SUMMARY
Hox genes control the anterior-posterior patterning of most metazoan embryos. Their sequential expression is initially established by the segmentation gene cascade in the early Drosophila embryo [1]. The maintenance of these patterns depends on the Polycomb group (PcG) and trithorax group (trxG) complexes during the remainder of the life cycle [2]. We provide both genetic and molecular evidence that the Hox genes are subject to an additional tier of regulation, i.e. at the level of transcription elongation. Both Ultrabithorax (Ubx) and Abdominal-B (Abd-B) genes contain stalled or paused RNA Polymerase II (Pol II) even when silent [3, 4]. The Pol II elongation factors Elongin-A and Cdk9 are essential for optimal Ubx and Abd-B expression. Mitotic recombination assays suggest that these elongation factors are also important for the regulation of Notch, EGF, and Dpp signaling genes. Stalled Pol II persists in tissues where Ubx and Abd-B are silenced by the PcG complex. We propose that stalling fosters both the rapid induction and precise silencing of Hox gene expression during development.
RESULTS AND DISCUSSION
Recent studies suggest that the regulation of Pol II elongation might be a common feature of developmental gene control in the Drosophila embryo. ChIP-chip assays in cultured cell lines suggest that a significant fraction of all protein coding genes contain stalled Pol II [3, 5]. As many as 10% of all protein coding genes in the early Drosophila embryo contain Pol II prior to their expression [3, 5]. Many of these genes are developmental control genes, such as those encoding components of cell signaling pathways including Wnt, FGF, and Dpp (TGFβ). Moreover, four of the eight Hox genes in Drosophila appear to contain stalled Pol II (lab, Antp, Ubx, and Abd-B) in the early embryo [3]. Here we investigate the role of Pol II elongation factors in Hox gene expression.
Stalled Pol II at the Ubx and Abd-B loci
To confirm the preliminary evidence for stalled Pol II at the Ubx and Abd-B loci (Fig. 1A), conventional ChIP assays were performed using different antibodies against Pol II, namely, 8WG16, which recognizes the CTD of Pol II, and H14, which recognizes the initiating form (Ser-5 phosphorylation) of Pol II [6]. Both of these antibodies have been used in earlier ChIP [7] as well as ChIP-chip [3] assays to elucidate and map distinct functions of the Pol II complex. Chromatin cross-linking was performed on 0–2 hr wild-type embryos, prior to the onset of Hox gene expression. The chromatin was sonicated, precipitated with anti-Pol II antibodies, and then the extracted DNA was used as a template for PCR amplification (Fig. 1B). Hsp70 was used as a control since it represents the prototypic example of paused Pol II [4, 8]. As expected, the hsp70 promoter region contains strong Pol II signals with both the 8WG16 and H14 antibodies, indicating that an initiated Pol II is bound to the hsp70 promoter prior to heat shock induction. The Ubx and Abd-B promoter regions also exhibit strong signals, whereas PCR amplification performed with exonic probes failed to detect Pol II binding within the main body of the transcription unit (Fig. 1B). The presence of the H14 signal at these promoters suggests that Ser5 of the Pol II CTD is phosphorylated (initiated Pol II) prior to the activation of Ubx and Abd-B expression. As predicted from the previous ChIP-chip assays (Fig. 1A; ref. 3), the abd-A promoter region lacks Pol II.
Figure 1. Ubx and Abd-B are paused in the early embryo.
(A) ChIP-chip assay revealed that Ubx and Abd-B genes have Pol II signal (shown by arrow) at their promoters in inactive conditions in the early 2–4h embryos. While abd-A does not display a Pol II signal at its promoter region. (B) Conventional ChIP assays, followed by PCR, using Pol II antibodies [8WG16 against CTD of Pol II and H14 against Ser5 phosphorylated CTD of Pol II (initiated Pol II)] were performed on 0–2h embryos and visualized by gel electrophoresis. Abd-B and Ubx genes are inactive in the early embryos (0–2h) but Pol II signals were observed at the promoter regions of both the genes (rows 1 and 3 respectively). The exonic regions of Abd-B and Ubx do not show any signal for Pol II binding (rows 2 and 4 respectively). Hsp70 promoter was used as a positive control and displayed signal for Pol II as expected (row5) while abd-A promoter which was shown to be non-paused in early embryos does not show any Pol II signal at its promoter (row 6).
