Abstract
A major issue of current virology concerns the characterization of cellular proteins that operate as functional components of the viral multiplication process. Here we describe a group of host factors designated as ‘NFAR proteins’ that are recruited by the replication machinery of bovine viral diarrhea virus, a close relative of the human pathogen hepatitis C virus. The NFAR proteins associate specifically with both the termini of the viral RNA genome involving regulatory elements in the 5′ and 3′ non-translated regions. Modification of the protein interaction sites in the 3′ non-translated region yielded viral RNAs that were replication deficient. Viral replication was also inhibited by RNAi approaches that reduced the concentration of RNA helicase A, a member of the NFAR group, in the host cell’s cytoplasm. Further experimental data suggest that NFAR proteins mediate a circular conformation of the viral genome that may be important for the coordination of translation and replication. Because NFAR proteins are presumed components of the antiviral response, we suspect that viral recruitment may also serve to weaken cellular defense mechanisms.
Keywords: hepatitis C virus/NF90/NFAR/pestivirus/RNA helicase A
Introduction
Bovine viral diarrhea virus (BVDV) belongs to the genus Pestivirus of widespread animal pathogens, which together with the genera Flavivirus and Hepacivirus [hepatitis C viruses (HCVs)] constitute the family Flaviviridae (Lindenbach and Rice, 2001). The pestiviruses have found particular attention due to their marked similarity to HCV, the major causative agent of liver cirrhosis and liver cancer in man. Because studies with the human pathogen are hampered by its low rate of replication and the lack of convenient cell culture systems, BVDV serves as a surrogate model for the life cycle of HCV.
The BVDV genome is a positive-strand RNA of length ∼13 kb. It consists of a long open reading frame (ORF) that is flanked by non-translated regions (NTRs) at the 5′ and 3′ ends. The viral RNA acts directly as a messenger in the host cell’s cytoplasm. Translation initiates through a type IV internal ribosomal entry site (IRES) in the 5′NTR and leads to the synthesis of a polyprotein NH2-Npro, C, Erns, E1, E2, p7, NS2-3, NS4A, NS4B, NS5A, NS5B-COOH, which is co- and post-translationally processed by cellular as well as viral proteases to yield the structural (C, Erns, E1, E2, p7) and non-structural (Npro, NS2-NS5B) viral proteins (Lindenbach and Rice, 2001) (Figure 1A).
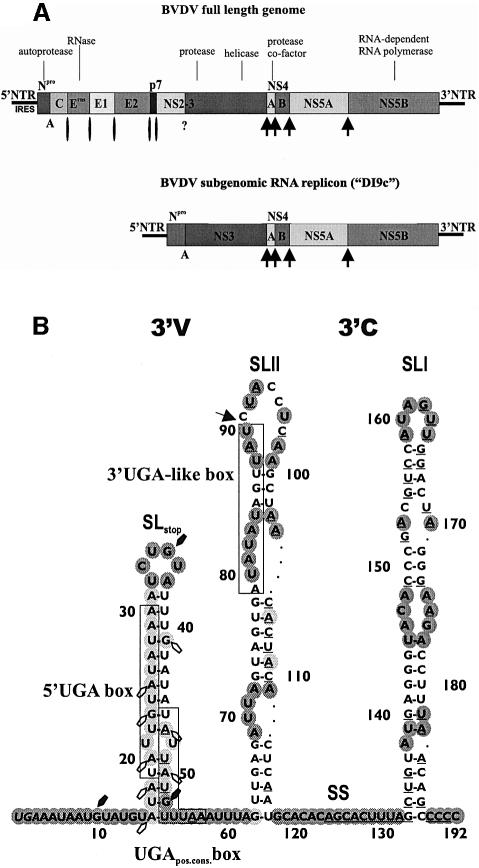
Fig. 1. Organization of genomic and subgenomic BVDV RNAs, and secondary structure of the BVDV DI9c 3′NTR. (A) Physical map of the full-length BVDV genome and the subgenomic replicon ‘DI9c’. The NTRs are indicated as single lines; the genetic units encoding the viral proteins are represented as shaded boxes. Enzymatic activities associated with the viral proteins are indicated (Lindenbach and Rice, 2001). The proteolytic cleavage sites in the polyprotein are marked as follows: arrow, cleavage by NS3/NS4A; circle, cleavage by signal peptidase; A, autoproteolytic activity; ?, uncertain. (B) Structure of the BVDV DI9c 3′NTR. The depicted region corresponds to nucleotides 12 090–12 281 of the genome of the DI9c ancestor BVDV CP7 (Meyers et al., 1996); numbering starts at the translational stop codon (italics). The UGA segments are boxed. An arrow marks the border between the variable 3′V and conserved 3′C region. Residues that are 100% conserved in all pestivirus strains are underlined. The depicted secondary structure summarizes the results of published experimental probing data on the structure of SLI, SS and SLII (Yu et al., 1999) and novel data on SLstop. They were superimposed on an mfold-aided structure prediction of the BVDV DI9c 3′NTR at lowest ΔGmax. Nucleotides that were highly exposed to single-strand specific RNases and/or chemical modification are indicated in dark gray; slightly exposed nucleotides are indicated in light gray. Residues in SLstop that were specifically accessible to RNases are marked by arrows: white (weak); black (strong). The partial accessibility of the A-U-rich stem of SLstop (especially its upstream portion) to nucleases and chemicals was explained by its low stability (see text; O.Isken, C.W.Grassmann, H.Yu and S.-E.Behrens, in preparation). Representative examples of chemical and enzymatic probing experiments of the BVDV 3′NTR are included in the Supplementary data.
As with other positive-strand RNA viruses, replication of the BVDV genome proceeds along a two-step pathway in the host cell. Following synthesis and maturation, the NS proteins and the viral RNA form ‘replication complexes’. These catalyze the transcription of negative-strand RNA intermediates from which, in turn, progeny positive-strand RNA molecules are generated. Hence, in addition to its function as mRNA, the viral genome acts as a template for RNA synthesis. The molecular features underlying the switch from translation to replication are poorly understood. Studies of poliovirus, a similarly organized positive-strand RNA virus, demonstrated that initiation of the replication cycle requires the clearance of ribosomes from the viral genome (Barton et al., 1999). Other poliovirus data suggest that translation is regulated in a feedback-like manner by nascent viral proteins and that the coordination of translation and RNA replication may involve a circular conformation of the viral genome (Gamarnik and Andino, 1998; Barton et al., 2001; Herold and Andino, 2001).
Studies of BVDV replication were significantly facilitated by the finding that subgenomic RNAs, which consist of the NTRs and the coding region of the proteins NS3–NS5B replicate autonomously in transfected host cells (Behrens et al., 1998) (Figure 1A). Focal points of interest investigating these replicons have been the NTRs, because these regions are supposed to encode the majority of features that orchestrate translation and RNA replication. Along this line, a stem–loop structure, ‘hairpin Ia’, at the immediate 5′ terminus of the BVDV RNA encodes different signals, some of which determine the efficiency of the IRES while others are essential for the initiation of the replication cycle (Yu et al., 2000). On the other hand, sequence and structural elements of the conserved ‘3′C’ portion of the 3′NTR were indicated to be components of the negative-strand promoter of the initial replication complex (Yu et al., 1999) (see Figure 1B). Recently, we found that the variable ‘3′V’ portion of the 3′NTR, which displays a surprising heterogeneity in size and sequence between different virus strains, modulates translation initiation as well as translation termination (O.Isken, C.W.Grassmann, H.Yu and S.-E.Behrens, in preparation). Thus hairpin Ia and 3′V appear to be crucial for the regulation of viral protein versus RNA synthesis.
There is much evidence suggesting that RNA viruses subvert cellular proteins to become part of their replication strategy (reviewed by Lai, 1998). In this article we identify a group of cellular proteins, including NF90/NFAR-1, NF45 and RNA helicase A, which by several lines of data were indicated to be required for BVDV replication. Because these proteins associate specifically with the 5′ as well as the 3′NTR of the viral genome involving hairpin Ia and 3′V, we suggest them as components of a circular conformation of the RNA necessary for the coordination of translation and replication. We consider these observations to be an important contribution toward an understanding of the role of cellular proteins in the life cycle of positive-strand RNA viruses.
Results
A defined set of cellular proteins binds specifically to the pestiviral 3′NTR
This study was originally aimed at searching for protein interaction partners of the diverse RNA motifs harbored by the pestiviral 3′NTR. For this purpose, we applied a classical UV cross-linking/label-transfer protocol to 32P-labeled transcripts of the 3′NTR of the BVDV replicon RNA ‘DI9c’ because this RNA was best characterized in terms of its functional elements and structure (Yu et al., 1999; see also Figure 1B). As a protein source, we used cytoplasmic extracts of cell types that support BVDV replication, i.e. MDBK (bovine kidney), BHK-21 (hamster kidney), Huh-7 (human hepatoma) or HeLa (human cervix) cells (Behrens et al., 1998). To detect potential RNA binding cellular as well as viral proteins, we performed the initial series of experiments with both naive and replicon-transfected cells (Figure 2A).
