Abstract
CCA-adding enzyme [ATP(CTP):tRNA nucleotidyltransferase], a template-independent RNA polymerase, adds the defined ‘cytidine–cytidine–adenosine’ sequence onto the 3′ end of tRNA. The archaeal CCA-adding enzyme (class I) and eubacterial/eukaryotic CCA-adding enzyme (class II) show little amino acid sequence homology, but catalyze the same reaction in a defined fashion. Here, we present the crystal structures of the class I archaeal CCA-adding enzyme from Archaeoglobus fulgidus, and its complexes with CTP and ATP at 2.0, 2.0 and 2.7 Å resolutions, respectively. The geometry of the catalytic carboxylates and the relative positions of CTP and ATP to a single catalytic site are well conserved in both classes of CCA-adding enzymes, whereas the overall architectures, except for the catalytic core, of the class I and class II CCA-adding enzymes are fundamentally different. Furthermore, the recognition mechanisms of substrate nucleotides and tRNA molecules are distinct between these two classes, suggesting that the catalytic domains of class I and class II enzymes share a common origin, and distinct substrate recognition domains have been appended to form the two presently divergent classes.
Keywords: archaeon/crystal structure/evolution/nucleotidyltransferase/tRNA
Introduction
The 3′-terminal CCA sequence (positions 74, 75 and 76) found in all mature tRNAs (Sprinzl and Cramer, 1979) is essential for various aspects of gene expression in all organisms. The CCA sequence is required for the aminoacylation by aminoacyl-tRNA synthetase, and for peptide-bond formation on ribosomes (Green and Noller, 1997; Nissen et al., 2000). The CCA sequence at the 3′ end of tRNAs is repaired and sometimes constructed de novo by a CCA-adding enzyme [ATP (CTP): tRNA nucleotidyltransferase] using CTP and ATP as substrates (Deutscher, 1982). The CCA-adding enzyme is present in all three primary kingdoms (Eubacteria, Eukarya and Archaea), indicating the conservation of its activity throughout evolution (Yue et al., 1996). The CCA-adding enzyme is indispensable in the many organisms in which some or all of the tRNA genes do not encode CCA (Aebi et al., 1990). Otherwise, the activity is advantageous for cell viability in the organisms, in which all of the tRNA genes encode CCA (Zhu and Deutscher, 1987).
The CCA-adding enzyme is a member of the nucleotidyltransferase (NT) family which encompasses enzymes as diverse as poly(A) polymerase (PAP), terminal deoxynucleotidyltransferase (TdT), DNA polymerase β (pol β), glutamine synthase adenylyltransferase and kanamycin nucleotidyltransferase (KNT) (Holm and Sander, 1995; Martin and Keller, 1996; Yue et al., 1996). The CCA-adding enzyme is a remarkable enzyme among the NT family; it synthesizes the ordered CCA sequence at the 3′ end of the tRNA primer without the aid of a nucleic acid template. Furthermore, the enzyme strictly monitors the status of the tRNA 3′ end to reconstruct the complete CCA terminus. Based on biochemical and biophysical studies, several models have been proposed to explain the specific activity of this remarkable RNA polymerase (Deutscher, 1982; Shi et al., 1998a; Yue et al., 1998; Hou, 2000; Li et al., 2000, 2002). However, the details of the CCA addition mechanism have remained elusive for three decades.
The CCA-adding enzymes are divided into two classes, the archaeal CCA-adding enzymes (class I) and the eubacterial/eukaryotic CCA-adding enzymes (class II), based on the sequence alignment (Yue et al., 1996). Although both classes of CCA-adding enzymes catalyze the same reaction in a defined fashion and cross-react with each other’s tRNA substrates, no significant amino acid sequence similarity can be readily identified, except in the regions around the active site signature motif. Recently, the structures of the class II eubacterial and eukaryotic CCA-adding enzymes and/or their complexes with CTP and ATP were reported, revealing the nucleotide recognition mechanism (Li et al., 2002; Augustin et al., 2003). In contrast, the structure of a class I archaeal CCA-adding enzyme has not been solved, and the question remains as to how the class I CCA-adding enzyme recognizes CTP and ATP to catalyze the same reaction performed by the class II CCA-adding enzymes. In the present study, we have determined the crystal structures of the class I archaeal CCA-adding enzyme and its complexes with CTP and ATP. The structures provide significant implications for the mechanism and evolution of the remarkable CCA-adding activity.
Results and discussion
Structure determination of Archaeoglobus fulgidus CCA-adding enzyme
The CCA-adding enzyme from the hyperthermophilic archaeon A.fulgidus (AfCCA) was overexpressed in Escherichia coli and crystallized. The apo structure was initially solved by multiwavelength anomalous dispersion (MAD) using the selenomethionine-labeled protein, and then the native structure was refined to an R factor of 19.8% (Rfree of 24.2%) using reflections up to 2.0 Å resolution. Subsequently, the model was refined against the datasets collected from crystals soaked in CTP or ATP. The structure of the CTP complex was refined to an R factor of 20.9% (Rfree of 24.9%) up to 2.0 Å resolution, while the structure of the ATP complex was refined to an R factor of 20.5% (Rfree of 26.0%) up to 2.7 Å resolution (Table I).
Table I. Summary of data collection and refinement statistics.
