completely block the function of TFIIF [15]. Therefore, these results together strongly suggest that there is a TFIIF resistance activity present in the extract but missing in the above defined system as recently described [15].
Figure 3. TFIIF activity is inhibited in the extract before P-TEFb functions.
A. Comparison of the function of TFIIF in a defined and a crude in vitro transcription system. Isolated EECs were left untreated or supplemented with indicated amounts of TFIIF and subsequent elongation was allowed for three minutes in the absence or presence of HNE and/or 1 μM of flavopiridol. B. Salt tolerance of the TFIIF resistance activity. EECs were isolated under the indicated salt conditions and then further elongated for three minutes in the presence of TFIIF. C. Analysis of HNE P-11 fractions for the TFIIF resistance activity. Isolated EECs were supplemented with TFIIF, 1 μM of flavopiridol, and indicated HNE P-11 fractions or the whole extract (“+”). Elongation was then carried out for seven minutes. Transcripts were analyzed as described in the legend to figure 2.
The elongation complexes were then isolated under less stringent conditions to look for the functional association of TFIIF resistance factor(s). Aliquots of the pulse labeled elongation complexes were isolated with buffers containing various concentrations of salt respectively, and the isolated EECs were further chased for three minutes in the presence of TFIIF (Fig. 3B). We found that when EECs were isolated by a low salt buffer containing 60 mM KCl, they were not able to respond to the stimulatory effect of TFIIF, indicating that the TFIIF resistance factor(s) remained associated (Fig. 3B, lanes 1 and 2). With increasing salt concentration, the TFIIF resistance activity was gradually eluted from the EECs during isolation (Fig. 3B, lanes 3-6). The EECs isolated with 240 mM salt still exhibited appreciable TFIIF resistance activity comparing to the regular high salt isolated EECs (1.6 M KCl), suggesting that the TFIIF resistance factor(s) bound relatively tightly with early elongation complexes formed in the presence of whole nuclear extract. Next, functional transcription assays were applied in conjunction with conventional chromatography to purify the factor(s) responsible (Fig. 3C). HNE was fractionated through a phosphocellulose (P11) column and the resultant four fractions (one flow-through “FT” fraction and three fractions eluting at 0.2, 0.4 and 0.8 M KCl) were analyzed in a functional transcription assay described below. Whole extract or each P11 fraction was added back to isolated EECs, together with TFIIF and flavopiridol. Elongation was then allowed to proceed for seven minutes and the lengths of the RNAs produced were analyzed. As shown in Figure 3C, among the four fractions only the 0.4 M step was able to block TFIIF function efficiently, indicating that the factor(s) that blocks TFIIF function eluted from phosphocellulose P11 between 0.2 and 0.4 M salt. To further purify this factor, the P11-0.4 fraction could be fractionated through another column and a similar strategy applied.
2.2.5. Examination of transcription termination and mRNA processing events
Besides examination of factors controlling RNAPII elongation, the in vitro transcription systems described here can also be applied to study the functional properties of factors involved in termination or mRNA processing.