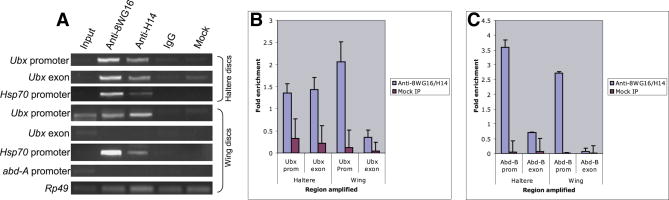
The preceding studies suggest that Ubx and Abd-B contain a stalled form of Pol II in early embryos. Additional assays were done to investigate Pol II binding in wing and haltere imaginal discs (Fig. 2A). The hsp70 promoter region contains strong Pol II signals in both wing and haltere discs, consistent with previous studies suggesting that the gene is stably paused in most or all tissues prior to induction by heat shock [9]. The ChIP assays also identify strong Pol II signals in the Ubx promoter region of wing discs (Fig. 2A, B), where the gene is silenced by the PcG complex [10]. In contrast, a probe directed against exon 1 failed to detect significant levels of Pol II within the main body of the transcription unit (Fig. 2A, B).
Figure 2. Ubx and Abd-B genes show promoter-proximal stalling of RNA Pol II in wing and imaginal discs tissues.
The Pol II ChIP was performed on wing and haltere imaginal discs, followed by conventional PCR and QPCR analysis. IP was done using a cocktail of antibody recognizing CTD of Pol II (8WG16) and Ser5 phosphorylated CTD of Pol II (H14) as well as each of them alone. (A) In the haltere discs Ubx gene is active and Pol II shows signals along the promoter (row 1) as well as exon regions (row 2), while in the wing discs it shows signal at the promoter region (row 4) and no signal at the exon (row5). Thus, supporting the fact that even when Ubx is inactive in the wing discs it still shows Pol II binding at the promoter regions and may be stalled/paused. The hsp70 gene shows Pol II signals at both haltere discs (row3) and wing discs (row 6). The abd-A promoter does not show Pol II signal in wing disc and was used as negative control (row 7) and rp49 was used as loading control (row8). (B) QPCRs using cocktail of Pol II antibody (8WG16 and H14) reveal signals of Pol II on Ubx promoter in wing discs, while haltere discs show similar signals of Pol II in promoter and exon regions. (C) Abd-B gene is known to be inactive in haltere as well as wing discs. QPCRs display enrichment of Pol II binding at the proximal-promoter region when compared to the exon in wing as well as haltere imaginal discs.
Very different results were obtained with haltere discs, where Ubx is strongly expressed and the resulting Ubx repressor inhibits wing development. In this case, strong Pol II signals are detected in both the promoter region and exon, as would be expected for an actively expressed gene (Fig. 2A). These findings were strengthened by the use of qPCR assays (Fig. 2B). For these experiments, ChIP assays were done with a cocktail of Pol II antibodies (both 8WG16 and H14), as described previously [3]. Pol II signals are detected in both the promoter region and exon of the Ubx locus in haltere discs, where the gene is active. In contrast, there are substantially higher levels of Pol II in the promoter region than exon in wing discs where Ubx is silent (Fig. 2B). Permanganate protection assays are consistent with the occurrence of paused Pol II located between +18 and +35 bp downstream of the Ubx transcription start site (see Supp. Fig. 1).
Abd-B also exhibits higher levels of Pol II binding in the promoter region as compared with exon 1 (Fig. 2C). However, unlike Ubx, Abd-B is silent in both the wing and haltere discs [11], so it is not surprising that Pol II is not significantly detected in exon 1 in either tissue. As seen in early embryos, the promoter region of abd-A lacks significant binding of Pol II in wing discs (Fig. 2A; compare with 1A).
Genetic interactions with Pol II elongation factors
Pol II stalling raises the possibility that Ubx might be regulated at the level of transcriptional elongation. A number of elongation factors have been identified in cell culture assays, including NELF (Negative Elongation Factors) A–E, ELONGIN-A (Elo-A), SPT (suppressor of termination) 4/5, and CDK9 (Cyclin dependant kinase 9) [4]. Reduced levels of Ubx+ activity cause a slight transformation of halteres into wings since Ubx functions as a repressor of wing development in the halteres [1]. We reasoned that if Ubx is regulated at the level of Pol II elongation, then reduced levels of critical elongation factors should enhance the patterning defects observed in weak Ubx mutants (Fig. 3).