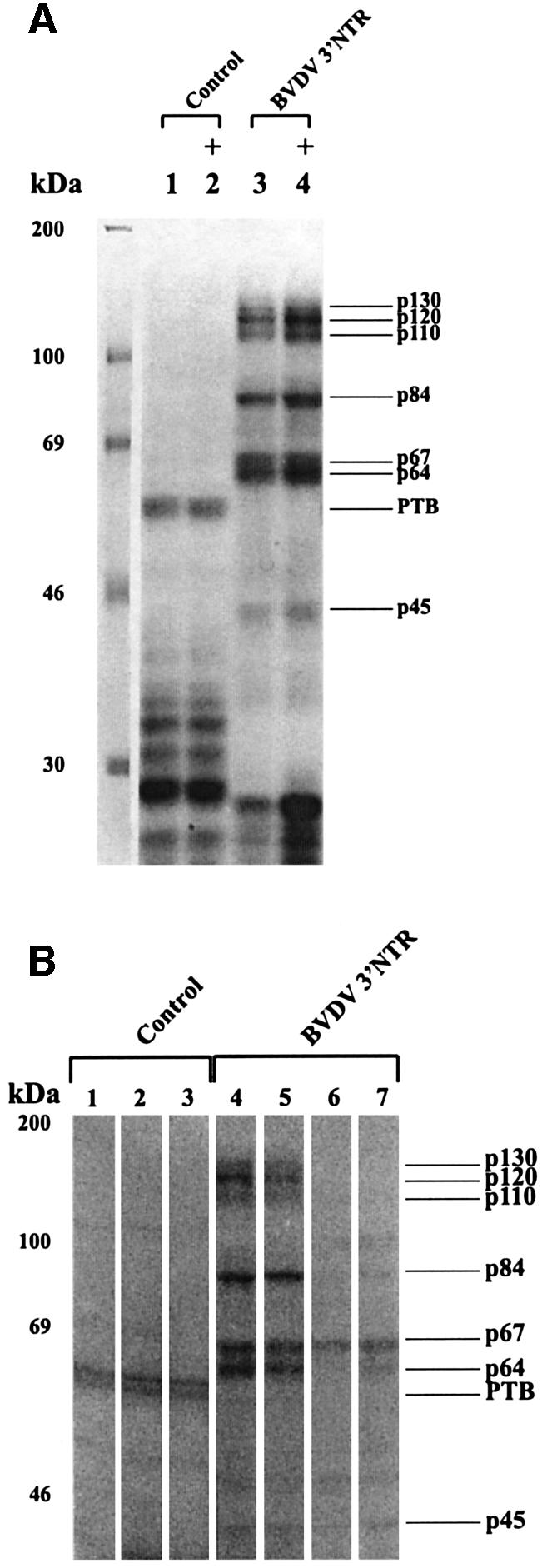
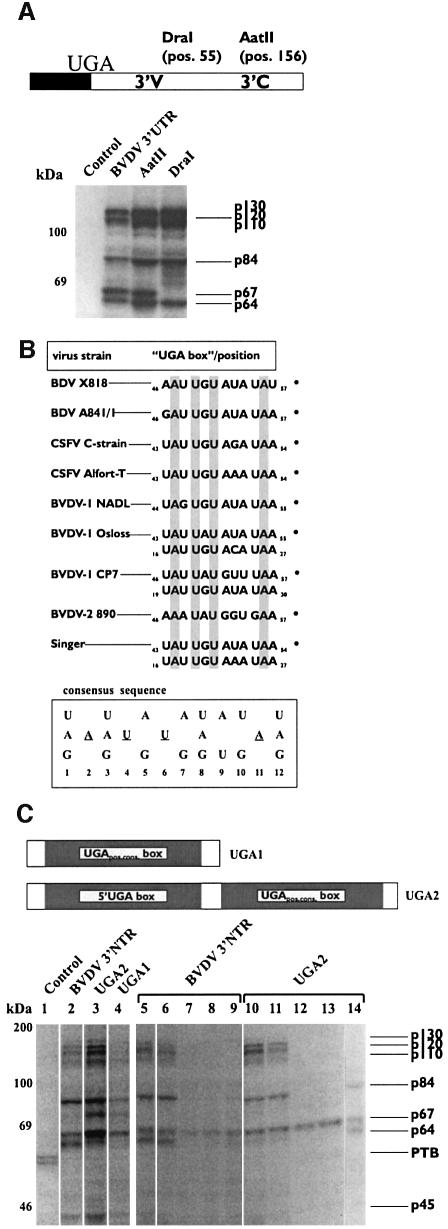
Fig. 2. A set of cellular proteins associates specifically with the pestiviral 3′NTR. (A) UV cross-linking/label transfer experiments performed with [32P]UTP-labeled RNA probes (∼10 ng) and extracts of BHK-21 cells. A non-related control RNA with the same specific activity was compared side-by-side with a transcript corresponding to the BVDV 3′NTR. The control RNA derived from transcription of the Bluescript polylinker; the BVDV transcript was described previously (Yu et al., 1999). Cytoplasmic extracts (20 µg protein/assay) of mock-transfected and BVDV DI9c transfected (+) BHK-21 cells were used as protein sources; all reactions were supplemented with 20 µg of tRNA. Proteins found to be charged with labeled RNA were named according to their molecular weight as they appeared on the analytical SDS–PAGE. PTB, which was found to interact with the control RNA, is also indicated; other control RNAs yielded a blank gel (not shown). Note that [32P]GTP-labeled transcripts yielded identical results (not shown). (B) Competition experiment with different pestiviral 3′NTR transcripts. UV cross-linking was performed as described above with labeled control RNA probe (lanes 1–3) and labeled BVDV 3′NTR probe (lanes 4–7) and in the absence or presence of unlabeled competitor RNA. Lanes 1 and 4, without competitor; lanes 2 and 5, in the presence of a 200-fold molar excess of unlabeled control RNA competitor; lanes 3 and 6, ∼200-fold molar excess of BVDV 3′NTR competitor; lane 7, ∼200-fold molar excess of CSFV 3′NTR competitor.
Apart from slight variations between independent experiments (see subsequent figures), which were explained by technical reasons, this assay gave rise to a distinct and reproducible pattern of proteins charged with RNA label (Figure 2A). While reactions with non-related RNAs often revealed a 60 kDa double band, which was identified as polypyrimidine-tract binding protein PTB (not shown), this signal was absent in experiments with the BVDV 3′NTR. Here, we detected three proteins with molecular masses of ∼130, 120 and 110 kDa and a further four proteins with masses of 84, 67, 64 and 45 kDa, respectively. As this pattern turned out to be the same with different cell types (see below) and to be indistinguishable in extracts of mock and replicon-transfected cells (Figure 2A), we deduced that these bands correspond to a set of ubiquitous cellular RNA binding proteins.
To determine the specificity of the detected RNA–protein interactions, we measured the efficiency of the UV cross-link in the presence of varying amounts of tRNA (up to a 2000-fold molar excess with respect to the viral RNA), at different ionic strengths (up to 1 M KCl) and in the presence of non-related or specific competitor RNA (up to a 200-fold molar excess with respect to the viral RNA). Thus RNA binding of the majority of proteins, i.e. p130, p120, p110, p84, p64 and p45, turned out to be highly specific because the cross-link was only chased by specific competitor RNA (Figure 2B) or high salt (not shown). In contrast, p67 was found to associate significantly less stringently; i.e. similarly to what was observed for the association of PTB with the control RNA, the cross-link of p67 to the BVDV 3′NTR remained largely unaffected even at a high excess of specific competitor (Figure 2B). Importantly, UV cross-linking experiments performed with 3′NTR transcripts of other pestiviral genomes, such as that of classical swine fever virus (CSFV), yielded the identical pattern of labeled cellular proteins as the BVDV RNA (data not shown). In agreement with this observation, CSFV 3′NTR transcripts competed with protein binding to the BVDV 3′NTR (Figure 2B). This suggested the existence of similar or even identical protein interaction sites in the different pestiviral 3′NTRs (see below).
Taking these data together, by applying a suitable UV cross-linking/label transfer protocol it was possible to detect a defined set of cytoplasmic cellular proteins (p130, p120, p110, p84, p64 and p45) which bind in a specific manner to the pestiviral 3′ non-translated region.