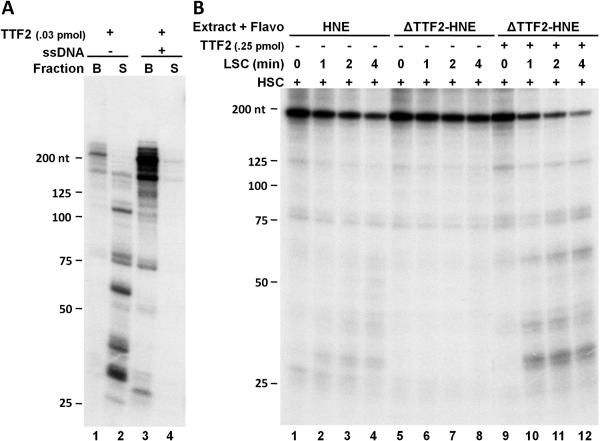
Figure 4A illustrates an example in which the transcription termination activity of TTF2 was analyzed. Using an immobilized template to form EECs makes it very easy to detect the termination activity because any transcript terminated would be released from the immobilized DNA into the supernatant. Elongation of isolated EECs was carried out for seven minutes in the presence of recombinant TTF2. Then the supernatants were separated from the beads and the transcripts in both fractions were extracted and analyzed. As shown in Figure 4A, most of the transcripts were present in the supernatant, indicating that they were terminated by TTF2 during the chase. We found earlier that the ATPase activity of TTF2 could be inhibited by ssDNA [35]. Here the expected result was obtained that further inclusion of ssDNA completely blocked the termination activity of TTF2, evidenced by the detection of most transcripts in the bead fraction (Fig. 4A, lanes 3 and 4). Normally elongation complexes that are unaffected by P-TEFb have propensities to terminate [34]. This can be detected using a modified crude system described below. Isolated EECs were allowed to elongate under standard conditions (60 mM salt) for indicated amounts of time in the presence of HNE and flavopiridol. During this time-course of low salt chase (LSC), some of the polymerases terminate. To better observe and quantify the termination activity, after the LSC, the salt was raised to 250 mM and the chase continued for another 7 minutes (high salt chase, HSC). During the HSC negative factors are blocked and all engaged polymerases reach the run-off site. In the complete reactions analyzed in Figure 4B, termination of the polymerases indeed occurred during the low salt chase, easily seen by the gradual loss of the run-off signal (lanes 1-4). As expected, termination during the low salt chase also reduced the accumulation of short terminated transcripts (mostly between 30 and 60 nt in length. The potential role of TTF2 in causing the transcription termination was then examined. Similar transcription assays were carried out to compare the termination activity of whole nuclear extract to that of an equivalent amount of the same extract with TTF2 depleted. As detected by examination of low salt run-off and short transcripts, depletion of TTF2 significantly reduced the termination activity in the extract (Fig. 4B, lanes 5-8), and in contrast, add-back of more recombinant TTF2 to the TTF2-depleted extract restored the termination activity (Fig. 4B, lanes 9-12). Taken together, our transcription system can be used as a powerful tool to study the transcription termination activity in vitro and the results from the above experiments demonstrate that TTF2 is the major RNAPII termination factor in HNE.
Figure 4. TTF2 is the main termination factor in HNE.
A. Termination activity of TTF2 in the defined system. Isolated EECs were left untreated or supplemented with recombinant TTF2 and elongation of isolated EECs was carried out for seven minutes in the absence or presence of 1 pmol of ssDNA (a 48-nt DNA oligo with random sequences). Then the supernatants were separated from the beads and the transcripts in both fractions were analyzed as described in the legend to Fig. 2. B. TTF2 causes transcription termination in the presence of HNE. Isolated EECs were supplemented with a whole or TTF2-depleted extract and elongation was first carried out under regular low salt conditions for indicated amounts of time (LSC) then the salt was raised to 250 mM and elongation was continued for another seven minutes (HSC). The template used in this experiment generates 183 nt run-off transcripts. Transcripts were analyzed as described in the legend to figure 2.
Detailed mechanisms of mRNA processing events can also be obtained from functional transcription assays using isolated elongation complexes. Using this system, we have shown that capping is functionally coupled to transcription [19]. More recently, we designed a template with the adenovirus-L3 poly(A) site integrated downstream of a transcribed region and further used it to study transcript cleavage and polyadenylation [17]. We have found that cleavage and polyadenylation of transcripts occur much more efficiently when the transcripts are present in elongation complexes than as free RNAs, demonstrating that cleavage and polyadenylation are also functionally coupled to transcription [17]. The add-back assays should be useful for further examining other mRNA processing events such as splicing.