| SeMet crystal |
Native | +CTP | +ATP | ||||
|---|---|---|---|---|---|---|---|
| Peak | Edge | Remote1 | Remote2 | ||||
| Data collection | |||||||
| Beam line | BL41XU | BL41XU | BL41XU | BL44XU | |||
| Wavelength (Å) | 0.9792 | 0.9795 | 0.9843 | 0.9753 | 0.9740 | 0.9704 | 1.0000 |
| Resolution (Å) | 50–3.0 | 50–3.0 | 50–3.0 | 50–3.0 | 50–2.0 | 50–2.0 | 50–2.7 |
| Unique reflections | 10 258 | 10 413 | 10 427 | 10 458 | 32 398 | 31 484 | 14 279 |
| Redundancy | 3.5 | 3.5 | 3.5 | 3.5 | 3.2 | 2.7 | 3.4 |
| Completeness (%) | 99.3 (98.5) | 99.0 (96.7) | 99.2 (97.7) | 99.2 (97.9) | 95.6 (87.3) | 90.5 (80.3) | 98.1 (80.7) |
| I/σ(I) | 20.1 (7.6) | 20.6 (7.2) | 20.4 (6.8) | 19.9 (6.1) | 20.0 (3.5) | 12.8 (1.8) | 13.8 (2.5) |
|
Rsym (%) |
6.3 (12.0) |
4.5 (12.1) |
4.2 (11.6) |
5.0 (13.5) |
3.9 (16.8) |
6.3 (35.6) |
5.8 (21.3) |
| Phasing statistics | |||||||
| No. of sites | 5 | 5 | 5 | 5 | |||
| Phasing power | |||||||
| Iso (acent/cent) | 0.42/0.31 | – | 0.72/0.52 | 0.94/0.61 | |||
| Rcullis | |||||||
| Iso (acent/cent) | 0.96/0.93 | – | 0.90/0.88 | 0.87/0.84 | |||
| Ano | 0.74 | 0.89 | 0.84 | 0.98 | |||
| FOM (acent/cent) |
|
0.47/0.41 |
|
|
|
|
|
| Refinement statistics | |||||||
| Resolution (Å) | 50–2.0 | 50–2.0 | 50–2.7 | ||||
| Reflections | 32 398 | 31 484 | 14 279 | ||||
| Protein atoms | 3560 | 3578 | 3578 | ||||
| Water oxygens | 305 | 287 | 121 | ||||
| Rwork (%) | 19.8 | 20.9 | 20.5 | ||||
| Rfree (%) | 24.2 | 24.9 | 26.0 | ||||
| R.m.s.d. bond lengths (Å) | 0.0061 | 0.0177 | 0.0065 | ||||
| R.m.s.d. bond angles (°) | 1.27 | 1.70 | 1.48 | ||||
| R.m.s.d. dihedrals (°) | 21.54 | 22.49 | 22.47 | ||||
| R.m.s.d. impropers (°) | 0.89 | 1.16 | 1.02 | ||||
The numbers in parentheses are for the last shell.
Rsym = Σ |Iavg – Ii|/ ΣIi
Rcullis = Σ||FPH + FP| – FH(calc)|/Σ|FPH|
Rwork = Σ|Fo – Fc|/ΣFo for reflections of work set.
Rfree = Σ|Fo – Fc|/ΣFo for reflections of test set (10% of total reflections).
Overall architecture of Archaeoglobus fulgidus CCA-adding enzyme
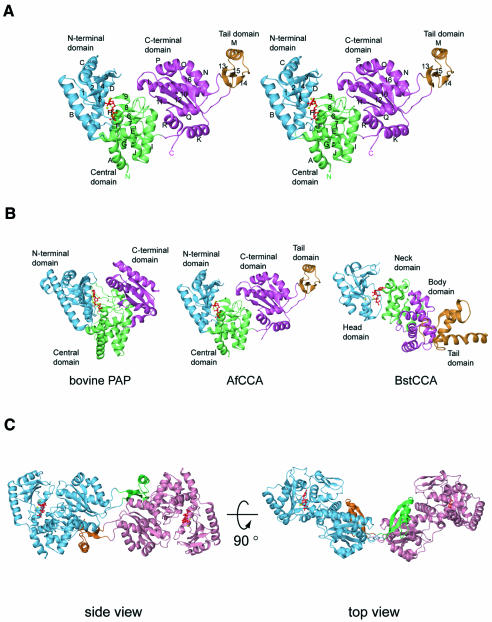
The AfCCA protein structure consists of three main domains: the N-terminal, central and C-terminal domains. An additional tail domain is inserted into, but spatially sequestered from, the C-terminal domain. The overall structure of the class I AfCCA has a U-shape, with a large cleft (∼20 × 16 × 18 Å) surrounded by the three main domains (Figure 1A). The ATP or CTP molecule sits in the interdomain cleft between the N-terminal and central domains (Figure 1A). Strikingly, the overall architecture of AfCCA is fundamentally distinct from the ‘sea-horse’ architecture of the class II CCA-adding enzymes from Bacillus stearothermophilus (BstCCA) (Figure 1B) and human mitochondria (HmtCCA), and is rather similar to that of eukaryotic PAP (Figure 1B). In contrast with PAP, AfCCA exists as a dimer in the crystal, with the subunits related by crystallographic symmetry (Figure 1C). The inserted tail domain of one subunit closely interacts with the tail, central and C-terminal domains of the other subunit, resulting in a buried surface area of 2.5 × 103 Å2. Sedimentation equilibrium and velocity experiments revealed that AfCCA exists as a dimer in solution (Supplementary figure S1A available at The EMBO Journal Online) like the Sulfolobus shibatae CCA-adding enzyme (Li et al., 2000). In contrast, although there are two molecules in the crystal asymmetric unit, class II BstCCA and HmtCCA are likely to exist as monomers, since the buried surface area is small (512 Å2 in BstCCA) and the dimeric arrangements of BstCCA and HmtCCA are completely different. Therefore the oligomeric state is likely to differ between the class I and class II CCA-adding enzymes. An amino acid sequence alignment of the archaeal class I CCA-adding enzymes is shown in Figure 2.
Fig. 1. (A) Stereo view of the A.fulgidus CCA-adding enzyme structure. The N-terminal domain (residues 16–143), the central domain (residues 2–15 and 144–259), the C-terminal domain (residues 260–339 and 384–437) and the tail domain (residues 340–383) are shown in cyan, green, magenta and orange, respectively. The CTP molecule bound to the active site is colored red and the two bound metal ions are shown as yellow spheres. The α-helices and β-strands are designated by letters and numerals, respectively. (B) Comparison of the structures of bovine poly(A) polymerase complexed with 3′-dATP (left), class I A.fulgidus CCA-adding enzyme complexed with CTP (middle) and class II B.stearothermophilus CCA-adding enzyme complexed with ATP (right). The orientations of the catalytic domains are adjusted as in (A). The bound NTPs are colored red. (C) Two views of the A.fulgidus CCA-adding enzyme dimer. The two subunits are colored cyan and pink, with the respective tail domains shown in green and orange, respectively. CTPs and metal ions bound to the active sites are shown as in (A).