Determining the RNA–protein interaction site
Next, we performed the cross-linking experiment with RNA transcripts that comprised different portions of the BVDV 3′NTR to specify the protein interaction site(s). Interestingly, these experiments revealed that the entire set of cellular proteins binds exclusively to the variable 3′V portion of the 3′NTR (Figure 3A; some data not shown). This was rather unexpected in view of our assumption that the protein interaction site should be somehow conserved. However, by performing experimental structure probing and alignment studies it became evident that the 3′V region, despite its sequence heterogeneity, encodes structure and sequence motifs that are common to all pestivirus strains. Thus, by determining the secondary structure of the 3′NTR of the BVDV DI9c RNA and comparing it with that of other pestiviral 3′NTRs (Deng and Brock, 1993, and data not shown), the 3′V region was shown to fold generally into two unstable stem–loop structures termed SLstop and SLII (see Figure 1B and Supplementary data available at The EMBO Journal Online). Moreover, all pestiviral 3′V regions were found to encode 12-nucleotide-long sequence stretches that correspond to a certain consensus sequence, four nucleotides of which are 100% conserved. Alluding to their sequence composition, we termed these elements ‘UGA boxes’. One motif, designated ‘UGApos.cons. box’, is present at nearly identical positions in all pestiviral genomes, i.e. 39–42 residues downstream of the ORF (see Figures 1B and 3B). The 3′V region of BVDV DI9c, which derived from strain BVDV CP7 (Meyers et al., 1996), contains two additional repeats. A second element, ‘5′UGA box’, corresponds exactly to the consensus and resides upstream of the UGApos.cons. box. A third sequence-stretch, termed ‘3′UGA-like box’, which largely resembles the consensus, locates further downstream (see Figure 1B).
Fig. 3. The cellular proteins bind to the 3′V portion of the 3′NTR. (A) UV cross-linking assay performed with shorter versions of the BVDV 3′NTR (conditions as described in Materials and methods and Figure 2). The upper panel shows the scheme of the BVDV 3′NTR transcript (Yu et al., 1999) (the black box represents a residual part of the ORF) indicating the positions of restriction sites (AatII, DraI) used to generate different run-off versions (for numbering see Figure 1B). The lower panel shows the cross-linking assay with the different transcripts. Note that experiments with transcripts that comprised only the 3′C portion of the 3′NTR yielded blank gels (not shown). (B) Conserved sequence elements (‘UGA boxes’) in the pestiviral 3′V region. The table shows an alignment of the sequence elements and their position in the genomic 3′V region of representative virus strains (for numbering see Figure 1B). The consensus sequence is depicted below. One hundred percent conserved nucleotides are underlined or marked in gray. Some strains contain several UGA box elements; in these cases, the UGApos.cons box is indicated by an asterisk (see text). (C) Viral RNA–cellular protein interaction studies with RNA probes encoding individual UGA box elements. The upper panel shows schemes of the UGA1 and UGA2 transcripts (for numbering see Figure 3B). The left lower panel shows cross-linking experiment performed with identical molar amounts of the following probes: control RNA (lane 1), BVDV 3′NTR (lane 2), UGA2 (lane 3) and UGA1 (lane 4). The right lower panel shows competition experiments carried out with the BVDV 3′NTR probe (lanes 5–9) and the UGA2 probe (lanes 10–14). Lanes 5 and 10, without competitor; lanes 6 and 11, in the presence of a 200-fold molar excess of unlabeled control RNA competitor; lanes 7 and 12, ∼200-fold excess of BVDV 3′NTR competitor; lanes 8 and 13, ∼200-fold excess of UGA2 competitor; lanes 9 and 14, ∼200-fold excess of UGA1 competitor.
To evaluate whether these motifs contribute to the interaction of the cellular proteins with the 3′V region, we repeated the cross-linking experiment with a transcript that consisted only of the individual UGApos.cons. box sequence (UGA1). Moreover, we tested an RNA that comprised both 5′-terminal UGA box elements of the DI9c 3′NTR (UGA2) and which was shown by structure probing to be capable of forming the SLstop motif (Figure 3C; probing data not shown). Remarkably, each of these transcripts yielded the familiar label-transfer pattern, demonstrating that protein interaction was already detectable with a single UGA box element. In keeping with this finding, the UGA1 transcript functioned as a specific competitor RNA (Figure 3C). However, taking the intensity of label transfer as a measure of the efficiency of RNA–protein interactions, the protein binding and competition experiments were significantly more effective with the long and structured UGA2 transcript (Figure 3).
Hence, we deduced that efficient association of the cellular proteins to the 3′V portion of the pestiviral 3′NTR involves both the presence of a common sequence motif, namely the UGA box consensus sequence, and the formation of the correct structure.
The same set of cellular proteins binds to the BVDV 5′NTR
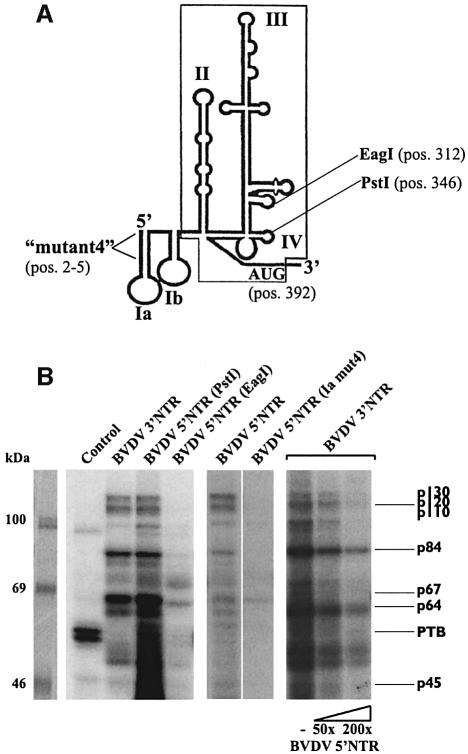
An important control for the specificity of the detected RNA–protein interactions was to test whether the cellular factors also bind to other regions of the BVDV genome. Accordingly, we performed the UV cross-linking assay with a range of transcripts that covered different portions of the non-structural ORF and each of the NTRs, respectively. While the ORF RNAs associated none or unrelated cellular proteins (not shown), we obtained strikingly similar label-transfer patterns when we probed transcripts of the 3′ and 5′NTR (Figure 4). The surprising observation that the same set of factors associates with the 3′ as well as the 5′ end of the viral RNA was confirmed by competition experiments showing that protein binding to the 3′NTR could be efficiently chased by 5′NTR transcripts and vice versa (Figure 4; 3′NTR competition not shown).
Fig. 4. The same set of cellular proteins binds specifically to each of the BVDV non-translated regions. (A) Scheme of the secondary/tertiary structure of the BVDV 5′NTR (Pestova and Hellen, 1999). The minimal IRES element is boxed. The position of ‘mutation 4’ which contains four nucleotide changes at positions 2–5 of the BVDV genome (Yu et al., 2000) is indicated. The positions of the AUG translation initiation codon and of restriction sites (EagI, PstI) used to generate different run-off transcripts are also indicated. (B) Cross-linking and competition experiments with BVDV 5′NTR transcripts (conditions as in Figures 2 and 3). The left panel shows cross-linking assays with the following probes: control RNA, BVDV 3′NTR, BVDV 5′NTR (PstI) and BVDV 5′NTR (EagI). The middle panel shows a side-by side experiment performed with wt 5′NTR probe and a 5′NTR probe (mut4) that does not form hairpin Ia. The right panel shows assays with the labeled BVDV 3′NTR probe performed in the absence and presence of a 50- and 200-fold molar excess of unlabeled 5′NTR competitor, respectively.
Sequence alignments revealed that the pestiviral 5′NTRs encode neither UGA box nor UGA-box-like elements (not shown). Hence, to gain again an idea on the mode of the RNA–protein interactions, we tested a cluster of different run-off transcripts of the BVDV DI9c 5′NTR. Interestingly, the association of the cellular proteins remained largely unaffected with transcripts that lacked only the 3′-terminal domain IV of the IRES. In comparison, protein binding was significantly reduced by deletions affecting domain III (Figure 4). Most remarkably, we also observed a substantial reduction of the RNA–protein interactions with 5′NTR transcripts that contained a set of point mutations in the immediate 5′-terminus (Figure 4) which inhibit the formation of hairpin Ia (Yu et al., 2000). Because transcripts encoding the individual hairpin Ia, domain III or domain IV motifs were negative in the assay (not shown), we concluded that the overall structure of the 5′NTR rather than single elements determines the association of the cellular proteins.
Thus the characteristics of the viral RNA–cellular protein interaction sites were found to surprisingly diverge between the 5′ and the 3′NTR. With the variable 3′V portion of the 3′NTR it was shown to involve primary sequence elements; with the highly conserved 5′NTR it was indicated to rely mainly on the correct folding of the complex RNA structure (see Discussion).