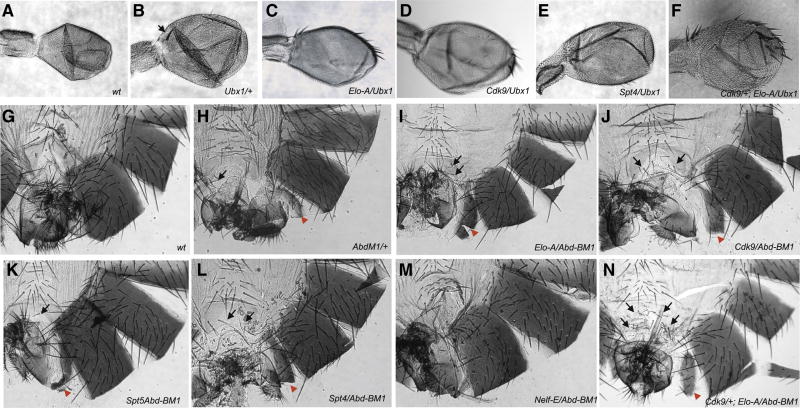
Figure 3. Enhancement and suppression of Ubx and Abd-B phenotypes by elongation factor mutants.
The dominant Ubx1 allele (B) which shows partial haltere to wing transformation [(slightly large halteres and presence of ectopic wing like britstle at the base of stalk (arrow) when compared to wt haltere (A)] was used for enhancement-suppression assay against the elongation mutants. In (C) Elo-A and (D) Cdk9 backgrounds the haltere to wing transformation was enhanced in 100% of transheterozygotes (as can be seen by increase in size and number of wing-like bristles in the haltere). The haltere to wing phenotype was partially suppressed in 100% of the transheterozygotes in (E) Spt4 background (as loss of prominent wing-like bristle along the leading edge of haltere occurs). To investigate if the elongation factors act together to control Ubx gene transcription a triple mutant fly was generated Cdk9/+; Elo-A/Ubx (F) and severe haltere to wing transformation (increased wing-like bristles and overall size of haltere) was observed in 100% of the triple mutants. The haplo-insufficient anteriorization phenotype of partial A7 to A6 (presence of small pigmented segmented below A6, shown by red arrow) and A6 to A5 transformation (presence of bristles in A6 sternite, shown by black arrows) in Abd-BM1 allele (H compare to G) was used as benchmark for looking for its interaction with the elongation factor mutants. Elongation factors like Elo-A (I), Cdk9 (J), Spt5 (K), Spt4 (L) all enhanced the partial A7 to A6 (increased the small pigmented segment size, shown by red arrows) and A6 to A5 (more bristles in A6 sternite, shown by black arrows) in 100% of the transheterozygotes suggesting its role in facilitating transcription of Abd-B gene. The Nelf-E mutation suppressed the dominant Abd-B phenotype (M compare to G) and made all the transheterozygotes look like wild type files (as seen by loss of A7 to A6 and A6 to A5 transformations) suggesting that it is bringing about negative regulation of Abd-B gene. The triple mutant Cdk9/+; Elo-A/Abd-BM1 (N) flies show severe enhancement of Abd-B phenotype [which is reflected by increased bristles in A6 sternite (black arrows) and stronger A7 to A6 transformation (red arrow)].
We specifically examined mutations in four different elongation factors, Elo-A, Cdk9, Spt4, and Spt5. Cdk9 has been shown to be a critical activator of paused Pol II at the hsp70 promoter [4]. Heterozygotes for each mutation were examined in a Ubx1/+ background, which displays a weak expansion of the halteres (Fig. 3B; compare with A). Elo-A/+; Ubx1/+ double heterozygotes display an enhanced transformation of halteres into wings (Fig. 3C). In particular, several wing-like bristles appear at the leading margin of the halteres. A similar phenotype was observed for Cdk9/+; Ubx1/+ double heterozygotes (Fig. 3D). Spt4 mutation cause a slight suppression of the Ubx1/+ phenotype, consistent with their dual activities in both attenuating and augmenting Pol II elongation (e.g., Fig. 3E).