Fig. 2. Sequence alignment of the CCA-adding enzymes from five archaea (A.fulgidus, Methanococcus jannaschii, Pyrococcus horikoshii, Methanobacterium thermoautotrophicum and S.shibatae). The sequences of the N-terminal and central domains of bovine poly(A) polymerase were also aligned according to their structural composition by using O (Jones et al., 1991). Blue bold numbers indicate the number of residues not displayed. Important conserved residues are highlighted in red for catalytic carboxylates and in yellow for residues interacting with the nucleotides. Highly conserved residues are in red. The signature sequence that forms the helical-turn motif is surrounded by a blue box. The secondary structure elements of A.fulgidus CCA-adding enzyme are indicated above the alignment, and those of bovine poly(A) polymerase are indicated below the alignment, as chains (α-helices), narrow chains (310 helices), arrows (β-strands) or TT (turns). The four domains in A.fulgidus CCA-adding enzyme are color coded as in Figure 1A. The putative tRNA-binding regions are shown with an orange bar and a blue bar just above the alignment in the subunit that catalyzes the CCA-addition and the other subunit, respectively.
The catalytic site of the nucleotidyltransfer reaction
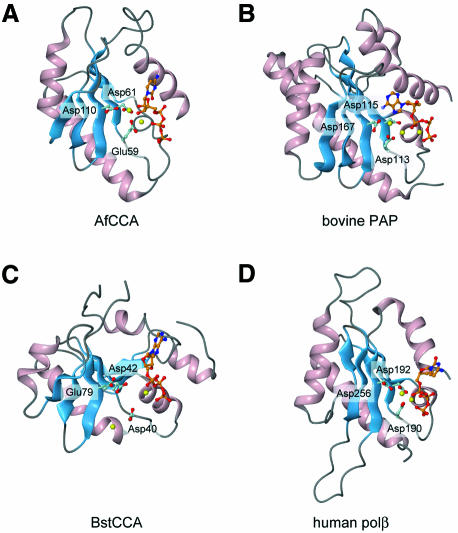
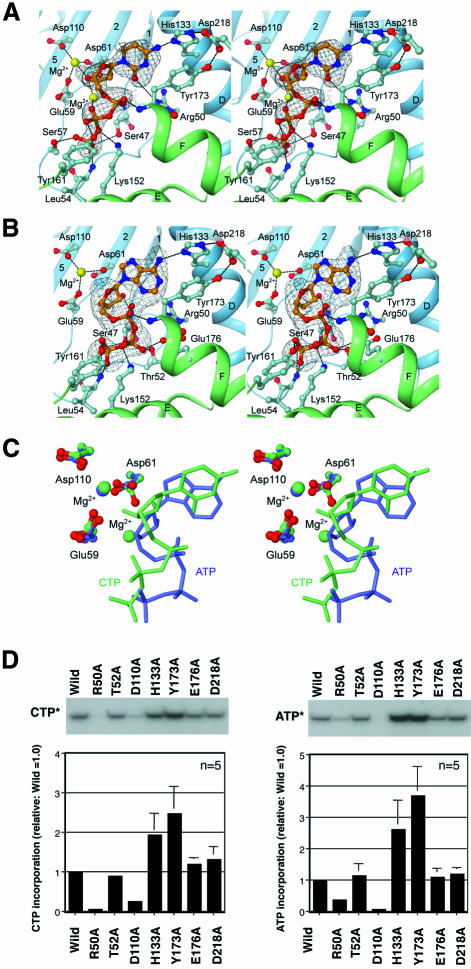
All NT family members, including pol β and other known polynucleotide polymerases, use the ‘two-metal-ion catalytic’ mechanism for the nucleotidyltransfer reaction (Brautigam and Steitz, 1998). In the structures of the known NTs, a metal ion(s) is coordinated in the active site (Figure 3) (Sakon et al., 1993; Pelletier et al., 1994; Bard et al., 2000; Martin et al., 2000). In the present structure of AfCCA, three carboxylates (Glu59, Asp61 and Asp110) are located in close proximity to each other and coordinate the catalytic Mg2+ ion(s) (Figures 3A and 4). In addition to Glu59 and Asp61, which were suggested to be catalytic carboxylates in the class I A.fulgidus CCA-adding enzyme (Asp53 and Asp55 in S.shibatae CCA-adding enzyme) (Yue et al., 1998), Asp110 (corresponding to Asp106 in S.shibatae CCA-adding enzyme) was now found to be the third carboxylate for the NT reaction (Figures 3A and 4). The mutation of Asp110 to Ala in the AfCCA enzyme resulted in a significant decrease in the rate of CMP and AMP incorporation into tRNA-DC (D is the discriminator nucleotide at position 73) and tRNA-DCC, respectively, in vitro (Figure 4D). In contrast, in the class II BstCCA, the third carboxylate has not yet been proved to be functionally important, and in the class II HmtCCA, it was not essential for catalysis (Augustin et al., 2003). In the CTP complex structure, the two Mg2+ ions interact with the three carboxylates in the active site (Figure 4A), while only one Mg2+ ion is bound to the active site in the ATP complex structure (Figure 4B). The three catalytic carboxylates of AfCCA are superimposable onto the corresponding three carboxylates of bovine PAP (Asp113, Asp115 and Asp167) (Figure 3B). The arrangements of the metal ions and the three carboxylates also correspond well to those observed in the structure of the pol β ternary complex (Figure 3D). Although the overall architectures of the class I and class II CCA-adding enzymes are different (Figure 1B), the class I N-terminal domain and the class II head domain have conserved amino acid residues and motifs. Between the two classes of CCA-adding enzymes, the five β-strands and two α-helices, which construct the catalytic core, can be superimposed with an r.m.s.d. of 2.1 Å for 62 corresponding Cα atoms. Moreover, the three carboxylates of BstCCA (Asp40, Asp42 and Glu79) are completely superimposable onto the three carboxylates of AfCCA (Glu59, Asp61 and Asp110) (Figure 3A and C). Therefore the catalytic cores of the class I and class II CCA-adding enzymes share a common ancestor, together with those of PAP and pol β, and catalyze the nucleotidyltransfer reaction by the same mechanism.
Fig. 3. Structural alignment of the conserved catalytic domains of (A) A.fulgidus CCA-adding enzyme (N-terminal domain), (B) bovine poly(A) polymerase (N-terminal domain), (C) B.stearothermophilus CCA-adding enzyme (Head domain) and (D) human pol β (Palm domain). The α-helices and β-strands are colored pink and blue, respectively. Three catalytic carboxylates are displayed. Metal ions are shown as yellow spheres.