Identification of the viral RNA binding proteins
Next, it was important to purify some of the above-determined cellular proteins and to define their identity. Considering the completion of the Human Genome Project and the fact that BVDV replicates efficiently in human cells, we employed cytoplasmic extracts of HeLa cells as a protein source. The purification was carried out following the scheme shown in Figure 5A; the process was monitored with the established cross-linking assay and BVDV 3′NTR as a probe (Figure 5B).
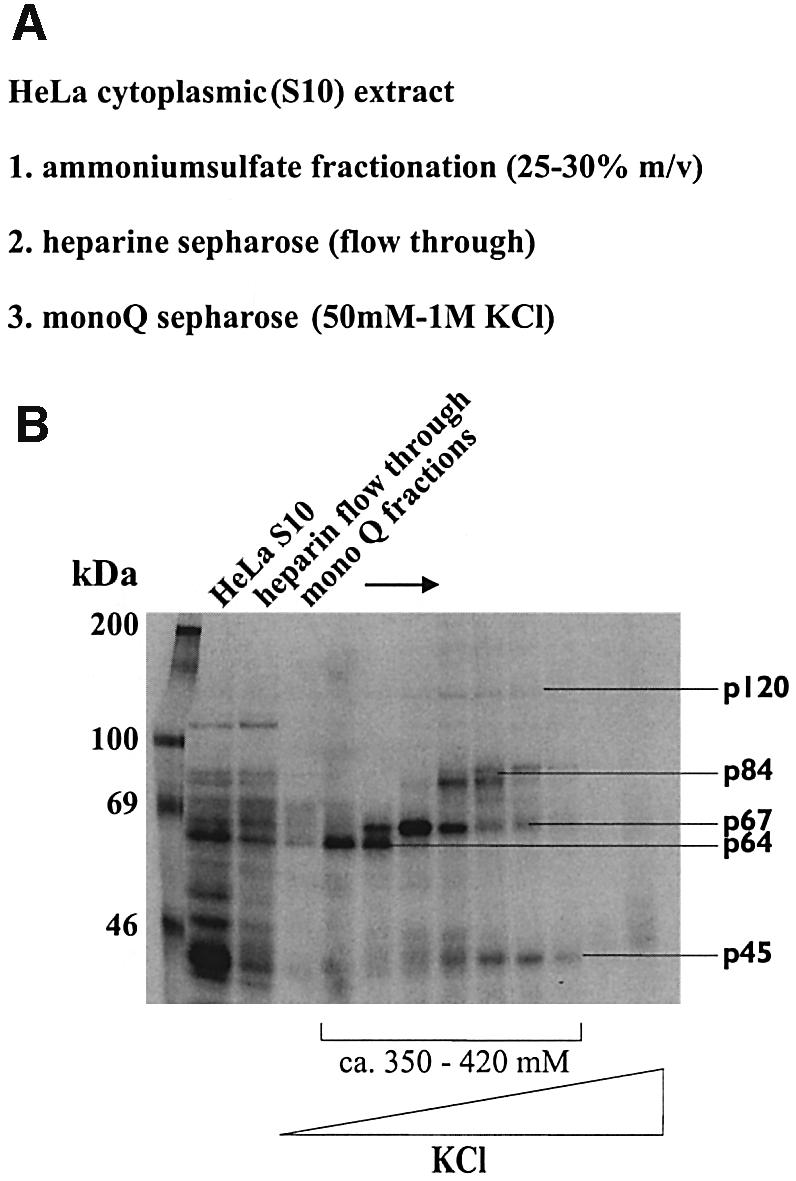
Fig. 5. Purification of the viral RNA binding proteins. (A) Purification scheme (for details, see Materials and methods). (B) Cross-link assays performed with the labeled BVDV 3′NTR probe and cytoplasmic extract, the heparin–Sepharose flowthrough and fractions that derived from the final MonoQ chromatography at ∼350–420 mM KCl, respectively. The viral RNA binding proteins are indicated.
Using this procedure, we obtained a reasonable degree of purification for p84. That is, silver staining of SDS gels of the final purification fractions revealed the presence of ∼20 protein bands; however, only one of these had the expected size of 84 kDa (data not shown). It was excised and trypsinized, and several peptides were analyzed by massspectrometry. All peptides derived from one protein, ‘nuclear factor 90’ (NF90), a predominantly nuclear protein which, however, is also present in the cytoplasm (Kao et al., 1994; Liao et al., 1998). A corrected C-terminal variant of NF90 was later termed NFAR-1 (Saunders et al., 2001). As expected, NF90/NFAR-1 represents an RNA binding protein with two double-strand RNA binding motifs (dsRBM) (St Johnston et al., 1992). Most interestingly, NF90/NFAR-1 recently came under scrutiny as a potential sensor of double-stranded RNA, for example in the course of a viral infection (see below).
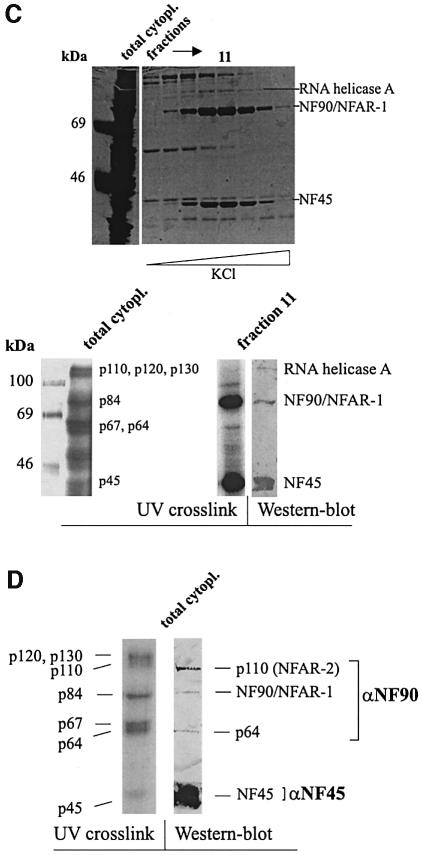
Several lines of evidence substantiated that NF90/NFAR-1 definitely corresponds to p84. Thus RNA gel-mobility shift assays (RMSA) carried out with HeLa cell extracts, BVDV 3′ (or 5′) NTR transcript and anti-NF90/NFAR-1 antiserum revealed the formation of specific antibody/protein/viral RNA complexes (Figure 6A). RNA–protein coprecipitation experiments with 35S-labeled in vitro translated protein confirmed that NF90/NFAR-1 binds at high priority to the BVDV RNA (Figure 6B). Other important data were derived when we purified NF90/NFAR-1 by poly(I):(C) affinity chromatography from cytoplasmic extracts. For this purpose, we applied a protocol (Liao et al., 1998) which yields the protein at high purity (see Figure 6C). Importantly, under these conditions NF90/NFAR-1 co-elutes with two other proteins, NF45 and, at a lower molar ratio, RNA helicase A (RHA), which have a size of 45 and 120 kDa, respectively (Liao et al., 1998) (Figure 6C). As this finding was strikingly reminiscent of our original purification scheme, where p84 was observed to cofractionate with p45 and p120 throughout all purification steps (see Figure 5B), it supported the notion that p84 represents NF90/NFAR-1. Moreover, it strongly suggested that p45 and p120 were NF45 and RHA. We confirmed this supposition by applying the poly(I):(C) fractions side-by-side with total cytoplasmic extract to UV cross-linking assays and immunoblots with antisera against the three proteins (Figure 6C).