Cdk9 and Elo-A are thought to regulate distinct aspects of Pol II elongation. The Cdk9 kinase phosphorylates Ser-2 of the Pol II CTD, which is critical for the release of Pol II from the pause site in the hsp70 promoter. Inhibition of Cdk9 activity causes a global reduction in Ser-2 phosphorylation [12]. In contrast, Elo-A appears to act at a later point of Pol II elongation, after release from the pause site [4]. Mutations in Cdk9 and Elo-A cause an additive enhancement in the Ubx1/+ phenotype (Fig. 3F). Triple heterozygotes display an expansion in the overall size of the haltere, and the anterior margin contains a series of bristles like those seen in wings. This phenotype suggests that diminished levels of Cdk9 and Elo-A cause significant reductions in Ubx+ activity.
ChIP-chip and conventional ChIP assays suggest that the Abd-B promoter region might also contain a stalled form of Pol II (see Figs. 1 and 2). As seen for Ubx, reduced levels of Cdk9 and Elo-A cause significant enhancements in the Abd-BM1/+ mutant phenotype (Fig. 3G–N). In particular, Abd-BM1/+ heterozygotes display a weak transformation of posterior abdominal segments into anterior segments, particularly the seventh abdominal segment (A7) into A6 (ectopic partial pigmentation) and A6 to A5 (ectopic bristles in A6 sternite) [13, 14] (Fig. 3H; compare with G). These phenotypes are augmented by reductions in either Cdk9 or Elo-A activity. Double heterozygotes display a more complete A7 to A6 transformation, as well as an increase in the number of bristles in A6 suggesting a more severe A6 to A5 transformation (Fig. 3I, J). These segmental transformations are weakly enhanced (not suppressed) by lower levels of Spt4 and Spt5 (Fig. 3K, L). In contrast, mutations in the negative elongation factor Nelf-E strongly suppress the Abd-BM1 phenotype (Fig. 3M), which is consistent with enhanced transcription of Abd-B. Triple heterozygotes, Abd-BM1/+; Cdk9/+; Elo-A/+, display an even more dramatic transformation of A7 to A6 and A6 to A5. Thus, as seen for Ubx, reduced levels of Cdk9 and Elo-A cause a significant diminishment in Abd-B+ gene activity (Fig. 3N).
Cdk9 and Elo-A mutations produce classical patterning defects
Stalled Pol II appears to be disproportionately associated with developmental control genes as compared with “housekeeping” genes that control cell metabolism and proliferation [e.g., 3, 5]. A substantial fraction of stalled genes exhibit localized patterns of expression during embryogenesis, such as Hox genes and genes encoding components of signaling pathways (e.g., Dpp, FGF, Notch, etc.) [3]. We therefore explored the possibility that elimination of Cdk9 and Elo-A activity via the production of mitotic clones might produce specific developmental defects in adult appendages. In these experiments there is no perturbation of Ubx or Abd-B activity. Cdk9 and Elo-A activities are disrupted in an otherwise wild-type background.
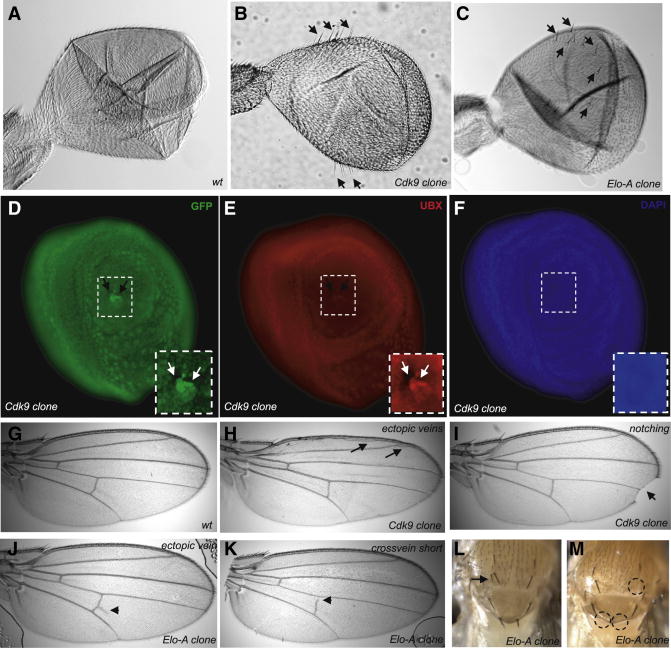
The localized loss of Cdk9 or Elo-A activity in the haltere discs leads to weak wing transformation phenotypes, similar to those seen for reductions in Ubx (Fig. 4B,C; compare with A). In particular, there is an expansion in the size of the halteres, and wing-like bristles appear at the margins. At least some of these phenotypes appear to arise from the specific loss of Ubx expression (Fig. 4D–F). Haltere discs containing clonal patches of Cdk9−/Cdk9− tissue (identified by the loss of GFP expression) display localized reductions in Ubx activity, as judged by the use of an anti-Ubx antibody (Fig. 4E). This observation suggests that Ubx transcription is particularly sensitive to diminished activities of Pol II elongation factors, which is consistent with the evidence that the Ubx promoter region contains stalled Pol II.