Fig. 4. CTP and ATP molecules bound to the active site of A.fulgidus CCA-adding enzyme. Interactions between (A) CTP or (B) ATP and conserved amino acid residues of A.fulgidus CCA-adding enzyme are shown. The secondary structure elements are color coded as in Figure 1A. Hydrogen bonds are shown as dashed black lines. The superimposed simulated annealing omit map (gray) is contoured at 3.0σ. (C) Superimposition of CTP (green) and ATP (blue) bound to the active site of A.fulgidus CCA-adding enzyme. Three conserved carboxylates in the CTP-bound and ATP-bound forms are displayed with green and blue backbones, respectively. (D) The activities of CTP (left) and ATP (right) incorporation into tRNA-DC and tRNA-DCC, respectively, by A.fulgidus CCA-adding enzyme mutants. The bars on the graphs indicate the SDs of five independent experiments.
Recognition of CTP and ATP
Both CTP and ATP are bound at the same pocket in the AfCCA complex structures (Figure 4A and C), underlying the utilization of a single active site pocket for the CCA addition by the class I CCA-adding enzyme (Yue et al., 1998). This was also observed in the structure of the class II CCA-adding enzyme complexed with NTP (Li et al., 2002), despite the different overall architectures between the two classes (Figure 1B). The relative position of the CTP or ATP to the active site in AfCCA is similar to that in BstCCA (Figure 3C). A conserved residue in the helical turn motif located in the N-terminal domain, Ser47, contacts the γ-phosphate of ATP and CTP in AfCCA (Figure 4A and B). This interaction between the Ser47 residue and the γ-phosphate of the incoming nucleotide is commonly observed in the structures of PAP, pol β and KNT. The main-chain amide group of Leu54 provides a hydrogen bond with the γ-phosphate moiety of both CTP and ATP (Figure 4A and B). In addition, Ser47 and Leu54 from the N-terminal domain and Lys152 and Tyr161 from the central domain are also involved in the recognition of the γ- and β-phosphate moieties of both ATP and CTP (Figure 4A and B). These two amino acid residues are strictly conserved among the class I archaeal CCA-adding enzymes and PAPs (Figure 2). The base moieties of CTP and ATP are recognized by the same amino acid residues (Arg50, His133 and Tyr173) (Figure 4A and B). His133, conserved among the class I CCA-adding enzymes (Figure 2), forms a hydrogen bond with either the 6-NH2 group of adenine or the 4-NH2 group of cytosine. The base moieties of CTP and ATP each provide a stacking interaction with Tyr173, which is also conserved among the class I CCA-adding enzymes (Figure 2). In both the ATP and CTP complex structures, Asp218 forms hydrogen bonds with the Nε group of His133 and the OH group of Tyr173 (Figure 4A and B), suggesting that Asp218 stabilizes the spatial arrangement of the two amino acid residues involved in the binding of both CTP and ATP. Another amino acid residue, Arg50, forms a weak hydrogen bond (distance of 3.69 Å) with the O2 atom of cytosine, and is in close enough proximity (distance of 3.91 Å) to interact with the N3 atom of adenine (Figure 4A and B). Among these amino acids interacting with base moieties, only Arg50 is crucial for the in vitro incorporation of CMP and AMP into tRNA-DC and tRNA-DCC, respectively (Figure 4D), while the mutation of His133, Tyr173 or Asp218 affects neither CMP nor AMP incorporation in vitro at low nucleotide concentrations (Figure 4D). The smaller effect of the R50A mutation on AMP incorporation than on CMP incorporation is probably due to the weaker interaction of ATP with Arg50 than that of CTP with Arg50. The Ala mutations of Thr52 and Glu176, which interact with the β phosphate of ATP (Figure 4B), but not CTP, did not affect the NT activity (Figure 4D). In the class II BstCCA, the base moiety of ATP or CTP is commonly recognized by Arg157 and Glu154, located on the neck domain (Li et al., 2002).
Although identical amino acids at a single active pocket recognize both ATP and CTP in class I AfCCA and class II BstCCA, the detailed mechanism of the nucleotide recognition significantly differs between the two enzyme families. First, the triphosphate moiety of ATP or CTP bound to class II BstCCA adopts a common extended conformation. The base moieties have the usual anti conformation. CTP bound to class I AfCCA adopts a similar extended triphosphate conformation, with the base moiety in an anti conformation (Figure 4A). In contrast, the ATP molecule bound to class I AfCCA adopts an unusual syn conformation, which is stabilized by a hydrogen bond between the α-phosphate group and the N3 atom of the adenine moiety (Figure 4B). As a result, the triphosphate moiety of ATP assumes a more stressed, kinked conformation, which may account for the exclusion of the second metal ion, as described above. These unusual conformations of the base and triphosphate moieties enable the 6-amino group and the N3 atom of adenine to be located at spatial positions quite similar to those of the 4-amino group and the O2 atom, respectively, of cytosine (only shifted by 1.01 Å and 0.25 Å, respectively), leading to the recognition of the functional groups by the common His133 and Arg50 residues (Figure 4C). Secondly, the nucleotide substrates are recognized by the N-terminal and central domains of class I AfCCA, while the nucleotides are recognized by only the neck domain of class II BstCCA. Thirdly, a comparison of the CTP-bound and ATP-bound structures of class I AfCCA reveals only slight conformational changes of the nucleotide-recognizing Arg50, His133, Tyr173, Ser47 and Tyr161 residues. This is probably due to the fact that the corresponding functional groups of CTP and ATP, which are recognized by AfCCA, are spatially located at almost the same position, as described above. In contrast, in the structures of the CTP-bound and ATP-bound class II BstCCAs, the 6-amino group and the N1 atom of adenine are shifted from the 4-amino group and the N3 atom of cytosine by 1.79 and 0.79 Å, respectively. Correspondingly, Arg157 and Asp154, which recognize the N1 (ATP) and N3 (CTP) atoms and the 6-amino (ATP) and 4-amino (CTP) groups, respectively, considerably change their conformations, thus exhibiting induced fit by nucleotide binding. On the other hand, as described above, the structures of the ATP and CTP triphosphate