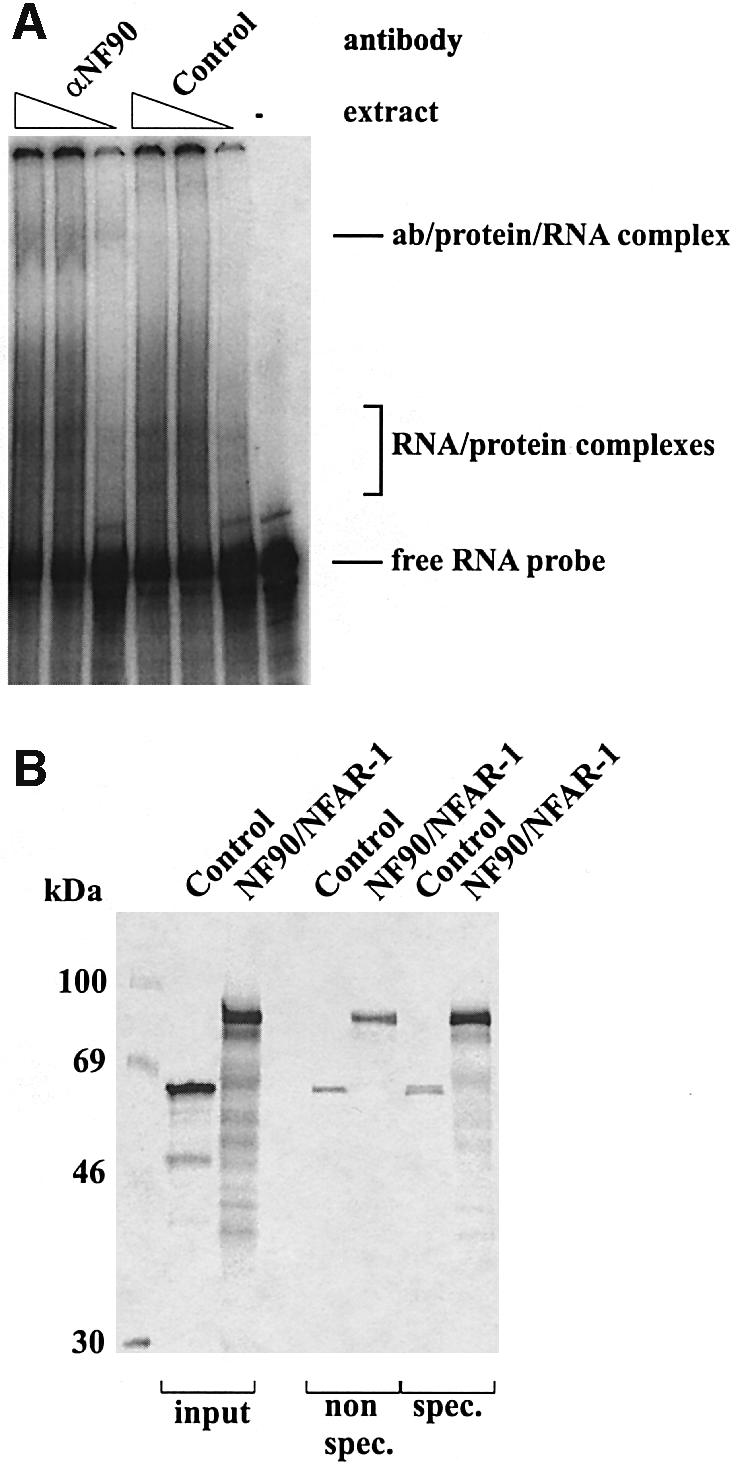
Fig. 6. The viral RNA binding proteins correspond to the ‘NFAR group of proteins’. (A) RMSA assay with αNF90. The experiment was performed as described in Materials and methods using 32P-labeled BVDV 3′NTR RNA. Except for the control lane (right), the RNA was incubated with increasing amounts of cytoplasmic extract (right to left: 5, 20 and 50 ng) and a constant amount of αNF90 or control antiserum. (B) PolyU pulldown experiment. The experiment was carried as described in Materials and methods using polyadenylated BVDV 3′NTR (spec.) and polyadenylated control RNA (non spec.). The binding behavior of in vitro translated [35S]-labeled NF90/NFAR-1 was compared with that of a control protein (luciferase). (C) Poly(I):(C) purification of the [NF90/NFAR-1, NF45, RHA] complex following the protocol described (Liao et al., 1998). The top panel shows aliquots of total HeLa cytoplasm and of fractions that eluted at high KCl (left to right ∼0.6–1 M) from poly(I):(C) analyzed by SDS–PAGE and silver staining. RHA, NF90/NFAR-1 and NF45 are indicated. The bottom panel (left and middle) shows the UV cross-linking assay with HeLa cytoplasmic extract (20 µg of protein) performed side-by-side with an aliquot (50 ng of protein) of the poly(I):(C) fraction 11. The autoradiograph of the experiment with fraction 11 was contrasted differently to reveal the under-represented band of RHA as well. The right bottom panel shows western blot analysis of fraction 11 performed with antisera against all three proteins. (D) αNF90 antiserum cross-reacts with proteins that exhibit essentially the same retardation factor as p110 and p64 on SDS–PAGE. The figure shows a side-by-side projection of a cross-link experiment (left) and a western blot experiment (right), each performed with total HeLa cytoplasmic extract. The western blot analysis was performed with a mixture of αNF45 and αNF90 (Kao et al., 1994); αNF90 alone stained the same pattern of proteins except for NF45 (not shown).
Western blot experiments also revealed preliminary information on the identity of the remaining viral RNA binding proteins that were originally detected by UV cross-linking. That is, when we tested the anti NF90/NFAR-1 antibody on total cytoplasmic extracts, it was found to stain p84 as well as two additional protein bands. Strikingly, these proteins had the same retardation factors on the SDS–PAGE as p110 and p64 (Figure 6D). This suggested that p110 corresponds to NFAR-2, i.e. a C-terminally extended alternatively spliced form of NFAR-1, which has a size of 110 kDa (Saunders et al., 2001). In addition, this result implied that p64 was another yet uncharacterized relative of NFAR-1.
Thus, although the latter data await final confirmation, we have accumulated convincing evidence that the majority of the viral RNA binding cellular proteins detected, hereinafter referred to as the ‘NFAR group of proteins’, represent either related forms or interaction partners of NF90/NFAR-1. In particular, it was confirmed that NF90/NFAR-1, NF45 and RHA form a stable higher-order protein complex (Liao et al., 1998).
Significance of the NFAR proteins for BVDV replication
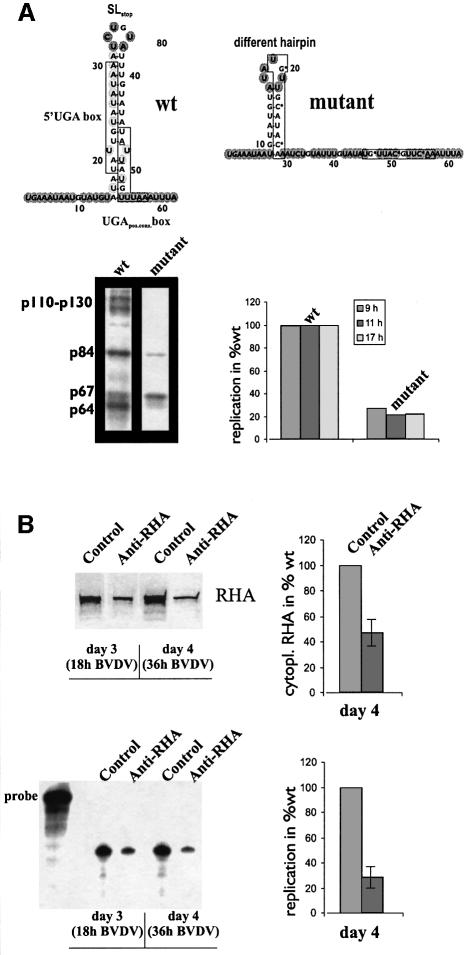
The above experiments demonstrated that the NFAR proteins associate specially with functional domains in each of the non-translated regions of the BVDV genome and gave rise to the supposition that these factors operate as functional components of the viral replication machinery. As an initial approach to test this hypothesis, we examined mutations in the 3′V region of the BVDV replicon in terms of their effect on the formation of the viral RNA/cellular protein complex and the RNA replication process, respectively. While this study is systematically described elsewhere (O.Isken, C.W.Grassmann, H.Yu and S.-E.Behrens, in preparation), it is important to note that nucleotide exchanges, which affected three of the four conserved positions in each of the 5′-terminal UGA boxes and which significantly modified the structure of the 3′V region, decreased protein binding and, concomitantly, impaired RNA replication (Figure 7A). In view of the fact that these mutations were shown to affect neither the stability of the viral RNA nor cis-encoded replica tion signals (O.Isken, C.W.Grassmann, H.Yu and S.-E.Behrens, in preparation; see also Discussion), we interpreted these results as a first important indication for the functional relevance of the viral/cellular ribonucleoprotein complex for viral replication.