Figure 4. Mitotic clones of Cdk9 and Elo-A display patterning phenotypes.
The mitotic clones of Cdk9 and Elo-A was generated using standard techniques and was screened for Ubx phenotype (haltere to wing transformation) in the adult flies. Mitotic clones of Cdk9 (B) and Elo-A (C) in adult haltere lead to transformation of haltere to wings (as seen by emergence of wing-like bristles) that is absent in wt haltere (A). Haltere imaginal discs containing Cdk9 clones [seen by using GFP as marker. Note the Cdk9−/− clone marked by no GFP and twin spot marked by GFP+/+ in a background of GFP+/−. (D)] lack Ubx expression [seen by anti-UBX staining in red. Note the loss of UBX staining from the Cdk9−/minus; clone and normal staining of Ubx in GFP+/+ clones (E)]. DAPI (F) was used to mark cell nuclei in the haltere imaginal discs. The adult mitotic clones of Cdk9 and Elo-A display phenotypes associated with signaling mutants. Cdk9 clones show ectopic wing veins (H), notched wings (I). Elo-A mitotic clones display similar signaling phenotypes of ectopic wing veins (J), short wing cross-veins (K), duplication of macrochaete in adult notum (L, shown by arrow) and loss of macrochaete from notum region (M, shown by dashed circles). All these phenotypes suggest perturbations in Notch, EGF, and Dpp (TGFβ) signaling
Cdk9 and Elo-A mitotic clones produce a variety of patterning defects in the wing and notum (Fig. 4G–M, Supp. Fig. 2C). Most notably, there is notching of the wing margins (Fig. 4I), ectopic wing veins (Fig. 4H, J), short crossveins (Fig 4K) and both losses and duplications of macrochaete in the notum (Fig. 4L, M). These phenotypes might arise from perturbations in Notch, EGF, and Dpp (TGFβ) signaling (also see Supp. Fig. 2C). Genes encoding components of each of these pathways appear to contain stalled Pol II in early embryos [3].
We presented evidence that the elongation factors Cdk9 and Elo-A are essential for optimal expression of at least a subset of Drosophila Hox genes, particularly Ubx+ activity in the developing halteres. Small patches of Elo-A−/Elo-A− or Cdk9−/Cdk9− mutant tissue also cause specific patterning defects in the wings and notum (Fig. 4). Both Pol II elongation factors are probably required for normal expression of a great number of genes in the Drosophila genome. Indeed, both elongation genes are essential and every attempt to create large mitotic clones resulted in larval lethality. Such lethality presumably reflects the general role of Elo-A and Cdk9 in gene expression. Previous studies have documented the general importance of the elongation factors ELL and Elo-A in Drosophila larval development and metamorphosis [15, 16]. Nonetheless, it would appear that a small number of patterning genes, including Ubx, are particularly sensitive to the loss of Elo-A and Cdk9 activity.
It has been extensively argued that Polycomb might mediate repression by propagating an inactive form of chromatin, for example, by methylation of H3K27 followed by recruitment of HP1 or other proteins that package chromatin in an inactive state [2]. However, the demonstration that TBP and Pol II are present in the Ubx proximal promoter in wing imaginal discs suggests that PcG silencing does not render the chromatin inaccessible for the binding of even large protein complexes [11]. Instead, we propose that paused Pol II could contribute to PcG silencing by excluding the binding of additional Pol II complexes. Such occlusion by steric hindrance might help reduce transcriptional noise and thereby maintain Ubx repression. Mutations in the elongation factor, ELL [Su(Tpl)], suppress Scr phenotypes caused by the Pc4 Polycomb mutant [15, 17], raising the possibility that Pol II elongation factors somehow communicate with the PcG silencing complex. We propose that stalling might serve the dual role of fostering both silencing and rapid induction, and thereby provide a sharp on/off switch in Hox regulation.