moieties significantly differ in AfCCA, such that the 3′OH group of the bound ATP excludes the second Mg2+ ion. In contrast, in the BstCCA structures, the triphosphate structures are almost the same between the ATP-bound and CTP-bound forms. As a result, in AfCCA, the carboxyl side chain of Asp61, which binds to the second Mg2+ ion and is directed toward the 3′OH of CTP (2.65 Å apart), rotates by about 90° to be 3.69 Å away from the 3′OH in the ATP-bound form. Therefore, in class I AfCCA, the 3′OH group of the bound CTP is likely to be more activated to attack nucleophilically the incoming NTP compared with the 3′OH group of the bound ATP, which may be related to the mechanism of the template-independent, but sequence-specific, addition of CCA by the enzyme. In class II BstCCA, this involvement of the active site metal was not identified. In the ATP-bound and CTP-bound forms of class II BstCCA, the 3′OH group of the NTP is more distant from the ‘catalytic triad’; the shortest distance is 5.83 Å, between the 3′OH of CTP and the carboxyl group of Asp42. Therefore, in AfCCA, the bound NTP is located in the catalytically favorable position for the phosphoryl transfer reaction, as compared with the corresponding positions in BstCCA and the eukaryotic PAPs. Thus the detailed mechanisms not only of nucleotide recognition but also of the specific polymerization reaction are likely to differ between the two classes of CCA-adding enzymes. From the structures presented here, the mechanism of switching from CMP incorporation to AMP incorporation, so as to complete the CCA synthesis, is still elusive. A previous biochemical analysis revealed that S.shibatae CCA-adding enzyme still catalyzes CA addition to a tRNA UV crosslinked to the enzyme, and the same tRNA phosphate groups remain in contact with the enzyme during the incorporation of C and A (Shi et al., 1998a). These results indicate that the tRNA is fixed on the enzyme surface during the CCA-adding reaction, and the 3′-end refolds successively to enable C, C and A addition. Considering that a single amino acid (Arg50) is involved in both CMP and AMP incorporation into tRNAs in vitro (Figure 4D), it is likely that either the refolding nucleotide at the 3′ end of the tRNA or the ribonucleoprotein composed of the tRNA 3′ end and enzyme together (Shi et al., 1998a; Li et al., 2002) is involved in switching the specificity via the network of Arg50, His133 and Tyr173. The details of the switching mechanism will await the structure determination of the enzyme, tRNA and nucleotide complexes.
tRNA-docking model
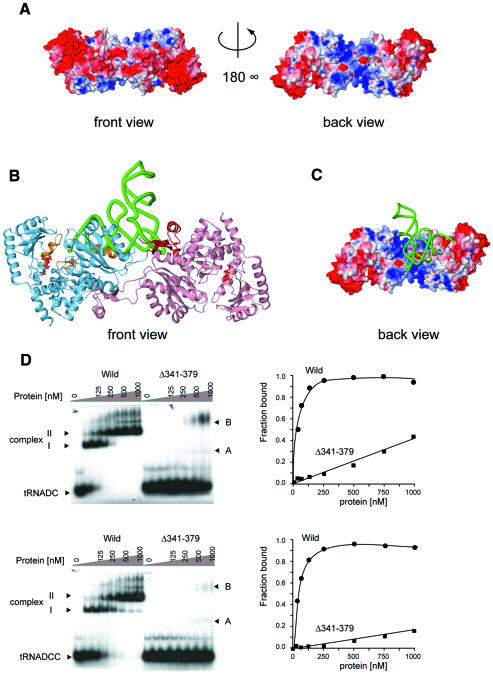
The C-terminal domain of AfCCA consists of a four-stranded antiparallel β-sheet, flanked by one long and six short helices on both sides (Figure 1A). A portion of this domain is topologically similar to the RNA-recognition motif (RRM) of several RNA-binding proteins, such as ribosomal protein S6 [Z score of 5.1 using the Dali server (Holm and Sander, 1998)], and the splicing factor U2AF 35 kDa subunit (Z score of 5.1). The electrostatic surface potential of the dimeric form of AfCCA shows a highly biased distribution of charged residues (Figure 5). The back surface of the central domain and large portions of the C-terminal and tail domains are positively charged. The dimeric form adopts a V-shaped structure (Figure 1C), the inside of which is positively charged and topologically suitable for accomodating tRNAs (Figure 5A). Therefore a plausible tRNA-docking model was built in which one tRNA molecule is bound to the AfCCA dimer, considering that the top-half region of the tRNA body is recognized by the class I CCA-adding enzyme (Shi et al., 1998b) (Figure 5B and C). Such a 2:1 complex of an archaeal CCA-adding enzyme and tRNA was biochemically detected previously (Li et al., 2000): S.shibatae CCA-adding enzyme was shown to form a stable dimer in solution and, upon binding of one tRNA per dimer, the two 2:1 complexes further assemble to form an active 4:2 complex. As described above, AfCCA forms a stable dimer through the interactions of the tail domains of each subunit (Figure 1C). A sedimentation velocity experiment revealed that the deletion of the tail domain (amino acid residues 341–379) mainly generates a monomeric enzyme (Supplementary figure S1), and results in less affinity toward tRNA primers in vitro (Figure 5D). A UV cross-linking experiment also indicated that the tail domain is involved in the tRNA binding (K.Tomita, unpublished data). Furthermore, the rates of CMP and AMP incorporation into tRNA-DC and tRNA-DCC, respectively, are significantly reduced in vitro; the Km values for tRNA in CMP and AMP incorporation are elevated by 30- and 64-fold, respectively (data not shown). Therefore AfCCA dimerization is prerequisite for tRNA binding. The gel shift analysis in Figure 5D showed two complexes, I and II. On the basis of the previous biophysical and biochemical experiment with S.shibatae (Li et al., 2000), complex I represents the 2:1 (enzyme:tRNA) complex and complex II represents the 4:2 complex.
Fig. 5. (A) Two views of the A.fulgidus CCA-adding enzyme dimer showing the surface colored according to its electrostatic potential (blue, positively charged; red, negatively charged). The back view displays the back side of the dimer. (B) Ribbon diagram of the docking model of yeast tRNAPhe and the A.fulgidus CCA-adding enzyme dimer. The tRNA phosphate backbone is displayed as a green tube. The ATP bound to each active site is colored red. The putative tRNA-binding regions are colored in orange and blue, as in Figure 2. The tail domains are colored in red. The orientation is the same as the front view in (A). (C) The back side of the A.fulgidus CCA-adding enzyme dimer, to which yeast tRNAPhe is docked, is shown as in (A). (D) The tRNA [tRNA-DC (upper) and tRNA-DCC (lower)] gel retardation assays of the A.fulgidus wild-type CCA-adding enzyme (circles) and the tail domain deletion mutant [Δ(341–379)] (squares). Two major complexes, denoted as complexes I and II for the wild-type, and complexes A and B for the Δ(341–379) mutant, could be detected. The Kd values of the wild-type CCA-adding enzyme toward tRNA-DC and tRNA-DCC were estimated to be 34 nM and 67 nM, respectively. Those of the Δ(341–379) mutant were estimated to be larger than 1 µM.