Fig. 7. Importance of the NFAR proteins for BVDV replication. (A) Mutations in the 3′V region inhibit protein binding and RNA replication. The top panel shows the structure of the upstream 62 residues of the wild type (wt) BVDV DI9c 3′V region compared with a mutant that contains nucleotide changes (marked by asterisks) at three conserved positions in the 5′UGA box and the UGApos.cons. box, respectively. The mutant RNA was shown by chemical modification (see Supplementary data) to form a different hairpin structure; the accessible residues are indicated as in Figure 1B. The structure of the residual 3′NTR was found unchanged (not shown). The bottom left panel shows a cross-linking assay performed with wt and mutant BVDV 3′NTR transcripts. The bottom right panel shows the replication capacity of the wt and the mutant replicon RNA. Replication was monitored by RNase protection of progeny positive-strand RNA at 9, 11 and 17 h post transfection into MDBK cells (for experimental details, see Behrens et al., 1998). The replication capacity of the mutant is compared with that of the wt (considered 100%). The diagram shows average values of three transcription/transfection experiments. (B) siRNA-mediated depletion of cytoplasmic RHA inhibits BVDV replication. The top left panel shows a representative western blot experiment analyzing the amount of RHA in the cytoplasm of Huh-7 cells at days 3 and 4 after the first round of transfection with a control siRNA and an anti-RHA siRNA, respectively. The top right panel shows average values of the concentration of RHA at day 4 as determined in three independent siRNA transfection experiments; the error bar indicates the mean deviation. Note that the level of depletion was found to be in the same range at days 3 and 4. The bottom left panel shows a representative RNase protection experiment measuring the replication capacity of BVDV DI9c RNA in control and anti-RHA siRNA transfected cells, respectively. Left, original RNA probe used for protection: lanes 1 and 2, protected progeny positive-strand RNA at 18 h post transfection of BVDV RNA (i.e. at day 3 after the first round of siRNA transfection); lanes 3 and 4, at ∼36 h post transfection of BVDV RNA (at day 4 after the first round of siRNA transfection). Transfection experiments with replication-defective BVDV RNA (Behrens et al., 1998) resulted in a blank gel (not shown). Right: diagram summarizing the data of three independent replication experiments; the error bar indicates the mean deviation. Note that analogous effects on the level of cytoplasmic RHA and viral replication were observed with two different anti-RHA siRNAs (see Materials and methods) and that, compatible with the idea of a limiting host factor, replication was inhibited in the same way with higher amounts of transfected BVDV RNA (e.g. 5 pmol instead of 1 pmol per 3 × 106 cells, not shown).
Further evidence came from experiments, where we investigated viral replication in cells that expressed lower levels of RHA due to a siRNA-directed depletion of RHA mRNA (Elbashir et al., 2001). We employed Huh-7 cells for this purpose, because these cells turned out to be most susceptible to siRNA transfections (transfection efficiency 80–90%; see Materials and methods). In pilot studies we determined that two subsequent transfections within two days using 600 pmol of anti-RHA siRNAs per 3 × 106 cells reduced the level of cytoplasmic RHA by ∼40–60% during the following two days (see Figure 7B). Importantly, the moderate decrease of the RHA concentration had no effect on the viability of the cells over this period (see below). The BVDV RNA was included in the second transfection round and viral replication was monitored 18 h and 36 h later, i.e. at days 3 and 4 after the first round of siRNA transfection. These time points were significantly prior to the onset of the cytopathic effect of BVDV replication in Huh-7 cells (Behrens et al., 1998) at ∼48 h post transfection of the viral RNA (day 5 of the total experiment). We assured the absence of cell-death up to day 5 by Annexin V tests and trypan blue exclusion (not shown). Most importantly, with anti RHA siRNA treated cells, we observed a significant reduction of viral replication by 60–85% compared to cells transfected with control siRNAs (Figure 7B). We attributed the inhibitory effect of RNAi on viral replication to the reduced amount of RHA at the sites of RNA replication in the cytoplasm.
In sum, these data indicated the NFAR group of proteins to be ‘real BVDV host-factors’, which participate actively in the viral RNA replication process (see Discussion).
Indications for a protein-mediated 5′–3′ interaction of the BVDV genome
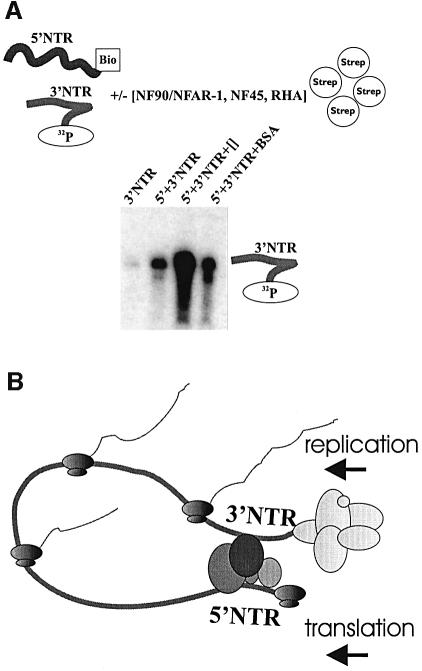
The final issue addressed in this work concerned the potential function of the NFAR proteins during viral replication. The scenario that p130, p120, p110, p84, p64 and p45 bind to the 3′ as well as to the 5′NTR of the BVDV genome fueled the idea that these proteins might act as a functional bond between the termini of the viral RNA. To evaluate this possibility, we designed a 5′–3′ co-precipitation assay applying streptavidin-beads covered with biotinylated transcripts of the viral 5′NTR and radioactively labeled transcripts of the 3′NTR. Interestingly, co-precipitation attempts performed exclusively with naked RNAs revealed a detectable degree of RNA–RNA interactions between both ends of the viral genome. That is, a small amount of 3′NTR was reproducibly pulled down by the 5′NTR-transcripts (Figure 8A). However, in experiments where the reaction mixture was complemented with poly(I):(C) fractions that contained the purified NF90/NFAR-1, NF45 and RHA (Figure 6C), we observed a significant increase of the level of 5′–3′ interactions (Figure 8A).
Fig. 8. The [NF90/NFAR-1, NF45, RHA] complex supports the interaction of both termini of the viral RNA genome. (A) 5′–3′coprecipitation assay. The top panel shows a schematic representation of the assay carried out with biotinylated 5′NTR and 32P-labeled 3′NTR (see Materials and methods). Bottom panel: precipitation of the 3′NTR (as indicated) was performed with the naked RNAs, in the presence of the [NF90/NFAR-1, NF45, RHA] complex ([]) or in the presence of bovine serum albumin (BSA) (20-fold molar excess with respect to the complex). (B) Model of a circularized viral RNA mediated by [NF90/NFAR-1, NF45, RHA]. The replication complex, which is supposed to form at the immediate 3′-terminus of the viral RNA, is depicted in light gray; translating ribosomes are represented in shaded black. The [NF90/NFAR-1, NF45, RHA] complex is schematized by four differently shaded circles (see text for details).
As outlined further below, these data supported the idea that NFAR proteins, in particular the complex consisting of NF90/NFAR-1, NF45 and RHA, are involved in the establishment of a functional bridge between the 5′ and the 3′-end of the viral genome.
Discussion
Pestiviruses such as BVDV are easy manageable positive-strand RNA viruses, studies of which are particularly important to serve as a potent model of the life cycle of the closely related human HCV. The characterization of BVDV replicons revealed that on the part of the virus only the non-structural proteins encoded by the 3–5B portion of the ORF are necessary and sufficient to catalyze both steps of the viral RNA replication pathway (Behrens et al., 1998; Grassmann et al., 2001). Hence, a major subsequent question concerned the identity and functional role of cellular proteins participating in viral replication.
Considering that formation of the viral replication complex involves direct interactions between cellular ‘host factors’ and the viral RNA genome, we based a search on a classical UV cross-linking protocol. Thus we defined a set of specifically viral RNA binding proteins, which during subsequent experiments were shown to correspond to what we have called the ‘NFAR group of proteins’.
The NFAR group essentially includes three types of proteins, namely NF90/NFAR-1 (nuclear factor 90/nuclear factor associated with RNA) and related forms, i.e. NF45 (nuclear factor 45) and RHA [RNA helicase A or nuclear DNA helicase II (NDHII)]. All members of this group belong to a family of proteins with widespread functional diversity including, for example, the Drosophila Staufen, the Vaccinia E3L, the Escherichia coli RNase III and the protein kinase PKR. The most conspicuous common feature of this family is the double-strand RNA binding motif (dsRBM) that exerts a variety of functions ranging from RNA recognition to catalytic activity (Fierro-Monti and Mathews, 2000).
We paid particular attention to rigorous controls, excluding the possibility that the association of the NFAR proteins with the viral RNA occurred just by chance. Along this line, we applied a broad range of non-viral and viral RNA probes in the cross-linking assay, performed stringent competition experiments and finally tested the individual proteins in coprecipitation experiments with non-related and viral RNAs (Figure 6). Based on these data we concluded that the NFAR proteins associate in a specific manner with the 3′ as well as the 5′NTR of the BVDV genome. The specificity of the viral RNA/NFAR protein interactions was further confirmed by the initial characterization of the protein binding sites. While protein binding to the 3′NTR was found to involve the UGA box sequence elements as well as the formation of the structure of the 3′V region (Figures 3C and 7A); the binding motif in the 5′NTR was implied as a complex structure that necessarily includes distant elements such as hairpin Ia and IRES domain III (Figure 4). Final evidence that we were not dealing with indiscriminate factors was derived from the observation that the binding behavior of the NFAR proteins clearly differed from that of other RNA-associating proteins such as PTB and p67 (see Figures 2 and 3).