EXPERIMENTAL PROCEDURES
Fly crosses
The flies were constantly raised at 25°C and trans-heterozygous crosses were made by crossing balanced stocks and scoring for flies without any balancers. The elongation factor mutations were obtained from Bloomington stock center and were generated either by the Exilexis or DrosDel projects. The stocks which were used in genetic studies are y1 w1118; Spt5MGE-3/SM1; Psn143/TM6B (Spt5), y[1] w[67c23]; P{w[+mC]=lacW}spt4[k05316]/CyO (Spt4), y[1] w[67c23]; y[1] w[67c23]; P{w[+mC] y[+mDint2]=EPgy2}EY07065/TM3, Sb[1] Ser[1] (Nelf-E), c00768/TM6b,Tb (Nelf-E), w[1118]; Df(3R)Exel6274, P{w[+mC]=XP-U}Exel6274/TM6B, Tb[1] (Elongin-A), w[1118]; PBac {w[+mC]=WH} Cdk9[f05537]/CyO (Cdk9). Ubx1/TM6B, Tb and Abd-BM1/TM6B, Tb was obtained from E.S. Herrero. The generation of mitotic clones was performed using standard techniques [18]. For generating Cdk9 clones w[1118]; P{ry[+t7.2]=neoFRT}42D P{w[+mC]=Ubi-GFP(S65T)nls}2R/CyO was used and for Elongin-A clones yw; P{neoFRT}82B, Sb/TM6 and w; P{neoFRT}82B, P{ubi-GFP} were used. The FLP line used for all the crosses was hs-flp/hs-flp,y; Dr/TM3,Sb.
Chromatin Immunoprecipitation
In brief, the embryos were collected for 0–2 hrs, overnight and dechorionated in bleach, collected in a mesh and washed thoroughly with water, rinsed with Triton-NaCl solution once and washed thoroughly with water again. The embryos were transferred to scintillation vials and fixed in formaldehyde saturated hexanes for 25 minutes [3]. The embryos were disrupted in 7ml Wheaton dounce homogenizer in homogenization buffer and washed several times in wash buffers. The chromatin was then sonicated to get desired fragment size and aliquoted for further immunoprecipitations (IP). IP was performed following ChIP protocol provided with Upstate Biotechnology ChIP Assay Kit. For IP, chromatin was incubated with Protein A Sepharose resin slurry (Upstate Biotechnology) for an hour at 4°C. Pre-cleared chromatin was incubated with pre-immune serum (IgG, IgM) or with α-8WG16 antibody (Covance), α-H14 (Covance) antibody overnight at 4 °C. Antibody bound chromatin was mixed with either 60 μl Protein A sepharose or Protein G sepharose resin slurry and incubated for 1 h at 4 °C. The protein A beads bound with anti mouse IgM (Sigma) was used for IP experiments using H14 antibody. Chromatin from chromatin–antibody–resin complex was recovered after treatment with RNase A, Proteinase K and column purified. Precipitated DNA was resuspended in equal volume analyzed by real time PCR or conventional PCR. Primer pairs were designed to amplify 100 bp–200 bp fragments at regular intervals along the regions of interest (Supp. Data 1). PCR was performed and monitored in 7300 Real-Time PCR System using Sybr Green master mix (Applied Biosystems). Dissociation curves were analyzed as a means to ensure quality of amplicon and to monitor primer dimers. Enrichment was determined based on the differences of the critical threshold (ΔCt) measurements and was calculated based on formulae described elsewhere [19].
For the wing and haltere imaginal disc ChIP assay the discs were dissected from late third instar larvae from yw stocks. Larvae were either maintained continuously at 18° C or shifted from 18°C to 25°C 48 hours prior dissection. Discs were dissected in serum free SL2 cell medium and stored on ice until ready for formaldehyde fixation. Group of 20 discs were fixed at a time in 1% Formaldehyde for 20 minutes on ice and then quenched in 5X Glycine. Washed thrice in cold 1X PBS and suspended in SDS Lysis buffer with protease inhibitor cocktail II (supplied by Upstate ChIP Kit). The discs were sonicated to get the chromatin and subjected to ChIP and samples were followed up by conventional PCR or Real time PCR as described above.