The residues conserved in the archaeal class I CCA-adding enzymes are concentrated in three regions: the loop between β3 and β4, the region from β8 to β9 and the N-terminal half of αD (Figures 1 and 2). These three regions are in close proximity to each other, and are not involved in the interactions with either ATP or CTP. In particular, residues 91–95 in the loop connecting β3 and β4 are disordered in the present structures, which implies that the loop between β3 and β4 is flexible but may be fixed upon tRNA binding. Our docking model implies that the loop between β3 and β4, as well as the N-terminal side of αD, interacts with the 3′ strand of the tRNA acceptor end. In the docking model, four other regions may contact the tRNA. The β8–β9 hairpin, with some sequence conservation (Figure 2), is located close to the discriminator base in the model. The long α-helix L in the C-terminal domain, which contains conserved residues on one face of the helix (Figure 2), may lie along and interact with the TΨC-stem of the tRNA. The tail domain may interact with the TΨC-loop so as to monitor the shape of the acceptor TΨC-helix. Intriguingly, it is possible that the region encompassing the end of αG to β7 from the other subunit interacts with the TΨC-loop and D-loop of the tRNA.
In the model, the tRNA molecule docks with the class I AfCCA (from the upper side in Figure 1B) such that the 3′-terminus enters the catalytic cleft from the reverse direction of the triphosphate moiety of the bound NTP. On the other hand, it was suggested that the monomeric eubacterial class II enzyme recognizes the top-half helix of the tRNA (Li et al., 1997) by the body and tail domains of the ‘sea-horse’ structure. Recently, we solved the crystal structure of the complex of the class II enzyme and tRNA (K.Tomita and O.Nureki, in preparation); in the complex, one tRNA molecule approaches the monomeric class II enzyme from the inner side of the ‘sea-horse’ structure (from the lower side in Figure 1B), such that the 3′-terminus enters the catalytic cleft from the same direction as the triphosphate moiety of the bound NTP. Therefore the directions in which tRNA approaches the polymerization domain are reversed between the class I and class II CCA-adding enzymes, which is similar to the situation where the tRNA approaches the catalytic site from the reverse directions between the class I and class II aminoacyl-tRNA synthetase (aaRS) families (Carter, 1993). Concomitantly, in the class I and class II aaRSs, the bound ATP adopts bent and extended conformations, respectively (Arnez and Moras, 1997), which may also be analogous to the present finding that the bound ATP adopts stressed and relaxed conformations, respectively, in the class I and class II CCA-adding enzymes.
Evolutionary implications of class I and class II CCA-adding enzymes
It is surprising that the structures of the two CCA-adding enzyme classes are completely different, except for the catalytic core (Figure 1B), and that the mechanisms of NTP, and probably tRNA, recognition are distinct between these two classes. The class I CCA-adding enzyme is closely related and structurally similar to eukaryotic PAP (Yue et al., 1996). Since phylogenetic analyses have suggested that Archaea are the ancestors of Eukarya, it is likely that the eukaryotic PAP might have descended from the archaeal CCA-adding enzyme. Alternatively, it might be possible that an archaeal PAP, if one exists, might be the origin of the eukaryotic PAP. It should be noted that the eukaryotic PAP is monomeric, with its C-terminal domain involved in RNA binding (Bard et al., 2000; Martin et al., 2000), while the class I CCA-adding enzyme exists as a dimer, which is essential for the recognition of tRNA molecules (Figure 5D) (Li et al., 2000). Therefore the class I CCA-adding enzyme might have evolved by a sophisticated strategy to specifically recognize tRNA molecules as primers. The class II eukaryotic/eubacterial CCA-adding enzyme and eubacterial PAP are closely related (Yue et al., 1996). Eukaryotic CCA-adding enzymes might have been derived from a eubacterial CCA-adding enzyme (Reichert et al., 2001). Recently, it was found that the CCA-adding activity in some ancient and slow-evolving eubacteria represents the CC- and A-adding enzymes (Tomita and Weiner, 2001, 2002). This suggests that the class II CCA-adding enzymes might have been derived from the CC- and A-adding enzymes. The local sequence homology and the structural similarity around the catalytic core between the class I and class II CCA-adding enzymes indicate that both classes of enzymes might have arisen from a common ancestor (Yue et al., 1996). However, the class I and class II CCA-adding enzymes might have undergone entirely independent and divergent evolutionary processes after their separation into their respective classes.
Materials and methods
Preparation and crystallization of the CCA-adding enzyme
The CCA-adding enzyme from A.fulgidus was overexpressed in E.coli strain BL21(DE3) Codon Plus (Stratagene) as a hexahistidine-tagged recombinant protein using the pET-22b expression system (Novagen). The recombinant protein was purified by heat treatment (65°C), followed by Ni2+-chelating, anion-exchange and affinity column chromatography. The protein solution was dialyzed against a buffer containing 20 mM Tris-HCl pH 7.8, 0.3 M KCl, 5 mM MgCl2 and 1 mM dithothreitol (DTT), and was concentrated to 3 mg/ml. The protein solution was mixed with the same volume of crystallization solution containing 45 mM Tris-HCl pH 7.8, 10.8% PEG4000, 135 mM (CH3COO)2Ca, 6.3% 1,6-hexanediol and 10 mM DTT. The drop solution was slowly equilibrated against 400 µl of reservoir solution (40 mM Tris-HCl pH 7.8, 9.6% PEG4000, 120 mM (CH3COO)2Ca, 5.6% 1,6-hexanediol and 0.4 M KCl). Crystals were grown at 20°C by the hanging-drop vapor diffusion method. The selenomethionine derivative was also overexpressed in the methionine auxotrophic E.coli strain B834 (DE3) Codon Plus and was purified as described above. Crystals of the selenomethionine derivative were grown under similar conditions as those for the native crystals.