Our findings immediately suggested that the NFAR proteins might act as a functional bridge between both ends of the viral genome (Figure 8B). This idea was considerably strengthened by precipitation assays conducted with RNA transcripts of both the termini of the BVDV genome and fractions of purified proteins (Figure 8A). First, these experiments revealed that there was also a weak but evident interaction of the BVDV 5′ and 3′NTR in the absence of protein, which could not be explained by obvious complementary sequence stretches as they exist in the termini of flavivirus genomes (Hahn et al., 1987). Most importantly, the RNA–RNA interactions were considerably augmented in the presence of NF90/NFAR-1, NF45 and RHA, i.e. those members of the NFAR group which were previously reported and confirmed here to form a protein complex (Liao et al., 1998). The molecular mechanisms underlying this remarkable observation remain to be clarified. It is conceivable that the [NF90/NFAR-1, NF45, RHA] complex stabilizes weak RNA–RNA bonds. Alternatively, the formation of the viral/cellular RNP could create entirely novel interactions. Current investigations are aimed at understanding the nature of the 5′–3′ RNA–RNA hybrid as well as how [NF90/NFAR-1, NF45, RHA] supports long-distance communication of the RNA termini and in this way circularization of the viral genome.
Several lines of evidence supported the notion that certain members or even the entire group of NFAR proteins are essentially involved in the viral RNA replication process. That is, mutations which changed the sequence of the UGA box elements as well as the structure of the 3′V region demonstrated an evident correlation between viral/cellular RNP formation and the replication capacity of the BVDV RNA (Figure 7A) (O.Isken, C.W.Grassmann, H.Yu and S.-E.Behrens, in preparation). Taking the well-characterized RNA helicase RHA (Zhang and Grosse, 1994) as the most promising target for a siRNA-mediated depletion approach, we found that even moderate reductions of the concentration of the protein in the cytoplasm, which had no detectable consequences for the viability of the cell, nevertheless yielded a significant negative effect on BVDV replication. Apart from proving RHA convincingly as a viral host-factor, these data implied circularization of the viral genome as a prerequisite for replication. Moreover, the fact that RHA unwinds nucleic acid double strands fuels the speculation that this activity is important for the formation and/or modulation of 5′–3′ RNA–RNA cross-talk.
Along the same lines, it is intriguing that the NFAR protein binding sites involve RNA elements in the BVDV NTRs, namely hairpin Ia at the genomic 5′ end and the 3′V portion of the 3′NTR, which are suggested regulators of translation and RNA replication (see Introduction). Thus hairpin Ia was reinforced to operate in a strikingly similar fashion to the ‘cloverleaf’ structure at the 5′ terminus of the poliovirus genome. Both motifs were demonstrated to be crucially involved in the first replication step and to be major constituents of a protein-mediated closed-loop conformation of the viral genome (Yu et al., 2000; Barton et al., 2001; Herold and Andino, 2001). In view of the fact that hairpin Ia and 3′V modulate the efficiency of translation initiation (Yu et al., 2000; Isken et al., in preparation), it is tempting to assume that the NFAR proteins are active parts of mechanisms that control IRES function, possibly during the switch from protein to RNA synthesis. In support of this hypothesis, almost the entire cytoplasmic fraction of the NFAR group cofractionates with ribosomal salt wash (Langland et al., 1999; our own observations) and an isoform of NF90/NFAR-1 has already been shown to act as a translational inhibitor of acid β-glucosidase mRNA, the enzyme deficient in Gaucher disease (Xu and Grabowski, 1999). Accordingly, a major topic of future investigations will be exploration of the NFAR proteins for a potential inhibitory action on IRES-mediated translation. Moreover, considering the feedback regulation model (see Introduction), it will be important to evaluate whether some of the viral non-structural proteins interact with the RNA/NFAR protein complex and potentially modulate its activity.
One of the most interesting aspects of this study relates to the nature of the NFAR proteins. NF90/NF45 was originally characterized as a heterodimeric complex that associates with the interleukin 2 promoter (Kao et al., 1994). Accordingly, these proteins gained prevailing attention as potential regulators of gene expression on the transcriptional or post-transcriptional level (Saunders et al., 2001; Reichman et al., 2002). Many lines of evidence indicate that RHA also exerts a pivotal role in gene expression (reviewed by Fierro-Monti and Mathews, 2000). Most interesting from the virology viewpoint are findings that in interferon-α-stimulated cells RHA translocates to promyelocytic leukemia nuclear bodies (PML NBs), which may indicate the enzyme as a potential transcriptional regulator of interferon-inducible genes attached to PML NBs (Regad and Celbi-Alix, 2001; Fuchsova et al., 2002). Accordingly, BVDV may act in two ways for its own needs because exploitation of RHA as a component of the viral replication machinery in the cytoplasm may simultaneously remove the enzyme from sites in the nucleus that stimulate the antiviral state of the cell. In other words, by operating as a trap for proteins involved in antiviral activities, the BVDV genome might undermine the cellular defense. In compelling agreement with this idea, NF90/NFAR-1 was reported to mediate the activation of the cellular antiviral expression cascade (Krasnoselskaya-Riz et al., 2002) and to modulate the activity of PKR, another major player in the cellular interferon response (Parker et al., 2001; Saunders et al., 2001). Thus the NFAR group of proteins, potential sensors or even opponents of a viral infection, may be subverted to being accomplices of the virus. It will be important to test next the role of these proteins in the life cycle of other positive-strand RNA viruses, in particular the human pathogen HCV.
Materials and methods
UV cross-linking assay
Fifteen microliters of cytoplasmic extract were preincubated for 10 min at 30°C with 20 µg of tRNA in 10 mM HEPES pH 7.6, 0.3 mM MgCl2, 40 mM KCl, 1 mM dithiothreitol (DTT), 5% glycerol and 40 U RNasin. Next, 500 000 c.p.m. (∼10 ng) of 32P-labeled RNA transcript were added and the incubation continued for 30 min. The samples were exposed to ultraviolet light for 15 min at 4°C followed by treatment with 7 µg RNase A and 20 U RNase T1 for 1 h. The proteins were separated by SDS–PAGE and radiolabeled bands were identified by autoradiography. In competition experiments, the indicated excess of unlabeled competitor RNA was included into the reaction prior to the treatment with UV.
Protein purification
The proteins were precipitated at 30–35% (NH4)2SO4 (m/v) from cytoplasmic extracts of HeLa S3 cells, collected by centrifugation (15 min, 10000 r.p.m., 4°C, JA-20 rotor, Beckman) and dialyzed against buffer A (2.5 mM Tris-HCl pH 7.5, 50 mM KCl, 0.025 mM EDTA). Buffer A was used for chromatography with heparin–Sepharose, during washing and for elution with a salt gradient of 0.05–1 M KCl. As shown in Figure 5, the viral RNA binding proteins remained in the flowthrough. Subsequent chromatography with MonoQ-Sepharose was performed under the same conditions; the proteins eluting at 300–400 mM KCl (Figure 5) were dialyzed to buffer A 0.05 M KCl for further use. Poly(I):(C) purification of NF45, NF90/NFAR-1 and RHA (Figure 6) was carried out essentially as described (Liao et al., 1998).
RNA gel-mobility shift assay (RMSA)
Indicated amounts of cytoplasmic extract were incubated for 10 min at 30°C in RMSA buffer (5 mM HEPES pH 7.6, 50 mM KCl, 2 mM MgCl2, 1.5 mM ATP, 1 mM DTT, 5% glycerol, 40 U RNasin, 10 µg tRNA). [32P]-labeled RNA probe (50 000 c.p.m., ∼1 ng RNA) and 1 µl of antiserum were added and incubation was continued for a further 20 min. The complexes were separated on non-denaturing 5% Tris-borate-EDTA acrylamide gels and analyzed by autoradiography.
5′–3′ coprecipitation assay
Two micrograms of biotinylated RNA obtained by in vitro transcription in the presence of 0.16 mM biotinylated CTP (Roche) was bound to 2 µl of streptavidin–Sepharose beads in buffer C [10 mM HEPES pH 7.6, 0.3 mM MgCl2, 40 mM KCl, 1 mM DTT, 5% glycerol, 40 U RNasin (Promega)]. In a total volume of 25 µl, the 32P-labeled RNA (∼10 ng) was preincubated with poly(I):(C) purified proteins (∼20 ng/assay) and 10 µg of tRNA (20 min at 30°C). The reaction mixtures were combined and incubated for an additional 20 min. The beads were collected by centrifugation and washed with buffer C containing 500 mM KCl (2×) and 150 mM KCl (2×), respectively, and the precipitated nucleic acids were analyzed on a denaturing 5% TBE acrylamide gel.
RNAi
siRNA duplexes anti-RHA (5′-aaccacacagguuccccagTT or 5′-acuccccauugagccucguTT) and control (anti GFP) siRNA (5′-gacacgugcugaagucaagTT) were purchased from Qiagen-Xeragon. They were electroporated (one pulse, 270 V, 950 µF) at 600 pmol per 3 × 106 Huh-7 cells. After 2 days, the transfection was repeated, this time including 1–5 pmol of BVDV replicon RNA transcript. The cells were harvested 18 and 36 h later and the cytoplasmic fraction was analyzed for the amount of RHA and progeny positive-strand viral RNA molecules (Figure 7B).