Antibody staining of imaginal discs
For antibody staining, imaginal discs were dissected out and fixed in 4% paraformaldehyde and then washed in PBS thoroughly. Discs were then incubated in PBTX [PBS+BSA (Sigma) (0.5%) + Triton X-100 (Sigma) (0.1%)] for blocking at room temperature. The discs were washed again with PBTX thoroughly and primary antibody was added (monoclonal α-UBX, gift from Rob White and Rabbit α-GFP, Invitrogen) and left for overnight at 4°C. After washing by PBTX 3 times, secondary antibody was added (anti-rabbit Alexa 488 and anti-mouse Alexa 555) and left for 2–3 hours at RT. The discs were washed in PBTX and PBS; the discs were mounted in Prolong Gold medium with DAPI (Invitrogen) and visualized by flourescent microscopy.
Supplementary Material
Supplementary Figure 1. The Permanganate mapping of open transcription bubbles reveals engaged Pol II between +18 to +35 and +51 at the Ubx locus in wing and haltere imaginal discs. The figure shows Adenine-Guanine and Cytosine-Thymine ladder used for position determination, the reactivity pattern for naked DNA as a control and the permanganate reactivity observed in haltere and wing discs.
Supplementary Figure 2. A) The enhancer suppression screen for Ubx with elongation factors Nelf-E and Spt5 result in suppression of the Ubx phenotype (i.e. the haltere to wing transformation). B) The CbxHM allele which displays a wing to haltere transformation (gain of function) due to ectopic expression of Ubx in the wings was also used in the enhancer suppressor screen with elongation factors. We saw a marginal suppression of dominant Cbx phenotype in Cdk9 background (wing to haltere transformation became more wing like) and enhancement of Cbx phenotype in Nelf-E background (wing to haltere transformation became more haltere like). C) The adult mitotic clones of Cdk9 and Elo-A display phenotypes reminiscent of Notch, EGFR and wingless signaling. Unstable and folded wing margins, cell death and necrotic patches, loss or wing margin bristles, notching of wings and occurrences of ectopic bristles in the notum region.
Acknowledgments
We would like to thank Ernesto-Sanchez Herrero for providing Ubx and Abd-B fly stocks, Ishwar Hariharan for FRT lines, Exilexis collection for EloA and Nelf-E fly stocks and Fred Biemar for double balancers and FLP lines. α-UBX antibody was a generous gift from Rob White. We also thank Preeti Paliwal and Levine Lab members for support and advice. This study was supported by an NIH grant to M.L. (GM34431).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Lewis EB. The bithorax complex: the first fifty years. Int J Dev Biol. 1998;42:403–415. [PubMed] [Google Scholar]
- 2.Ringrose L, Paro R. Epigenetic regulation of cellular memory by the Polycomb and Trithorax group proteins. Annu Rev Genet. 2004;38:413–443. doi: 10.1146/annurev.genet.38.072902.091907. [DOI] [PubMed] [Google Scholar]
- 3.Zeitlinger J, Stark A, Kellis M, Hong JW, Nechaev S, Adelman K, Levine M, Young RA. RNA polymerase stalling at developmental control genes in the Drosophila melanogaster embryo. Nat Genet. 2007;39:1512–1516. doi: 10.1038/ng.2007.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Saunders A, Core LJ, Lis JT. Breaking barriers to transcription elongation. Nat Rev Mol Cell Biol. 2006;7:557–567. doi: 10.1038/nrm1981. [DOI] [PubMed] [Google Scholar]
- 5.Muse GW, Gilchrist DA, Nechaev S, Shah R, Parker JS, Grissom SF, Zeitlinger J, Adelman K. RNA polymerase is poised for activation across the genome. Nat Genet. 2007;39:1507–1511. doi: 10.1038/ng.2007.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Patturajan M, Schulte RJ, Sefton BM, Berezney R, Vincent M, Bensaude O, Warren SL, Corden JL. Growth-related changes in phosphorylation of yeast RNA polymerase II. J Biol Chem. 1998;273:4689–4694. doi: 10.1074/jbc.273.8.4689. [DOI] [PubMed] [Google Scholar]