2.3. Examination of physical interactions of factor(s) with RNAPII elongation complexes using EMSA
Because the expected functional consequences of many elongation factors can be extracted using our transcription systems, we reason that these factors must be associating with the elongation complexes in an appropriate way. To obtain valuable information concerning the physical associations of these factors with active elongation complexes, we recently established a new experimental system to directly analyze factor interactions to isolated elongation complexes on native gels, namely elongation complex-electrophoretic mobility shift assay (ECEMSA). The detailed protocol for EC-EMSA was documented in our recent paper [36]. Briefly, RNAPII elongation complexes are generated and isolated using the same protocol as in the transcription assays described in the previous section. Isolated elongation complexes are released from the beads through restriction enzyme digestion of the DNA template and then incubated with purified transcription factor(s) and analyzed on a low-percentage native gel. The capability of the factor(s) of interest to interact with ECs is evaluated by monitoring the gel shift of these elongation complexes. Using this EC-EMSA system, we were able to detect the interactions of TFIIF, TTF2, TFIIS, DSIF, and P-TEFb with isolated elongation complexes [36].
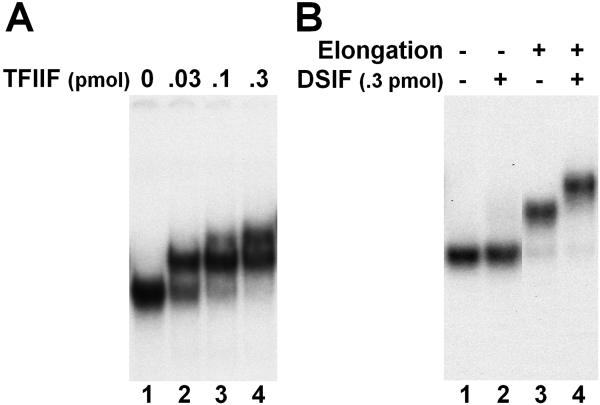
Figure 5A illustrates an example of analyzing the association of TFIIF with isolated EECs. A strong radioactive signal with a distinct migration was detected with isolated EECs alone (Fig. 5A, lane 1). This signal is truly due to intact elongation complexes since it disappeared upon the disruption of the complexes by different means [36]. Incubation of TFIIF with elongation complexes resulted in a mobility shift of the complexes to a higher position on the native gel and the completion of the shift was dependent on the amount of factor used (Fig. 5A, lanes 2 and 3).
Figure 5. Examination of EC•factor interaction using EC-EMSA.
A. Association of TFIIF with ECs. Isolated EECs were released from the beads by SacI digestion (crestriction site is 17 bp upstream of transcription start site). Then the complexes were left untreated or incubated with indicated amounts of TFIIF before analysis on a native gel. B. Nascent RNA facilitates the EC•DSIF interaction. Isolated EECs were left untreated or further elongated for three minutes before being released by SacI. Then the elongation complexes were incubated without or with 0.3 pmol of DSIF and further analyzed on a native gel.
The association of DSIF with elongation complexes was also characterized. Initially we were surprised to find that an extra amount of DSIF (over the amount of EECs used) was not able to cause an appreciable shift of the elongation complexes since DSIF was known to function during early elongation (Fig. 5B, lanes 1 and 2). Later we discovered that the nascent transcripts facilitated the binding of DSIF with ECs. When the nascent transcripts in isolated EECs were further extended to 35 nt or longer, the association of DSIF with ECs became very efficient (Fig. 5B, lanes 3 and 4) and the ECs could be completely shifted even when much less DSIF was used [36]. In addition, we have determined that the mobility shift of ECs upon transcript extension (Fig. 5B, compare lanes 3 and 1) is mainly due to the change of the relative position of the polymerase on the short DNA template and this can be eliminated through a mild DNase treatment to remove the DNA content extruded from ECs [36]. Furthermore, this system was applied to investigate the influence of phosphorylation of RNAPII-CTD and DSIF by P-TEFb on the interaction between DSIF and ECs [36].
Using isolated RNAPII elongation complexes to study factor binding makes the EC-EMSA system described here a potentially useful method for analyzing detailed mechanisms of EC•factor interactions. First of all, the elongation complexes used in these binding assays have been proven to be functionally active in transcription assays. Second, these elongation complexes isolated by stringent conditions are highly purified, which makes the binding reactions very reproducible and allows stronger interpretations of the results obtained. Third, the DNA and RNA content of the elongation complexes can be easily manipulated (eg. trimmed back by nuclease digestion) so that it can be determined if a EC•factor interaction is due to the direct association of the factor with polymerase, DNA template or nascent RNA. Fourth, the impact of RNAPII-CTD on factor binding can be directly tested by modifying the CTD prior to factor addition or by removal of the CTD through chymotrypsin treatment [17, 36, 37]. In addition, this system can be used to examine the effect of specific mutations or modifications on the binding of individual factors with ECs.