Data collection, structure determination and refinement
For the structural determination, native and selenomethionine derivative datasets were collected at station BL41XU at SPring-8 (Harima, Japan). Flash-frozen crystals were cryoprotected with 20% (v/v) ±-2-methyl-2,4-pentanediol. All the datasets were processed with the programs DENZO and SCALEPACK (Otwinowski and Minor, 1997). The native crystal belongs to the space group C2, with a = 87.4 Å, b = 76.5 Å, c = 76.8 Å and β = 98.5°. X-ray fluorescence spectra of the selenomethionine derivative crystal were also measured at station BL41XU at SPring-8. The scaled dataset at the peak wavelength of the MAD experiment up to 3.0 Å was used to locate the selenium atoms, using the program SnB (Weeks and Miller, 1999). After several trials of SnB, 5 consistent peaks were picked out of the six atoms expected in the asymmetric unit. Subsequent phase refinements and calculations were carried out with the program MLPHARE (CCP4, 1994) using reflections up to 3.0 Å. The resultant phases were further improved and extended up to 2.6 Å resolution by solvent flattening using the RESOLVE (Terwilliger, 2000) or DM (CCP4, 1994) program. In the resultant electron density maps, we can locate one CCA-adding enzyme molecule in one asymmetric unit. The model was built using the program O (Jones et al., 1991), and was refined against the reflections from the native crystal up to 2.0 Å resolution using the program CNS (Brunger et al., 1998) by rigid-body refinement, energy minimization and simulated annealing. The Ramachandran plot analysis of the final model with the program PROCHECK (Laskowski et al., 1993) showed that 98.9% of the residues in the present structure are in the most favored and additionally allowed regions.
Preparation of mutant AfCCA proteins and in vitro CCA-adding assay
Each mutation (R50A, T52A, E110A, H133R, Y173A, E176A, D218A) was introduced into the AfCCA overexpression plasmid by using the Quickchange Mutagenesis kit (Stratagene) according to the manufacturer’s instructions. The deletion mutant plasmid (Δ341–379) was also constructed in the same way. The mutant AfCCA proteins were overexpressed and purified as described above. Substrate tRNAs lacking the terminal CA or A (tRNA-DC or tRNA-DCC; D is the discriminator nucleotide at position 73) were prepared from the linearized pmBSDCCA plasmid as described (Oh and Pace, 1994). The CCA-adding assay was carried out under the challenging conditions (Yue et al., 1998) in a buffer containing 50 mM glycine–NaOH pH 8.5, 10 mM MgCl2, 2 mM DTT, 4 µM CTP (or ATP), 100 nM [α-32P]CTP (or [α-32P]ATP) (3000 Ci/mmol), 0.25 µM tRNA-DC (or tRNA-DCC) and 5 nM enzyme at 50°C for 5 min. The Km values for CTP and ATP were 50 and 240 µM, respectively. Therefore the assay conditions using a nucleotide concentration of 4 µM were considered to be sensitive enough for the evaluation of mutant activities. Under these conditions, the reactions are in the linear range. The reaction was stopped by adding an equal volume of stop solution (9 M urea, 0.002% BPB, 0.002% XC), and the products were separated on a 12% polyacrylamide gel containing 7 M urea. The 32P-labeled products were quantified by a BAS-2000 Image analyzer (Fuji-film).
Gel-retardation assay
tRNA-DC or tRNA-DCC uniformly labeled with [α-32P]UTP (3000 Ci/mmol) was prepared from the linearized pmBSDCCA plasmid by using an in vitro transcription kit (Promega) according to the manufacturer’s instructions. Aliquots (10 µl) of solutions containing 50 mM glycine–NaOH pH 8.5, 10 mM MgCl2, 2 mM DTT, 1.25 fmol 32P-labeled tRNA-DC (or tRNA-DCC) and various amounts of protein (wild or Δ341–379; 0–10 pmol) were incubated at room temperature for 10 min and then cooled on ice. After 1 µl of 0.02% BPB was added, the solution was separated by 5% native polyacrylamide gel electrophoresis (1 × TBE) in a cold room (4°C). The fraction of the total shifted band was quantified by a BAS-2000 Image analyzer.
Accession numbers
The coordinates and structure factors have been deposited in the Protein Data Bank (accession codes 1UET, 1UEU and 1UEV)
Supplementary data
Supplementary data are available at The EMBO Journal Online.
Acknowledgments
Acknowledgements
We thank Alan M.Weiner for the AfCCA overexpression plasmid. We thank M.Kawamoto and H.Sakai (JASRI) for their help in data collection at SPring-8. This work was supported by a PRESTO Program grant from JST (Japan Science and Technology) to O.N., the Kurata Memorial Hitachi Science and Technology Foundation, the Takeda Science Foundation and a Grant-in-Aid for Scientific Research for Young Scientists (K.T.). K.T. is a research associate supported by a grant from the Ministry of Education, Culture, Sports, Science and Technology to N.T.