Poly(U) pulldown
Polyadenylated versions of the BVDV 3′NTR and of non-related RNA were obtained by in vitro transcription of respective plasmid constructs. The 35S-labeled proteins were obtained by in vitro translation. Two micrograms of the RNA transcripts were incubated for 60 min at 30°C with 50µl of the in vitro translation reaction in incubation buffer (10 mM HEPES pH 7.4, 1.5 mM MgCl2, 90 mM KCl, 2.5 mM DTT, 0.05% NP-40) and in the presence of 10 µg of tRNA. Subsequently, the samples were adsorbed to poly(U)–agarose (Sigma). After washing with incubation buffer containing 250 mM KCl, the resin-bound proteins were released by boiling in standard SDS-sample buffer and analyzed by SDS–PAGE and autoradiography.
Information on the following procedures is given in the Supplementary data: construction of plasmids; preparation of cytoplasmic extracts; probing of the RNA structure; transcription, purification and translation of RNA; transfection of RNA; western blots and antibodies; protein sequence analysis; RNase protection assay (RPA); cell viability assays.
Supplementary data
Supplementary data are available at The EMBO Journal Online.
Acknowledgments
Acknowledgements
We are grateful to M.Niepmann (Giessen) for purified PTB and α-PTB serum and R.Lührmann (Göttingen) for providing HeLa extracts. This study was supported by the SFB 535, by a grant of GSK for C.G. and by a PA Tobacco Formula Grant for S.E.B. O.I. was supported by the ‘Kompetenznetz Hepatitis’ (Projekt 13.3).
References
- Barton D.J., Morasco,B.J. and Flanegan,J.B. (1999) Translating ribosomes inhibit poliovirus negative-strand RNA synthesis. J. Virol., 73, 10104–10112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barton D.J., O’Donnell,B. and Flanegan,J.B. (2001) 5′ cloverleaf in poliovirus RNA is a cis-acting replication element required for negative-strand synthesis. EMBO J., 20, 1439–1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behrens S.-E., Grassmann,C.W., Thiel,H.-J., Meyers,G. and Tautz,N. (1998) Characterization of an autonomous subgenomic pestivirus RNA replicon. J. Virol., 72, 2364–2372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng R. and Brock,K.V. (1993) 5′ and 3′ untranslated regions of pestivirus genome: primary and secondary structure analysis. Nucleic Acids Res., 21, 1949–1957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elbashir S.M., Harborth,J., Lendeckel,W., Yalcin,A., Weber,K. and Tuschl,T. (2001) Duplexes of 21-nucleotide RNAs mediate RNA interference in mammalian cell culture. Nature, 411, 494–498. [DOI] [PubMed] [Google Scholar]
- Fierro-Monti I. and Mathews,M.B. (2000). Proteins binding to duplexed RNA: one motif, multiple functions. Trends Biochem., 25, 241–246. [DOI] [PubMed] [Google Scholar]
- Fuchsova B., Novak,P., Kafkova,J. and Hozak,P. (2002) Nuclear DNA helicase II is recruited to IFN-alpha-activated transcription sites at PML nuclear bodies. J. Cell Biol., 158, 463–473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamarnik A. and Andino,R. (1998) Switch from translation to replication in a positive-stranded RNA virus. Genes Dev., 12, 2293–2304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grassmann C.W., Isken,O., Tautz,N. and Behrens,S.-E. (2001) Genetic analysis of the pestivirus nonstructural coding region: defects in the NS5A unit can be complemented in trans. J. Virol., 75, 7791–7802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn C.S., Hahn,Y.S., Rice,C.M., Lee,E., Dalgarno,L., Strauss,E.G. and Strauss,J.H. (1987) Conserved elements in the 3′ untranslated region of flavivirus RNAs and potential cyclization sequences. J. Mol. Biol., 198, 33–41. [DOI] [PubMed] [Google Scholar]
- Herold J. and Andino,R. (2001) Poliovirus RNA replication requires genome circularization through a protein–protein bridge. Mol. Cell, 7, 581–591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kao P.N., Chen,L., Brock,G., Ng,J., Kenny,J., Smith,A.J. and Courthesy,B. (1994) Cloning and expression of cyclosporine A- and FK506-sensitive nuclear factor of activated T-cells: NF45 and NF90. J. Biol. Chem., 269, 20691–20699. [PubMed] [Google Scholar]
- Krasnoselskaya-Riz I., Spruill,A., Chen,Y.W., Schuster,D., Teslovich,T., Baker,C., Kumar,A. and Stephan,D.A. (2002) Nuclear factor 90 mediates activation of the cellular antiviral expression cascade. AIDS Res. Hum. Retroviruses, 18, 591–604. [DOI] [PubMed] [Google Scholar]
- Lai M.M. (1998). Cellular factors in the transcription and replication of viral RNA genomes: a parallel to DNA-dependent RNA transcription. Virology, 244, 1–12. [DOI] [PubMed] [Google Scholar]
- Langland J.O., Kao,P.N. and Jacobs,B.L. (1999) Nuclear factor-90 of activated T-cells: a double-stranded RNA-binding protein and substrate for the double-stranded RNA-dependent protein kinase, PKR. Biochemistry, 38, 6361–6368. [DOI] [PubMed] [Google Scholar]
- Liao H.J., Kobayashi,R. and Mathews,M.B. (1998) Activities of adenovirus virus-associated RNAs: purification and characterization of RNA binding proteins. Proc. Natl Acad. Sci. USA, 95, 8514–8519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindenbach B.D. and Rice,C.M. (2001). Flaviviridae: the viruses and their replication. In Fields,B.N. (ed.), Virology, 4th edn. Lippincott–Raven, Philadelphia, PA, pp. 991–1042. [Google Scholar]
- Meyers G., Tautz,N., Becher,P., Thiel,H.-J. and Kümmerer,B.M. (1996) Recovery of cytopathogenic and noncytopathogenic bovine viral diarrhea viruses from cDNA constructs. J. Virol., 70, 8606–8613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker L.M., Fierro-Monti,I. and Mathews,M.B. (2001) Nuclear factor 90 is a substrate and regulator of the eukaryotic initiation factor 2 kinase double-stranded RNA-activated protein kinase. J. Biol. Chem., 276, 32522–32530. [DOI] [PubMed] [Google Scholar]
- Pestova T.V. and Hellen,C.U. (1999) Internal translation of bovine viral diarrhea virus RNA. Virology, 258, 249–256. [DOI] [PubMed] [Google Scholar]
- Regad T. and Chelbi-Alix,M.K. (2001). Role and fate of PML nuclear bodies in response to interferon and viral infections. Oncogene, 20, 7274–7286. [DOI] [PubMed] [Google Scholar]
- Reichman T.W., Muniz,L.C. and Mathews,M.B. (2002) The RNA binding protein nuclear factor 90 functions as both a positive and negative regulator of gene expression in mammalian cells. Mol. Cell. Biol., 22, 343–356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saunders L.R., Perkins,D.J., Balachandran,S., Michaels,R., Ford,R., Mayeda,A. and Barber,G.N. (2001) Characterization of two evolutionarily conserved, alternatively spliced nuclear phosphoproteins, NFAR-1 and -2, that function in mRNA processing and interact with the double-stranded RNA-dependent protein kinase, PKR. J. Biol. Chem., 276, 32300–32312. [DOI] [PubMed] [Google Scholar]
- St Johnston D., Brown,N.H., Gall,J.G. and Jantsch,M. (1992) A conserved double-stranded RNA-binding domain. Proc. Natl Acad. Sci. USA, 89, 10979–10983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y.H. and Grabowski,G.A. (1999) Molecular cloning and characterization of a translational inhibitory protein that binds to coding sequences of human acid beta-glucosidase and other mRNAs. Mol. Genet. Metab., 68, 441–454. [DOI] [PubMed] [Google Scholar]
- Yu H., Grassmann,C.W. and Behrens,S.-E. (1999) Sequence and structural elements at the 3′ terminus of bovine viral diarrhea virus genomic RNA: functional role during RNA replication. J. Virol., 73, 3638–3648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H., Isken,O., Grassmann,C.W. and Behrens,S.-E. (2000) A stem–loop motif formed by the immediate 5′-terminus of the bovine viral diarrhea virus genome modulates translation as well as replication of the viral RNA. J. Virol., 74, 5825–5835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S. and Grosse,F. (1994) Nuclear DNA helicase II unwinds both DNA and RNA. Biochemistry, 33, 3906–3912. [DOI] [PubMed] [Google Scholar]