3. Concluding remarks
The experimental systems described here have provided important insights into some of the mechanisms by which various elongation, termination, and processing factors act on RNAPII elongation complexes. Most significantly, they have helped to reconstitute the fundamental RNAPII elongation control process in vitro that requires carefully orchestrated actions of RNAPII, DSIF, NELF, P-TEFb, and TFIIF. Importantly, the systems have been used to discover new activities involved. As the emphasis in the transcription field shifts to the investigation of controlling gene expression during elongation phase, it is likely that these methods will be increasingly used to provide information about the molecular mechanisms behind the RNAPII elongation control machinery. In addition, the combination of our in vitro transcription systems with EC-EMSA has made it possible to correlate binding with functional consequences of the transcription factors of interest. These methods will be further applied to examine the biochemical properties of any newly discovered elongation factors and other termination or processing factors and ultimately a better defined system will be created that more faithfully mimics the properties of the crude extract.
Supplementary Material
Acknowledgment
This work was supported by NIH grant GM35500 for D.H.P.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Marshall NF, Price DH. Mol Cell Biol. 1992;12:2078–90. doi: 10.1128/mcb.12.5.2078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yamaguchi Y, Takagi T, Wada T, Yano K, Furuya A, Sugimoto S, Hasegawa J, Handa H. Cell. 1999;97:41–51. doi: 10.1016/s0092-8674(00)80713-8. [DOI] [PubMed] [Google Scholar]
- 3.Wada T, Takagi T, Yamaguchi Y, Watanabe D, Handa H. Embo J. 1998;17:7395–403. doi: 10.1093/emboj/17.24.7395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wada T, Takagi T, Yamaguchi Y, Ferdous A, Imai T, Hirose S, Sugimoto S, Yano K, Hartzog GA, Winston F, Buratowski S, Handa H. Genes Dev. 1998;12:343–56. doi: 10.1101/gad.12.3.343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Marshall NF, Price DH. J Biol Chem. 1995;270:12335–8. doi: 10.1074/jbc.270.21.12335. [DOI] [PubMed] [Google Scholar]
- 6.Guenther MG, Levine SS, Boyer LA, Jaenisch R, Young RA. Cell. 2007;130:77–88. doi: 10.1016/j.cell.2007.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Muse GW, Gilchrist DA, Nechaev S, Shah R, Parker JS, Grissom SF, Zeitlinger J, Adelman K. Nat Genet. 2007;39:1507–11. doi: 10.1038/ng.2007.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zeitlinger J, Stark A, Kellis M, Hong JW, Nechaev S, Adelman K, Levine M, Young RA. Nat Genet. 2007;39:1512–6. doi: 10.1038/ng.2007.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Price DH. Mol Cell. 2008;30:7–10. doi: 10.1016/j.molcel.2008.03.001. [DOI] [PubMed] [Google Scholar]
- 10.Peterlin BM, Price DH. Mol Cell. 2006;23:297–305. doi: 10.1016/j.molcel.2006.06.014. [DOI] [PubMed] [Google Scholar]
- 11.Renner DB, Yamaguchi Y, Wada T, Handa H, Price DH. J Biol Chem. 2001;276:42601–9. doi: 10.1074/jbc.M104967200. [DOI] [PubMed] [Google Scholar]