- 7.Wang X, Lee C, Gilmour DS, Gergen JP. Transcription elongation controls cell fate specification in the Drosophila embryo. Genes Dev. 2007;21:1031–1036. doi: 10.1101/gad.1521207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rougvie AE, Lis JT. Postinitiation transcriptional control in Drosophila melanogaster. Mol Cell Biol. 1990;10:6041–6045. doi: 10.1128/mcb.10.11.6041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rougvie AE, Lis JT. The RNA polymerase II molecule at the 5′ end of the uninduced hsp70 gene of D. melanogaster is transcriptionally engaged. Cell. 1988;54:795–804. doi: 10.1016/s0092-8674(88)91087-2. [DOI] [PubMed] [Google Scholar]
- 10.Beuchle D, Struhl G, Muller J. Polycomb group proteins and heritable silencing of Drosophila Hox genes. Development. 2001;128:993–1004. doi: 10.1242/dev.128.6.993. [DOI] [PubMed] [Google Scholar]
- 11.Papp B, Muller J. Histone trimethylation and the maintenance of transcriptional ON and OFF states by trxG and PcG proteins. Genes Dev. 2006;20:2041–2054. doi: 10.1101/gad.388706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ni Z, Schwartz BE, Werner J, Suarez JR, Lis JT. Coordination of transcription, RNA processing, and surveillance by P-TEFb kinase on heat shock genes. Mol Cell. 2004;13:55–65. doi: 10.1016/s1097-2765(03)00526-4. [DOI] [PubMed] [Google Scholar]
- 13.Casanova J, Sanchez-Herrero E, Morata G. Identification and characterization of a parasegment specific regulatory element of the abdominal-B gene of Drosophila. Cell. 1986;47:627–636. doi: 10.1016/0092-8674(86)90627-6. [DOI] [PubMed] [Google Scholar]
- 14.Sanchez-Herrero E, Vernos I, Marco R, Morata G. Genetic organization of Drosophila bithorax complex. Nature. 1985;313:108–113. doi: 10.1038/313108a0. [DOI] [PubMed] [Google Scholar]
- 15.Eissenberg JC, Ma J, Gerber MA, Christensen A, Kennison JA, Shilatifard A. dELL is an essential RNA polymerase II elongation factor with a general role in development. Proc Natl Acad Sci U S A. 2002;99:9894–9899. doi: 10.1073/pnas.152193699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Smith ER, Winter B, Eissenberg JC, Shilatifard A. Regulation of the transcriptional activity of poised RNA polymerase II by the elongation factor ELL. Proc Natl Acad Sci U S A. 2008;105:8575–8579. doi: 10.1073/pnas.0804379105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kehle J, Beuchle D, Treuheit S, Christen B, Kennison JA, Bienz M, Muller J. dMi-2, a hunchback-interacting protein that functions in polycomb repression. Science. 1998;282:1897–1900. doi: 10.1126/science.282.5395.1897. [DOI] [PubMed] [Google Scholar]
- 18.Xu T, Rubin GM. Analysis of genetic mosaics in developing and adult Drosophila tissues. Development. 1993;117:1223–1237. doi: 10.1242/dev.117.4.1223. [DOI] [PubMed] [Google Scholar]
- 19.Mukhopadhyay A, Deplancke B, Walhout AJ, Tissenbaum HA. Chromatin immunoprecipitation (ChIP) coupled to detection by quantitative real-time PCR to study transcription factor binding to DNA in Caenorhabditis elegans. Nat Protoc. 2008;3:698–709. doi: 10.1038/nprot.2008.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1. The Permanganate mapping of open transcription bubbles reveals engaged Pol II between +18 to +35 and +51 at the Ubx locus in wing and haltere imaginal discs. The figure shows Adenine-Guanine and Cytosine-Thymine ladder used for position determination, the reactivity pattern for naked DNA as a control and the permanganate reactivity observed in haltere and wing discs.
Supplementary Figure 2. A) The enhancer suppression screen for Ubx with elongation factors Nelf-E and Spt5 result in suppression of the Ubx phenotype (i.e. the haltere to wing transformation). B) The CbxHM allele which displays a wing to haltere transformation (gain of function) due to ectopic expression of Ubx in the wings was also used in the enhancer suppressor screen with elongation factors. We saw a marginal suppression of dominant Cbx phenotype in Cdk9 background (wing to haltere transformation became more wing like) and enhancement of Cbx phenotype in Nelf-E background (wing to haltere transformation became more haltere like). C) The adult mitotic clones of Cdk9 and Elo-A display phenotypes reminiscent of Notch, EGFR and wingless signaling. Unstable and folded wing margins, cell death and necrotic patches, loss or wing margin bristles, notching of wings and occurrences of ectopic bristles in the notum region.