References
- Aebi M., Kirchner,G., Chen,J.Y., Vijayraghavan,U., Jacobson,A., Martin,N.C. and Abelson,J. (1990) Isolation of a temperature-sensitive mutant with an altered tRNA nucleotidyltransferase and cloning of the gene encoding tRNA nucleotidyltransferase in the yeast Saccharomyces cerevisiae. J. Biol. Chem., 265, 16216–16220. [PubMed] [Google Scholar]
- Arnez J.G. and Moras,D. (1997) Structural and functional considerations of the aminoacylation reaction. Trends Biochem. Sci., 22, 211–216. [DOI] [PubMed] [Google Scholar]
- Augustin M.A., Reichert,A.S., Betat,H., Huber,R., Mörl,M. and Steegborn,C. (2003) Crystal structure of the human CCA-adding enzyme: insights into template-independent polymerization. J. Mol. Biol., 328, 985–994. [DOI] [PubMed] [Google Scholar]
- Bard J., Zhelkovsky,A.M., Helmling,S., Earnest,T.N., Moore,C.L. and Bohm,A. (2000) Structure of yeast poly(A) polymerase alone and in complex with 3′-dATP. Science, 289, 1346–1349. [DOI] [PubMed] [Google Scholar]
- Brautigam C.A. and Steitz,T.A. (1998) Structural and functional insights provided by crystal structures of DNA polymerases and their substrate complexes. Curr. Opin. Struct. Biol., 8, 54–63. [DOI] [PubMed] [Google Scholar]
- Brunger A.T. et al. (1998) Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr. D, 54, 905–921. [DOI] [PubMed] [Google Scholar]
- Carter C.W. Jr (1993) Cognition, mechanism and evolutionary relationships in aminoacyl-tRNA synthetases. Annu. Rev. Biochem., 62, 715–748. [DOI] [PubMed] [Google Scholar]
- CCP4 (1994) The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D, 50, 760–763. [DOI] [PubMed] [Google Scholar]
- Deutscher M.P. (1982) tRNA nucleotidyltransferase. Enzymes, 15, 183–215. [Google Scholar]
- Green R. and Noller,H.F. (1997) Ribosomes and translation. Annu. Rev. Biochem., 66, 679–716. [DOI] [PubMed] [Google Scholar]
- Holm L. and Sander,C. (1995) DNA polymerase beta belongs to an ancient nucleotidyltransferase superfamily. Trends Biochem. Sci., 20, 345–347. [DOI] [PubMed] [Google Scholar]
- Holm L. and Sander,C. (1998) Touring protein fold space with Dali/FSSP. Nucleic Acids Res., 26, 316–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou Y.M. (2000) Unusual synthesis by the Escherichia coli CCA-adding enzyme. RNA, 6, 1031–1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones T.A., Zou,J.Y., Cowan,S.W. and Kjeldgaard. (1991) Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A, 47, 110–119. [DOI] [PubMed] [Google Scholar]
- Laskowski R.A., MacArthur,M.W., Moss,A.L. and Thornton,J.M. (1993) PROCHECK:a program to check the stereochemical quality of protein structures J. Appl. Crystallogr., 26, 283–291. [Google Scholar]
- Li F., Wang,J. and Steitz,T.A. (2000) Sulfolobus shibatae CCA-adding enzyme forms a tetramer upon binding two tRNA molecules: a scrunching-shuttling model of CCA specificity. J. Mol. Biol., 304, 483–492. [DOI] [PubMed] [Google Scholar]
- Li F., Xiong,Y., Wang,J., Cho,H.D., Tomita,K., Weiner,A.M. and Steitz,T.A. (2002) Crystal structures of the Bacillus stearothermophilus CCA-adding enzyme and its complexes with ATP or CTP. Cell, 111, 815–824. [DOI] [PubMed] [Google Scholar]
- Li Z., Sun,Y. and Thurlow,D.L. (1997) RNA minihelices as model substrates for ATP/CTP:tRNA nucleotidyltransferase. Biochem. J., 327, 847–851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin G. and Keller,W. (1996) Mutational analysis of mammalian poly(A) polymerase identifies a region for primer binding and catalytic domain, homologous to the family X polymerases, and to other nucleotidyltransferases. EMBO J., 15, 2593–2603. [PMC free article] [PubMed] [Google Scholar]
- Martin G., Keller,W. and Doublie,S. (2000) Crystal structure of mammalian poly(A) polymerase in complex with an analog of ATP. EMBO J., 19, 4193–4203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nissen P., Hansen,J., Ban,N., Moore,P.B. and Steitz,T.A. (2000) The structural basis of ribosome activity in peptide bond synthesis. Science, 289, 920–930. [DOI] [PubMed] [Google Scholar]
- Oh B.K. and Pace,N.R. (1994) Interaction of the 3′-end of tRNA with ribonuclease P RNA. Nucleic Acids Res., 22, 4087–4094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otwinowski Z. and Minor,W. (1997) Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol., 276, 307–326. [DOI] [PubMed] [Google Scholar]
- Pelletier H., Sawaya,M.R., Kumar,A., Wilson,S.H. and Kraut,J. (1994) Crystal structure of rat DNA polymerase beta: evidence for a common polymerase mechanism. Science, 264, 1930–1935. [DOI] [PubMed] [Google Scholar]
- Reichert A.S., Thurlow,D.L. and Mörl,M. (2001) A eubacterial origin for the human tRNA nucleotidyltransferase? Biol. Chem., 382, 1431–1438. [DOI] [PubMed] [Google Scholar]
- Sakon J., Liao,H.H., Kanikula,A.M., Benning,M.M., Rayment,I. and Holden,H.M. (1993) Molecular structure of kanamycin nucleotidyltransferase determined to 3.0-Å resolution. Biochemistry, 32, 11977–11984. [DOI] [PubMed] [Google Scholar]
- Shi P.Y., Maizels,N. and Weiner,A.M. (1998a) CCA addition by tRNA nucleotidyltransferase: polymerization without translocation? EMBO J., 17, 3197–3206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi P.Y., Weiner,A.M. and Maizels,N. (1998b) A top-half tDNA minihelix is a good substrate for the eubacterial CCA-adding enzyme. RNA, 4, 276–284. [PMC free article] [PubMed] [Google Scholar]
- Sprinzl M. and Cramer,F. (1979) The –C-C-A end of tRNA and its role in protein biosynthesis. Prog. Nucleic Acid Res. Mol. Biol., 22, 1–69. [DOI] [PubMed] [Google Scholar]
- Terwilliger T.C. (2000) Maximum-likelihood density modification. Acta Crystallogr. D, 56, 965–972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomita K. and Weiner,A.M. (2001) Collaboration between CC- and A-adding enzymes to build and repair the 3′-terminal CCA of tRNA in Aquifex aeolicus. Science, 294, 1334–1336. [DOI] [PubMed] [Google Scholar]
- Tomita K. and Weiner,A.M. (2002) Closely related CC- and A-adding enzymes collaborate to construct and repair the 3′-terminal CCA of tRNA in Synechocystis sp. and Deinococcus radiodurans. J. Biol. Chem., 277, 48192–48198. [DOI] [PubMed] [Google Scholar]
- Weeks C.M. and Miller,R. (1999) The design and implementation of SnB version 2.0. J. Appl. Crystallogr., 32, 120–124. [Google Scholar]
- Yue D., Maizels,N. and Weiner,A.M. (1996) CCA-adding enzymes and poly(A) polymerases are all members of the same nucleotidyltransferase superfamily: characterization of the CCA-adding enzyme from the archaeal hyperthermophile Sulfolobus shibatae. RNA, 2, 895–908. [PMC free article] [PubMed] [Google Scholar]
- Yue D., Weiner,A.M. and Maizels,N. (1998) The CCA-adding enzyme has a single active site. J. Biol. Chem., 273, 29693–29700. [DOI] [PubMed] [Google Scholar]
- Zhu L. and Deutscher,M.P. (1987) tRNA nucleotidyltransferase is not essential for Escherichia coli viability. EMBO J., 6, 2473–2477. [DOI] [PMC free article] [PubMed] [Google Scholar]