- 12.Gilmour DS, Lis JT. Mol Cell Biol. 1986;6:3984–9. doi: 10.1128/mcb.6.11.3984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rougvie AE, Lis JT. Cell. 1988;54:795–804. doi: 10.1016/s0092-8674(88)91087-2. [DOI] [PubMed] [Google Scholar]
- 14.Marshall NF, Peng J, Xie Z, Price DH. J Biol Chem. 1996;271:27176–83. doi: 10.1074/jbc.271.43.27176. [DOI] [PubMed] [Google Scholar]
- 15.Cheng B, Price DH. J Biol Chem. 2007;282:21901–12. doi: 10.1074/jbc.M702936200. [DOI] [PubMed] [Google Scholar]
- 16.Bentley DL. Curr Opin Cell Biol. 2005;17:251–6. doi: 10.1016/j.ceb.2005.04.006. [DOI] [PubMed] [Google Scholar]
- 17.Adamson TE, Shutt DC, Price DH. J Biol Chem. 2005;280:32262–71. doi: 10.1074/jbc.M505532200. [DOI] [PubMed] [Google Scholar]
- 18.Adamson TE, Price DH. Mol Cell Biol. 2003;23:4046–55. doi: 10.1128/MCB.23.12.4046-4055.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Moteki S, Price D. Mol Cell. 2002;10:599–609. doi: 10.1016/s1097-2765(02)00660-3. [DOI] [PubMed] [Google Scholar]
- 20.Sluder AE, Price DH, Greenleaf AL. J Biol Chem. 1988;263:9917–25. [PubMed] [Google Scholar]
- 21.Kerppola TK, Kane CM. Mol Cell Biol. 1988;8:4389–94. doi: 10.1128/mcb.8.10.4389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kane CM, Chamberlin MJ. Biochemistry. 1985;24:2254–62. doi: 10.1021/bi00330a020. [DOI] [PubMed] [Google Scholar]
- 23.Kadesch TR, Chamberlin MJ. J Biol Chem. 1982;257:5286–95. [PubMed] [Google Scholar]
- 24.Kephart DD, Marshall NF, Price DH. Mol Cell Biol. 1992;12:2067–77. doi: 10.1128/mcb.12.5.2067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Price DH, Sluder AE, Greenleaf AL. Mol Cell Biol. 1989;9:1465–75. doi: 10.1128/mcb.9.4.1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sluder AE, Greenleaf AL, Price DH. J Biol Chem. 1989;264:8963–9. [PubMed] [Google Scholar]
- 27.Xie Z, Price DH. J Biol Chem. 1996;271:11043–6. doi: 10.1074/jbc.271.19.11043. [DOI] [PubMed] [Google Scholar]
- 28.Aso T, Lane WS, Conaway JW, Conaway RC. Science. 1995;269:1439–43. doi: 10.1126/science.7660129. [DOI] [PubMed] [Google Scholar]
- 29.Adamson TE, Shore SM, Price DH. Methods Enzymol. 2003;371:264–75. doi: 10.1016/S0076-6879(03)71019-2. [DOI] [PubMed] [Google Scholar]
- 30.Price DH, Sluder AE, Greenleaf AL. J Biol Chem. 1987;262:3244–55. [PubMed] [Google Scholar]
- 31.Peng J, Zhu Y, Milton JT, Price DH. Genes Dev. 1998;12:755–62. doi: 10.1101/gad.12.5.755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Peng J, Liu M, Marion J, Zhu Y, Price DH. Cold Spring Harb Symp Quant Biol. 1998;63:365–70. doi: 10.1101/sqb.1998.63.365. [DOI] [PubMed] [Google Scholar]
- 33.Palangat M, Renner DB, Price DH, Landick R. Proc Natl Acad Sci U S A. 2005;102:15036–41. doi: 10.1073/pnas.0409405102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jiang Y, Liu M, Spencer CA, Price DH. Mol Cell. 2004;14:375–85. doi: 10.1016/s1097-2765(04)00234-5. [DOI] [PubMed] [Google Scholar]
- 35.Xie Z, Price DH. J Biol Chem. 1998;273:3771–7. doi: 10.1074/jbc.273.6.3771. [DOI] [PubMed] [Google Scholar]
- 36.Cheng B, Price DH. Nucleic Acids Res. 2008;36:e135. doi: 10.1093/nar/gkn630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zehring WA, Lee JM, Weeks JR, Jokerst RS, Greenleaf AL. Proc Natl Acad Sci U S A. 1988;85:3698–702. doi: 10.1073/pnas.85.11.3698. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.